The Recombinant Fragment of Human κ-Casein Induces Cell Death by Targeting the Proteins of Mitochondrial Import in Breast Cancer Cells
Abstract
1. Introduction
2. Results
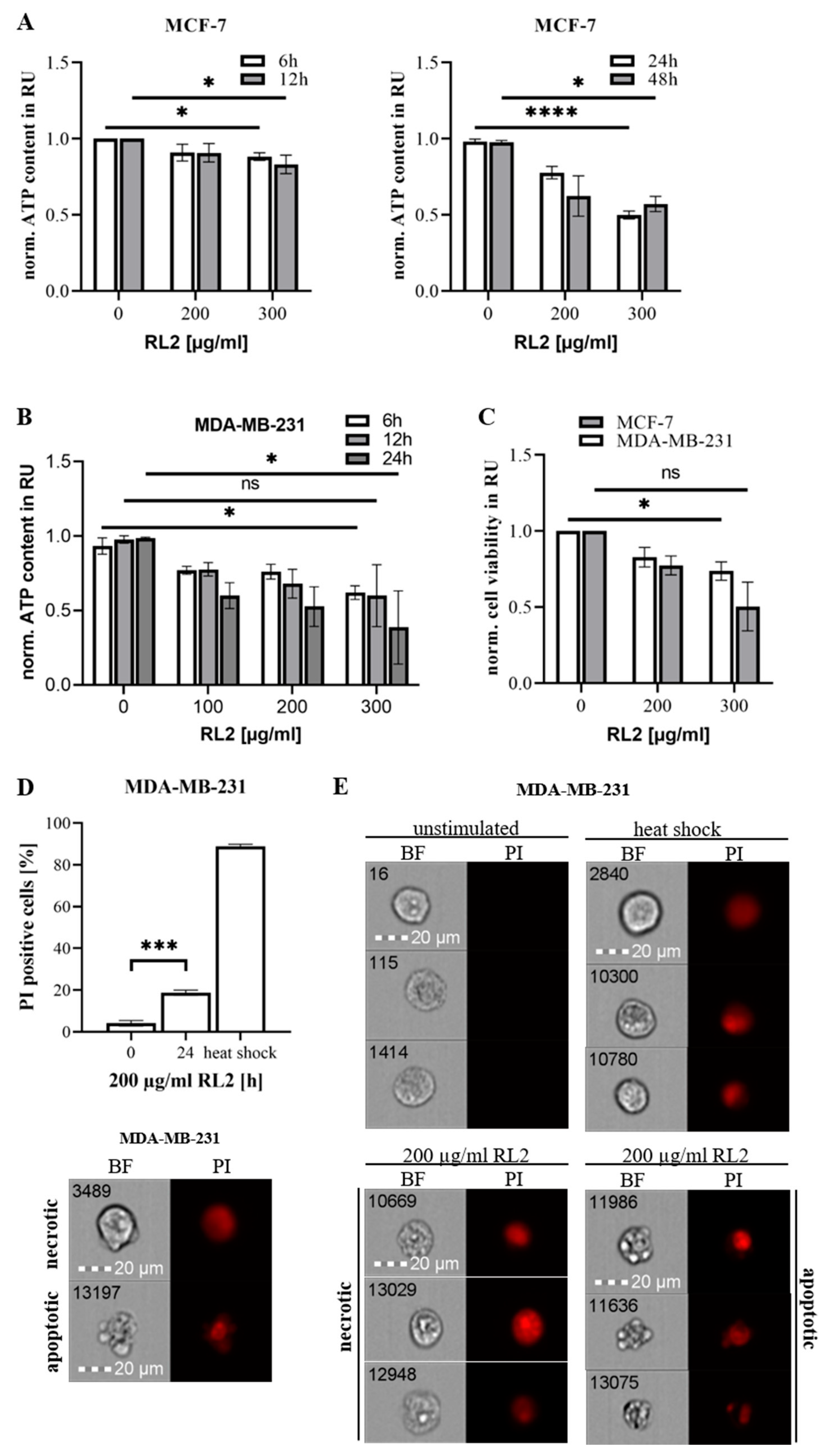
2.1. RL2 Induces ATP Loss and Cell Death in Breast Cancer Cells
2.2. RL2 Induces Caspase Activity in Breast Cancer Cells
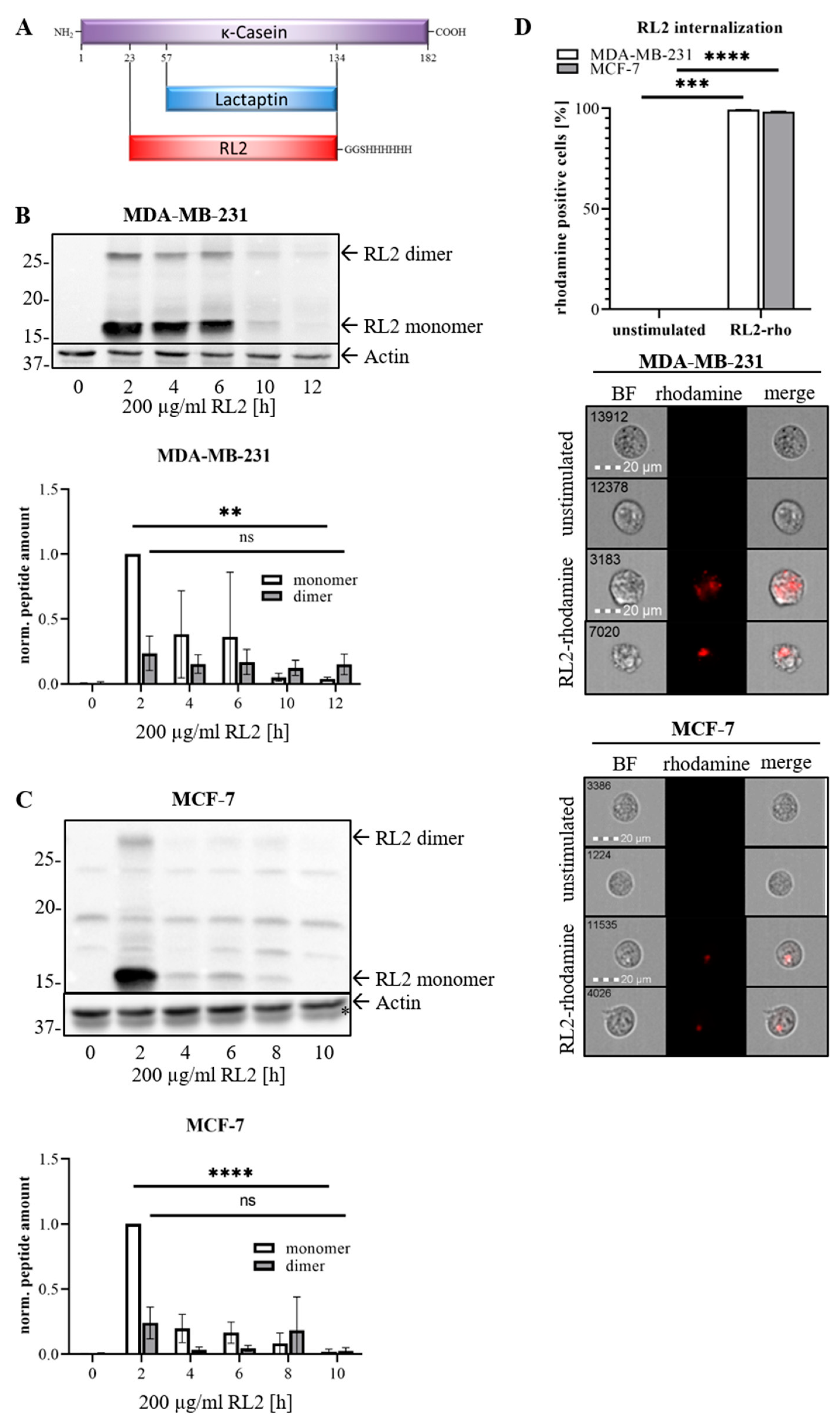
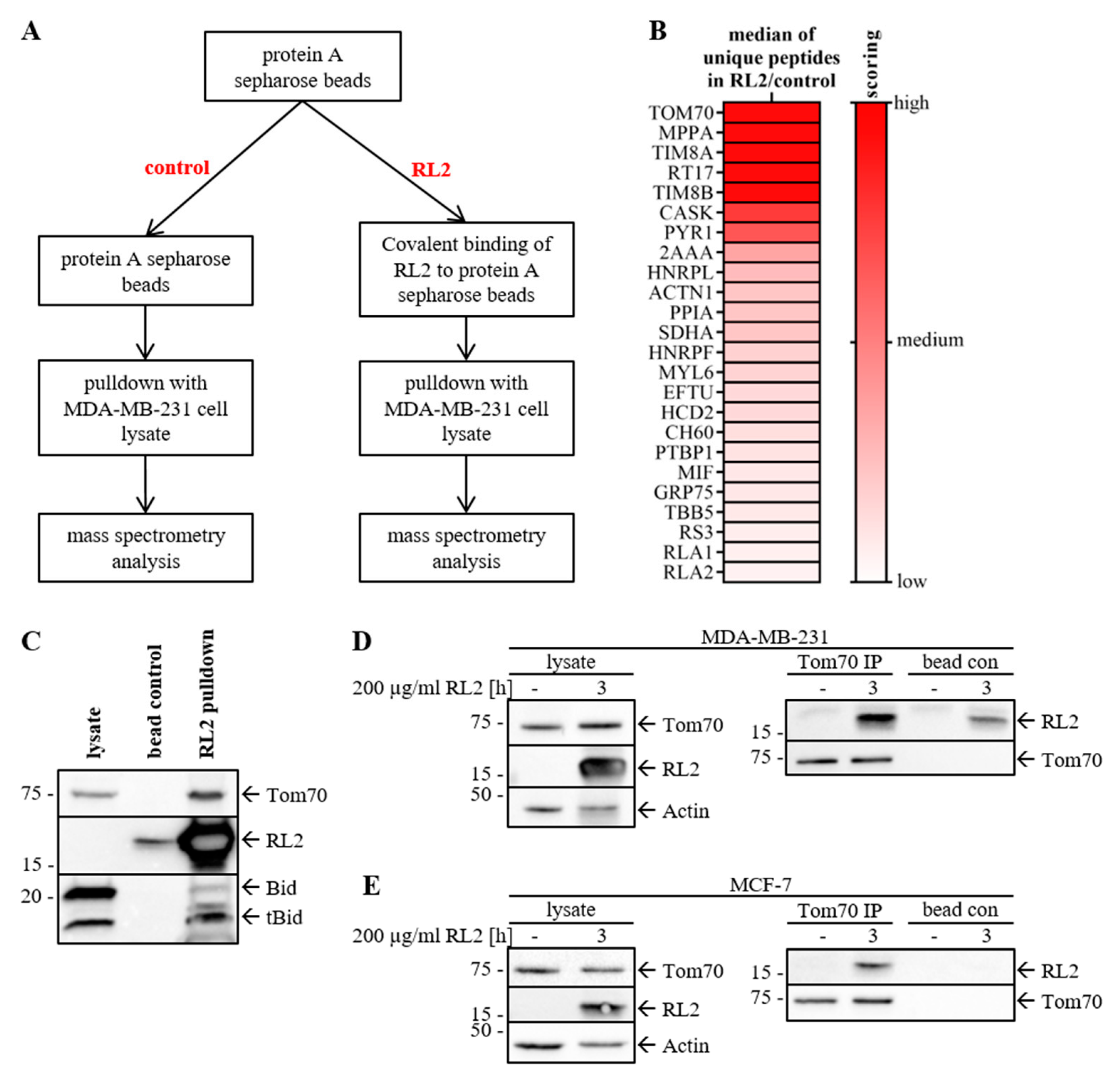
2.3. RL2 Interacts with the Mitochondrial Import Protein TOM70
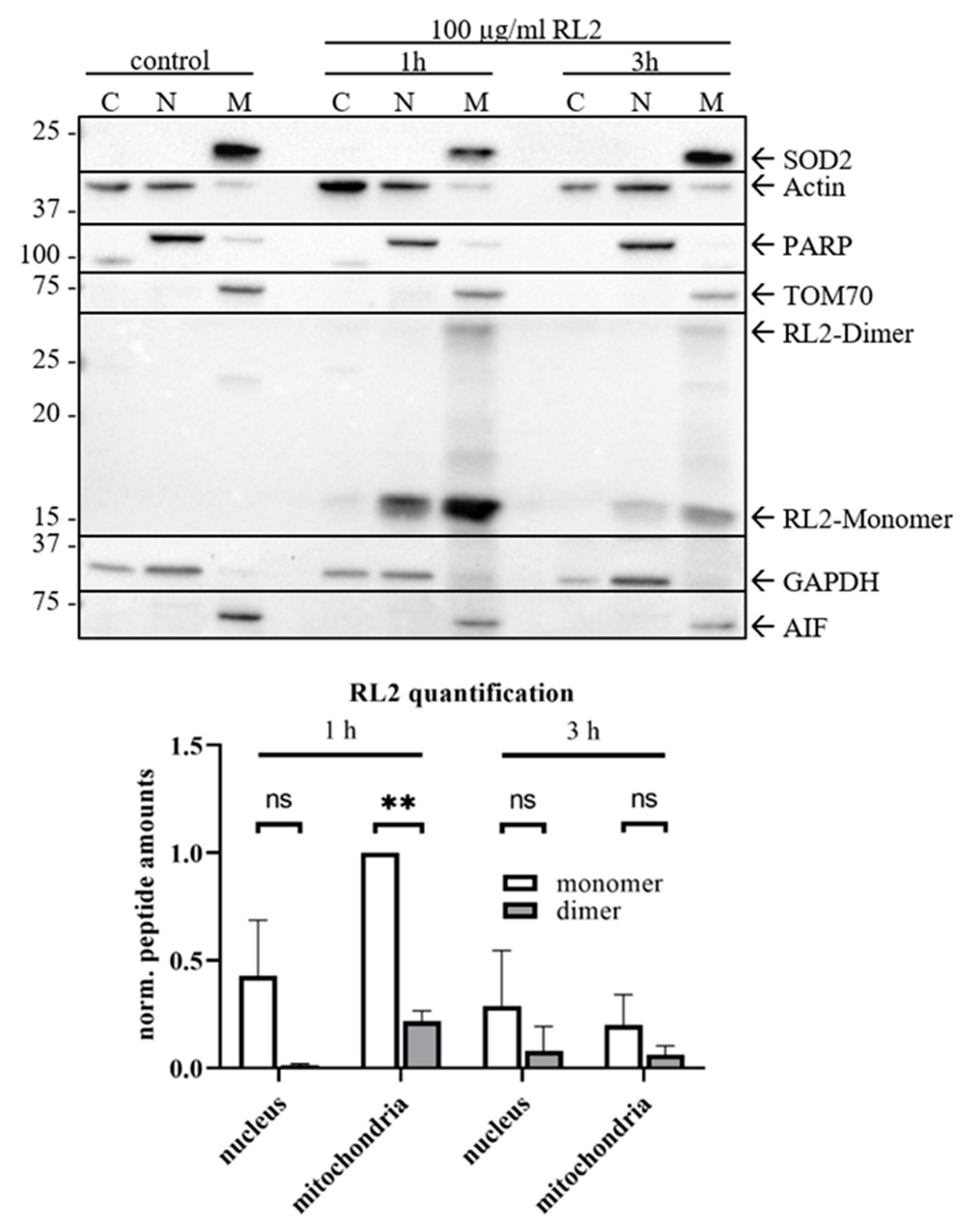
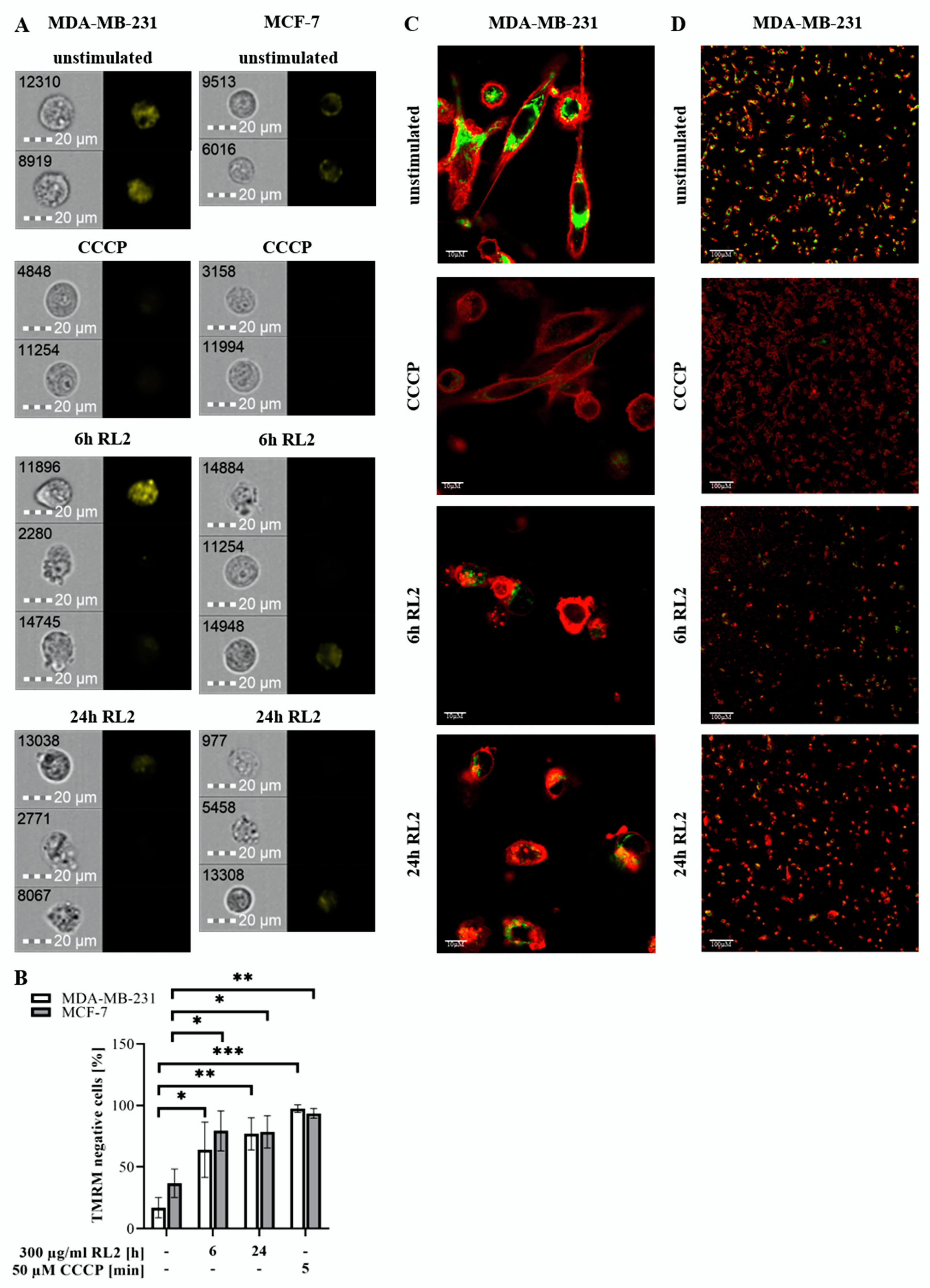
2.4. RL2 Is Targeted to the Mitochondria and Induces Loss of Mitochondrial Membrane Potential
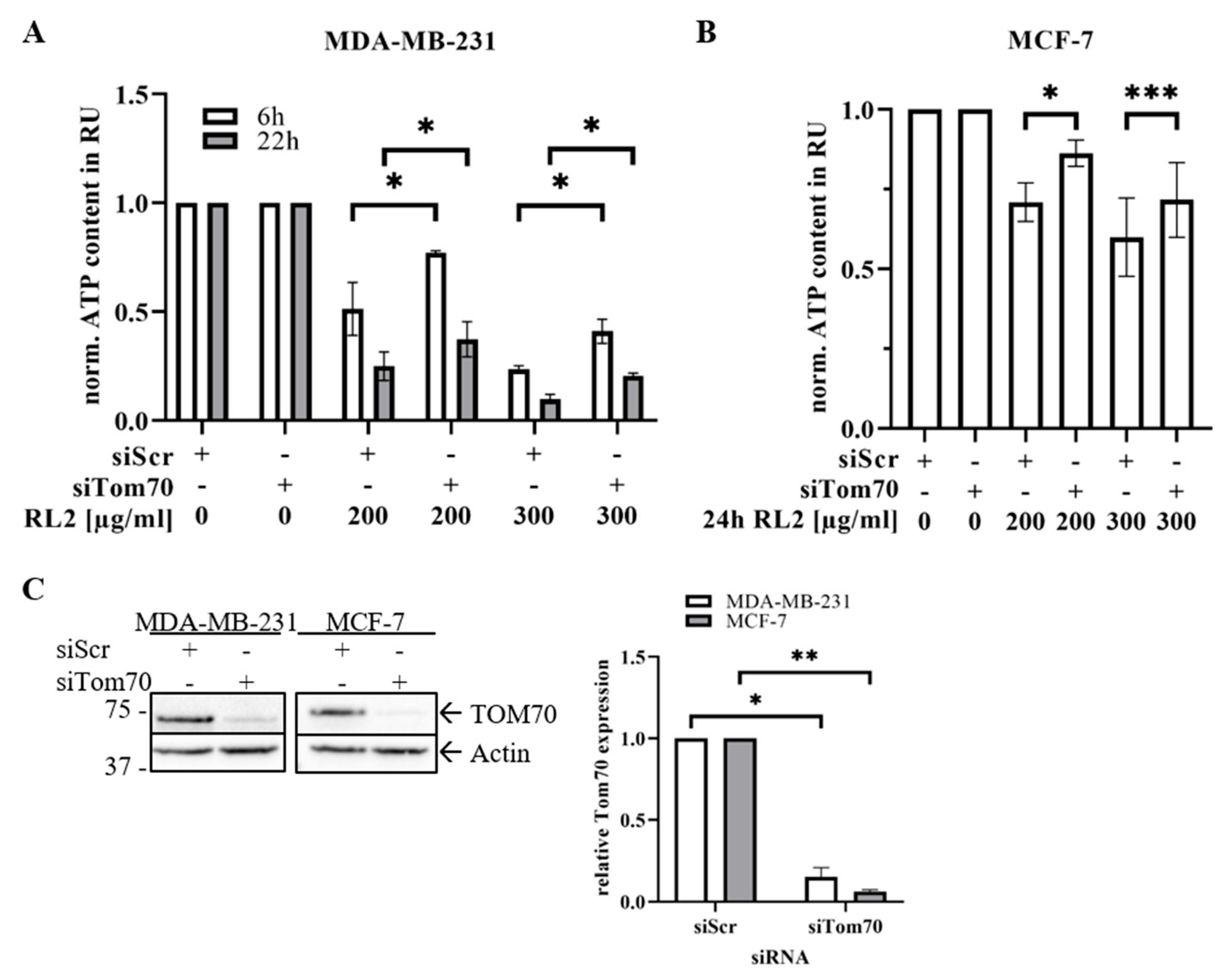
2.5. TOM70 Downregulation Partially Rescues RL2-Induced ATP Loss
3. Discussion
4. Material and Methods
4.1. Antibodies and Reagents
4.2. Cell Culture
4.3. ATP Measurement Assay
4.4. Cell Viability Measurements by Metabolic (MTT) Assay
4.5. Caspase-3/7 Activity Assay
4.6. Cell Death and Internalization Measurements by Imaging Flow Cytometry
4.7. Western Blot Analysis and Immunoprecipitation
4.8. Cellular Fractionation
4.9. Protein Knock-Down
4.10. Protein Pull-Down and Mass Spectrometry Analysis
4.11. Mitochondrial Membrane Potential Analysis by Confocal Laser Scanning Microscopy and Imaging Flow Cytometry
4.12. Statistics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| ACN | Acetonitrile |
| ACTN1 | Actin |
| AIF | apoptosis inducing factor |
| CCCP | Carbonylcyanid-m-chlorphenylhydrazon |
| Cyt c | Cytochrome c |
| DISC | death inducing signaling complex |
| DR | death receptor |
| EndoG | Endonuclease G |
| FA | Formic acid |
| IP | Immunoprecipitation |
| MOMP | mitochondrial outer membrane permeabilization |
| OMM | outer mitochondrial membrane |
| RL2 | recombinant Lactaptin 2 |
| RU | relative units |
| TFA | Trifluoroacetic acid |
| TIM/TOM | translocase of the inner/outer membrane |
| TMRM | Tetramethylrhodamin-methylester |
| TBB5 | Tubulin |
References
- Spector, D.; Deroo, L.A.; Sandler, D.P. Lifestyle behaviors in black and white women with a family history of breast cancer. Prev. Med. 2011, 52, 394–397. [Google Scholar] [CrossRef] [PubMed]
- Lord, S.R.; Cheng, W.-C.; Liu, D.; Gaude, E.; Haider, S.; Metcalf, T.; Patel, N.; Teoh, E.J.; Gleeson, F.; Bradley, K.; et al. Integrated Pharmacodynamic Analysis Identifies Two Metabolic Adaption Pathways to Metformin in Breast Cancer. Cell Metab. 2018, 28, 679–688. [Google Scholar] [CrossRef] [PubMed]
- Greer, Y.E.; Porat-Shliom, N.; Nagashima, K.; Stuelten, C.; Crooks, D.; Koparde, V.N.; Gilbert, S.F.; Islam, C.; Ubaldini, A.; Ji, Y.; et al. ONC201 kills breast cancer cells in vitro by targeting mitochondria. Oncotarget 2018, 9, 18454–18479. [Google Scholar] [CrossRef] [PubMed]
- Ho, J.C.S.; Nadeem, A.; Svanborg, C. HAMLET—A protein-lipid complex with broad tumoricidal activity. Biochem. Biophys. Res. Commun. 2017, 482, 454–458. [Google Scholar] [CrossRef] [PubMed]
- Pereira, C.S.; Guedes, J.P.; Gonçalves, M.; Loureiro, L.; Castro, L.; Gerós, H.; Rodrigues, L.R.; Côrte-Real, M. Lactoferrin selectively triggers apoptosis in highly metastatic breast cancer cells through inhibition of plasmalemmal V-H+-ATPase. Oncotarget 2016, 7, 62144–62158. [Google Scholar] [CrossRef] [PubMed]
- Semenov, D.V.; Fomin, A.S.; Kuligina, E.V.; Koval, O.A.; Matveeva, V.A.; Babkina, I.N.; Tikunova, N.V.; Richter, V.A. Recombinant Analogs of a Novel Milk Pro-Apoptotic Peptide, Lactaptin, and Their Effect on Cultured Human Cells. Protein J. 2010, 29, 174–180. [Google Scholar] [CrossRef] [PubMed]
- Koval, O.A.; Fomin, A.S.; Kaledin, V.I.; Semenov, D.V.; Potapenko, M.O.; Kuligina, E.V.; Nikolinb, V.P.; Nikitenkoc, E.V.; Richter, V.A. A novel pro-apoptotic effector lactaptin inhibits tumor growth in mice models. Biochimie 2012, 94, 2467–2474. [Google Scholar] [CrossRef]
- Koval, O.A.; Tkachenko, A.V.; Fomin, A.S.; Semenov, D.V.; Nushtaeva, A.A.; Kuligina, E.V.; Zavjalov, E.L.; Richter, V.A. Lactaptin induces p53-independent cell death associated with features of apoptosis and autophagy and delays growth of breast cancer cells in mouse xenografts. PLoS ONE 2014, 9, e93921. [Google Scholar] [CrossRef]
- Kaledin, V.I.; Koval, O.A.; Kuligina, E.V.; Lushnikova, E.L.; Nikolin, V.P.; Popova, N.A.; Pyshnaya, I.A.; Richter, V.A. Antimetastatic Effect of Liposomal Recombinant Lactaptin. Bull. Exp. Biol. Med. 2018, 164, 762–765. [Google Scholar] [CrossRef]
- Koval, O.A.; Sakaeva, G.R.; Fomin, A.S.; Nushtaeva, A.A.; Semenov, D.V.; Kuligina, E.V.; Gulyaeva, L.F.; Gerasimov, A.V.; Richter, V.A. Sensitivity of endometrial cancer cells from primary human tumor samples to new potential anticancer peptide lactaptin. J. Cancer Res. Ther. 2015, 11, 345–351. [Google Scholar] [CrossRef]
- Chinak, O.A.; Shernyukov, A.V.; Ovcherenko, S.S.; Sviridov, E.A.; Golyshev, V.M.; Fomin, A.S.; Pyshnaya, I.A.; Kuligina, E.V.; Richter, V.A.; Bagryanskaya, E.G. Structural and aggregation features of a human κ-Casein fragment with antitumor and cell-penetrating properties. Molecules 2019, 24, 2919. [Google Scholar] [CrossRef] [PubMed]
- Chinak, O.; Golubitskaya, E.; Pyshnaya, I.; Stepanov, G.; Zhuravlev, E.; Richter, V.; Koval, O. Nucleic acids delivery into the cells using pro-apoptotic protein lactaptin. Front. Pharmacol. 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Trauth, B.C.; Klas, C.; Peters, A.M.; Matzku, S.; Möller, P.; Falk, W.; Debatin, K.M.; Krammer, P.H. Monoclonal antibody-mediated tumor regression by induction of apoptosis. Science 1989, 245, 301–305. [Google Scholar] [CrossRef] [PubMed]
- Yonehara, S.; Ishii, A.; Yonehara, M. A cell-killing monoclonal antibody (anti-Fas) to a cell surface antigen co-downregulated with the receptor of tumor necrosis factor. J. Exp. Med. 1989, 169, 1747–1756. [Google Scholar] [CrossRef]
- Wiley, S.R.; Schooley, K.; Smolak, P.J.; Din, W.S.; Huang, C.P.; Nicholl, J.K.; Sutherland, G.R.; Smith, T.D.; Rauch, C.; Smith, C.A.; et al. Identification and characterization of a new member of the TNF family that induces apoptosis. Immunity 1995, 3, 673–682. [Google Scholar] [CrossRef]
- Kischkel, F.C.; Hellbardt, S.; Behrmann, I.; Germer, M.; Pawlita, M.; Krammer, P.H.; Peter, M.E. Cytotoxicity-dependent APO-1 (Fas/CD95)-associated proteins form a death-inducing signaling complex (DISC) with the receptor. EMBO J. 1995, 14, 5579–5588. [Google Scholar] [CrossRef]
- Sprick, M.R.; Weigand, M.A.; Rieser, E.; Rauch, C.T.; Juo, P.; Blenis, J.; Krammer, P.H.; Walczak, H. FADD/MORT1 and caspase-8 are recruited to TRAIL receptors 1 and 2 and are essential for apoptosis mediated by TRAIL receptor 2. Immunity 2000, 12, 599–609. [Google Scholar] [CrossRef]
- Chipuk, J.E.; Bouchier-Hayes, L.; Green, D.R. Mitochondrial outer membrane permeabilization during apoptosis: The innocent bystander scenario. Cell Death Differ. 2006, 13, 1396–1402. [Google Scholar] [CrossRef]
- Bender, T.; Martinou, J.-C. Where killers meet–permeabilization of the outer mitochondrial membrane during apoptosis. Cold Spring Harb. Perspect. Biol. 2013, 5, a011106. [Google Scholar] [CrossRef]
- Dave, Z.; Byfield, M.; Bossy-Wetzel, E. Assessing mitochondrial outer membrane permeabilization during apoptosis. Methods 2008, 46, 319–323. [Google Scholar] [CrossRef]
- Korsmeyer, S.J.; Wei, M.C.; Saito, M.; Weiler, S.; Oh, K.J.; Schlesinger, P.H. Pro-apoptotic cascade activates BID, which oligomerizes BAK or BAX into pores that result in the release of cytochrome c. Cell Death Differ. 2000, 7, 1166–1173. [Google Scholar] [CrossRef] [PubMed]
- Wiedemann, N.; Pfanner, N.; Ryan, M.T. The three modules of ADP/ATP carrier cooperate in receptor recruitment and translocation into mitochondria. EMBO J. 2001, 20, 951–960. [Google Scholar] [CrossRef] [PubMed]
- Alberts, B.; Johnson, A.; Lewis, J.; Raff, M.; Roberts, K.; Walter, P. Molecular Biology of the Cell; Garland Science: New York, NY, USA, 2002. [Google Scholar]
- Ahting, U.; Thieffry, M.; Engelhardt, H.; Hegerl, R.; Neupert, W.; Nussberger, S. Tom40, the pore-forming component of the protein-conducting TOM channel in the outer membrane of mitochondria. J. Cell Biol. 2001, 153, 1151–1160. [Google Scholar] [CrossRef] [PubMed]
- Chou, C.-H.; Lee, R.-S.; Yang-Yen, H.-F. An internal EELD domain facilitates mitochondrial targeting of Mcl-1 via a Tom70-dependent pathway. Shaw J., editor. Mol. Biol. Cell 2006, 17, 3952–3963. [Google Scholar] [CrossRef][Green Version]
- Yano, M.; Hoogenraad, N.; Terada, K.; Mori, M. Identification and functional analysis of human Tom22 for protein import into mitochondria. Mol. Cell. Biol. 2000, 20, 7205–7213. [Google Scholar] [CrossRef]
- Baker, M.J.; Frazier, A.E.; Gulbis, J.M.; Ryan, M.T. Mitochondrial protein-import machinery: Correlating structure with function. Trends Cell Biol. 2007, 17, 456–464. [Google Scholar] [CrossRef]
- Chinak, O.A.; Fomin, A.S.; Nushtaeva, A.A.; Koval, O.A.; Savelyeva, A.V.; Kuligina, E.V.; Richter, V. Penetration of the peptide lactaptin into human cancer cells. Russ. J. Bioorg. Chem. 2016, 42, 361–371. [Google Scholar] [CrossRef]
- Kochneva, G.; Sivolobova, G.; Tkacheva, A.; Grazhdantseva, A.; Troitskaya, O.; Nushtaeva, A.; Kuligina, E.; Richter, V.; Koval, O.A. Engineering of double recombinant vaccinia virus with enhanced oncolytic potential for solid tumor virotherapy. Oncotarget 2016, 7, 74171–74188. [Google Scholar] [CrossRef]
- Belovezhets, T.N.; Matvienko, D.A.; Volkova, O.Y.; Koval, O.A.; Tkachenko, A.V.; Kuligina, E.V.; Taranin, A.V.; Richter, V. Analysis of in vitro cytotoxicity of human NK cell line co-expressing a PSMA-specific CAR and an antitumor agent lactaptin. Genes Cells 2018, 3, 89–93. [Google Scholar] [CrossRef]
- Söllner, T.; Pfaller, R.; Griffiths, G.; Pfanner, N.; Neupert, W. A mitochondrial import receptor for the ADP/ATP carrier. Cell 1990, 62, 107–115. [Google Scholar] [CrossRef]
- Ryan, M.T.; Müller, H.; Pfanner, N. Functional staging of ADP/ATP carrier translocation across the outer mitochondrial membrane. J. Biol. Chem. 1999, 274, 20619–20627. [Google Scholar] [CrossRef] [PubMed]
- Filadi, R.; Leal, N.S.; Schreiner, B.; Rossi, A.; Dentoni, G.; Pinho, C.M.; Wiehager, B.; Cieri, D.; Cali, T.; Pizzo, P.; et al. TOM70 Sustains Cell Bioenergetics by Promoting IP3R3-Mediated ER to Mitochondria Ca2+ Transfer. Curr. Biol. 2018, 28, 369–382. [Google Scholar] [CrossRef] [PubMed]
- Tucker, K.; Park, E. Cryo-EM structure of the mitochondrial protein-import channel TOM complex at near-atomic resolution. Nat. Struct. Mol. Biol. 2019, 26, 1158–1166. [Google Scholar] [CrossRef] [PubMed]
- Galluzzi, L.; Vitale, I.; Aaronson, S.A.; Abrams, J.M.; Adam, D.; Agostinis, P.; Alnemri, E.S.; Altucci, L.; Amelio, I.; Andrews, D.W.; et al. Molecular mechanisms of cell death: Recommendations of the Nomenclature Committee on Cell Death 2018. Cell Death Differ. 2018, 25, 486–541. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Outschoorn, U.E.; Peiris-Pagés, M.; Pestell, R.G.; Sotgia, F.; Lisanti, M.P. Cancer metabolism: A therapeutic perspective. In Volume 14, Nature Reviews Clinical Oncology; Nature Publishing Group: New York, NY, USA, 2017; pp. 11–31. [Google Scholar]
- De Francesco, E.M.; Sotgia, F.; Lisanti, M.P. Cancer stem cells (CSCs): Metabolic strategies for their identification and eradication. Biochem. J. 2018, 475, 1611–1634. [Google Scholar] [CrossRef]
- Singan, V.R.; Simpson, J.C. Implementation of the Rank-Weighted Co-localization (RWC) algorithm in multiple image analysis platforms for quantitative analysis of microscopy images. Source Code Biol. Med. 2016, 11, 2. [Google Scholar] [CrossRef]


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Richter, M.; Wohlfromm, F.; Kähne, T.; Bongartz, H.; Seyrek, K.; Kit, Y.; Chinak, O.; Richter, V.A.; Koval, O.A.; Lavrik, I.N. The Recombinant Fragment of Human κ-Casein Induces Cell Death by Targeting the Proteins of Mitochondrial Import in Breast Cancer Cells. Cancers 2020, 12, 1427. https://doi.org/10.3390/cancers12061427
Richter M, Wohlfromm F, Kähne T, Bongartz H, Seyrek K, Kit Y, Chinak O, Richter VA, Koval OA, Lavrik IN. The Recombinant Fragment of Human κ-Casein Induces Cell Death by Targeting the Proteins of Mitochondrial Import in Breast Cancer Cells. Cancers. 2020; 12(6):1427. https://doi.org/10.3390/cancers12061427
Chicago/Turabian StyleRichter, Max, Fabian Wohlfromm, Thilo Kähne, Hannes Bongartz, Kamil Seyrek, Yuriy Kit, Olga Chinak, Vladimir A. Richter, Olga A. Koval, and Inna N. Lavrik. 2020. "The Recombinant Fragment of Human κ-Casein Induces Cell Death by Targeting the Proteins of Mitochondrial Import in Breast Cancer Cells" Cancers 12, no. 6: 1427. https://doi.org/10.3390/cancers12061427
APA StyleRichter, M., Wohlfromm, F., Kähne, T., Bongartz, H., Seyrek, K., Kit, Y., Chinak, O., Richter, V. A., Koval, O. A., & Lavrik, I. N. (2020). The Recombinant Fragment of Human κ-Casein Induces Cell Death by Targeting the Proteins of Mitochondrial Import in Breast Cancer Cells. Cancers, 12(6), 1427. https://doi.org/10.3390/cancers12061427







