Conditional Gene Targeting Reveals Cell Type-Specific Roles of the Lysosomal Protease Cathepsin L in Mammary Tumor Progression
Abstract
1. Introduction
2. Results
2.1. Generation and Characterization of Conditional Ctsl Knockout Mice
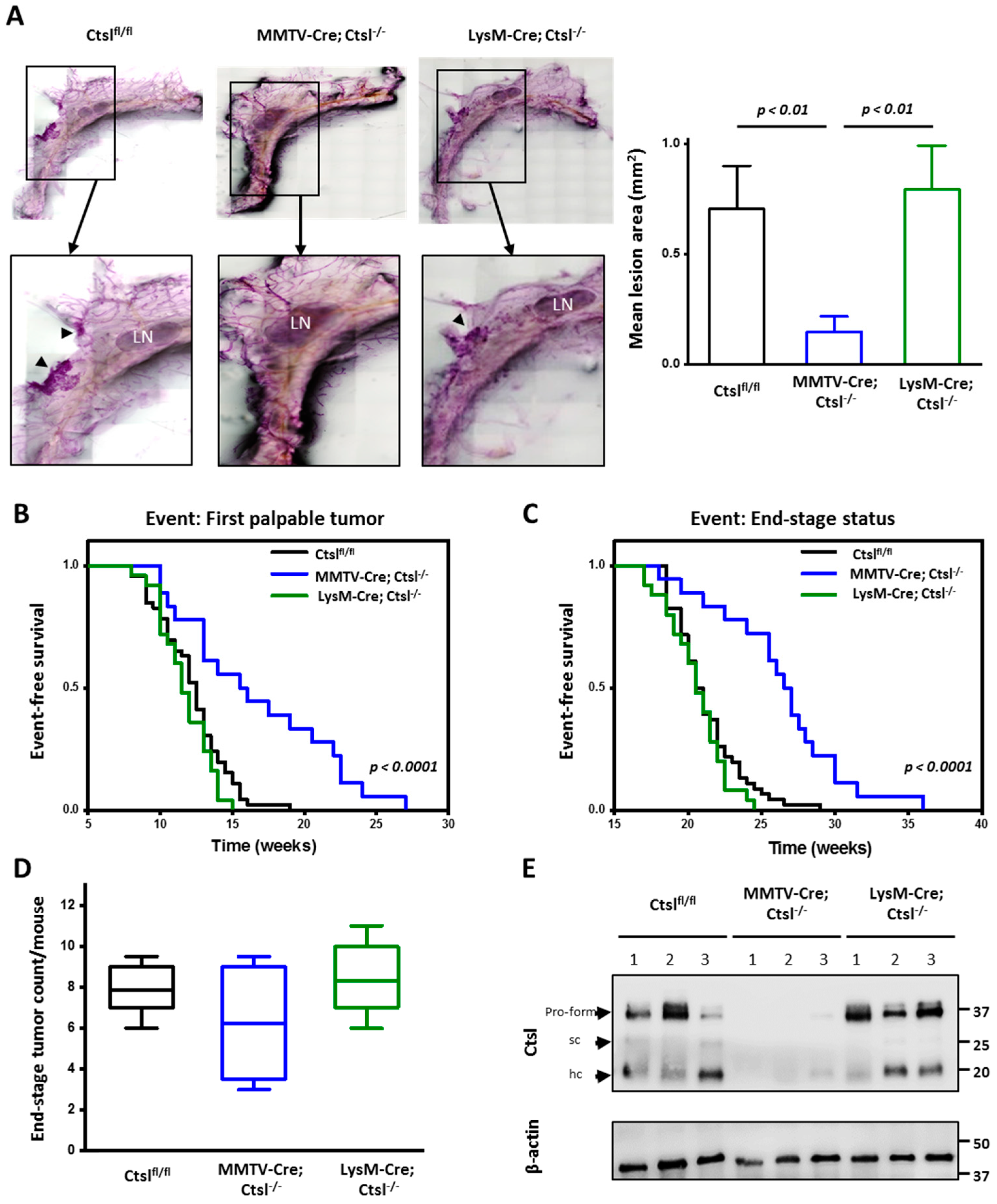
2.2. Ctsl Deletion in Mammary Epithelial Cells Delays Tumor Onset
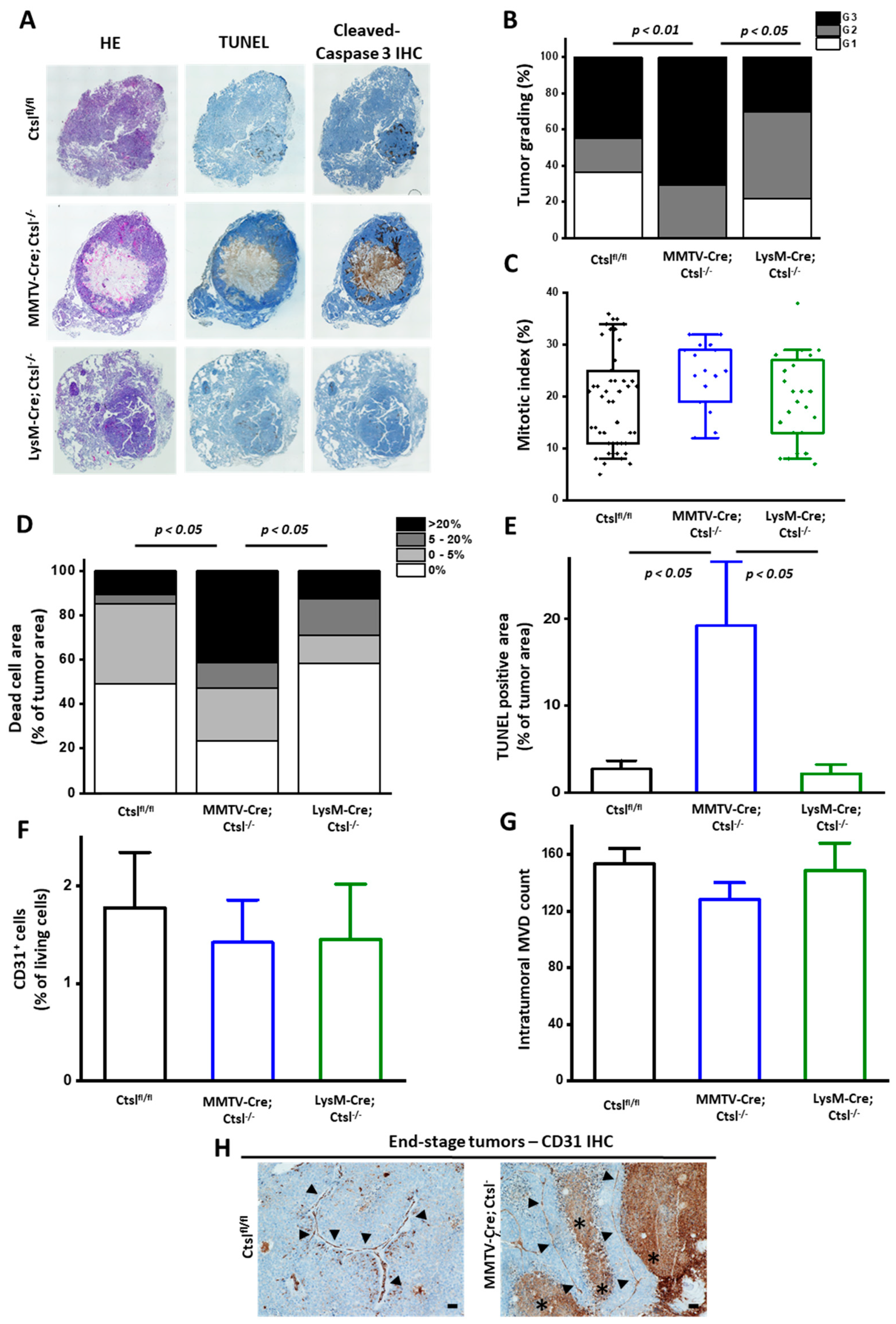
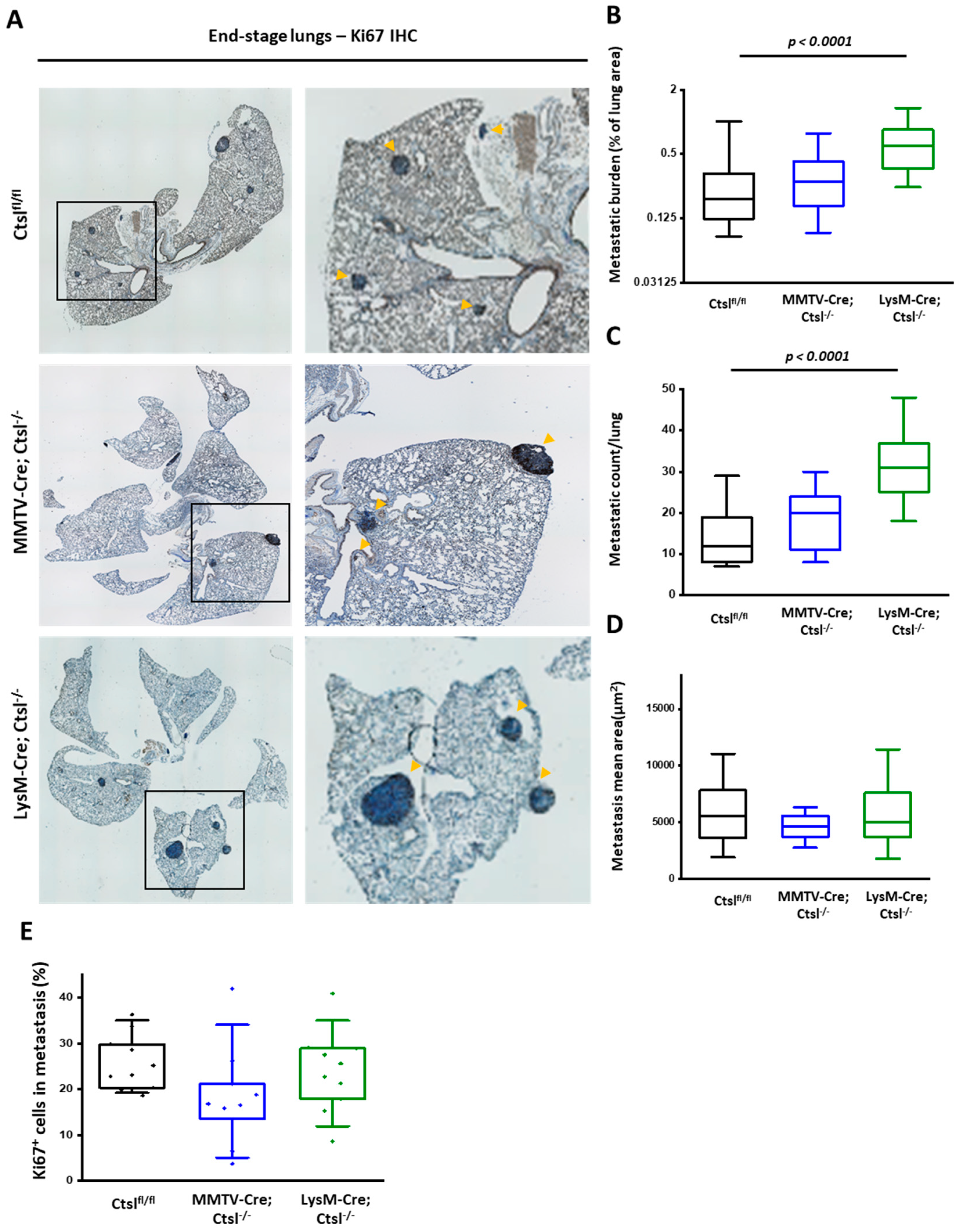
2.3. Distinct Effects of Cell Specific Ctsl-Deficiency on Histological Appearance and Metastasis of MMTV-PyMT Breast Cancers
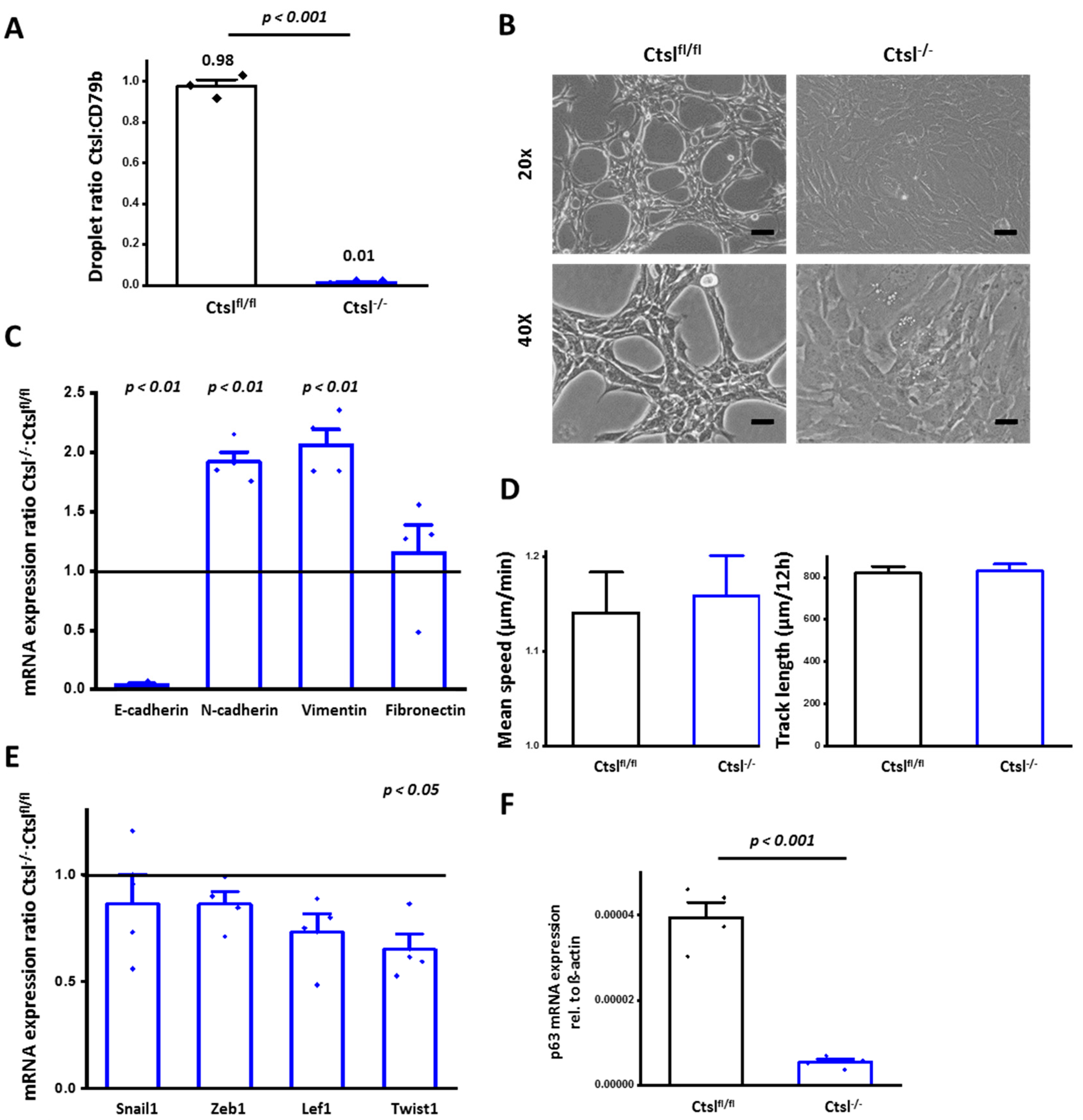
2.4. Ctsl−/− Mammary Epithelial Cancer Cells De-Differentiate In Vitro
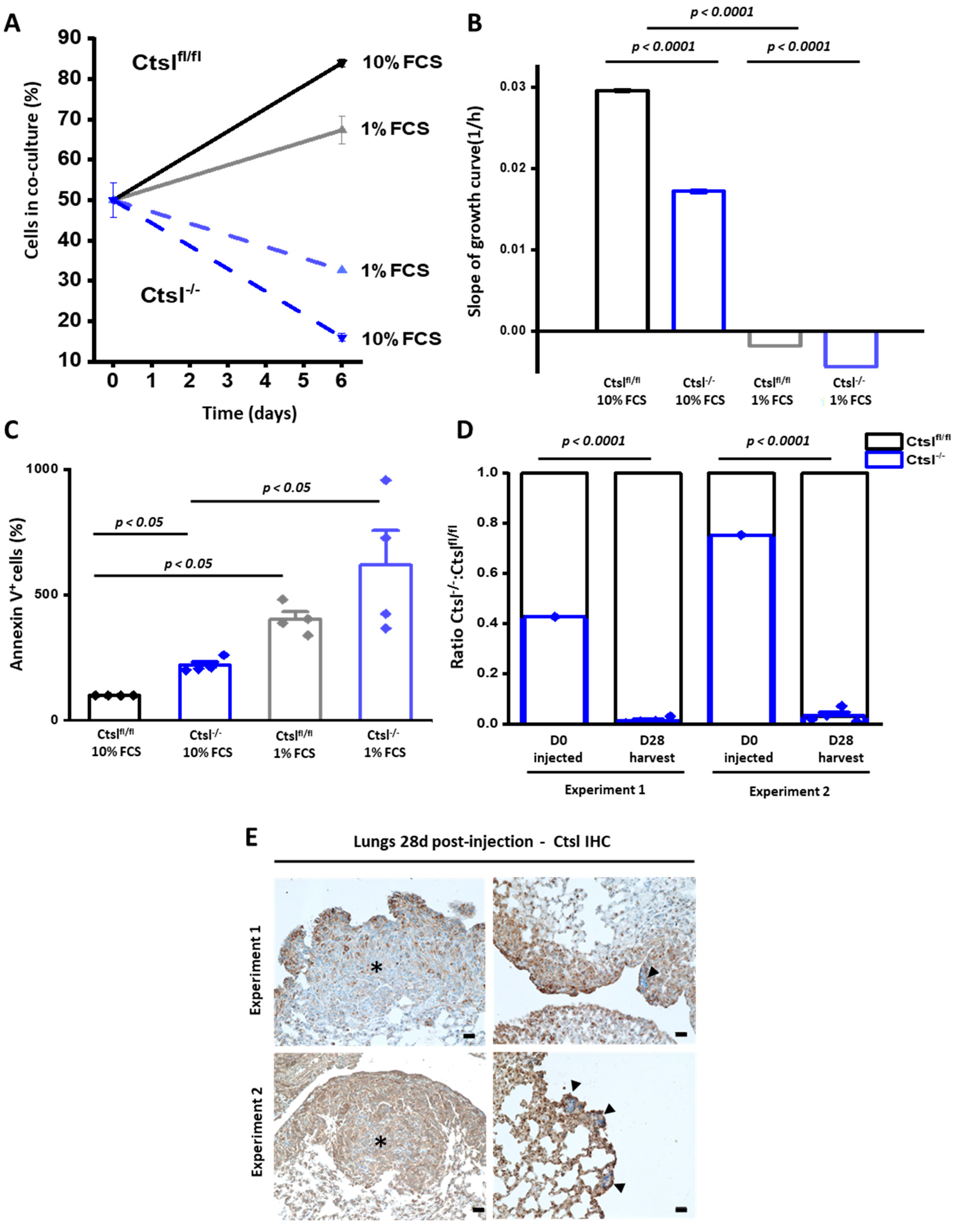
2.5. Ctsl−/− Mammary Epithelial Cancer Cells Are Growth Defective
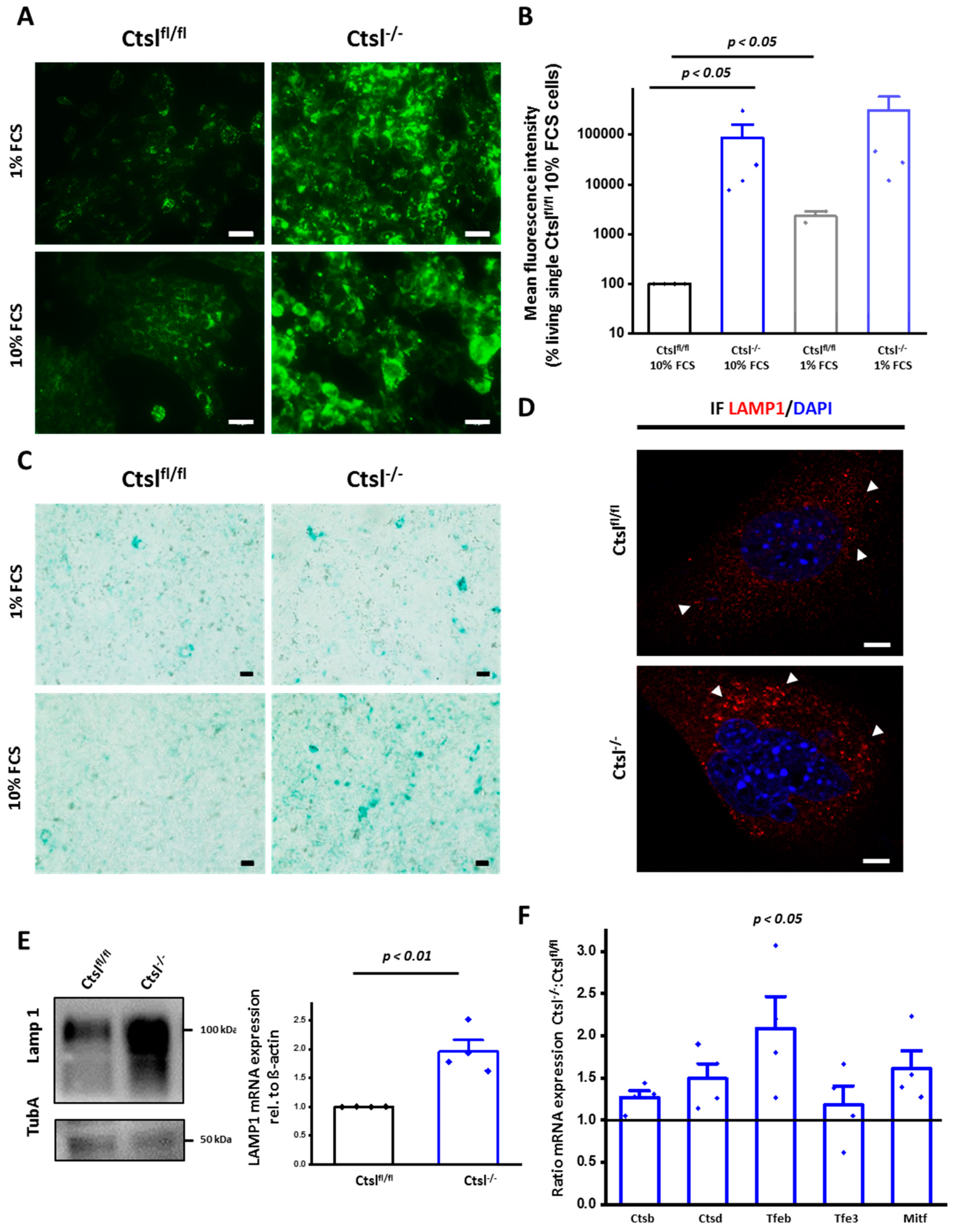
2.6. Expansion of the Lysosomal Compartment of Ctsl−/− Mammary Epithelial Cancer Cells
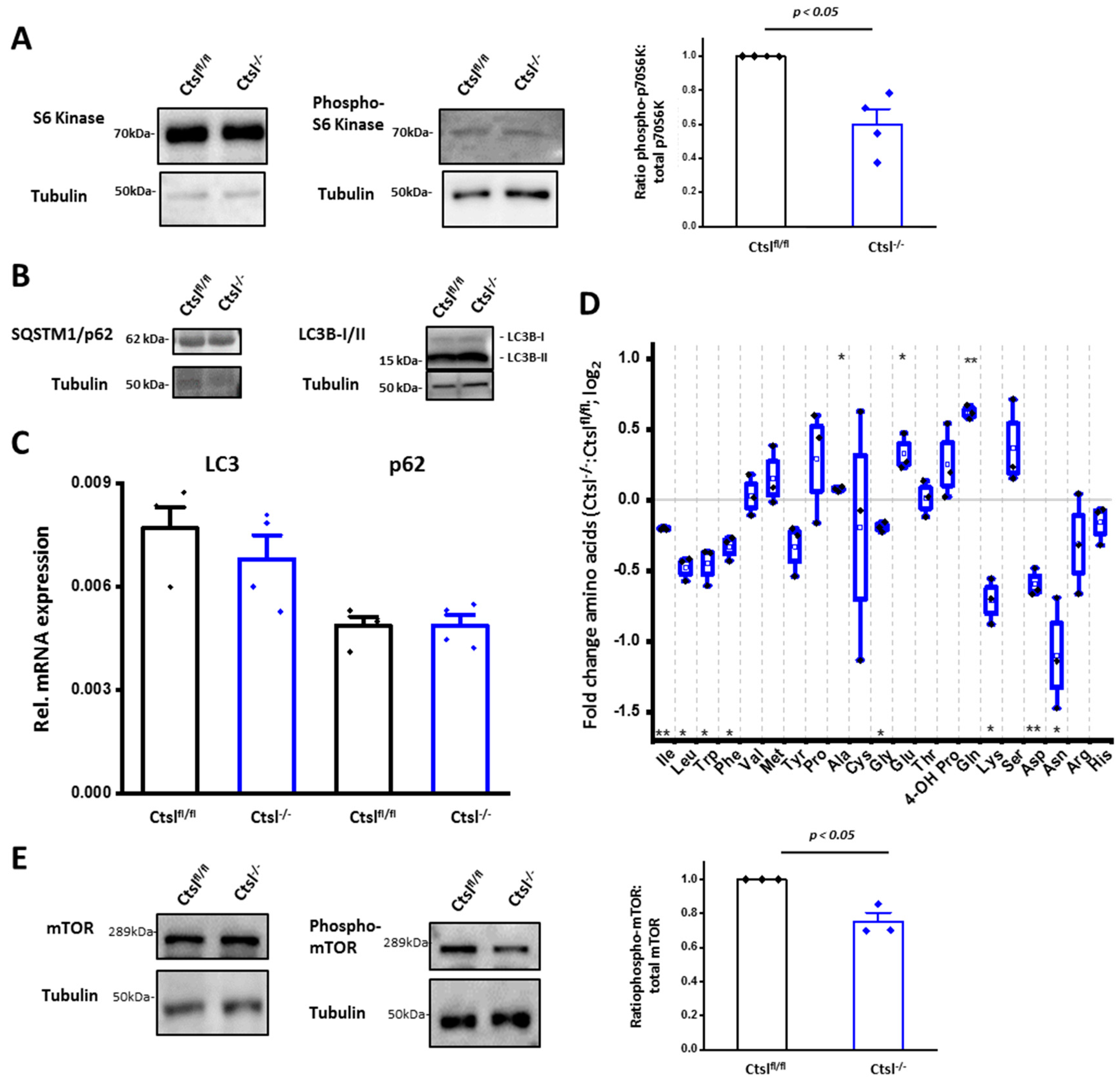
2.7. Impaired mTORC1 Activity in Ctsl−/− Cancer cells
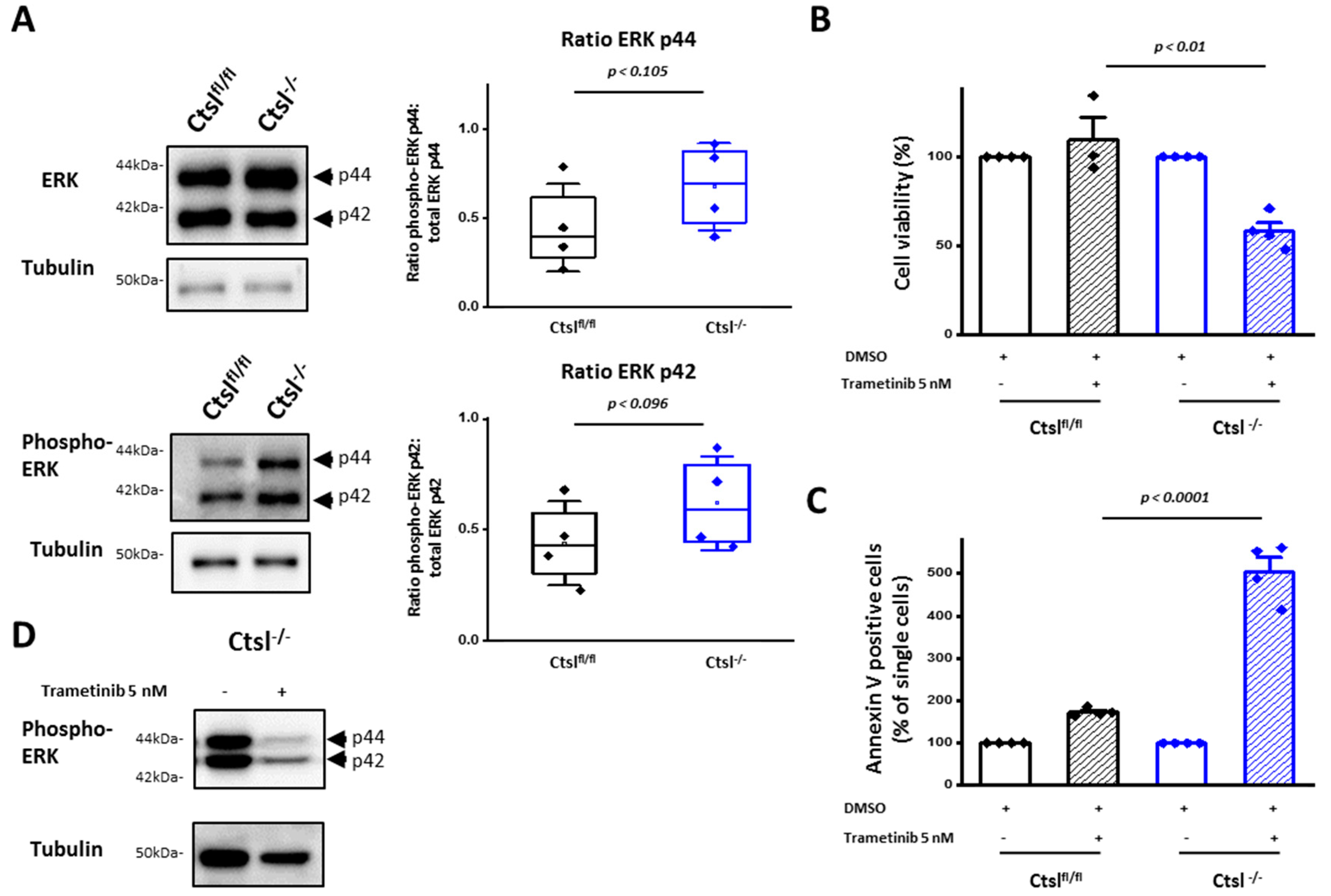
2.8. ERK 1/2 Signaling Is Crucial for Viability and Survival of Ctsl−/− Mammary Cancer Cells
3. Discussion
4. Materials and Methods
4.1. Mice
4.2. Genotyping
4.3. Bone Marrow Isolation and Macrophage Differentiation
4.4. Flow Cytometry Analysis of Macrophage Differentiation Stages
4.5. Mammary Whole Mounts
4.6. Immunohistochemistry
4.7. Histological Grading of Tumors
4.8. Quantification of Metastasis
4.9. Flow Cytometry of Tissues and Cells
4.10. Microvessel Density (MVD) Measurement
4.11. Generation and Culture of Breast Cancer Primary Cell Lines
4.12. RT-Cell Proliferation Monitoring
4.13. Droplet Digital PCR, Co-Culture- and Experimental Metastasis Assay
4.14. Cell Movement Tracking Assay
4.15. Immunofluorescence
4.16. β-Galactosidase Assay and LysoTracker™ Stainings
4.17. Preparation of Protein Lysates
4.18. SDS-Page and Western Blotting
4.19. RNA Isolation and RT-PCR
4.20. Amino Acid Profiling by Metabolomics
4.21. MEK Inhibitor (Trametinib) Treatment
4.22. MTT Cell Viability Assay
4.23. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Sevenich, L.; Joyce, J.A. Pericellular proteolysis in cancer. Genes Dev. 2014, 28, 2331–2347. [Google Scholar] [CrossRef] [PubMed]
- Hillebrand, L.E.; Wickberg, S.M.; Gomez-Auli, A.; Follo, M.; Maurer, J.; Busch, H.; Boerries, M.; Reinheckel, T. MMP14 empowers tumor-initiating breast cancer cells under hypoxic nutrient-depleted conditions. FASEB J. 2019, 33, 4124–4140. [Google Scholar] [CrossRef] [PubMed]
- Mason, S.D.; Joyce, J. A Proteolytic networks in cancer. Trends Cell Biol. 2011, 21, 228–237. [Google Scholar] [CrossRef] [PubMed]
- Osuala, K.O.; Ji, K.; Mattingly, R.R.; Sloane, B.F. Breast Cancer: Proteolysis and Migration. Adv. Exp. Med. Biol. 2019, 1152, 401–411. [Google Scholar] [CrossRef] [PubMed]
- Joyce, J.A.; Baruch, A.; Chehade, K.; Meyer-Morse, N.; Giraudo, E.; Tsai, F.-Y.; Greenbaum, D.C.; Hager, J.H.; Bogyo, M.; Hanahan, D. Cathepsin cysteine proteases are effectors of invasive growth and angiogenesis during multistage tumorigenesis. Cancer Cell 2004, 5, 443–453. [Google Scholar] [CrossRef]
- Vasiljeva, O.; Papazoglou, A.; Krüger, A.; Brodoefel, H.; Korovin, M.; Deussing, J.; Augustin, N.; Nielsen, B.S.; Almholt, K.; Bogyo, M.; et al. Tumor Cell–Derived and Macrophage-Derived Cathepsin B Promotes Progression and Lung Metastasis of Mammary Cancer. Cancer Res. 2006, 66, 5242–5250. [Google Scholar] [CrossRef]
- Akkari, L.; Gocheva, V.; Quick, M.L.; Kester, J.C.; Spencer, A.K.; Garfall, A.L.; Bowman, R.L.; Joyce, J.A. Combined deletion of cathepsin protease family members reveals compensatory mechanisms in cancer. Genes Dev. 2016, 30, 220–232. [Google Scholar] [CrossRef]
- Fonović, M.; Turk, B. Cysteine cathepsins and extracellular matrix degradation. Biochim. Biophys. Acta Gen. Subj. 2014, 1840, 2560–2570. [Google Scholar] [CrossRef]
- Vidak, E.; Javoršek, U.; Vizovišek, M.; Turk, B. Cysteine Cathepsins and their Extracellular Roles: Shaping the Microenvironment. Cells 2019, 8, 264. [Google Scholar] [CrossRef]
- Rudzińska, M.; Parodi, A.; Soond, S.M.; Vinarov, A.Z.; Korolev, D.O.; Morozov, A.O.; Daglioglu, C.; Tutar, Y.; Zamyatnin, A.A. The role of cysteine cathepsins in cancer progression and drug resistance. Int. J. Mol. Sci. 2019, 20, 3602. [Google Scholar] [CrossRef]
- Thomssen, C.; Schmitt, M.; Goretzki, L.; Oppelt, P.; Pache, L.; Dettmar, P.; Jänicke, F.; Graeff, H. Prognostic value of the cysteine proteases cathepsins B and cathepsin L in human breast cancer. Clin. Cancer Res. 1995, 1, 741–746. [Google Scholar] [PubMed]
- Foekens, J.A.; Kos, J.; Peters, H.A.; Krasovec, M.; Look, M.P.; Cimerman, N.; Meijer-van Gelder, M.E.; Henzen-Logmans, S.C.; van Putten, W.L.; Klijn, J.G. Prognostic significance of cathepsins B and L in primary human breast cancer. J. Clin. Oncol. 1998, 16, 1013–1021. [Google Scholar] [CrossRef] [PubMed]
- Harbeck, N.; Alt, U.; Berger, U.; Krüger, A.; Thomssen, C.; Jänicke, F.; Höfler, H.; Kates, R.E.; Schmitt, M. Prognostic impact of proteolytic factors (urokinase-type plasminogen activator, plasminogen activator inhibitor 1, and cathepsins B, D, and L) in primary breast cancer reflects effects of adjuvant systemic therapy. Clin. Cancer Res. 2001, 7, 2757–2764. [Google Scholar]
- Nakagawa, T.; Roth, W.; Wong, P.; Nelson, A.; Farr, A.; Deussing, J.; Villadangos, J.A.; Ploegh, H.; Peters, C.; Rudensky, A.Y. Cathepsin L: Critical role in Ii degradation and CD4 T cell selection in the thymus. Science 1998, 280, 450–453. [Google Scholar] [CrossRef]
- Roth, W.; Deussing, J.; Botchkarev, V.A.; Pauly-Evers, M.; Saftig, P.; Hafner, A.; Schmidt, P.; Schmahl, M.; Scherer, J.; Anton-Lamprecht, I.; et al. Cathepsin L deficiency as molecular defect of furless: Hyperproliferation of keratinocytes and pertubation of hair follicle cycling. FASEB J. 2000, 14, 2075–2086. [Google Scholar] [CrossRef]
- Stypmann, J.; Glaser, K.; Roth, W.; Tobin, D.J.; Petermann, I.; Matthias, R.; Monnig, G.; Haverkamp, W.; Breithardt, G.; Schmahl, W.; et al. Dilated cardiomyopathy in mice deficient for the lysosomal cysteine peptidase cathepsin L. Proc. Natl. Acad. Sci. USA 2002, 99, 6234–6239. [Google Scholar] [CrossRef] [PubMed]
- Yasothornsrikul, S.; Greenbaum, D.; Medzihradszky, K.F.; Toneff, T.; Bundey, R.; Miller, R.; Schilling, B.; Petermann, I.; Dehnert, J.; Logvinova, A.; et al. Cathepsin L in secretory vesicles functions as a prohormone-processing enzyme for production of the enkephalin peptide neurotransmitter. Proc. Natl. Acad. Sci. USA 2003, 100, 9590–9595. [Google Scholar] [CrossRef]
- Hook, V.; Yasothornsrikul, S.; Greenbaum, D.; Medzihradszky, K.F.; Troutner, K.; Toneff, T.; Bundey, R.; Logrinova, A.; Reinheckel, T.; Peters, C.; et al. Cathepsin L and Arg/Lys aminopeptidase: A distinct prohormone processing pathway for the biosynthesis of peptide neurotransmitters and hormones. Biol. Chem. 2004, 385, 473–480. [Google Scholar] [CrossRef]
- Funkelstein, L.; Beinfeld, M.; Minokadeh, A.; Zadina, J.; Hook, V. Unique biological function of cathepsin L in secretory vesicles for biosynthesis of neuropeptides. Neuropeptides 2010, 44, 457–466. [Google Scholar] [CrossRef]
- Gocheva, V.; Zeng, W.; Ke, D.; Klimstra, D.; Reinheckel, T.; Peters, C.; Hanahan, D.; Joyce, J.A. Distinct roles for cysteine cathepsin genes in multistage tumorigenesis. Genes Dev. 2006, 20, 543–556. [Google Scholar] [CrossRef]
- Brindle, N.R.; Joyce, J.A.; Rostker, F.; Lawlor, E.R.; Swigart-Brown, L.; Evan, G.; Hanahan, D.; Shchors, K. Deficiency for the Cysteine Protease Cathepsin L Impairs Myc-Induced Tumorigenesis in a Mouse Model of Pancreatic Neuroendocrine Cancer. PLoS ONE 2015, 10, e0120348. [Google Scholar] [CrossRef] [PubMed]
- Boudreau, F.; Lussier, C.R.; Mongrain, S.; Darsigny, M.; Drouin, J.L.; Doyon, G.; Suh, E.R.; Beaulieu, J.-F.; Rivard, N.; Perreault, N. Loss of cathepsin L activity promotes claudin-1 overexpression and intestinal neoplasia. FASEB J. 2007, 21, 3853–3865. [Google Scholar] [CrossRef] [PubMed]
- Dennemärker, J.; Lohmüller, T.; Mayerle, J.; Tacke, M.; Lerch, M.M.; Coussens, L.M.; Peters, C.; Reinheckel, T. Deficiency for the cysteine protease cathepsin L promotes tumor progression in mouse epidermis. Oncogene 2010, 29, 1611–1621. [Google Scholar] [CrossRef] [PubMed]
- Benavides, F.; Perez, C.; Blando, J.; Contreras, O.; Shen, J.; Coussens, L.M.; Fischer, S.M.; Kusewitt, D.F.; DiGiovanni, J.; Conti, C.J. Protective role of cathepsin L in mouse skin carcinogenesis. Mol. Carcinog. 2012, 51, 352–361. [Google Scholar] [CrossRef]
- Tholen, M.; Wolanski, J.; Stolze, B.; Chiabudini, M.; Gajda, M.; Bronsert, P.; Stickeler, E.; Rospert, S.; Reinheckel, T. Stress-resistant Translation of Cathepsin L mRNA in Breast Cancer Progression. J. Biol. Chem. 2015, 290, 15758–15769. [Google Scholar] [CrossRef]
- Gocheva, V.; Wang, H.-W.; Gadea, B.B.; Shree, T.; Hunter, K.E.; Garfall, A.L.; Berman, T.; Joyce, J.A. IL-4 induces cathepsin protease activity in tumor-associated macrophages to promote cancer growth and invasion. Genes Dev. 2010, 24, 241–255. [Google Scholar] [CrossRef]
- Clausen, B.E.; Burkhardt, C.; Reith, W.; Renkawitz, R.; Förster, I. Conditional gene targeting in macrophages and granulocytes using LysMcre mice. Transgenic Res. 1999, 8, 265–277. [Google Scholar] [CrossRef]
- Wagner, K.-U.; Wall, R.J.; St-Onge, L.; Gruss, P.; Wynshaw-Boris, A.; Garrett, L.; Li, M.; Furth, P.A.; Hennighausen, L. Cre-mediated gene deletion in the mammary gland. Nucleic Acids Res. 1997, 25, 4323–4330. [Google Scholar] [CrossRef]
- Guy, C.T.; Cardiff, R.D.; Muller, W.J. Induction of mammary tumors by expression of polyomavirus middle T oncogene: A transgenic mouse model for metastatic disease. Mol. Cell. Biol. 1992, 12, 954–961. [Google Scholar] [CrossRef]
- Davie, S.A.; Maglione, J.E.; Manner, C.K.; Young, D.; Cardiff, R.D.; MacLeod, C.L.; Ellies, L.G. Effects of FVB/NJ and C57Bl/6J strain backgrounds on mammary tumor phenotype in inducible nitric oxide synthase deficient mice. Transgenic Res. 2007, 16, 193–201. [Google Scholar] [CrossRef]
- Elston, C.W.; Ellis, I.O. Pathological prognostic factors in breast cancer. I. The value of histological grade in breast cancer: Experience from a large study with long-term follow-up. Histopathology 1991, 19, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Weidner, N.; Semple, J.P.; Welch, W.R.; Folkman, J. Tumor Angiogenesis and Metastasis—Correlation in Invasive Breast Carcinoma. N. Engl. J. Med. 1991, 324, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Ogura, M.; Bridgeman, V.L.; Malanchi, I. Macrophages unlock progression of breast cancer cells experiencing matrigel-segregation in transplantation models. Sci. Rep. 2017, 7, 11028. [Google Scholar] [CrossRef] [PubMed]
- Qian, B.-Z.; Zhang, H.; Li, J.; He, T.; Yeo, E.-J.; Soong, D.Y.H.; Carragher, N.O.; Munro, A.; Chang, A.; Bresnick, A.R.; et al. FLT1 signaling in metastasis-associated macrophages activates an inflammatory signature that promotes breast cancer metastasis. J. Exp. Med. 2015, 212, 1433–1448. [Google Scholar] [CrossRef] [PubMed]
- Wculek, S.K.; Malanchi, I. Neutrophils support lung colonization of metastasis-initiating breast cancer cells. Nature 2015, 528, 413–417. [Google Scholar] [CrossRef] [PubMed]
- Gatti, V.; Bongiorno-Borbone, L.; Fierro, C.; Annicchiarico-Petruzzelli, M.; Melino, G.; Peschiaroli, A. p63 at the Crossroads between Stemness and Metastasis in Breast Cancer. Int. J. Mol. Sci. 2019, 20, 2683. [Google Scholar] [CrossRef]
- Lawrence, R.E.; Zoncu, R. The lysosome as a cellular centre for signalling, metabolism and quality control. Nat. Cell Biol. 2019, 21, 133–142. [Google Scholar] [CrossRef]
- Bar-Peled, L.; Sabatini, D.M. Regulation of mTORC1 by amino acids. Trends Cell Biol. 2014, 24, 400–406. [Google Scholar] [CrossRef] [PubMed]
- Abe, H.; Kikuchi, S.; Hayakawa, K.; Iida, T.; Nagahashi, N.; Maeda, K.; Sakamoto, J.; Matsumoto, N.; Miura, T.; Matsumura, K.; et al. Discovery of a Highly Potent and Selective MEK Inhibitor: GSK1120212 (JTP-74057 DMSO Solvate). ACS Med. Chem. Lett. 2011, 2, 320–324. [Google Scholar] [CrossRef]
- Koboldt, D.C.; Fulton, R.S.; McLellan, M.D.; Schmidt, H.; Kalicki-Veizer, J.; McMichael, J.F.; Fulton, L.L.; Dooling, D.J.; Ding, L.; Mardis, E.R.; et al. Comprehensive molecular portraits of human breast tumours. Nature 2012, 490, 61–70. [Google Scholar] [CrossRef]
- Fluck, M.M.; Schaffhausen, B.S. Lessons in Signaling and Tumorigenesis from Polyomavirus Middle T Antigen. Microbiol. Mol. Biol. Rev. 2009, 73, 542–563. [Google Scholar] [CrossRef] [PubMed]
- Lin, E.Y.; Nguyen, A.V.; Russell, R.G.; Pollard, J.W. Colony-stimulating factor 1 promotes progression of mammary tumors to malignancy. J. Exp. Med. 2001, 193, 727–739. [Google Scholar] [CrossRef]
- Hollern, D.P.; Swiatnicki, M.R.; Andrechek, E.R. Histological subtypes of mouse mammary tumors reveal conserved relationships to human cancers. PLoS Genet. 2018, 14, e1007135. [Google Scholar] [CrossRef] [PubMed]
- Rothberg, J.M.; Bailey, K.M.; Wojtkowiak, J.W.; Ben-Nun, Y.; Bogyo, M.; Weber, E.; Moin, K.; Blum, G.; Mattingly, R.R.; Gillies, R.J.; et al. Acid-mediated tumor proteolysis: Contribution of cysteine cathepsins. Neoplasia 2013, 15, 1125–1137. [Google Scholar] [CrossRef] [PubMed]
- Sevenich, L.; Bowman, R.L.; Mason, S.D.; Quail, D.F.; Rapaport, F.; Elie, B.T.; Brogi, E.; Brastianos, P.K.; Hahn, W.C.; Holsinger, L.J.; et al. Analysis of tumour- and stroma-supplied proteolytic networks reveals a brain-metastasis-promoting role for cathepsin S. Nat. Cell Biol. 2014, 16, 876–888. [Google Scholar] [CrossRef] [PubMed]
- Small, D.M.; Burden, R.E.; Jaworski, J.; Hegarty, S.M.; Spence, S.; Burrows, J.F.; McFarlane, C.; Kissenpfennig, A.; McCarthy, H.O.; Johnston, J.A.; et al. Cathepsin S from both tumor and tumor-associated cells promote cancer growth and neovascularization. Int. J. Cancer 2013, 133, 2102–2112. [Google Scholar] [CrossRef]
- Akkari, L.; Gocheva, V.; Kester, J.C.; Hunter, K.E.; Quick, M.L.; Sevenich, L.; Wang, H.-W.; Peters, C.; Tang, L.H.; Klimstra, D.S.; et al. Distinct functions of macrophage-derived and cancer cell-derived cathepsin Z combine to promote tumor malignancy via interactions with the extracellular matrix. Genes Dev. 2014, 28, 2134–2150. [Google Scholar] [CrossRef]
- Tobin, D.J.; Foitzik, K.; Reinheckel, T.; Mecklenburg, L.; Botchkarev, V.A.; Peters, C.; Paus, R. The Lysosomal Protease Cathepsin L Is an Important Regulator of Keratinocyte and Melanocyte Differentiation During Hair Follicle Morphogenesis and Cycling. Am. J. Pathol. 2002, 160, 1807–1821. [Google Scholar] [CrossRef]
- Petermann, I.; Mayer, C.; Stypmann, J.; Biniossek, M.L.; Tobin, D.J.; Engelen, M.A.; Dandekar, T.; Grune, T.; Schild, L.; Peters, C.; et al. Lysosomal, cytoskeletal, and metabolic alterations in cardiomyopathy of cathepsin L knockout mice. FASEB J. 2006, 20, 1266–1268. [Google Scholar] [CrossRef]
- Rabanal-Ruiz Y1, K.V. mTORC1 and Nutrient Homeostasis: The Central Role of the Lysosome. Int. J. Mol. Sci. 2018, 19, 818. [Google Scholar] [CrossRef]
- Rogala, K.B.; Gu, X.; Kedir, J.F.; Abu-Remaileh, M.; Bianchi, L.F.; Bottino, A.M.S.; Dueholm, R.; Niehaus, A.; Overwijn, D.; Fils, A.-C.P.; et al. Structural basis for the docking of mTORC1 on the lysosomal surface. Science 2019, 366, 468–475. [Google Scholar] [CrossRef] [PubMed]
- Holen, I.; Speirs, V.; Morrissey, B.; Blyth, K. In vivo models in breast cancer research: Progress, challenges and future directions. Dis. Models Mech. 2017, 10, 359–371. [Google Scholar] [CrossRef] [PubMed]
- Klarenbeek, S.; van Miltenburg, M.H.; Jonkers, J. Genetically engineered mouse models of PI3K signaling in breast cancer. Mol. Oncol. 2013, 7, 146–164. [Google Scholar] [CrossRef] [PubMed]
- Reinheckel, T.; Hagemann, S.; Dollwet-Mack, S.; Martinez, E.; Lohmüller, T.; Zlatkovic, G.; Tobin, D.J.; Maas-Szabowski, N.; Peters, C. The lysosomal cysteine protease cathepsin L regulates keratinocyte proliferation by control of growth factor recycling. J. Cell Sci. 2005, 118, 3387–3395. [Google Scholar] [CrossRef]
- Köhler, M.; Ehrenfeld, S.; Halbach, S.; Lauinger, M.; Burk, U.; Reischmann, N.; Cheng, S.; Spohr, C.; Uhl, F.M.; Köhler, N.; et al. B-Raf deficiency impairs tumor initiation and progression in a murine breast cancer model. Oncogene 2019, 38, 1324–1339. [Google Scholar] [CrossRef]
- Schlimpert, M.; Lagies, S.; Budnyk, V.; Müller, B.; Walz, G.; Kammerer, B. Metabolic Phenotyping of Anks3 Depletion in mIMCD-3 cells-a Putative Nephronophthisis Candidate. Sci. Rep. 2018, 8, 9022. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Parigiani, M.A.; Ketscher, A.; Timme, S.; Bronsert, P.; Schlimpert, M.; Kammerer, B.; Jacquel, A.; Chaintreuil, P.; Reinheckel, T. Conditional Gene Targeting Reveals Cell Type-Specific Roles of the Lysosomal Protease Cathepsin L in Mammary Tumor Progression. Cancers 2020, 12, 2004. https://doi.org/10.3390/cancers12082004
Parigiani MA, Ketscher A, Timme S, Bronsert P, Schlimpert M, Kammerer B, Jacquel A, Chaintreuil P, Reinheckel T. Conditional Gene Targeting Reveals Cell Type-Specific Roles of the Lysosomal Protease Cathepsin L in Mammary Tumor Progression. Cancers. 2020; 12(8):2004. https://doi.org/10.3390/cancers12082004
Chicago/Turabian StyleParigiani, María Alejandra, Anett Ketscher, Sylvia Timme, Peter Bronsert, Manuel Schlimpert, Bernd Kammerer, Arnaud Jacquel, Paul Chaintreuil, and Thomas Reinheckel. 2020. "Conditional Gene Targeting Reveals Cell Type-Specific Roles of the Lysosomal Protease Cathepsin L in Mammary Tumor Progression" Cancers 12, no. 8: 2004. https://doi.org/10.3390/cancers12082004
APA StyleParigiani, M. A., Ketscher, A., Timme, S., Bronsert, P., Schlimpert, M., Kammerer, B., Jacquel, A., Chaintreuil, P., & Reinheckel, T. (2020). Conditional Gene Targeting Reveals Cell Type-Specific Roles of the Lysosomal Protease Cathepsin L in Mammary Tumor Progression. Cancers, 12(8), 2004. https://doi.org/10.3390/cancers12082004





