Single-Cell RNA-seq Reveals Obesity-Induced Alterations in the Brca1-Mutated Mammary Gland Microenvironment
Abstract
1. Introduction
2. Results
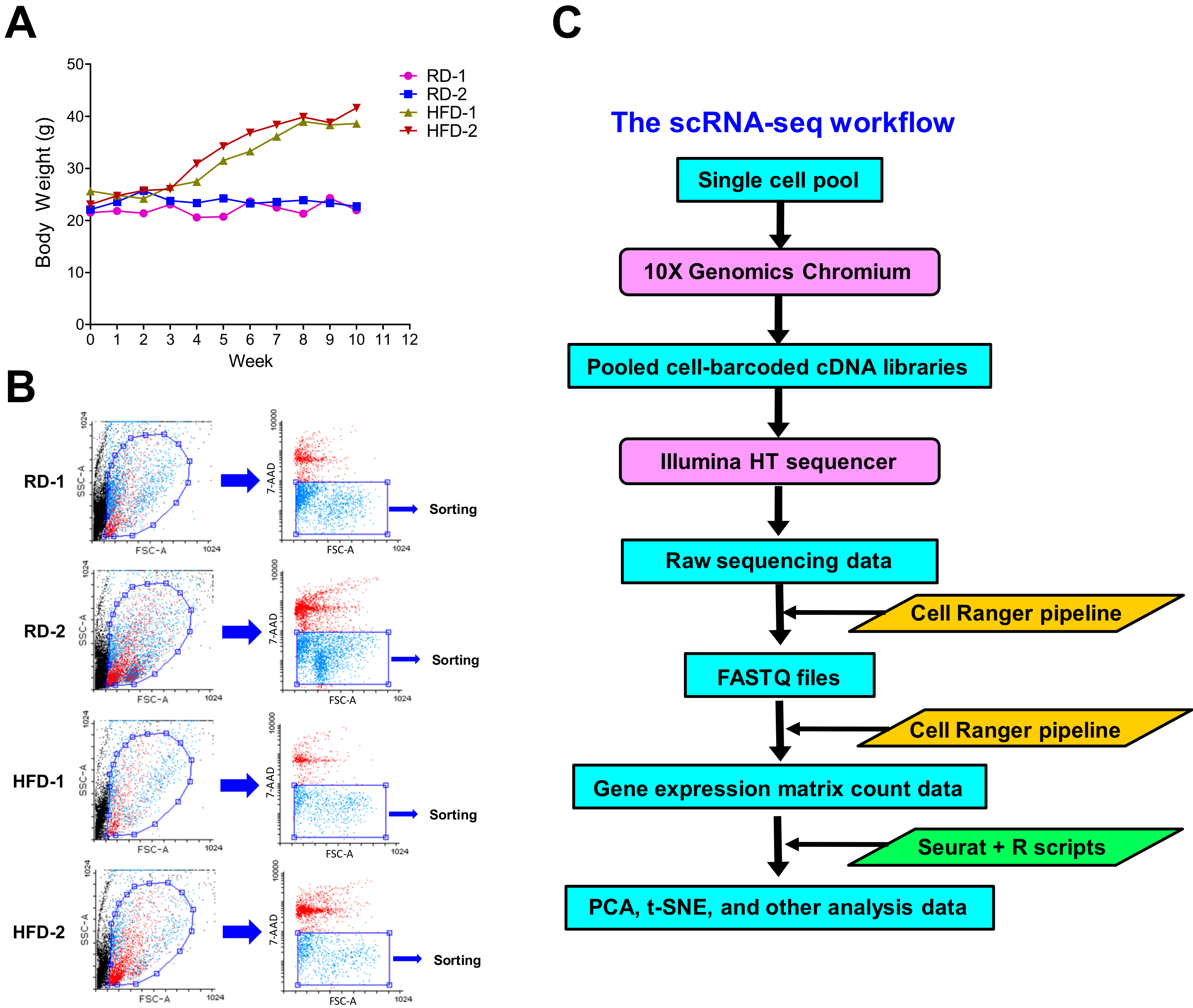
2.1. High-Fat Diets Induce Obesity in Brca1−/−; p53+/− Mice for scRNA-seq Analysis
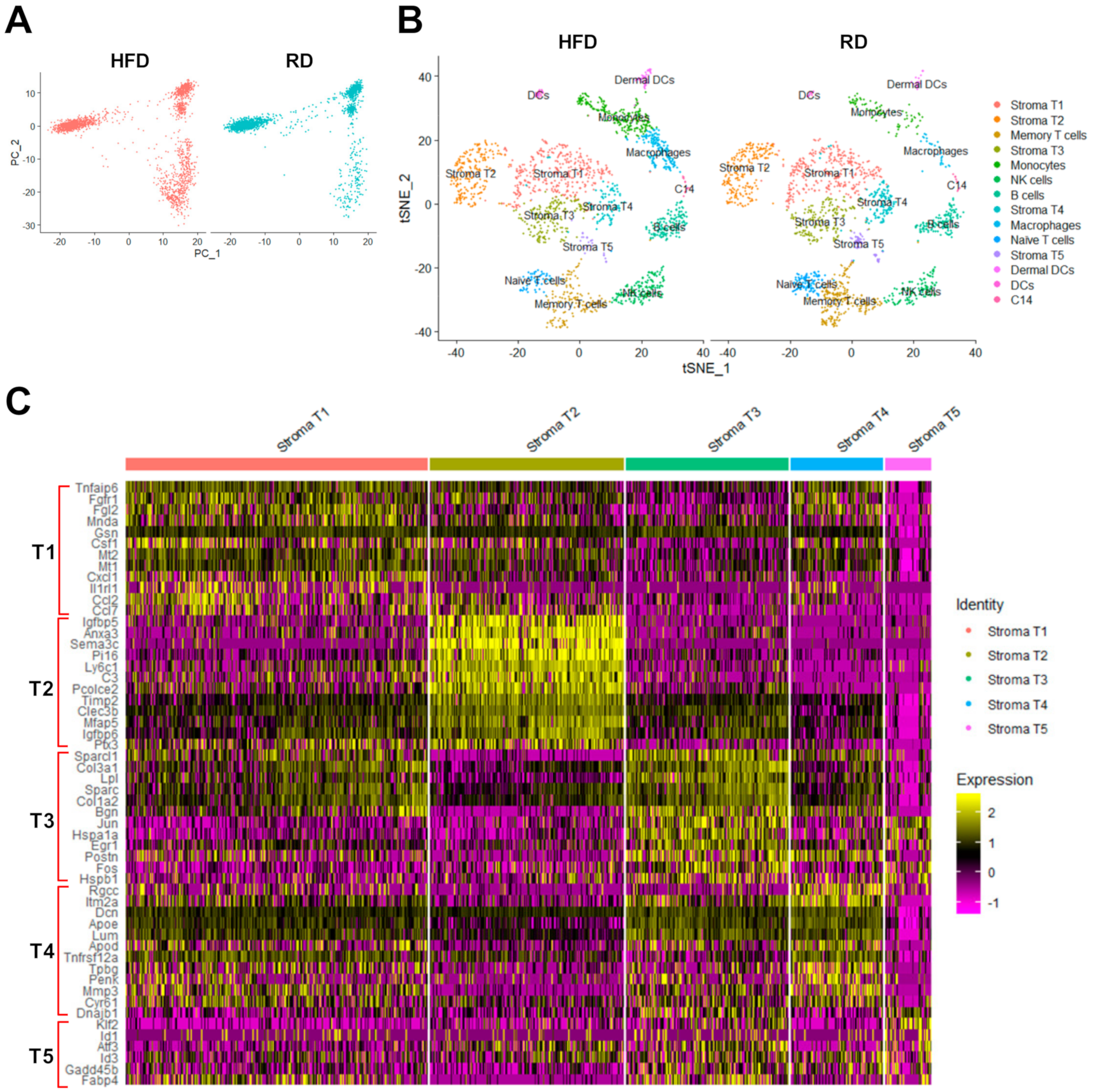
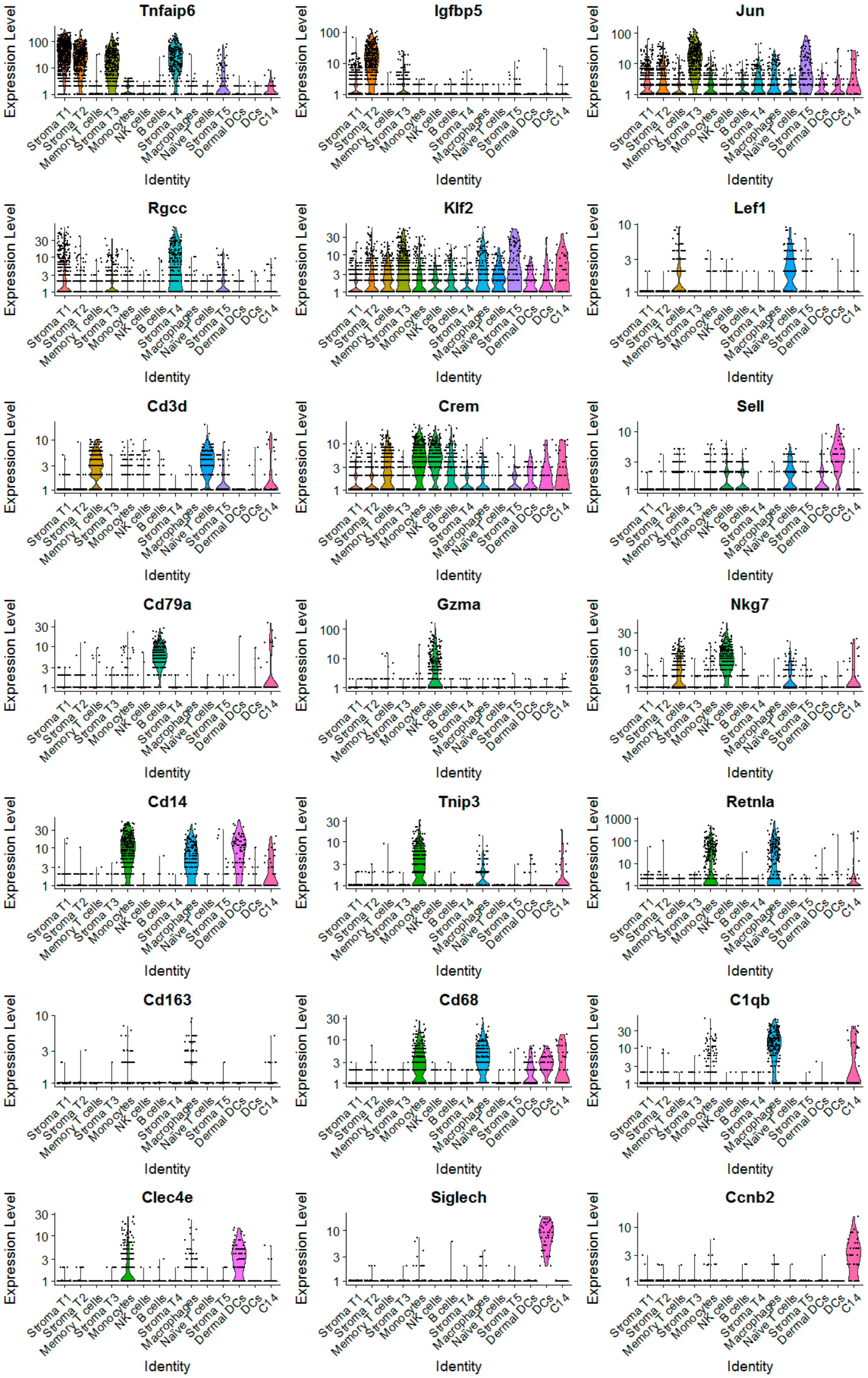
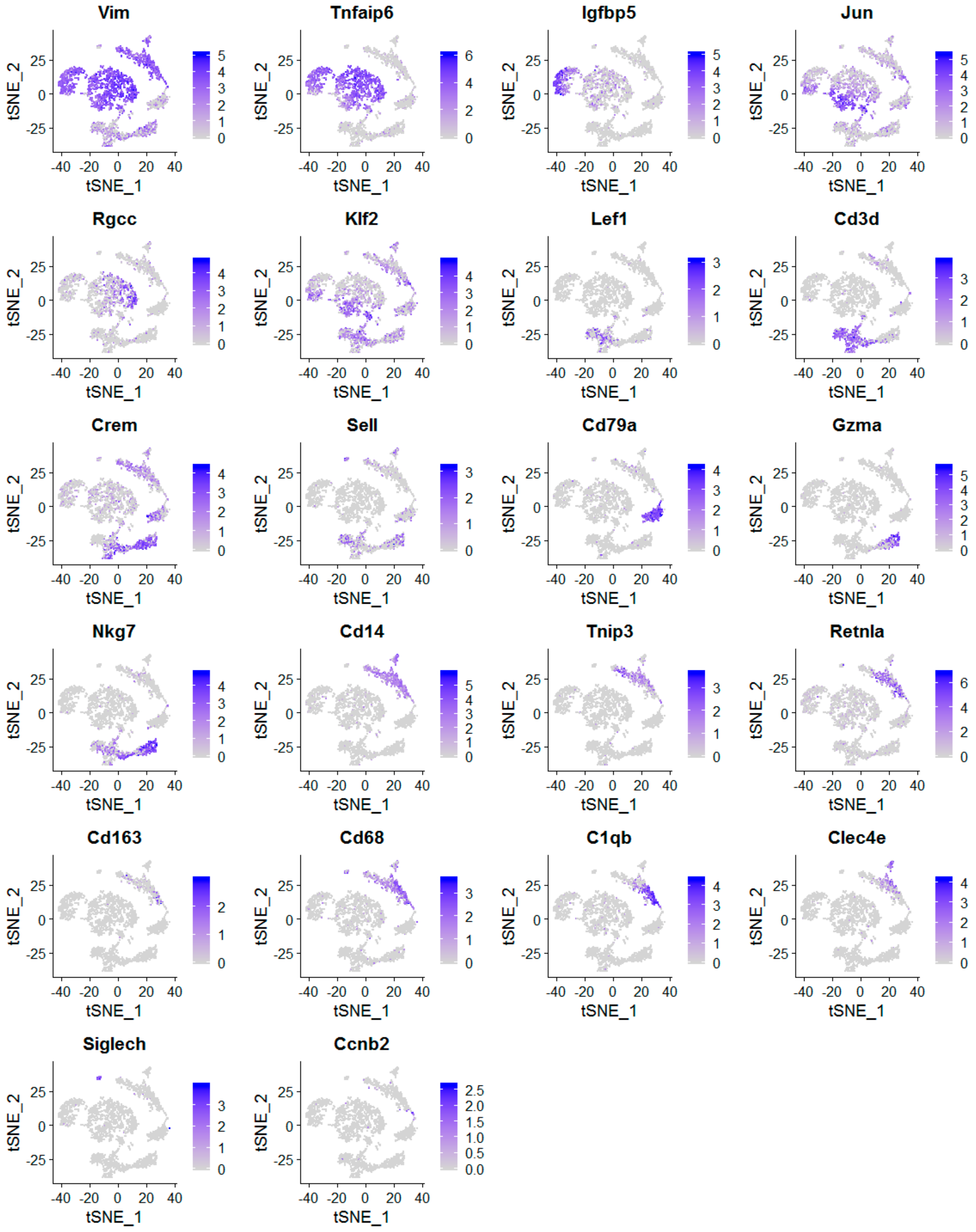
2.2. The scRNA-Seq Analysis Identifies Various Molecular Subtypes of Mammary Stromal Fibroblasts in Brca1−/−; p53+/− mice
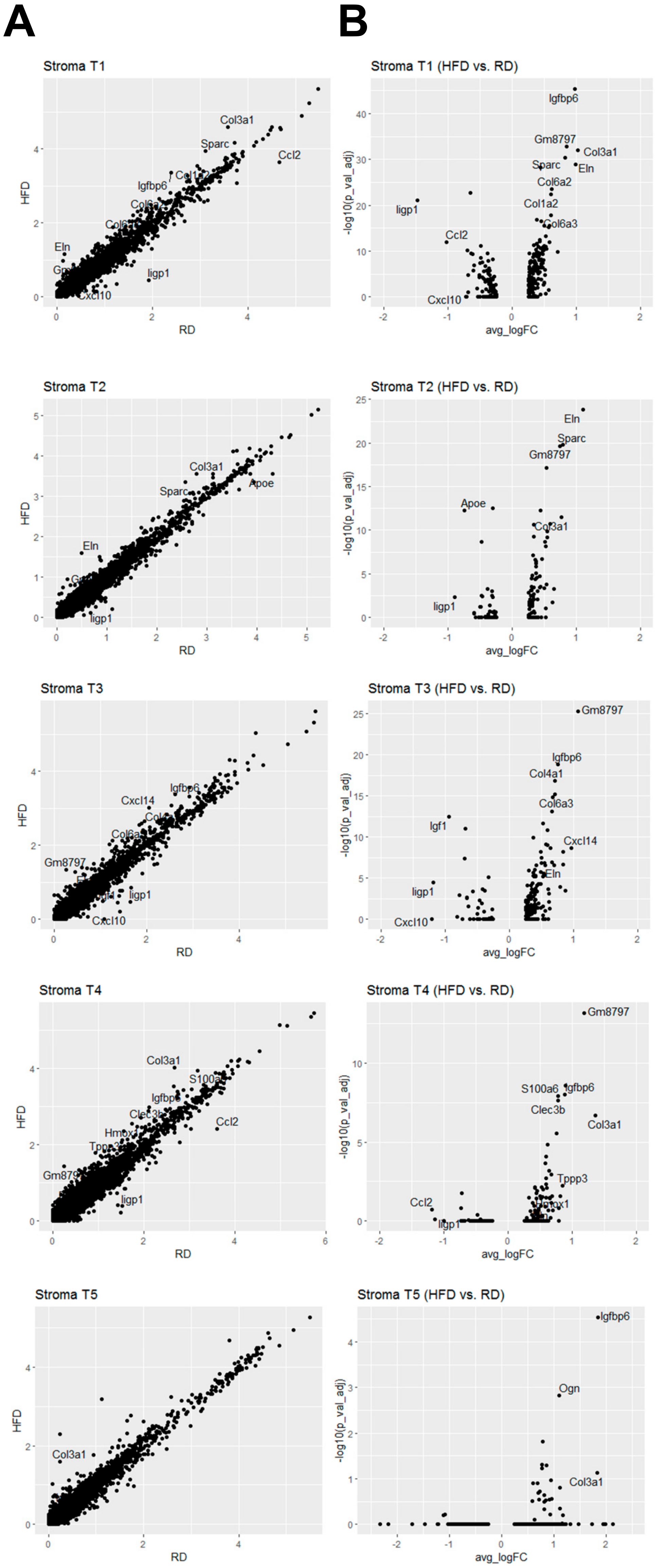
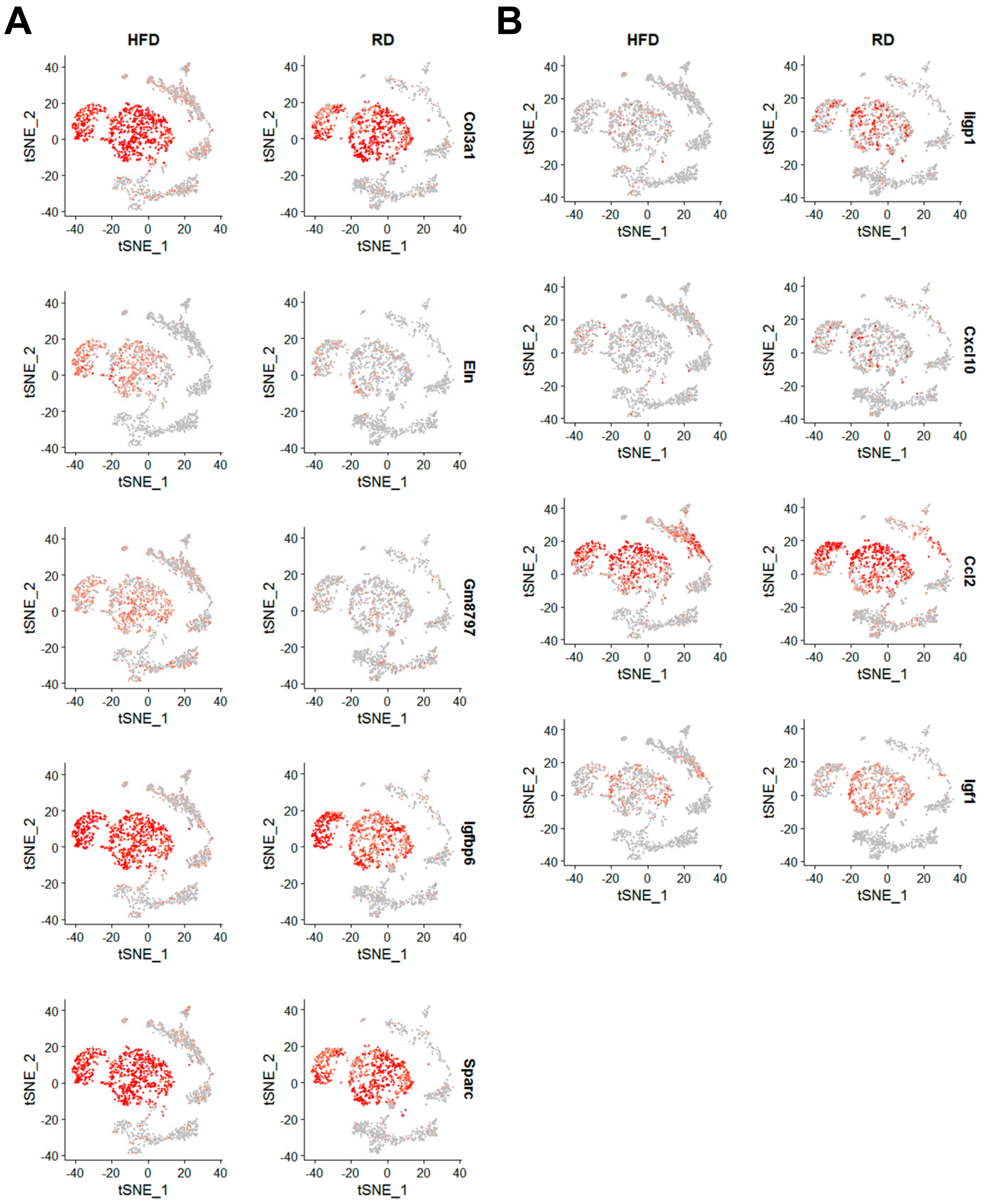
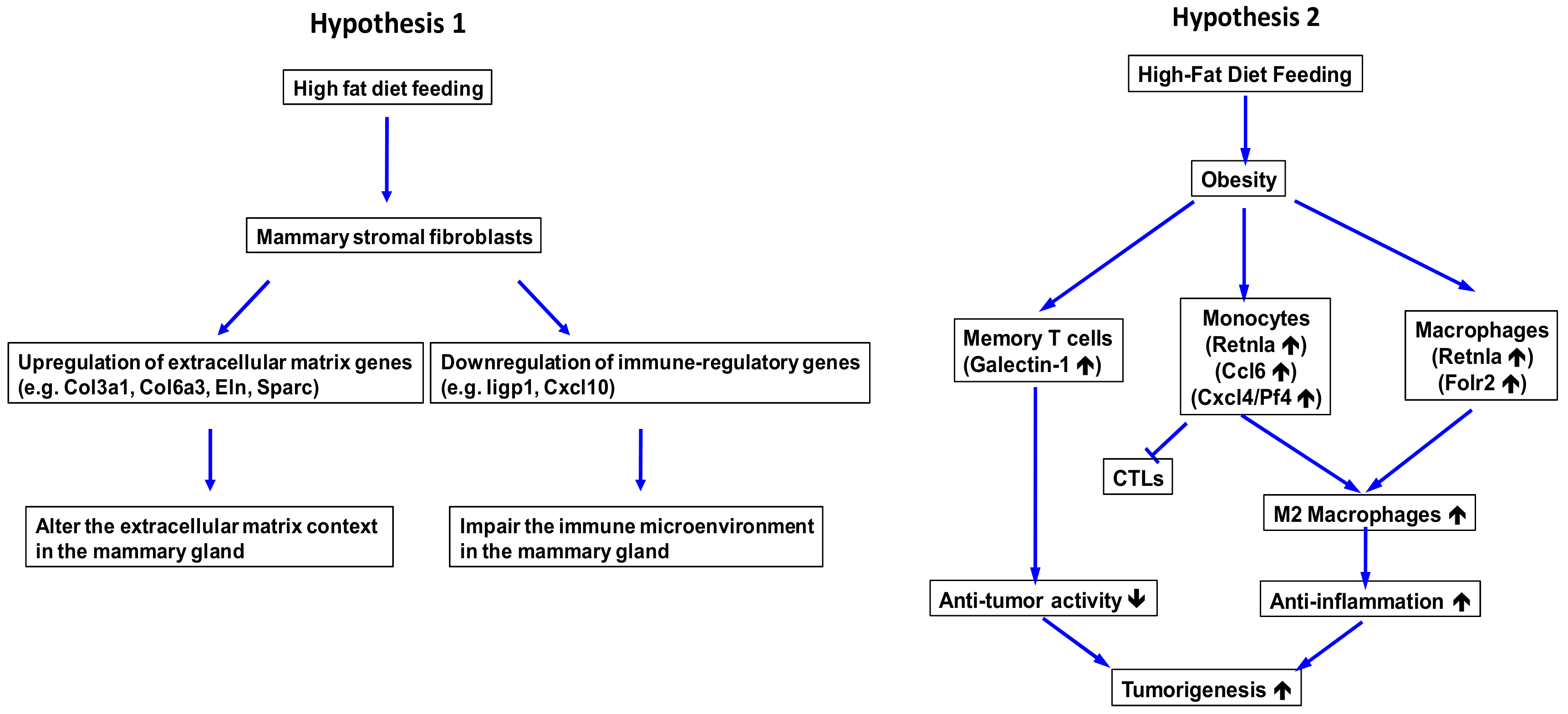
2.3. Obesity Aberrantly Modulates the Transcriptomic Landscape of Brca1−/−; p53+/− Mammary Stromal Fibroblasts to Alter their Functions in the Extracellular Matrix and Immunological Regulation
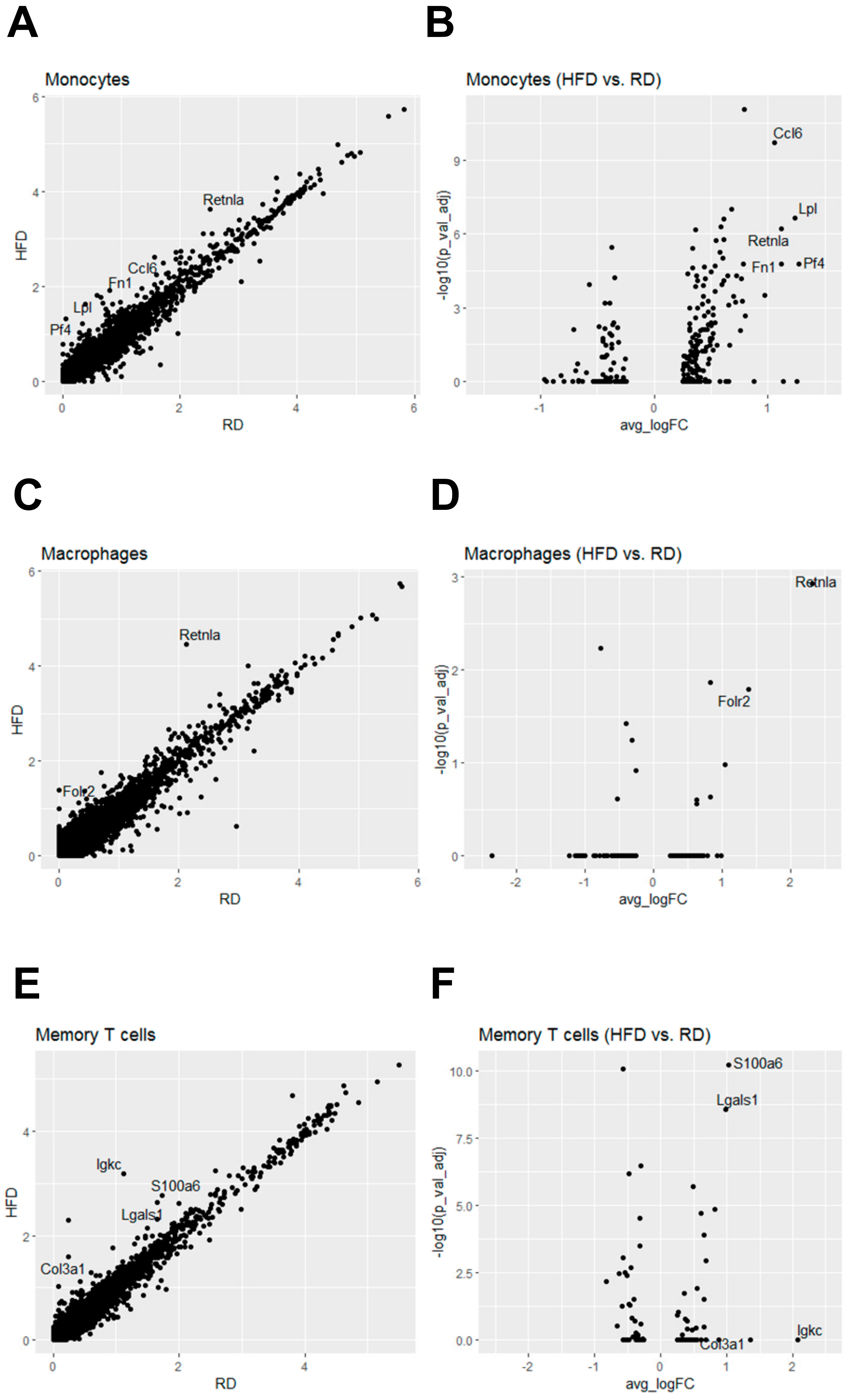
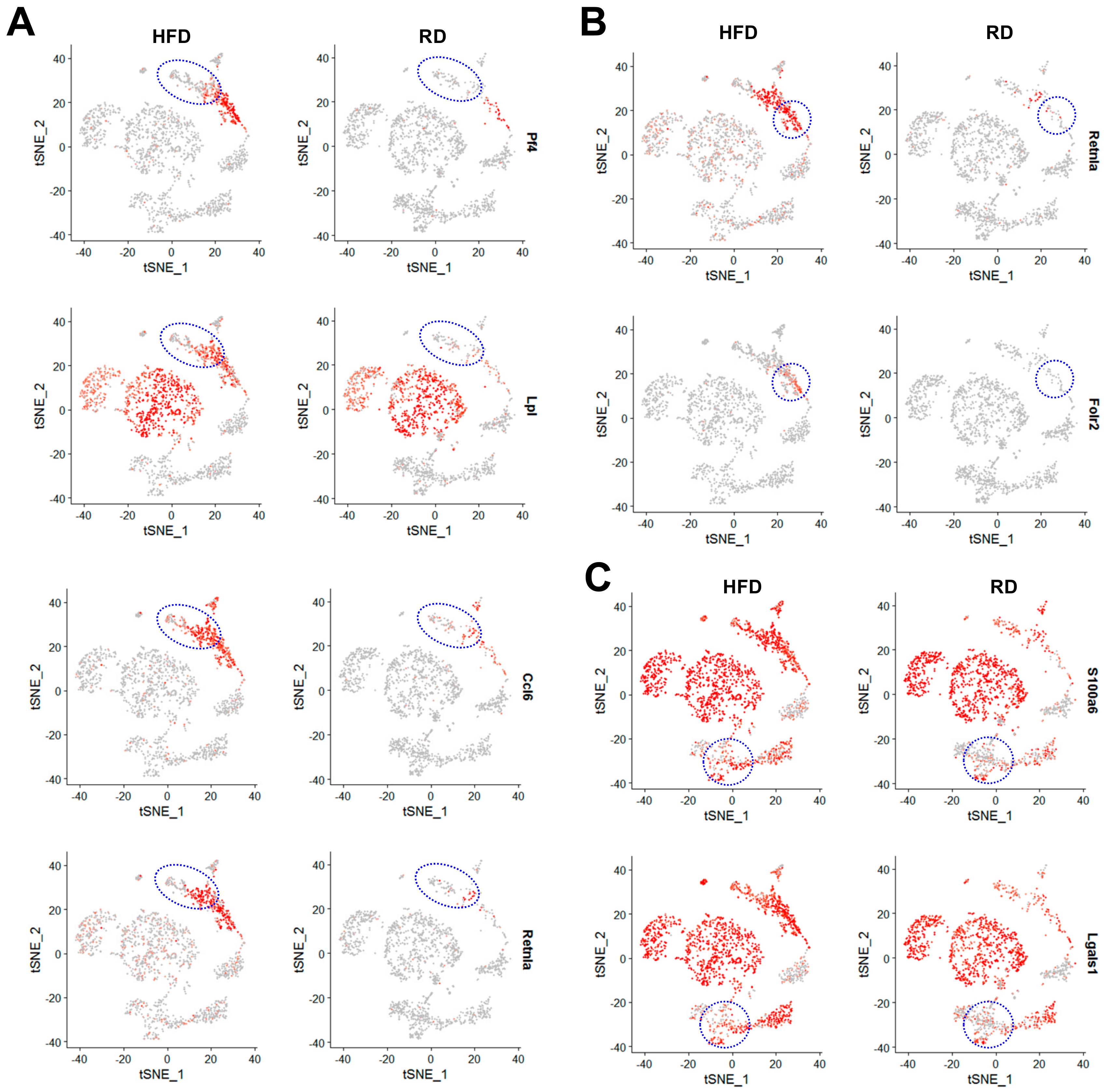
2.4. Obesity Aberrantly Regulates the Immune Ecosystem within Brca1−/−; p53+/− Mammary Glands to Establish a Tumor-Favorable Tissue Microenvironment
3. Discussion
4. Materials and Methods
4.1. HFD-Induced Obesity in Brca1−/−; p53+/− Mice
4.2. Single-Cell Isolation of Mammary Stromal and Immune Cells
4.3. Single-Cell RNA Sequencing and Data Analysis
4.4. Statistics and Reproducibility
5. Conclusions
DVA/US Government Disclaimer
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Goodwin, P.J.; Stambolic, V. Impact of the obesity epidemic on cancer. Annu. Rev. Med. 2015, 66, 281–296. [Google Scholar] [CrossRef]
- Sung, H.; Siegel, R.L.; Torre, L.A.; Pearson-Stuttard, J.; Islami, F.; Fedewa, S.A.; Goding Sauer, A.; Shuval, K.; Gapstur, S.M.; Jacobs, E.J.; et al. Global patterns in excess body weight and the associated cancer burden. CA Cancer J. Clin. 2019, 69, 88–112. [Google Scholar] [CrossRef] [PubMed]
- Khandekar, M.J.; Cohen, P.; Spiegelman, B.M. Molecular mechanisms of cancer development in obesity. Nat. Rev. Cancer 2011, 11, 886–895. [Google Scholar] [CrossRef] [PubMed]
- Carmichael, A.R. Obesity and prognosis of breast cancer. Obes. Rev. 2006, 7, 333–340. [Google Scholar] [CrossRef]
- Brown, K.A.; Simpson, E.R. Obesity and breast cancer: Progress to understanding the relationship. Cancer Res. 2010, 70, 4–7. [Google Scholar] [CrossRef] [PubMed]
- Jain, R.; Strickler, H.D.; Fine, E.; Sparano, J.A. Clinical studies examining the impact of obesity on breast cancer risk and prognosis. J. Mammary Gland Biol. Neoplasia 2013, 18, 257–266. [Google Scholar] [CrossRef] [PubMed]
- James, F.R.; Wootton, S.; Jackson, A.; Wiseman, M.; Copson, E.R.; Cutress, R.I. Obesity in breast cancer—What is the risk factor? Eur. J. Cancer 2015, 51, 705–720. [Google Scholar] [CrossRef]
- Agurs-Collins, T.; Ross, S.A.; Dunn, B.K. The Many Faces of Obesity and Its Influence on Breast Cancer Risk. Front. Oncol. 2019, 9, 765. [Google Scholar] [CrossRef]
- Dirat, B.A.; Bochet, L.; Escourrou, G.; Valet, P.; Muller, C. Unraveling the obesity and breast cancer links: A role for cancer-associated adipocytes? Endocr. Dev. 2010, 19, 45–52. [Google Scholar]
- Simpson, E.R.; Brown, K.A. Minireview: Obesity and breast cancer: A tale of inflammation and dysregulated metabolism. Mol. Endocrinol. 2013, 27, 715–725. [Google Scholar] [CrossRef]
- Bertolini, F.; Orecchioni, S.; Petit, J.Y.; Kolonin, M.G. Obesity, proinflammatory mediators, adipose tissue progenitors, and breast cancer. Curr. Opin. Oncol. 2014, 26, 545–550. [Google Scholar] [CrossRef] [PubMed]
- Rose, D.P.; Gracheck, P.J.; Vona-Davis, L. The Interactions of Obesity, Inflammation and Insulin Resistance in Breast Cancer. Cancers 2015, 7, 2147–2168. [Google Scholar] [CrossRef] [PubMed]
- Feola, A.; Ricci, S.; Kouidhi, S.; Rizzo, A.; Penon, A.; Formisano, P.; Giordano, A.; Di Carlo, A.; Di Domenico, M. Multifaceted Breast Cancer: The Molecular Connection With Obesity. J. Cell Physiol. 2017, 232, 69–77. [Google Scholar] [CrossRef] [PubMed]
- Place, A.E.; Jin Huh, S.; Polyak, K. The microenvironment in breast cancer progression: Biology and implications for treatment. Breast Cancer Res. 2011, 13, 227. [Google Scholar] [CrossRef] [PubMed]
- Artacho-Cordón, A.; Artacho-Cordón, F.; Ríos-Arrabal, S.; Calvente, I.; Núñez, M.I. Tumor microenvironment and breast cancer progression: A complex scenario. Cancer Biol. Ther. 2012, 13, 14–24. [Google Scholar] [CrossRef]
- Mitra, S.; Stemke-Hale, K.; Mills, G.B.; Claerhout, S. Interactions between tumor cells and microenvironment in breast cancer: A new opportunity for targeted therapy. Cancer Sci. 2012, 103, 400–407. [Google Scholar] [CrossRef]
- Lim, B.; Woodward, W.A.; Wang, X.; Reuben, J.M.; Ueno, N.T. Inflammatory breast cancer biology: The tumour microenvironment is key. Nat. Rev. Cancer 2018, 18, 485–499. [Google Scholar] [CrossRef]
- Korkaya, H.; Liu, S.; Wicha, M.S. Breast cancer stem cells, cytokine networks, and the tumor microenvironment. J. Clin. Investig. 2011, 121, 3804–3809. [Google Scholar] [CrossRef]
- Mao, Y.; Keller, E.T.; Garfield, D.H.; Shen, K.; Wang, J. Stromal cells in tumor microenvironment and breast cancer. Cancer Metastasis Rev. 2013, 32, 303–315. [Google Scholar] [CrossRef]
- Houthuijzen, J.M.; Jonkers, J. Cancer-associated fibroblasts as key regulators of the breast cancer tumor microenvironment. Cancer Metastasis Rev. 2018, 37, 577–597. [Google Scholar] [CrossRef]
- Bhat, V.; Allan, A.L.; Raouf, A. Role of the Microenvironment in Regulating Normal and Cancer Stem Cell Activity: Implications for Breast Cancer Progression and Therapy Response. Cancers 2019, 11, 1240. [Google Scholar] [CrossRef] [PubMed]
- Tower, H.; Ruppert, M.; Britt, K. The Immune Microenvironment of Breast Cancer Progression. Cancers 2019, 11, 1375. [Google Scholar] [CrossRef] [PubMed]
- Sundaram, S.; Johnson, A.R.; Makowski, L. Obesity, metabolism and the microenvironment: Links to cancer. J. Carcinog 2013, 12, 19. [Google Scholar] [PubMed]
- Iyengar, N.M.; Gucalp, A.; Dannenberg, A.J.; Hudis, C.A. Obesity and Cancer Mechanisms: Tumor Microenvironment and Inflammation. J. Clin. Oncol. 2016, 34, 4270–4276. [Google Scholar] [CrossRef]
- Olson, O.C.; Quail, D.F.; Joyce, J.A. Obesity and the tumor microenvironment. Science 2017, 358, 1130–1131. [Google Scholar] [CrossRef]
- Huh, J.Y.; Park, Y.J.; Ham, M.; Kim, J.B. Crosstalk between adipocytes and immune cells in adipose tissue inflammation and metabolic dysregulation in obesity. Mol. Cells 2014, 37, 365–371. [Google Scholar] [CrossRef]
- Maurizi, G.; Della Guardia, L.; Maurizi, A.; Poloni, A. Adipocytes properties and crosstalk with immune system in obesity-related inflammation. J. Cell Physiol. 2018, 233, 88–97. [Google Scholar] [CrossRef]
- Xu, X.; Wagner, K.U.; Larson, D.; Weaver, Z.; Li, C.; Ried, T.; Hennighausen, L.; Wynshaw-Boris, A.; Deng, C.X. Conditional mutation of Brca1 in mammary epithelial cells results in blunted ductal morphogenesis and tumour formation. Nat. Genet. 1999, 22, 37–43. [Google Scholar] [CrossRef]
- Brodie, S.G.; Xu, X.; Qiao, W.; Li, W.M.; Cao, L.; Deng, C.X. Multiple genetic changes are associated with mammary tumorigenesis in Brca1 conditional knockout mice. Oncogene 2001, 20, 7514–7523. [Google Scholar] [CrossRef]
- Zhang, X.; Li, T.; Liu, F.; Chen, Y.; Yao, J.; Li, Z.; Huang, Y.; Wang, J. Comparative Analysis of Droplet-Based Ultra-High-Throughput Single-Cell RNA-Seq Systems. Mol. Cells 2019, 73, 130–142. [Google Scholar] [CrossRef]
- Ferguson, A.; Chen, K. Analysis of Transcriptional Profiling of Immune Cells at the Single-Cell Level. Methods Mol. Biol. 2020, 2111, 47–57. [Google Scholar] [PubMed]
- Satija, R.; Farrell, J.A.; Gennert, D.; Schier, A.F.; Regev, A. Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 2015, 33, 495–502. [Google Scholar] [CrossRef]
- Butler, A.; Hoffman, P.; Smibert, P.; Papalexi, E.; Satija, R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 2018, 36, 411–420. [Google Scholar] [CrossRef] [PubMed]
- Harman, A.N.; Bye, C.R.; Nasr, N.; Sandgren, K.J.; Kim, M.; Mercier, S.K.; Botting, R.A.; Lewin, S.R.; Cunningham, A.L.; Cameron, P.U. Identification of lineage relationships and novel markers of blood and skin human dendritic cells. J. Immunol. 2013, 190, 66–79. [Google Scholar] [CrossRef] [PubMed]
- Takagi, H.; Fukaya, T.; Eizumi, K.; Sato, Y.; Sato, K.; Shibazaki, A.; Otsuka, H.; Hijikata, A.; Watanabe, T.; Ohara, O.; et al. Plasmacytoid dendritic cells are crucial for the initiation of inflammation and T cell immunity in vivo. Immunity 2011, 35, 958–971. [Google Scholar] [CrossRef] [PubMed]
- Naba, A.; Clauser, K.R.; Hoersch, S.; Liu, H.; Carr, S.A.; Hynes, R.O. The matrisome: In silico definition and in vivo characterization by proteomics of normal and tumor extracellular matrices. Mol. Cell Proteom. 2012, 11, M111.014647. [Google Scholar] [CrossRef]
- Fabregat, A.; Sidiropoulos, K.; Viteri, G.; Marin-Garcia, P.; Ping, P.; Stein, L.; D’Eustachio, P.; Hermjakob, H. Reactome diagram viewer: Data structures and strategies to boost performance. Bioinformatics 2018, 34, 1208–1214. [Google Scholar] [CrossRef]
- Eden, E.; Navon, R.; Steinfeld, I.; Lipson, D.; Yakhini, Z. GOrilla: A tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinform. 2009, 10, 48. [Google Scholar] [CrossRef]
- Uthaiah, R.C.; Praefcke, G.J.; Howard, J.C.; Herrmann, C. IIGP1, an interferon-gamma-inducible 47-kDa GTPase of the mouse, showing cooperative enzymatic activity and GTP-dependent multimerization. J. Biol. Chem. 2003, 278, 29336–29343. [Google Scholar] [CrossRef]
- Tiwari, S.; Macmicking, J.D. Bacterial phagosome acidification within IFN-gamma-activated macrophages: Role of host p47 immunity-related GTPases IRGs). Methods Mol. Biol. 2008, 445, 407–415. [Google Scholar]
- Conti, I.; Rollins, B.J. CCL2 (monocyte chemoattractant protein-1) and cancer. Semin. Cancer Biol. 2004, 14, 149–154. [Google Scholar] [CrossRef] [PubMed]
- Arendt, L.M.; McCready, J.; Keller, P.J.; Baker, D.D.; Naber, S.P.; Seewaldt, V.; Kuperwasser, C. Obesity promotes breast cancer by CCL2-mediated macrophage recruitment and angiogenesis. Cancer Res. 2013, 73, 6080–6093. [Google Scholar] [CrossRef] [PubMed]
- Bousquenaud, M.; Fico, F.; Solinas, G.; Rüegg, C.; Santamaria-Martínez, A. Obesity promotes the expansion of metastasis-initiating cells in breast cancer. Breast Cancer Res. 2018, 20, 104. [Google Scholar] [CrossRef] [PubMed]
- Karin, N.; Razon, H. Chemokines beyond chemo-attraction: CXCL10 and its significant role in cancer and autoimmunity. Cytokine 2018, 109, 24–28. [Google Scholar] [CrossRef]
- Raes, G.; Noël, W.; Beschin, A.; Brys, L.; de Baetselier, P.; Hassanzadeh, G.H. FIZZ1 and Ym as tools to discriminate between differentially activated macrophages. Dev. Immunol. 2002, 9, 151–159. [Google Scholar] [CrossRef]
- Puig-Kröger, A.; Sierra-Filardi, E.; Domínguez-Soto, A.; Samaniego, R.; Corcuera, M.T.; Gómez-Aguado, F.; Ratnam, M.; Sánchez-Mateos, P.; Corbí, A.L. Folate receptor beta is expressed by tumor-associated macrophages and constitutes a marker for M2 anti-inflammatory/regulatory macrophages. Cancer Res. 2009, 69, 9395–9403. [Google Scholar] [CrossRef]
- Italiani, P.; Boraschi, D. From Monocytes to M1/M2 Macrophages: Phenotypical vs. Functional Differentiation. Front. Immunol. 2014, 5, 514. [Google Scholar] [CrossRef]
- Mills, C.D. Anatomy of a discovery: m1 and m2 macrophages. Front. Immunol. 2015, 6, 212. [Google Scholar] [CrossRef]
- Laviron, M.; Boissonnas, A. Ontogeny of Tumor-Associated Macrophages. Front. Immunol. 2019, 10, 1799. [Google Scholar] [CrossRef]
- Chanmee, T.; Ontong, P.; Konno, K.; Itano, N. Tumor-associated macrophages as major players in the tumor microenvironment. Cancers 2014, 6, 1670–1690. [Google Scholar] [CrossRef]
- Pesce, J.T.; Ramalingam, T.R.; Wilson, M.S.; Mentink-Kane, M.M.; Thompson, R.W.; Cheever, A.W.; Urban, J.F., Jr.; Wynn, T.A. Retnla (relmalpha/fizz1) suppresses helminth-induced Th2-type immunity. PLoS Pathog. 2009, 5, e1000393. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Dhanasekaran, S.M.; Jin, H.; Hu, B.; Tomlins, S.A.; Chinnaiyan, A.M.; Phan, S.H. FIZZ1 stimulation of myofibroblast differentiation. Am. J. Pathol. 2004, 164, 1315–1326. [Google Scholar] [CrossRef]
- Yabluchanskiy, A.; Ma, Y.; DeLeon-Pennell, K.Y.; Altara, R.; Halade, G.V.; Voorhees, A.P.; Nguyen, N.T.; Jin, Y.F.; Winniford, M.D.; Hall, M.E.; et al. Myocardial Infarction Superimposed on Aging: MMP-9 Deletion Promotes M2 Macrophage Polarization. J. Gerontol. Ser. A Biol. Sci. Med. Sci. 2016, 71, 475–483. [Google Scholar] [CrossRef] [PubMed]
- Deng, S.; Deng, Q.; Zhang, Y.; Ye, H.; Yu, X.; Zhang, Y.; Han, G.Y.; Luo, P.; Wu, M.; Yu, Y.; et al. Non-platelet-derived CXCL4 differentially regulates cytotoxic and regulatory T cells through CXCR3 to suppress the immune response to colon cancer. Cancer Lett. 2019, 443, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Ito, K.; Stannard, K.; Gabutero, E.; Clark, A.M.; Neo, S.Y.; Onturk, S.; Blanchard, H.; Ralph, S.J. Galectin-1 as a potent target for cancer therapy: Role in the tumor microenvironment. Cancer Metastasis Rev. 2012, 31, 763–778. [Google Scholar] [CrossRef]
- Sadeghi, M.; Ordway, B.; Rafiei, I.; Borad, P.; Fang, B.; Koomen, J.L.; Zhang, C.; Yoder, S.; Johnson, J.; Damaghi, M. Integrative Analysis of Breast Cancer Cells Reveals an Epithelial-Mesenchymal Transition Role in Adaptation to Acidic Microenvironment. Front. Oncol. 2020, 10, 304. [Google Scholar] [CrossRef]
- Leśniak, W.; Słomnicki, Ł.P.; Filipek, A. S100A6—New facts and features. Biochem. Biophys. Res. Commun. 2009, 390, 1087–1092. [Google Scholar] [CrossRef]
- Henegar, C.; Tordjman, J.; Achard, V.; Lacasa, D.; Cremer, I.; Guerre-Millo, M.; Poitou, C.; Basdevant, A.; Stich, V.; Viguerie, N.; et al. Adipose tissue transcriptomic signature highlights the pathological relevance of extracellular matrix in human obesity. Genome Biol. 2008, 9, R14. [Google Scholar] [CrossRef]
- Park, J.; Scherer, P.E. Adipocyte-derived endotrophin promotes malignant tumor progression. J. Clin. Investig. 2012, 122, 4243–4256. [Google Scholar] [CrossRef]
- Egeblad, M.; Rasch, M.G.; Weaver, V.M. Dynamic interplay between the collagen scaffold and tumor evolution. Curr. Opin. Cell Biol. 2010, 22, 697–706. [Google Scholar] [CrossRef]
- Schrader, J.; Gordon-Walker, T.T.; Aucott, R.L.; van Deemter, M.; Quaas, A.; Walsh, S.; Benten, D.; Forbes, S.J.; Wells, R.G.; Iredale, J.P. Matrix stiffness modulates proliferation, chemotherapeutic response, and dormancy in hepatocellular carcinoma cells. Hepatology 2011, 53, 1192–1205. [Google Scholar] [CrossRef]
- Paszek, M.J.; Zahir, N.; Johnson, K.R.; Lakins, J.N.; Rozenberg, G.I.; Gefen, A.; Reinhart-King, C.A.; Margulies, S.S.; Dembo, M.; Boettiger, D.; et al. Tensional homeostasis and the malignant phenotype. Cancer Cell 2005, 8, 241–254. [Google Scholar] [CrossRef] [PubMed]
- Engler, A.J.; Sen, S.; Sweeney, H.L.; Discher, D.E. Matrix elasticity directs stem cell lineage specification. Cell 2006, 126, 677–689. [Google Scholar] [CrossRef] [PubMed]
- Mammoto, A.; Connor, K.M.; Mammoto, T.; Yung, C.W.; Huh, D.; Aderman, C.M.; Mostoslavsky, G.; Smith, L.E.; Ingber, D.E. A mechanosensitive transcriptional mechanism that controls angiogenesis. Nature 2009, 457, 1103–1108. [Google Scholar] [CrossRef]
- Provenzano, P.P.; Inman, D.R.; Eliceiri, K.W.; Keely, P.J. Matrix density-induced mechano-regulation of breast cell phenotype, signaling and gene expression through a FAK-ERK linkage. Oncogene 2009, 28, 4326–4343. [Google Scholar] [CrossRef] [PubMed]
- Chandler, E.M.; Seo, B.R.; Califano, J.P.; Andresen Eguiluz, R.C.; Lee, J.S.; Yoon, C.J.; Tims, D.T.; Wang, J.X.; Cheng, L.; Mohanan, S.; et al. Implanted adipose progenitor cells as physicochemical regulators of breast cancer. Proc. Natl. Acad. Sci. USA 2012, 109, 9786–9791. [Google Scholar] [CrossRef] [PubMed]
- Kanneganti, T.D.; Dixit, V.D. Immunological complications of obesity. Nat. Immunol. 2012, 13, 707–712. [Google Scholar] [CrossRef] [PubMed]
- Gerriets, V.A.; MacIver, N.J. Role of T cells in malnutrition and obesity. Front. Immunol. 2014, 5, 379. [Google Scholar] [CrossRef]
- Grant, R.W.; Dixit, V.D. Adipose tissue as an immunological organ. Obesity 2015, 23, 512–518. [Google Scholar] [CrossRef]
- Wensveen, F.M.; Valentić, S.; Šestan, M.; Turk Wensveen, T.; Polić, B. The “Big Bang” in obese fat: Events initiating obesity-induced adipose tissue inflammation. Eur. J. Immunol. 2015, 45, 2446–2456. [Google Scholar] [CrossRef]
- Saltiel, A.R.; Olefsky, J.M. Inflammatory mechanisms linking obesity and metabolic disease. J. Clin. Investig. 2017, 127, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Haniffa, M.; Bigley, V.; Collin, M. Human mononuclear phagocyte system reunited. Semin. Cell Dev. Biol. 2015, 41, 59–69. [Google Scholar] [CrossRef] [PubMed]
- Yona, S.; Gordon, S. From the Reticuloendothelial to Mononuclear Phagocyte System—The Unaccounted Years. Front. Immunol. 2015, 6, 328. [Google Scholar] [CrossRef] [PubMed]
- Gordon, S.; Taylor, P.R. Monocyte and macrophage heterogeneity. Nat. Rev. Immunol. 2005, 5, 953–964. [Google Scholar] [CrossRef]
- Yona, S.; Kim, K.W.; Wolf, Y.; Mildner, A.; Varol, D.; Breker, M.; Strauss-Ayali, D.; Viukov, S.; Guilliams, M.; Misharin, A.; et al. Fate mapping reveals origins and dynamics of monocytes and tissue macrophages under homeostasis. Immunity 2013, 38, 79–91. [Google Scholar] [CrossRef]
- Qiu, S.Q.; Waaijer, S.; Zwager, M.C.; de Vries, E.; van der Vegt, B.; Schröder, C.P. Tumor-associated macrophages in breast cancer: Innocent bystander or important player? Cancer Treat. Rev. 2018, 70, 178–189. [Google Scholar] [CrossRef]
- Hou, J.; Shi, J.; Chen, L.; Lv, Z.; Chen, X.; Cao, H.; Xiang, Z.; Han, X. M2 macrophages promote myofibroblast differentiation of LR-MSCs and are associated with pulmonary fibrogenesis. Cell Commun. Signal. 2018, 16, 89. [Google Scholar] [CrossRef]
- Bility, M.T.; Nio, K.; Li, F.; McGivern, D.R.; Lemon, S.M.; Feeney, E.R.; Chung, R.T.; Su, L. Chronic hepatitis C infection-induced liver fibrogenesis is associated with M2 macrophage activation. Sci. Rep. 2016, 6, 39520. [Google Scholar] [CrossRef]
- Chawla, A.; Nguyen, K.D.; Goh, Y.P. Macrophage-mediated inflammation in metabolic disease. Nat. Rev. Immunol. 2011, 11, 738–749. [Google Scholar] [CrossRef]
- Sun, H.; Miao, Z.; Zhang, X.; Chan, U.I.; Su, S.M.; Guo, S.; Wong, C.K.H.; Xu, X.; Deng, C.X. Single-cell RNA-Seq reveals cell heterogeneity and hierarchy within mouse mammary epithelia. J. Biol. Chem. 2018, 293, 8315–8329. [Google Scholar] [CrossRef]
- Chung, W.; Eum, H.H.; Lee, H.-O.; Lee, K.-M.; Lee, H.-B.; Kim, K.-T.; Ryu, H.S.; Kim, S.; Lee, J.E.; Park, Y.H.; et al. Single-cell RNA-seq enables comprehensive tumour and immune cell profiling in primary breast cancer. Nat. Commun. 2017, 8, 15081. [Google Scholar] [CrossRef] [PubMed]
- Karaayvaz, M.; Cristea, S.; Gillespie, S.M.; Patel, A.P.; Mylvaganam, R.; Luo, C.C.; Specht, M.C.; Bernstein, B.E.; Michor, F.; Ellisen, L.W. Unravelling subclonal heterogeneity and aggressive disease states in TNBC through single-cell RNA-seq. Nat. Commun. 2018, 9, 3588. [Google Scholar] [CrossRef] [PubMed]
- Sebastian, A.; Hum, N.R.; Martin, K.A.; Gilmore, S.F.; Peran, I.; Byers, S.W.; Wheeler, E.K.; Coleman, M.A.; Loots, G.G. Single-Cell Transcriptomic Analysis of Tumor-Derived Fibroblasts and Normal Tissue-Resident Fibroblasts Reveals Fibroblast Heterogeneity in Breast Cancer. Cancers 2020, 12, 1307. [Google Scholar] [CrossRef] [PubMed]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lo, P.-K.; Yao, Y.; Zhou, Q. Single-Cell RNA-seq Reveals Obesity-Induced Alterations in the Brca1-Mutated Mammary Gland Microenvironment. Cancers 2020, 12, 2235. https://doi.org/10.3390/cancers12082235
Lo P-K, Yao Y, Zhou Q. Single-Cell RNA-seq Reveals Obesity-Induced Alterations in the Brca1-Mutated Mammary Gland Microenvironment. Cancers. 2020; 12(8):2235. https://doi.org/10.3390/cancers12082235
Chicago/Turabian StyleLo, Pang-Kuo, Yuan Yao, and Qun Zhou. 2020. "Single-Cell RNA-seq Reveals Obesity-Induced Alterations in the Brca1-Mutated Mammary Gland Microenvironment" Cancers 12, no. 8: 2235. https://doi.org/10.3390/cancers12082235
APA StyleLo, P.-K., Yao, Y., & Zhou, Q. (2020). Single-Cell RNA-seq Reveals Obesity-Induced Alterations in the Brca1-Mutated Mammary Gland Microenvironment. Cancers, 12(8), 2235. https://doi.org/10.3390/cancers12082235



