Rare Tumor-Normal Matched Whole Exome Sequencing Identifies Novel Genomic Pathogenic Germline and Somatic Aberrations
Abstract
1. Introduction
2. Results
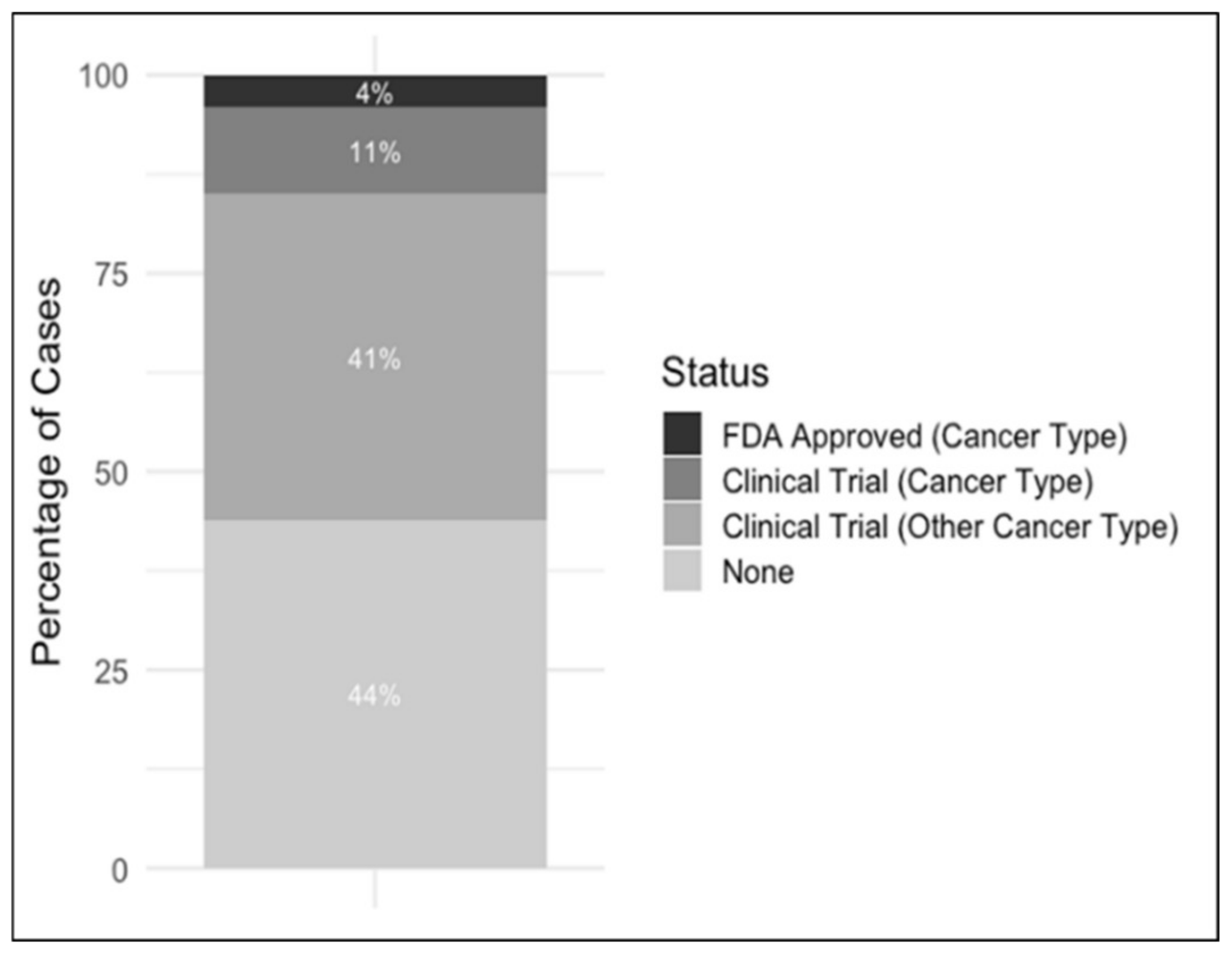
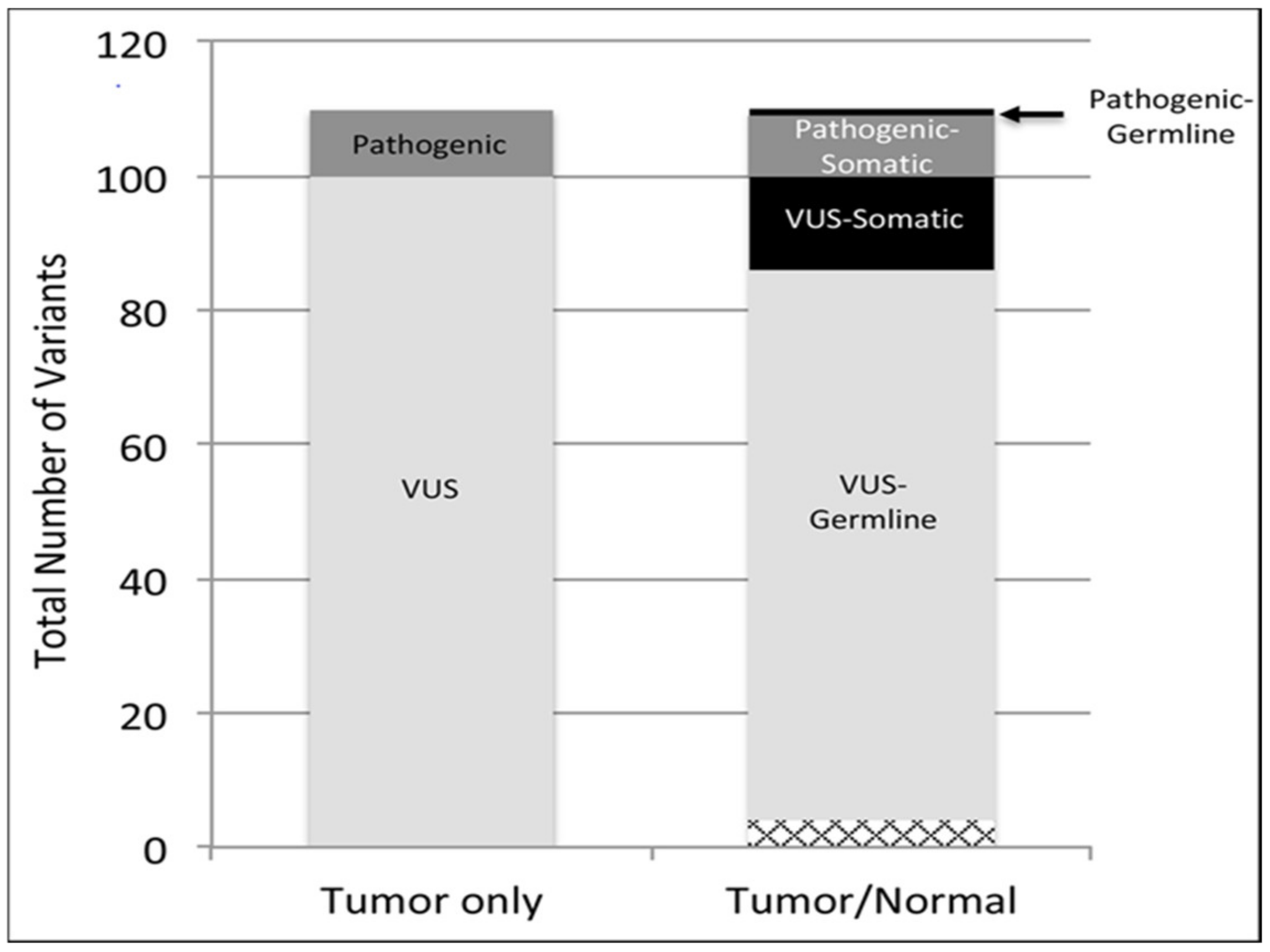
2.1. Somatic and Pathogenic Germline Variants Detected in the Rare Tumor Cohort
2.2. Germline Pathogenic Variants Are More Common in Rare Cancers
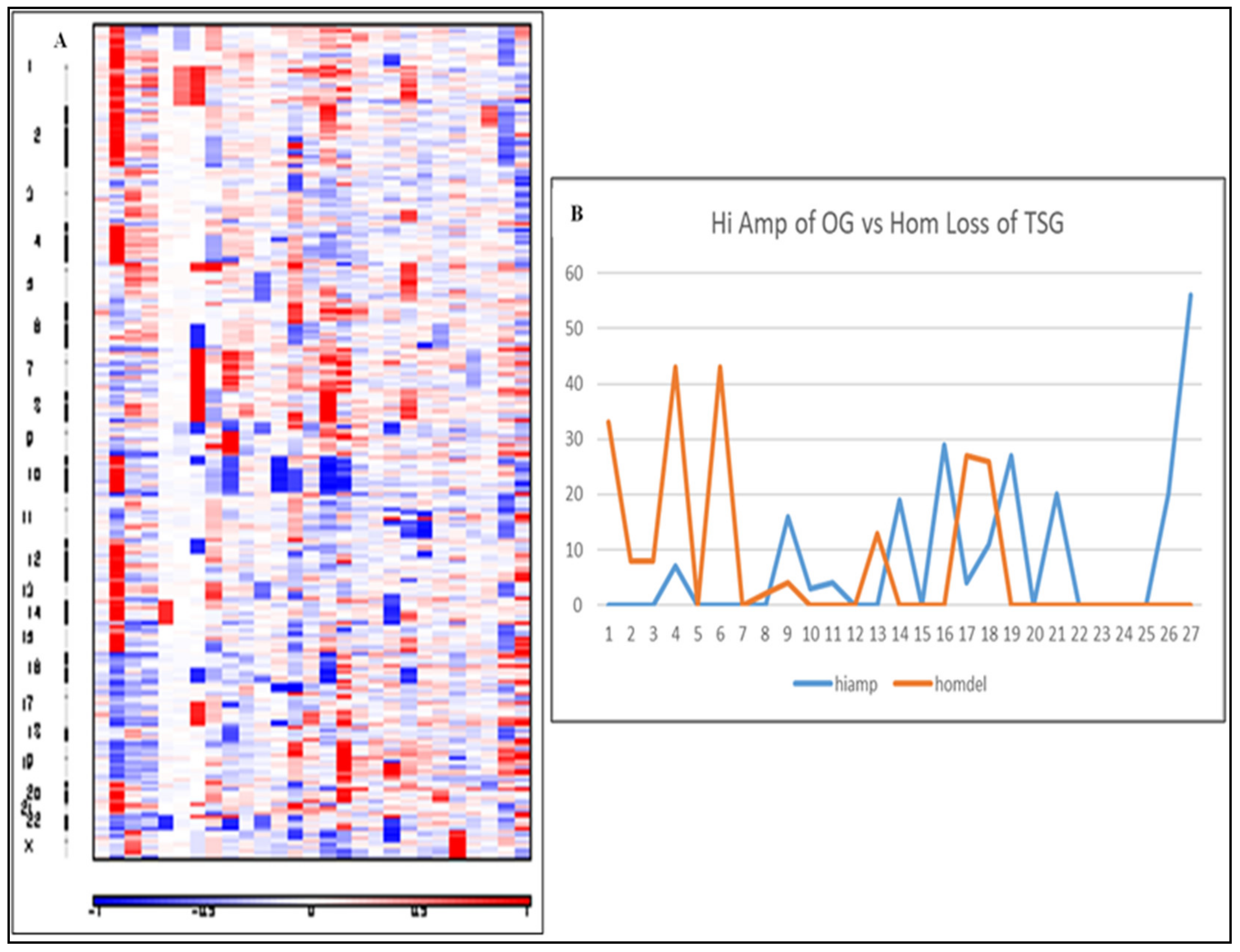
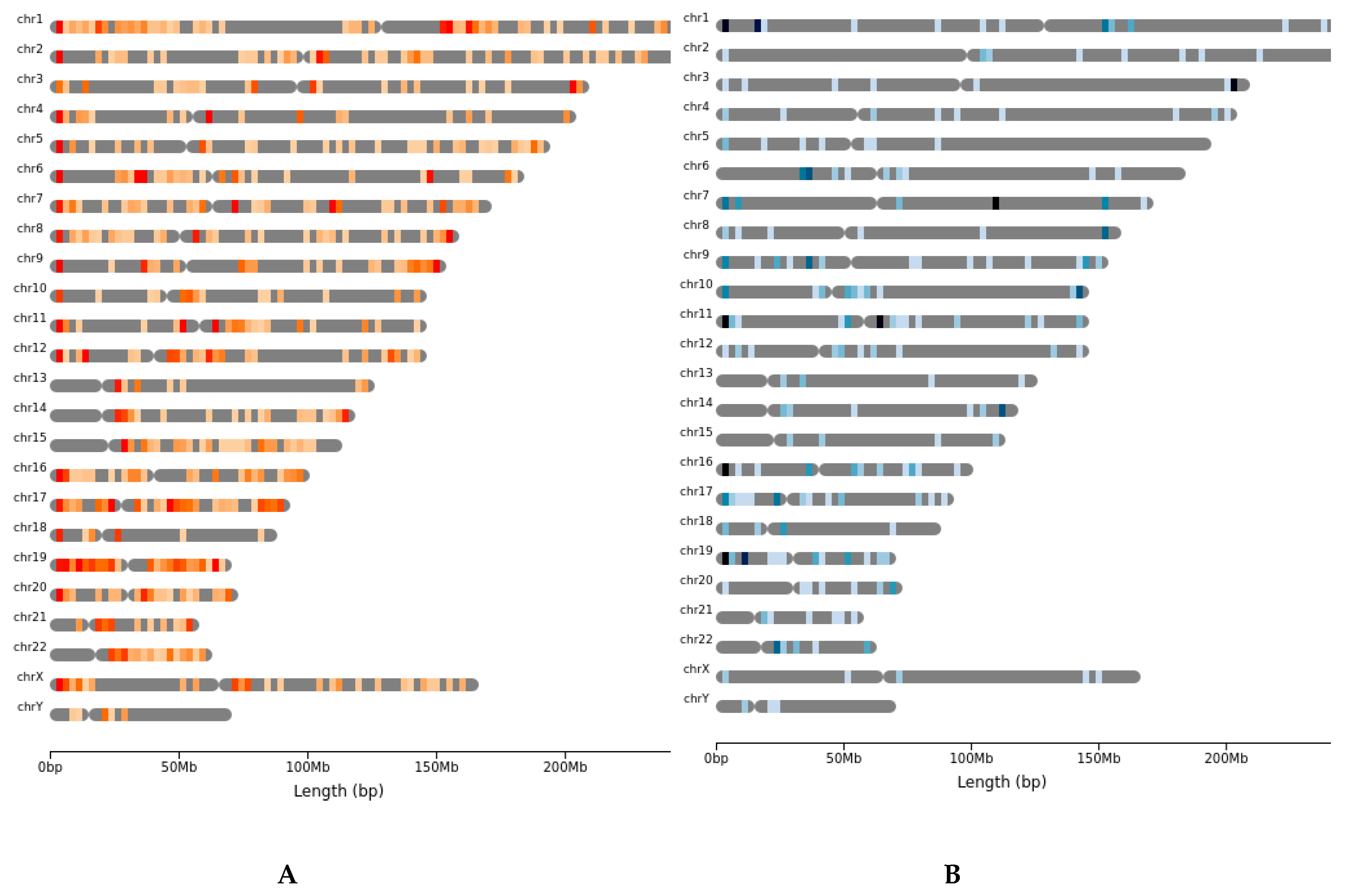
2.3. Copy Number Alterations Are Variable and Depend on the Rare Tumor Type
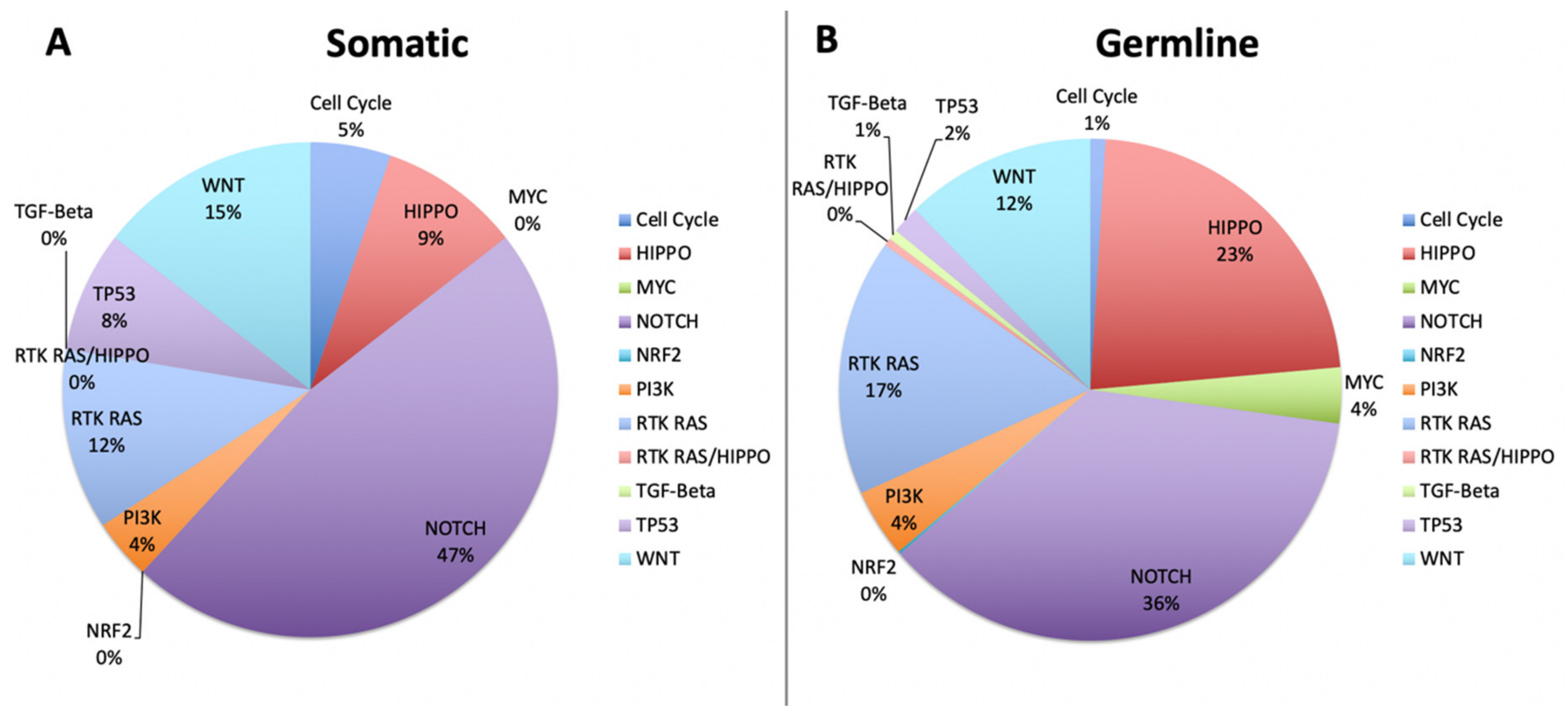
2.4. Cell Fate Pathways and TP53 Mutations Dominate Pathogenesis in Rare Cancers
2.5. Double Hit Analysis Shows Driver Oncogenes Correlate to CNAs
3. Discussion
4. Materials and Methods
4.1. Patients with Rare Cancers
4.2. Tumor and Germline Whole Exome and Gene Panel Sequencing
4.3. Sequence Analysis
4.4. Pathway and Double Hit Analysis
4.5. Copy Number Alteration Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| ACC | Adrenocortical carcinoma |
| CHOL | Cholangiocarcinoma |
| GBM | Glioblastoma multiforme |
| LGG | Brain Lower Grade Glioma |
| MESO | Mesothelioma |
| PCPG | Pheochromocytoma and Paraganglioma |
| SARC | Sarcoma |
| TGCT | Testicular Germ Cell Tumors |
| THYM | Thymoma |
| UCS | Uterine carcinoma |
| UVM | Uveal melanoma |
| BRCA | Breast invasive carcinoma |
| COAD | Colon adenocarcinoma |
| LUAD | Lung adenocarcinoma |
| LUSC | Lung squamous cell carcinoma |
| PRAD | Prostate adenocarcinoma |
| READ | Rectum adenocarcinoma |
| ACMG | American College of Medical Genetics |
References
- Dienstmann, R.; Rodon, J.; Barretina, J.; Tabernero, J. Genomic medicine frontier in human solid tumors: Prospects and challenges. J. Clin. Oncol. 2013, 31, 1874–1884. [Google Scholar] [CrossRef] [PubMed]
- Bradford, D.; Reilly, K.M.; Widemann, B.C.; Sandler, A.; Kummar, S. Developing therapies for rare tumors: Opportunities, challenges and progress. Exp. Opin. Orphan Drugs 2015, 4, 93–103. [Google Scholar] [CrossRef]
- Kato, S.; Kurasaki, K.; Ikeda, S.; Kurzrock, R. Rare tumor clinic: The university of california san diego moores cancer center experience with a precision therapy approach. Oncologist 2018, 23, 171–178. [Google Scholar] [CrossRef] [PubMed]
- Gatta, G.; van der Zwan, J.M.; Casali, P.G.; Siesling, S.; Dei Tos, A.P.; Kunkler, I.; Otter, R.; Licitra, L.; Mallone, S.; Tavilla, A.; et al. Rare cancers are not so rare: The rare cancer burden in Europe. Eur. J. Cancer 2011, 47, 2493–2511. [Google Scholar] [CrossRef] [PubMed]
- Tap, W.D.; Wainberg, Z.A.; Anthony, S.P.; Ibrahim, P.N.; Zhang, C.; Healey, J.H.; Chmielowski, B.; Staddon, A.P.; Cohn, A.L.; Shapiro, G.I.; et al. Structure-guided blockade of csf1r kinase in tenosynovial giant-cell tumor. N. Engl. J. Med. 2015, 373, 428–437. [Google Scholar] [CrossRef] [PubMed]
- Ballinger, M.L.; Goode, D.L.; Ray-Coquard, I.; James, P.A.; Mitchell, G.; Niedermayr, E.; Puri, A.; Schiffman, J.D.; Dite, G.S.; Cipponi, A.; et al. Monogenic and polygenic determinants of sarcoma risk: An international genetic study. Lancet Oncol. 2016, 17, 1261–1271. [Google Scholar] [CrossRef]
- Fishbein, L.; Nathanson, K.L. Pheochromocytoma and paraganglioma: Understanding the complexities of the genetic background. Cancer Genet. 2012, 205, 1–11. [Google Scholar] [CrossRef]
- Huang, K.L.; Mashl, R.J.; Wu, Y.; Ritter, D.I.; Wang, J.; Oh, C.; Paczkowska, M.; Reynolds, S.; Wyczalkowski, M.A.; Oak, N.; et al. Pathogenic germline variants in 10,389 adult cancers. Cell 2018. [Google Scholar] [CrossRef]
- Solomon, S.; Das, S.; Brand, R.; Whitcomb, D.C. Inherited pancreatic cancer syndromes. Cancer J. 2012, 18, 485–491. [Google Scholar] [CrossRef]
- Urbini, M.; Nannini, M.; Astolfi, A.; Indio, V.; Vicennati, V.; De Luca, M.; Tarantino, G.; Corso, F.; Saponara, M.; Gatto, L.; et al. Whole exome sequencing uncovers germline variants of cancer-related genes in sporadic pheochromocytoma. Int. J. Genom. 2018. [Google Scholar] [CrossRef]
- Larson, K.; Kannaiyan, R.; Pandey, R.; Chen, Y.; Babiker, H.M.; Mahadevan, D. A Comparative Analysis of Tumors and Plasma Circulating Tumor DNA in 145 Advanced Cancer Patients Annotated by 3 Core Cellular Processes. Cancers 2020, 12, 701. [Google Scholar] [CrossRef] [PubMed]
- National Cancer Registration and Analysis Service. Available online: http://www.ncin.org (accessed on 6 April 2020).
- Seifert, G.; Brocheriou, C.; Cardesa, A.; Everson, J.W. WHO international classification of tumours. Tentative histological classification of salivary gland tumours. Pathol. Res. Pract. 1990, 186, 555–581. [Google Scholar] [CrossRef]
- Duggan, M.A.; Anderson, W.F.; Altekruse, S.; Penberthy, L.; Sherman, M.E. The surveillance, epidemiology, and end results (SEER) program and pathology: Toward strengthening the critical relationship. Am. J. Surg. Pathol. 2016, 40, e94–e102. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, R.M.; Steenstrup Jensen, S.; Juel, J. Clear cell sarcoma—A review. J. Orthop. 2018, 15, 963–966. [Google Scholar] [CrossRef] [PubMed]
- Moten, A.S.; Jayarajan, S.N.; Willis, A.I. Spindle cell carcinoma of the breast: A comprehensive analysis. Am. J. Surg. 2016, 211, 716–721. [Google Scholar] [CrossRef] [PubMed]
- Søreide, K.; Sandvik, O.M.; Søreide, J.A.; Giljaca, V.; Jureckova, A.; Bulusu, V.R. Global epidemiology of gastrointestinal stromal tumours (gist): A systematic review of population-based cohort studies. Cancer Epidemiol. 2016, 40, 39–46. [Google Scholar] [CrossRef]
- Starrett, G.J.; Marcelus, C.; Cantalupo, P.G.; Katz, J.P.; Cheng, J.; Akagi, K.; Thakuria, M.; Rabinowits, G.; Wang, L.C.; Symer, D.E.; et al. Merkel cell polyomavirus exhibits dominant control of the tumor genome and transcriptome in virus-associated Merkel cell carcinoma. mBio 2017. [Google Scholar] [CrossRef]
- Collins, R.R.J.; Patel, K.; Putnam, W.C.; Kapur, P.; Rakheja, D. Oncometabolites: A New Paradigm for Oncology, Metabolism, and the Clinical Laboratory. Clin. Chem. 2017, 63, 1812–1820. [Google Scholar] [CrossRef]
- Postow, M.A.; Robson, M.E. Inherited gastrointestinal stromal tumor syndromes: Mutations, clinical features, and therapeutic implications. Clin. Sarcoma Res. 2012, 2, 16. [Google Scholar] [CrossRef]
- Bertelsen, B.; Tuxen, I.V.; Yde, C.W.; Gabrielaite, M.; Torp, M.H.; Kinalis, S.; Oestrup, O.; Rohrberg, K.; Spangaard, I.; Santoni-Rugiu, E.; et al. High frequency of pathogenic germline variants within homologous recombination repair in patients with advanced cancer. NPJ Genom. Med. 2019, 4, 13. [Google Scholar] [CrossRef]
- Favero, F.; Joshi, T.; Marquard, A.M.; Birkbak, N.J.; Krzystanek, M.; Li, Q.; Szallasi, Z.; Eklund, A.C. Sequenza: Allele-specific copy number and mutation profiles from tumor sequencing data. Ann. Oncol. 2015, 26, 64–70. [Google Scholar] [CrossRef] [PubMed]
- Mermel, C.H.; Schumacher, S.E.; Hill, B.; Meyerson, M.L.; Beroukhim, R.; Getz, G. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 2011, 12. [Google Scholar] [CrossRef] [PubMed]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio Cancer Genomics Portal: An Open Platform for Exploring Multidimensional Cancer Genomics Data. Cancer Discov. 2012, 2, 401. [Google Scholar] [CrossRef] [PubMed]
- Zack, T.I.; Schumacher, S.E.; Carter, S.L.; Cherniack, A.D.; Saksena, G.; Tabak, B.; Lawrence, M.S.; Zhang, C.-Z.; Wala, J.; Mermel, C.H.; et al. Pan-cancer patterns of somatic copy number alteration. Nat. Genet. 2013, 45, 1134–1140. [Google Scholar] [CrossRef]
- Hieronymus, H.; Murali, R.; Tin, A.; Yadav, K.; Abida, W.; Moller, H.; Berney, D.; Scher, H.; Carver, B.; Scardino, P.; et al. Tumor copy number alteration burden is a pan-cancer prognostic factor associated with recurrence and death. eLife 2018, 7, e37294. [Google Scholar] [CrossRef]
- Shlien, A.; Malkin, D. Copy number variations and cancer. Genome Med. 2009, 1, 62. [Google Scholar] [CrossRef]
- Sanchez-Vega, F.; Mina, M.; Armenia, J.; Chatila, W.K.; Luna, A.; La, K.C.; Dimitradoy, S.; Liu, D.L.; Kantheti, H.S.; Saghafinia, S.; et al. Oncogenic Signaling Pathways in The Cancer Genome Atlas. Cell 2018, 173, 321–337. [Google Scholar] [CrossRef]
- Yewale, C.; Baradia, D.; Vhora, I.; Patil, S.; Misra, A. Epidermal growth factor receptor targeting in cancer: A review of trends and strategies. Biomaterials 2013, 34, 8690–8707. [Google Scholar] [CrossRef]
- Charpidou, A.; Blatza, D.; Anagnostou, V.; Syrigos, K.N. EGFR mutations in non-small cell lung cancer-clinical implications. In Vivo 2008, 22, 529–536. [Google Scholar]
- Groisberg, R.; Hong, D.S.; Roszik, J.; Janku, F.; Tsimberidou, A.M.; Javle, M.; Meric-Bernstam, F.; Subbiah, V. Clinical next-generation sequencing for precision oncology in rare cancers. Mol. Cancer Ther. 2018, 17, 1595–1601. [Google Scholar] [CrossRef]
- Jones, S.; Anagnostou, V.; Lytle, K.; Parpart-Li, S.; Nesselbush, M.; Riley, D.R.; Shukla, M.; Chesnick, B.; Kadan, M.; Papp, E.; et al. Personalized genomic analyses for cancer mutation discovery and interpretation. Sci. Transl. Med. 2015, 7, 283ra253. [Google Scholar] [CrossRef] [PubMed]
- Rahman, N. Realizing the promise of cancer predisposition genes. Nature 2014, 505, 302–308. [Google Scholar] [CrossRef] [PubMed]
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hegde, M.; Lyon, E.; Spector, E.; et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the american college of medical genetics and genomics and the association for molecular pathology. Genet. Med. 2015, 17, 405–424. [Google Scholar] [CrossRef] [PubMed]
- Carro, M.S.; Lim, W.K.; Alvarez, M.J.; Bollo, R.J.; Zhao, X.; Snyder, E.Y.; Sulman, E.P.; Anne, S.L.; Doetsch, F.; Colman, H.; et al. The transcriptional network for mesenchymal transformation of brain tumours. Nature 2010, 463, 318–325. [Google Scholar] [CrossRef]
- Bogoch, Y.; Friedlander-Malik, G.; Lupu, L.; Bondar, E.; Zohar, N.; Langier, S.; Ram, Z.; Nachmany, I.; Klausner, J.M.; Pencovich, N. Augmented expression of runx1 deregulates the global gene expression of u87 glioblastoma multiforme cells and inhibits tumor growth in mice. Tumour Biol. 2017. [Google Scholar] [CrossRef]
- Zhang, J.; Walsh, M.F.; Wu, G.; Edmonson, M.N.; Gruber, T.A.; Easton, J.; Hedges, D.; Ma, X.; Zhou, X.; Yergeau, D.A.; et al. Germline mutations in predisposition genes in pediatric cancer. N. Engl. J. Med. 2015, 373, 2336–2346. [Google Scholar] [CrossRef]
- Cheng, H.; Powers, J.; Schaffer, K.; Sartor, O. Practical methods for integrating genetic testing into clinical practice for advanced prostate cancer. Am. Soc. Clin. Oncol. Educ. Book 2018, 38, 372–381. [Google Scholar] [CrossRef]
- Abiria, S.A.; Williams, T.V.; Munden, A.L.; Grover, V.K.; Wallace, A.; Lundberg, C.J.; Valadez, J.G.; Cooper, M.K. Expression of Hedgehog ligand and signal transduction components in mutually distinct isocitrate dehydrogenase mutant glioma cells supports a role for paracrine signaling. J. Neurooncol. 2014, 119, 243–251. [Google Scholar] [CrossRef][Green Version]
- Infante, P.; Faedda, R.; Bernardi, F.; Bufalieri, F.; Lospinoso Severini, L.; Alfonsi, R.; Mazzà, D.; Siler, M.; Coni, S.; Po, A.; et al. Itch/beta-arrestin2-dependent non-proteolytic ubiquitylation of sufu controls hedgehog signalling and medulloblastoma tumorigenesis. Nat. Commun. 2018, 9, 976. [Google Scholar] [CrossRef]
- Hinsch, A.; Brolund, M.; Hube-Magg, C.; Kluth, M.; Simon, R.; Möller-Koop, C.; Sauter, G.; Steurer, S.; Luebke, A.; Angerer, A.; et al. Immunohistochemically detected idh1(r132h) mutation is rare and mostly heterogeneous in prostate cancer. World J. Urol. 2018, 36, 877–882. [Google Scholar] [CrossRef]
- Huang, A.; Garraway, L.A.; Ashworth, A.; Weber, B. Synthetic lethality as an engine for cancer drug target discovery. Nat. Rev. Drug Discov. 2020, 19, 23–38. [Google Scholar] [CrossRef] [PubMed]
- Shah, S.P.; Köbel, M.; Senz, J.; Morin, R.D.; Clarke, B.A.; Wiegand, K.C.; Leung, G.; Zayed, A.; Mehl, E.; Kalloger, S.E.; et al. Mutation of FOXL2 in granulosa-cell tumors of the ovary. N. Engl. J. Med. 2009, 360, 2719–2729. [Google Scholar] [CrossRef] [PubMed]
- Ghadban, T.; Schmidt-Yang, M.; Uzunoglu, F.G.; Perez, D.R.; El Gammal, A.T.; Miro, J.T.; Wellner, U.; Pantel, K.; Izbicki, J.R.; Vashist, Y.K. Evaluation of the germline single nucleotide polymorphism rs583522 in the tnfaip3 gene as a prognostic marker in esophageal cancer. Cancer Genet. 2015, 208, 595–601. [Google Scholar] [CrossRef]
- Liu, C.; Fu, X.; Zhong, Z.; Zhang, J.; Mou, H.; Wu, Q.; Sheng, T.; Huang, B.; Zou, Y. CHD1L Expression Increases Tumor Progression and Acts as a Predictive Biomarker for Poor Prognosis in Pancreatic Cancer. Dig. Dis. Sci. 2017, 62, 2376–2385. [Google Scholar] [CrossRef] [PubMed]
- Marshall, A.D.; Bailey, C.G.; Champ, K.; Vellozzi, M.; O’Young, P.; Metierre, C.; Feng, Y.; Thoeng, A.; Richards, A.M.; Schmitz, U.; et al. CTCF genetic alterations in endometrial carcinoma are pro-tumorigenic. Oncogene 2017, 36, 4100–4110. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.E.; Tzen, C.Y.; Wang, S.Y.; Yeh, C.N. Clinical Diagnosis of Gastrointestinal Stromal Tumor (GIST): From the Molecular Genetic Point of View. Cancers 2019, 16, 11. [Google Scholar] [CrossRef]
- Flavahan, W.A.; Drier, Y.; Johnstone, S.E.; Hemming, M.L.; Tarjan, D.R.; Hegazi, E.; Shareef, S.J.; Javed, N.M.; Raut, C.P.; Eschle, B.K.; et al. Altered chromosomal topology drives oncogenic programs in SDH-deficient GISTs. Nature 2019, 575, 229–233. [Google Scholar] [CrossRef]
- Ock, C.-Y.; Hwang, J.-E.; Keam, B.; Kim, S.-B.; Shim, J.-J.; Jang, H.-J.; Park, S.; Sohn, B.H.; Cha, M.; Ajani, J.A.; et al. Genomic landscape associated with potential response to anti-CTLA-4 treatment in cancers. Nat. Commun. 2017, 8, 1050. [Google Scholar] [CrossRef]
- Davis, L.E.; Nicholls, L.A.; Babiker, H.M.; Liau, J.; Mahadevan, D. PD-1 Inhibition Achieves a Complete Metabolic Response in a Patient with Malignant Peripheral Nerve Sheath Tumor. Cancer Immunol. Res. 2019, 7, 1396–1400. [Google Scholar] [CrossRef]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The genome analysis toolkit: A mapreduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef]
- Cibulskis, K.; Lawrence, M.S.; Carter, S.L.; Sivachenko, A.; Jaffe, D.; Sougnez, C.; Gabriel, S.; Meyerson, M.; Lander, E.S.; Getz, G. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat. Biotechnol. 2013, 31, 213–219. [Google Scholar] [CrossRef] [PubMed]
- Garrison, E.; Marth, G. Haplotype-based variant detection from short-read sequencing. arXiv 2012, arXiv:1207.3907. [Google Scholar]
- Wang, K.; Li, M.; Hakonarson, H. ANNOVAR: Functional annotation of genetic variants from next-generation sequencing data. Nucleic Acids Res. 2010, 38, e164. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Sun, J.; Zhao, M. ONGene, a literature-based database for human oncogenes. J. Genet. Genom. 2017, 44, 119–121. [Google Scholar] [CrossRef] [PubMed]
- TSGene 2.0. Available online: https://bioinfo.uth.edu/TSGene/ (accessed on 20 November 2019).
- Tamborero, D.; Rubio-Perez, C.; Deu-Pons, J.; Schroeder, M.P.; Vivancos, A.; Rovira, A.; Tusquets, I.; Albanell, J.; Rodon, J.; Tabernero, J.; et al. Cancer Genome Interpreter annotates the biological and clinical relevance of tumor alterations. Genome Med. 2018, 10, 25. [Google Scholar] [CrossRef] [PubMed]
- Lawrence, M.; Huber, W.; Pagès, H.; Aboyoun, P.; Carlson, M.; Gentleman, R.; Morgan, M.T.; Carey, V.J. Software for Computing and Annotating Genomic Ranges. PLoS Comput. Biol. 2013. [Google Scholar] [CrossRef]
- Cavalcante, R.G.; Sartor, M.A. annotatr: Genomic regions in context. Bioinformatics 2017, 33, 2381–2383. [Google Scholar] [CrossRef]
- Carlson, M. org.Hs.eg.db: Genome Wide Annotation for Human. R Package Version 3.8.2. 2019. Available online: https://bioconductor.org/packages/release/data/annotation/html/org.Hs.eg.db.html (accessed on 24 May 2019).
- Durinck, S.; Spellman, P.T.; Birney, E.; Huber, W. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat. Protoc. 2009, 4, 1184–1191. [Google Scholar] [CrossRef]
- Anand, L. chromoMap: Interactive Visualization and Mapping of Chromosomes. R Package Version 0.2. 2019. Available online: https://CRAN.R-project.org/package=chromoMap (accessed on 9 September 2019).
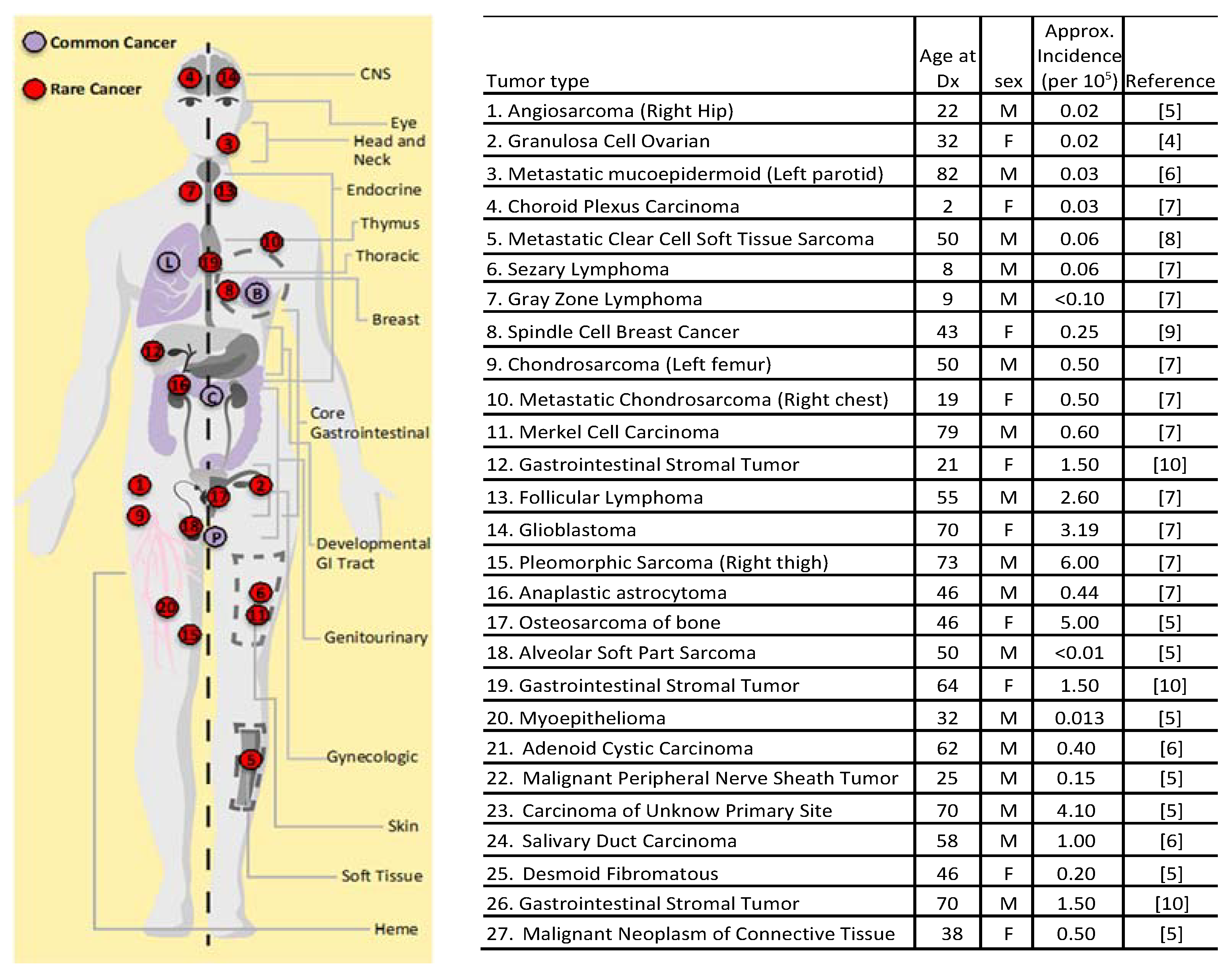

| Germline TSG Drivers | ||||||||
|---|---|---|---|---|---|---|---|---|
| Case | Ref | Alt | Gene | ExonicFunc.refGene | avsnp150 | gnomAD Freq. | Polyphen2 | PhyloP |
| 4 | G | A | MAD1L1 | nonsyn. SNV | rs121908982 | 0.0036 | Prob. Damaging | 5.785 |
| 7 | C | G | IGF2R | nonsyn. SNV | rs8191844 | 0.0102 | Prob. Damaging | 4.24 |
| 7 | C | - | BRCA2 | frameshift deletion | rs80359718 | N/A | N/A | N/A |
| 13 | G | A | NUP98 | nonsyn. SNV | rs61751338 | 0.0014 | Prob. Damaging | 6.327 |
| 13 | T | C | TET2 | nonsyn. SNV | rs144386291 | 0.0055 | Prob. Damaging | 6.829 |
| 2 | C | T | EPHA2 | nonsyn. SNV | rs34192549 | 0.0123 | Benign | 0.073 |
| 12 | TGGTGAAGAACATTCAGGCAA | - | BARD1 | inframe deletion | rs28997575 | 0.024 | N/A | N/A |
| 5 | C | T | EPHA2 | nonsyn. SNV | rs34192549 | 0.0123 | Benign | 0.073 |
| 5 | G | A | IGF2R | nonsyn. SNV | rs8191753 | 0.0023 | Prob. Damaging | 3.63 |
| 1 | C | T | EXT2 | nonsyn. SNV | rs138495222 | 0.0006 | Prob. Damaging | 7.813 |
| 14 | G | A | AXIN2 | nonsyn. SNV | rs138287857 | 0.0012 | Poss. Damaging | 7.551 |
| 15 | G | C | LZTS1 | nonsyn. SNV | rs148775156 | 0.001 | Prob. Damaging | 2.853 |
| 15 | G | A | PTCH1 | nonsyn. SNV | rs138911275 | 0.0007 | Poss. Damaging | 7.645 |
| 11 | G | A | ARID2 | nonsyn. SNV | rs200040222 | 0.0001 | Prob. Damaging | 9.317 |
| 10 | C | A | DLC1 | nonsyn. SNV | . | N/A | Prob. Damaging | 2.753 |
| 3 | TGGTGAAGAACATTCAGGCAA | - | BARD1 | inframe deletion | rs28997575 | 0.024 | N/A | N/A |
| 3 | C | T | CTNND1 | nonsyn. SNV | rs199813020 | 0.0003 | Prob. Damaging | 2.101 |
| 8 | G | A | STARD13 | nonsyn. SNV | rs144801804 | 0.0008 | Prob. Damaging | 9.889 |
| 8 | C | T | SDHA | stopgain | rs781764920 | 6.37E-05 | N/A | 2.131 |
| 8 | - | AGG | FBXW7 | inframe insertion | rs541979458 | 0.0014 | N/A | N/A |
| 8 | C | T | ATM | nonsyn. SNV | rs28904919 | 0.0014 | Poss. Damaging | 3.491 |
| 16 | A | C | NOTCH2 | nonsyn. SNV | rs147223770 | 0.0032 | Poss. Damaging | 8.947 |
| 16 | C | T | ATM | nonsyn. SNV | rs56009889 | 0.0004 | Prob. Damaging | 3.766 |
| 17 | G | A | CHEK2 | nonsyn. SNV | rs730881690 | N/A | Prob. Damaging | 8.668 |
| 18 | C | T | EPHA2 | nonsyn. SNV | rs34192549 | 0.0123 | Benign | 0.073 |
| 18 | - | GCGGGT | CEBPA | inframe insertion | rs762459325 | 0.0416 | N/A | N/A |
| 20 | G | A | BRCA2 | nonsyn. SNV | rs41293503 | N/A | Prob. Damaging | 3.408 |
| 20 | C | G | KMT2C | nonsyn. SNV | rs138119145 | 0.0065 | Prob. Damaging | 6.729 |
| 20 | C | T | ATR | nonsyn. SNV | . | N/A | Poss. Damaging | 6.179 |
| 21 | T | C | DICER1 | nonsyn. SNV | rs747510783 | N/A | Prob. Damaging | 7.502 |
| 21 | T | C | ATM | nonsyn. SNV | . | N/A | Poss. Damaging | 6.589 |
| 21 | A | G | MSH2 | nonsyn. SNV | rs773177076 | N/A | Prob. Damaging | 9.147 |
| 25 | T | C | TET2 | nonsyn. SNV | rs144386291 | 0.0055 | Prob. Damaging | 6.829 |
| 25 | T | C | NUP98 | nonsyn. SNV | rs201011075 | 0.0005 | Prob. Damaging | 4.516 |
| 22 | A | G | LZTS1 | nonsyn. SNV | rs149140637 | 0.0031 | Prob. Damaging | 7.441 |
| 22 | C | T | LATS1 | nonsyn. SNV | rs148506316 | 0.0002 | Benign | 5.885 |
| 23 | G | T | PPP2R5C | nonsyn. SNV | rs147942579 | 0.0003 | Prob. Damaging | 9.014 |
| 24 | A | G | SDHB | nonsyn. SNV | rs771004483 | N/A | Poss. Damaging | 8.563 |
| 24 | C | T | EPHA2 | nonsyn. SNV | rs139787163 | 0.0003 | Prob. Damaging | 1.668 |
| 24 | C | T | GPC3 | nonsyn. SNV | rs11539789 | 0.003 | Benign | 2.917 |
| 24 | A | G | SDHD | nonsyn. SNV | rs11214077 | 0.0048 | Prob. Damaging | 2.521 |
| 26 | C | T | SMARCA4 | nonsyn. SNV | rs763471007 | N/A | Prob. Damaging | 7.813 |
| 26 | G | A | TSC1 | nonsyn. SNV | rs878853968 | N/A | Prob. Damaging | 8.039 |
| 27 | C | T | EPHA2 | nonsyn. SNV | rs34192549 | 0.0123 | Benign | 0.073 |
| 27 | - | GCGGGT | CEBPA | inframe insertion | rs762459325 | 0.0416 | N/A | N/A |
| 19 | T | A | MCC | nonsyn. SNV | rs17313892 | 0.0076 | Prob. Damaging | 4.64 |
| 19 | C | T | EPHA2 | nonsyn. SNV | rs34192549 | 0.0123 | Benign | 0.073 |
| 13 | G | A | TXNIP | nonsyn. SNV | rs781868836 | N/A | N/A | 4.673 |
| Germline Oncogene Drivers | ||||||||
| Case | Ref | Alt | Gene | ExonicFunc.refGene | avsnp150 | gnomAD Freq. | Polyphen2 | PhyloP |
| 13 | G | A | NUP98 | nonsyn. SNV | rs61751338 | 0.0014 | Prob. Damaging | 6.327 |
| 13 | C | T | CSF1R | nonsyn. SNV | rs138432536 | 0.003 | Poss. Damaging | 3.03 |
| 2 | C | T | EPHA2 | nonsyn. SNV | rs34192549 | 0.0123 | Benign | 0.073 |
| 12 | TGGTGAAGAACATTCAGGCAA | - | BARD1 | inframe deletion | rs28997575 | 0.024 | N/A | N/A |
| 5 | C | T | EPHA2 | nonsyn. SNV | rs34192549 | 0.0123 | Benign | 0.073 |
| 1 | G | C | MUC4 | nonsyn. SNV | rs369326402 | 0.0001 | Poss. Damaging | −1.2 |
| 14 | C | T | MET | nonsyn. SNV | rs34589476 | 0.0025 | Benign | 3.567 |
| 15 | G | A | PTCH1 | nonsyn. SNV | rs138911275 | 0.0007 | Poss. Damaging | 7.645 |
| 9 | G | A | NSD2 | nonsyn. SNV | rs758343111 | N/A | Poss. Damaging | 3.202 |
| 10 | T | - | CHD1L | stopgain | rs781989601 | N/A | N/A | N/A |
| 3 | TGGTGAAGAACATTCAGGCAA | - | BARD1 | inframe deletion | rs28997575 | 0.024 | N/A | N/A |
| 3 | GGAGCTCCATCC | - | TRIO | inframe deletion | rs140308852 | 0.0076 | N/A | N/A |
| 18 | T | G | PDGFRA | nonsyn. SNV | rs148654387 | 0.0004 | Poss. Damaging | 6.111 |
| 18 | C | T | EPHA2 | nonsyn. SNV | rs34192549 | 0.0123 | Benign | 0.073 |
| 18 | C | T | MYC | nonsyn. SNV | rs200431478 | 0.0002 | Prob. Damaging | 4.735 |
| 25 | G | C | MUC4 | nonsyn. SNV | rs369326402 | 0.0001 | Poss. Damaging | −1.2 |
| 25 | T | C | NUP98 | nonsyn. SNV | rs201011075 | 0.0005 | Prob. Damaging | 4.516 |
| 24 | G | A | FIP1L1 | nonsyn. SNV | rs777738679 | N/A | Prob. Damaging | 7.569 |
| 24 | C | T | EPHA2 | nonsyn. SNV | rs139787163 | 0.0003 | Prob. Damaging | 1.668 |
| 26 | G | C | MUC4 | nonsyn. SNV | rs369326402 | 0.0001 | Poss. Damaging | −1.2 |
| 27 | C | T | EPHA2 | nonsyn. SNV | rs34192549 | 0.0123 | Benign | 0.073 |
| 19 | T | A | MCC | nonsyn. SNV | rs17313892 | 0.0076 | Prob. Damaging | 4.64 |
| 19 | G | C | MUC4 | nonsyn. SNV | rs369326402 | 0.0001 | Poss. Damaging | -1.2 |
| 19 | C | T | EPHA2 | nonsyn. SNV | rs34192549 | 0.0123 | Benign | 0.073 |
| Somatic TSG Drivers | ||||||||
| Case | Ref | Alt | Gene | ExonicFunc.refGene | avsnp150 | gnomAD Freq. | Polyphen2 | PhyloP |
| 4 | G | A | TP53 | nonsyn. SNV | rs876659802 | N/A | Prob. damaging | 10.003 |
| 13 | C | - | ARID1A | frameshift deletion | . | N/A | N/A | N/A |
| 13 | G | A | CREBBP | stopgain | . | N/A | N/A | 6.795 |
| 2 | G | C | FOXL2 | nonsyn. SNV | rs1057519865 | N/A | Prob. damaging | 2.22 |
| 12 | C | T | KMT2C | nonsyn. SNV | rs145833795 | 9.55E-05 | Prob. damaging | 4.525 |
| 1 | A | - | RB1 | frameshift deletion | . | N/A | N/A | N/A |
| 14 | G | A | TP53 | nonsyn. SNV | rs587780070 | N/A | Prob. damaging | 10.003 |
| 14 | A | T | TP53 | nonsyn. SNV | rs1057519982 | N/A | Prob. damaging | 9.325 |
| 15 | G | A | CDKN2A | stopgain | rs121913388 | N/A | Benign | 1.26 |
| 15 | G | C | IDH1 | nonsyn. SNV | rs121913499 | N/A | Poss. damaging | 7.103 |
| 9 | G | A | IDH1 | nonsyn. SNV | rs121913499 | N/A | Benign | 7.103 |
| 10 | C | A | TP53 | stopgain | rs201744589 | N/A | N/A | −0.746 |
| 19 | - | T | SETD2 | frameshift insertion | . | N/A | N/A | N/A |
| 19 | AAG | - | CTCF | inframe deletion | . | N/A | N/A | N/A |
| 7 | TCATCTCA | - | TNFAIP3 | frameshift deletion | . | N/A | N/A | N/A |
| Somatic Oncogene Drivers | ||||||||
| Case | Ref | Alt | Gene | ExonicFunc.refGene | avsnp150 | gnomAD Freq. | Polyphen2 | PhyloP |
| 7 | GGAG | - | CHD1L | frameshift deletion | rs782573713 | N/A | N/A | N/A |
| 2 | G | C | MUC4 | nonsyn. SNV | rs369326402 | 0.0001 | Poss. damaging | −1.2 |
| 15 | G | C | IDH1 | nonsyn. SNV | rs121913499 | N/A | Poss. damaging | 7.103 |
| 9 | G | A | IDH1 | nonsyn. SNV | rs121913499 | N/A | Benign | 7.103 |
| 3 | G | T | HRAS | nonsyn. SNV | rs28933406 | N/A | Benign | 9.821 |
| 3 | G | A | PIK3CA | nonsyn. SNV | rs121913273 | N/A | Prob. damaging | 9.602 |
| 16 | C | T | EGFR | nonsyn. SNV | rs149840192 | N/A | Prob. damaging | 7.882 |
| 20 | G | A | PIK3CA | nonsyn. SNV | rs121913273 | N/A | Prob. damaging | 9.602 |
| 19 | AGTGGA | - | KIT | inframe deletion | rs869025568 | N/A | N/A | N/A |
| Case ID | Cancer Type | Gene | Mutation Type | Coding Change | Protein Change | gnomAD Freq | Polyphen2 | PhyloP | ACMG Classification |
|---|---|---|---|---|---|---|---|---|---|
| 7 | Gray Zone Lymphoma | BRCA2 | Frameshift | NM_000059.3: c.8575delC | NP_000050.2: p.Gln2859Lysfs | N/A | N/A | N/A | Pathogenic |
| 8 | Breast Cancer, Spindle Cell | SDHA | Nonsense | NM_004168.3: c.223C>T | NP_004159.2: p.Arg75Ter | 6.47E-05 | N/A | 2.131 | Pathogenic |
| 12 | GIST | SDHC | Frameshift | NM_003001.3: c.6delT | NP_002992.1: p.Ala3Argfs | N/A | N/A | N/A | Pathogenic |
| 14 | Glioblastoma | RUNX1 | Missense | NM_001754.4: c.451A>T | NP_001745.2: p.Met151Leu | N/A | N/A | N/A | Likely Pathogenic |
| 16 | Glioma (Anaplastic Astrocytoma) | FANCC | Splice Region (Exon Skipping) | NM_000136.2: c.456+4A>T | N/A | 0.0001 | N/A | N/A | Pathogenic |
| 18 | Aveolar Soft Part Sarcoma | MUTYH | Missense | NM_001128425.1: c.1187G>A | NP_001121897.1: p.Gly396Asp | 0.0032 | Prob. damaging | 4.511 | Pathogenic |
| Cancer Case Set | Total Cases | Pathogenic/Likely Pathogenic Germline Variant | No Pathogenic Germline Variant |
|---|---|---|---|
| Rare Cancer (UAGC) | 27 | 6 (22.2%) | 21 (77.8%) |
| Common Cancer (Total) | 3451 | 274 (7.9%) | 3177 (92.1%) |
| Fisher Exact Test p-value, UAGC vs. Total Common: 0.01800 | |||
| Common Cancers | Total Cases | Pathogenic/Likely Pathogenic Germline Variant | No Pathogenic Germline Variant |
| CRC [21] | 141 | 27 | 114 |
| Breast [21] | 85 | 16 | 69 |
| Prostate [21] | 26 | 3 | 23 |
| NSCLC [21] | 33 | 4 | 29 |
| SCLC [21] | 11 | 0 | 11 |
| COAD (colon) [14] | 419 | 25 | 394 |
| READ (rectal) [14] | 145 | 6 | 139 |
| BRCA (breast) [14] | 1076 | 106 | 970 |
| PRAD (prostate) [14] | 498 | 27 | 471 |
| LUAD (lung adeno) [14] | 518 | 33 | 485 |
| LUSC (lung squamous) [14] | 499 | 27 | 472 |
| Common - Total | 3451 | 274 | 3177 |
| Rare Cancers | Total Cases | Pathogenic/Likely Pathogenic Germline Variant | No Pathogenic Germline Variant | ||
|---|---|---|---|---|---|
| UAGC Rare Cancer | 27 | 6 (22.2%) | 21 (77.8%) | ||
| Huang et al. [14] Rare Cancer | 1955 | 163 (8.3%) | 1792 (91.7%) | ||
| Bertelsen et al. [21] Rare Cancer | 122 | 22 (18.0%) | 100 (82.0%) | ||
| [14] + [21] Rare Cancer | 2077 | 185 (8.9%) | 1892 (91.1%) | ||
| Fisher Exact Test p-value, UAGC vs. [14] + [21]: 0.03035 | |||||
| Huang et al. [14] Rare Cancers: | Total Cases | Pathogenic/Likely Pathogenic Germline Variant | No Pathogenic Germline Variant | ||
| Adrenocortical Carcinoma | 92 | 4 | 88 | ||
| Cholangiocarcinoma | 45 | 1 | 44 | ||
| Glioblastoma Multiforme | 393 | 23 | 370 | ||
| Brain Lower Grade Glioma | 515 | 31 | 484 | ||
| Mesothelioma | 82 | 7 | 75 | ||
| Pheochromocytoma and Paraganglioma | 179 | 41 | 138 | ||
| Sarcoma | 255 | 32 | 223 | ||
| Thymoma | 123 | 6 | 117 | ||
| Uterine Carcinosarcoma | 57 | 2 | 55 | ||
| Uveal Melanoma | 80 | 4 | 76 | ||
| Testicular Germ Cell Tumors | 134 | 12 | 122 | ||
| Sum | 1955 | 163 | 1792 | ||
| Bertelsen et al. [21] Rare Cancers: | Total Cases | Pathogenic/Likely Pathogenic Germline Variant | No Pathogenic Germline Variant | ||
| Bile Duct Cancer | 47 | 8 | 39 | ||
| Sarcoma | 14 | 3 | 11 | ||
| Neuroendocrine Cancer | 13 | 1 | 12 | ||
| Malignant Mesothelioma | 12 | 7 | 5 | ||
| Adrenocortical Cancer | 8 | 0 | 8 | ||
| Thymoma | 8 | 1 | 7 | ||
| Adenoid Cystic Carcinoma | 5 | 1 | 4 | ||
| Myoepithelial Carcinoma | 4 | 0 | 4 | ||
| Glioblastoma | 4 | 0 | 4 | ||
| Anogenital Cancer | 3 | 0 | 3 | ||
| Vulvovaginal Cancer | 2 | 1 | 1 | ||
| Germ Cell Cancer | 2 | 0 | 2 | ||
| Sum | 122 | 22 | 100 | ||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sprissler, R.; Perkins, B.; Johnstone, L.; Babiker, H.M.; Chalasani, P.; Lau, B.; Hammer, M.; Mahadevan, D. Rare Tumor-Normal Matched Whole Exome Sequencing Identifies Novel Genomic Pathogenic Germline and Somatic Aberrations. Cancers 2020, 12, 1618. https://doi.org/10.3390/cancers12061618
Sprissler R, Perkins B, Johnstone L, Babiker HM, Chalasani P, Lau B, Hammer M, Mahadevan D. Rare Tumor-Normal Matched Whole Exome Sequencing Identifies Novel Genomic Pathogenic Germline and Somatic Aberrations. Cancers. 2020; 12(6):1618. https://doi.org/10.3390/cancers12061618
Chicago/Turabian StyleSprissler, Ryan, Bryce Perkins, Laurel Johnstone, Hani M. Babiker, Pavani Chalasani, Branden Lau, Michael Hammer, and Daruka Mahadevan. 2020. "Rare Tumor-Normal Matched Whole Exome Sequencing Identifies Novel Genomic Pathogenic Germline and Somatic Aberrations" Cancers 12, no. 6: 1618. https://doi.org/10.3390/cancers12061618
APA StyleSprissler, R., Perkins, B., Johnstone, L., Babiker, H. M., Chalasani, P., Lau, B., Hammer, M., & Mahadevan, D. (2020). Rare Tumor-Normal Matched Whole Exome Sequencing Identifies Novel Genomic Pathogenic Germline and Somatic Aberrations. Cancers, 12(6), 1618. https://doi.org/10.3390/cancers12061618







