The miR-26b-5p/KPNA2 Axis Is an Important Regulator of Burkitt Lymphoma Cell Growth
Abstract
1. Introduction
2. Results
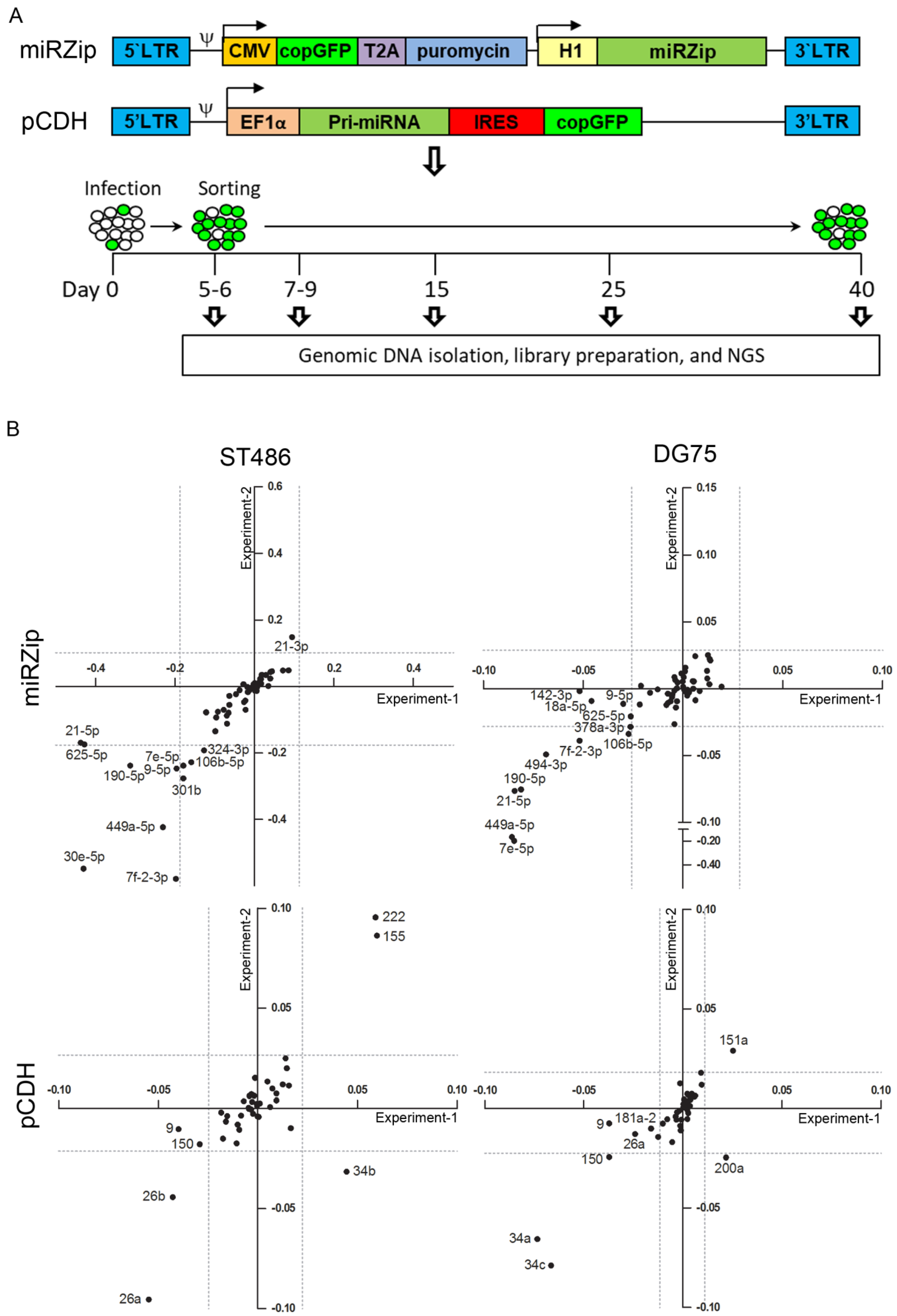
2.1. Validation of the Efficiency of the miRNA Inhibition and Overexpression Pools
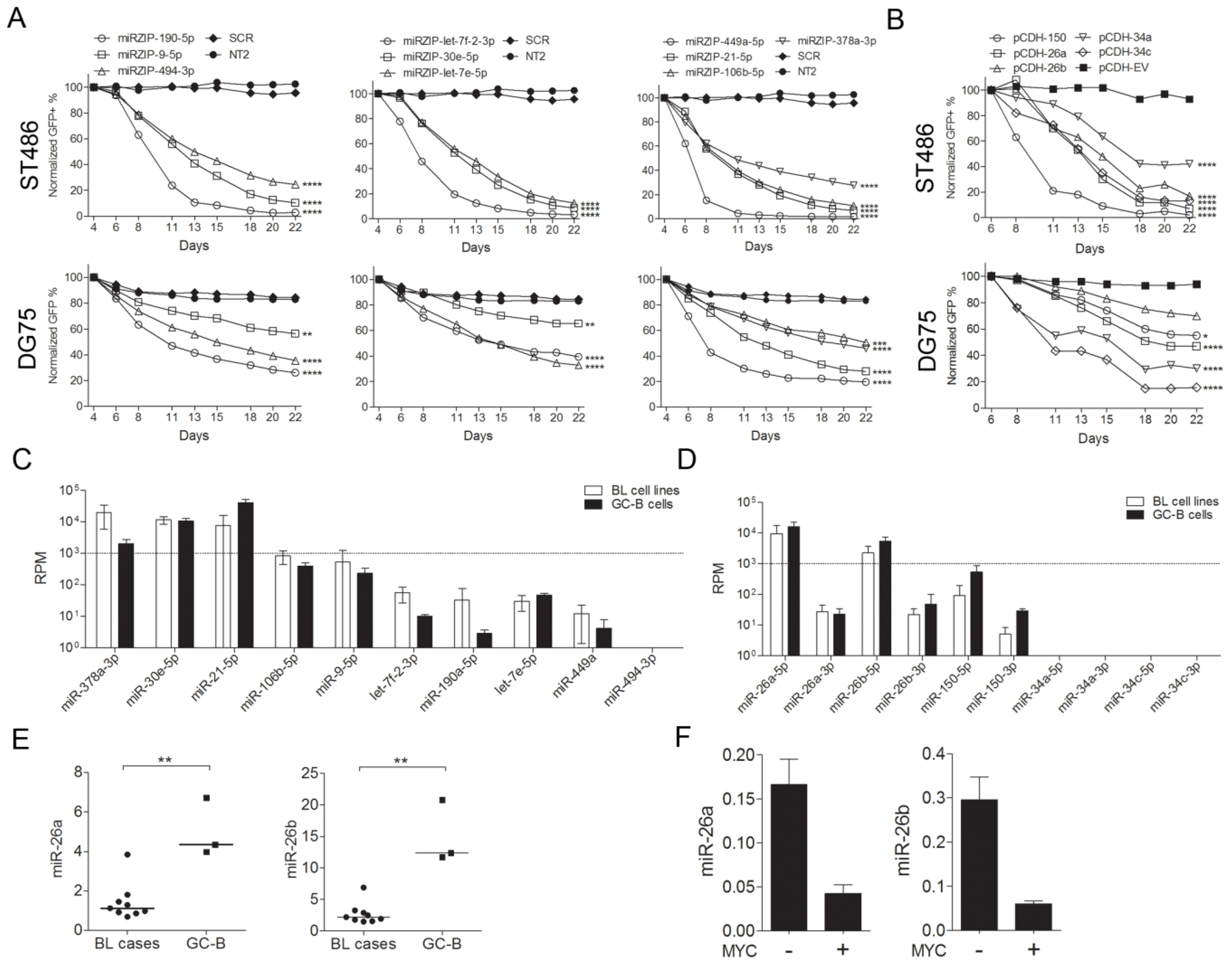
2.2. Identification of miRNAs Affecting the Growth of BL Cells
2.3. Selection of MiRNA Candidates for Functional Studies
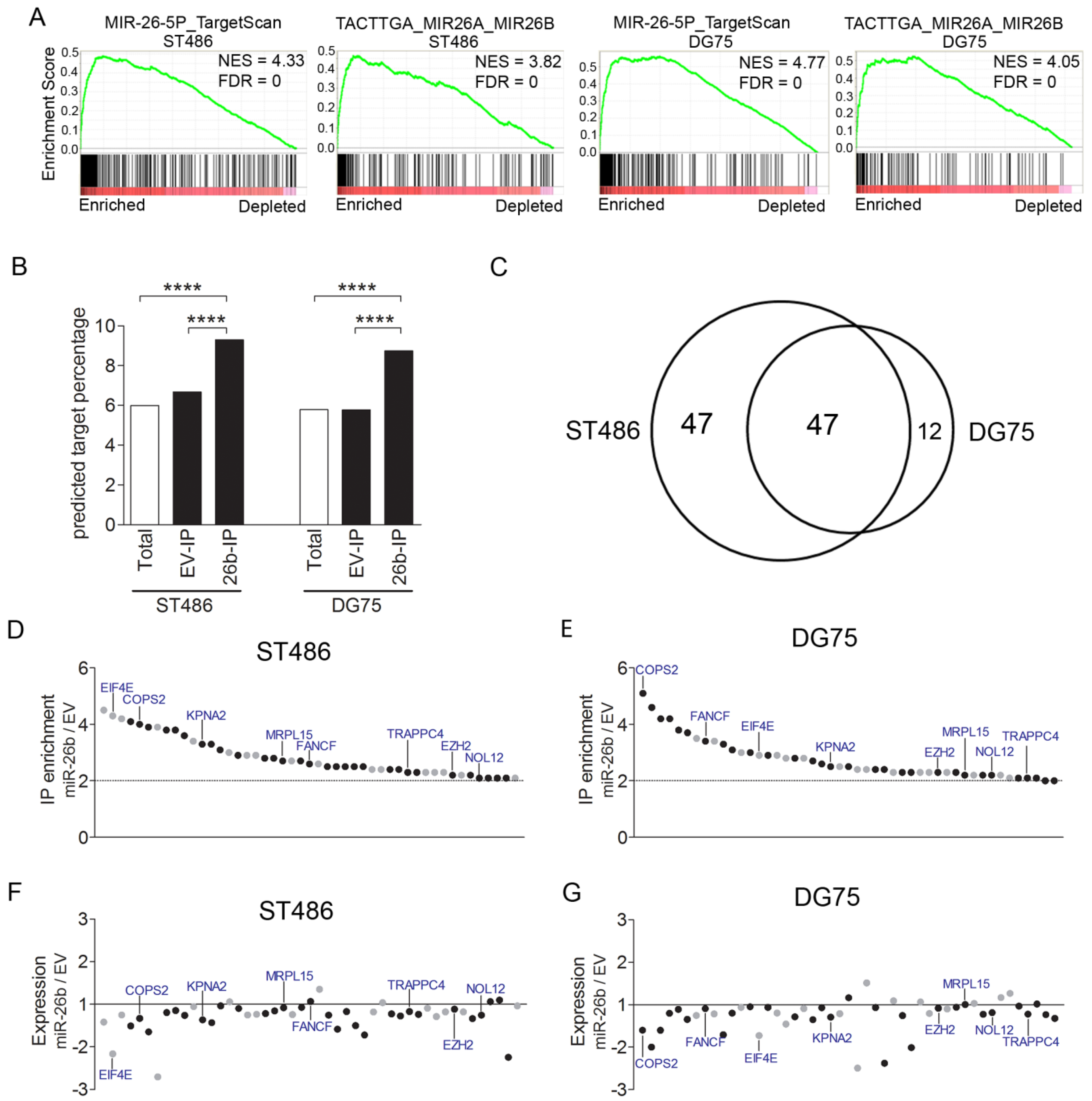
2.4. Identification of Targets of the miR-26-5p Family by Ago2-RIP-Chip
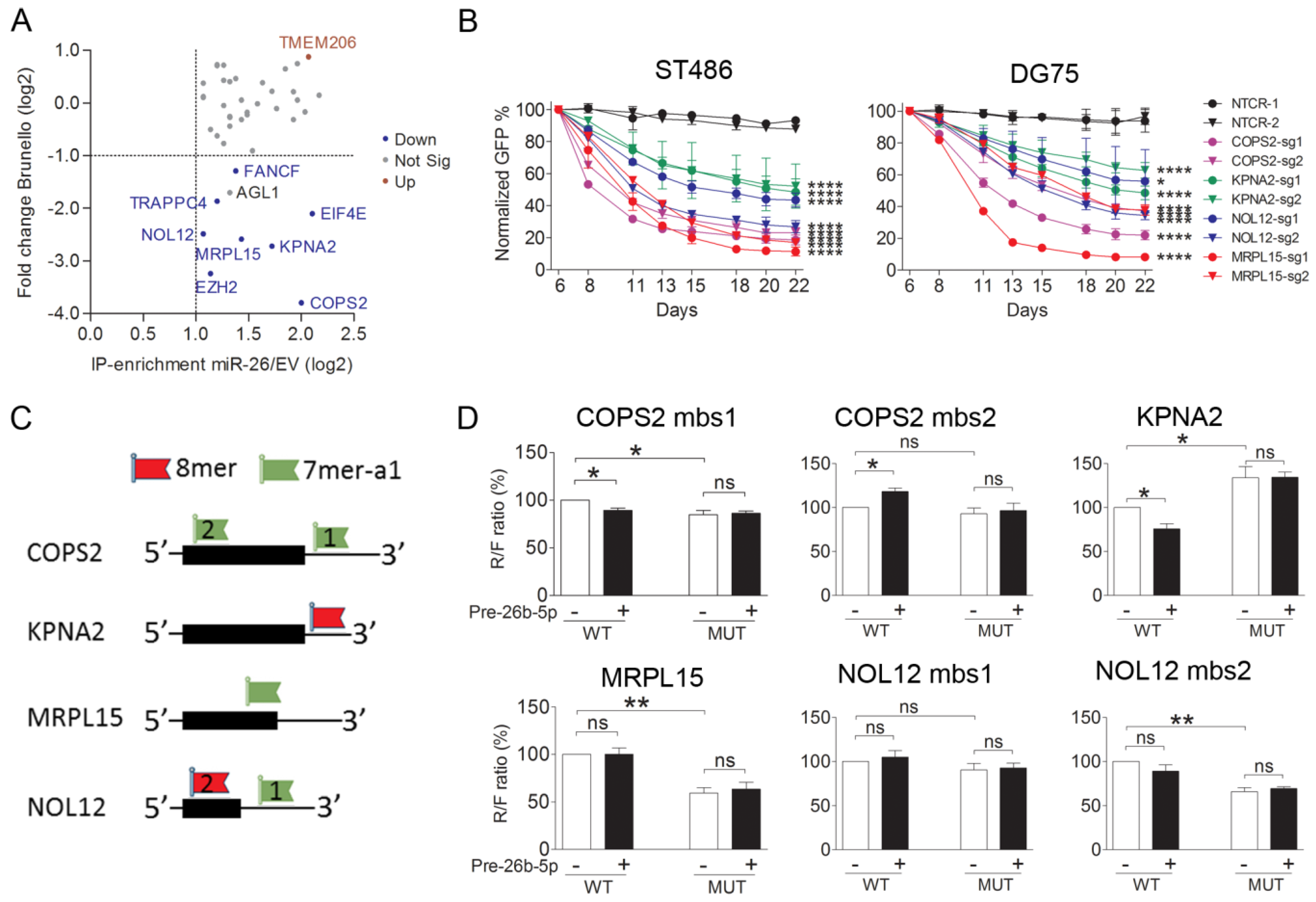
2.5. COPS2, NOL12, MRPL15, and KPNA2 Are Potential Targets of miR-26b-5p and the Essential Genes for the BL Cells
3. Discussion
4. Materials and Methods
4.1. Tissue Samples and Cell Lines
4.2. Generation of the Construct Pools for miRNA Inhibition and Overexpression
4.3. Production of Lentiviral Particles
4.4. Quality Control of miRNA Inhibition and Overexpression Pools
4.5. High Throughput Screen
4.6. DNA Isolation and Amplification of the Inserts
4.7. Library Preparation, Next Generation Sequencing, and Data Analysis
4.8. Green Fluorescent Protein (GFP) Growth Competition Assay
4.9. MiRNA qRT-PCR
4.10. Ago2-RIP-Chip
4.11. Prediction of MiRNA Binding Sites and Gene set Enrichment Analysis
4.12. Genome-Wide CRISPR/Cas9 Knockdown Screen for Essential Genes in BL
4.13. Validation of MiR-26b-5p Binding to the Predicted Binding Sites
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Swerdlow, S.H.; Campo, E.; Harris, N.L.; Jaffe, E.S.; Pileri, S.A.; Stein, H.; Thiele, J. WHO Classification of Tumours of the Haematopoietic and Lymphoid Tissues, Revised 4th ed; IARC: Lyon, France, 2017. [Google Scholar]
- Burkitt, D. A Sarcoma Involving the Jaws in African Children. Br. J. Surg. 1958, 46, 218–223. [Google Scholar] [CrossRef]
- Victora, G.D.; Dominguez-Sola, D.; Holmes, A.B.; Deroubaix, S.; Dalla-Favera, R.; Nussenzweig, M.C. Identification of Human Germinal Center Light and Dark Zone Cells and Their Relationship to Human B-Cell Lymphomas. Blood 2012, 120, 2240–2248. [Google Scholar] [CrossRef]
- Schmitz, R.; Ceribelli, M.; Pittaluga, S.; Wright, G.; Staudt, L.M. Oncogenic Mechanisms in Burkitt Lymphoma. Cold Spring Harb. Perspect. Med. 2014, 4. [Google Scholar] [CrossRef]
- Burkitt, D.P. Etiology of Burkitt’s Lymphoma--An Alternative Hypothesis to a Vectored Virus. J. Natl. Cancer Inst. 1969, 42, 19–28. [Google Scholar]
- Zech, L.; Haglund, U.; Nilsson, K.; Klein, G. Characteristic Chromosomal Abnormalities in Biopsies and Lymphoid-Cell Lines from Patients with Burkitt and Non-Burkitt Lymphomas. Int. J. Cancer 1976, 17, 47–56. [Google Scholar] [CrossRef]
- Hausser, J.; Zavolan, M. Identification and Consequences of miRNA-Target Interactions--Beyond Repression of Gene Expression. Nat. Rev. Genet. 2014, 15, 599–612. [Google Scholar] [CrossRef]
- Musilova, K.; Mraz, M. MicroRNAs in B-Cell Lymphomas: How a Complex Biology Gets More Complex. Leukemia 2015, 29, 1004–1017. [Google Scholar] [CrossRef]
- Di Lisio, L.; Sanchez-Beato, M.; Gomez-Lopez, G.; Rodriguez, M.E.; Montes-Moreno, S.; Mollejo, M.; Menarguez, J.; Martinez, M.A.; Alves, F.J.; Pisano, D.G.; et al. MicroRNA Signatures in B-Cell Lymphomas. Blood Cancer J. 2012, 2, e57. [Google Scholar] [CrossRef]
- Hezaveh, K.; Kloetgen, A.; Bernhart, S.H.; Mahapatra, K.D.; Lenze, D.; Richter, J.; Haake, A.; Bergmann, A.K.; Brors, B.; Burkhardt, B.; et al. Alterations of microRNA and microRNA-Regulated Messenger RNA Expression in Germinal Center B-Cell Lymphomas Determined by Integrative Sequencing Analysis. Haematologica 2016, 101, 1380–1389. [Google Scholar] [CrossRef]
- Lenze, D.; Leoncini, L.; Hummel, M.; Volinia, S.; Liu, C.G.; Amato, T.; De Falco, G.; Githanga, J.; Horn, H.; Nyagol, J.; et al. The Different Epidemiologic Subtypes of Burkitt Lymphoma Share a Homogenous Micro RNA Profile Distinct from Diffuse Large B-Cell Lymphoma. Leukemia 2011, 25, 1869–1876. [Google Scholar] [CrossRef]
- Oduor, C.I.; Kaymaz, Y.; Chelimo, K.; Otieno, J.A.; Ong’echa, J.M.; Moormann, A.M.; Bailey, J.A. Integrative microRNA and mRNA Deep-Sequencing Expression Profiling in Endemic Burkitt Lymphoma. BMC Cancer 2017, 17, 761. [Google Scholar] [CrossRef]
- Robertus, J.L.; Kluiver, J.; Weggemans, C.; Harms, G.; Reijmers, R.M.; Swart, Y.; Kok, K.; Rosati, S.; Schuuring, E.; van Imhoff, G.; et al. MiRNA Profiling in B non-Hodgkin Lymphoma: A MYC-Related miRNA Profile Characterizes Burkitt Lymphoma. Br. J. Haematol. 2010, 149, 896–899. [Google Scholar] [CrossRef]
- Bartolome-Izquierdo, N.; de Yebenes, V.G.; Alvarez-Prado, A.F.; Mur, S.M.; Lopez Del Olmo, J.A.; Roa, S.; Vazquez, J.; Ramiro, A.R. miR-28 Regulates the Germinal Center Reaction and Blocks Tumor Growth in Preclinical Models of non-Hodgkin Lymphoma. Blood 2017, 129, 2408–2419. [Google Scholar] [CrossRef]
- Chen, S.; Wang, Z.; Dai, X.; Pan, J.; Ge, J.; Han, X.; Wu, Z.; Zhou, X.; Zhao, T. Re-Expression of microRNA-150 induces EBV-Positive Burkitt Lymphoma Differentiation by Modulating c-Myb in Vitro. Cancer Sci. 2013, 104, 826–834. [Google Scholar] [CrossRef]
- Dzikiewicz-Krawczyk, A.; Kok, K.; Slezak-Prochazka, I.; Robertus, J.L.; Bruining, J.; Tayari, M.M.; Rutgers, B.; de Jong, D.; Koerts, J.; Seitz, A.; et al. ZDHHC11 and ZDHHC11B are Critical Novel Components of the Oncogenic MYC-miR-150-MYB Network in Burkitt Lymphoma. Leukemia 2017, 31, 1470–1473. [Google Scholar] [CrossRef]
- Zhang, X.; Zhao, X.; Fiskus, W.; Lin, J.; Lwin, T.; Rao, R.; Zhang, Y.; Chan, J.C.; Fu, K.; Marquez, V.E.; et al. Coordinated Silencing of MYC-Mediated miR-29 by HDAC3 and EZH2 as a Therapeutic Target of Histone Modification in Aggressive B-Cell Lymphomas. Cancer Cell 2012, 22, 506–523. [Google Scholar] [CrossRef]
- Robaina, M.C.; Faccion, R.S.; Mazzoccoli, L.; Rezende, L.M.; Queiroga, E.; Bacchi, C.E.; Thomas-Tikhonenko, A.; Klumb, C.E. miR-17-92 Cluster Components Analysis in Burkitt Lymphoma: Overexpression of miR-17 is Associated with Poor Prognosis. Ann. Hematol. 2016, 95, 881–891. [Google Scholar] [CrossRef]
- Slezak-Prochazka, I.; Kluiver, J.; de Jong, D.; Smigielska-Czepiel, K.; Kortman, G.; Winkle, M.; Rutgers, B.; Koerts, J.; Visser, L.; Diepstra, A.; et al. Inhibition of the miR-155 Target NIAM Phenocopies the Growth Promoting Effect of miR-155 in B-Cell Lymphoma. Oncotarget 2016, 7, 2391–2400. [Google Scholar] [CrossRef]
- Niu, F.; Dzikiewicz-Krawczyk, A.; Koerts, J.; de Jong, D.; Wijenberg, L.; Fernandez Hernandez1, M.; Slezak-Prochazka, I.; Winkle1, M.; Kooistra, W.; van der Sluis, T.; et al. MiR-378a-3p is Critical for Burkitt Lymphoma Cell Growth. Cancers. (under review).
- Mullokandov, G.; Baccarini, A.; Ruzo, A.; Jayaprakash, A.D.; Tung, N.; Israelow, B.; Evans, M.J.; Sachidanandam, R.; Brown, B.D. High-Throughput Assessment of microRNA Activity and Function Using microRNA Sensor and Decoy Libraries. Nat. Methods 2012, 9, 840–846. [Google Scholar] [CrossRef]
- Chang, T.C.; Yu, D.; Lee, Y.S.; Wentzel, E.A.; Arking, D.E.; West, K.M.; Dang, C.V.; Thomas-Tikhonenko, A.; Mendell, J.T. Widespread microRNA Repression by Myc Contributes to Tumorigenesis. Nat. Genet. 2008, 40, 43–50. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Zhang, L.; Huang, H.; Huang, Y.; Huang, L.; Wang, J.; Huang, S.; He, L.; Zhou, Y.; Jia, W.; et al. MiR-26b/KPNA2 Axis Inhibits Epithelial Ovarian Carcinoma Proliferation and Metastasis through Downregulating OCT4. Oncotarget 2015, 6, 23793–23806. [Google Scholar] [CrossRef] [PubMed]
- Tan, S.; Ding, K.; Li, R.; Zhang, W.; Li, G.; Kong, X.; Qian, P.; Lobie, P.E.; Zhu, T. Identification of miR-26 as a Key Mediator of Estrogen Stimulated Cell Proliferation by Targeting CHD1, GREB1 and KPNA2. Breast Cancer Res. 2014, 16, R40. [Google Scholar] [CrossRef]
- Deng, M.; Zhang, R.; He, Z.; Qiu, Q.; Lu, X.; Yin, J.; Liu, H.; Jia, X.; He, Z. TET-Mediated Sequestration of miR-26 Drives EZH2 Expression and Gastric Carcinogenesis. Cancer Res. 2017, 77, 6069–6082. [Google Scholar] [CrossRef]
- Gentilin, E.; Tagliati, F.; Filieri, C.; Mole, D.; Minoia, M.; Rosaria Ambrosio, M.; Degli Uberti, E.C.; Zatelli, M.C. miR-26a Plays an Important Role in Cell Cycle Regulation in ACTH-Secreting Pituitary Adenomas by Modulating Protein Kinase Cdelta. Endocrinology 2013, 154, 1690–1700. [Google Scholar] [CrossRef]
- Wei, K.; Pan, C.; Yao, G.; Liu, B.; Ma, T.; Xia, Y.; Jiang, W.; Chen, L.; Chen, Y. MiR-106b-5p Promotes Proliferation and Inhibits Apoptosis by Regulating BTG3 in Non-Small Cell Lung Cancer. Cell. Physiol. Biochem. 2017, 44, 1545–1558. [Google Scholar] [CrossRef]
- Kazimierska, M.; Zurawek, M.; Wozniak, T.; Podralska, M.; Kluiver, J.; Van Den Berg, A.; Rozwadowska, N.; Dzikiewicz-Krawczyk, A. MYC-Dependent Vulnerabilities in Cancer Cells. manuscript in preparation.
- Sander, S.; Bullinger, L.; Klapproth, K.; Fiedler, K.; Kestler, H.A.; Barth, T.F.E.; Moller, P.; Stilgenbauer, S.; Pollack, J.R.; Wirth, T. MYC Stimulates EZH2 Expression by Repression of Its Negative Regulator miR-26a. Blood 2008, 112, 4202–4212. [Google Scholar] [CrossRef]
- Yuan, Y.; Niu, F.; Nolte, I.M.; Koerts, J.; De Jong, D.; Rutgers, B.; Osinga, J.; Azkanaz, M.; Terpstra, M.; Bystrykh, L.; et al. MicroRNA High Throughput Loss-of-Function Screening Reveals an Oncogenic Role for miR-21-5p in Hodgkin Lymphoma. Cell. Physiol. Biochem. 2018, 49, 144–159. [Google Scholar] [CrossRef]
- Laudato, S.; Patil, N.; Abba, M.L.; Leupold, J.H.; Benner, A.; Gaiser, T.; Marx, A.; Allgayer, H. P53-Induced miR-30e-5p Inhibits Colorectal Cancer Invasion and Metastasis by Targeting ITGA6 and ITGB1. Int. J. Cancer 2017, 141, 1879–1890. [Google Scholar] [CrossRef]
- Xu, G.; Cai, J.; Wang, L.; Jiang, L.; Huang, J.; Hu, R.; Ding, F. MicroRNA-30e-5p Suppresses Non-Small Cell Lung Cancer Tumorigenesis by Regulating USP22-Mediated Sirt1/JAK/STAT3 Signaling. Exp. Cell Res. 2018, 362, 268–278. [Google Scholar] [CrossRef]
- Zhao, J.J.; Lin, J.; Zhu, D.; Wang, X.; Brooks, D.; Chen, M.; Chu, Z.B.; Takada, K.; Ciccarelli, B.; Admin, S.; et al. miR-30-5p Functions as a Tumor Suppressor and Novel Therapeutic Tool by Targeting the Oncogenic Wnt/beta-catenin/BCL9 Pathway. Cancer Res. 2014, 74, 1801–1813. [Google Scholar] [CrossRef]
- Lu, J.; Wei, J.H.; Feng, Z.H.; Chen, Z.H.; Wang, Y.Q.; Huang, Y.; Fang, Y.; Liang, Y.P.; Cen, J.J.; Pan, Y.H.; et al. miR-106b-5p Promotes Renal Cell Carcinoma Aggressiveness and Stem-Cell-Like Phenotype by Activating Wnt/beta-Catenin Signalling. Oncotarget 2017, 8, 21461–21471. [Google Scholar] [CrossRef]
- Liu, F.; Gong, J.; Huang, W.; Wang, Z.; Wang, M.; Yang, J.; Wu, C.; Wu, Z.; Han, B. MicroRNA-106b-5p Boosts Glioma Tumorigensis by Targeting Multiple Tumor Suppressor Genes. Oncogene 2014, 33, 4813–4822. [Google Scholar] [CrossRef][Green Version]
- Guo, F.; Hou, X.; Sun, Q. MicroRNA-9-5p Functions as a Tumor Suppressor in Papillary Thyroid Cancer via Targeting BRAF. Oncol. Lett. 2018, 16, 6815–6821. [Google Scholar] [CrossRef]
- Li, G.; Wu, F.; Yang, H.; Deng, X.; Yuan, Y. MiR-9-5p Promotes Cell Growth and Metastasis in Non-Small Cell Lung Cancer through the Repression of TGFBR2. Biomed. Pharmacother. 2017, 96, 1170–1178. [Google Scholar] [CrossRef]
- Leucci, E.; Zriwil, A.; Gregersen, L.H.; Jensen, K.T.; Obad, S.; Bellan, C.; Leoncini, L.; Kauppinen, S.; Lund, A.H. Inhibition of miR-9 De-Represses HuR and DICER1 and Impairs Hodgkin Lymphoma Tumour Outgrowth in Vivo. Oncogene 2012, 31, 5081–5089. [Google Scholar] [CrossRef]
- Liu, H.; Cheng, Y.; Xu, Y.; Xu, H.; Lin, Z.; Fan, J.; Lang, J. The Inhibition of Tumor Protein p53 by microRNA-151a-3p Induced Cell Proliferation, Migration and Invasion in Nasopharyngeal Carcinoma. Biosci. Rep. 2019, 39. [Google Scholar] [CrossRef]
- Sun, S.; Wang, H.; Ji, M. Overexpression of miR-222-3p Promotes the Proliferation and Inhibits the Apoptosis of Diffuse Large B-Cell Lymphoma Cells via Suppressing PPP2R2A. Technol. Cancer Res. Treat. 2019, 18. [Google Scholar] [CrossRef]
- Gao, J.; Liu, Q.G. The Role of miR-26 in Tumors and Normal Tissues (Review). Oncol. Lett. 2011, 2, 1019–1023. [Google Scholar] [CrossRef]
- Koh, C.M.; Iwata, T.; Zheng, Q.; Bethel, C.; Yegnasubramanian, S.; De Marzo, A.M. Myc Enforces Overexpression of EZH2 in Early Prostatic Neoplasia via Transcriptional and Post-Transcriptional Mechanisms. Oncotarget 2011, 2, 669–683. [Google Scholar] [CrossRef]
- Bialopiotrowicz, E.; Noyszewska-Kania, M.; Kachamakova-Trojanowska, N.; Loboda, A.; Cybulska, M.; Grochowska, A.; Kopczynski, M.; Mikula, M.; Prochorec-Sobieszek, M.; Firczuk, M.; et al. Serine Biosynthesis Pathway Supports MYC-miR-494-EZH2 Feed-Forward Circuit Necessary to Maintain Metabolic and Epigenetic Reprogramming of Burkitt Lymphoma Cells. Cancers 2020, 12, 580. [Google Scholar] [CrossRef]
- Li, P.; Ding, N.; Zhang, W.; Chen, L. COPS2 Antagonizes OCT4 to Accelerate the G2/M Transition of Mouse Embryonic Stem Cells. Stem Cell Rep. 2018, 11, 317–324. [Google Scholar] [CrossRef]
- Scott, D.D.; Trahan, C.; Zindy, P.J.; Aguilar, L.C.; Delubac, M.Y.; Van Nostrand, E.L.; Adivarahan, S.; Wei, K.E.; Yeo, G.W.; Zenklusen, D.; et al. Nol12 is a multifunctional RNA binding protein at the nexus of RNA and DNA metabolism. Nucleic Acids Res. 2017, 45, 12509–12528. [Google Scholar] [CrossRef]
- Zhang, W.Y.; Ni, P.L.; Mou, C.L.; Zhang, Y.Q.; Guo, H.C.; Zhao, T.; Loh, Y.H.; Chen, L.Y. Cops2 Promotes Pluripotency Maintenance by Stabilizing Nanog Protein and Repressing Transcription. Sci. Rep. UK 2016, 6. [Google Scholar] [CrossRef]
- Sotgia, F.; Fiorillo, M.; Lisanti, M.P. Mitochondrial Markers Predict Recurrence, Metastasis and Tamoxifen-Resistance in Breast Cancer Patients: Early Detection of Treatment Failure with Companion Diagnostics. Oncotarget 2017, 8, 68730–68745. [Google Scholar] [CrossRef]
- Xiang, S.; Wang, Z.; Ye, Y.; Zhang, F.; Li, H.; Yang, Y.; Miao, H.; Liang, H.; Zhang, Y.; Jiang, L.; et al. E2F1 and E2F7 Differentially Regulate KPNA2 to Promote the Development of Gallbladder Cancer. Oncogene 2019, 38, 1269–1281. [Google Scholar] [CrossRef]
- Tsai, M.M.; Huang, H.W.; Wang, C.S.; Lee, K.F.; Tsai, C.Y.; Lu, P.H.; Chi, H.C.; Lin, Y.H.; Kuo, L.M.; Lin, K.H. MicroRNA-26b Inhibits Tumor Metastasis by Targeting the KPNA2/c-jun Pathway in Human Gastric Cancer. Oncotarget 2016, 7, 39511–39526. [Google Scholar] [CrossRef]
- Guo, X.; Wang, Z.; Zhang, J.; Xu, Q.; Hou, G.; Yang, Y.; Dong, C.; Liu, G.; Liang, C.; Liu, L.; et al. Upregulated KPNA2 Promotes Hepatocellular Carcinoma Progression and Indicates Prognostic Significance across Human Cancer Types. Acta Biochim. Biophys. Sin. (Shanghai) 2019, 51, 285–292. [Google Scholar] [CrossRef]
- Wang, P.; Zhao, Y.; Liu, K.; Liu, X.; Liang, J.; Zhou, H.; Wang, Z.; Zhou, Z.; Xu, N. Wip1 Cooperates with KPNA2 to Modulate the Cell Proliferation and Migration of Colorectal Cancer via a p53-Dependent Manner. J. Cell. Biochem. 2019, 120, 15709–15718. [Google Scholar] [CrossRef]
- Huang, L.; Wang, H.Y.; Li, J.D.; Wang, J.H.; Zhou, Y.; Luo, R.Z.; Yun, J.P.; Zhang, Y.; Jia, W.H.; Zheng, M. KPNA2 Promotes Cell Proliferation and Tumorigenicity in Epithelial Ovarian Carcinoma through Upregulation of c-Myc and Downregulation of FOXO3a. Cell Death Dis. 2013, 4, e745. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Liu, Q.; Liu, Z.; Xia, Q.; Zhang, Z.; Zhang, R.; Gao, T.; Gu, G.; Wang, Y.; Wang, D.; et al. KPNA2 Promotes Metabolic Reprogramming in Glioblastomas by Regulation of c-Myc. J. Exp. Clin. Cancer Res. 2018, 37, 194. [Google Scholar] [CrossRef] [PubMed]
- Winkle, M.; Van Den Berg, A.; Tayari, M.; Sietzema, J.; Terpstra, M.; Kortman, G.; De Jong, D.; Visser, L.; Diepstra, A.; Kok, K.; et al. Long Noncoding RNAs as a Novel Component of the Myc Transcriptional Network. FASEB J. 2015, 29, 2338–2346. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Kluiver, J.; Koerts, J.; De Jong, D.; Rutgers, B.; Razak, F.R.A.; Terpstra, M.; Plaat, B.E.; Nolte, I.M.; Diepstra, A.; et al. miR-24-3p Is Overexpressed in Hodgkin Lymphoma and Protects Hodgkin and Reed-Sternberg Cells from Apoptosis. Am. J. Pathol. 2017, 187, 1343–1355. [Google Scholar] [CrossRef]
- Kluiver, J.; Slezak-Prochazka, I.; Van Den Berg, A. Studying microRNAs in Lymphoma. Methods Mol. Biol. 2013, 971, 265–276. [Google Scholar] [CrossRef]
- Kluiver, J.; Niu, F.; Yuan, Y.; Kok, K.; Van Den Berg, A.; Dzikiewicz-Krawczyk, A. NGS-Based High-Throughput Screen to Identify MicroRNAs Regulating Growth of B-Cell Lymphoma. Methods Mol. Biol. 2019, 1956, 269–282. [Google Scholar] [CrossRef]
- Kluiver, J.; Gibcus, J.H.; Hettinga, C.; Adema, A.; Richter, M.K.; Halsema, N.; Slezak-Prochazka, I.; Ding, Y.; Kroesen, B.J.; Van Den Berg, A. Rapid Generation of microRNA Sponges for microRNA Inhibition. PLoS ONE 2012, 7, e29275. [Google Scholar] [CrossRef]
- Tan, L.P.; Seinen, E.; Duns, G.; De Jong, D.; Sibon, O.C.M.; Poppema, S.; Kroesen, B.J.; Kok, K.; Van Den Berg, A. A High throughput Experimental Approach to Identify miRNA Targets in Human Cells. Nucleic Acids Res. 2009, 37. [Google Scholar] [CrossRef]
- Agarwal, V.; Bell, G.W.; Nam, J.W.; Bartel, D.P. Predicting Effective microRNA Target Sites in Mammalian mRNAs. Elife 2015, 4. [Google Scholar] [CrossRef]
- Tsai, S.Q.; Nguyen, N.T.; Malagon-Lopez, J.; Topkar, V.V.; Aryee, M.J.; Joung, J.K. CIRCLE-seq: A Highly Sensitive in Vitro Screen for Genome-Wide CRISPR Cas9 Nuclease Off-Targets. Nat. Methods 2017, 14, 607–614. [Google Scholar] [CrossRef]
| IP/T Ratio | ST486 (n = 12,286) | DG75 (n = 9206) | ||||
| pCDH-miR-26b | EV | pCDH-miR-26b/EV | pCDH-miR-26b | EV | pCDH-miR-26b/EV | |
| ≥2 | 1587 | 1524 | 94 | 1236 | 1140 | 59 |
| ≥4 | 560 | 573 | 10 | 462 | 412 | 5 |
| ≥8 | 169 | 205 | 0 | 141 | 143 | 0 |
| Gene | Transcript ID | FC of IP/T Ratio | Brunello Screen | miR-26b-5p Binding Site ** | |||
|---|---|---|---|---|---|---|---|
| DG75 | ST486 | FC | p-Value * | CDS | 3′UTR | ||
| COPS2 | ENST00000388901 | 5.1 | 4.0 | −13.9 | 4.97 × 10−86 | 7mA1 | 7mA1 |
| EZH2 | ENST00000320356 | 2.3 | 2.2 | −9.4 | 3.87 × 10−45 | 7mA1 | 8m |
| KPNA2 | ENST00000330459 | 2.5 | 3.3 | −6.6 | 1.16 × 10−6 | 8m | |
| MRPL15 | ENST00000260102 | 2.2 | 2.7 | −6 | 5.03 × 10−43 | 7mA1 | |
| NOL12 | ENST00000359114 | 2.2 | 2.1 | −5.6 | 2.67 × 10−47 | 8m | 7mA1 |
| EIF4E | ENST00000505992 | 2.9 | 4.3 | −4.3 | 5.76 × 10−12 | ||
| TRAPPC4 | ENST00000533632 | 2.1 | 2.3 | −3.7 | 1.0 × 10−8 | 7mA1 | |
| ALG1 | ENST00000262374 | 2.3 | 2.5 | −3.2 | 0.042 | 7m8 | |
| FANCF | ENST00000327470 | 3.4 | 2.6 | −2.4 | 3.52 × 10−4 | 7mA1/8m | |
| B3GNT2 | ENST00000301998 | 3.1 | 2.9 | −1.9 | 1 | 8m | |
| MT2A | ENST00000245185 | 3.4 | 2.4 | −1.7 | 0.045 | ||
| NXT1 | ENST00000254998 | 2.3 | 2.3 | −1.5 | 1 | ||
| MT1B | ENST00000334346 | 2.3 | 2.2 | −1.4 | 1 | ||
| MSMO1 | ENST00000261507 | 2.3 | 2.5 | −1.4 | 1 | 8m | |
| PPP1CC | ENST00000335007 | 2.9 | 3.8 | −1.2 | 1 | 7mA1 | |
| MT1E | ENST00000306061 | 2.2 | 2.7 | −1.2 | 1 | ||
| MPV17L2 | ENST00000599612 | 2.6 | 2.5 | −1.2 | 1 | 8m | |
| RHOQ | ENST00000238738 | 2.8 | 2.5 | −1.2 | 1 | 7mA1/7m8/8m | |
| TBC1D7 | ENST00000379300 | 2.0 | 2.8 | −1.2 | 1 | 8m | |
| TMEM156 | ENST00000381938 | 3.0 | 2.8 | −1.1 | 1 | 8m | 7mA1 |
| REEP4 | ENST00000306306 | 4.6 | 4.1 | −1.1 | 1 | 7m8 | |
| ASB10 | ENST00000420175 | 2.1 | 3.4 | −1.1 | 1 | ||
| OSCP1 | ENST00000235532 | 2.9 | 2.4 | -1 | 1 | ||
| H3F3C | ENST00000340398 | 2.3 | 3.0 | -1 | 1 | ||
| PRMT3 | ENST00000331079 | 2.1 | 2.7 | 1 | 1 | 7mA1 | |
| ZDHHC6 | ENST00000369405 | 4.2 | 3.9 | 1 | 1 | 8m | |
| DIABLO | ENST00000650715 | 2.1 | 2.1 | 1.1 | 1 | 7mA1 | |
| TXNDC17 | ENST00000250101 | 2.5 | 2.1 | 1.1 | 1 | 7mA1 | |
| MGST1 | ENST00000396209 | 3.5 | 4.5 | 1.1 | 1 | ||
| MSRB2 | ENST00000376510 | 3.8 | 3.8 | 1.2 | 1 | 7mA1 | |
| FRAT2 | ENST00000371019 | 3.7 | 3.3 | 1.2 | 1 | 8m | |
| ADAM19 | ENST00000257527 | 2.4 | 2.1 | 1.3 | 1 | 8m | |
| PRKCD | ENST00000330452 | 4.2 | 3.1 | 1.3 | 1 | 8m | |
| ACBD5 | ENST00000396271 | 3.3 | 2.5 | 1.3 | 1 | 7mA1/8m | |
| ZNF410 | ENST00000555044 | 2.4 | 2.4 | 1.3 | 1 | 8m | |
| ACYP2 | ENST00000394666 | 2.4 | 2.6 | 1.4 | 1 | ||
| BID | ENST00000317361 | 2.7 | 3.6 | 1.6 | 1 | 8m | |
| SLC25A36 | ENST00000324194 | 2.3 | 2.3 | 1.6 | 1 | ||
| CRADD | ENST00000332896 | 2.0 | 2.4 | 1.6 | 1 | 8m | |
| POLR3G | ENST00000369314 | 2.4 | 3.9 | 1.7 | 1 | ||
| SAAL1 | ENST00000524803 | 2.2 | 2.3 | 1.7 | 0.984 | 7mA1/8m | |
| TMEM206 | ENST00000261455 | 3.0 | 4.2 | 1.8 | 6.96 × 10−4 | ||
| LINC00847 | ENST00000501855 | 2.8 | 2.9 | ||||
| LOC100294145 | N/A | 2.2 | 2.1 | ||||
| MT1L | ENST00000565768 | 2.8 | 2.3 | ||||
| XLOC_l2_008009 | TCONS_l2_00014564 | 2.5 | 2.9 | ||||
| lnc-C2orf81-2 | lnc-C2orf81-2:1–2 | 2.3 | 2.2 | 7m8/8m *** | |||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Niu, F.; Kazimierska, M.; Nolte, I.M.; Terpstra, M.M.; de Jong, D.; Koerts, J.; van der Sluis, T.; Rutgers, B.; O’Connell, R.M.; Kok, K.; et al. The miR-26b-5p/KPNA2 Axis Is an Important Regulator of Burkitt Lymphoma Cell Growth. Cancers 2020, 12, 1464. https://doi.org/10.3390/cancers12061464
Niu F, Kazimierska M, Nolte IM, Terpstra MM, de Jong D, Koerts J, van der Sluis T, Rutgers B, O’Connell RM, Kok K, et al. The miR-26b-5p/KPNA2 Axis Is an Important Regulator of Burkitt Lymphoma Cell Growth. Cancers. 2020; 12(6):1464. https://doi.org/10.3390/cancers12061464
Chicago/Turabian StyleNiu, Fubiao, Marta Kazimierska, Ilja M. Nolte, Miente Martijn Terpstra, Debora de Jong, Jasper Koerts, Tineke van der Sluis, Bea Rutgers, Ryan M. O’Connell, Klaas Kok, and et al. 2020. "The miR-26b-5p/KPNA2 Axis Is an Important Regulator of Burkitt Lymphoma Cell Growth" Cancers 12, no. 6: 1464. https://doi.org/10.3390/cancers12061464
APA StyleNiu, F., Kazimierska, M., Nolte, I. M., Terpstra, M. M., de Jong, D., Koerts, J., van der Sluis, T., Rutgers, B., O’Connell, R. M., Kok, K., van den Berg, A., Dzikiewicz-Krawczyk, A., & Kluiver, J. (2020). The miR-26b-5p/KPNA2 Axis Is an Important Regulator of Burkitt Lymphoma Cell Growth. Cancers, 12(6), 1464. https://doi.org/10.3390/cancers12061464








