HMGA Proteins in Hematological Malignancies
Abstract
:1. Introduction
2. HMGA Proteins in Oncology
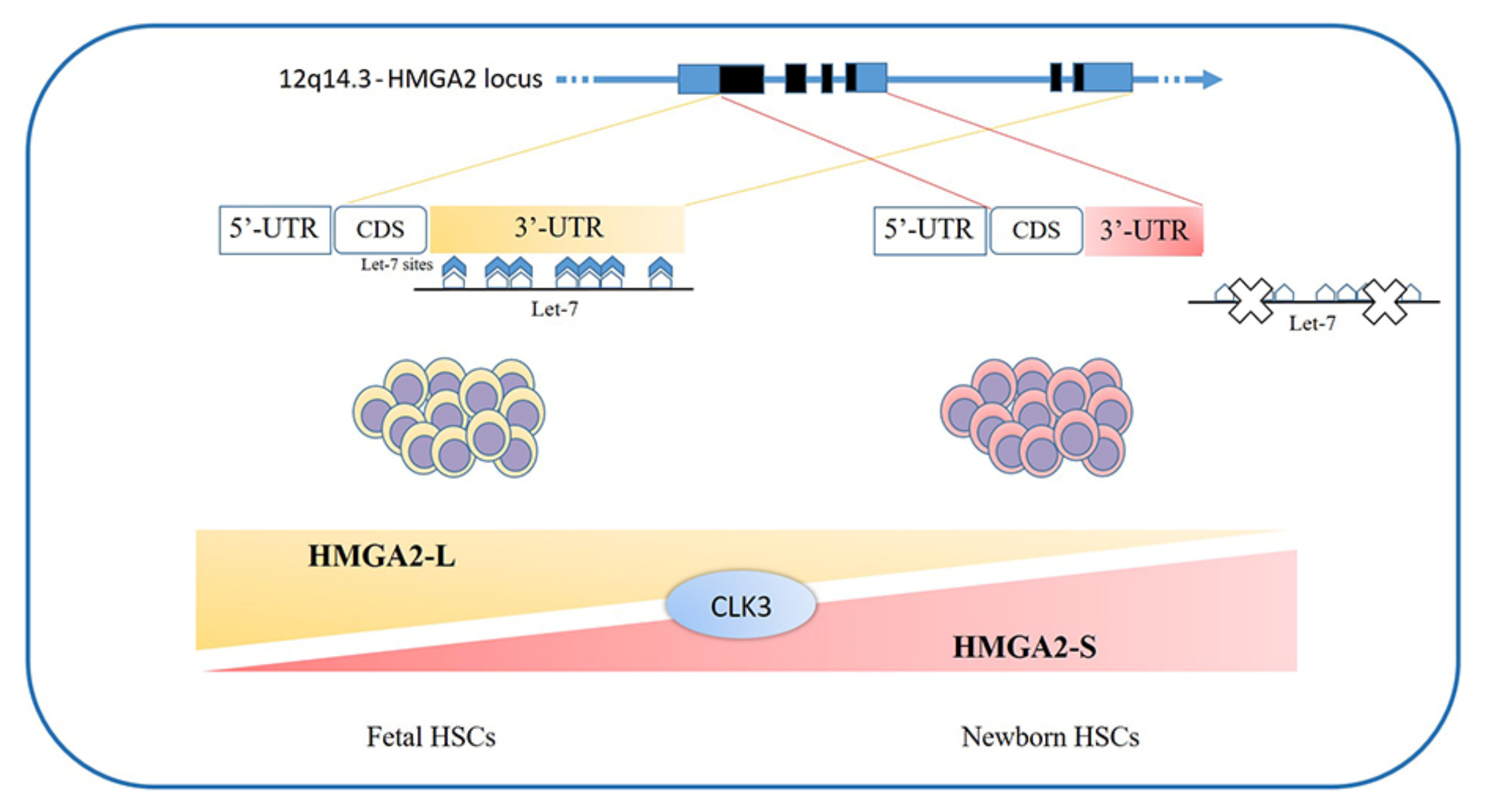
3. HMGA Proteins in Hematopoiesis
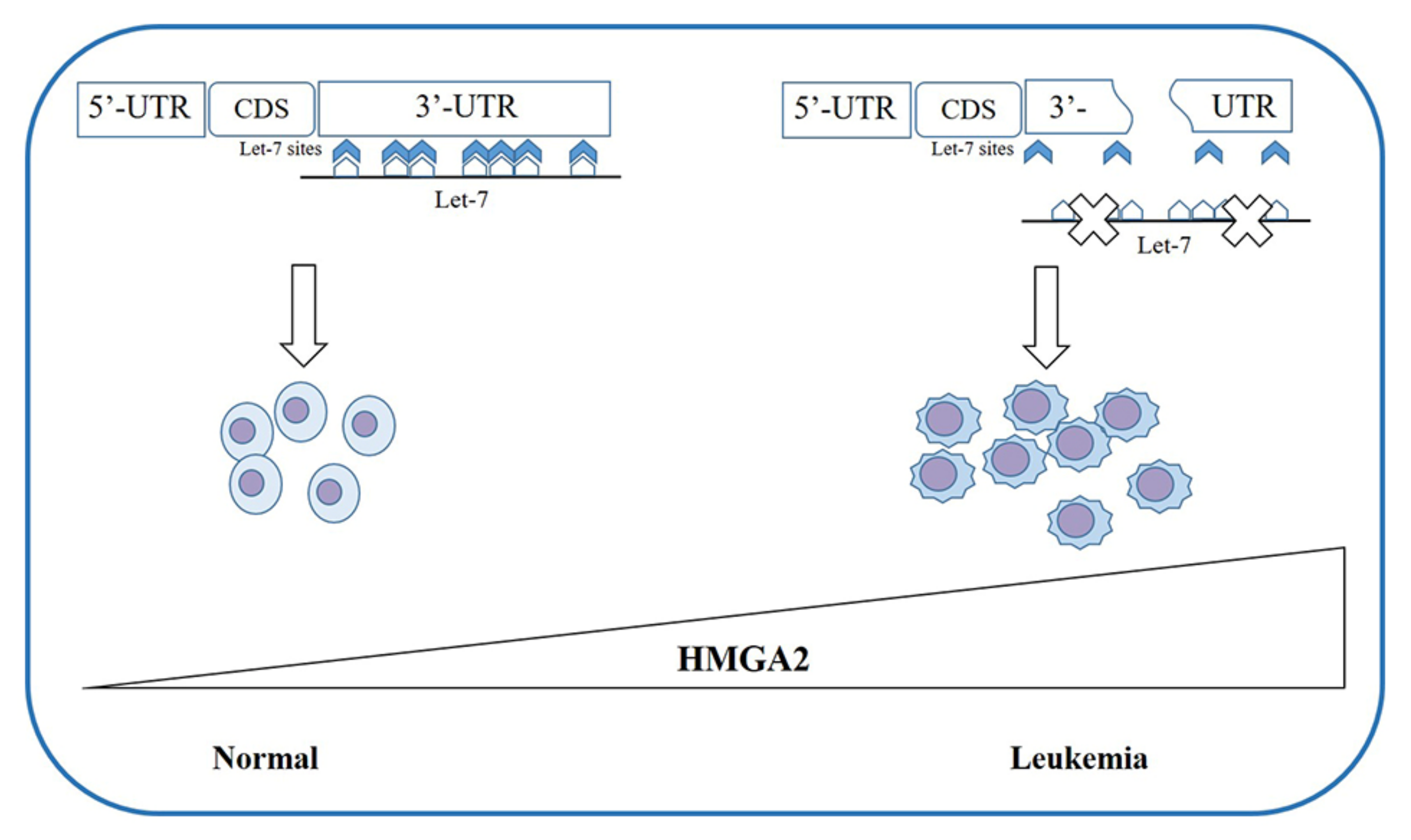
4. HMGAs Involvement in Myeloid Malignancies
4.1. HMGAs Involvement in Myeloproliferative Neoplasms
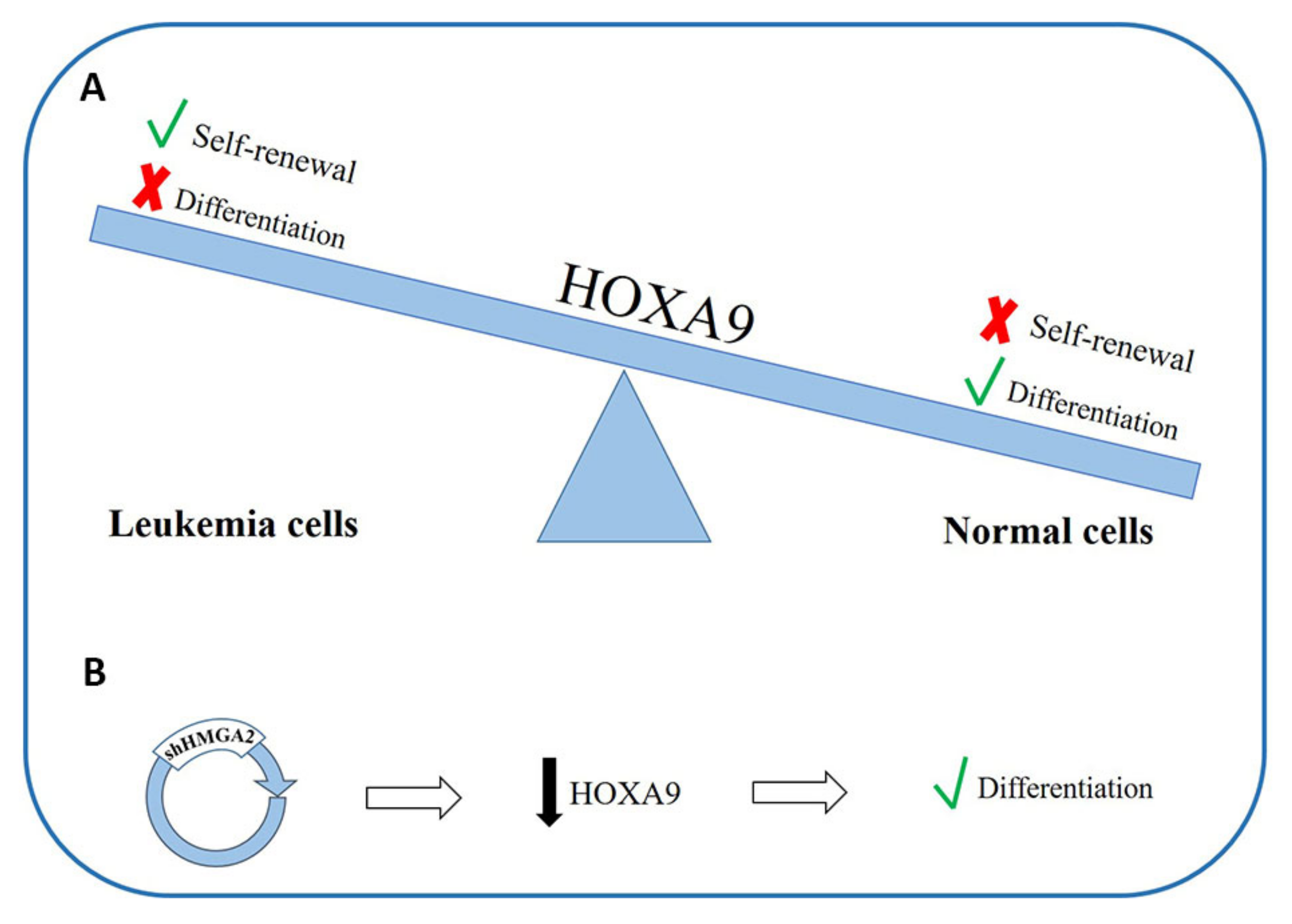
4.2. HMGAs Involvement in Acute Myeloid Leukemia
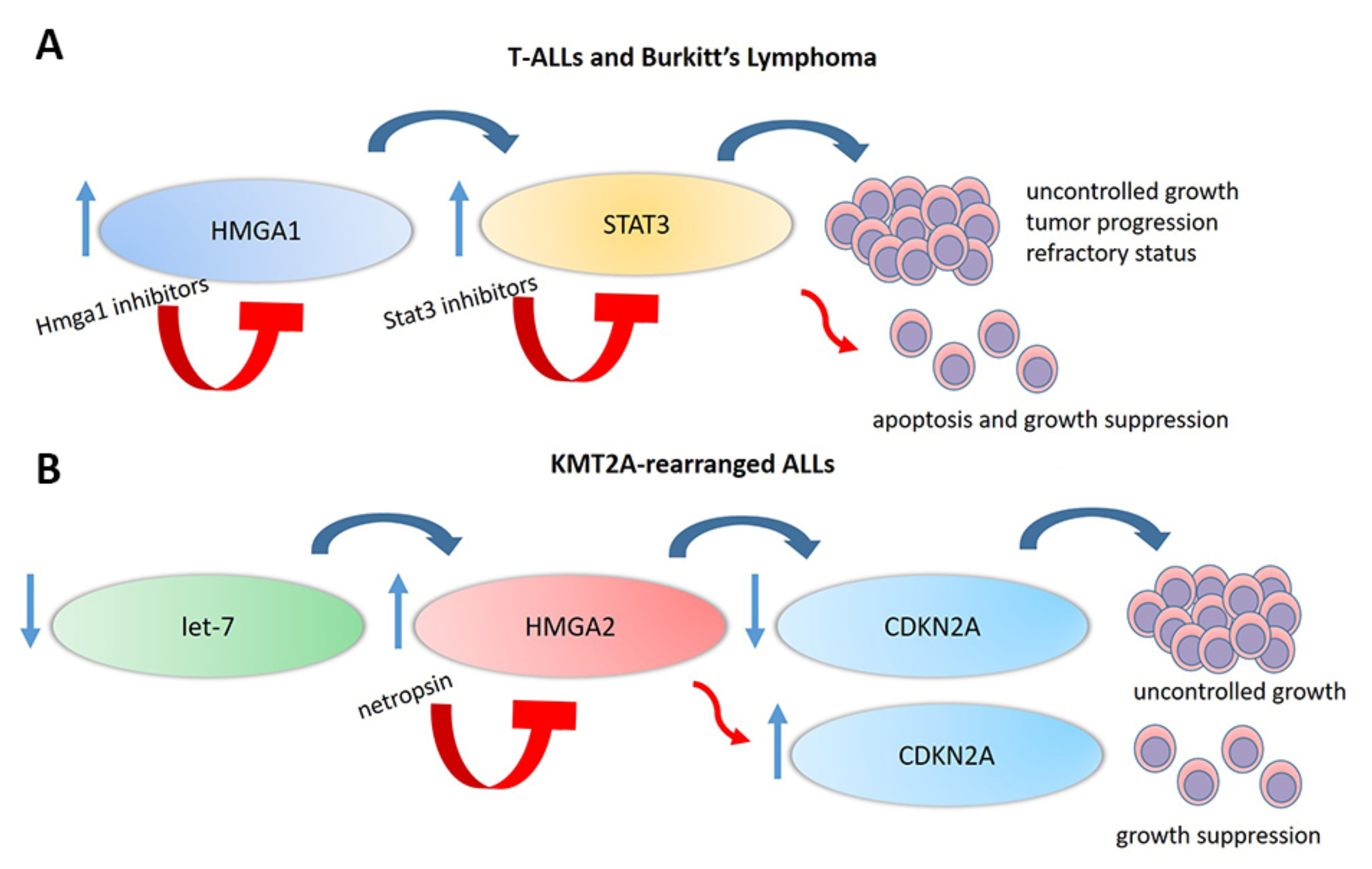
5. HMGAs Involvement in Lymphoid Malignancies
6. HMGA Proteins as Targetable Markers
7. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Reeves, R. Molecular biology of HMGA proteins: Hubs of nuclear function. Gene 2001, 277, 63–81. [Google Scholar] [CrossRef]
- Fusco, A.; Fedele, M. Roles of HMGA proteins in cancer. Nat. Rev. Cancer 2007, 7, 899–910. [Google Scholar] [CrossRef] [PubMed]
- Resar, L.M.S. The high mobility group A1 gene: Transforming inflammatory signals into cancer? Cancer Res. 2010, 70, 436–439. [Google Scholar] [CrossRef] [Green Version]
- Shah, S.N.; Resar, L.M.S. High mobility group A1 and cancer: Potential biomarker and therapeutic target. Histol. Histopathol. 2012, 27, 567–579. [Google Scholar]
- Friedmann, M.; Holth, L.T.; Zoghbi, H.Y.; Reeves, R. Organization, inducible-expression and chromosome localization of the human HMG-I(Y) nonhistone protein gene. Nucleic Acids Res. 1993, 21, 4259–4267. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cleynen, I.; Van de Ven, W.J.M. The HMGA proteins: A myriad of functions (Review). Int. J. Oncol. 2008, 32, 289–305. [Google Scholar] [CrossRef] [Green Version]
- Nagpal, S.; Ghosn, C.; DiSepio, D.; Molina, Y.; Sutter, M.; Klein, E.S.; Chandraratna, R.A. Retinoid-dependent recruitment of a histone H1 displacement activity by retinoic acid receptor. J. Biol. Chem. 1999, 274, 22563–22568. [Google Scholar] [CrossRef] [Green Version]
- Cesana, M.; Guo, M.H.; Cacchiarelli, D.; Wahlster, L.; Barragan, J.; Doulatov, S.; Vo, L.T.; Salvatori, B.; Trapnell, C.; Clement, K.; et al. A CLK3-HMGA2 Alternative Splicing Axis Impacts Human Hematopoietic Stem Cell Molecular Identity throughout Development. Cell Stem Cell 2018, 22, 575–588.e7. [Google Scholar] [CrossRef] [Green Version]
- Zhang, S.; Mo, Q.; Wang, X. Oncological role of HMGA2 (Review). Int. J. Oncol. 2019, 55, 775–778. [Google Scholar] [CrossRef]
- Reeves, R.; Wolffe, A.P. Substrate structure influences binding of the non-histone protein HMG-I(Y) to free and nucleosomal DNA. Biochemistry 1996, 35, 5063–5074. [Google Scholar] [CrossRef]
- Nissen, M.S.; Reeves, R. Changes in superhelicity are introduced into closed circular DNA by binding of high mobility group protein I/Y. J. Biol. Chem. 1995, 270, 4355–4360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hill, D.A.; Pedulla, M.L.; Reeves, R. Directional binding of HMG-I(Y) on four-way junction DNA and the molecular basis for competitive binding with HMG-1 and histone H1. Nucleic Acids Res. 1999, 27, 2135–2144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reeves, R.; Beckerbauer, L. HMGI/Y proteins: Flexible regulators of transcription and chromatin structure. Biochim. Biophys. Acta Gene Struct. Expr. 2001, 1519, 13–29. [Google Scholar] [CrossRef]
- Frank, O.; Schwanbeck, R.; Wiśniewski, J.R. Protein footprinting reveals specific binding modes of a high mobility group protein I to DNAs of different conformation. J. Biol. Chem. 1998, 273, 20015–20020. [Google Scholar] [CrossRef] [Green Version]
- Thanos, D.; Maniatis, T. Virus induction of human IFNβ gene expression requires the assembly of an enhanceosome. Cell 1995, 83, 1091–1100. [Google Scholar] [CrossRef] [Green Version]
- Thanos, D.; Du, W.; Maniatis, T. The high mobility group protein HMG I(Y) is an essential structural component of a virus-inducible enhancer complex. In Cold Spring Harbor Symposia on Quantitative Biology; Cold Spring Harbor Laboratory Press: New York, NY, USA, 1993; Volume 58, pp. 73–81. [Google Scholar] [CrossRef] [PubMed]
- Thanos, D.; Maniatis, T. The High Mobility Group protein HMG I(Y) is required for NF-κB-dependent virus induction of the human IFN-β gene. Cell 1992, 71, 777–789. [Google Scholar] [CrossRef]
- Wolffe, A.P. Architectural transcription factors. Science 1994, 264, 1100–1101. [Google Scholar] [CrossRef]
- Carey, M. The enhanceosome and transcriptional synergy. Cell 1998, 92, 5–8. [Google Scholar] [CrossRef] [Green Version]
- Radic, M.Z.; Saghbini, M.; Elton, T.S.; Reeves, R.; Hamkalo, B.A. Hoechst 33258, distamycin A, and high mobility group protein I (HMG-I) compete for binding to mouse satellite DNA. Chromosoma 1992, 101, 602–608. [Google Scholar] [CrossRef]
- Käs, E.; Poljak, L.; Adachi, Y.; Laemmli, U.K. A model for chromatin opening: Stimulation of topoisomerase II and restriction enzyme cleavage of chromatin by distamycin. EMBO J. 1993, 12, 115–126. [Google Scholar] [CrossRef] [PubMed]
- Baldassarre, G.; Battista, S.; Belletti, B.; Thakur, S.; Pentimalli, F.; Trapasso, F.; Fedele, M.; Pierantoni, G.; Croce, C.M.; Fusco, A. Negative Regulation of BRCA1 Gene Expression by HMGA1 Proteins Accounts for the Reduced BRCA1 Protein Levels in Sporadic Breast Carcinoma. Mol. Cell. Biol. 2003, 23, 2225–2238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hammond, S.M.; Sharpless, N.E. HMGA2, MicroRNAs, and Stem Cell Aging. Cell 2008, 135, 1013–1016. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nishino, J.; Kim, I.; Chada, K.; Morrison, S.J. Hmga2 promotes neural stem cell self-renewal in young but not olNishino, J., Kim, I., Chada, K., & Morrison, S. J. (2008). Hmga2 promotes neural stem cell self-renewal in young but not old mice by reducing p16Ink4a and p19Arf Expression. Cell 2008, 135, 227–239. [Google Scholar]
- Tzatsos, A.; Bardeesy, N. Ink4a/Arf regulation by let-7b and Hmga2: A genetic pathway governing stem cell aging. Cell Stem Cell 2008, 3, 469–470. [Google Scholar] [CrossRef] [Green Version]
- Zhou, X.; Benson, K.F.; Przybysz, K.; Liu, J.; Hou, Y.; Cherath, L.; Chada, K. Genomic structure and expression of the murine Hmgi-c gene. Nucleic Acids Res. 1996, 24, 4071–4077. [Google Scholar] [CrossRef]
- Rogalla, P.; Drechsler, K.; Frey, G.; Hennig, Y.; Helmke, B.; Bonk, U.; Bullerdiek, J. HMGI-C expression patterns in human tissues. Implications for the genesis of frequent mesenchymal tumors. Am. J. Pathol. 1996, 149, 775–779. [Google Scholar]
- Rommel, B.; Rogalla, P.; Jox, A.; Kalle, C.V.; Kazmierczak, B.; Wolf, J.; Bullerdiek, J. HMGI-C, a member of the high mobility group family of proteins, is expressed in hematopoietic stem cells and in leukemic cells. Leuk. Lymphoma 1997, 26, 603–607. [Google Scholar] [CrossRef]
- Gattas, G.J.; Quade, B.J.; Nowak, R.A.; Morton, C.C. HMGIC expression in human adult and fetal tissues and in uterine leiomyomata. Genes. Chromosomes Cancer 1999, 25, 316–322. [Google Scholar] [CrossRef]
- Somervaille, T.C.P.; Matheny, C.J.; Spencer, G.J.; Iwasaki, M.; Rinn, J.L.; Witten, D.M.; Chang, H.Y.; Shurtleff, S.A.; Downing, J.R.; Cleary, M.L. Hierarchical maintenance of MLL myeloid leukemia stem cells employs a transcriptional program shared with embryonic rather than adult stem cells. Cell Stem Cell 2009, 4, 129–140. [Google Scholar] [CrossRef] [Green Version]
- Chiappetta, G.; Avantaggiato, V.; Visconti, R.; Fedele, M.; Battista, S.; Trapasso, F.; Merciai, B.M.; Fidanza, V.; Giancotti, V.; Santoro, M.; et al. High level expression of the HMGI (Y) gene during embryonic development. Oncogene 1996, 13, 2439–2446. [Google Scholar] [PubMed]
- Zhou, X.; Benson, K.F.; Ashar, H.R.; Chada, K. Mutation responsible for the mouse pygmy phenotype in the developmentally regulated factor HMGI-C. Nature 1995, 376, 771–774. [Google Scholar] [CrossRef] [PubMed]
- Battista, S.; Fidanza, V.; Fedele, M.; Klein-Szanto, A.J.; Outwater, E.; Brunner, H.; Santoro, M.; Croce, C.M.; Fusco, A. The expression of a truncated HMGI-C gene induces gigantism associated with lipomatosis. Cancer Res. 1999, 59, 4793–4797. [Google Scholar] [PubMed]
- Melillo, R.M.; Pierantoni, G.M.; Scala, S.; Battista, S.; Fedele, M.; Stella, A.; De Biasio, M.C.; Chiappetta, G.; Fidanza, V.; Condorelli, G.; et al. Critical Role of the HMGI(Y) Proteins in Adipocytic Cell Growth and Differentiation. Mol. Cell. Biol. 2001, 21, 2485–2495. [Google Scholar] [CrossRef] [Green Version]
- Tallini, G.; Dal Cin, P. HMGI(Y) and HMGI-C dysregulation: A common occurrence in human tumors. Adv. Anat. Pathol. 1999, 6, 237–246. [Google Scholar] [CrossRef]
- Harada-Shirado, K.; Ikeda, K.; Ogawa, K.; Ohkawara, H.; Kimura, H.; Kai, T.; Noji, H.; Morishita, S.; Komatsu, N.; Takeishi, Y. Dysregulation of the MIRLET7/HMGA2 axis with methylation of the CDKN2A promoter in myeloproliferative neoplasms. Br. J. Haematol. 2015, 168, 338–349. [Google Scholar] [CrossRef] [Green Version]
- Huang, M.L.; Chen, C.C.; Chang, L.C. Gene expressions of HMGI-C and HMGI(Y) are associated with stage and metastasis in colorectal cancer. Int. J. Colorectal Dis. 2009, 24, 1281–1286. [Google Scholar] [CrossRef]
- Wang, X.; Liu, X.; Li, A.Y.-J.; Chen, L.; Lai, L.; Lin, H.H.; Hu, S.; Yao, L.; Peng, J.; Loera, S.; et al. Overexpression of HMGA2 promotes metastasis and impacts survival of colorectal cancers. Clin. Cancer Res. 2011, 17, 2570–2580. [Google Scholar] [CrossRef] [Green Version]
- Pallante, P.; Sepe, R.; Puca, F.; Fusco, A. High mobility group a proteins as tumor markers. Front. Med. 2015, 2, 15. [Google Scholar] [CrossRef] [Green Version]
- Berlingieri, M.T.; Pierantoni, G.M.; Giancotti, V.; Santoro, M.; Fusco, A. Thyroid cell transformation requires the expression of the HMGA1 proteins. Oncogene 2002, 21, 2971–2980. [Google Scholar] [CrossRef] [Green Version]
- Scala, S.; Portella, G.; Fedele, M.; Chiappetta, G.; Fusco, A. Adenovirus-mediated suppression of HMGI(Y) protein synthesis as potential therapy of human malignant neoplasias. Proc. Natl. Acad. Sci. USA 2000, 97, 4256–4261. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berlingieri, M.T.; Manfioletti, G.; Santoro, M.; Bandiera, A.; Visconti, R.; Giancotti, V.; Fusco, A. Inhibition of HMGI-C protein synthesis suppresses retrovirally induced neoplastic transformation of rat thyroid cells. Mol. Cell. Biol. 1995, 15, 1545–1553. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fedele, M.; Berlingieri, M.T.; Scala, S.; Chiariotti, L.; Viglietto, G.; Rippel, V.; Bullerdiek, J.; Santoro, M.; Fusco, A. Truncated and chimeric HMGI-C genes induce neoplastic transformation of NIH3T3 murine fibroblasts. Oncogene 1998, 17, 413–418. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wood, L.J.; Maher, J.F.; Bunton, T.E.; Resar, L.M. The oncogenic properties of the HMG-I gene family. Cancer Res. 2000, 60, 4256–4261. [Google Scholar]
- Baldassarre, G.; Fedele, M.; Battista, S.; Vecchione, A.; Klein-Szanto, A.J.P.; Santoro, M.; Waldmann, T.A.; Azimi, N.; Croce, C.M.; Fusco, A. Onset of natural killer cell lymphomas in transgenic mice carrying a truncated HMGI-C gene by the chronic stimulation of the IL-2 and IL-15 pathway. Proc. Natl. Acad. Sci. USA 2001, 98, 7970–7975. [Google Scholar] [CrossRef] [Green Version]
- Xu, Y.; Sumter, T.F.; Bhattacharya, R.; Tesfaye, A.; Fuchs, E.J.; Wood, L.J.; Huso, D.L.; Resar, L.M.S. The HMG-I oncogene causes highly penetrant, aggressive lymphoid malignancy in transgenic mice and is overexpressed in human leukemia. Cancer Res. 2004, 64, 3371–3375. [Google Scholar] [CrossRef] [Green Version]
- Fedele, M.; Pentimalli, F.; Baldassarre, G.; Battista, S.; Klein-Szanto, A.J.P.; Kenyon, L.; Visone, R.; De Martino, I.; Ciarmiello, A.; Arra, C.; et al. Transgenic mice overexpressing the wild-type form of the HMGA1 gene develop mixed growth hormone/prolactin cell pituitary adenomas and natural killer cell lymphomas. Oncogene 2005, 24, 3427–3435. [Google Scholar] [CrossRef] [Green Version]
- Fedele, M.; Battista, S.; Kenyon, L.; Baldassarre, G.; Fidanza, V.; Klein-Szanto, A.J.P.; Parlow, A.F.; Visone, R.; Pierantoni, G.M.; Outwater, E.; et al. Overexpression of the HMGA2 gene in transgenic mice leads to the onset of pituitary adenomas. Oncogene 2002, 21, 3190–3198. [Google Scholar] [CrossRef] [Green Version]
- Ben-Porath, I.; Thomson, M.W.; Carey, V.J.; Ge, R.; Bell, G.W.; Regev, A.; Weinberg, R.A. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat. Genet. 2008, 40, 499–507. [Google Scholar] [CrossRef]
- Chou, B.-K.; Mali, P.; Huang, X.; Ye, Z.; Dowey, S.N.; Resar, L.M.; Zou, C.; Zhang, Y.A.; Tong, J.; Cheng, L. Efficient human iPS cell derivation by a non-integrating plasmid from blood cells with unique epigenetic and gene expression signatures. Cell Res. 2011, 21, 518–529. [Google Scholar] [CrossRef]
- Karp, J.E.; Smith, B.D.; Resar, L.S.; Greer, J.M.; Blackford, A.; Zhao, M.; Moton-Nelson, D.; Alino, K.; Levis, M.J.; Gore, S.D.; et al. Phase 1 and pharmacokinetic study of bolus-infusion flavopiridol followed by cytosine arabinoside and mitoxantrone for acute leukemias. Blood 2011, 117, 3302–3310. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nelson, D.M.; Joseph, B.; Hillion, J.; Segal, J.; Karp, J.E.; Resar, L.M.S. Flavopiridol induces BCL-2 expression and represses oncogenic transcription factors in leukemic blasts from adults with refractory acute myeloid leukemia. Leuk. Lymphoma 2011, 52, 1999–2006. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, G.; Chen, J.; Lee, S.; Clark, T.; Rowley, J.D.; Wang, S.M. The pattern of gene expression in human CD34(+) stem/progenitor cells. Proc. Natl. Acad. Sci. USA 2001, 98, 13966–13971. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dolde, C.E.; Mukherjee, M.; Cho, C.; Resar, L.M.S. HMG-I/Y in human breast cancer cell lines. Breast Cancer Res. Treat. 2002, 71, 181–191. [Google Scholar] [CrossRef] [PubMed]
- Tesfaye, A.; Di Cello, F.; Hillion, J.; Ronnett, B.M.; Elbahloul, O.; Ashfaq, R.; Dhara, S.; Prochownik, E.; Tworkoski, K.; Reeves, R.; et al. The high-mobility group A1 gene up-regulates cyclooxygenase 2 expression in uterine tumorigenesis. Cancer Res. 2007, 67, 3998–4004. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Di Cello, F.; Hillion, J.; Hristov, A.; Wood, L.J.; Mukherjee, M.; Schuldenfrei, A.; Kowalski, J.; Bhattacharya, R.; Ashfaq, R.; Resar, L.M.S. HMGA2 participates in transformation in human lung cancer. Mol. Cancer Res. 2008, 6, 743–750. [Google Scholar] [CrossRef] [Green Version]
- Di Cello, F.; Hillion, J.; Kowalski, J.; Ronnett, B.M.; Aderinto, A.; Huso, D.L.; Resar, L.M.S. Cyclooxygenase inhibitors block uterine tumorigenesis in HMGA1a transgenic mice and human xenografts. Mol. Cancer Ther. 2008, 7, 2090–2095. [Google Scholar] [CrossRef] [Green Version]
- Hillion, J.; Dhara, S.; Sumter, T.F.; Mukherjee, M.; Di Cello, F.; Belton, A.; Turkson, J.; Jaganathan, S.; Cheng, L.; Ye, Z.; et al. The high-mobility group A1a/signal transducer and activator of transcription-3 axis: An achilles heel for hematopoietic malignancies? Cancer Res. 2008, 68, 10121–10127. [Google Scholar] [CrossRef] [Green Version]
- Hristov, A.C.; Cope, L.; Di Cello, F.; Reyes, M.D.; Singh, M.; Hillion, J.A.; Belton, A.; Joseph, B.; Schuldenfrei, A.; Iacobuzio-Donahue, C.A.; et al. HMGA1 correlates with advanced tumor grade and decreased survival in pancreatic ductal adenocarcinoma. Mod. Pathol. 2010, 23, 98–104. [Google Scholar] [CrossRef]
- Belton, A.; Gabrovsky, A.; Bae, Y.K.; Reeves, R.; Iacobuzio-Donahue, C.; Huso, D.L.; Resar, L.M.S. HMGA1 induces intestinal polyposis in transgenic mice and drives tumor progression and stem cell properties in colon cancer cells. PLoS ONE 2012, 7, e30034. [Google Scholar] [CrossRef] [Green Version]
- Pomeroy, S.L.; Tamayo, P.; Gaasenbeek, M.; Sturla, L.M.; Angelo, M.; McLaughlin, M.E.; Kim, J.Y.H.; Goumnerova, L.C.; Black, P.M.; Lau, C.; et al. Prediction of central nervous system embryonal tumour outcome based on gene expression. Nature 2002, 415, 436–442. [Google Scholar] [CrossRef] [PubMed]
- Wood, L.J.; Mukherjee, M.; Dolde, C.E.; Xu, Y.; Maher, J.F.; Bunton, T.E.; Williams, J.B.; Resar, L.M. HMG-I/Y, a new c-Myc target gene and potential oncogene. Mol. Cell. Biol. 2000, 20, 5490–5502. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schuldenfrei, A.; Belton, A.; Kowalski, J.; Talbot, C.C.; Di Cello, F.; Poh, W.; Tsai, H.-L.; Shah, S.N.; Huso, T.H.; Huso, D.L.; et al. HMGA1 drives stem cell, inflammatory pathway, and cell cycle progression genes during lymphoid tumorigenesis. BMC Genomics 2011, 12, 549. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Efanov, A.; Zanesi, N.; Coppola, V.; Nuovo, G.; Bolon, B.; Wernicle-Jameson, D.; Lagana, A.; Hansjuerg, A.; Pichiorri, F.; Croce, C.M. Human HMGA2 protein overexpressed in mice induces precursor T-cell lymphoblastic leukemia. Blood Cancer J. 2014, 4, e227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Di Cello, F.; Dhara, S.; Hristov, A.C.; Kowalski, J.; Elbahloul, O.; Hillion, J.; Roy, S.; Meijerink, J.P.P.; Winter, S.S.; Larson, R.S.; et al. Inactivation of the Cdkn2a locus cooperates with HMGA1 to drive T-cell leukemogenesis. Leuk. Lymphoma 2013, 54, 1762–1768. [Google Scholar] [CrossRef] [PubMed]
- Petit, M.M.R.; Mols, R.; Schoenmakers, E.F.P.M.; Mandahl, N.; Van De Ven, W.J.M. LPP, the preferred fusion partner gene of HMGIC in lipomas, is a novel member of the LIM protein gene family. Genomics 1996, 36, 118–129. [Google Scholar] [CrossRef]
- Schoenmakers, E.F.; Huysmans, C.; Van de Ven, W.J. Allelic knockout of novel splice variants of human recombination repair gene RAD51B in t(12;14) uterine leiomyomas. Cancer Res. 1999, 59, 19–23. [Google Scholar]
- Schoenmakers, E.F.; Wanschura, S.; Mols, R.; Bullerdiek, J.; Van den Berghe, H.; Van de Ven, W.J. Recurrent rearrangements in the high mobility group protein gene, HMGI-C, in benign mesenchymal tumours. Nat. Genet. 1995, 10, 436–444. [Google Scholar] [CrossRef]
- Kazmierczak, B.; Meyer-Bolte, K.; Tran, K.H.; Wöckel, W.; Breightman, I.; Rosigkeit, J.; Bartnitzke, S.; Bullerdiek, J. A high frequency of tumors with rearrangements of genes of the HMGI(Y) family in a series of 191 pulmonary chondroid hamartomas. Genes. Chromosomes Cancer 1999, 26, 125–133. [Google Scholar] [CrossRef]
- Odero, M.D.; Grand, F.H.; Iqbal, S.; Ross, F.; Roman, J.P.; Vizmanos, J.L.; Andrieux, J.; Laï, J.L.; Calasanz, M.J.; Cross, N.C.P. Disruption and aberrant expression of HMGA2 as a consequence of diverse chromosomal translocations in myeloid malignancies. Leukemia 2005, 19, 245–252. [Google Scholar] [CrossRef]
- Pasquinelli, A.E.; Reinhart, B.J.; Slack, F.; Martindale, M.Q.; Kuroda, M.I.; Maller, B.; Hayward, D.C.; Ball, E.E.; Degnan, B.; Müller, P.; et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature 2000, 408, 86–89. [Google Scholar] [CrossRef] [PubMed]
- Geurts, J.M.W.; Schoenmakers, E.F.P.M.; Van De Ven, W.J.M. Molecular characterization of a complex chromosomal rearrangement in a pleomorphic salivary gland adenoma involving the 3′-UTR of HMGIC. Cancer Genet. Cytogenet. 1997, 95, 198–205. [Google Scholar] [CrossRef]
- Inoue, N.; Izui-Sarumaru, T.; Murakami, Y.; Endo, Y.; Nishimura, J.I.; Kurokawa, K.; Kuwayama, M.; Shime, H.; Machii, T.; Kanakura, Y.; et al. Molecular basis of clonal expansion of hematopoiesis in 2 patients with paroxysmal nocturnal hemoglobinuria (PNH). Blood 2006, 108, 4232–4236. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, Y.S.; Dutta, A. The tumor suppressor microRNA let-7 represses the HMGA2 oncogene Email alerting service The tumor suppressor microRNA let-7 represses the HMGA2 oncogene. Genes Dev. 2007, 1025–1030. [Google Scholar] [CrossRef] [Green Version]
- Hebert, C.; Norris, K.; Scheper, M.A.; Nikitakis, N.; Sauk, J.J. High mobility group A2 is a target for miRNA-98 in head and neck squamous cell carcinoma. Mol. Cancer 2007, 6. [Google Scholar] [CrossRef] [Green Version]
- Baba, O.; Horie, T.; Nakao, T.; Hakuno, D.; Nakashima, Y.; Nishi, H.; Kuwabara, Y.; Nishiga, M.; Nishino, T.; Ide, Y.; et al. MicroRNA 33 Regulates the Population of Peripheral Inflammatory Ly6C high Monocytes through Dual Pathways. Mol. Cell. Biol. 2018, 38. [Google Scholar] [CrossRef] [Green Version]
- Reinhart, B.J.; Slack, F.J.; Basson, M.; Pasquienelll, A.E.; Bettlnger, J.C.; Rougvle, A.E.; Horvitz, H.R.; Ruvkun, G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 2000, 403, 901–906. [Google Scholar] [CrossRef]
- Johnson, S.M.; Grosshans, H.; Shingara, J.; Byrom, M.; Jarvis, R.; Cheng, A.; Labourier, E.; Reinert, K.L.; Brown, D.; Slack, F.J. RAS is regulated by the let-7 microRNA family. Cell 2005, 120, 635–647. [Google Scholar] [CrossRef] [Green Version]
- Mayr, C.; Hemann, M.T.; Bartel, D.P. Disrupting the pairing between let-7 and Hmga2 enhances oncogenic transformation. Science 2007, 315, 1576–1579. [Google Scholar] [CrossRef] [Green Version]
- Wang, T.; Zhang, X.; Obijuru, L.; Laser, J.; Aris, V.; Lee, P.; Mittal, K.; Soteropoulos, P.; Wei, J.-J. A micro-RNA signature associated with race, tumor size, and target gene activity in human uterine leiomyomas. Genes. Chromosomes Cancer 2007, 46, 336–347. [Google Scholar] [CrossRef]
- Grosveld, F.; van Assendelft, G.B.; Greaves, D.R.; Kollias, G. Position-independent, high-level expression of the human β-globin gene in transgenic mice. Cell 1987, 51, 975–985. [Google Scholar] [CrossRef] [Green Version]
- Kottickal, L.V.; Sarada, B.; Ashar, H.; Chada, K.; Nagarajan, L. Preferential expression of HMGI-C isoforms lacking the acidic carboxy terminal in human leukemia. Biochem. Biophys. Res. Commun. 1998, 242, 452–456. [Google Scholar] [CrossRef] [PubMed]
- Santulli, B.; Kazmierczak, B.; Napolitano, R.; Caliendo, I.; Chiappetta, G.; Rippe, V.; Bullerdiek, J.; Fusco, A. A 12q13 translocation involving the HMGI-C gene in Richter transformation of a chronic lymphocytic leukemia. Cancer Genet. Cytogenet. 2000, 119, 70–73. [Google Scholar] [CrossRef]
- Cirera-Salinas, D.; Pauta, M.; Allen, R.M.; Salerno, A.G.; Ramírez, C.M.; Chamorro-Jorganes, A.; Wanschel, A.C.; Lasuncion, M.A.; Morales-Ruiz, M.; Suarez, Y.; et al. Mir-33 regulates cell proliferation and cell cycle progression. Cell Cycle 2012, 11, 922–933. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thomas, M.; Lange-Grünweller, K.; Weirauch, U.; Gutsch, D.; Aigner, A.; Grünweller, A.; Hartmann, R.K. The proto-oncogene Pim-1 is a target of miR-33a. Oncogene 2012, 31, 918–928. [Google Scholar] [CrossRef] [Green Version]
- Rice, S.J.; Lai, S.-C.; Wood, L.W.; Helsley, K.R.; Runkle, E.A.; Winslow, M.M.; Mu, D. MicroRNA-33a mediates the regulation of high mobility group AT-hook 2 gene (HMGA2) by thyroid transcription factor 1 (TTF-1/NKX2-1). J. Biol. Chem. 2013, 288, 16348–16360. [Google Scholar] [CrossRef] [Green Version]
- Wiemer, E.A.C. The role of microRNAs in cancer: No small matter. Eur. J. Cancer 2007, 43, 1529–1544. [Google Scholar] [CrossRef]
- Godley, L.A. HMGA2 levels in CML: Reflective of miRNA gene regulation in a hematopoietic tumor? Leuk. Lymphoma 2007, 48, 1898–1899. [Google Scholar] [CrossRef]
- Lu, J.; Getz, G.; Miska, E.A.; Alvarez-Saavedra, E.; Lamb, J.; Peck, D.; Sweet-Cordero, A.; Ebert, B.L.; Mak, R.H.; Ferrando, A.A.; et al. MicroRNA expression profiles classify human cancers. Nature 2005, 435, 834–838. [Google Scholar] [CrossRef]
- Kumar, M.S.; Lu, J.; Mercer, K.L.; Golub, T.R.; Jacks, T. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat. Genet. 2007, 39, 673–677. [Google Scholar] [CrossRef]
- Sgarra, R.; Pegoraro, S.; D’Angelo, D.; Ros, G.; Zanin, R.; Sgubin, M.; Petrosino, S.; Battista, S.; Manfioletti, A.G. High Mobility Group A (HMGA): Chromatin Nodes Controlled by a Knotty miRNA Network. Int. J. Mol. Sci. 2020, 21, 717. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boyerinas, B.; Park, S.-M.; Shomron, N.; Hedegaard, M.M.; Vinther, J.; Andersen, J.S.; Feig, C.; Xu, J.; Burge, C.B.; Peter, M.E. Identification of let-7-regulated oncofetal genes. Cancer Res. 2008, 68, 2587–2591. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Copley, M.R.; Babovic, S.; Benz, C.; Knapp, D.J.H.F.; Beer, P.A.; Kent, D.G.; Wohrer, S.; Treloar, D.Q.; Day, C.; Rowe, K.; et al. The Lin28b-let-7-Hmga2 axis determines the higher self-renewal potential of fetal haematopoietic stem cells. Nat. Cell Biol. 2013, 15, 916–925. [Google Scholar] [CrossRef] [PubMed]
- Cavazzana-Calvo, M.; Payen, E.; Negre, O.; Wang, G.; Hehir, K.; Fusil, F.; Down, J.; Denaro, M.; Brady, T.; Westerman, K.; et al. Transfusion independence and HMGA2 activation after gene therapy of human β-thalassaemia. Nature 2010, 467, 318–322. [Google Scholar] [CrossRef]
- Wang, G.P.; Berry, C.C.; Malani, N.; Leboulch, P.; Fischer, A.; Hacein-Bey-Abina, S.; Cavazzana-Calvo, M.; Bushman, F.D. Dynamics of gene-modified progenitor cells analyzed by tracking retroviral integration sites in a human SCID-X1 gene therapy trial. Blood 2010, 115, 4356–4366. [Google Scholar] [CrossRef]
- De Vasconcellos, J.F.; Lee, Y.T.; Byrnes, C.; Tumburu, L.; Rabel, A.; Miller, J.L. HMGA2 Moderately Increases Fetal Hemoglobin Expression in Human Adult Erythroblasts. PLoS ONE 2016, 11, e0166928. [Google Scholar] [CrossRef]
- De Vasconcellos, J.F.; Byrnes, C.; Lee, Y.T.; Allwardt, J.M.; Kaushal, M.; Rabel, A.; Miller, J.L. Tough decoy targeting of predominant let-7 miRNA species in adult human hematopoietic cells. J. Transl. Med. 2017, 15, 169. [Google Scholar] [CrossRef]
- Bluteau, O.; Langlois, T.; Rivera-Munoz, P.; Favale, F.; Rameau, P.; Meurice, G.; Dessen, P.; Solary, E.; Raslova, H.; Mercher, T.; et al. Developmental changes in human megakaryopoiesis. J. Thromb. Haemost. 2013, 11, 1730–1741. [Google Scholar] [CrossRef]
- Stolla, M.C.; Catherman, S.C.; Kingsley, P.D.; Rowe, R.G.; Koniski, A.D.; Fegan, K.; Vit, L.; McGrath, K.E.; Daley, G.Q.; Palis, J. Lin28b regulates age-dependent differences in murine platelet function. Blood Adv. 2019, 3, 72–82. [Google Scholar] [CrossRef] [Green Version]
- Battista, S.; Pentimalli, F.; Baldassarre, G.; Fedele, M.; Fidanza, V.; Croce, C.M.; Fusco, A. Loss of Hmga1 gene function affects embryonic stem cell lympho-hematopoietic differentiation. FASEB J. 2003, 17, 1496–1498. [Google Scholar] [CrossRef]
- Pierantoni, G.M.; Agosti, V.; Fedele, M.; Bond, H.; Caliendo, I.; Chiappetta, G.; Lo Coco, F.; Pane, F.; Turco, M.C.; Morrone, G.; et al. High-mobility group A1 proteins are overexpressed in human leukaemias. Biochem. J. 2003, 372, 145–150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berent-Maoz, B.; Montecino-Rodriguez, E.; Fice, M.; Casero, D.; Seet, C.S.; Crooks, G.M.; Lowry, W.; Dorshkind, K. The expansion of thymopoiesis in neonatal mice is dependent on expression of High mobility group A 2 protein (Hmga2). PLoS ONE 2015, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xi, Y.; Watanabe, S.; Hino, Y.; Sakamoto, C.; Nakatsu, Y.; Okada, S.; Nakao, M. Hmga1 is differentially expressed and mediates silencing of the CD4/CD8 loci in T cell lineages and leukemic cells. Cancer Sci. 2012, 103, 439–447. [Google Scholar] [CrossRef] [PubMed]
- Nikolajczyk, B.S.; Nelsen, B.; Sen, R. Precise alignment of sites required for mu enhancer activation in B cells. Mol. Cell. Biol. 1996, 16, 4544–4554. [Google Scholar] [CrossRef] [Green Version]
- Nikolajczyk, B.S.; Cortes, M.; Feinman, R.; Sen, R. Combinatorial determinants of tissue-specific transcription in B cells and macrophages. Mol. Cell. Biol. 1997, 17, 3527–3535. [Google Scholar] [CrossRef] [Green Version]
- Blackwell, T.K.; Moore, M.W.; Yancopoulos, G.D.; Suh, H.; Lutzker, S.; Selsing, E.; Alt, F.W. Recombination between immunoglobulin variable region gene segments is enhanced by transcription. Nature 1986, 324, 585–589. [Google Scholar] [CrossRef]
- Sakai, E.; Bottaro, A.; Davidson, L.; Sleckman, B.P.; Alt, F.W. Recombination and transcription of the endogenous Ig heavy chain locus is effected by the Ig heavy chain intronic enhancer core region in the absence of the matrix attachment regions. Proc. Natl. Acad. Sci. USA 1999, 96, 1526–1531. [Google Scholar] [CrossRef] [Green Version]
- Lewis, R.T.; Andreucci, A.; Nikolajczyk, B.S. PU. 1-mediated transcription is enhanced by HMG-I(Y)-dependent structural mechanisms. J. Biol. Chem. 2001, 276, 9550–9557. [Google Scholar] [CrossRef] [Green Version]
- Nagulapalli, S.; Pongubala, J.M.; Atchison, M.L. Multiple proteins physically interact with PU.1. Transcriptional synergy with NF-IL6 beta (C/EBP delta, CRP3). J. Immunol. 1995, 155, 4330–4338. [Google Scholar]
- McCarthy, K.M.; McDevit, D.; Andreucci, A.; Reeves, R.; Nikolajczyk, B.S. HMGA1 co-activates transcription in B cells through indirect association with DNA. J. Biol. Chem. 2003, 278, 42106–42114. [Google Scholar] [CrossRef] [Green Version]
- Vardiman, J.W.; Thiele, J.; Arber, D.A.; Brunning, R.D.; Borowitz, M.J.; Porwit, A.; Harris, N.L.; Le Beau, M.M.; Hellström-Lindberg, E.; Tefferi, A.; et al. The 2008 revision of the World Health Organization (WHO) classification of myeloid neoplasms and acute leukemia: Rationale and important changes. Blood 2009, 114, 937–951. [Google Scholar] [CrossRef] [Green Version]
- Dameshek, W. Some speculations on the myeloproliferative syndromes [editorial]. Blood. 1951;6(4):372-375. Blood 2016, 127, 663. [Google Scholar]
- Nowell, P.C.; Hungerford, D.A. Chromosome studies on normal and leukemic human leukocytes. J. Natl. Cancer Inst. 1960, 25, 85–109. [Google Scholar]
- Rowley, J.D. A new consistent chromosomal abnormality in chronic myelogenous leukaemia identified by quinacrine fluorescence and Giemsa staining. Nature 1973, 243, 290–293. [Google Scholar] [CrossRef]
- Chen, C.C.; You, J.Y.; Lung, J.; Huang, C.E.; Chen, Y.Y.; Leu, Y.W.; Ho, H.Y.; Li, C.P.; Lu, C.H.; Lee, K.D.; et al. Aberrant let7a/HMGA2 signaling activity with unique clinical phenotype in JAK2-mutated myeloproliferative neoplasms. Haematologica 2017, 102, 509–518. [Google Scholar] [CrossRef] [Green Version]
- James, C.; Ugo, V.; Le Couédic, J.P.; Staerk, J.; Delhommeau, F.; Lacout, C.; Garçon, L.; Raslova, H.; Berger, R.; Bennaceur-Griscelli, A.; et al. A unique clonal JAK2 mutation leading to constitutive signalling causes polycythaemia vera. Nature 2005, 434, 1144–1148. [Google Scholar] [CrossRef]
- Kralovics, R.; Passamonti, F.; Buser, A.S.; Teo, S.S.; Tiedt, R.; Passweg, J.R.; Tichelli, A.; Cazzola, M.; Skoda, R.C. A gain-of-function mutation of JAK2 in myeloproliferative disorders. N. Engl. J. Med. 2005, 352, 1779–1790. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Levine, R.L.; Wadleigh, M.; Cools, J.; Ebert, B.L.; Wernig, G.; Huntly, B.J.P.; Boggon, T.J.; Wlodarska, I.; Clark, J.J.; Moore, S.; et al. Activating mutation in the tyrosine kinase JAK2 in polycythemia vera, essential thrombocythemia, and myeloid metaplasia with myelofibrosis. Cancer Cell 2005, 7, 387–397. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baxter, E.J.; Scott, L.M.; Campbell, P.J.; East, C.; Fourouclas, N.; Swanton, S.; Vassiliou, G.S.; Bench, A.J.; Boyd, E.M.; Curtin, N.; et al. Acquired mutation of the tyrosine kinase JAK2 in human myeloproliferative disorders. Lancet 2005, 365, 1054–1061. [Google Scholar] [CrossRef]
- Vainchenker, W.; Delhommeau, F.; Constantinescu, S.N.; Bernard, O.A. New mutations and pathogenesis of myeloproliferative neoplasms. Blood 2011, 118, 1723–1735. [Google Scholar] [CrossRef] [PubMed]
- Aliano, S.; Cirmena, G.; Garuti, A.; Fugazza, G.; Bruzzone, R.; Rocco, I.; Malacarne, M.; Ballestrero, A.; Sessarego, M. HMGA2 overexpression in polycythemia vera with t(12;21)(q14;q22). Cancer Genet. Cytogenet. 2007, 177, 115–119. [Google Scholar] [CrossRef] [PubMed]
- Storlazzi, C.T.; Albano, F.; Locunsolo, C.; Lonoce, A.; Funes, S.; Guastadisegni, M.C.; Cimarosto, L.; Impera, L.; D’Addabbo, P.; Panagopoulos, I.; et al. t(3;12)(q26;q14) in polycythemia vera is associated with upregulation of the HMGA2 gene. Leukemia 2006, 20, 2190–2192. [Google Scholar] [CrossRef] [Green Version]
- Etienne, A.; Carbuccia, N.; Adélaïde, J.; Bekhouche, I.; Rémy, V.; Sohn, C.; Sainty, D.; Gastaut, J.A.; Olschwang, S.; Birnbaum, D.; et al. Rearrangements involving 12q in myeloproliferative disorders: Possible role of HMGA2 and SOCS2 genes. Cancer Genet. Cytogenet. 2007, 176, 80–88. [Google Scholar] [CrossRef]
- Andrieux, J.; Demory, J.L.; Dupriez, B.; Quief, S.; Plantier, I.; Roumier, C.; Bauters, F.; Laï, J.L.; Kerckaert, J.P. Dysregulation and Overexpression of HMGA2 in Myelofibrosis with Myeloid Metaplasia. Genes Chromosom. Cancer 2004, 39, 82–87. [Google Scholar] [CrossRef] [PubMed]
- Guglielmelli, P.; Zini, R.; Bogani, C.; Salati, S.; Pancrazzi, A.; Bianchi, E.; Mannelli, F.; Ferrari, S.; Le Bousse-Kerdilès, M.-C.; Bosi, A.; et al. Molecular Profiling of CD34 + Cells in Idiopathic Myelofibrosis Identifies a Set of Disease-Associated Genes and Reveals the Clinical Significance of Wilms’ Tumor Gene 1 ( WT1 ). Stem Cells 2007, 25, 165–173. [Google Scholar] [CrossRef] [PubMed]
- Young, A.R.J.; Narita, M. Oncogenic HMGA2: Short or small? Genes Dev. 2007, 21, 1005–1009. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ikeda, K.; Ogawa, K.; Takeishi, Y. The role of HMGA2 in the proliferation and expansion of a hematopoietic cell in myeloproliferative neoplasms. Fukushima J. Med. Sci. 2012, 58, 91–100. [Google Scholar] [CrossRef] [Green Version]
- James, C.; Mazurier, F.; Dupont, S.; Chaligne, R.; Lamrissi-Garcia, I.; Tulliez, M.; Lippert, E.; Marion, F.X.; Pasquet, J.M.; Etienne, G.; et al. The hematopoietic stem cell compartment of JAK2V617F-positive myeloproliferative disorders is a reflection of disease heterogeneity. Blood 2008, 112, 2429–2438. [Google Scholar] [CrossRef] [Green Version]
- Ikeda, K.; Mason, P.J.; Bessler, M. 3′UTR-truncated Hmga2 cDNA causes MPN-like hematopoiesis by conferring a clonal growth advantage at the level of HSC in mice. Blood 2011, 117, 5860–5869. [Google Scholar] [CrossRef] [Green Version]
- Li, O.; Li, J.; Dröge, P. DNA architectural factor and proto-oncogene HMGA2 regulates key developmental genes in pluripotent human embryonic stem cells. FEBS Lett. 2007, 581, 3533–3537. [Google Scholar] [CrossRef] [Green Version]
- Yu, F.; Yao, H.; Zhu, P.; Zhang, X.; Pan, Q.; Gong, C.; Huang, Y.; Hu, X.; Su, F.; Lieberman, J.; et al. let-7 Regulates Self Renewal and Tumorigenicity of Breast Cancer Cells. Cell 2007, 131, 1109–1123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ueda, K.; Ikeda, K.; Ikezoe, T.; Harada-Shirado, K.; Ogawa, K.; Hashimoto, Y.; Sano, T.; Ohkawara, H.; Kimura, S.; Shichishima-Nakamura, A.; et al. Hmga2 collaborates with JAK2V617F in the development of myeloproliferative neoplasms. Blood Adv. 2017, 1, 1001–1015. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oguro, H.; Yuan, J.; Tanaka, S.; Miyagi, S.; Mochizuki-Kashio, M.; Ichikawa, H.; Yamazaki, S.; Koseki, H.; Nakauchi, H.; Iwama, A. Lethal myelofibrosis induced by Bmi1-deficient hematopoietic cells unveils a tumor suppressor function of the polycomb group genes. J. Exp. Med. 2012, 209, 445–454. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Y.; Akada, H.; Nath, D.; Hutchison, R.E.; Mohi, G. Loss of Ezh2 cooperates with Jak2V617F in the development of myelofibrosis in a mouse model of myeloproliferative neoplasm. Blood 2016, 127, 3410–3423. [Google Scholar] [CrossRef] [Green Version]
- Martin, S.E.; Sausen, M.; Joseph, A.; Kingham, B.F.; Martin, E.S. Identification of a HMGA2-EFCAB6 gene rearrangement following next-generation sequencing in a patient with a t(12;22)(q14.3;q13.2) and JAK2V617F-positive myeloproliferative neoplasm. Cancer Genet. 2012, 205, 295–303. [Google Scholar] [CrossRef]
- Meyer, B.; Krisponeit, D.; Junghanss, C.; Escobar, H.M.; Bullerdiek, J. Quantitative expression analysis in peripheral blood of patients with chronic myeloid leukaemia: Correlation between HMGA2 expression and white blood cell count. Leuk. Lymphoma 2007, 48, 2008–2013. [Google Scholar] [CrossRef]
- Wei, J.; Li, H.; Wang, S.; Li, T.; Fan, J.; Liang, X.; Li, J.; Han, Q.; Zhu, L.; Fan, L.; et al. let-7 enhances osteogenesis and bone formation while repressing adipogenesis of human stromal/mesenchymal stem cells by regulating HMGA2. Stem Cells Dev. 2014, 23, 1452–1463. [Google Scholar] [CrossRef] [Green Version]
- Bruchova, H.; Merkerova, M.; Prchal, J.T. Aberrant expression of microRNA in polycythemia vera. Haematologica 2008, 93, 1009–1016. [Google Scholar] [CrossRef] [Green Version]
- Guglielmelli, P.; Tozzi, L.; Pancrazzi, A.; Bogani, C.; Antonioli, E.; Ponziani, V.; Poli, G.; Zini, R.; Ferrari, S.; Manfredini, R.; et al. MicroRNA expression profile in granulocytes from primary myelofibrosis patients. Exp. Hematol. 2007, 35, 1708.e1–1708.e12. [Google Scholar] [CrossRef]
- Zhan, H.; Cardozo, C.; Yu, W.; Wang, A.; Moliterno, A.R.; Dang, C.V.; Spivak, J.L. MicroRNA deregulation in polycythemia vera and essential thrombocythemia patients. Blood Cells Mol. Dis. 2013, 50, 190–195. [Google Scholar] [CrossRef] [Green Version]
- Albano, F.; Anelli, L.; Zagaria, A.; Coccaro, N.; Casieri, P.; Minervini, A.; Specchia, G. SETBP1 and miR-4319 dysregulation in primary myelofibrosis progression to acute myeloid leukemia. J. Hematol. Oncol. 2012, 5, 48. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kota, J.; Caceres, N.; Constantinescu, S.N. Aberrant signal transduction pathways in myeloproliferative neoplasms. Leukemia 2008, 22, 1828–1840. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murakami, Y.; Inoue, N.; Shichishima, T.; Ohta, R.; Noji, H.; Maeda, Y.; Nishimura, J.I.; Kanakura, Y.; Kinoshita, T. Deregulated expression of HMGA2 is implicated in clonal expansion of PIGA deficient cells in paroxysmal nocturnal haemoglobinuria. Br. J. Haematol. 2012, 156, 383–387. [Google Scholar] [CrossRef] [PubMed]
- Serio, B.; Selleri, C.; Maciejewski, J.P. Impact of immunogenetic polymorphisms in bone marrow failure syndromes. Mini Rev. Med. Chem. 2011, 11, 544–552. [Google Scholar] [CrossRef] [PubMed]
- Shichishima, T.; Okamoto, M.; Ikeda, K.; Kaneshige, T.; Sugiyama, H.; Terasawa, T.; Osumi, K.; Maruyama, Y. HLA class II haplotype and quantitation of WT1 RNA in Japanese patients with paroxysmal nocturnal hemoglobinuria. Blood 2002, 100, 22–28. [Google Scholar] [CrossRef]
- Shichishima, T.; Noji, H.; Ikeda, K.; Akutsu, K.; Maruyama, Y. Bone marrow failure: The frequency of HLA class I alleles in Japanese patients with bone marrow failure. Haematologica 2006, 91, 856–857. [Google Scholar]
- Ikeda, K.; Shichishima, T.; Yasukawa, M.; Nakamura-Shichishima, A.; Noji, H.; Akutsu, K.; Osumi, K.; Maruyama, Y. The role of Wilms’ tumor gene peptide-specific cytotoxic T lymphocytes in immunologic selection of a paroxysmal nocturnal hemoglobinuria clone. Exp. Hematol. 2007, 35, 618–626. [Google Scholar] [CrossRef]
- Luzzatto, L.; Bessler, M. The dual pathogenesis of paroxysmal nocturnal hemoglobinuria. Curr. Opin. Hematol. 1996, 3, 101–110. [Google Scholar] [CrossRef]
- Fleischman, A.G.; Aichberger, K.J.; Luty, S.B.; Bumm, T.G.; Petersen, C.L.; Doratotaj, S.; Vasudevan, K.B.; LaTocha, D.H.; Yang, F.; Press, R.D.; et al. TNFα facilitates clonal expansion of JAK2V617F positive cells in myeloproliferative neoplasms. Blood 2011, 118, 6392–6398. [Google Scholar] [CrossRef]
- Verstovsek, S.; Kantarjian, H.; Mesa, R.A.; Pardanani, A.D.; Cortes-Franco, J.; Thomas, D.A.; Estrov, Z.; Fridman, J.S.; Bradley, E.C.; Erickson-Viitanen, S.; et al. Safety and efficacy of INCB018424, a JAK1 and JAK2 inhibitor, in myelofibrosis. N. Engl. J. Med. 2010, 363, 1117–1127. [Google Scholar] [CrossRef] [Green Version]
- Miller, K.D.; Siegel, R.L.; Lin, C.C.; Mariotto, A.B.; Kramer, J.L.; Rowland, J.H.; Stein, K.D.; Alteri, R.; Jemal, A. Cancer treatment and survivorship statistics, 2016. CA. Cancer J. Clin. 2016, 66, 271–289. [Google Scholar] [CrossRef] [Green Version]
- Döhner, H.; Weisdorf, D.J.; Bloomfield, C.D. Acute myeloid leukemia. N. Engl. J. Med. 2015, 373, 1136–1152. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mrózek, K.; Heerema, N.A.; Bloomfield, C.D. Cytogenetics in acute leukemia. Blood Rev. 2004, 18, 115–136. [Google Scholar] [CrossRef]
- Rowley, J.D. Chromosomal translocations: Revisited yet again. Blood 2008, 112, 2183–2189. [Google Scholar] [CrossRef]
- Jin, J.; Yu, M.; Hu, C.; Ye, L.; Xie, L.; Jin, J.; Chen, F.; Tong, H. Pesticide exposure as a risk factor for myelodysplastic syndromes: A meta-analysis based on 1942 cases and 5359 controls. PLoS ONE 2014, 9. [Google Scholar] [CrossRef] [PubMed]
- Döhner, H.; Estey, E.; Grimwade, D.; Amadori, S.; Appelbaum, F.R.; Büchner, T.; Dombret, H.; Ebert, B.L.; Fenaux, P.; Larson, R.A.; et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood 2017, 129, 424–447. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cumbo, C.; Minervini, C.F.; Orsini, P.; Anelli, L.; Zagaria, A.; Minervini, A.; Coccaro, N.; Impera, L.; Tota, G.; Parciante, E.; et al. Nanopore targeted sequencing for rapid gene mutations detection in acute myeloid leukemia. Genes (Basel) 2019, 10, 1026. [Google Scholar] [CrossRef] [Green Version]
- Minervini, C.F.; Cumbo, C.; Orsini, P.; Brunetti, C.; Anelli, L.; Zagaria, A.; Minervini, A.; Casieri, P.; Coccaro, N.; Tota, G.; et al. TP53 gene mutation analysis in chronic lymphocytic leukemia by nanopore MinION sequencing. Diagn. Pathol. 2016, 11, 96. [Google Scholar] [CrossRef] [Green Version]
- Rockova, V.; Abbas, S.; Wouters, B.J.; Erpelinck, C.A.J.; Beverloo, H.B.; Delwel, R.; Van Putten, W.L.J.; Löwenberg, B.; Valk, P.J.M. Risk stratification of intermediate-risk acute myeloid leukemia: Integrative analysis of a multitude of gene mutation and gene expression markers. Blood 2011, 118, 1069–1076. [Google Scholar] [CrossRef] [Green Version]
- Mroźek, K.; Marcucci, G.; Nicolet, D.; Maharry, K.S.; Becker, H.; Whitman, S.P.; Metzeler, K.H.; Schwind, S.; Wu, Y.Z.; Kohlschmidt, J.; et al. Prognostic significance of the European LeukemiaNet standardized system for reporting cytogenetic and molecular alterations in adults with acute myeloid leukemia. J. Clin. Oncol. 2012, 30, 4515–4523. [Google Scholar] [CrossRef]
- Grimwade, D.; Walker, H.; Oliver, F.; Wheatley, K.; Harrison, C.; Harrison, G.; Rees, J.; Hann, I.; Stevens, R.; Burnett, A.; et al. The importance of diagnostic cytogenetics on outcome in AML: Analysis of 1612 patients entered into the MRC AML 10 trial. The Medical Research Council Adult and Children’s Leukaemia Working Parties. Blood 1998, 92, 2322–2333. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marquis, M.; Beaubois, C.; Lavallée, V.P.; Abrahamowicz, M.; Danieli, C.; Lemieux, S.; Ahmad, I.; Wei, A.; Ting, S.B.; Fleming, S.; et al. High expression of HMGA2 independently predicts poor clinical outcomes in acute myeloid leukemia. Blood Cancer J. 2018, 8. [Google Scholar] [CrossRef] [PubMed]
- Nyquist, K.B.; Panagopoulos, I.; Thorsen, J.; Roberto, R.; Wik, H.S.; Tierens, A.; Heim, S.; Micci, F. t(12;13)(q14;q31) leading to HMGA2 upregulation in acute myeloid leukaemia. Br. J. Haematol. 2012, 157, 769–771. [Google Scholar] [CrossRef] [PubMed]
- Tan, L.; Wei, X.; Zheng, L.; Zeng, J.; Liu, H.; Yang, S.; Tan, H. Amplified HMGA2 promotes cell growth by regulating Akt pathway in AML. J. Cancer Res. Clin. Oncol. 2016, 142, 389–399. [Google Scholar] [CrossRef]
- Ma, S.; Yang, L.L.; Niu, T.; Cheng, C.; Zhong, L.; Zheng, M.W.; Xiong, Y.; Li, L.L.; Xiang, R.; Chen, L.J.; et al. SKLB-677, an FLT3 and Wnt/β-catenin signaling inhibitor, displays potent activity in models of FLT3-driven AML. Sci. Rep. 2015, 5. [Google Scholar] [CrossRef]
- Sokolowski, K.M.; Koprowski, S.; Kunnimalaiyaan, S.; Balamurugan, M.; Gamblin, T.C.; Kunnimalaiyaan, M. Potential Molecular Targeted Therapeutics: Role of PI3-K/Akt/mTOR Inhibition in Cancer. Anticancer Agents Med. Chem. 2016, 16, 29–37. [Google Scholar] [CrossRef]
- Li, J.; Volk, A.; Zhang, J.; Cannova, J.; Dai, S.; Hao, C.; Hu, C.; Sun, J.; Xu, Y.; Wei, W.; et al. Sensitizing leukemia stem cells to NF-κB inhibitor treatment in vivo by inactivation of both TNF and IL-1 signaling. Oncotarget 2017, 8, 8420–8435. [Google Scholar]
- Cook, A.M.; Li, L.; Ho, Y.; Lin, A.; Li, L.; Stein, A.; Forman, S.; Perrotti, D.; Jove, R.; Bhatia, R. Role of altered growth factor receptor-mediated JAK2 signaling in growth and maintenance of human acute myeloid leukemia stem cells. Blood 2014, 123, 2826–2837. [Google Scholar] [CrossRef] [Green Version]
- Arcaro, A.; Guerreiro, A. The Phosphoinositide 3-Kinase Pathway in Human Cancer: Genetic Alterations and Therapeutic Implications. Curr. Genomics 2007, 8, 271–306. [Google Scholar] [CrossRef]
- Yuan, T.L.; Cantley, L.C. PI3K pathway alterations in cancer: Variations on a theme. Oncogene 2008, 27, 5497–5510. [Google Scholar] [CrossRef] [Green Version]
- Tan, L.; Xu, H.; Chen, G.; Wei, X.; Yu, B.; Ye, J.; Xu, L.; Tan, H. Silencing of HMGA2 reverses retardance of cell differentiation in human myeloid leukaemia. Br. J. Cancer 2018, 118, 405–415. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, S.; Gu, Y.; Wang, G.; Hu, Q.; Chen, S.; Wang, Y.; Zhao, M. HMGA2 regulates acute myeloid leukemia progression and sensitivity to daunorubicin via Wnt/β-catenin signaling. Int. J. Mol. Med. 2019, 44, 427–436. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zha, L.; Zhang, J.; Tang, W.; Zhang, N.; He, M.; Guo, Y.; Wang, Z. HMGA2 elicits EMT by activating the Wnt/β-catenin pathway in gastric cancer. Dig. Dis. Sci. 2013, 58, 724–733. [Google Scholar] [CrossRef] [PubMed]
- Wend, P.; Runke, S.; Wend, K.; Anchondo, B.; Yesayan, M.; Jardon, M.; Hardie, N.; Loddenkemper, C.; Ulasov, I.; Lesniak, M.S.; et al. WNT10B/β-catenin signalling induces HMGA2 and proliferation in metastatic triple-negative breast cancer. EMBO Mol. Med. 2013, 5, 264–279. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Donnell, M.R.; Abboud, C.N.; Altman, J.; Appelbaum, F.R.; Arber, D.A.; Attar, E.; Borate, U.; Coutre, S.E.; Damon, L.E.; Goorha, S.; et al. NCCN Clinical Practice Guidelines Acute myeloid leukemia. J. Natl. Compr. Canc. Netw. 2012, 10, 984–1021. [Google Scholar] [CrossRef] [Green Version]
- Quiney, C.; Billard, C.; Faussat, A.M.; Salanoubat, C.; Kolb, J.P. Hyperforin inhibits P-gp and BCRP activities in chronic lymphocytic leukaemia cells and myeloid cells. Leuk. Lymphoma 2007, 48, 1587–1599. [Google Scholar] [CrossRef]
- Andrieux, J.; Bilhou-Nabera, C.; Lippert, E.; Le Bousse-Kerdiles, M.C.; Dupriez, B.; Grardel, N.; Pierre-Louis, O.; Desterke, C.; Praloran, V.; Luc Laï, J.; et al. Expression of HMGA2 in PB leukocytes and purified CD34+ cells from controls and patients with Myelofibrosis and myeloid metaplasia. Leuk. Lymphoma 2006, 47, 1956–1959. [Google Scholar] [CrossRef]
- Thorsteinsdottir, U.; Mamo, A.; Kroon, E.; Jerome, L.; Bijl, J.; Lawrence, H.J.; Humphries, K.; Sauvageau, G. Overexpression of the myeloid leukemia-associated Hoxa9 gene in bone marrow cells induces stem cell expansion. Blood 2002, 99, 121–129. [Google Scholar] [CrossRef]
- Faber, J.; Krivtsov, A.V.; Stubbs, M.C.; Wright, R.; Davis, T.N.; Van Den Heuvel-Eibrink, M.; Zwaan, C.M.; Kung, A.L.; Armstrong, S.A. HOXA9 is required for survival in human MLL-rearranged acute leukemias. Blood 2009, 113, 2375–2385. [Google Scholar] [CrossRef] [Green Version]
- Sauvageau, G.; Lansdorp, P.M.; Eaves, C.J.; Hogge, D.E.; Dragowska, W.H.; Reid, D.S.; Largman, C.; Lawrence, H.J.; Humphries, R.K. Differential expression of homeobox genes in functionally distinct CD34+ subpopulations of human bone marrow cells. Proc. Natl. Acad. Sci. USA 1994, 91, 12223–12227. [Google Scholar] [CrossRef] [Green Version]
- Sun, M.; Song, C.-X.; Huang, H.; Frankenberger, C.A.; Sankarasharma, D.; Gomes, S.; Chen, P.; Chen, J.; Chada, K.K.; He, C.; et al. HMGA2/TET1/HOXA9 signaling pathway regulates breast cancer growth and metastasis. Proc. Natl. Acad. Sci. USA 2013, 110, 9920–9925. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Golub, T.R.; Slonim, D.K.; Tamayo, P.; Huard, C.; Gaasenbeek, M.; Mesirov, J.P.; Coller, H.; Loh, M.L.; Downing, J.R.; Caligiuri, M.A.; et al. Molecular classification of cancer: Class discovery and class prediction by gene expression monitoring. Science 1999, 286, 531–537. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thorsteinsdottir, U.; Kroon, E.; Jerome, L.; Blasi, F.; Sauvageau, G. Defining Roles for HOX and MEIS1 Genes in Induction of Acute Myeloid Leukemia. Mol. Cell. Biol. 2001, 21, 224–234. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Emmrich, S.; Katsman-Kuipers, J.E.; Henke, K.; Khatib, M.E.; Jammal, R.; Engeland, F.; Dasci, F.; Zwaan, C.M.; Den Boer, M.L.; Verboon, L.; et al. MiR-9 is a tumor suppressor in pediatric AML with t(8;21). Leukemia 2014, 28, 1022–1032. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Vodyanik, M.A.; Smuga-Otto, K.; Antosiewicz-Bourget, J.; Frane, J.L.; Tian, S.; Nie, J.; Jonsdottir, G.A.; Ruotti, V.; Stewart, R.; et al. Induced pluripotent stem cell lines derived from human somatic cells. Science 2007, 318, 1917–1920. [Google Scholar] [CrossRef] [PubMed]
- Piskounova, E.; Polytarchou, C.; Thornton, J.E.; Lapierre, R.J.; Pothoulakis, C.; Hagan, J.P.; Iliopoulos, D.; Gregory, R.I. Lin28A and Lin28B inhibit let-7 MicroRNA biogenesis by distinct mechanisms. Cell 2011, 147, 1066–1079. [Google Scholar] [CrossRef] [Green Version]
- Heo, I.; Joo, C.; Cho, J.; Ha, M.; Han, J.; Kim, V.N. Lin28 Mediates the Terminal Uridylation of let-7 Precursor MicroRNA. Mol. Cell 2008, 32, 276–284. [Google Scholar] [CrossRef]
- Viswanathan, S.R.; Daley, G.Q.; Gregory, R.I. Selective blockade of microRNA processing by Lin28. Science 2008, 320, 97–100. [Google Scholar] [CrossRef] [Green Version]
- Rybak, A.; Fuchs, H.; Smirnova, L.; Brandt, C.; Pohl, E.E.; Nitsch, R.; Wulczyn, F.G. A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nat. Cell Biol. 2008, 10, 987–993. [Google Scholar] [CrossRef]
- Newman, M.A.; Thomson, J.M.; Hammond, S.M. Lin-28 interaction with the Let-7 precursor loop mediates regulated microRNA processing. RNA 2008, 14, 1539–1549. [Google Scholar] [CrossRef] [Green Version]
- Büssing, I.; Slack, F.J.; Großhans, H. let-7 microRNAs in development, stem cells and cancer. Trends Mol. Med. 2008, 14, 400–409. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Qin, Y.Z.; Yang, S.; Wang, Y.; Chang, Y.J.; Zhao, T.; Jiang, Q.; Huang, X.J. Meis1 is critical to the maintenance of human acute myeloid leukemia cells independent of MLL rearrangements. Ann. Hematol. 2017, 96, 567–574. [Google Scholar] [CrossRef] [PubMed]
- In’t Hout, F.E.M.; van der Reijden, B.A.; Monteferrario, D.; Jansen, J.H.; Huls, G. High expression of transcription factor 4 (TCF4) is an independent adverse prognostic factor in acute myeloid leukemia that could guide treatment decisions. Haematologica 2014, 99, e257–e259. [Google Scholar] [CrossRef] [PubMed]
- Lam, K.; Muselman, A.; Du, R.; Harada, Y.; Scholl, A.G.; Yan, M.; Matsuura, S.; Weng, S.; Harada, H.; Zhang, D.E. Hmga2 is a direct target gene of RUNX1 and regulates expansion of myeloid progenitors in mice. Blood 2014, 124, 2203–2212. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gerritsen, M.; Yi, G.; Tijchon, E.; Kuster, J.; Schuringa, J.J.; Martens, J.H.A.; Vellenga, E. RUNX1 mutations enhance self-renewal and block granulocytic differentiation in human in vitro models and primary AMLs. Blood Adv. 2019, 3, 320–332. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tremblay, G.; Rousseau, B.; Marquis, M.; Beaubois, C.; Sauvageau, G.; Hébert, J. Cost-Effectiveness Analysis of a HMGA2 Prognostic Test for Acute Myeloid Leukemia in a Canadian Setting. Appl. Health Econ. Health Policy 2019, 17, 827–839. [Google Scholar] [CrossRef] [Green Version]
- Hunger, S.P.; Loh, M.L.; Whitlock, J.A.; Winick, N.J.; Carroll, W.L.; Devidas, M.; Raetz, E.A. Children’s oncology Group’s 2013 blueprint for research: Acute lymphoblastic leukemia. Pediatr. Blood Cancer 2013, 60, 957–963. [Google Scholar] [CrossRef] [Green Version]
- Pui, C.H.; Robison, L.L.; Look, A.T. Acute lymphoblastic leukaemia. Lancet 2008, 371, 1030–1043. [Google Scholar] [CrossRef]
- Coccaro, N.; Anelli, L.; Zagaria, A.; Specchia, G.; Albano, F. Next-generation sequencing in acute lymphoblastic Leukemia. Int. J. Mol. Sci. 2019, 20, 2929. [Google Scholar] [CrossRef] [Green Version]
- Van Vlierberghe, P.; Ferrando, A. The molecular basis of T cell acute lymphoblastic leukemia. J. Clin. Investig. 2012, 122, 3398–3406. [Google Scholar] [CrossRef] [Green Version]
- Weng, A.P.; Ferrando, A.A.; Lee, W.; Morris IV, J.P.; Silverman, L.B.; Sanchez-Irizarry, C.; Blacklow, S.C.; Look, A.T.; Aster, J.C. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science 2004, 306, 269–271. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fedele, M.; Fidanza, V.; Battista, S.; Pentimalli, F.; Klein-Szanto, A.J.P.; Visone, R.; De Martino, I.; Curcio, A.; Morisco, C.; Del Vecchio, L.; et al. Haploinsufficiency of the Hmga1 gene causes cardiac hypertrophy and myelo-lymphoproliferative disorders in mice. Cancer Res. 2006, 66, 2536–2543. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xi, Y.; Li, Y.-S.; Tang, H.-B. High mobility group A1 protein acts as a new target of Notch1 signaling and regulates cell proliferation in T leukemia cells. Mol. Cell. Biochem. 2013, 374, 173–180. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, S.; Bartram, C.R.; Seriu, T.; Miller, C.W.; Tobler, A.; Janssen, J.W.; Reiter, A.; Ludwig, W.D.; Zimmermann, M.; Schwaller, J. Analysis of a family of cyclin-dependent kinase inhibitors: P15/MTS2/INK4B, p16/MTS1/INK4A, and p18 genes in acute lymphoblastic leukemia of childhood. Blood 1995, 86, 755–760. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lowe, S.W.; Sherr, C.J. Tumor suppression by Ink4a-Arf: Progress and puzzles. Curr. Opin. Genet. Dev. 2003, 13, 77–83. [Google Scholar] [CrossRef]
- Roy, S.; Di Cello, F.; Kowalski, J.; Hristov, A.C.; Tsai, H.L.; Bhojwani, D.; Meyer, J.A.; Carroll, W.L.; Belton, A.; Resar, L.M.S. HMGA1 overexpression correlates with relapse in childhood B-lineage acute lymphoblastic leukemia. Leuk. Lymphoma 2013, 54, 2565–2567. [Google Scholar] [CrossRef] [Green Version]
- Maia, S.; Haining, W.N.; Ansén, S.; Xia, Z.; Armstrong, S.A.; Seth, N.P.; Ghia, P.; den Boer, M.L.; Pieters, R.; Sallan, S.E.; et al. Gene expression profiling identifies BAX-delta as a novel tumor antigen in acute lymphoblastic leukemia. Cancer Res. 2005, 65, 10050–10058. [Google Scholar] [CrossRef] [Green Version]
- Andersson, A.; Olofsson, T.; Lindgren, D.; Nilsson, B.; Ritz, C.; Edén, P.; Lassen, C.; Råde, J.; Fontes, M.; Mörse, H.; et al. Molecular signatures in childhood acute leukemia and their correlations to expression patterns in normal hematopoietic subpopulations. Proc. Natl. Acad. Sci. USA 2005, 102, 19069–19074. [Google Scholar] [CrossRef] [Green Version]
- Shah, S.N.; Kerr, C.; Cope, L.; Zambidis, E.; Liu, C.; Hillion, J.; Belton, A.; Huso, D.L.; Resar, L.M.S. HMGA1 Reprograms Somatic Cells into Pluripotent Stem Cells by Inducing Stem Cell Transcriptional Networks. PLoS ONE 2012, 7. [Google Scholar] [CrossRef] [Green Version]
- Nagel, S.; Scherr, M.; Kel, A.; Hornischer, K.; Crawford, G.E.; Kaufmann, M.; Meyer, C.; Drexler, H.G.; MacLeod, R.A.F. Activation of TLX3 and NKX2-5 in t(5;14)(q35;q32) T-cell acute lymphoblastic leukemia by remote 3′-BCL11B enhancers and coregulation by PU.1 and HMGA1. Cancer Res. 2007, 67, 1461–1471. [Google Scholar] [CrossRef] [Green Version]
- Hillion, J.; Belton, A.M.; Shah, S.N.; Turkson, J.; Jing, N.; Tweardy, D.J.; Di Cello, F.; Huso, D.L.; Resar, L.M.S. Nanoparticle delivery of inhibitory signal transducer and activator of transcription 3 G-quartet oligonucleotides blocks tumor growth in HMGA1 transgenic model of T-cell leukemia. Leuk. Lymphoma 2014, 55, 1194–1197. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.; Yue, P.; Page, B.D.G.; Li, T.; Zhao, W.; Namanja, A.T.; Paladino, D.; Zhao, J.; Chen, Y.; Gunning, P.T.; et al. Orally bioavailable small-molecule inhibitor of transcription factor Stat3 regresses human breast and lung cancer xenografts. Proc. Natl. Acad. Sci. USA 2012, 109, 9623–9628. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jing, N.; Li, Y.; Xiong, W.; Sha, W.; Jing, L.; Tweardy, D.J. G-quartet oligonucleotides: A new class of signal transducer and activator of transcription 3 inhibitors that suppresses growth of prostate and breast tumors through induction of apoptosis. Cancer Res. 2004, 64, 6603–6609. [Google Scholar] [CrossRef] [Green Version]
- Jing, N.; Zhu, Q.; Yuan, P.; Li, Y.; Mao, L.; Tweardy, D.J. Targeting signal transducer and activator of transcription 3 with G-quartet oligonucleotides: A potential novel therapy for head and neck cancer. Mol. Cancer Ther. 2006, 5, 279–286. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patel, H.S.; Kantarjian, H.M.; Bueso-Ramos, C.E.; Medeiros, L.J.; Haidar, M.A. Frequent deletions at 12q14.3 chromosomal locus in adult acute lymphoblastic leukemia. Genes. Chromosomes Cancer 2005, 42, 87–94. [Google Scholar] [CrossRef]
- Nishi, M.; Eguchi-Ishimae, M.; Wu, Z.; Gao, W.; Iwabuki, H.; Kawakami, S.; Tauchi, H.; Inukai, T.; Sugita, K.; Hamasaki, Y.; et al. Suppression of the let-7b microRNA pathway by DNA hypermethylation in infant acute lymphoblastic leukemia with MLL gene rearrangements. Leukemia 2013, 27, 389–397. [Google Scholar] [CrossRef] [Green Version]
- Nakamura, M.; Sugita, K.; Inukai, T.; Goi, K.; Iijima, K.; Tezuka, T.; Kojika, S.; Shiraishi, K.; Miyamoto, N.; Karakida, N.; et al. P16/MTS1/INK4A gene is frequently inactivated by hypermethylation in childhood acute lymphoblastic leukemia with 11q23 translocation. Leukemia 1999, 13, 884–890. [Google Scholar] [CrossRef] [Green Version]
- Wu, Z.; Eguchi-Ishimae, M.; Yagi, C.; Iwabuki, H.; Gao, W.; Tauchi, H.; Inukai, T.; Sugita, K.; Ishii, E.; Eguchi, M. HMGA2 as a potential molecular target in KMT2A-AFF1-positive infant acute lymphoblastic leukaemia. Br. J. Haematol. 2015, 171, 818–829. [Google Scholar] [CrossRef]
- Miao, Y.; Cui, T.; Leng, F.; Wilson, W.D. Inhibition of high-mobility-group A2 protein binding to DNA by netropsin: A biosensor-surface plasmon resonance assay. Anal. Biochem. 2008, 374, 7–15. [Google Scholar] [CrossRef] [Green Version]
- Fedele, M.; Fusco, A. HMGA and Cancer. Biochim. Biophys. Acta Gene Regul. Mech. 2010, 1799, 48–54. [Google Scholar] [CrossRef]
- Beckerbauer, L.; Tepe, J.J.; Cullison, J.; Reeves, R.; Williams, R.M. FR900482 class of anti-tumor drugs cross-links oncoprotein HMG I/Y to DNA in vivo. Chem. Biol. 2000, 7, 805–812. [Google Scholar] [CrossRef] [Green Version]
- Belton, A.; Xian, L.; Huso, T.; Koo, M.; Luo, L.Z.; Turkson, J.; Page, B.D.G.; Gunning, P.T.; Liu, G.; Huso, D.L.; et al. STAT3 inhibitor has potent antitumor activity in B-lineage acute lymphoblastic leukemia cells overexpressing the high mobility group A1 (HMGA1)–STAT3 pathway. Leuk. Lymphoma 2016, 57, 2681–2684. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Böll, B.; Hansen, H.; Heuck, F.; Reiners, K.; Borchmann, P.; Rothe, A.; Engert, A.; Pogge von Strandmann, E. The fully human anti-CD30 antibody 5F11 activates NF-{kappa}B and sensitizes lymphoma cells to bortezomib-induced apoptosis. Blood 2005, 106, 1839–1842. [Google Scholar]
- Jia, L.; Gopinathan, G.; Sukumar, J.T.; Gribben, J.G. Blocking autophagy prevents bortezomib-induced NF-κB activation by reducing I-κBα degradation in lymphoma cells. PLoS ONE 2012, 7. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.-P.; Ding, D.-Z.; Shi, B.; Zhang, S.-Q.; Gu, L.-L.; Wang, Y.-C.; Cheng, C. Expression of TRIM28 correlates with proliferation and Bortezomib-induced apoptosis in B-cell non-Hodgkin lymphoma. Leuk. Lymphoma 2018, 59, 2639–2649. [Google Scholar] [CrossRef] [PubMed]
- Pongas, G.N.; Annunziata, C.M.; Staudt, L.M. PI3Kδ inhibition causes feedback activation of PI3Kα in the ABC subtype of diffuse large B-cell lymphoma. Oncotarget 2017, 8, 81794–81802. [Google Scholar] [CrossRef] [Green Version]
- Song, J.; Shao, Q.; Li, C.; Liu, H.; Li, J.; Wang, Y.; Song, W.; Li, L.; Wang, G.; Shao, Z.; et al. Effects of microRNA-21 on apoptosis by regulating the expression of PTEN in diffuse large B-cell lymphoma. Medicine (Baltimore) 2017, 96, e7952. [Google Scholar] [CrossRef]
- Coccaro, N.; Anelli, L.; Zagaria, A.; Perrone, T.; Specchia, G.; Albano, F. Molecular Complexity of Diffuse Large B-Cell Lymphoma: Can It Be a Roadmap for Precision Medicine? Cancers (Basel) 2020, 12, 185. [Google Scholar] [CrossRef] [Green Version]
- Wang, R.; Shen, J.; Wang, Q.; Zhang, M. Bortezomib inhibited the progression of diffuse large B-cell lymphoma via targeting miR-198. Biomed. Pharmacother. 2018, 108, 43–49. [Google Scholar] [CrossRef]
- Shen, Y.; Lu, L.; Xu, J.; Meng, W.; Qing, Y.; Liu, Y.; Zhang, B.; Hu, H. Bortezomib induces apoptosis of endometrial cancer cells through microRNA-17-5p by targeting p21. Cell Biol. Int. 2013, 37, 1114–1121. [Google Scholar] [CrossRef]
- Lin, Y.; Chen, H.; Hu, Z.; Mao, Y.; Xu, X.; Zhu, Y.; Xu, X.; Wu, J.; Li, S.; Mao, Q.; et al. miR-26a inhibits proliferation and motility in bladder cancer by targeting HMGA1. FEBS Lett. 2013, 587, 2467–2473. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.; Tao, T.; Liu, C.; Guan, H.; Huang, Y.; Xu, B.; Chen, M. Downregulation of miR-195 promotes prostate cancer progression by targeting HMGA1. Oncol. Rep. 2016, 36, 376–382. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.; Wang, J.; Jia, Y.; Shen, F.; Han, W.; Kang, Y. MiR-142-3p functions as a potential tumor suppressor in human osteosarcoma by targeting HMGA1. Cell. Physiol. Biochem. 2014, 33, 1329–1339. [Google Scholar] [CrossRef] [PubMed]
- Cinkornpumin, J.; Roos, M.; Nguyen, L.; Liu, X.; Gaeta, X.; Lin, S.; Chan, D.N.; Liu, A.; Gregory, R.I.; Jung, M.; et al. A small molecule screen to identify regulators of let-7 targets. Sci. Rep. 2017, 7, 15973. [Google Scholar] [CrossRef]
- Manley, P.W.; Cowan-Jacob, S.W.; Mestan, J. Advances in the structural biology, design and clinical development of Bcr-Abl kinase inhibitors for the treatment of chronic myeloid leukaemia. Biochim. Biophys. Acta 2005, 1754, 3–13. [Google Scholar] [CrossRef]
- O’Brien, S.G.; Guilhot, F.; Larson, R.A.; Gathmann, I.; Baccarani, M.; Cervantes, F.; Cornelissen, J.J.; Fischer, T.; Hochhaus, A.; Hughes, T.; et al. Imatinib compared with interferon and low-dose cytarabine for newly diagnosed chronic-phase chronic myeloid leukemia. N. Engl. J. Med. 2003, 348, 994–1004. [Google Scholar] [CrossRef] [Green Version]
- Baccarani, M.; Deininger, M.W.; Rosti, G.; Hochhaus, A.; Soverini, S.; Apperley, J.F.; Cervantes, F.; Clark, R.E.; Cortes, J.E.; Guilhot, F.; et al. European LeukemiaNet recommendations for the management of chronic myeloid leukemia: 2013. Blood 2013, 122, 872–884. [Google Scholar] [CrossRef]
- Rossi, A.R.; Breccia, M.; Abruzzese, E.; Castagnetti, F.; Luciano, L.; Gozzini, A.; Annunziata, M.; Martino, B.; Stagno, F.; Cavazzini, F.; et al. Outcome of 82 chronic myeloid leukemia patients treated with nilotinib or dasatinib after failure of two prior tyrosine kinase inhibitors. Haematologica 2013, 98, 399–403. [Google Scholar] [CrossRef]
- Ahuja, N.; Sharma, A.R.; Baylin, S.B. Epigenetic Therapeutics: A New Weapon in the War Against Cancer. Annu. Rev. Med. 2016, 67, 73–89. [Google Scholar] [CrossRef] [Green Version]
- Vitkeviciene, A.; Baksiene, S.; Borutinskaite, V.; Navakauskiene, R. Epigallocatechin-3-gallate and BIX-01294 have different impact on epigenetics and senescence modulation in acute and chronic myeloid leukemia cells. Eur. J. Pharmacol. 2018, 838, 32–40. [Google Scholar] [CrossRef]
- Gan, R.-Y.; Li, H.-B.; Sui, Z.-Q.; Corke, H. Absorption, metabolism, anti-cancer effect and molecular targets of epigallocatechin gallate (EGCG): An updated review. Crit. Rev. Food Sci. Nutr. 2018, 58, 924–941. [Google Scholar] [CrossRef]
- Granja, A.; Pinheiro, M.; Reis, S. Epigallocatechin gallate nanodelivery systems for cancer therapy. Nutrients 2016, 8, 307. [Google Scholar] [CrossRef]
- Huang, J.; Dorsey, J.; Chuikov, S.; Pérez-Burgos, L.; Zhang, X.; Jenuwein, T.; Reinberg, D.; Berger, S.L. G9a and Glp methylate lysine 373 in the tumor suppressor p53. J. Biol. Chem. 2010, 285, 9636–9641. [Google Scholar] [CrossRef] [Green Version]
- Kubicek, S.; O’Sullivan, R.J.; August, E.M.; Hickey, E.R.; Zhang, Q.; Teodoro, M.L.; Rea, S.; Mechtler, K.; Kowalski, J.A.; Homon, C.A.; et al. Reversal of H3K9me2 by a small-molecule inhibitor for the G9a histone methyltransferase. Mol. Cell 2007, 25, 473–481. [Google Scholar] [CrossRef]
- Nardella, C.; Clohessy, J.G.; Alimonti, A.; Pandolfi, P.P. Pro-senescence therapy for cancer treatment. Nat. Rev. Cancer 2011, 11, 503–511. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Minervini, A.; Coccaro, N.; Anelli, L.; Zagaria, A.; Specchia, G.; Albano, F. HMGA Proteins in Hematological Malignancies. Cancers 2020, 12, 1456. https://doi.org/10.3390/cancers12061456
Minervini A, Coccaro N, Anelli L, Zagaria A, Specchia G, Albano F. HMGA Proteins in Hematological Malignancies. Cancers. 2020; 12(6):1456. https://doi.org/10.3390/cancers12061456
Chicago/Turabian StyleMinervini, Angela, Nicoletta Coccaro, Luisa Anelli, Antonella Zagaria, Giorgina Specchia, and Francesco Albano. 2020. "HMGA Proteins in Hematological Malignancies" Cancers 12, no. 6: 1456. https://doi.org/10.3390/cancers12061456
APA StyleMinervini, A., Coccaro, N., Anelli, L., Zagaria, A., Specchia, G., & Albano, F. (2020). HMGA Proteins in Hematological Malignancies. Cancers, 12(6), 1456. https://doi.org/10.3390/cancers12061456







