The Role of p53 Dysfunction in Colorectal Cancer and Its Implication for Therapy
Abstract
Simple Summary
Abstract
1. Introduction
1.1. The Physiological Role of p53 in CRC
1.2. p53 Mutations in CRC
1.3. Treatment of CRC
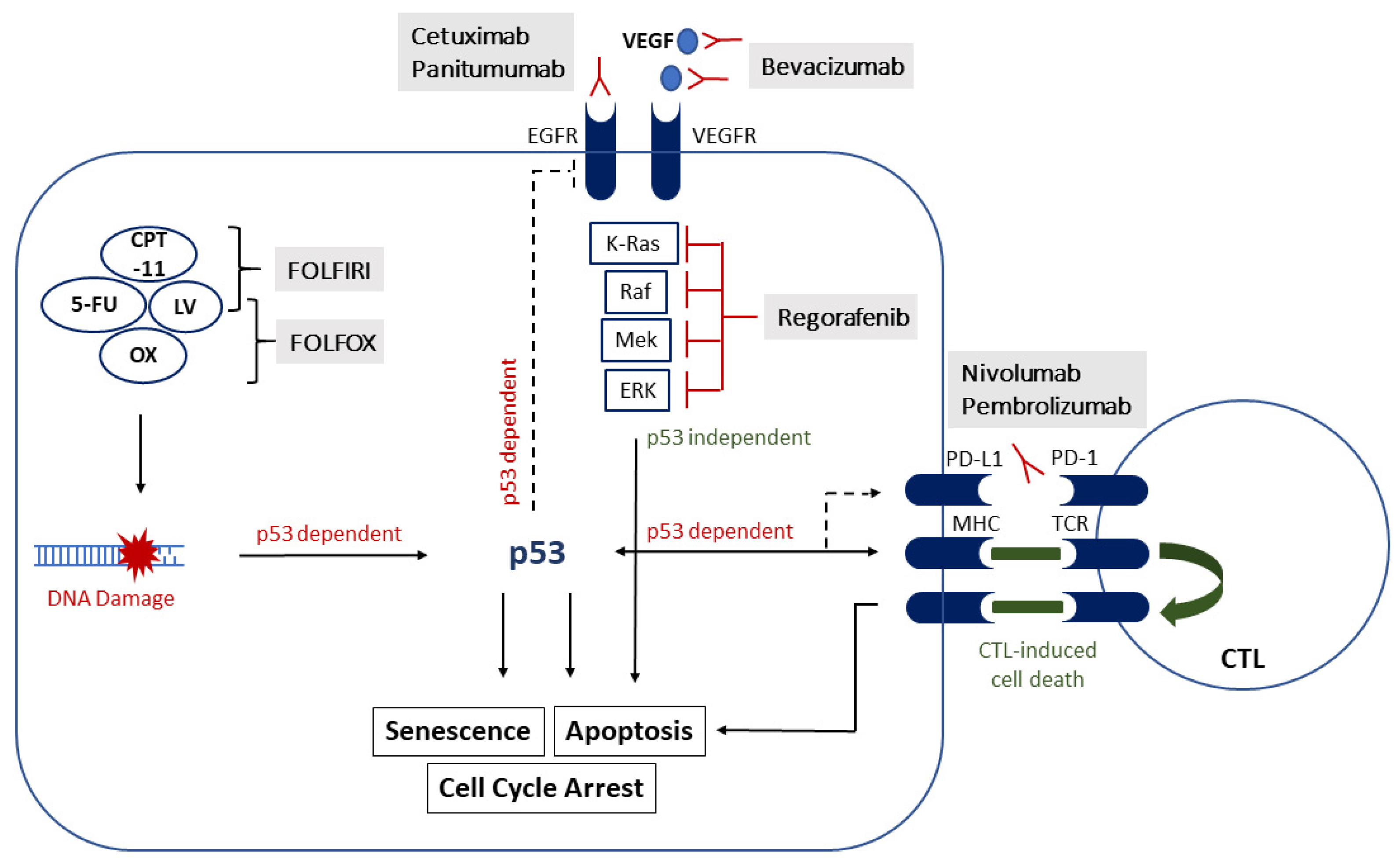
2. Treatment Response to Systemic Therapy
2.1. Chemotherapy
2.1.1. 5-Fluoruracil
2.1.2. Oxaliplatin
2.1.3. Irinotecan
2.2. VEGF- and EGFR-Targeted Therapies
2.2.1. Bevacizumab (Anti-VEGF)
2.2.2. Cetuximab, Panitumumab (Anti-EGFR)
2.3. Tyrosine Kinase Inhibitors
3. Immunotherapy
3.1. Immune Checkpoint Inhibitors
3.2. Individualized Immunotherapy
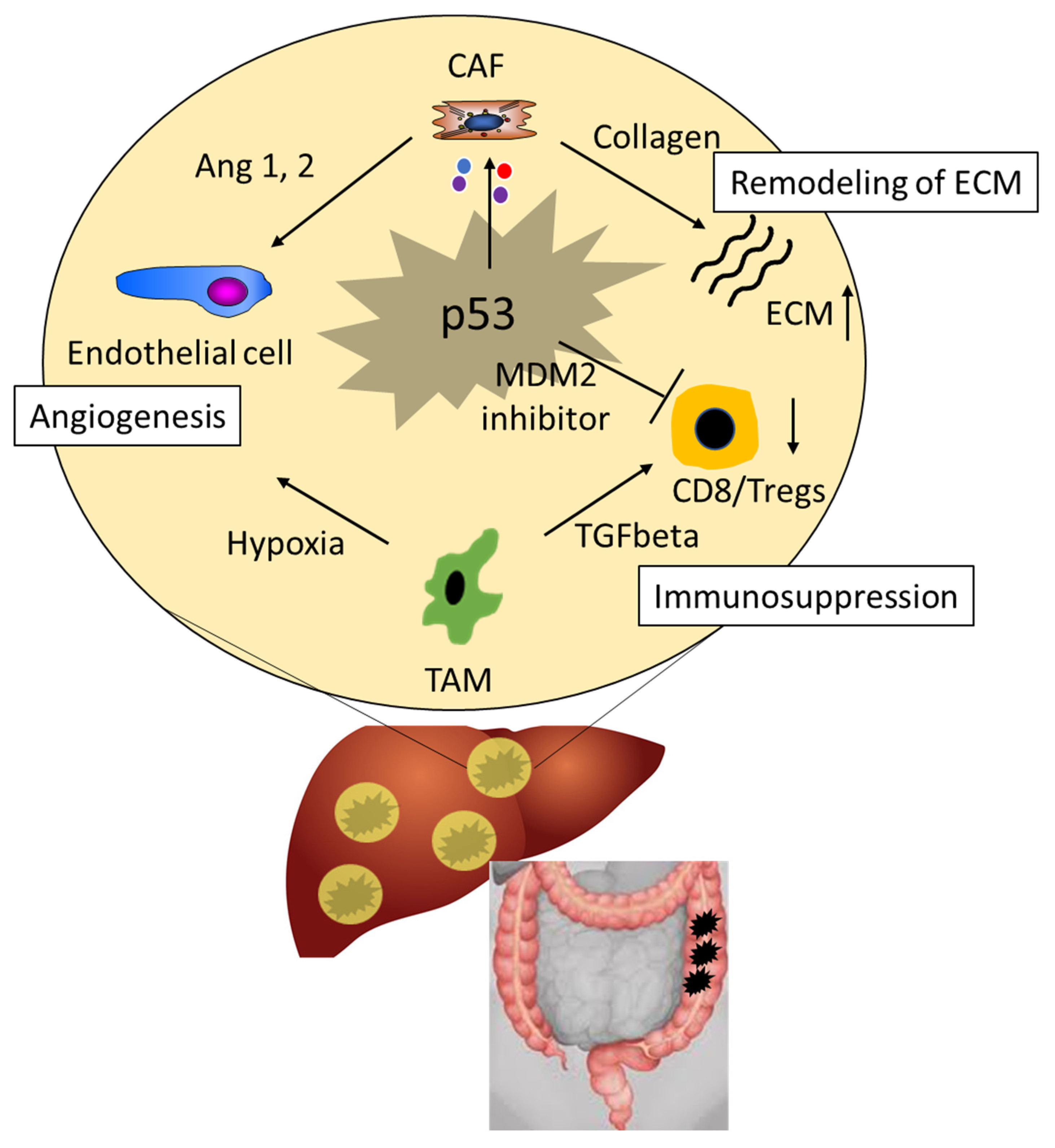
4. The Impact of p53 on Treatment Outcomes of Colorectal Liver Metastasis (CRLM)
5. The Role of p53 in the Tumor Microenvironment
6. p53 as a Druggable Target
7. Conclusions and Outlook
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| 5-FU | 5-Fluoruracil |
| Ang | Angiopoietin |
| CAFs | Cancer-associated fibroblasts |
| CCL | Chemokine ligand |
| CDK | Cyclin-dependent kinase |
| CPT-11 | Irinotecan |
| CRC | Colorectal cancer |
| CRLM | Colorectal liver metastasis |
| CTL | Cytotoxic T-lymphocyte |
| CTLA-4 | Cytotoxic T-lymphocyte antigen-4 |
| DBD | DNA-binding domain |
| DFS | Disease-free survival |
| dMMR | Deficient mismatch repair |
| dTMP | Deoxythymidine monophosphate |
| ECM | Extracellular matrix |
| EGFR | Epidermal growth factor receptor |
| FAP | Fibroblast activation protein |
| FdUMP | Fluorodeoxyuridine monophosphate |
| FP | Fluoropyrimidine |
| FSP-1 | Fibroblast-specific protein, S100A4 |
| GOF | Gain-of-function |
| HAI | Hepatic arterial infusion |
| HDI | Human Development Index |
| HMG-CoA reductase | 3-hydroxy-3-methylglutaryl coenzyme A reductase |
| Hsp | Heat shock protein |
| ICI | Immune checkpoint inhibitor |
| LOF | Loss-of-function |
| LV | Leucovorin |
| mCRC | Metastatic CRC |
| MDM2 | E3 ubiquitin-protein ligase Mdm2 |
| MHC I | Major histocompatibility complex |
| micro RNA | miRNA |
| MMP | Metalloproteinases |
| MSI-H | High microsatellite instability |
| MSS/I | Microsatellite stable/instable |
| NF-κB | Nuclear factor ‘kappa-light-chain-enhancer’ of activated B-cells |
| OS | Overall survival |
| OX | Oxaliplatin |
| PD-1 | Programmed death-1 |
| PDGFR α/β | Platelet derived growth factor receptor-alpha/beta |
| PD-L1 | Programmed death ligand 1 |
| pMMR | Proficient mismatch repair |
| POLR2A | DNA-directed RNA polymerase II subunit RPB1 |
| PUMA | P53 Upregulated Modulator of Apoptosis |
| ROS | Reactive oxygen species |
| siRNA | silencing RNA |
| SREBP | Sterol regulatory element-binding protein |
| TAMs | Tumor-associated macrophages |
| TCR | T-cell receptor |
| TME | Tumor microenvironment |
| Tregs | Regulatory T cells |
| TNF-α | Tumor necrosis factor-alpha |
| TS | Thymidylate synthase |
| UICC | Union for International Cancer Control |
| VEGF | Vascular endothelial growth factor |
| wt | Wild-type |
| ZMC-1 | Zinc metallochaperone-1 |
| α-SMA | α-smooth muscle actin |
References
- Global Cancer Observatory. Available online: https://gco.iarc.fr (accessed on 30 December 2020).
- Ferlay, J.; Colombet, M.; Soerjomataram, I.; Dyba, T.; Randi, G.; Bettio, M.; Gavin, A.; Visser, O.; Bray, F. Cancer incidence and mortality patterns in Europe: Estimates for 40 countries and 25 major cancers in 2018. Eur. J. Cancer 2018, 103, 356–387. [Google Scholar] [CrossRef]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef]
- Rawla, P.; Sunkara, T.; Barsouk, A. Epidemiology of colorectal cancer: Incidence, mortality, survival, and risk factors. Gastroenterol. Rev. 2019, 14, 89–103. [Google Scholar] [CrossRef] [PubMed]
- Taylor, D.P.; Burt, R.W.; Williams, M.S.; Haug, P.J.; Cannon–Albright, L.A. Population-Based Family History–Specific Risks for Colorectal Cancer: A Constellation Approach. Gastroenterology 2010, 138, 877–885. [Google Scholar] [CrossRef]
- A Eaden, J.; Abrams, K.R.; Mayberry, J.F. The risk of colorectal cancer in ulcerative colitis: A meta-analysis. Gut 2001, 48, 526–535. [Google Scholar] [CrossRef] [PubMed]
- Liang, P.S.; Chen, T.-Y.; Giovannucci, E. Cigarette smoking and colorectal cancer incidence and mortality: Systematic review and meta-analysis. Int. J. Cancer 2009, 124, 2406–2415. [Google Scholar] [CrossRef] [PubMed]
- Fedirko, V.; Tramacere, I.; Bagnardi, V.; Rota, M.; Scotti, L.; Islami, F.; Negri, E.; Straif, K.; Romieu, I.; La Vecchia, C.; et al. Alcohol drinking and colorectal cancer risk: An overall and dose–response meta-analysis of published studies. Ann. Oncol. 2011, 22, 1958–1972. [Google Scholar] [CrossRef] [PubMed]
- Chan, D.S.M.; Lau, R.; Aune, D.; Vieira, R.; Greenwood, D.C.; Kampman, E.; Norat, T. Red and Processed Meat and Colorectal Cancer Incidence: Meta-Analysis of Prospective Studies. PLoS ONE 2011, 6, e20456. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Yang, Y.; Wang, F.; Zhang, P.; Shi, C.; Zou, Y.; Qin, H. Obesity and Risk of Colorectal Cancer: A Systematic Review of Prospective Studies. PLoS ONE 2013, 8, e53916. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Ben, Q.; Shen, H.; Lu, W.; Zhang, Y.; Zhu, J. Diabetes mellitus and incidence and mortality of colorectal cancer: A systematic review and meta-analysis of cohort studies. Eur. J. Epidemiol. 2011, 26, 863–876. [Google Scholar] [CrossRef] [PubMed]
- Brenner, H.; Kloor, M.; Pox, C.P. Colorectal cancer. Lancet 2014, 383, 1490–1502. [Google Scholar] [CrossRef]
- Brenner, H.; Chang-Claude, J.; Seiler, C.M.; Rickert, A.; Hoffmeister, M. Protection From Colorectal Cancer After Colonoscopy. Ann. Intern. Med. 2011, 154, 22–30. [Google Scholar] [CrossRef] [PubMed]
- Aune, D.; Lau, R.; Chan, D.S.; Vieira, R.; Greenwood, D.C.; Kampman, E.; Norat, T. Nonlinear Reduction in Risk for Colorectal Cancer by Fruit and Vegetable Intake Based on Meta-analysis of Prospective Studies. Gastroenterology 2011, 141, 106–118. [Google Scholar] [CrossRef]
- Aune, D.; Chan, D.S.M.; Lau, R.; Vieira, R.; Greenwood, D.C.; Kampman, E.; Norat, T. Dietary fibre, whole grains, and risk of colorectal cancer: Systematic review and dose-response meta-analysis of prospective studies. BMJ 2011, 343, d6617. [Google Scholar] [CrossRef] [PubMed]
- Aune, D.; Lau, R.; Chan, D.S.M.; Vieira, R.; Greenwood, D.C.; Kampman, E.; Norat, T. Dairy products and colorectal cancer risk: A systematic review and meta-analysis of cohort studies. Ann. Oncol. 2012, 23, 37–45. [Google Scholar] [CrossRef]
- Wu, S.; Feng, B.; Li, K.; Zhu, X.; Liang, S.; Liu, X.; Han, S.; Wang, B.; Wu, K.; Miao, D.; et al. Fish Consumption and Colorectal Cancer Risk in Humans: A Systematic Review and Me-ta-analysis. Am. J. Med. 2012, 125, 551–559.e5. [Google Scholar] [CrossRef] [PubMed]
- Demierre, M.-F.; Higgins, P.D.R.; Gruber, S.B.; Hawk, E.T.; Lippman, S.M. Statins and cancer prevention. Nat. Rev. Cancer 2005, 5, 930–942. [Google Scholar] [CrossRef] [PubMed]
- Elmunzer, B.J.; Hayward, R.A.; Schoenfeld, P.S.; Saini, S.D.; Deshpande, A.; Waljee, A.K. Effect of Flexible Sigmoidoscopy-Based Screening on In-cidence and Mortality of Colorectal Cancer: A Systematic Review and Meta-Analysis of Randomized Controlled Trials. PLoS Med. 2012, 9, e1001352. [Google Scholar] [CrossRef] [PubMed]
- Nakayama, M.; Oshima, M. Mutant p53 in colon cancer. J. Mol. Cell Biol. 2019, 11, 267–276. [Google Scholar] [CrossRef]
- Fearon, E.R.; Vogelstein, B. A genetic model for colorectal tumorigenesis. Cell 1990, 61, 759–767. [Google Scholar] [CrossRef]
- Baran, B.; Ozupek, N.M.; Tetik, N.Y.; Acar, E.; Bekcioglu, O.; Baskin, Y. Difference Between Left-Sided and Right-Sided Colorectal Cancer: A Focused Review of Literature. Gastroenterol. Res. 2018, 11, 264–273. [Google Scholar] [CrossRef]
- Giannakis, M.; Mu, X.J.; Shukla, S.A.; Qian, Z.R.; Cohen, O.; Nishihara, R.; Bahl, S.; Cao, Y.; Amin-Mansour, A.; Yamauchi, M.; et al. Genomic Correlates of Immune-Cell Infiltrates in Colorectal Car-cinoma. Cell Rep. 2016, 15, 857–865. [Google Scholar] [CrossRef]
- Robles, A.I.; Jen, J.; Harris, C.C. Clinical Outcomes of TP53 Mutations in Cancers. Cold Spring Harb. Perspect. Med. 2016, 6, a026294. [Google Scholar] [CrossRef] [PubMed]
- Cho, Y.; Gorina, S.; Jeffrey, P.; Pavletich, N. Crystal structure of a p53 tumor suppressor-DNA complex: Understanding tumorigenic muta-tions. Science 1994, 265, 346–355. [Google Scholar] [CrossRef]
- Li, H.; Zhang, J.; Tong, J.H.M.; Chan, A.W.H.; Yu, J.; Kang, W.; To, K.F. Targeting the Oncogenic p53 Mutants in Colorectal Cancer and Other Solid Tumors. Int. J. Mol. Sci. 2019, 20, 5999. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Lozano, G. Molecular Pathways: Targeting Mdm2 and Mdm4 in Cancer Therapy. Clin. Cancer Res. 2013, 19, 34–41. [Google Scholar] [CrossRef] [PubMed]
- Li, X.-L.; Zhou, J.; Chen, Z.-R.; Chng, W.-J. p53mutations in colorectal cancer- molecular pathogenesis and pharmacological reactivation. World J. Gastroenterol. 2015, 21, 84–93. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Ma, W.; Benchimol, S. Pidd, a new death-domain–containing protein, is induced by p53 and promotes apoptosis. Nat. Genet. 2000, 26, 122–127. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Kon, N.; Jiang, L.; Tan, M.; Ludwig, T.; Zhao, Y.; Baer, R.; Gu, W. Tumor Suppression in the Absence of p53-Mediated Cell-Cycle Arrest, Apoptosis, and Senescence. Cell 2012, 149, 1269–1283. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, S.; Tanaka, T.; Poyurovsky, M.V.; Nagano, H.; Mayama, T.; Ohkubo, S.; Lokshin, M.; Hosokawa, H.; Nakayama, T.; Suzuki, Y.; et al. Phosphate-activated glutaminase (GLS2), a p53-inducible regulator of glutamine metabolism and reactive oxygen species. Proc. Natl. Acad. Sci. USA 2010, 107, 7461–7466. [Google Scholar] [CrossRef] [PubMed]
- A Sablina, A.; Budanov, A.V.; Ilyinskaya, G.V.; Agapova, L.S.; E Kravchenko, J.; Chumakov, P.M. The antioxidant function of the p53 tumor suppressor. Nat. Med. 2005, 11, 1306–1313. [Google Scholar] [CrossRef] [PubMed]
- Smit, W.L.; Spaan, C.N.; de Boer, R.J.; Ramesh, P.; Garcia, T.M.; Meijer, B.J.; Vermeulen, J.L.M.; Lezzerini, M.; MacInnes, A.W.; Koster, J.; et al. Driver mutations of the adenoma-carcinoma sequence govern the intestinal epithelial global translational capacity. Proc. Natl. Acad. Sci.USA 2020, 117, 25560–25570. [Google Scholar] [CrossRef] [PubMed]
- Ryan, K.M.; Phillips, A.C.; Vousden, K.H. Regulation and function of the p53 tumor suppressor protein. Curr. Opin. Cell Biol. 2001, 13, 332–337. [Google Scholar] [CrossRef]
- Russo, A.; Bazan, V.; Iacopetta, B.; Kerr, D.; Soussi, T.; Gebbia, N. The TP53 Colorectal Cancer International Collaborative Study on the Prog-nostic and Predictive Significance of p53 Mutation: Influence of Tumor Site, Type of Mutation, and Adjuvant Treatment. J. Clin. Oncol. 2005, 23, 7518–7528. [Google Scholar] [CrossRef]
- Willis, A.; Jung, E.J.; Wakefield, T.; Chen, X. Mutant p53 exerts a dominant negative effect by preventing wild-type p53 from binding to the promoter of its target genes. Oncogene 2004, 23, 2330–2338. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Yang, J.; Lu, G.; Liu, P.C.; Chen, Q.; Xie, Z.; Wu, C. One Stone Kills Three Birds: Novel Boron-Containing Vesicles for Potential BNCT, Controlled Drug Release, and Diagnostic Imaging. Mol. Pharm. 2014, 11, 3291–3299. [Google Scholar] [CrossRef] [PubMed]
- Argilés, G.; Tabernero, J.; Labianca, R.; Hochhauser, D.; Salazar, R.; Iveson, T.; Laurent-Puig, P.; Quirke, P.; Yoshino, T.; Taieb, J.; et al. Localised colon cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2020, 31, 1291–1305. [Google Scholar] [CrossRef]
- Costas-Chavarri, A.; Nandakumar, G.; Temin, S.; Lopes, G.; Cervantes, A.; Cruz Correa, M.; Engineer, R.; Hamashima, C.; Fuang Ho, G.; David Huitzil, F.; et al. Treatment of Patients with Early-Stage Colorectal Cancer: ASCO Resource-Stratified Guideline. J. Glob. Oncol. 2019, 5, 1–19. [Google Scholar] [PubMed]
- Popat, S.; Hubner, R.; Houlston, R. Systematic Review of Microsatellite Instability and Colorectal Cancer Prognosis. J. Clin. Oncol. 2005, 23, 609–618. [Google Scholar] [CrossRef]
- Chiorean, E.G.; Nandakumar, G.; Fadelu, T.; Temin, S.; Alarcon-Rozas, A.E.; Bejarano, S.; Croitoru, A.-E.; Grover, S.; Lohar, P.V.; Odhiambo, A.; et al. Treatment of Patients with Late-Stage Colorectal Cancer: ASCO Resource-Stratified Guideline. Jco Glob. Oncol. 2020, 6, 414–438. [Google Scholar] [CrossRef]
- Cassidy, J.; Saltz, L.; Twelves, C.; Van Cutsem, E.; Hoff, P.; Kang, Y.; Saini, J.P.; Gilberg, F.; Cunningham, D. Efficacy of capecitabine versus 5-fluorouracil in colorectal and gastric cancers: A meta-analysis of individual data from 6171 patients. Ann. Oncol. 2011, 22, 2604–2609. [Google Scholar] [CrossRef]
- Van Cutsem, E.; Cervantes, A.; Nordlinger, B.; Arnold, D. Metastatic colorectal cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2014, 25, iii1–iii9. [Google Scholar] [CrossRef] [PubMed]
- Douillard, J.; Cunningham, D.; Roth, A.; Navarro, M.; James, R.; Karasek, P.; Jandik, P.; Iveson, T.; Carmichael, J.; Alakl, M.; et al. Irinotecan combined with fluorouracil compared with fluorouracil alone as first-line treatment for metastatic colorectal cancer: A multicentre randomised trial. Lancet 2000, 355, 1041–1047. [Google Scholar] [CrossRef]
- André, T.; Boni, C.; Navarro, M.; Tabernero, J.; Hickish, T.; Topham, C.; Bonetti, A.; Clingan, P.; Bridgewater, J.; Rivera, F.; et al. Improved overall survival with oxaliplatin, fluorouracil, and leu-covorin as adjuvant treatment in stage II or III colon cancer in the MOSAIC trial. J. Clin. Oncol. 2009, 27, 3109–3116. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2020. CA. Cancer J. Clin. 2020, 70, 7–30. [Google Scholar] [CrossRef]
- Longley, D.B.; Allen, W.L.; McDermott, U.; Wilson, T.R.; Latif, T.; Boyer, J.; Lynch, M.; Johnston, P.G. The Roles of Thymidylate Synthase and p53 in Regulating Fas-Mediated Apoptosis in Response to Antimetabolites. Clin. Cancer Res. 2004, 10, 3562–3571. [Google Scholar] [CrossRef][Green Version]
- Thirion, P.; Michiels, S.; Pignon, J.P.; Buyse, M.; Braud, A.C.; Carlson, R.W.; O’Connell, M.; Sargent, P.; Piedbois, P. Meta-Analysis Group in Cancer. Modulation of fluorouracil by leucovorin in patients with advanced colorectal cancer: An updated meta-analysis. J. Clin. Oncol. 2004, 22, 3766–3775. [Google Scholar]
- Yang, B.; Eshleman, J.R.; Berger, N.A.; Markowitz, S.D. Wild-type p53 protein potentiates cytotoxicity of therapeutic agents in human colon cancer cells. Clin. Cancer Res. 1996, 2, 1649–1657. [Google Scholar] [PubMed]
- Liang, J.-T.; Huang, K.-C.; Cheng, Y.-M.; Hsu, H.-C.; Cheng, A.-L.; Hsu, C.-H.; Yeh, K.-H.; Wang, S.-M.; Chang, K.-J. P53 overexpression predicts poor chemosensitivity to high-dose 5-fluorouracil plus leucovorin chemotherapy for stage IV colorectal cancers after palliative bowel resection. Int. J. Cancer 2001, 97, 451–457. [Google Scholar] [CrossRef]
- Ahnen, D.J.; Feigl, P.; Quan, G.; Fenoglio-Preiser, C.; Lovato, L.C.; A Bunn, P.; Stemmerman, G.; Wells, J.D.; Macdonald, J.S.; Meyskens, F.L. Ki-ras mutation and p53 overexpression predict the clinical behavior of colorectal cancer: A Southwest Oncology Group study. Cancer Res. 1998, 58, 1149–1158. [Google Scholar] [PubMed]
- Pugacheva, E.N.; Ivanov, A.V.; Kravchenko, J.E.; Kopnin, B.P.; Levine, A.J.; Chumakov, P.M. Novel gain of function activity of p53 mutants: Activation of the dUTPase gene expression leading to resistance to 5-fluorouracil. Oncogene 2002, 21, 4595–4600. [Google Scholar] [CrossRef] [PubMed]
- Zaanan, A.; Cuilliere-Dartigues, P.; Guilloux, A.; Parc, Y.; Louvet, C.; de Gramont, A.; Tiret, E.; Dumont, S.; Gayet, B.; Validire, P.; et al. Impact of p53 expression and microsatellite instability on stage III colon cancer disease-free survival in patients treated by 5-fluorouracil and leucovorin with or without oxaliplatin. Ann. Oncol. 2009, 21, 772–780. [Google Scholar] [CrossRef]
- Boyer, J.; McLean, E.G.; Aroori, S.; Wilson, P.; McCulla, A.; Carey, P.D.; Longley, D.B.; Johnston, P.G. Characterization of p53 Wild-Type and Null Isogenic Colorectal Cancer Cell Lines Resistant to 5-Fluorouracil, Oxaliplatin, and Irinotecan. Clin. Cancer Res. 2004, 10, 2158–2167. [Google Scholar] [CrossRef] [PubMed]
- Dominijanni, A.; Gmeiner, W.H. Improved potency of F10 relative to 5-fluorouracil in colorectal cancer cells with p53 mutations. Cancer Drug Resist. 2018, 1, 48–58. [Google Scholar] [CrossRef] [PubMed]
- Bunz, F.; Hwang, P.M.; Torrance, C.; Waldman, T.; Zhang, Y.; Dillehay, L.; Williams, J.; Lengauer, C.; Kinzler, K.W.; Vogelstein, B. Disruption of p53 in human cancer cells alters the responses to therapeutic agents. J. Clin. Investig. 1999, 104, 263–269. [Google Scholar] [CrossRef]
- Issaeva, N.; Bozko, P.; Enge, M.; Protopopova, M.; Verhoef, L.G.G.C.; Masucci, M.G.; Pramanik, A.; Selivanova, G. Small molecule RITA binds to p53, blocks p53–HDM-2 interaction and activates p53 function in tumors. Nat. Med. 2004, 10, 1321–1328. [Google Scholar] [CrossRef] [PubMed]
- Wiegering, A.; Matthes, N.; Mühling, B.; Koospal, M.; Quenzer, A.; Peter, S.; Germer, C.T.; Linnebacher, M.; Otto, C. Reactivating p53 and Inducing Tumor Apoptosis (RITA) Enhances the Response of RITA-Sensitive Colorectal Cancer Cells to Chemotherapeutic Agents 5-Fluorouracil and Oxaliplatin. Neoplasia 2017, 19, 301–309. [Google Scholar] [CrossRef] [PubMed]
- Pothuraju, R.; Rachagani, S.; Krishn, S.R.; Chaudhary, S.; Nimmakayala, R.K.; Siddiqui, J.A.; Ganguly, K.; Lakshmanan, I.; Cox, J.L.; Mallya, K.; et al. Molecular implications of MUC5AC-CD44 axis in colorectal cancer progression and chemoresistance. Mol. Cancer 2020, 19, 37. [Google Scholar] [CrossRef] [PubMed]
- Ferris, J.; Espona-Fiedler, M.; Hamilton, C.; Holohan, C.; Crawford, N.; McIntyre, A.J.; Roberts, J.Z.; Wappett, M.; McDade, S.S.; Longley, D.B.; et al. Pevonedistat (MLN4924): Mechanism of cell death induction and therapeutic potential in colorectal cancer. Cell Death Discov. 2020, 6, 1–14. [Google Scholar] [CrossRef]
- Liang, Y.; Hou, L.; Li, L.; Li, L.; Zhu, L.; Wang, Y.; Huang, X.; Hou, Y.; Zhu, D.; Zou, H.; et al. Dichloroacetate restores colorectal cancer chemosensitivity through the p53/miR-149-3p/PDK2-mediated glucose metabolic pathway. Oncogene 2020, 39, 469–485. [Google Scholar] [CrossRef]
- Wang, H.; Zhang, L.; Yang, X.; Jin, Y.; Pei, S.; Zhang, D.; Zhang, H.; Zhou, B.; Zhang, Y.; Lin, D. PUMA mediates the combinational therapy of 5-FU and NVP-BEZ235 in colon cancer. Oncotarget 2015, 6, 14385–14398. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Liu, N.; Liu, J.; Liu, Y.; Zhang, C.; Long, S.; Luo, G.; Zhang, L.; Zhang, Y. Mutant p53 drives cancer chemotherapy resistance due to loss of function on acti-vating transcription of PUMA. Cell Cycle. 2019, 18, 3442–3455. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Pan, T.; Xiang, Y.; Zhang, M.; Feng, J.; Liu, S.; Duan, T.; Chen, P.; Zhai, B.; Chen, X.; et al. β-Elemene Reverses the Resistance of p53-Deficient Colorectal Cancer Cells to 5-Fluorouracil by Inducing Pro-death Autophagy and Cyclin D3-Dependent Cycle Arrest. Front. Bioeng. Biotechnol. 2020, 8. [Google Scholar] [CrossRef] [PubMed]
- Oh, H.J.; Bae, J.M.; Wen, X.; Jung, S.; Kim, Y.; Kim, K.J.; Cho, N.-Y.; Kim, J.H.; Han, S.-W.; Kim, T.-Y.; et al. p53 expression status is associated with cancer-specific survival in stage III and high-risk stage II colorectal cancer patients treated with oxaliplatin-based adjuvant chemotherapy. Br. J. Cancer 2019, 120, 797–805. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, T.; Wu, T.-T.; Catalano, P.J.; Ueki, T.; Satriano, R.; Haller, D.G.; Benson, A.B.; Hamilton, S.R. Molecular Predictors of Survival after Adjuvant Chemotherapy for Colon Cancer. N. Engl. J. Med. 2001, 344, 1196–1206. [Google Scholar] [CrossRef] [PubMed]
- Popat, S.; Chen, Z.; Zhao, D.; Pan, H.; Hearle, N.; Chandler, I.; Shao, Y.; Aherne, W.; Houlston, R.S. A prospective, blinded analysis of thymidylate synthase and p53 expressi-on as prognostic markers in the adjuvant treatment of colorectal cancer. Ann. Oncol. 2006, 17, 1810–1817. [Google Scholar] [CrossRef] [PubMed]
- Li, A.J.; Li, H.G.; Tang, E.J.; Wu, W.; Chen, Y.; Jiang, H.H.; Lin, M.B.; Yin, L. PIK3CA and TP53 mutations predict overall survival of stage II/III colorectal cancer patients. World J. Gastroenterol. 2018, 24, 631–640. [Google Scholar] [CrossRef]
- He, L.; Zhu, H.; Zhou, S.; Wu, T.; Wu, H.; Yang, H.; Mao, H.; SekharKathera, C.; Janardhan, A.; Edick, A.M.; et al. Wnt pathway is involved in 5-FU drug resistance of colorectal cancer cells. Exp. Mol. Med. 2018, 50, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhang, X.; Han, C.; Wan, G.; Huang, X.; Ivan, C.; Jiang, D.; Rodriguez-Aguayo, C.; Lopez-Berestein, G.; Rao, P.H.; et al. TP53 loss creates therapeutic vulnerability in colorectal cancer. Nat. Cell Biol. 2015, 520, 697–701. [Google Scholar] [CrossRef]
- Manic, S.; Gatti, L.; Carenini, N.; Fumagalli, G.; Zunino, F.; Perego, P. Mechanisms controlling sensitivity to platinum complexes: Role of p53 and DNA mismatch repair. Curr. Cancer Drug Targets 2003, 3, 21–29. [Google Scholar] [CrossRef] [PubMed]
- Arango, D.; Wilson, A.J.; Shi, Q.; A Corner, G.; Aranes, M.J.; Nicholas, C.J.; Lesser, M.; Mariadason, J.M.; Augenlicht, L.H. Molecular mechanisms of action and prediction of response to oxaliplatin in colorectal cancer cells. Br. J. Cancer 2004, 91, 1931–1946. [Google Scholar] [CrossRef] [PubMed]
- Therachiyil, L.; Haroon, J.; Sahir, F.; Siveen, K.S.; Uddin, S.; Kulinski, M.; Buddenkotte, J.; Steinhoff, M.; Krishnankutty, R. Dysregulated Phosphorylation of p53, Autophagy and Stemness Attributes the Mutant p53 Harboring Colon Cancer Cells Impaired Sensitivity to Oxaliplatin. Front. Oncol. 2020, 10. [Google Scholar] [CrossRef] [PubMed]
- Brachtendorf, S.; Wanger, R.A.; Birod, K.; Thomas, D.; Trautmann, S.; Wegner, M.S.; Fuhrmann, D.C.; Brüne, B.; Geisslinger, G.; Grösch, S. Chemosensitivity of human colon cancer cells is in-fluenced by a p53-dependent enhancement of ceramide synthase 5 and induction of autophagy. Biochim. Biophys. Acta-Mol. Cell Biol. Lipids. 2018, 1863, 1214–1227. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.-Y.; Islam, A.; Yuan, T.-M.; Chen, S.-W.; Liu, P.-F.; Chueh, P.J. Regulation of tNOX expression through the ROS-p53-POU3F2 axis contri-butes to cellular responses against oxaliplatin in human colon cancer cells. J. Exp. Clin. Cancer Res. 2018, 37, 161. [Google Scholar] [CrossRef] [PubMed]
- Marine, J.-C.; Lozano, G. Mdm2-mediated ubiquitylation: p53 and beyond. Cell Death Differ. 2009, 17, 93–102. [Google Scholar] [CrossRef] [PubMed]
- Azmi, A.S.; Banerjee, S.; Ali, S.; Wang, Z.; Bao, B.; Beck, F.W.J.; Maitah, M.; Choi, M.; Shields, T.F.; Philip, P.A.; et al. Network modeling of MDM2 inhibitor-oxaliplatin combination reveals bio-logical synergy in wt-p53 solid tumors. Oncotarget 2011, 2, 378–392. [Google Scholar] [CrossRef]
- Poel, D.; Boyd, L.N.; Beekhof, R.; Schelfhorst, T.; Pham, T.V.; Piersma, S.R.; Jaco, C.K.; Connie, R.J.; Henk, M.W.V.; Tineke, E.B. Proteomic Analysis of miR-195 and miR-497 Replacement Reveals Potential Candidates that Increase Sensitivity to Oxaliplatin in MSI/P53wt Colorectal Cancer Cells. Cells 2019, 8, 1111. [Google Scholar] [CrossRef] [PubMed]
- Pilat, N.; Grünberger, T.; Längle, F.; Mittlböck, M.; Perisanidis, B.; Kappel, S.; Wolf, B.; Starlinger, P.; Kührer, I.; Mühlbacher, F.; et al. Assessing the TP53 marker type in patients treated with or without neoadjuvant chemotherapy for resectable colorectal liver metastases: A p53 Research Group study. Eur. J. Surg. Oncol. 2015, 41, 683–689. [Google Scholar] [CrossRef]
- Netter, J.; Lehmann-Che, J.; Lambert, J.; Tallet, A.; Lourenco, N.; Soliman, H.; Bertheau, P.; Pariente, B.; Chirica, M.; Pocard, M.; et al. Functional TP53 mutations have no impact on response to cytotoxic agents in metastatic colon cancer. Bull Cancer 2015, 102, 117–125. [Google Scholar] [CrossRef]
- Bras-Gonçalves, A.R.; Rosty, C.; Laurent-Puig, P.; Soulié, P.; Dutrillaux, B.; Poupon, M.-F. Sensitivity to CPT-11 of xenografted human colorectal cancers as a function of microsatellite instability and p53 status. Br. J. Cancer 2000, 82, 913–923. [Google Scholar] [CrossRef] [PubMed]
- Weekes, J.; Ho, Y.-H.; Sebesan, S.; Ong, K.; Lam, A.K.-Y. Irinotecan and colorectal cancer: The role of p53, VEGF-C and α-B-crystallin expression. Int. J. Color. Dis. 2009, 25, 907. [Google Scholar] [CrossRef][Green Version]
- Warren, R.S.; Atreya, C.E.; Niedzwiecki, D.; Weinberg, V.K.; Donner, D.B.; Mayer, R.J.; Goldberg, R.M.; Compton, C.C.; Zuraek, M.B.; Ye, C.; et al. Association of TP53 Mutational Status and Gender with Survival after Adjuvant Treatment for Stage III Colon Cancer: Results of CALGB 89803. Clin. Cancer Res. 2013, 19, 5777–5787. [Google Scholar] [CrossRef]
- Hurwitz, H.; Fehrenbacher, L.; Novotny, W.; Cartwright, T.; Hainsworth, J.; Heim, W.; Berlin, J.; Baron, A.; Griffing, S.; Holmgren, E.; et al. Bevacizumab plus Irinotecan, Fluorouracil, and Leucovorin for Metastatic Colorectal Cancer. N. Engl. J. Med. 2004, 350, 2335–2342. [Google Scholar] [CrossRef]
- Bouvet, M.; Ellis, L.M.; Nishizaki, M.; Fujiwara, T.; Liu, W.; Bucana, C.D.; Fang, B.; Lee, J.J.; A Roth, J. Adenovirus-mediated wild-type p53 gene transfer down-regulates vascular endothelial growth factor expression and inhibits angiogenesis in human colon cancer. Cancer Res. 1998, 58, 2288–2292. [Google Scholar]
- Huemer, F.; Thaler, J.; Piringer, G.; Hackl, H.; Pleyer, L.; Hufnagl, C.; Weiss, L.; Greil, R. Sidedness and TP53 mutations impact OS in anti-EGFR but not anti-VEGF treated mCRC-An analysis of the KRAS registry of the AGMT (Arbeitsgemeinschaft Medikamentöse Tumortherapie). BMC Cancer 2018, 18, 11. [Google Scholar] [CrossRef] [PubMed]
- Ince, W.L.; Jubb, A.M.; Holden, S.N.; Holmgren, E.B.; Tobin, P.; Sridhar, M.; Hurwitz, H.I.; Kabbinavar, F.; Novotny, W.F.; Hillan, K.J.; et al. Association of k-ras, b-raf, and p53 Status with the Treatment Effect of Bevacizumab. J. Natl. Cancer Inst. 2005, 97, 981–989. [Google Scholar] [CrossRef] [PubMed]
- De Bruijn, M.T.; Raats, D.A.E.; Tol, J.; Hinrichs, J.; Teerenstra, S.; Punt, C.J.A.; Borel, R.I.H.M.; Kranenburg, O. Combined KRAS and TP53 mutation status is not predictive in CAPOX-treated metastatic colorectal cancer. Anticancer Res. 2011, 31, 1379–1385. [Google Scholar] [PubMed]
- Hasan, M.R.; Ho, S.H.Y.; Owen, D.A.; Tai, I.T. Inhibition of VEGF induces cellular senescence in colorectal cancer cells. Int. J. Cancer 2011, 129, 2115–2123. [Google Scholar] [CrossRef] [PubMed]
- Bardelli, A.; Siena, S. Molecular Mechanisms of Resistance to Cetuximab and Panitumumab in Colorectal Cancer. J. Clin. Oncol. 2010, 28, 1254–1261. [Google Scholar] [CrossRef] [PubMed]
- Lo Nigro, C.; Ricci, V.; Vivenza, D.; Granetto, C.; Fabozzi, T.; Miraglio, E.; Merlano, M.C. Prognostic and predictive biomarkers in metastatic colorectal cancer anti-EGFR therapy. World J. Gastroenterol. 2016, 22, 6944. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Jin, M.; Ye, S.; Yan, S. CHI3L1 promotes proliferation and improves sensitivity to cetuximab in colon cancer cells by down-regulating p53. J. Clin. Lab. Anal. 2019, 34, e23026. [Google Scholar] [CrossRef]
- Yang, M.; Schell, M.J.; Loboda, A.; Nebozhyn, M.; Li, J.; Teer, J.K.; Pledger, W.J.; Yeatman, T.J. Repurposing EGFR Inhibitor Utility in Colorectal Cancer in Mutant APC and TP53 Subpopulations. Cancer Epidemiol. Biomark. Prev. 2019, 28, 1141–1152. [Google Scholar] [CrossRef]
- Oden-Gangloff, A.; Di Fiore, F.; Bibeau, F.; Lamy, A.; Bougeard, G.; Charbonnier, F.; Blanchard, F.; Tougeron, D.; Ychou, M.; Boissière, F.; et al. TP53 mutations predict disease control in metastatic colorectal cancer treated with cetuximab-based chemotherapy. Br. J. Cancer 2009, 100, 1330–1335. [Google Scholar] [CrossRef] [PubMed]
- Stintzing, S.; Wirapati, P.; Lenz, H.-J.; Neureiter, D.; Fischer von Weikersthal, L.; Decker, T.; Kiani, A.; Kaiser, F.; Al-Batran, S.; Heintges, T.; et al. Consensus molecular subgroups (CMS) of co-lorectal cancer (CRC) and first-line efficacy of FOLFIRI plus cetuximab or bevacizumab in the FIRE3 (AIO KRK-0306) trial. Ann. Oncol. 2019, 30, 1796–1803. [Google Scholar] [CrossRef] [PubMed]
- Di Fiore, F.; Lamy, A.; Blanchard, F.; Oden-Gangloff, A.; Sesboüé, R.; Sabourin, J.; Frébourg, T.; Michel, P.; Laurent-Puig, P. TP53 mutations in irinotecan-refractory KRAS wt-BRAF wt metastatic colorectal cancer patients treated with cetuximab-based chemotherapy. J. Clin. Oncol. 2011, 29, 426. [Google Scholar] [CrossRef]
- Papaxoinis, G.; Kotoula, V.; Giannoulatou, E.; Koliou, G.A.; Karavasilis, V.; Lakis, S.; Koureas, A.; Bobos, M.; Chalaralambous, E.; Daskalaki, E. Phase II study of panitumumab combined with cape-citabine and oxaliplatin as first-line treatment in metastatic colorectal cancer patients: Clinical results including extended tumor geno-typing. Med. Oncol. 2018, 35, 16. [Google Scholar] [CrossRef] [PubMed]
- Di Bartolomeo, M.; Pietrantonio, F.; Perrone, F.; Dotti, K.F.; Lampis, A.; Bertan, C.; Beretta, E.; Rimassa, L.; Carbone, C.; Biondani, P.; et al. Lack of KRAS, NRAS, BRAF and TP53 mutations impro-ves outcome of elderly metastatic colorectal cancer patients treated with cetuximab, oxaliplatin and UFT. Target. Oncol. 2014, 9, 155–162. [Google Scholar] [CrossRef] [PubMed]
- Ciardiello, F.; Normanno, N.; Maiello, E.; Martinelli, E.; Troiani, T.; Pisconti, S.; Giuliani, F.; Barone, C.; Cartenì, G.; Rachiglio, A.M.; et al. Clinical activity of FOLFIRI plus cetuximab according to ex-tended gene mutation status by next-generation sequencing: Findings from the CAPRI-GOIM trial. Ann. Oncol. 2014, 25, 1756–1761. [Google Scholar] [CrossRef]
- Grothey, A.; Van Cutsem, E.; Sobrero, A.; Siena, S.; Falcone, A.; Ychou, M.; Humblet, Y.; Bouché, O.; Mineur, L.; Barone, C.; et al. Regorafenib monotherapy for previously treated metastatic colorectal cancer (CORRECT): An international, multicentre, randomised, placebo-controlled, phase 3 trial. Lancet 2013, 381, 303–312. [Google Scholar] [CrossRef]
- Tong, J.; Tan, S.; Zou, F.; Yu, J.; Zhang, L. FBW7 mutations mediate resistance of colorectal cancer to targeted therapies by blocking Mcl-1 degradation. Oncogene 2017, 36, 787–796. [Google Scholar] [CrossRef]
- Marks, E.I.; Tan, C.; Zhang, J.; Zhou, L.; Yang, Z.; Scicchitano, A.; El-Deiry, W.S. Regorafenib with a fluoropyrimidine for metastatic colorectal cancer after progression on multiple 5-FU-containing combination therapies and regorafenib monotherapy. Cancer Biol. Ther. 2015, 16, 1710–1719. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Wei, L.; Yu, J.; Zhang, L. Regorafenib Inhibits Colorectal Tumor Growth through PUMA-Mediated Apoptosis. Clin. Cancer Res. 2014, 20, 3472–3484. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Zhu, Z.; Zhang, X.; Zhang, N.; Yao, Z. Idelalisib induces PUMA-dependent apoptosis in colon cancer cells. Oncotarget 2016, 8, 6102–6113. [Google Scholar] [CrossRef] [PubMed]
- Iwai, Y.; Ishida, M.; Tanaka, Y.; Okazaki, T.; Honjo, T.; Minato, N. Involvement of PD-L1 on tumor cells in the escape from host immune system and tumor immunotherapy by PD-L1 blockade. Proc. Natl. Acad. Sci. USA 2002, 99, 12293–12297. [Google Scholar] [CrossRef] [PubMed]
- Overman, M.J.; McDermott, R.; Leach, J.L.; Lonardi, S.; Lenz, H.J.; Morse, M.A.; Desai, J.; Hill, A.; Axelson, M.; Moss, R.A.; et al. Nivolumab in patients with metastatic DNA mismatch repair-deficient or microsatellite instability-high colorectal cancer (CheckMate 142): An open-label, multicentre, phase 2 study. Lancet Oncol. 2017, 18, 1182–1191. [Google Scholar] [CrossRef]
- André, T.; Shiu, K.-K.; Kim, T.W.; Jensen, B.V.; Jensen, L.H.; Punt, C.; Smith, D.; Garcia-Carbonero, R.; Benavides, M.; Gibbs, P.; et al. Pembrolizumab in Microsatellite-Instability–High Advanced Colorectal Cancer. N. Engl. J. Med. 2020, 383, 2207–2218. [Google Scholar] [CrossRef] [PubMed]
- Cohen, R.; Hain, E.; Buhard, O.; Guilloux, A.; Bardier, A.; Kaci, R.; Bertheau, P.; Renaud, F.; Bibeau, F.; Fléjou, J.-F.; et al. Association of Primary Resistance to Immune Checkpoint Inhibitors in Metastatic Colorectal Cancer with Misdiagnosis of Microsatellite Instability or Mismatch Repair Deficiency Status. JAMA Oncol. 2019, 5, 551–555. [Google Scholar] [CrossRef]
- Le, D.T.; Durham, J.N.; Smith, K.N.; Wang, H.; Bartlett, B.R.; Aulakh, L.K.; Lu, S.; Kemberling, H.; Wilt, C.; Luber, B.S.; et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science 2017, 357, 409–413. [Google Scholar] [CrossRef] [PubMed]
- Webber, E.M.; Kauffman, T.L.; O’Connor, E.; Goddard, K.A.B. Systematic review of the predictive effect of MSI status in colorectal cancer pa-tients undergoing 5FU-based chemotherapy. BMC Cancer 2015, 15, 156. [Google Scholar] [CrossRef]
- Jass, J.R.; Do, K.-A.; Simms, L.A.; Iino, H.; Wynter, C.; Pillay, S.P.; Searle, J.; Radford-Smith, G.; Young, J.; Leggett, B. Morphology of sporadic colorectal cancer with DNA replication errors. Gut 1998, 42, 673–679. [Google Scholar] [CrossRef]
- Lin, E.I.; Tseng, L.-H.; Gocke, C.D.; Reil, S.; Le, D.T.; Azad, N.S.; Eshleman, J.R. Mutational profiling of colorectal cancers with microsatellite instability. Oncotarget 2015, 6, 42334–42344. [Google Scholar] [CrossRef] [PubMed]
- Braun, M.W.; Iwakuma, T. Regulation of cytotoxic T-cell responses by p53 in cancer. Transl. Cancer Res. 2016, 5, 692–697. [Google Scholar] [CrossRef]
- Zhu, K.; Wang, J.; Zhu, J.; Jiang, J.; Shou, J.; Chen, X. p53 induces TAP1 and enhances the transport of MHC class I peptides. Oncogene 1999, 18, 7740–7747. [Google Scholar] [CrossRef]
- Thiery, J.; Dorothée, G.; Haddada, H.; Echchakir, H.; Richon, C.; Stancou, R.; Vergnon, I.; Benard, J.; Mami-Chouaib, F.; Chouaib, S. Potentiation of a tumor cell susceptibility to autologous CTL killing by restoration of wild-type p53 function. J. Immunol. 2003, 170, 5919–5926. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Niu, D.; Lai, L.; Ren, E.C. p53 increases MHC class I expression by upregulating the endoplasmic reticulum aminopeptidase ERAP1. Nat. Commun. 2013, 4, 2359. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Li, M.; Wang, X. Cancer type-dependent correlations between TP53 mutations and antitumor immunity. DNA Repair 2020, 88, 102785. [Google Scholar] [CrossRef] [PubMed]
- Correale, P.; Aquino, A.; Giuliani, A.; Pellegrini, M.; Micheli, L.; Cusi, M.G.; Nencini, C.; Petrioli, R.; Prete, S.; De Vecchis, L.; et al. Treatment of colon and breast carcinoma cells with 5-fluorouracil enhances expression of carcinoembryonic antigen and susceptibility to HLA-A(*)02.01 restricted, CEA-peptide-specific cytotoxic T cellsin vitro. Int. J. Cancer 2003, 104, 437–445. [Google Scholar] [CrossRef]
- Helleday, T. Making immunotherapy “cold” tumours “hot” by chemotherapy-induced mutations-A misconception. Ann. Oncol. 2019, 30, 360–361. [Google Scholar] [CrossRef] [PubMed]
- Yoon, K.W.; Byun, S.; Kwon, E.; Hwang, S.-Y.; Chu, K.; Hiraki, M.; Jo, S.-H.; Weins, A.; Hakroush, S.; Cebulla, A.; et al. Control of signaling-mediated clearance of apoptotic cells by the tumor suppressor p53. Science 2015, 349, 1261669. [Google Scholar] [CrossRef]
- Agupitan, A.D.; Neeson, P.; Williams, S.; Howitt, J.; Haupt, S.; Haupt, Y. P53: A Guardian of Immunity Becomes Its Saboteur through Mutation. Int. J. Mol. Sci. 2020, 21, 3452. [Google Scholar] [CrossRef] [PubMed]
- Cortez, M.A.; Ivan, C.; Valdecanas, D.; Wang, X.; Peltier, H.J.; Ye, Y.; Araujo, L.; Carbone, D.P.; Shilo, K.; Giri, D.K.; et al. PDL1 Regulation by p53 via miR-34. J. Natl. Cancer Inst. 2016, 108. [Google Scholar] [CrossRef]
- Kim, D.W.; Haydu, L.E.; Joon, A.Y.; Bassett, R.L.; Siroy, A.E.; Tetzlaff, M.T.; Routbort, M.J.; Amaria, R.N.; Wargo, J.A.; McQuade, J.L.; et al. Clinicopathological features and clinical outcomes associated with TP53 and BRAFNon-V600 mutations in cutaneous melanoma patients. Cancer 2017, 123, 1372–1381. [Google Scholar] [CrossRef]
- Breakstone, R. Colon cancer and immunotherapy—Can we go beyond microsatellite instability? Transl. Gastroenterol. Hepatol. 2021, 6, 12. [Google Scholar] [CrossRef] [PubMed]
- Schumacher, T.N.; Schreiber, R.D. Neoantigens in cancer immunotherapy. Science 2015, 348, 69–74. [Google Scholar] [CrossRef]
- Brown, S.D.; Warren, R.L.; Gibb, E.A.; Martin, S.D.; Spinelli, J.J.; Nelson, B.H.; Holt, R.A. Neo-antigens predicted by tumor genome meta-analysis cor-relate with increased patient survival. Genome Res. 2014, 24, 743–750. [Google Scholar] [CrossRef] [PubMed]
- Malekzadeh, P.; Pasetto, A.; Robbins, P.F.; Parkhurst, M.R.; Paria, B.C.; Jia, L.; Gartner, J.J.; Hill, V.; Yu, Z.; Restifo, N.P.; et al. Neoantigen screening identifies broad TP53 mutant immu-nogenicity in patients with epithelial cancers. J. Clin. Investig. 2019, 129, 1109–1114. [Google Scholar] [CrossRef] [PubMed]
- Ciernik, I.F.; A Berzofsky, J.; Carbone, D.P. Human lung cancer cells endogenously expressing mutant p53 process and present the mutant epitope and are lysed by mutant-specific cytotoxic T lymphocytes. Clin. Cancer Res. 1996, 2, 877–882. [Google Scholar] [PubMed]
- Mandal, R.; Chan, T.A. Personalized Oncology Meets Immunology: The Path toward Precision Immunotherapy. Cancer Discov. 2016, 6, 703–713. [Google Scholar] [CrossRef]
- Kaur, H.B.; Lu, J.; Guedes, L.B.; Maldonado, L.; Reitz, L.; Barber, J.R.; De Marzo, A.M.; Tomlins, S.A.; Sfanos, K.S.; Eisenberger, M.; et al. TP53 missense mutation is associated with increased tumor-infiltrating T cells in primary prostate cancer. Hum. Pathol. 2019, 87, 95–102. [Google Scholar] [CrossRef] [PubMed]
- Lo, W.; Parkhurst, M.; Robbins, P.F.; Tran, E.; Lu, Y.-C.; Jia, L.; Gartner, J.J.; Pasetto, A.; Deniger, D.; Malekzadeh, P.; et al. Immunologic Recognition of a Shared p53 Mutated Neoantigen in a Patient with Metastatic Colorectal Cancer. Cancer Immunol. Res. 2019, 7, 534–543. [Google Scholar] [CrossRef] [PubMed]
- Sur, D.; Havasi, A.; Cainap, C.; Samasca, G.; Burz, C.; Balacescu, O.; Lupan, I.; Deleanu, D.; Irimie, A. Chimeric Antigen Receptor T-Cell Therapy for Colorectal Cancer. J. Clin. Med. 2020, 9, 182. [Google Scholar] [CrossRef] [PubMed]
- Jin, K.-T.; Chen, B.; Liu, Y.-Y.; Lan, H.U.-R.; Yan, J.-P. Monoclonal antibodies and chimeric antigen receptor (CAR) T cells in the treatment of colorectal cancer. Cancer Cell Int. 2021, 21, 1–15. [Google Scholar] [CrossRef] [PubMed]
- John, L.B.; Devaud, C.; Duong, C.P.; Yong, C.S.; Beavis, P.A.; Haynes, N.M.; Chow, M.T.; Smyth, M.J.; Kershaw, M.H.; Darcy, P.K. Anti-PD-1 Antibody Therapy Potently Enhances the Eradication of Established Tumors By Gene-Modified T Cells. Clin. Cancer Res. 2013, 19, 5636–5646. [Google Scholar] [CrossRef] [PubMed]
- Lang, H.; Baumgart, J.; Heinrich, S.; Tripke, V.; Passalaqua, M.; Maderer, A.; Galle, P.R.; Roth, W.; Kloth, M.; Moehler, M. Extended Molecular Profiling Improves Stratification and Prediction of Survival After Resection of Colorectal Liver Metastases. Ann. Surg. 2019, 270, 799–805. [Google Scholar] [CrossRef] [PubMed]
- Lang, H. ALPPS for Colorectal Liver Metastases. J. Gastrointest. Surg. 2016, 21, 190–192. [Google Scholar] [CrossRef] [PubMed]
- Chakedis, J.; Squires, M.H.; Beal, E.W.; Hughes, T.; Lewis, H.; Paredes, A.; Al-Mansour, M.; Sun, S.; Cloyd, J.M.; Pawlik, T.M. Update on current problems in colorectal liver metastasis. Curr. Probl. Surg. 2017, 54, 554–602. [Google Scholar] [CrossRef] [PubMed]
- Nordlinger, B.; Sorbye, H.; Glimelius, B.; Poston, G.J.; Schlag, P.M.; Rougier, P.; O Bechstein, W.; Primrose, J.N.; Walpole, E.T.; Finch-Jones, M.; et al. Perioperative chemotherapy with FOLFOX4 and surgery versus surgery alone for resectable liver metastases from colorectal cancer (EORTC Intergroup trial 40983): A randomised controlled trial. Lancet 2008, 371, 1007–1016. [Google Scholar] [CrossRef]
- Jones, R.P.; Jackson, R.; Dunne, D.F.J.; Malik, H.Z.; Fenwick, S.W.; Poston, G.J.; Ghaneh, P. Systematic review and meta-analysis of follow-up after hepatectomy for colorectal liver metastases. Br. J. Surg. 2012, 99, 477–486. [Google Scholar] [CrossRef]
- Tsilimigras, D.I.; Ntanasis-Stathopoulos, I.; Bagante, F.; Moris, D.; Cloyd, J.; Spartalis, E.; Pawlik, T.M. Clinical significance and prognostic relevance of KRAS, BRAF, PI3K and TP53 genetic mutation analysis for resectable and unresectable colorectal liver metastases: A systematic review of the current evidence. Surg. Oncol. 2018, 27, 280–288. [Google Scholar] [CrossRef]
- Chun, Y.S.; Passot, G.; Yamashita, S.; Nusrat, M.; Katsonis, P.; Loree, J.M.; Conrad, C.; Tzeng, C.W.D.; Xiao, L.; Aloia, T.A.; et al. Deleterious Effect of RAS and Evolutionary High-risk TP53 Double Mutation in Colorectal Liver Metastases. Ann. Surg. 2019, 269, 917–923. [Google Scholar] [CrossRef]
- Leung, M.; Gholami, S. The state of hepatic artery infusion chemotherapy in the management of metastatic colorectal cancer to the liver. Chin. Clin. Oncol. 2019, 8, 10. [Google Scholar] [CrossRef] [PubMed]
- Koerkamp, B.G.; Sadot, E.; Kemeny, N.E.; Gönen, M.; Leal, J.N.; Allen, P.J.; Cercek, A.; DeMatteo, R.P.; Kingham, T.P.; Jarnagin, W.R.; et al. Perioperative Hepatic Arterial Infusion Pump Chemotherapy Is Associated with Longer Survival After Resection of Colorectal Liver Metastases: A Propensity Score Analysis. J. Clin. Oncol. 2017, 35, 1938–1944. [Google Scholar] [CrossRef]
- Khan, Z.; Jonas, S.; Feldmann, K.; Patel, H.; Wharton, R.; Tarragona, A.; Ivison, A.; Allen-Mersh, T. p53 mutation and response to hepatic arterial floxuridine in patients with colorectal liver metastases. J. Cancer Res. Clin. Oncol. 2001, 127, 675–680. [Google Scholar] [CrossRef] [PubMed]
- Backus, H.H.J.; Van Riel, J.M.G.H.; Van Groeningen, C.J.; Vos, W.; Dukers, D.F.; Bloemena, E.; Wouters, D.; Pinedo, H.M.; Peters, G.J. Rb, mc1-1 and p53 expression correlate with clinical outcome in patients with liver metastases from colorectal cancer. Ann. Oncol. 2001, 12, 779–785. [Google Scholar] [CrossRef] [PubMed]
- Yamagishi, S.; Shimada, H.; Ishikawa, T.; Fujii, S.; Tanaka, K.; Masui, H.; Yamaguchi, S.; Ichikawa, Y.; Togo, S.; Ike, H. Expression of dihydropyrimidine dehydrogenase, thymidylate synthase, p53 and p21 in metastatic liver tumor from colorectal cancer after 5-fluorouracil-based chemotherapy. Anticancer. Res. 2005, 25, 1237–1242. [Google Scholar] [PubMed]
- Smith, J.J.; Chatila, W.K.; Sanchez-Vega, F.; Datta, J.; Connell, L.C.; Szeglin, B.C.; Basunia, A.; Boucher, T.M.; Hauser, H.; Wasserman, I.; et al. Genomic stratification beyond Ras/B-Raf in colorectal liver metastasis patients treated with hepatic arterial infusion. Cancer Med. 2019, 8, 6538–6548. [Google Scholar] [CrossRef] [PubMed]
- Warren, R.S.; Kirn, D.H. Liver-directed viral therapy for cancer p53-targeted adenoviruses and beyond. Surg. Oncol. Clin. N. Am. 2002, 11, 571–588. [Google Scholar] [CrossRef]
- Atencio, I.A.; Grace, M.; Bordens, R.; Fritz, M.; A Horowitz, J.; Hutchins, B.; Indelicato, S.; Jacobs, S.; Kolz, K.; Maneval, D.; et al. Biological activities of a recombinant adenovirus p53 (SCH 58500) administered by hepatic arterial infusion in a Phase 1 colorectal cancer trial. Cancer Gene. 2005, 13, 169–181. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, M.; Saha, S.; Bettke, J.; Nagar, R.; Parrales, A.; Iwakuma, T.; van der Velden, A.W.M.; Martinez, L.A. Mutant p53 suppresses innate immune signaling to promote tumo-rigenesis. Cancer Cell. 2021, 39, 494–508. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.Q.; Mulford, I.J.; Sharp, F.; Liang, J.; Kurtulus, S.; Trabucco, G.; Quinn, D.S.; A Longmire, T.; Patel, N.; Patil, R.; et al. Inhibition of MDM2 promotes anti-tumor responses in p53 wild-type cancer cells through their interaction with the immune and stromal microenvironment. Cancer Res. 2021. [Google Scholar] [CrossRef] [PubMed]
- Capaci, V.; Mantovani, F.; Del Sal, G. Amplifying Tumor–Stroma Communication: An Emerging Oncogenic Function of Mutant Pfront. Front. Oncol. 2021, 10. [Google Scholar] [CrossRef]
- Cordani, M.; Pacchiana, R.; Butera, G.; D’Orazi, G.; Scarpa, A.; Donadelli, M. Mutant p53 proteins alter cancer cell secretome and tumour microenvironment: Involvement in cancer invasion and metastasis. Cancer Lett. 2016, 376, 303–309. [Google Scholar] [CrossRef]
- Yeudall, W.; Vaughan, C.A.; Miyazaki, H.; Ramamoorthy, M.; Choi, M.-Y.; Chapman, C.G.; Wang, H.; Black, E.; Bulysheva, A.A.; Deb, S.P.; et al. Gain-of-function mutant p53 upregulates CXC chemokines and enhances cell migration. Carcinog. 2011, 33, 442–451. [Google Scholar] [CrossRef] [PubMed]
- Cooks, T.; Pateras, I.S.; Tarcic, O.; Solomon, H.; Schetter, A.J.; Wilder, S.; Lozano, G.; Pikarsky, E.; Forshew, T.; Rozenfeld, N.; et al. Mutant p53 Prolongs NF-κB Activation and Promotes Chronic In-flammation and Inflammation-Associated Colorectal Cancer. Cancer Cell 2013, 23, 634–646. [Google Scholar] [CrossRef] [PubMed]
- Shakya, R.; Tarulli, G.A.; Sheng, L.; A Lokman, N.; Ricciardelli, C.; I Pishas, K.; I Selinger, C.; Kohonen-Corish, M.R.J.; A Cooper, W.; Turner, A.G.; et al. Mutant p53 upregulates alpha-1 antitrypsin expression and promotes invasion in lung cancer. Oncogene 2017, 36, 4469–4480. [Google Scholar] [CrossRef]
- Neilsen, P.M.; Noll, J.E.; Suetani, R.J.; Schulz, R.B.; Al-Ejeh, F.; Evdokiou, A.; Lane, D.P.; Callen, D.F. Mutant p53 uses p63 as a molecular chaperone to alter gene expression and induce a pro-invasive secretome. Oncotarget 2011, 2, 1203–1217. [Google Scholar] [CrossRef]
- Fitzgerald, A.A.; Weiner, L.M. The role of fibroblast activation protein in health and malignancy. Cancer Metastasis Rev. 2020, 39, 783–803. [Google Scholar] [CrossRef]
- Kaps, L.; Schuppan, D. Targeting Cancer Associated Fibroblasts in Liver Fibrosis and Liver Cancer Using Nanocarriers. Cells 2020, 9, 2027. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Han, C.; Wang, S.; Fang, P.; Ma, Z.; Xu, L.; Yin, R. Cancer-associated fibroblasts: An emerging target of anti-cancer immunotherapy. J. Hematol. Oncol. 2019, 12, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Hirata, E.; Sahai, E. Tumor Microenvironment and Differential Responses to Therapy. Cold Spring Harb. Perspect. Med. 2017, 7, a026781. [Google Scholar] [CrossRef]
- Bhowmick, N.A.; Neilson, E.G.; Moses, H.L. Stromal fibroblasts in cancer initiation and progression. Nat. Cell Biol. 2004, 432, 332–337. [Google Scholar] [CrossRef]
- Piersma, B.; Hayward, M.; Weaver, V.M. Fibrosis and cancer: A strained relationship. Biochim. Biophys. Acta (BBA) Rev. Cancer 2020, 1873, 188356. [Google Scholar] [CrossRef]
- Liu, F.; Zhang, W.; Yang, F.; Feng, T.; Zhou, M.; Yu, Y.; Yu, X.; Zhao, W.; Yi, F.; Tang, W.; et al. Interleukin-6-stimulated progranulin expression contributes to the malignancy of hepatocellular carcinoma cells by activating mTOR signaling. Sci. Rep. 2016, 6, 21260. [Google Scholar] [CrossRef]
- Blagih, J.; Buck, M.D.; Vousden, K.H. p53, cancer and the immune response. J. Cell Sci. 2020, 133. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.; Lu, Z.; Takwi, A.A.L.; Chen, W.; Callander, N.S.; Ramos, K.S.; Young, K.H.; Li, Y. Negative regulation of the tumor suppressor p53 gene by microR-NAs. Oncogene 2011, 30, 843–853. [Google Scholar] [CrossRef]
- Capaci, V.; Bascetta, L.; Fantuz, M.; Beznoussenko, G.V.; Sommaggio, R.; Cancila, V.; Bisso, A.; Campaner, E.; Mironov, A.A.; Wiśniewski, J.R.; et al. Mutant p53 induces Golgi tubulo-vesiculation dri-ving a prometastatic secretome. Nat. Commun. 2020, 11, 3945. [Google Scholar] [CrossRef] [PubMed]
- Frum, R.A.; Grossman, S.R. Mechanisms of mutant p53 stabilization in cancer. In Mutant p53 and MDM2 in Cancer; Deb, S.P., Deb, S., Eds.; Springer: Dordrecht, The Netherlands, 2014; pp. 187–197. [Google Scholar]
- Bockamp, E.; Rosigkeit, S.; Siegl, D.; Schuppan, D. Nano-Enhanced Cancer Immunotherapy: Immunology Encounters Nanotechnology. Cells 2020, 9, 2102. [Google Scholar] [CrossRef]
- Mamrot, J.; Balachandran, S.; Steele, E.J.; Lindley, R.A. Molecular model linking Th2 polarized M2 tumour-associated macrophages with deaminase-mediated cancer progression mutation signatures. Scand. J. Immunol. 2019, 89, e12760. [Google Scholar] [CrossRef]
- Riabov, V.; Gudima, A.; Wang, N.; Mickley, A.; Orekhov, A.; Kzhyshkowska, J. Role of tumor associated macrophages in tumor angiogenesis and lymphangiogenesis. Front. Physiol. 2014, 5, 75. [Google Scholar] [CrossRef] [PubMed]
- Haist, M.; Stege, H.; Grabbe, S.; Bros, M. The Functional Crosstalk between Myeloid-Derived Suppressor Cells and Regulatory T Cells within the Immunosuppressive Tumor Microenvironment. Cancers 2021, 13, 210. [Google Scholar] [CrossRef]
- Taniyama, D.; Taniyama, K.; Kuraoka, K.; Yamamoto, H.; Zaitsu, J.; Saito, A.; Sakamoto, N.; Sentani, K.; Oue, N.; Yasui, W. CD204-Positive Tumor-associated Macrophages Relate to Malignant Transformation of Colorectal Adenoma. Anticancer. Res. 2019, 39, 2767–2775. [Google Scholar] [CrossRef]
- Kang, J.-C.; Chen, J.-S.; Lee, C.-H.; Chang, J.-J.; Shieh, Y.-S. Intratumoral macrophage counts correlate with tumor progression in colorectal cancer. J. Surg. Oncol. 2010, 102, 242–248. [Google Scholar] [CrossRef] [PubMed]
- Edin, S.; Wikberg, M.L.; Oldenborg, P.-A.; Palmqvist, R. Macrophages. Oncoimmunology 2013, 2, e23038. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.; Zhu, Y.; Han, L.; Kim, A.L.; Kopelovich, L.; Bickers, D.R.; Athar, M. CP-31398 restores mutant p53 tumor suppressor function and inhibits UVB-induced skin carcinogenesis in mice. J. Clin. Investig. 2007, 117, 3753–3764. [Google Scholar] [CrossRef] [PubMed]
- Bykov, V.J.N.; Issaeva, N.; Shilov, A.; Hultcrantz, M.; Pugacheva, E.; Chumakov, P.; Bergman, J.; Wiman, K.G.; Selivanova, G. Restoration of the tumor suppressor function to mu-tant p53 by a low-molecular-weight compound. Nat. Med. 2002, 8, 282–288. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Bykov, V.J.N.; Wiman, K.G.; Zawacka-Pankau, J. Correction: APR-246 reactivates mutant p53 by targeting cysteines 124 and 277. Cell Death Dis. 2019, 10, 1–2. [Google Scholar] [CrossRef] [PubMed]
- Zhu, G.; Pan, C.; Bei, J.-X.; Li, B.; Liang, C.; Xu, Y.; Fu, X. Mutant p53 in Cancer Progression and Targeted Therapies. Front. Oncol. 2020, 10. [Google Scholar] [CrossRef] [PubMed]
- Boeckler, F.M.; Joerger, A.C.; Jaggi, G.; Rutherford, T.J.; Veprintsev, D.B.; Fersht, A.R. Targeted rescue of a destabilized mutant of p53 by an in si-lico screened drug. Proc. Natl. Acad. Sci. USA 2008, 105, 10360–10365. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Wilcken, R.; Joerger, A.C.; Chuckowree, I.S.; Amin, J.; Spencer, J.; Fersht, A.R. Small molecule induced reactivation of mutant p53 in cancer cells. Nucleic Acids Res. 2013, 41, 6034–6044. [Google Scholar] [CrossRef]
- Puca, R.; Nardinocchi, L.; Porru, M.; Simon, A.J.; Rechavi, G.; Leonetti, C.; Givol, D.; D’Orazi, G. Restoring p53 active conformation by zinc increases the response of mutant p53 tumor cells to anticancer drugs. Cell Cycle 2011, 10, 1679–1689. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Vazquez, A.; Levine, A.J.; Carpizo, D.R. Allele-Specific p53 Mutant Reactivation. Cancer Cell. 2012, 21, 614–625. [Google Scholar] [CrossRef] [PubMed]
- Salim, K.Y.; Maleki Vareki, S.; Danter, W.R.; San-Marina, S.; Koropatnick, J. COTI-2, a novel small molecule that is active against multiple hu-man cancer cell lines in vitro and in vivo. Oncotarget 2016, 7, 41363–41379. [Google Scholar] [CrossRef] [PubMed]
- Weinmann, L.; Wischhusen, J.; Demma, M.J.; Naumann, U.; Roth, P.; DasMahapatra, B.; Weller, M. A novel p53 rescue compound induces p53-dependent growth arrest and sensitises glioma cells to Apo2L/TRAIL-induced apoptosis. Cell Death Differ. 2008, 15, 718–729. [Google Scholar] [CrossRef] [PubMed]
- Demma, M.; Maxwell, E.; Ramos, R.; Liang, L.; Li, C.; Hesk, D.; Rossman, R.; Mallams, A.; Doll, R.; Liu, M.; et al. SCH529074, a Small Molecule Activator of Mutant p53, Which Binds p53 DNA Binding Domain (DBD), Restores Growth-suppressive Function to Mutant p53 and Interrupts HDM2-mediated Ubiquitination of Wild Type p53. J. Biol. Chem. 2010, 285, 10198–10212. [Google Scholar] [CrossRef] [PubMed]
- Schulz-Heddergott, R.; Stark, N.; Edmunds, S.J.; Li, J.; Conradi, L.-C.; Bohnenberger, H.; Ceteci, F.; Greten, F.R.; Dobbelstein, M.; Moll, U.M. Therapeutic Ablation of Gain-of-Function Mutant p53 in Colorectal Cancer Inhibits Stat3-Mediated Tumor Growth and Invasion. Cancer Cell 2018, 34, 298–314.e7. [Google Scholar] [CrossRef] [PubMed]
- Alexandrova, E.M.; Yallowitz, A.R.; Li, D.; Xu, S.; Schulz, R.; A Proia, D.; Lozano, G.; Dobbelstein, M.; Moll, U.M. Improving survival by exploiting tumour dependence on stabilized mutant p53 for treatment. Nat. Cell Biol. 2015, 523, 352–356. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Marchenko, N.; Moll, U.M. SAHA shows preferential cytotoxicity in mutant p53 cancer cells by destabilizing mutant p53 through inhibition of the HDAC6-Hsp90 chaperone axis. Cell Death Differ. 2011, 18, 1904–1913. [Google Scholar] [CrossRef]
- Pushpakom, S.; Iorio, F.; Eyers, P.A.; Escott, K.J.; Hopper, S.; Wells, A.; Doig, A.; Guilliams, T.; Latimer, J.; McNamee, C.; et al. Drug repurposing: Progress, challenges and recommendations. Nat. Rev. Drug Discov. 2019, 18, 41–58. [Google Scholar] [CrossRef]
- Freed-Pastor, W.A.; Mizuno, H.; Zhao, X.; Langerød, A.; Moon, S.-H.; Rodriguez-Barrueco, R.; Barsotti, A.; Chicas, A.; Li, W.; Polotskaia, A.; et al. Mutant p53 Disrupts Mammary Tissue Architecture via the Mevalonate Pathway. Cell 2012, 148, 244–258. [Google Scholar] [CrossRef]
- Sorrentino, G.; Ruggeri, N.; Specchia, V.; Cordenonsi, M.; Mano, M.; Dupont, S.; Manfrin, A.; Ingallina, E.; Sommaggio, R.; Piazza, S.; et al. Metabolic control of YAP and TAZ by the mevalonate pathway. Nat. Cell Biol. 2014, 16, 357–366. [Google Scholar] [CrossRef]
- Kaps, L.; Nuhn, L.; Aslam, M.; Brose, A.; Foerster, F.; Rosigkeit, S.; Renz, P.; Heck, R.; Kim, Y.O.; Lieberwirth, I.; et al. In Vivo Gene-Silencing in Fibrotic Liver by siRNA-Loaded Cationic Nanohydrogel Particles. Adv. Healthc. Mater. 2015, 4, 2809–2815. [Google Scholar] [CrossRef] [PubMed]
- Leber, N.; Kaps, L.; Aslam, M.; Schupp, J.; Brose, A.; Schäffel, D.; Fischer, K.; Diken, M.; Strand, D.; Koynov, K.; et al. SiRNA-mediated in vivo gene knockdown by acid-degradable cationic nanohydrogel particles. J. Control. Release. 2017, 248, 10–23. [Google Scholar] [CrossRef]
- Leber, N.; Kaps, L.; Yang, A.; Aslam, M.; Giardino, M.; Klefenz, A.; Choteschovsky, N.; Rosigkeit, S.; Mostafa, A.; Nuhn, L.; et al. α-Mannosyl-Functionalized Cationic Nanohydrogel Particles for Targeted Gene Knockdown in Immunosuppressive Macrophages. Macromol. Biosci. 2019, 19, 1900162. [Google Scholar] [CrossRef] [PubMed]
- Kaps, L.; Leber, N.; Klefenz, A.; Choteschovsky, N.; Zentel, R.; Nuhn, L.; Schuppan, D. In Vivo siRNA Delivery to Immunosuppressive Liver Macropha-ges by α-Mannosyl-Functionalized Cationic Nanohydrogel Particles. Cells 2020, 9, 1905. [Google Scholar] [CrossRef] [PubMed]
- Hsiue, E.H.-C.; Wright, K.M.; Douglass, J.; Hwang, M.S.; Mog, B.J.; Pearlman, A.H.; Paul, S.; DiNapoli, S.R.; Konig, M.F.; Wang, Q.; et al. Targeting a neoantigen derived from a common TP53 mu-tation. Science 2021, 371, eabc8697. [Google Scholar] [CrossRef] [PubMed]
| Drug | Type of Drug | Mechanism | Stage of Development | Reference | |
|---|---|---|---|---|---|
| 1. | APR-246 | Small molecule | Restores wild-type conformation | Clinical phase II | [177,178,179] |
| CP-31398 | Small molecule | Restores wild-type conformation | Preclinical | [26,180] | |
| 2. | PK083 | Small molecule | Restores wild-type conformation | Preclinical | [26,180] |
| 3. | PK7088 | Small molecule | Restores wild-type conformation | Preclinical | [26,184] |
| 4. | Zinc | Cofactor | Restores wild-type conformation | Preclinical | [26] |
| 5. | ZMC-1 | Zn2+-chelating compounds | Restores wild-type conformation | Preclinical | [183] |
| 6. | COTI-2 | Zn2+-chelating compounds | Restores wild-type conformation | Clinical phase I | [184] |
| 7. | P53R3 | Small molecule | Restores DNA-binding ability | Preclinical | [26] |
| 8. | SCH529074 | Small molecule | Restores DNA-binding ability and prevents degradation of p53 | Preclinical | [186] |
| 9. | Hsp90 inhibitor | Small molecule | Depletion of p53 mutants | Preclinical | [187] |
| 10. | Ganetespib (potent Hsp90 inhibitor) | Small molecule | Depletion of p53 mutants | Discontinuation of clinical phase I | [188] |
| SAHA | Small molecule | Depletion of p53 mutants | Preclinical | [189] | |
| 11. | Statins | Small molecule | Inhibition of p53 mutant related downstream targets | Preclinical | [191,192] |
| 12. | siRNA | RNA based therapy | Specific knockdown of p53 mutants | No data yet available | |
| 13. | H2 | Bispecific antibody | Specific targeting of TP53R175H | Preclinical | [197] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Michel, M.; Kaps, L.; Maderer, A.; Galle, P.R.; Moehler, M. The Role of p53 Dysfunction in Colorectal Cancer and Its Implication for Therapy. Cancers 2021, 13, 2296. https://doi.org/10.3390/cancers13102296
Michel M, Kaps L, Maderer A, Galle PR, Moehler M. The Role of p53 Dysfunction in Colorectal Cancer and Its Implication for Therapy. Cancers. 2021; 13(10):2296. https://doi.org/10.3390/cancers13102296
Chicago/Turabian StyleMichel, Maurice, Leonard Kaps, Annett Maderer, Peter R. Galle, and Markus Moehler. 2021. "The Role of p53 Dysfunction in Colorectal Cancer and Its Implication for Therapy" Cancers 13, no. 10: 2296. https://doi.org/10.3390/cancers13102296
APA StyleMichel, M., Kaps, L., Maderer, A., Galle, P. R., & Moehler, M. (2021). The Role of p53 Dysfunction in Colorectal Cancer and Its Implication for Therapy. Cancers, 13(10), 2296. https://doi.org/10.3390/cancers13102296





