KLK4 Induces Anti-Tumor Effects in Human Xenograft Mouse Models of Orthotopic and Metastatic Prostate Cancer
Abstract
:Simple Summary
Abstract
1. Introduction
2. Results
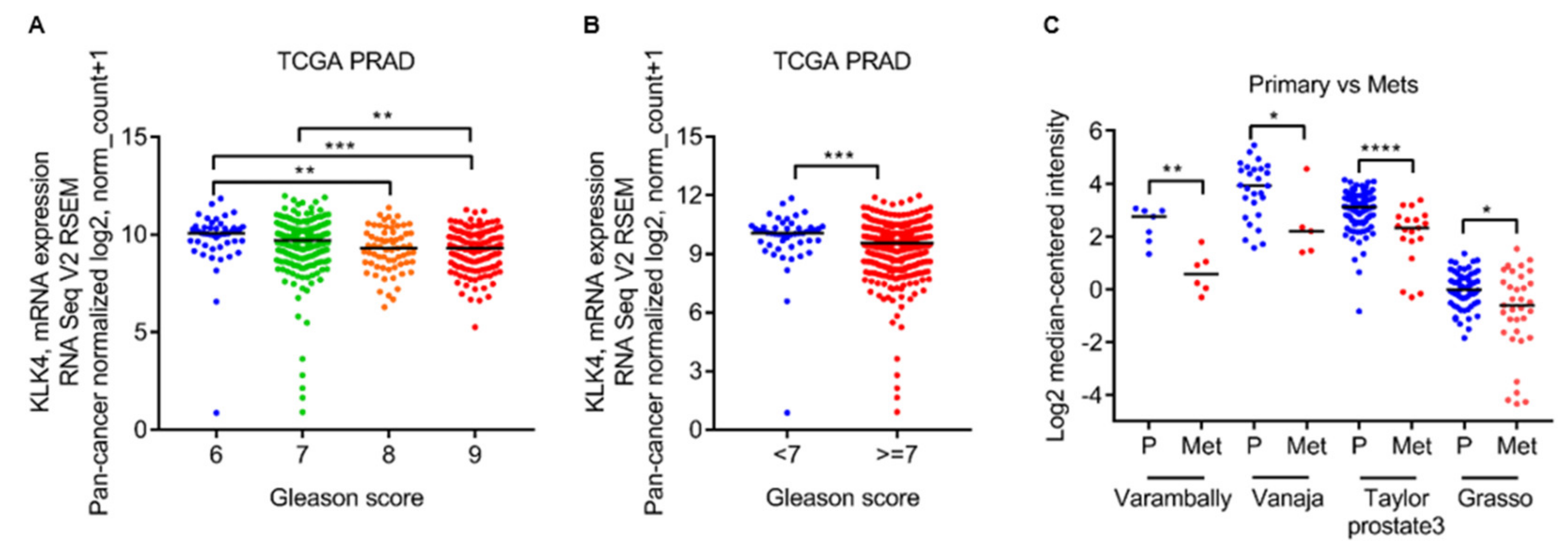
2.1. Low Kallikrein-Related Peptidase 4 (KLK4) mRNA Levels Are Associated with Gleason Grade and Metastatic Lesions
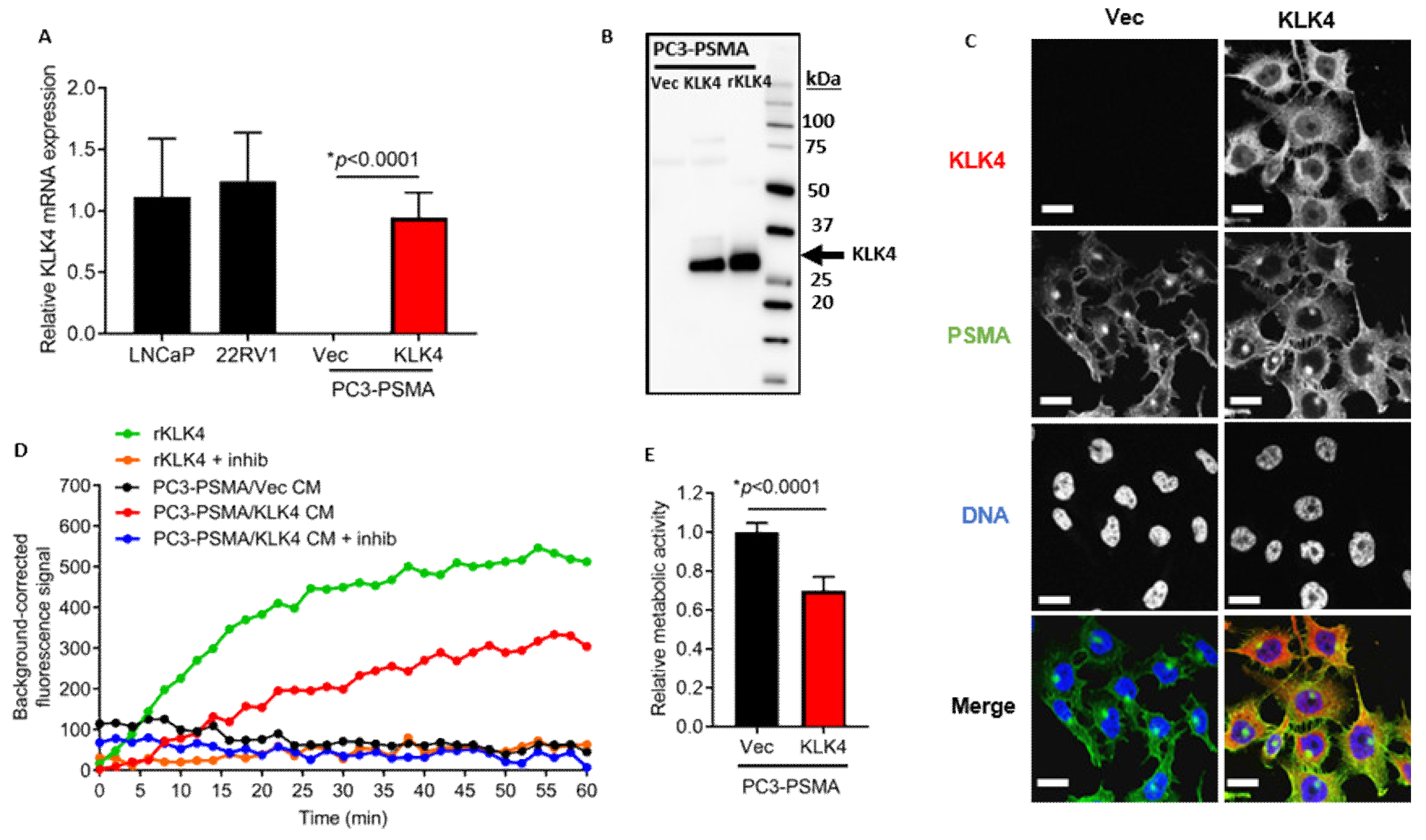
2.2. KLK4-Transfected PC3-PSMA Cells Secrete Enzymatically Active KLK4 Protein, and Exhibit Reduced Proliferative Rate In Vitro
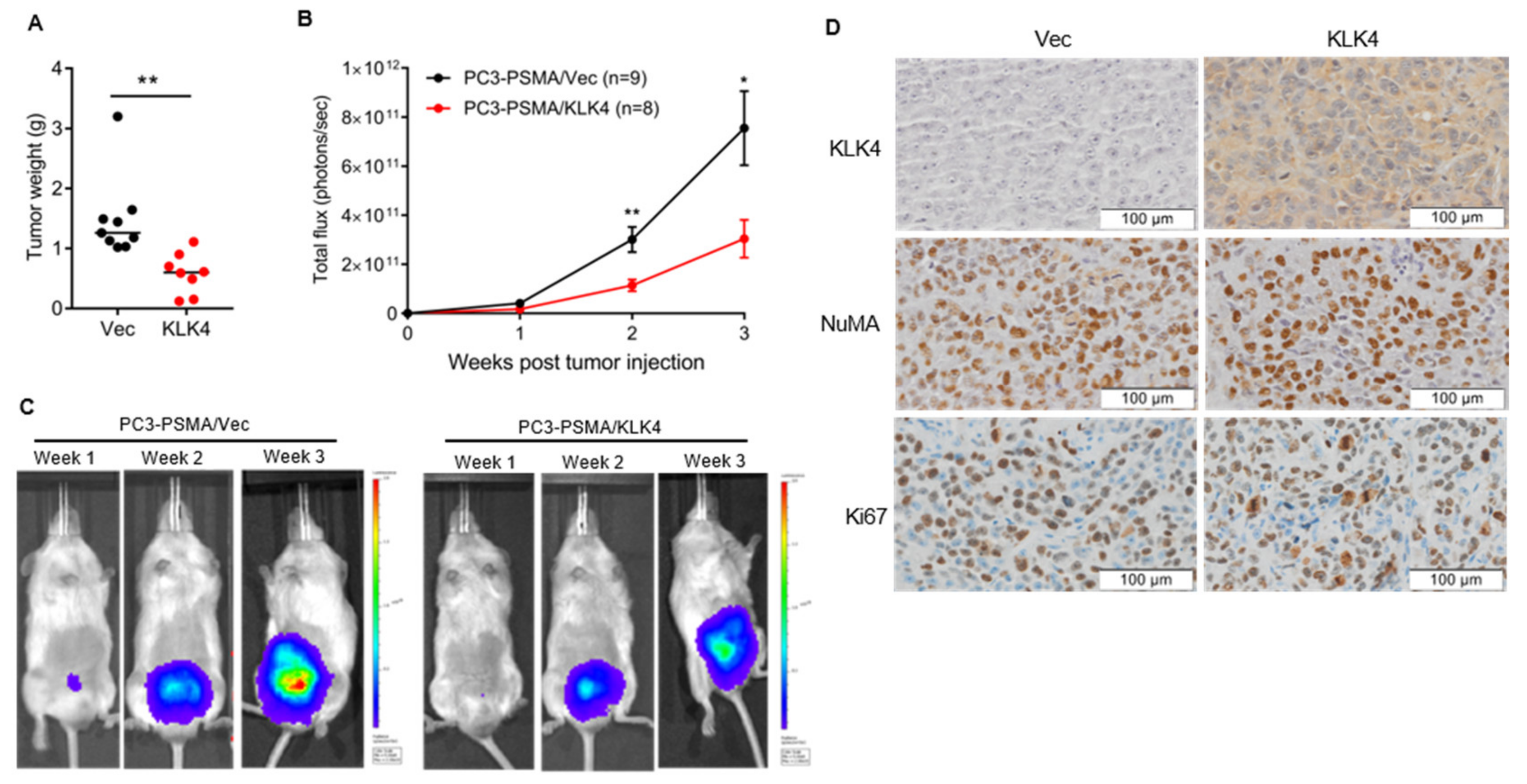
2.3. Tumor Secretion of KLK4 Inhibits the Growth of Orthotopically Implanted Prostate Tumors in Mice
2.4. Tumor Secretion of KLK4 Is Associated with Significantly Lower Metastatic Tumor Burden in Mice
3. Discussion
4. Materials and Methods
4.1. Evaluation of KLK4 Expression in Clinical Samples
4.2. Cell Culture
4.3. Generation of KLK4 Transfected Cell Lines
4.4. Quantitative PCR
4.5. Western Blotting
4.6. Enzyme Activity Assay
4.7. Fluorescence Microscopy
4.8. Cell Proliferation Assay
4.9. Animal Ethics Statement
4.10. Mice and Housing Conditions
4.11. In Vivo Tumorigenesis Studies
4.12. Tumor Bioluminescence Imaging
4.13. PET-CT Imaging
4.14. High Resolution microCT (Ex Vivo)
4.15. Immunohistochemical Staining of Xenograft Tumors
4.16. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [Green Version]
- Lawrence, M.G.; Lai, J.; Clements, J.A. Kallikreins on steroids: Structure, function, and hormonal regulation of prostate-specific antigen and the extended kallikrein locus. Endocr. Rev. 2010, 31, 407–446. [Google Scholar] [CrossRef] [PubMed]
- Kryza, T.; Silva, M.L.; Loessner, D.; Heuze-Vourc’h, N.; Clements, J.A. The kallikrein-related peptidase family: Dysregulation and functions during cancer progression. Biochimie 2016, 122, 283–299. [Google Scholar] [CrossRef] [PubMed]
- Obiezu, C.V.; Shan, S.J.; Soosaipillai, A.; Luo, L.Y.; Grass, L.; Sotiropoulou, G.; Petraki, C.D.; Papanastasiou, P.A.; Levesque, M.A.; Diamandis, E.P. Human kallikrein 4: Quantitative study in tissues and evidence for its secretion into biological fluids. Clin. Chem. 2005, 51, 1432–1442. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kryza, T.; Silva, L.M.; Bock, N.; Fuhrman-Luck, R.A.; Stephens, C.R.; Gao, J.; Samaratunga, H.; Australian Prostate Cancer, B.; Lawrence, M.G.; Hooper, J.D.; et al. Kallikrein-related peptidase 4 induces cancer-associated fibroblast features in prostate-derived stromal cells. Mol. Oncol. 2017, 11, 1307–1329. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Veveris-Lowe, T.L.; Lawrence, M.G.; Collard, R.L.; Bui, L.; Herington, A.C.; Nicol, D.L.; Clements, J.A. Kallikrein 4 (hK4) and prostate-specific antigen (PSA) are associated with the loss of E-cadherin and an epithelial-mesenchymal transition (EMT)-like effect in prostate cancer cells. Endocr. Relat. Cancer 2005, 12, 631–643. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fuhrman-Luck, R.A.; Stansfield, S.H.; Stephens, C.R.; Loessner, D.; Clements, J.A. Prostate cancer-associated kallikrein-related peptidase 4 activates matrix Metalloproteinase-1 and Thrombospondin-1. J. Proteome Res. 2016, 15, 2466–2478. [Google Scholar] [CrossRef]
- Gao, J.; Collard, R.L.; Bui, L.; Herington, A.C.; Nicol, D.L.; Clements, J.A. Kallikrein 4 is a potential mediator of cellular interactions between cancer cells and osteoblasts in metastatic prostate cancer. Prostate 2007, 67, 348–360. [Google Scholar] [CrossRef]
- Weinstein, J.N.; Collisson, E.A.; Mills, G.B.; Shaw, K.R.; Ozenberger, B.A.; Ellrott, K.; Shmulevich, I.; Sander, C.; Stuart, J.M.; The Cancer Genome Atlas Research Network. The cancer genome atlas pan-cancer analysis project. Nat. Genet. 2013, 45, 1113–1120. [Google Scholar] [CrossRef]
- Varambally, S.; Yu, J.; Laxman, B.; Rhodes, D.R.; Mehra, R.; Tomlins, S.A.; Shah, R.B.; Chandran, U.; Monzon, F.A.; Becich, M.J.; et al. Integrative genomic and proteomic analysis of prostate cancer reveals signatures of metastatic progression. Cancer Cell 2005, 8, 393–406. [Google Scholar] [CrossRef] [Green Version]
- Vanaja, D.K.; Cheville, J.C.; Iturria, S.J.; Young, C.Y. Transcriptional silencing of zinc finger protein 185 identified by expression profiling is associated with prostate cancer progression. Cancer Res. 2003, 63, 3877–3882. [Google Scholar] [PubMed]
- Taylor, B.S.; Schultz, N.; Hieronymus, H.; Gopalan, A.; Xiao, Y.; Carver, B.S.; Arora, V.K.; Kaushik, P.; Cerami, E.; Reva, B.; et al. Integrative genomic profiling of human prostate cancer. Cancer Cell 2010, 18, 11–22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grasso, C.S.; Wu, Y.M.; Robinson, D.R.; Cao, X.; Dhanasekaran, S.M.; Khan, A.P.; Quist, M.J.; Jing, X.; Lonigro, R.J.; Brenner, J.C.; et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature 2012, 487, 239–243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Avgeris, M.; Stravodimos, K.; Scorilas, A. Kallikrein-related peptidase 4 gene (KLK4) in prostate tumors: Quantitative expression analysis and evaluation of its clinical significance. Prostate 2011, 71, 1780–1789. [Google Scholar] [CrossRef] [PubMed]
- Klokk, T.I.; Kilander, A.; Xi, Z.; Waehre, H.; Risberg, B.; Danielsen, H.E.; Saatcioglu, F. Kallikrein 4 is a proliferative factor that is overexpressed in prostate cancer. Cancer Res. 2007, 67, 5221–5230. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mukai, S.; Yorita, K.; Yamasaki, K.; Nagai, T.; Kamibeppu, T.; Sugie, S.; Kida, K.; Onizuka, C.; Tsukino, H.; Kamimura, T.; et al. Expression of human kallikrein 1-related peptidase 4 (KLK4) and MET phosphorylation in prostate cancer tissue: Immunohistochemical analysis. Hum. Cell 2015, 28, 133–142. [Google Scholar] [CrossRef] [PubMed]
- Seiz, L.; Kotzsch, M.; Grebenchtchikov, N.I.; Geurts-Moespot, A.J.; Fuessel, S.; Goettig, P.; Gkazepis, A.; Wirth, M.P.; Schmitt, M.; Lossnitzer, A.; et al. Polyclonal antibodies against kallikrein-related peptidase 4 (KLK4): Immunohistochemical assessment of KLK4 expression in healthy tissues and prostate cancer. Biol. Chem. 2010, 391, 391–401. [Google Scholar] [CrossRef]
- Whitbread, A.K.; Veveris-Lowe, T.L.; Lawrence, M.G.; Nicol, D.L.; Clements, J.A. The role of kallikrein-related peptidases in prostate cancer: Potential involvement in an epithelial to mesenchymal transition. Biol. Chem. 2006, 387, 707–714. [Google Scholar] [CrossRef]
- Ramsay, A.J.; Dong, Y.; Hunt, M.L.; Linn, M.; Samaratunga, H.; Clements, J.A.; Hooper, J.D. Kallikrein-related peptidase 4 (KLK4) initiates intracellular signaling via protease-activated receptors (PARs). KLK4 and PAR-2 are co-expressed during prostate cancer progression. J. Biol. Chem. 2008, 283, 12293–12304. [Google Scholar] [CrossRef] [Green Version]
- Wang, W.; Mize, G.J.; Zhang, X.; Takayama, T.K. Kallikrein-related peptidase-4 initiates tumor-stroma interactions in prostate cancer through protease-activated receptor-1. Int. J. Cancer 2010, 126, 599–610. [Google Scholar] [CrossRef]
- Wilson, S.R.; Gallagher, S.; Warpeha, K.; Hawthorne, S.J. Amplification of MMP-2 and MMP-9 production by prostate cancer cell lines via activation of protease-activated receptors. Prostate 2004, 60, 168–174. [Google Scholar] [CrossRef] [PubMed]
- LeBleu, V.S.; Kalluri, R. A peek into cancer-associated fibroblasts: Origins, functions and translational impact. Dis. Model Mech. 2018, 11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, T.; Zhou, L.; Li, D.; Andl, T.; Zhang, Y. Cancer-Associated Fibroblasts Build and Secure the Tumor Microenvironment. Front Cell Dev. Biol. 2019, 7, 60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moradi, A.; Srinivasan, S.; Clements, J.; Batra, J. Beyond the biomarker role: Prostate-specific antigen (PSA) in the prostate cancer microenvironment. Cancer Metastasis Rev. 2019, 38, 333–346. [Google Scholar] [CrossRef] [PubMed]
- Costanza, B.; Umelo, I.A.; Bellier, J.; Castronovo, V.; Turtoi, A. Stromal Modulators of TGF-beta in Cancer. J. Clin. Med. 2017, 6. [Google Scholar] [CrossRef] [Green Version]
- Drabsch, Y.; ten Dijke, P. TGF-beta signalling and its role in cancer progression and metastasis. Cancer Metastasis Rev. 2012, 31, 553–568. [Google Scholar] [CrossRef]
- Dallas, S.L.; Zhao, S.; Cramer, S.D.; Chen, Z.; Peehl, D.M.; Bonewald, L.F. Preferential production of latent transforming growth factor beta-2 by primary prostatic epithelial cells and its activation by prostate-specific antigen. J. Cell Physiol. 2005, 202, 361–370. [Google Scholar] [CrossRef]
- Killian, C.S.; Corral, D.A.; Kawinski, E.; Constantine, R.I. Mitogenic response of osteoblast cells to prostate-specific antigen suggests an activation of latent TGF-beta and a proteolytic modulation of cell adhesion receptors. Biochem. Biophys. Res. Commun. 1993, 192, 940–947. [Google Scholar] [CrossRef]
- Konrad, L.; Scheiber, J.A.; Schwarz, L.; Schrader, A.J.; Hofmann, R. TGF-beta1 and TGF-beta2 strongly enhance the secretion of plasminogen activator inhibitor-1 and matrix metalloproteinase-9 of the human prostate cancer cell line PC-3. Regul. Pept. 2009, 155, 28–32. [Google Scholar] [CrossRef]
- Sung, E.; Kwon, O.K.; Lee, J.M.; Lee, S. Proteomics approach to identify novel metastatic bone markers from the secretome of PC-3 prostate cancer cells. Electrophoresis 2017, 38, 2638–2645. [Google Scholar] [CrossRef]
- Goettig, P.; Magdolen, V.; Brandstetter, H. Natural and synthetic inhibitors of kallikrein-related peptidases (KLKs). Biochimie 2010, 92, 1546–1567. [Google Scholar] [CrossRef] [PubMed]
- Shahinian, H.; Loessner, D.; Biniossek, M.L.; Kizhakkedathu, J.N.; Clements, J.A.; Magdolen, V.; Schilling, O. Secretome and degradome profiling shows that Kallikrein-related peptidases 4, 5, 6, and 7 induce TGFbeta-1 signaling in ovarian cancer cells. Mol. Oncol. 2014, 8, 68–82. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kobayashi-Kinoshita, S.; Yamakoshi, Y.; Onuma, K.; Yamamoto, R.; Asada, Y. TGF-beta1 autocrine signalling and enamel matrix components. Sci. Rep. 2016, 6, 33644. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanchez, W.Y.; de Veer, S.J.; Swedberg, J.E.; Hong, E.J.; Reid, J.C.; Walsh, T.P.; Hooper, J.D.; Hammond, G.L.; Clements, J.A.; Harris, J.M. Selective cleavage of human sex hormone-binding globulin by kallikrein-related peptidases and effects on androgen action in LNCaP prostate cancer cells. Endocrinology 2012, 153, 3179–3189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Korkmaz, K.S.; Korkmaz, C.G.; Pretlow, T.G.; Saatcioglu, F. Distinctly different gene structure of KLK4/KLK-L1/prostase/ARM1 compared with other members of the kallikrein family: Intracellular localization, alternative cDNA forms, and Regulation by multiple hormones. DNA Cell Biol. 2001, 20, 435–445. [Google Scholar] [CrossRef] [PubMed]
- Stephenson, S.A.; Verity, K.; Ashworth, L.K.; Clements, J.A. Localization of a new prostate-specific antigen-related serine protease gene, KLK4, is evidence for an expanded human kallikrein gene family cluster on chromosome 19q13.3-13.4. J. Biol. Chem. 1999, 274, 23210–23214. [Google Scholar] [CrossRef] [Green Version]
- Lai, J.; An, J.; Nelson, C.C.; Lehman, M.L.; Batra, J.; Clements, J.A. Analysis of androgen and anti-androgen regulation of KLK-related peptidase 2, 3, and 4 alternative transcripts in prostate cancer. Biol. Chem. 2014, 395, 1127–1132. [Google Scholar] [CrossRef] [Green Version]
- Dong, Y.; Bui, L.T.; Odorico, D.M.; Tan, O.L.; Myers, S.A.; Samaratunga, H.; Gardiner, R.A.; Clements, J.A. Compartmentalized expression of kallikrein 4 (KLK4/hK4) isoforms in prostate cancer: Nuclear, cytoplasmic and secreted forms. Endocr. Relat. Cancer 2005, 12, 875–889. [Google Scholar] [CrossRef]
- Jin, Y.; Qu, S.; Tesikova, M.; Wang, L.; Kristian, A.; Maelandsmo, G.M.; Kong, H.; Zhang, T.; Jeronimo, C.; Teixeira, M.R.; et al. Molecular circuit involving KLK4 integrates androgen and mTOR signaling in prostate cancer. Proc. Natl. Acad. Sci. USA 2013, 110, E2572–E2581. [Google Scholar] [CrossRef] [Green Version]
- Xi, Z.; Klokk, T.I.; Korkmaz, K.; Kurys, P.; Elbi, C.; Risberg, B.; Danielsen, H.; Loda, M.; Saatcioglu, F. Kallikrein 4 is a predominantly nuclear protein and is overexpressed in prostate cancer. Cancer Res. 2004, 64, 2365–2370. [Google Scholar] [CrossRef] [Green Version]
- Karakosta, T.D.; Soosaipillai, A.; Diamandis, E.P.; Batruch, I.; Drabovich, A.P. Quantification of Human Kallikrein-Related Peptidases in Biological Fluids by Multiplatform Targeted Mass Spectrometry Assays. Mol. Cell Proteom. 2016, 15, 2863–2876. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Obiezu, C.V.; Soosaipillai, A.; Jung, K.; Stephan, C.; Scorilas, A.; Howarth, D.H.; Diamandis, E.P. Detection of human kallikrein 4 in healthy and cancerous prostatic tissues by immunofluorometry and immunohistochemistry. Clin. Chem. 2002, 48, 1232–1240. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fuchs, A.V.; Tse, B.W.; Pearce, A.K.; Yeh, M.C.; Fletcher, N.L.; Huang, S.S.; Heston, W.D.; Whittaker, A.K.; Russell, P.J.; Thurecht, K.J. Evaluation of polymeric nanomedicines targeted to PSMA: Effect of ligand on targeting efficiency. Biomacromolecules 2015, 16, 3235–3247. [Google Scholar] [CrossRef]
- Prezas, P.; Arlt, M.J.; Viktorov, P.; Soosaipillai, A.; Holzscheiter, L.; Schmitt, M.; Talieri, M.; Diamandis, E.P.; Kruger, A.; Magdolen, V. Overexpression of the human tissue kallikrein genes KLK4, 5, 6, and 7 increases the malignant phenotype of ovarian cancer cells. Biol. Chem. 2006, 387, 807–811. [Google Scholar] [CrossRef] [PubMed]
- Tse, B.W.C.; Volpert, M.; Ratther, E.; Stylianou, N.; Nouri, M.; McGowan, K.; Lehman, M.L.; McPherson, S.J.; Roshan-Moniri, M.; Butler, M.S.; et al. Neuropilin-1 is upregulated in the adaptive response of prostate tumors to androgen-targeted therapies and is prognostic of metastatic progression and patient mortality. Oncogene 2017, 36, 3417–3427. [Google Scholar] [CrossRef] [Green Version]
- Swedberg, J.E.; Nigon, L.V.; Reid, J.C.; de Veer, S.J.; Walpole, C.M.; Stephens, C.R.; Walsh, T.P.; Takayama, T.K.; Hooper, J.D.; Clements, J.A.; et al. Substrate-guided design of a potent and selective kallikrein-related peptidase inhibitor for kallikrein 4. Chem. Biol. 2009, 16, 633–643. [Google Scholar] [CrossRef]
- Tse, B.W.; Cowin, G.J.; Soekmadji, C.; Jovanovic, L.; Vasireddy, R.S.; Ling, M.T.; Khatri, A.; Liu, T.; Thierry, B.; Russell, P.J. PSMA-targeting iron oxide magnetic nanoparticles enhance MRI of preclinical prostate cancer. Nanomedicine (Lond) 2015, 10, 375–386. [Google Scholar] [CrossRef] [Green Version]
- Tse, B.W.; Russell, P.J.; Lochner, M.; Forster, I.; Power, C.A. IL-18 inhibits growth of murine orthotopic prostate carcinomas via both adaptive and innate immune mechanisms. PLoS ONE 2011, 6, e24241. [Google Scholar] [CrossRef] [Green Version]
- Zhou, H.; Zhao, D. Ultrasound imaging-guided intracardiac injection to develop a mouse model of breast cancer brain metastases followed by longitudinal MRI. J. Vis. Exp. 2014. [Google Scholar] [CrossRef] [Green Version]
- Kryza, T.; Khan, T.; Puttick, S.; Li, C.; Sokolowski, K.A.; Tse, B.W.; Cuda, T.; Lyons, N.; Gough, M.; Yin, J.; et al. Effective targeting of intact and proteolysed CDCP1 for imaging and treatment of pancreatic ductal adenocarcinoma. Theranostics 2020, 10, 4116–4133. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tse, B.W.-C.; Kryza, T.; Yeh, M.-C.; Dong, Y.; Sokolowski, K.A.; Walpole, C.; Dreyer, T.; Felber, J.; Harris, J.; Magdolen, V.; et al. KLK4 Induces Anti-Tumor Effects in Human Xenograft Mouse Models of Orthotopic and Metastatic Prostate Cancer. Cancers 2020, 12, 3501. https://doi.org/10.3390/cancers12123501
Tse BW-C, Kryza T, Yeh M-C, Dong Y, Sokolowski KA, Walpole C, Dreyer T, Felber J, Harris J, Magdolen V, et al. KLK4 Induces Anti-Tumor Effects in Human Xenograft Mouse Models of Orthotopic and Metastatic Prostate Cancer. Cancers. 2020; 12(12):3501. https://doi.org/10.3390/cancers12123501
Chicago/Turabian StyleTse, Brian W.-C., Thomas Kryza, Mei-Chun Yeh, Ying Dong, Kamil A. Sokolowski, Carina Walpole, Tobias Dreyer, Johanna Felber, Jonathan Harris, Viktor Magdolen, and et al. 2020. "KLK4 Induces Anti-Tumor Effects in Human Xenograft Mouse Models of Orthotopic and Metastatic Prostate Cancer" Cancers 12, no. 12: 3501. https://doi.org/10.3390/cancers12123501
APA StyleTse, B. W.-C., Kryza, T., Yeh, M.-C., Dong, Y., Sokolowski, K. A., Walpole, C., Dreyer, T., Felber, J., Harris, J., Magdolen, V., Russell, P. J., & Clements, J. A. (2020). KLK4 Induces Anti-Tumor Effects in Human Xenograft Mouse Models of Orthotopic and Metastatic Prostate Cancer. Cancers, 12(12), 3501. https://doi.org/10.3390/cancers12123501




