Abstract
Secondary resistant mutations in cancer cells arise in response to certain small molecule inhibitors. These mutations inevitably cause recurrence and often progression to a more aggressive form. Resistant mutations may manifest in various forms. For example, some mutations decrease or abrogate the affinity of the drug for the protein. Others restore the function of the enzyme even in the presence of the inhibitor. In some cases, resistance is acquired through activation of a parallel pathway which bypasses the function of the drug targeted pathway. The Catalogue of Somatic Mutations in Cancer (COSMIC) produced a compendium of resistant mutations to small molecule inhibitors reported in the literature. Here, we build on these data and provide a comprehensive review of resistant mutations in cancers. We also discuss mechanistic parallels of resistance.
1. Introduction
Paul Ehrlich who is famous for discovering a treatment for syphilis as well as making impactful contributions to the treatment of other bacterial infections was the first to use the word “chemotherapy” when referring to treatment of diseases with chemicals [1]. However, the use of drugs to treat cancer was pioneered by Sydney Farber during his work on childhood leukemia [2]. Because of Farber’s foundational contribution to the development of various anti-cancer drugs, today we think of chemotherapy as a cancer treatment.
Farber’s drug of choice was aminopterin, a competitive inhibitor of dihydrofolate reductase [3]. The drug blocks nucleotide synthesis which affects both DNA replication and transcription. It targeted the fast dividing leukemia cells in children leading to remissions in about 30% of the patients [4]. Farber and his colleagues were careful not to use the word “cured” when discussing these patients. The remission patients eventually developed resistance to the drug and ultimately succumbed to the disease. One answer was to administer a higher dose of the drug, but Farber was reluctant to do so because he was deeply affected by his patients’ suffering. Emil Freireich and Emil Frei working at the National Cancer Institute (NCI) proposed the use of combination chemotherapy. Using randomized studies, the NCI has been central in the discovery of a plethora of chemotherapeutic drugs, both synthetic and natural plant extracts [5]. In Siddhartha Mukherjee’s book, The Emperor of All Maladies: A biography of Cancer, the historical timeline of these events are outlined [6].
Advances in chemical synthesis combined with a deeper understanding of protein three dimensional structures have allowed identification of druggable targets [7,8]. Importantly, not all factors involved in a pathway can be druggable. For example, in a signal transduction pathway only certain key enzymes (e.g., tyrosine kinases) may be druggable [9]. Inhibiting these key enzymes is sufficient to inactivate the entire pathway. Most modern drug discovery focuses on identifying key targets that are druggable and synthesizing small molecule inhibitors [10,11,12]. This type of treatment with small molecule inhibitors is known as “targeted therapy.” We now use the word “chemotherapy” to refer only to general cell cycle inhibitors.
Cellular adaptation to targeted therapy has been an issue in cancer treatment from the very beginning and is the reason for Farber’s only temporary remissions. One form of adaptation is to compensate for the inhibition of one pathway by modulating other pathways and escape the drug’s inhibitory effects. Combination therapy is, therefore, more efficient in killing the cells because it targets several pathways at once. Another form is to acquire a mutation in the target gene making the protein resistant to the drug. To overcome the resistance, it is often possible to synthesize a second generation, or even third or higher generation drug that is potent even against the resistant mutations.
Unlike cancer driver mutations which generally appear before drug treatment [13,14,15], resistant mutations are secondary mutations that appear in response to adaptation to the drug [16]. Here we used the Catalogue of Somatic Mutations in Cancer (COSMIC) database (cancer.sanger.ac.uk) [17] to retrieve targeted therapy resistant mutations. We map these data and discuss the implication of these resistant mutations. Although some of these mutations have been reviewed before, this is a comprehensive review of all cancers with the aim of pointing out parallels between the various resistant mechanisms.
2. Data Acquisition and Manipulation
The “COSMIC resistance mutations” file was downloaded from the version 90 COSMIC database (https://cancer.sanger.ac.uk/cosmic/download). This file categorizes all the reported mutations and provides the PubMed ID for all queries. The information in this file was used to plot all the mutations in Figure 1, Figure 2, Figure 3, Figure 4, Figure 5 and Figure 6. Lollipop figures were made as previously described [18]. The Lollipops software uses Uniprot and Pfam to gather the protein information. Next to each mutation we indicated the number of times it appears in the file (e.g., X4). In our figures we restrict our diagrams of the targeted pathways to only those relevant to the drugs discussed. All references curated by COSMIC and listed in the file are provided in Supplementary Tables S1–S7. Some references provided in this file were also used in this report. Because the literature on this topic is vast, we could not include most of the references. Although we tried to cite primary research papers when referring to specific mutations, we often had to cite reviews when referring to broader aspects of the field. The goal of this review is to give an overview of the most reported resistance mutations.
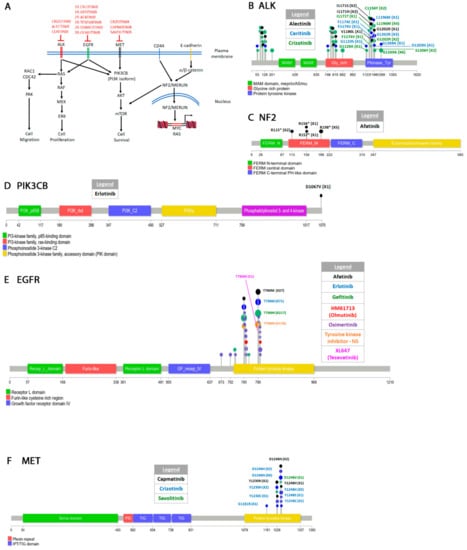
Figure 1.
Signal transduction pathways in non-small cell lung cancers (NSCLC) and drug resistant mutations. (A) Some of the signaling pathways targeted by drugs that are discussed in this paper. Three receptor tyrosine kinases (ALK, EGFR, and MET) acquire resistant mutations to the drugs shown in red. For EGFR 1st, 2nd, and 3rd generation drugs are indicated as 1R, 2R, and 3R respectively. The CD44-cadherin-NF2/Merlin pathway that activates transcription of the MYC and RAS proto-oncogenes is also shown. Activating mutations in the NF2/merlin arise in response to drugs that target ALK, EGFR, and MET. (B–F) Diagrams showing the position of the mutations in several genes shown in (A). Note, that in order to not over-clutter the diagrams only some of the mutations are shown for certain genes. A comprehensive list is found in the supplementary material.
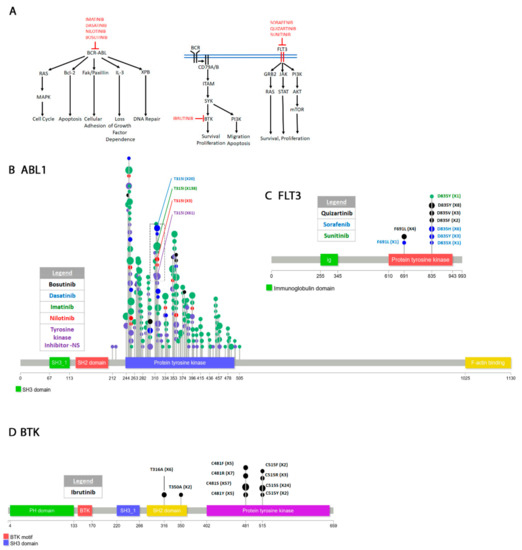
Figure 2.
Pathways targeted in cancers of the hematopoietic and lymphoid tissue and drug resistant mutations. (A) The fused BCR-ABL protein signals through several pathways to activate cell cycle, apoptosis, adhesion, DNA repair and loss of growth factor dependence. The drugs that target the BCR-ABL fusion are shown. Some of these drugs also target the ABL1 wild type protein. Please see text for discussion. The pathways involving BTK and the FLT3 receptor tyrosine kinase are also shown because they are discussed in the text. (B) Mutations in the ABL1 gene cluster in the protein tyrosine domain. Only a few of the mutations are labeled to prevent clutter. Please see Supplementary Figure S1 for all mutations. (C,D) Resistant mutations in FLT3 and BTK.
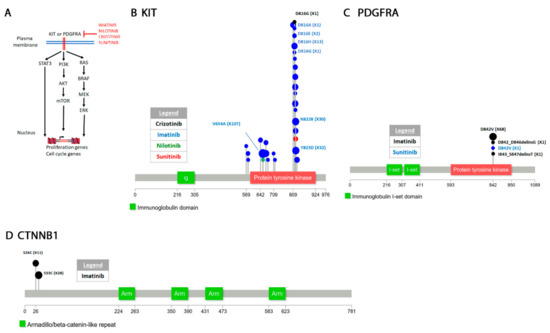
Figure 3.
Signaling and mutations arising in gastrointestinal stromal tumors (GIST) soft tissue. (A) The KIT or PDGFRA receptor tyrosine kinases signal to at least three pathways to activate transcription of proliferation and cell cycle genes. The two receptor tyrosine kinases (RTKs) acquire resistance in response to the drugs shown. (B–D) Diagrams of resistant mutations in some of the genes targeted by the drugs shown in (A).
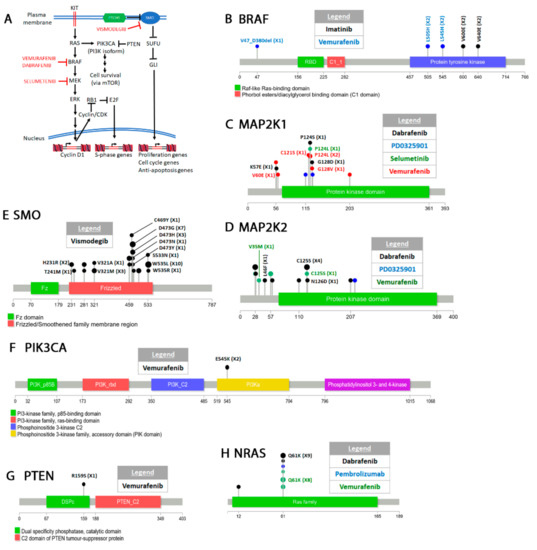
Figure 4.
Drugs that target melanoma pathways. (A) Some of the signal transduction pathways discussed here and the drugs that target them. Please see text for details. (B) Drug resistant point mutations arising in BRAF. Also shown is one structural alteration. Other resistant structural alterations of BRAF are listed in Supplementary Table S4. (C–F) Secondary mutations acquired in the other genes involved in the pathways shown in (A).

Figure 5.
ERalpha (ESR1) in breast cancer. (A) Upon binding to its ligand the estrogen receptor translocates to the nucleus where it interacts with estrogen response elements to activate survival and proliferation genes. Estrogen receptor (ER) phosphorylation also appears to be required for activation. Not discussed here are the roles of the ER receptor in activating cytoplasmic signal transduction pathways through interaction with plasma membrane receptors [496]. (B) Resistant mutations in ESR1 arising in response to endocrine therapy.

Figure 6.
The androgen receptor (AR) signaling and drug resistant mutations. (A) Diagram of the signal transduction pathway for AR signaling. Testosterone is synthesized from a precursor in several epistatic biochemical steps. CYP17A is an enzyme involved in one of these steps. Testosterone crosses the plasma membrane of the target cell and is converted to 5α-dihydrotestosterone (DHT). DHT interacts with the AR receptor and the complex translocates to the nucleus where it binds androgen response elements (ARE) and activates gene transcription. The drugs that inhibit either CYP17A or AR are shown. (B) Diagram of resistant mutations in the androgen receptor. Only the amino acids discussed are shown.
3. Overview of Resistance Mutations
Using the COSMIC database, we retrieved resistance mutations to 37 therapies that target 23 proteins (Table 1). Some drugs are not specified (e.g., Tyrosine kinase inhibitors-NS). The most represented tissues were lung and hematopoietic and lymphoid while adenocarcinoma was the most likely tumor type to develop resistance. Resistance mutations arise most often in protein tyrosine kinase domains. The properties of different small molecule protein kinase inhibitors types have been discussed elsewhere [19,20,21]. We focus this review on secondary resistant mutations that arise in response to the therapy. Thus, we do not discuss all therapeutic drugs but only those that acquire secondary resistant mutations.

Table 1.
Summary of secondary mutations acquired by various genes in response to small molecule inhibitors.
4. Resistant Mutations Appear in Several Cancers in Response to Small Molecule Inhibitors
4.1. Non-Small Cell Lung Cancer
4.1.1. Classification
Lung cancer is classified into small cell lung cancer (SCLC) and non-small cell lung cancer (NSCLC). NSCLC is further sub-classified into adenocarcinoma, large cell carcinoma, and squamous cell carcinoma [23]. Bronchioalveolar (BAC) and acinar carcinoma are two forms of adenocarcinoma. Under the new classification of the Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society (IASLC/ATS/ERS), the term “bronchioalveolar carcinoma (BAC)” has been proposed to be discontinued. BAC is currently re-classified as adenocarcinoma in situ, lepidic predominant adenocarcinoma, and minimally invasive adenocarcinoma [24]. However, since some COSMIC cases precede this classification, we will still use the term bronchioalveolar in this report. Squamous cell carcinoma and adenosquamous carcinoma are the two other forms of lung cancer [25] reported on COSMIC. “Non-small cell carcinoma” refers to non-classified NSCLC or unknown origin.
4.1.2. Epidermal Growth Factor Receptor (EGFR)
Most NSCLC drug therapies target tyrosine kinases in several signal transduction pathways [26,27,28]. Several drugs have been approved for treatment of NSCLC [29,30]. We discuss the ones that affect the pathways presented in Figure 1A because resistance mutations have been reported. The epidermal growth factor receptor (EGFR or ErbB1) is a receptor tyrosine kinase from the ErbB family RTKs. This family also includes ERBB2 or HER2, ERBB3 or HER3, and ERBB4 or HER4 [31]. EGFR signals primarily through the PI3K-AKT-mTOR [32,33] and the RAS-RAF-MEK-ERK [34,35] pathways. Physiologically, the function of EGFR is to promote survival and proliferation. EGFR also activates CDC42 which is involved in cell migration [36,37]. JAK and STAT3 (Signal transducer and activator of transcription 3) are two other pathways activated by EGFR not shown in Figure 1A [31]. Most NSCLCs are characterized by EGFR overexpression [38,39] or activating mutations also known as “common mutations” (e.g., L858R, delL747-P753ins, G719S, several exon 19 deletions and exon 20 insertions) [40,41,42,43]. These mutations are found in the P-loop or activation loop of the kinase which are essential for function and are universally conserved in kinases [44,45,46]. Some of these mutations sensitize the kinase to first generation tyrosine inhibitors erlotinib and gefitinib (see next paragraph).
ERLOTINIB and GEFITINIB are first generation reversible tyrosine kinase inhibitors. Erlotinib (CP-358,774) is an ATP analog that inhibits EGFR tyrosine autophosphorylation [47]. Gefitinib (ZD1839) is an independently synthesized ATP-competitive inhibitor of EGFR [48,49]. When compared to the WT enzyme, the L858R mutation decreases the affinity of enzyme for ATP (higher Km) while simultaneously increasing its affinity for the drugs (lower Ki) [50,51]. Continuous treatment with either erlotinib or gefitinib leads to the acquired EGFR T790M gatekeeper mutation (Figure 1E) [52,53]. The T790M mutation appears to compensate for the loss of ATP affinity caused by the L858R mutation. Double L858R T790M mutants show the same ATP affinity as WT cells [54,55,56]. The T790M resistant mutation occurs even when treating patients with EGFR activating mutations other than L858R. In at least one instance, erlotinib treatment of an EGFR (G719A L861Q) patient resulted in the T790M mutation. The patient also acquired the D1067V mutation in PIK3CB (Figure 1D) which leads to oncogenic transformation of this kinase [57]. The PIK3CB mutation which is in the kinase domain activates the PIK3 pathway in response to EGFR inhibition and leads to cellular transformation both in vitro and in vivo [57,58].
AFATINIB (BIBW2992) is a second-generation inhibitor that targets the entire ErbB receptor family [59,60]. Preclinical studies found that it inhibits the activity of WT enzyme as well as that of L858R single or L858R T790M double mutants [61]. Afatinib is an irreversible inhibitor because it forms a covalent interaction with C797 of EGFR and blocks the enzymatic activity of the kinase [62]. Remarkably, patients treated with afatinib that harbored EGFR common activating mutations also acquired the T790M resistant secondary mutation (Figure 1E, Supplementary Table S1) [63,64]. A clinical study also found that the T790M mutation was resistant to afatinib [65]. Some L858R afatinib treated patients also acquired a C797S mutation. Both the T790M and the C797S mutations rendered cells resistant to the drug [64]. Mapping the C797S mutation on the crystal structure of EGFR shows that it disrupts the ability of the drug to make a covalent bond with the kinase [64]. The L792F was another mutation identified by the same study to arise after treatment with afatinib. The L792F mutation causes steric hindrance decreasing the drug’s ability to interact with the enzyme.
Combination therapy of afatinib and the EGFR antibody cetuximab given to mice harboring the T790M mutation led to activation of the mTOR pathway and acquired resistance [66]. Two mutations were identified in the mTOR modulating protein NF2 (R198*, a truncating mutation and 811-2A > T, a splicing mutant) (Figure 1C). NF2 encodes the protein Merlin which regulates contact inhibition by several mechanisms including neutralizing growth factor receptors [67,68], and promoting cadherin cell attachments [69]. Contact inhibition is primarily mediated through interaction with CD44 [70,71] or E-cadherin [72] (Figure 1A). In mouse embryonic fibroblasts, loss of NF2 function decreases the strength of cadherin mediated adherens junctions and increases confluency [73]. Merlin is also involved in signaling through the Hippo pathway to restrict growth of tissues [74] and the mTOR pathway [75]. Finally, translocation of merlin to the nucleus indirectly modulates expression of several oncogenes including MYC and RAS [76,77]. Some early reports identified biallelic inactivation of NF2 in mesotheliomas but not lung cancers [78,79] and has been singled as a therapeutic target in these cancers [75]. The NF2 resistant mutations identified in afatinib and cetuximab combination therapy patients [66] show that modulating the mTOR pathway is one mechanism to escape EGFR TK inhibition.
TESEVATINIB (XL647) is another second generation inhibitor [80] that targets both EGFR T790M gatekeeper mutation as well vascular endothelial growth factor (VEGFR) [81], the ephrin receptor B2 [82], and human epidermal growth factor receptor 2 (HER-2) [83]. Patients with Exon19 deletion or the L858R mutation responded well to tesevatinib though its efficacy may be increased because it also targets VEGFR which promotes angiogenesis. However, a subsequent study found that patients who had acquired the T790M mutation after erlotinib and gefitinib treatment had only very modest response to tesevatinib and questioned its efficacy in treatment of patients that acquire this resistant mutation [84]. Patients who only harbored the EGFR activating mutations but were not previously treated with any drug (naïve) fared better [85].
OSIMERTINIB (AZD9291) was developed as a third-generation drug by AstraZeneca to treat patients with the T790M mutation resistant to generation I and II drugs [86,87]. Like afatinib, osimertinib is an irreversible inhibitor that covalently interacts with C797. Unlike afatinib, osimertinib is highly specific for EGFR and does not target the other members of the ErbB1 family. Osimertinib works best against the L858R/T790M EGFR double mutant but not as well as afatinib against L858R single mutant, the Exon 19del mutant or WT EGFR. At least one report found that osimertinib may not be efficient for patients that harbor both the T790M and the exon 19 deletion [88]. As expected, resistant mutations in response to osimertinib treatment arise by inactivating cysteine 797 (C797G or C797S) [89,90] (Figure 1E). A G796D mutation has been reported in a Chinese patient to also be resistant to osimertinib [90]. Molecular modeling shows that this mutation causes steric hindrance that prevents interaction of osmiertinib with C797. Two other mutations G796S/R and L792F/H also interfere with the drug’s ability to interact with the kinase domain [91]. Recently a L718Q mutation was reported to render EGFR resistant to osimertinib [92]. This patient, positive for L858R, acquired the T790M mutation after initial treatment with gefitinib and was subsequently started on osimertinib. Within eight months the patient acquired the L718Q mutation. Remarkably, in vitro analysis suggests that the L718Q T790M double mutant re-sensitizes the receptor to afatinib [93]. Molecular modeling shows that the L718Q mutation appears to block osimertinib from interacting covalently with C797 [94]. Finally, a L747P mutation [95] also renders cells resistant to osimertinib while sensitizing cells to afatinib [96,97]. The L747P single mutation appears to be sufficient to confer resistance to erlotinib or gefitinib [98,99,100] if it occurs spontaneously but does not appear to arise in response to gefitinib/erlotinib treatment. Other mechanisms of resistance including mutations or upregulation of other pathways have also been documented [101,102] but we do not discuss them here.
OLMUTINIB (HM61713) is another third-generation drug [103] that selectively inhibits EGFR T790M, L858R and exon 19 deletions but works poorly against WT EGFR [104,105]. In addition, olmutinib also inhibits the function of several ABC (ATP-binding cassette) transporters [106], which are usually overexpressed in cancer cells showing multidrug resistance [107]. A T790M mutant patient treated with olmutinib acquired the C797S mutation suggesting that the mechanism of action of olmutinib is similar to osimertinib [108]. Several other small molecule EGFR inhibitors have also been synthesized and they are reviewed elsewhere [109,110,111,112].
4.1.3. Hepatocyte Growth Factor Receptor (HFGR, c-Met)
The hepatocyte growth factor receptor HGFR encoded by c-Met is a receptor tyrosine kinase activated by several ligands including the hepatocyte growth factor [113,114]. The signal is transduced through PI3K [115], AKT [116], and mTOR which promotes cell survival [117] (Figure 1A). There is also crosstalk between MET and EFGR or ALK (see Section 4.1.4 and Section 4.3.1) that activates the RAS-RAF-MEK-ERK pathway which promotes cell proliferation [118]. Finally, there are several other MET downstream targets [118] including STAT3 [119].
MET amplification and overexpression occurs often in NSCLC EGFR TK-inhibitors treated patients [120,121,122,123,124,125,126,127]. A mutation that causes alternative splicing of exon 14 known as “exon skipping” leads to an in-frame deletion of a MET negative regulatory domain creating a gain of function allele [128,129] Several other activating mutations have also been identified most importantly in the juxtamembrane domain [130].
Small molecule inhibitors of MET have been generated that target the ATP binding pocket [131,132]. These molecules are known as Class I and Class II. Some but not all Class I inhibitors are highly selective for MET while Class II usually also target other kinases [133,134,135]. Class III inhibitors have also been developed that are non-ATP competitive. Tivatinib is such a molecule that appears to stabilize MET in an inhibitory conformation [136,137,138]. Below we discuss a few examples of resistant mutations that arise against Class I inhibitors.
CRIZOTINIB (PF-02341066) synthesized by Pfizer is a non-selective Class I inhibitor that targets ALK, ROS1 and RON or MST1R (Macrophage stimulating protein receptor) in addition to MET [139] (please see Section 4.1.4 for a discussion on ALK resistant mutations). When bound to MET, crizotinib forms interactions with the activation loop blocking the enzyme in an autoinhibitory conformation [140]. Treatment of patients with crizotinib eventually leads to resistant mutations (Figure 1F). Two mutations in the A loop D1246H/N [141,142,143,144,145] and Y1248H/C/S [141,144,145,146] stabilize the active conformation of the A loop and decreases the efficacy of the drug [147]. At least one study identified the G1181R in addition to the Y1248 and D1246 mutations when an exon 14 skipping patient was treated with crizotinib [145]. G1181R also affects the ATP pocket and is presumed to lead to resistance by the same mechanism.
CAPMATINIB (INC280, INCB28060) was synthesized by Incyte Corporation as a reversible ATP competitive inhibitor highly selective for c-MET [148]. It works best against the exon 14 deletion mutant or the R988C mutant but is also highly potent against the non-mutated allele in cells with MET amplification or over-expression. It interacts with MET through the Y1230 residue. A salt bridge between D1246 and Y1248 further stabilizes the inhibitor [149]. Resistance mutations to capmatinib arise that disrupt the interaction of the drug with the kinase. Patients with MET amplification treated with gefitinib and capmatinib developed the D1246N and the Y1248H resistant mutations [144]. Molecular modeling showed that the mutant c-MET was sufficiently distorted to prohibit the binding of the drug.
SAVOLITINIB (volitinib, AZD6094) was synthesized by AstraZeneca as a Type I specific c-MET inhibitor [150] and shown initially to be active against gastric and renal cancers xenograft models [151,152]. The drug was subsequently shown to be active against NSCLC models both in vivo and in vitro where it also targets PI3K and MAPK [153]. Upon treatment with savolitinib, the D1246V/H/N mutation arises and renders cells resistant to the drug [153] (Figure 1E). Even though savolitinib does not form a molecular interaction with D1246, molecular modeling shows that substitution of the charged Asp with uncharged Val, His, or Asn induces a conformation shift in the catalytic A-loop that significantly decreases the affinity of the drug for the enzyme [154,155]. Although we do not discuss Type II or Type III inhibitors in this review, it is worth noting that the D1246V/H/N mutation renders the kinase sensitive to the Type II inhibitors [154]. Savolitinib resistance also arises by over-expression of c-MYC or constitutive expression of mTOR [153].
4.1.4. Anaplastic Lymphoma Kinase (ALK)
ALK is a receptor tyrosine kinase that promotes cell proliferation, survival and migration involved in gut and neuronal development [156,157] (Figure 1A). Although long considered an orphan receptor in vertebrates, ligands have been recently identified in the nervous system [158]. The signal for cell proliferation is transduced through RAS-RAF-MEK-ERK but there is also crosstalk with the PI3K-AKT-mTOR and RAC1/CDC42-PAK pathways [156]. Signal can also be transduced through the JAK3-STAT3 and PLCγ-PIP2-IP3 pathways [159] (not shown in Figure 1). To our knowledge a ligand in lung tissue has not been identified.
ALK disfunction was identified in anaplastic large cell lymphoma as a fusion between the catalytic domain and nucleophosmin (NPM) amino terminus [160]. NPM is a nuclear protein with pleiotropic functions including genome stability and chromatin remodeling [161]. The ALK-NPM fusion appears to allow expression and activity of ALK in lymphatic tissues which contributes to the development of lymphomas. Another fusion between ALK and tropomyosin (TPM) with similar effect as the ALK-NPM fusion has also been described in lymphomas [162]. Fusions between ALK and other proteins also appear to activate ALK in NSCLC [163,164,165,166,167,168]. Most unique to lung cancer is a fusion between ALK and the microtubule associated protein like 4 EML4 [169,170]. Several of these EML4-ALK fusions have been identified in which various EML4 exons are always fused with ALK exons 20–29. All of these fusions lead to ALK gain of function. ALK inhibitors show some efficacy but eventually most acquire resistance [171].
Crizotinib, mentioned above, was one of the first drug used to target NSCLCs characterized by ALK fusions [172]. Crizotinib interacts with ALK similarly to MET through the L1196 gatekeeper residue (L1158 in MET), as well as the G1202 and G1269 residues [173,174]. Other interacting residues are also conserved between MET and ALK but no corresponding tyrosine residue (Y1248 MET) interaction is found in ALK. This loss of interaction appears to be the reason for the higher affinity of the drug for MET than ALK. The drug is effective for about 10 months [172] but eventually resistance develops [175] (Figure 1A). As expected, the most prevalent ALK resistant mutations are L1196M [176,177,178,179] and G1269A [178,179] which disrupt the interaction of the drug with the enzyme and decreases the efficacy of the drug for the EML4-ALK target almost ten-fold [174]. At least two reports have identified the G1202R mutation [177,180] as well as another S1206Y mutation [177,181] which decrease the affinity of the drug for the kinase. Other structural mutations are likely to have the same effect (Supplementary Table S1). Two other mutations in residues somewhat distal from the contact residues (C1156Y and F1174V) [179,180,182] show a lower degree of resistance. A G1128A mutation [183] in EML4-ALK lung cancers does not appear to affect the drug’s affinity for the enzyme but rather induces a conformational change that increases the kinase activity [184]. A I1171T/N mutation has also been identified that shows resistance to both crizotinib and the second generation alectinib drug in NSCLC [185].
ALECTINIB (CH5424802) is a second generation selective ALK inhibitor designed to target secondary resistant mutations resulting from crizotinib treatment [186]. Alectinib is very efficient in inhibiting the activity of EML4-ALK L1196M gatekeeper mutant in NSCLC xenograft models [186,187]. An additional advantage of alectinib is that it can penetrate more tissues including the central nervous system (CNS) which is impenetrable to crizotinib [188]. This makes alectinib appealing for treatment of NSCLC which have a tendency to metastasize to the CNS as well as other MET positive tumors. Resistant mutations to alectinib in NSCLC are primarily in the I1171S/N residue mentioned above [185,189,190,191] which cause fluctuations in the A-loop and destabilize the drug ALK interaction [192]. A V1180L resistant mutant [193] has the same properties as I1171S/N. The G1202R resistant mutation resulting from crizotinib treatment also arises in alectinib treatment [171,190].
CERITINIB (LDK378) is another second generation selective ALK inhibitor produced by Novartis with high activity against the ALK L1196M, I1171T, S1206Y and G1269A crizotinib resistance mutations [181]. Similarly to crizotinib, it interacts with the ATP binding pocket of ALK. It can also penetrate the blood brain barrier [194]. Some resistant mutations that decrease the efficacy of ceritinib are similar to those that arise for crizotinib and alectinib [181]. A resistant mutation unique to ceritinib is the G1123S. This mutation and the F1174C/V mutation which also arises in response to crizotinib treatment [181,195] induce a conformation change in the structure of the P-loop to decrease the affinity of the drug [196]. Another unique D1203N mutation was also shown to be resistant in both cell lines and in vivo models [197] although it is currently not clear how it leads to resistance. Due to its proximity to the G1202 residue it can be speculated that it destabilizes the interaction of the drug with the kinase.
4.2. Hematopoietic and Lymphoid Tissue
4.2.1. Classification
The World Health Organization (WHO) convenes from time to time to classify tumors of the hematopoietic and lymphoid tissues [198,199]. Both morphology and genetics are considered when classifying hematopoietic and lymphoid tumors. Here we briefly summarize the classification of the tumors discussed in this review.
Chronic myeloid leukemias (CMLs) are generally characterized by the presence of BCR-ABL1 fusions [200]. CMLs can progress to more aggressive forms such as acute myeloid leukemia (AML) or acute lymphoblastic leukemia (ALL). AML and CML are characterized by acquisition of additional mutations to the BCR-ABL1 fusion. Chronic lymphocytic leukemia (CLL) usually develops later in life and is characterized by an increase in lymphocytic count [201]. Genetically, this disease is associated with BCR mutations and several chromosomal deletions including 13q14, 11q22-23 (associated with ATM: ataxia telangiectasia mutated, a checkpoint gene), SF3B1 (splicing factor 3b subunit 1), and BIRC3 (baculoviral IAP repeat containing 3) mutations [202,203,204] and 17p13 (associated with TP53 mutations) as well as trisomy 12. Genomic sequencing identified several other altered pathways including NOTCH1, apoptosis, and cell cycle defects [205]. Lymphoplasmacytic lymphoma (LPL) is an elderly rare disorder, a subtype of non-Hodgkin’s lymphoma, genetically characterized by mutations in MYD88 and CXCR4 [206]. Another form of non-Hodgkin’s lymphoma is mantle cell lymphoma (MCL) which overexpresses Cyclin D1 and also shows defects in TP53, NOTCH1/2 PI3K, mTOR and NF-kB in more aggressive forms [207].
4.2.2. Abelson Murine Leukemia Viral Oncogene Homolog 1 (ABL1)
ABL1 is an ubiquitously expressed non-receptor tyrosine kinase but with highly controlled enzymatic activity [208] with several functions including cell differentiation, division and adhesion [209,210,211,212] (Figure 2A).
Interaction between the SH3 domain and the kinase domain forms an inhibitory conformation [213]. SH3 domain mutations serve as activating c-Abl1 mutations in various hematopoietic and lymphoid cancers. However, a hallmark of many leukemias is the presence of the Philadelphia chromosome, a reciprocal translocation between chromosome 22 and 9 [214,215,216,217]. The break in chromosome 22 occurs within the BCR gene at one of three different locations: just after the first exon, around exon 13 and after exon 19. Nearly the entire Abl1 gene is fused to one of these three fragments of BCR creating three different variations (p190Bcr-Abl, exon 18 BCR break; p210Bcr-Abl, exon 13 BCR break; and p230Bcl-Abl, exon 1 break) [218]. WT BCR and ABL1 can interact with each other and it appears that BCR1 negatively regulates ABL1 [219]. Remarkably, the BCR-ABL fusion permanently activates the ABL tyrosine kinase activity [220]. Pathways destabilized by the BCR-ABL fusion include cell cycle regulation through Ras signaling, apoptosis [221,222,223], cell adhesion [224,225], loss of growth factor interaction [226,227], and DNA repair (NER and DSB repair) (Figure 2A) [228,229].
IMATINIB MESYLATE (STI571) is the first-generation drug designed by Novartis Pharma as a specific PDGF-R inhibitor. The subsequent realization that it selectively inhibits both Abl1 and Bcr-Abl kinase activity facilitated FDA approval for CML treatment [230,231]. Imatinib is a strong specific inhibitor of ATP binding to Bcr-Abl [232]. Crystal structure of the Abl kinase domain and imatinib complex reveals that a hydrogen bond between the drug and T315 (the gatekeeper residue) stabilizes the drug in a conformation that blocks ATP interaction with the enzyme active site [233,234,235].
Resistance to imatinib eventually develops [236,237]. The T315I gatekeeper mutation is by far the most commonly occurring mutation that renders cells resistant to Imatinib [238,239] (Figure 2B, Supplementary Figure S1). Substitution of threonine for isoleucine abrogates the hydrogen bond required for Imatinib interaction and drastically reduces the affinity of the drug for the enzyme [240]. The F311I/L and F317L/V also affect the drug affinity, but they are not as drastic because they do not completely abrogate the interaction (Supplementary Figure S1, Supplementary Table S2).
Remarkably several other mutations have also been reported to render resistance to imatinib. In order of incidence reported on COSMIC they are E255K/V > Y253F/H > G250E > M351T > F359A/C/I/V/X > M244V > F317L/V (mentioned above) > E355A/G/X > Q252E/H/K/R > E459G/K/Q, H396P/R. Others arise at much lower frequency (Supplementary Figure S1, Supplementary Table S2). The Y253F/H, E255K/V, G250E, M244V, and Q252E/E/H/K/R detected in the BCR-ABL patients are all found in the P-loop of the kinase [241]. The P-loop is the ATP binding site and these mutations generally reduces the drug’s affinity for the enzyme but does not affect the kinase activity [242,243,244]. It has been demonstrated that the sensitivity of the kinase to imatinib is decreased in patients harboring these mutations by 70%–100% [245]. At least one of the P-loop mutations (Y253F) appears to also act as an activating mutation because it increases the enzyme activity compared to WT [246]. The M351T, F359A/C/I/V/X, and E355A/G/X are in the SH2 contact region (Supplementary Figure S1). The SH2 and SH3 interactions with the kinase domain promote an inhibitory conformation that inactivate the enzyme [247,248,249]. These and other mutations in the regions where the SH2 or SH3 domains contact the kinase domain (Supplementary Figure S1, Supplementary Table S2) affect the autoinhibitory conformation form of the enzyme and decrease the drug’s potency. The H396P/R mutation is in the activation (A)-loop of the enzyme. It appears to stabilize the loop from a closed to an open conformation causing the enzyme to remain activated for longer periods of time [250].
DASATINIB, nilotinib and bosutinib are second generation BCR-ABL inhibitors [251,252,253,254]. Dasatinib (BMS-354825) was designed by Bristol-Myers Squibb and Otskuka Pharmaceutical Co., Ltd. to inhibit imatinib resistant BCR-ABL mutations in CML but it also works against Src family kinases [255,256,257,258]. The crystal structure of the kinase domain + dasatinib complex shows that the drug makes hydrogen bonds with T315 and M318. It efficiently inhibits most imatinib resistant mutants except the T315I gatekeeper mutation. In fact, if patients without the T315I mutation are treated with dasatinib they also acquire this resistant mutation as well as the neighboring F317L/R (Supplementary Figure S1, Supplementary Table S2). A few other mutations most importantly in the P-loop (E255K Y253H and G250E) also eventually confer resistance to the drug. These mutations are similar to the imatinib resistant mutations and confer resistance by a similar mechanism (Supplementary Figure S1, Supplementary Table S2) [259].
NILOTINIB (AMN107) was designed by Novartis Pharmaceuticals to inhibit Abl kinase activity in imatinib-resistant cells [260,261,262,263]. The crystal structure of the Abl-Nilotinib complex shows that although it occupies the same hydrophobic pocket as imatinib, nilotinib fits better in this pocket and therefore has higher affinity for the enzyme making it a more potent inhibitor [263]. However, the drug is not very efficient against P-loop mutations (Y253F/H, E255K/V) that affect the inhibitory conformation of the enzyme or the T315I gatekeeper mutation with which it makes a hydrogen bond similarly to imatinib and dasatinib. In fact, nilotinib resistant secondary mutations arise in the same amino acids (P-loop, T315, SH2 contact site) as for imatinib and dasatinib [264] (Supplementary Figure S1, Supplementary Table S2)
An advantage of nilotinib is that it also inhibits KIT, both WT and the D816G/V mutant resistant to crizotinib (see Section 4.3.1), making it useful for treatment of other c-Kit driven tumors [263,265,266]. Thus, nilotinib is used for treatment of advanced gastro-intestinal tumors (GISTs) [267]. A resistant KIT N655T mutation was identified in a treated GIST patient [268]. This mutation also appears to arise spontaneously [269].
BOSUTINIB (SKI-606) was subsequently produced by Wyeth (now Pfizer Inc., NY, USA) as a Bcr-Abl ATP competitive inhibitor with some activity against Src [270,271]. Bosutinib can inhibit BCR-ABL at nanomolar range and is less toxic than imatinib, nilotinib and dasatinib [272,273,274,275,276] making it a more desirable drug. The drug’s therapeutic potential is also enhanced by its dual inhibition of Src and Abl [277]. The crystal structure of bosutinib complexed with the enzyme shows that its interaction is similar to that of dasatinib with the exception that it only forms a hydrogen bond with M318 and only Van der Waals interactions with T315 [278]. In addition, bosutinib also does not interact with the P-loop making it good for treating P-loop resistant mutations compared to the other drugs. Further, unlike nilotinib and dasatinib, bosutinib can inhibit all but the T315I and V299L imatinib resistant mutants [271,277,279]. Even though the drug forms no interaction with the T315 and V299 residues, the T315I and V299L substitutions causes sufficient steric hindrance that drastically decreases the affinity of the drug for the enzyme [280]. An L248R mutation in the P-loop which was previously shown in vitro to cause imatinib resistance [281] was clinically isolated from a F359I imatinib resistant patient subsequently treated with bosutinib [282]. In vitro analysis showed that although the L248R single mutation confers only moderate resistance than the L248R F359I double mutant it is highly resistant to bosutinib as well as the other drugs. Because bosutinib does not interact with the P-loop, the L248R resistant mutation is surprising and to our knowledge no in silico modeling with this mutation has been performed. Except for the Y253F mutation, most P-loop mutations decrease the affinity of the drug for the enzyme. Mutations in the F359 residue alter the inhibitory conformation of the enzyme. Therefore, it is possible that the L248R F359I double mutant may cause resistance through a combination of decreased drug affinity and increase enzymatic activity.
4.2.3. Bruton’s Tyrosine Kinase (BTK)
The cytoplasmic Bruton’s tyrosine kinase (BTK) is a member of a complex B-cell signal transduction relay required for survival, proliferation, migration and apoptosis of B-cells [283,284]. Interaction of an antigen with the B-cell receptor (BCR) activates a cascade of signal transduction pathways (only a simplified form shown in Figure 2A) that eventually leads to activation of BTK [285,286,287,288]. BTK phosphorylation at Y551 in response to BCR signaling significantly increases the kinase activity of the enzyme [289,290,291,292]. BTK may also autoactivate by self-phosphorylation of Tyr223 in the SH3 domain which appears to increase its affinity for downstream targets [289,293,294,295,296,297]. Activated BTK promotes B-cell maturation.
Mutations in BTK that affect its enzymatic activity have initially been identified in inherited X-linked agammaglobulinemia (XLA) [298]. XLA patients have a deficiency in B-cell maturation resulting in very low plasma B-cell counts [299]. Constitutive BTK activation is essential for proliferation of abnormal B-cells in several types of blood malignancies including chronic lymphocytic leukemia (CLL) [300,301], mantle cell lymphoma [302,303], and lymphoplasmacytic lymphoma [304]. Remarkably, activating oncogenic BTK mutations have not been clinically identified. The kinase appears to be activated by other mutations in the components of the BCR signaling pathway. Mutations in ITAM, CD79A, CD79B [305] as well as other members of the BCR signal transduction pathway [283] have been reported in various cancers. Nevertheless, inhibition of BTK or other components of the BCR signaling pathway drastically reduces B-cell proliferation in vitro [305] making BTK an attractive druggable target.
IBRUTINIB (PCI-32765) was the first generation targeted BTK inhibitor with subsequent development of others (Acalabrutinib, Tirabrutinib, Zanabrutinib, Spebrutinib) [306]. A team at Celera Genomics synthesized the irreversible inhibitor ibrutinib in 2006 [307]. The inhibitor forms a covalent bond with C481 and inhibits the kinase activity of BTK and autophosphorylation at Y223 [308,309,310].
Resistance to ibrutinib develops almost always by amino acid substitution of C481S which abrogates the covalent bond of the inhibitor with the kinase (Figure 2D). This mutation restores the BTK Y233 autophosphorylation and the kinase activity for downstream targets [311,312,313]. Remarkably, ibrutinib may still inhibit the C481S mutant form but only reversibly and about 100 times less efficiently. The Abl inhibitor, dasatinib which is a less effective WT BTK inhibitor than ibrutinib inhibits both the mutant and non-mutant form with the same effectiveness [312]. C481F/R/Y substitutions have also been reported (Figure 2D, Supplementary Table S2). A clinically identified T316A resistant mutation in the SH2 domain has the same effect as the C481S mutation [314]. Molecular modeling shows that this mutation does not affect ibrutinib interaction with the enzyme. Nevertheless, both Y223 autophosphorylation and activation of downstream targets were comparably decreased in C481S and T316A.
4.2.4. FMS-like Tyrosine Kinase 3 (FLT3)
FLT3 is a receptor tyrosine kinase that appears to be expressed mainly in hematopoietic cells [315,316]. It is a Type III receptor tyrosine kinase (RTK) that has a distinct activation mechanism [317]. Interaction of the juxtamembrane domain with the kinase domain leads to an inactive conformation state [318]. Upon ligand binding, a conformation shift occurs that cross-phosphorylates and activates the kinase. This activation mechanism is unlike other RTKs where ligand binding dimerizes the receptor and causes cross-phosphorylation of activation loop [319]. Consequently, FLT3 mutations or alterations of the juxtamembrane domain constitutively activate the receptor [320]. One such activating change identified in acute myeloid leukemia is a tandem duplication of the juxtamembrane domain that affects the inhibitory conformation of the enzyme and leads to constitutive activation [321,322,323,324,325]. Further, in vitro analysis has shown that shortening of the juxtamembrane domain also leads to constitutive activation of the enzyme [320]. Less frequently, two other activation mutations in D835 and I836 have been clinically identified in AML patients [326]. These mutations are in the activation loop of the enzyme and are able to induce an active conformation change [327,328]. Physiologically, the function of FLT3 is to promote survival and proliferation which is accomplished by activating several parallel pathways (Figure 2A) [329]. Several anti-FLT3 drugs have been developed for the treatment of AML patients [329,330]. Here we only discuss resistant mutations to sorafenib, quizartinib, and sunitinib in AML patients.
SORAFENIB (BAY 43-9006) was initially identified through a collaboration of Bayer and Onyx Pharmaceuticals in a screen for Raf1 inhibitors. Subsequent structural analysis allowed minor modifications of the chemical compound to increase its Raf1 inhibitory potency [331,332,333,334]. In addition to Raf1, sorafenib can also inhibit b-Raf, BEGFR1/2/3, PDGFR, FGFR1, c-kit, FLT3, and RET making it a broad-spectrum drug [335,336]. Following clinical trials, the drug was approved to treat various cancers including AML [337]. The most often detected resistant mutation to sorafenib is in the D835 residue mentioned above which is often substituted for other residues including H, Y, and X [338,339] (Figure 2C, Supplementary Table S2). Molecular modeling shows that D835 which is in the activation loop is crucial for facilitating docking of the drug with the enzyme [327,340]. Any mutation in this amino acid severely affects the ability of the drug to inhibit FLT3. An F691L gatekeeper mutation has also been identified in sorafenib treated patients (Figure 2C, Supplementary Table S2) [339]. Molecular modeling shows that sorafenib makes contact with this residue and the F691L change prohibits this contact [339].
QUIZARTINIB (AC220) was identified in a screen for specific FLT3 inhibitors [341,342]. The drug has high selectivity for FLT3 with minor effects on other kinases and has been used for treatment of AML [343,344]. Crystal structure of the FLT3 catalytic domain bound to the drug shows that similarly to sorafenib it makes interactions with F691 but not D835 [345]. Predictably, the F691L resistant mutation affects the drug’s ability to interact with the enzyme while the D835 mutations cause a hyperactive kinase form [327,339,346,347].
SUNITINIB (SU011248) is another drug that targets several receptor tyrosine kinases including KIT and FLT3 [348]. It was initially approved for treatment of gastrointestinal tumors [349]. Unlike quizartinib and sorafenib, sunitinib does not make interactions with F691 [339] and consequently F691L resistant mutations have not been identified. However, a D835Y mutation affects the ability of the drug to interact with the enzyme similarly to sorafenib and quizartinib.
For a more in depth reading of skin cancer resistance to targeted therapies there are several recent reviews [350,351].
4.3. Gastrointestinal Stromal Tumors (GISTs)
Gastrointestinal stromal tumors (GISTs) occur in the cells of Cajal which coordinate smooth muscle contractions in the gastrointestinal tract [352,353,354,355]. GISTs show c-KIT and PDGFRA gain of function, two receptor tyrosine kinases that promote the cell cycle and cell proliferation (Figure 3A) [356,357].
4.3.1. Tyrosine-Protein Kinase KIT
KIT is a receptor tyrosine kinase which appears to function primarily in stem cells [358]. It has also been characterized as a dependence receptor which can function in the absence of a ligand to trigger apoptosis [359]. KIT signals through the PI3K-AKT-mTOR pathway to promote anti-apoptosis and through the RAS-RAF-MEK-MAPK pathway to activate proliferation (Figure 3A) [360,361]. Activating c-KIT oncogenic mutations have been first identified in gastrointestinal and leukemia cancers [362,363,364] as well as a variety of other hematopoietic cancers [365,366]. Exon 11 encodes the autoinhibitory juxtamembrane domain where many KIT mutations are found [360,367]. Another common D816V mutation occurs in the activation loop [368,369]. Caspase dependent cleavage of c-KIT at D816 is required for its pro-apoptotic activity and the D816V prevents cleavage [359].
4.3.2. Platelet Derived Growth Factor Receptor Alpha (PDGFRA)
PDGFRA is a receptor tyrosine kinase that signals through the same pathways as KIT (Figure 3A) [370]. About 15% of GISTs patients have activating mutations in PGFRA and not KIT. PDGFRA mutations occur in the juxtamembrane domain but also in the activation loop and ATP binding domain [357]. The D842V mutation is the most common which occurs in one of the tyrosine kinase domains [371].
Imatinib, nilotininb and sunitinib are all used for treatment of GISTs. Imatinib is able to inhibit KIT [372] and PDGFR [373] receptors. KIT D816 mutations (D816A/E/G/H) also appear as secondary mutations in response to the drug (Figure 3B, Supplementary Table S3). The two other most reported KIT resistant mutation to imatinib are V654A and Y823D. Molecular modeling shows that these mutations decrease the affinity of the drug for KIT [374]. In PDGFRA, the D842V substitution occurs most often (Figure 3C, Supplementary Table S3) [375]. Structural analysis shows that the D842V is an activating mutation that increases PDGFRA affinity for ATP [376]. A melanoma patient treated with imatinib developed both a L576P KIT mutation and a CTNNB1 S33C mutation (Figure 3D) [377]. Modeling simulations show that the L576P KIT mutation which also appears in GIST [378] is an activating mutation [379]. CTNNB1 encodes beta-catenin which is part of the Wnt signaling pathway that has diverse functions including cell proliferation and migration [380,381]. The S33C mutation knocks out a phosphorylation site and appears to stabilize the protein [382,383].
A D816G KIT mutation has also been recently identified in a NSCLC patient treated with crizotinib [384]. This patient also harbored a ROS1 chromosomal re-arrangement. The ROS1 protooncogene is a receptor tyrosine kinase similar to ALK [385,386] and re-arrangements of ROS1 have been described as “driver mutations” in lung cancers [387,388,389,390,391]. Tyrosine Kinase inhibitors including crizotinib and ceritinib have been used for treatment of lung cancers harboring ROS1 re-arrangements [392,393,394]. Treatment of this ROS1 re-arranged patient with crizotinib for 15 months led to acquired resistance to the drug. Subsequent sequencing revealed that the KIT D816G mutation causes the resistant phenotype. In vitro assays showed that this mutation induces KIT autophosphorylation and activation. These data suggest that activation of the KIT pathway bypasses the inhibition of the ROS pathway.
4.4. Melanoma
Classification
Melanomas are malignant melanocytic neoplasms. Although malignancies can occur in any tissue where melanocytes are found, skin melanomas are the most prevalent in sun exposed white people [395,396]. Skin melanomas are generally classified as chronically sun damaged (CSD) and non-chronically sun damaged (non-CSD) [397]. CSD melanomas are characterized by high mutation burden that leads to disruption of several pathways [396]. The serine/threonine kinase BRAFV600E mutation that disrupts the KIT-NRAS-BRAF-MEK-ERK-CDK4/6-RB pathway (Figure 4) is almost always present in both CSD and non-CSD melanomas [398]. The mutation also appears in NSCLC [399]. The CSD melanomas often have additional KIT, NRAS and NF1 mutations [400]. The BRAFV600E is a driver mutation in melanomas because it constitutively activates BRAF and the ERK pathway [401,402].
The KIT receptor tyrosine kinase described above is also mutated in melanoma [400,403]. Shain and colleagues [404] sequenced 293 relevant genes in melanoma tissues and delineated the progression of mutations in these malignancies. They showed that BRAFV600E is almost always the first mutation to occur followed by NRAS and TERT (telomerase). Homozygous CDKN2A, PTEN and TP53 deletions are associated with invasive and metastatic melanoma. Mutations in the neurofibromatosis factor NF1 are also associated with invasive and metastatic melanomas [404,405]. One study showed that NF1 mutations suppress BRAF targeted therapy making BRAF inhibitors less efficient against BRAF and NF1 double mutants [406]. The therapeutic agents used for melanoma treatment have been reviewed recently [407]. Below we discuss resistant mutations that arise in response to some of these agents.
VEMURAFENIB (PLX4032) is a BRAFV600E inhibitor discovered by researchers at Plexxikon [408,409]. Drug synthesis was informed through a crystallography and molecular modeling approach. Because previous BRAF drugs were not very specific [410,411], the investigators wanted a compound that targeted BRAFV600E but had little activity against WT BRAF or other kinases. This method allowed synthesis of highly selective compounds including PLX4720, another BRAFV600E not discussed here [408]. Vemurafenib forms a hydrogen bond with Asp594. A salt bridge between E600 and L507 prevents the activation loop from interacting with the ATP binding site, thus inhibiting enzyme activation. Preclinical trials showed that vemurafenib is a very effective inhibitor [409,412]. Subsequent clinical trials showed that vemurafenib induces efficient tumor regression in BRAFV600E melanoma patients as well as other cancers [413,414,415,416,417]. Vemurafenib and the related drug dabrafenib activate WT BRAF and are only indicated for BRAFV600E patients [418].
Vemurafenib resistant mutations arise in several genes in the MAPK pathway including MEK1 (MAP2K1), MEK2 (MAP2K2), NRAS, PIK3CA, and PTEN as well as various alterations of the BRAF gene itself (Figure 4, Supplementary Table S4) [419]. The MEK1 C121S mutation leads to a hyperactive form of the enzyme which remains active in the absence of an upstream signal [420]. A screen for BRAF inhibitor resistant mutations identified the MEK1 G128V, P124L/S, and V60E mutations as well as other MEK2 and NRAS [421]. Various BRAF splice variants also confer resistance to vemurafenib [422] (Supplementary Table S4). These variants usually lack the RAS binding site which resides between exons 4–8 and allows BRAF activation in the absence of RAS interaction [423,424,425,426]. A BRAF L505H mutation [427,428] appears to affect the ability of the drug to prohibit activation loop interaction with the binding site (Figure 4B). Remarkably, molecular modeling showed that the double V600E L505H mutant may still be sensitive to sorafenib [427]. Finally activating mutations in PIK3CA [429,430] or complete inactivation of PTEN [423] also arise in response to vemurafenib.
DABRAFENIB (GSK2118436) is an inhibitor that interacts with the inactive conformation of BRAFV600E [431,432,433,434,435,436]. It was synthesized by GlaxoSmithKline [437]. X-ray structure of dabrafenib complexed with BRAFV600E shows that the drug is stabilized in the active site of the enzyme by hydrogen bonds with C532, K483, and F595 [433]. Several other hydrophobic interactions are required for drug interaction with the active site of BRAFV600E. In addition to BRAFV600E, dabrafenib also inhibits MEK (MAP2K1) and MEK2 (MAP2K2) [438] and NRAS [431].
BRAFV600E resistance to dabrafenib has been reported. In one case, a splicing variant missing exons 2–10 was isolated from dabrafenib treated patients (Supplementary Table S4) [422,423,439]. This splicing variant was initially described as a resistant mutation to vemurafenib treated patients [426]. Deletion of this region causes BRAF dimerization and activation in the absence of RAS interaction. Other BRAF structural alterations that confer resistance to vemurafenib and dabrafenib have also been identified (Supplementary Table S4). BRAF amplification, MEK1/2 and NRAS mutations are other forms of BRAFV600E acquired resistance to dabrafenib [440]. Identified MEK1 resistant mutations are V60E, P124S, C121S, G128V, K57E, and G128D (Figure 4C) [421,422,441]. Most of these mutations (with the exception of V60E and K57E) map in the protein kinase domain. Western blotting shows that the V60E and G128V mutations cause hyperphosphorylation of MEK1/2 and ERK1/2. Therefore, these mutations appear to constitutively activate the MEK/ERK pathway. Dabrafenib MEK2 resistant mutations arise in amino acids V35M, L46F, C125S, and N126D (Figure 4D) [421]. Two of these mutations (V35M and L46F) occur in the N-terminus while two others (C125S and N126D) in the kinase domain. The mutations flank the ATP binding pocket and all cause hyperphosphorylation of MEK1/2 and ERK1/2. Combination therapy of dabrafenib and other inhibitors of the MAPK pathway (e.g., trametinib a MEK inhibitor) [442] leads to mutation in other genes in the pathway such as MEK2 or NRAS [443]. For a more comprehensive review of MAPK pathway resistant mutations please see [444].
SELUMETINIB (ARRY-142889, AZD6244) is a second generation MEK1/2 inhibitor synthesized by AstraZeneca [445,446]. The drug effectively inhibits purified MEK1’s ability to phosphorylate ERK1 as well as tumor growth in mouse xenografts [445]. It is also better at inhibiting tumor cell line growth that harbor BRAF or RAS mutations [446]. Following clinical trials, the drug has been used for the treatment of melanoma [447,448]. The drug is also used for the treatment of NSCLC [449,450] where it appears to work best in combination with PIK3CA inhibitors [451,452] as well as other cancers not discussed here. Cells resistant to selumetinib also show resistance to vemurafenib suggesting that resistance is acquired by the same mechanism [453]. The MAP2K1 P124L mutation was identified in one patient with advanced melanoma treated with selumetinib (Figure 4C) [454].
PEMBROLIZUMAB (MK-3475), designed by Merck &Co, is a monoclonal antibody that targets the program cell death-1 (PD-1) receptor [455]. It was approved in the US for the treatment of NSCLC, breast cancer, lymphoma, melanoma, as well as other forms of cancers [456]. PD-1 is expressed in the T-cells and interacts with the PD-L1 and PD-L2 ligands on the antigen-presenting cells [457]. This interaction suppresses the immune system. Thus PD-1 and other receptors like it act as an immune checkpoint to prevents an autoimmune response [458,459]. Binding of pembrolizumab to PD-1 blocks the receptor–ligand interaction and allows the immune system to attack cancer cells [460]. Although we do not discuss them here, nivolumab is another anti PD-1 antibody, while atezolizumab, avelumab, and durvalumab are anti PD-L1 antibodies that have been FDA approved [461,462]. Ipilimumab [463] is an antibody against CTLA-4 another inhibitory receptor [464,465]. Resistance to pembrolizumab arises by inactivating the function of the JAK1 and JAK2 genes [466]. JAK1/2 are involved in interferon signaling [467,468]. An in-depth review of PD-1/PD-L1 mechanisms of resistance mutations has been recently published [469].
VISMODEGIB (GDC-0449) was discovered by scientists at Genetech in a screen for inhibitors of the hedgehog signaling pathway [470]. It targets SMO (smoothened), a transmembrane protein that transduces signals to the cytoplasm upon binding to cholesterol [471,472,473]. The hedgehog pathway facilitates embryonic development [474] but is also activated in cancer where it contributes to cellular proliferation and differentiation (Figure 4A) [475,476,477]. Hedgehog pathway signaling activates the glioblastoma Gli1/2/3 genes (named so because they were initially identified in glioblastomas [478]) through a complex epistatic pathway of negative regulators [479]. Targeting of the hedgehog pathway in cancer cells has been an efficient way to inhibit tumor growth [480,481,482,483].
Vismodegib has been useful for treating metastatic tumors [484,485,486] including advanced basal cell carcinoma [486,487,488]. Resistance can be inherited or acquired de novo. Whole exome sequencing of vismodegib resistant basal cell carcinomas from patients with Gorlin syndrome, an autosomal dominant disease that causes early onset basal cell carcinoma, identified several mutations in SMO [489,490]. Several other mutations were identified in basal cell carcinoma patients not previously diagnosed with Gorlin syndrome (Figure 4E) [490]. Molecular modeling and docking on a crystal structure of the SMO transmembrane domain [491] showed that resistant mutations fall into two categories, those that affect drug interaction with SMO and those that lead to constitutive activation of SMO. Most mutations occur in the transmembrane domain of the protein.
4.5. ESR1 in Breast Cancer
Classification
Breast cancer is classified genetically by the status of three receptors: Estrogen receptor (ER), human epidermal growth factor receptor (HER2), and progesterone receptor (PR) [492,493]. Cancers that do not express any of the receptors are known as “triple negative”. Around 70% of all breast cancers are ER-positive and are generally treated with anti-hormone (endocrine) therapy [493]. Two isoforms of the estrogen receptor exist: ERalpha (ESR1) and ERBeta (ESR2) [494]. Upon binding to estrogen (the ligand), the receptor dimerizes and translocate to the nucleus [495] (Figure 5A).
The receptor functions as a homodimer and while ERα/ERβ heterodimers have been reported in vitro, they have not been identified in vivo. In the nucleus, the dimers bind to the estrogen response elements (EREs) [497,498]. Binding of the ligand does not appear to be required for ESR1-ERE interaction in vitro but it strongly increases its affinity in vivo [499,500]. ESR1 phosphorylation further increases its affinity for ERE [501]. In the nucleus, it activates estrogen responsive genes including TP53 [502], BRCA1 [503], CDK4/6 [504], as well as many others [505,506,507,508,509]. These genes promote cellular proliferation and survival [506].
ENDOCRINE THERAPY refers to modulation of hormone activity or complete hormone removal to treat breast cancer [510]. This can be accomplished either by preventing the ovaries from making hormones or by interfering with the function of estrogen and progesterone. Pre-menopausal patients who still produce estrogen in the ovaries have traditionally been treated by surgically removing the ovaries though endocrine therapy has decreased this practice in recent years [511,512,513,514]. Resistant mutations arising from endocrine therapy treatment have been thoroughly reviewed recently [515,516]. Generally, activating mutations in ESR1 renders the receptor resistant to endocrine therapy [517].
TAMOXIFEN. The selective estrogen receptor modulator (SERM) Tamoxifen (ICI46,474) was discovered by ICI Pharmaceuticals Division and has quickly been adopted as a treatment option [518,519] for both pre- and post-menopausal women. Tamoxifen is metabolized to 4-hydroxytamoxifen (OHT) which is the active antagonist that interreacts with the ligand binding domain (LBD) of ERα [520]. The crystal structure of the LBD-OHD complex shows that the drug blocks interaction of ESR1 with its co-activator [521,522]. Helix 12 of the ligand binding domain is required for interaction of ESR1 with co-activators and tamoxifen induces a conformation change that prevents this interaction. Molecular modeling shows that a D538G amino acid substitution frequently identified in tamoxifen treated patients is an ESR1 activating mutation that makes the ligand dispensable (Figure 5B, Supplementary Table S5) [523]. A Y537S/N/C mutation increases the flexibility of helix 12 by restricting the number of stabilizing hydrogen bonds formed and also allows interaction of ESR1 with its co-activators in the absence of the ligand [523,524]. Combination therapy is suggested for patients with ESR1 activating mutations which include CDK4/6 or mTOR inhibitors [525,526,527,528].
Aromatase (CYP19A1) is the enzyme responsible for synthesizing estrogen from androgen precursors [529]. Aromatase inhibitors and gonadotropin releasing hormone agonists suppress estrogen production and are often prescribed as adjuvant therapy to post-menopausal patients [530,531,532]. The ESR1 D538G, Y537S/C/N, and to a lesser degree L536H/R activating mutations also appear in aromatase inhibitor treated patients (Figure 5B, Supplementary Table S5) [529,533,534,535]. Two other mutations E380Q and S436P arise at significant rate in aromatase inhibitor treated patients that cause constitutive ER activity in the absence of ligand [536,537,538,539]. Finally, several other mutations have also been identified more recently [536,537,540,541]. An E380Q mutation found in helix 12 next to the estrogen binding site increases the flexibility of this helix and decreases the affinity of the protein for the drug [542]. Not discussed here are resistant mutations in aromatase (CYP19A1).
4.6. Androgen Receptor in Prostate Cancer
The androgen receptor (AR) is a transcription factor that controls male developmental genes [543,544,545]. Upon interaction with its ligand, the testosterone derivative 5-alpha-dihydrotestosterone (DHT), the androgen receptor binds androgen-responsive elements (AREs) [546,547] (Figure 6A).
The prostate specific antigen (PSA) is an AR target. For full activation of PSA transcription, the AR must interact with both the promoter region and an enhancer regulatory cassette [548]. The proliferation of prostate cancer cells requires constitutive AR activation and one of the treatments is androgen ablation which constitutes blocking production of AR binding ligands (male hormones) [549,550]. For example, luteinizing hormone releasing hormone (LHRH) agonists decrease the levels of androgen receptors produced by the testicles and are used as a form of treatment [551]. Surgical castration has also been shown to be effective because AR receptors originate in the testis [552]. However, ablation provides only temporal relief because prostate cancer cells often respond by over-expressing AR [552,553,554,555,556]. Mutations have also been identified which increase the transcription factor activation potential making it more likely to promote the same transcriptional levels even when ligands are depleted. Some of these mutations appear only after androgen ablation while others after subsequent treatment with therapeutic agents [553,557,558,559,560,561,562,563,564].
Prostate cancer resistant to castration eventually develops in almost 100% of the cases. Therefore, therapeutic agents must be administered for efficient treatment [565]. The two major types of treatments for castration resistant prostate cancer are inhibitors of synthesis of androgen ligand precursor (ketoconazole and abiraterone) and AR antagonists (enzalutamide and flutamide). Even in castrated patients, androgen ligands may still be synthesized by other pathways or de novo. Both ketoconazole and abiraterone are inhibitors of CYP17A [566,567] (Figure 6A), a hydroxylase/lyase involved in steroid synthesis that appears to be responsible for continual androgen synthesis in castrated patients [568]. Ketoconazole is significantly more toxic that abiraterone [569,570]. Abiraterone was developed later by modification of esters of pyridyl acetic acid [571,572], a potent inhibitor of CYP17A [573]. It is a more selective inhibitor than ketoconazole [571,574,575]. The antagonists enzalutamide and flutamide block androgen binding to AR significantly reducing ARs ability to activate transcription [576,577,578,579,580].
Resistance mutations to the AR drugs are generally restricted to two amino acids, 877 and 878 (Figure 6B, Supplementary Table S6). The most frequent mutation is T878A which can confer resistance to all drugs mentioned above [581,582,583,584]. T878S was identified in at least one abiraterone resistant cancer [581,584] and F877L in one enzalutamide treated cancer [583,585]. Modeling data of the AR ligand binding domain shows that these mutations are selected for because they convert the antagonists enzalutamide and to a lesser degree flutamide into agonists [586]. Treatment with ketoconazole and abiraterone causes upregulation of AR synthesis but with remarkable selection for the mutant form (T878A) [582]. Importantly, none of the patients that upregulate AR T878A were previously treated with flutamide or enzalutamide. Thus, the mutant can occur directly in response to ketoconazole and abiraterone.
4.7. Other Resistant Mutations
INFIGRATINIB (BGJ 398) is a drug that can inhibit the function of fibroblast growth factor receptors 1–4 (FGFR 1–4) [587,588]. The FGFR receptors are good targets because they are involved in a cascade of signaling pathways that control proliferation, cell survival, and angiogenesis [589,590,591]. Infigratinib is able to inhibit FGFR function in sorafenib resistant cells [592] and is efficient in treatment of biliary tract cancer [593,594]. A secondary resistant mutation (V564) to infigratinib has been identified in the FGFR2 gene [595]. Molecular modeling shows that the V564F is a gatekeeper mutation that blocks the drug interaction with the kinase while retaining the activity of the enzyme (Supplementary Table S7).
EVEROLIMUS (RAD001) is a rapamycin analog that targets mTOR [596]. Everolimus is used as a chemotherapeutic agent for a variety of cancers including thyroid cancer. A patient with anaplastic thyroid cancer treated with everolimus acquired the mTOR F2108L secondary resistant mutation (Supplementary Table S7) [597]. This mutation is similar to a rapamycin resistant mutation identified in fission yeast tor2 (F2049L) [598]. Structural modeling showed that the human F2108L occurs in the rapamycin binding domain. Subsequent in vitro studies showed that this mutation also affects the effect of rapamycin on mTOR activity. In at least one report, a breast cancer cell line treated with the mTOR inhibitor rapamycin led to the F2108L resistant mutation [527]. Molecular modeling showed that this mutation inhibits drug interaction with the protein [597]. Thus, the mechanism of everolimus resistance is likely to be similar to that of rapamycin.
PF-04217903 is a c-Met ATP competitive inhibitor that can slow or stop tumor progression both in vitro and in vivo [599]. A patient with a kidney tumor was treated with PF-04217903 in a Phase I clinical trial acquired a M1268T secondary resistant mutation (Supplementary Table S7) [600]. The significance of this mutation remains to be investigated.
PD0325901, developed by Pfizer, is a non-competitive ATP inhibitor of MAP2K1/MAP2K2 [601,602]. PD0325901 and similar MAP2K specific inhibitors can be used to target the ERK pathway in a variety of cancers [603,604]. F129L and L115P MAP2K1 resistant mutations arise after drug treatment (Supplementary Table S7) [605]. These mutations appear to affect drug affinity for the kinase. The F129L also seems to function as an activating mutation [606]. A V215E MAP2K2 mutation has the same effect as the F129L MAP2K1 mutation.
5. Conclusions and Future Directions
Resistance to small molecule inhibitors highlights the plasticity of cellular processes that drive tumorigenesis. Drug synthesis has been a cat and mouse game and it is likely, that this will continue at least for the foreseeable future. Nevertheless, this should not be interpreted as a failure to produce efficient treatments. On the contrary, our understanding of protein structure has in many cases allowed development of targeted small molecule drugs that are more efficient and selective than chemotherapy.
Understanding resistance mutations requires knowledge of organic chemistry and protein structure. In silico molecular modeling has also been helpful but often specialized software and computer programing knowledge is required. However, recently more user-friendly platforms are being developed. For example, COSMIC launched a platform known as COSMIC-3D to understand “cancer mutations in the context of 3D protein structure” (https://cancer.sanger.ac.uk/cosmic3d/) [607]. Interfaces such as this, will undoubtedly allow other scientists not necessarily trained as organic or protein biochemists to provide input and generate hypothesis that will be useful for drug development.
As seen in this review, many resistant mechanisms are similar. For example, when targeting kinases, certain “gatekeeper” residues are essential for facilitating drug–enzyme interaction. Mutations in these residues almost always affect the efficacy of the drug. Although each kinase is unique, patients have benefited from faster drug discovery when scientists compared different resistance mechanisms [608]. Comparisons of resistance mechanisms will accelerate next generation drug development.
Immunotherapy (briefly mentioned in this review) and modulation of the function of various non-coding RNAs are other forms of cancer therapies that we did not have time to address here [609,610,611,612,613,614]. The immune system function in slowing tumor progression has been well documented [615,616]. Non-coding RNAs have a plethora of functions in the cell including regulation of gene expression and protein localization [611]. Non-coding RNAs also regulate gene expression in the immune cells allowing tumors to evade the immune system [617]. One challenge has been the delivery of these therapies. However, with technical advances such as CRISPR [618,619] and nanotechnology [620,621,622,623,624] it is clear that in the near future we will see much more complex forms of cancer treatments.
Supplementary Materials
The following are available online at https://www.mdpi.com/2072-6694/12/4/927/s1, Figure S1: Resistant mutations in the ABL 1 tyrosine kinase domain, Table S1: Resistant mutations in non-small cell lung cancer (NSCLC), Table S2: Resistant mutations detected in cancers of the hematopoietic and lymphoid tissues, Table S3: Resistant mutations in gastrointestinal stromal tumors (GIST) soft tissue, Table S4: Resistant mutations in melanoma, Table S5: Resistant mutations in breast cancer, Table S6: Androgen receptor (AR) resistant mutations in prostate cancer, Table S7: Resistant mutations detected in other tissues.
Author Contributions
A.B.H. created all figures and R.C.P. wrote the first draft. Both A.B.H. and R.C.P. revised subsequent drafts. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by Pelotonia funds to R.C.P. from the Ohio State University Comprehensive Cancer Center.
Acknowledgments
We thank James and Ellen Bazzoli for their generous donation to sponsor our laboratory.
Conflicts of Interest
The authors declare no conflict of interest.
References
- DeVita, V.T., Jr.; Chu, E. A history of cancer chemotherapy. Cancer Res. 2008, 68, 8643–8653. [Google Scholar] [CrossRef] [PubMed]
- Farber, S. Chemotherapy in the treatment of leukemia and Wilms’ tumor. JAMA 1966, 198, 826–836. [Google Scholar] [CrossRef] [PubMed]
- Ribatti, D. Sidney Farber and the treatment of childhood acute lymphoblastic leukemia with a chemotherapeutic agent. Pediatr. Hematol. Oncol. 2012, 29, 299–302. [Google Scholar] [CrossRef] [PubMed]
- Farber, S.; Diamond, L.K. Temporary remissions in acute leukemia in children produced by folic acid antagonist, 4-aminopteroyl-glutamic acid. N. Engl. J. Med. 1948, 238, 787–793. [Google Scholar] [CrossRef] [PubMed]
- Grever, M.R.; Schepartz, S.A.; Chabner, B.A. The National Cancer Institute: Cancer drug discovery and development program. Semin. Oncol. 1992, 19, 622–638. [Google Scholar] [PubMed]
- Mukherjee, S. The Emperor of all Maladies: A Biography of Cancer; Scribner: New York, NY, USA, 2010. [Google Scholar]
- Meisner, N.C.; Hintersteiner, M.; Uhl, V.; Weidemann, T.; Schmied, M.; Gstach, H.; Auer, M. The chemical hunt for the identification of drugable targets. Curr. Opin. Chem. Biol. 2004, 8, 424–431. [Google Scholar] [CrossRef] [PubMed]
- Piggott, A.M.; Karuso, P. Quality, not quantity: The role of natural products and chemical proteomics in modern drug discovery. Comb. Chem. High. Throughput Scr. 2004, 7, 607–630. [Google Scholar] [CrossRef]
- Sioud, M.; Leirdal, M. Druggable signaling proteins. Methods Mol. Biol. 2007, 361, 1–24. [Google Scholar] [CrossRef]
- Yuan, Y.X.; Pei, J.F.; Lai, L.H. Binding site detection and druggability prediction of protein targets for structure-based drug design. Curr. Pharm. Des. 2013, 19, 2326–2333. [Google Scholar] [CrossRef]
- Perola, E.; Herman, L.; Weiss, J. Development of a rule-based method for the assessment of protein druggability. J. Chem. Inf. Modeling 2012, 52, 1027–1038. [Google Scholar] [CrossRef]
- Sledz, P.; Caflisch, A. Protein structure-based drug design: From docking to molecular dynamics. Curr. Opin. Struc. Biol. 2018, 48, 93–102. [Google Scholar] [CrossRef] [PubMed]
- Bianchi, J.J.; Zhao, X.; Mays, J.C.; Davoli, T. Not all cancers are created equal: Tissue specificity in cancer genes and pathways. Curr. Opin. Cell. Biol. 2020, 63, 135–143. [Google Scholar] [CrossRef] [PubMed]
- Bailey, M.H.; Tokheim, C.; Porta-Pardo, E.; Sengupta, S.; Bertrand, D.; Weerasinghe, A.; Colaprico, A.; Wendl, M.C.; Kim, J.; Reardon, B.; et al. Comprehensive characterization of cancer driver genes and mutations. Cell 2018, 174, 1034–1035. [Google Scholar] [CrossRef] [PubMed]
- Greenman, C.; Stephens, P.; Smith, R.; Dalgliesh, G.L.; Hunter, C.; Bignell, G.; Davies, H.; Teague, J.; Butler, A.; Stevens, C.; et al. Patterns of somatic mutation in human cancer genomes. Nature 2007, 446, 153–158. [Google Scholar] [CrossRef]
- Boumahdi, S.; de Sauvage, F.J. The great escape: Tumour cell plasticity in resistance to targeted therapy. Nat. Rev. Drug Discov. 2020, 19, 39–56. [Google Scholar] [CrossRef]
- Tate, J.G.; Bamford, S.; Jubb, H.C.; Sondka, Z.; Beare, D.M.; Bindal, N.; Boutselakis, H.; Cole, C.G.; Creatore, C.; Dawson, E.; et al. COSMIC: The catalogue of somatic mutations in cancer. Nucleic Acids Res. 2019, 47, D941–D947. [Google Scholar] [CrossRef]
- Jay, J.J.; Brouwer, C. Lollipops in the clinic: Information dense mutation plots for precision medicine. PLoS ONE 2016, 11, e0160519. [Google Scholar] [CrossRef]
- Roskoski, R. Classification of small molecule protein kinase inhibitors based upon the structures of their drug-enzyme complexes. Pharmacol. Res. 2016, 103, 26–48. [Google Scholar] [CrossRef]
- Roskoski, R., Jr. Properties of FDA-approved small molecule protein kinase inhibitors: A 2020 update. Pharmacol. Res. 2020, 152, 104609. [Google Scholar] [CrossRef]
- Roskoski, R. A historical overview of protein kinases and their targeted small molecule inhibitors. Pharmacol. Res. 2015, 100, 1–23. [Google Scholar] [CrossRef]
- Helbig, G. Imatinib for the treatment of hypereosinophilic syndromes. Expert Rev. Clin. Immunol. 2018, 14, 163–170. [Google Scholar] [CrossRef] [PubMed]
- Duma, N.; Santana-Davila, R.; Molina, J.R. Non-small cell lung cancer: Epidemiology, screening, diagnosis, and treatment. Mayo Clin. Proc. 2019, 94, 1623–1640. [Google Scholar] [CrossRef] [PubMed]
- Gardiner, N.; Jogai, S.; Wallis, A. The revised lung adenocarcinoma classification-an imaging guide. J. Thorac. Dis 2014, 6, 537–546. [Google Scholar] [CrossRef]
- Li, C.; Lu, H. Adenosquamous carcinoma of the lung. Onco Targets Ther. 2018, 11, 4829–4835. [Google Scholar] [CrossRef]
- Zhu, C.; Zhuang, W.; Chen, L.; Yang, W.; Ou, W.B. Frontiers of ctDNA, targeted therapies, and immunotherapy in non-small-cell lung cancer. Transl. Lung Cancer Res. 2020, 9, 111–138. [Google Scholar] [CrossRef]
- Ruiz-Cordero, R.; Devine, W.P. Targeted therapy and checkpoint immunotherapy in lung cancer. Surg. Pathol. Clin. 2020, 13, 17–33. [Google Scholar] [CrossRef]
- Lim, Z.F.; Ma, P.C. Emerging insights of tumor heterogeneity and drug resistance mechanisms in lung cancer targeted therapy. J. Hematol. Oncol. 2019, 12, 134. [Google Scholar] [CrossRef]
- Hirsch, F.R.; Scagliotti, G.V.; Mulshine, J.L.; Kwon, R.; Curran, W.J.; Wu, Y.L.; Paz-Ares, L. Lung cancer: Current therapies and new targeted treatments. Lancet 2017, 389, 299–311. [Google Scholar] [CrossRef]
- Xu, Z.Y.; Li, J.L. Comparative review of drug-drug interactions with epidermal growth factor receptor tyrosine kinase inhibitors for the treatment of non-small-cell lung cancer. Oncotargets Ther. 2019, 12, 5467–5484. [Google Scholar] [CrossRef]
- Herbst, R.S. Review of epidermal growth factor receptor biology. Int. J. Radiat. Oncol. 2004, 59, 21–26. [Google Scholar] [CrossRef]
- Burgering, B.M.T.; Coffer, P.J. Protein-Kinase-B (C-Akt) in Phosphatidylinositol-3-Oh Inase signal-transduction. Nature 1995, 376, 599–602. [Google Scholar] [CrossRef] [PubMed]
- Mayer, I.A.; Arteaga, C.L. The PI3K/AKT pathway as a target for cancer treatment. Annu. Rev. Med. 2016, 67, 11–28. [Google Scholar] [CrossRef] [PubMed]
- Alroy, I.; Yarden, Y. The ErbB signaling network in embryogenesis and oncogenesis: Signal diversification through combinatorial ligand-receptor interactions. FEBS Lett. 1997, 410, 83–86. [Google Scholar] [CrossRef]
- Wee, P.; Wang, Z. Epidermal growth factor receptor cell proliferation signaling pathways. Cancers 2017, 9, 52. [Google Scholar] [CrossRef]
- Bishop, A.L.; Hall, A. Rho GTPases and their effector proteins. Biochem. J. 2000, 348, 241–255. [Google Scholar] [CrossRef] [PubMed]
- Aguilar, B.J.; Zhou, H.; Lu, Q. Cdc42 signaling pathway inhibition as a therapeutic target in ras- related cancers. Curr. Med. Chem. 2017, 24, 3485–3507. [Google Scholar] [CrossRef]
- Nicholson, R.I.; Gee, J.M.; Harper, M.E. EGFR and cancer prognosis. Eur. J. Cancer 2001, 37, 9–15. [Google Scholar] [CrossRef]
- Hirsch, F.R.; Varella-Garcia, M.; Bunn, P.A., Jr.; Di Maria, M.V.; Veve, R.; Bremmes, R.M.; Baron, A.E.; Zeng, C.; Franklin, W.A. Epidermal growth factor receptor in non-small-cell lung carcinomas: Correlation between gene copy number and protein expression and impact on prognosis. J. Clin. Oncol. 2003, 21, 3798–3807. [Google Scholar] [CrossRef]
- Hsu, P.C.; Jablons, D.M.; Yang, C.T.; You, L. Epidermal Growth Factor Receptor (EGFR) pathway, Yes-Associated Protein (YAP) and the Regulation of Programmed Death-Ligand 1 (PD-L1) in Non-Small Cell Lung Cancer (NSCLC). Int. J. Mol. Sci. 2019, 20, 3821. [Google Scholar] [CrossRef]
- Lynch, T.J.; Bell, D.W.; Sordella, R.; Gurubhagavatula, S.; Okimoto, R.A.; Brannigan, B.W.; Harris, P.L.; Haserlat, S.M.; Supko, J.G.; Haluska, F.G.; et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N. Engl. J. Med. 2004, 350, 2129–2139. [Google Scholar] [CrossRef]
- Paez, J.G.; Janne, P.A.; Lee, J.C.; Tracy, S.; Greulich, H.; Gabriel, S.; Herman, P.; Kaye, F.J.; Lindeman, N.; Boggon, T.J.; et al. EGFR mutations in lung cancer: Correlation with clinical response to gefitinib therapy. Science 2004, 304, 1497–1500. [Google Scholar] [CrossRef] [PubMed]
- Pao, W.; Miller, V.; Zakowski, M.; Doherty, J.; Politi, K.; Sarkaria, I.; Singh, B.; Heelan, R.; Rusch, V.; Fulton, L.; et al. EGF receptor gene mutations are common in lung cancers from "never smokers" and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc. Natl. Acad. Sci. USA 2004, 101, 13306–13311. [Google Scholar] [CrossRef] [PubMed]
- Huse, M.; Kuriyan, J. The conformational plasticity of protein kinases. Cell 2002, 109, 275–282. [Google Scholar] [CrossRef]
- Vyse, S.; Huang, P.H. Targeting EGFR exon 20 insertion mutations in non-small cell lung cancer. Signal Transduct. Target. Ther. 2019, 4, 5. [Google Scholar] [CrossRef] [PubMed]
- Ward, R.A.; Anderton, M.J.; Ashton, S.; Bethel, P.A.; Box, M.; Butterworth, S.; Colclough, N.; Chorley, C.G.; Chuaqui, C.; Cross, D.A.; et al. Structure- and reactivity-based development of covalent inhibitors of the activating and gatekeeper mutant forms of the epidermal growth factor receptor (EGFR). J. Med. Chem. 2013, 56, 7025–7048. [Google Scholar] [CrossRef]
- Pollack, V.A.; Savage, D.M.; Baker, D.A.; Tsaparikos, K.E.; Sloan, D.E.; Moyer, J.D.; Barbacci, E.G.; Pustilnik, L.R.; Smolarek, T.A.; Davis, J.A.; et al. Inhibition of epidermal growth factor receptor-associated tyrosine phosphorylation in human carcinomas with CP-358,774: Dynamics of receptor inhibition in situ and antitumor effects in athymic mice. J. Pharmacol. Exp. Ther. 1999, 291, 739–748. [Google Scholar]
- Wakeling, A.E.; Barker, A.J.; Davies, D.H.; Brown, D.S.; Green, L.R.; Cartlidge, S.A.; Woodburn, J.R. Specific inhibition of epidermal growth factor receptor tyrosine kinase by 4-anilinoquinazolines. Breast Cancer Res. Treat. 1996, 38, 67–73. [Google Scholar] [CrossRef]
- Ciardiello, F.; Caputo, R.; Bianco, R.; Damiano, V.; Pomatico, G.; De Placido, S.; Bianco, A.R.; Tortora, G. Antitumor effect and potentiation of cytotoxic drugs activity in human cancer cells by ZD-1839 (Iressa), an epidermal growth factor receptor-selective tyrosine kinase inhibitor. Clin. Cancer Res. 2000, 6, 2053–2063. [Google Scholar]
- Mulloy, R.; Ferrand, A.; Kim, Y.; Sordella, R.; Bell, D.W.; Haber, D.A.; Anderson, K.S.; Settleman, J. Epidermal growth factor receptor mutants from human lung cancers exhibit enhanced catalytic activity and increased sensitivity to gefitinib. Cancer Res. 2007, 67, 2325–2330. [Google Scholar] [CrossRef]
- Carey, K.D.; Garton, A.J.; Romero, M.S.; Kahler, J.; Thomson, S.; Ross, S.; Park, F.; Haley, J.D.; Gibson, N.; Sliwkowski, M.X. Kinetic analysis of epidermal growth factor receptor somatic mutant proteins shows increased sensitivity to the epidermal growth factor receptor tyrosine kinase inhibitor, erlotinib. Cancer Res. 2006, 66, 8163–8171. [Google Scholar] [CrossRef]
- Kobayashi, S.; Boggon, T.J.; Dayaram, T.; Janne, P.A.; Kocher, O.; Meyerson, M.; Johnson, B.E.; Eck, M.J.; Tenen, D.G.; Halmos, B. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N. Engl. J. Med. 2005, 352, 786–792. [Google Scholar] [CrossRef] [PubMed]
- Pao, W.; Miller, V.A.; Politi, K.A.; Riely, G.J.; Somwar, R.; Zakowski, M.F.; Kris, M.G.; Varmus, H. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med. 2005, 2, e73. [Google Scholar] [CrossRef] [PubMed]
- Eck, M.J.; Yun, C.H. Structural and mechanistic underpinnings of the differential drug sensitivity of EGFR mutations in non-small cell lung cancer. Biochim. Biophys. Acta 2010, 1804, 559–566. [Google Scholar] [CrossRef] [PubMed]
- Yun, C.H.; Mengwasser, K.E.; Toms, A.V.; Woo, M.S.; Greulich, H.; Wong, K.K.; Meyerson, M.; Eck, M.J. The T790M mutation in EGFR kinase causes drug resistance by increasing the affinity for ATP. Proc. Natl. Acad. Sci. USA 2008, 105, 2070–2075. [Google Scholar] [CrossRef]
- Yun, C.H.; Boggon, T.J.; Li, Y.; Woo, M.S.; Greulich, H.; Meyerson, M.; Eck, M.J. Structures of lung cancer-derived EGFR mutants and inhibitor complexes: Mechanism of activation and insights into differential inhibitor sensitivity. Cancer Cell 2007, 11, 217–227. [Google Scholar] [CrossRef]
- Pazarentzos, E.; Giannikopoulos, P.; Hrustanovic, G.; St John, J.; Olivas, V.R.; Gubens, M.A.; Balassanian, R.; Weissman, J.; Polkinghorn, W.; Bivona, T.G. Oncogenic activation of the PI3-kinase p110beta isoform via the tumor-derived PIK3Cbeta(D1067V) kinase domain mutation. Oncogene 2016, 35, 1198–1205. [Google Scholar] [CrossRef]
- Dbouk, H.A.; Khalil, B.D.; Wu, H.; Shymanets, A.; Nurnberg, B.; Backer, J.M. Characterization of a tumor-associated activating mutation of the p110beta PI 3-kinase. PLoS ONE 2013, 8, e63833. [Google Scholar] [CrossRef]
- Hirsh, V. New developments in the treatment of advanced squamous cell lung cancer: Focus on afatinib. Onco Targets Ther. 2017, 10, 2513–2526. [Google Scholar] [CrossRef]
- Li, D.; Ambrogio, L.; Shimamura, T.; Kubo, S.; Takahashi, M.; Chirieac, L.R.; Padera, R.F.; Shapiro, G.I.; Baum, A.; Himmelsbach, F.; et al. BIBW2992, an irreversible EGFR/HER2 inhibitor highly effective in preclinical lung cancer models. Oncogene 2008, 27, 4702–4711. [Google Scholar] [CrossRef]
- Kim, Y.; Ko, J.; Cui, Z.; Abolhoda, A.; Ahn, J.S.; Ou, S.H.; Ahn, M.J.; Park, K. The EGFR T790M mutation in acquired resistance to an irreversible second-generation EGFR inhibitor. Mol. Cancer Ther. 2012, 11, 784–791. [Google Scholar] [CrossRef]
- Solca, F.; Dahl, G.; Zoephel, A.; Bader, G.; Sanderson, M.; Klein, C.; Kraemer, O.; Himmelsbach, F.; Haaksma, E.; Adolf, G.R. Target binding properties and cellular activity of afatinib (BIBW 2992), an irreversible ErbB family blocker. J. Pharmacol. Exp. Ther. 2012, 343, 342–350. [Google Scholar] [CrossRef]
- Wu, S.G.; Liu, Y.N.; Tsai, M.F.; Chang, Y.L.; Yu, C.J.; Yang, P.C.; Yang, J.C.; Wen, Y.F.; Shih, J.Y. The mechanism of acquired resistance to irreversible EGFR tyrosine kinase inhibitor-afatinib in lung adenocarcinoma patients. Oncotarget 2016, 7, 12404–12413. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, Y.; Azuma, K.; Nagai, H.; Kim, Y.H.; Togashi, Y.; Sesumi, Y.; Chiba, M.; Shimoji, M.; Sato, K.; Tomizawa, K.; et al. Characterization of EGFR T790M, L792F, and C797S mutations as mechanisms of acquired resistance to afatinib in lung cancer. Mol. Cancer Ther. 2017, 16, 357–364. [Google Scholar] [CrossRef] [PubMed]
- Miller, V.A.; Hirsh, V.; Cadranel, J.; Chen, Y.M.; Park, K.; Kim, S.W.; Zhou, C.; Su, W.C.; Wang, M.; Sun, Y.; et al. Afatinib versus placebo for patients with advanced, metastatic non-small-cell lung cancer after failure of erlotinib, gefitinib, or both, and one or two lines of chemotherapy (LUX-Lung 1): A phase 2b/3 randomised trial. Lancet Oncol. 2012, 13, 528–538. [Google Scholar] [CrossRef]
- Pirazzoli, V.; Nebhan, C.; Song, X.; Wurtz, A.; Walther, Z.; Cai, G.; Zhao, Z.; Jia, P.; de Stanchina, E.; Shapiro, E.M.; et al. Acquired resistance of EGFR-mutant lung adenocarcinomas to afatinib plus cetuximab is associated with activation of mTORC1. Cell Rep. 2014, 7, 999–1008. [Google Scholar] [CrossRef]
- Curto, M.; Cole, B.K.; Lallemand, D.; Liu, C.H.; McClatchey, A.I. Contact-dependent inhibition of EGFR signaling by Nf2/Merlin. J. Cell Biol. 2007, 177, 893–903. [Google Scholar] [CrossRef] [PubMed]
- Okada, T.; Lopez-Lago, M.; Giancotti, F.G. Merlin/NF-2 mediates contact inhibition of growth by suppressing recruitment of Rac to the plasma membrane. J. Cell Biol. 2005, 171, 361–371. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Morrison, H.; Sherman, L.S.; Legg, J.; Banine, F.; Isacke, C.; Haipek, C.A.; Gutmann, D.H.; Ponta, H.; Herrlich, P. The NF2 tumor suppressor gene product, merlin, mediates contact inhibition of growth through interactions with CD44. Genes Dev. 2001, 15, 968–980. [Google Scholar] [CrossRef]
- Sainio, M.; Zhao, F.; Heiska, L.; Turunen, O.; den Bakker, M.; Zwarthoff, E.; Lutchman, M.; Rouleau, G.A.; Jaaskelainen, J.; Vaheri, A.; et al. Neurofibromatosis 2 tumor suppressor protein colocalizes with ezrin and CD44 and associates with actin-containing cytoskeleton. J. Cell Sci. 1997, 110, 2249–2260. [Google Scholar] [PubMed]
- Gladden, A.B.; Hebert, A.M.; Schneeberger, E.E.; McClatchey, A.I. The NF2 tumor suppressor, Merlin, regulates epidermal development through the establishment of a junctional polarity complex. Dev. Cell 2010, 19, 727–739. [Google Scholar] [CrossRef] [PubMed]
- Lallemand, D.; Curto, M.; Saotome, I.; Giovannini, M.; McClatchey, A.I. NF2 deficiency promotes tumorigenesis and metastasis by destabilizing adherens junctions. Genes Dev. 2003, 17, 1090–1100. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Pan, D. The hippo signaling pathway in development and disease. Dev. Cell 2019, 50, 264–282. [Google Scholar] [CrossRef] [PubMed]
- Sato, T.; Sekido, Y. NF2/Merlin inactivation and potential therapeutic targets in mesothelioma. Int. J. Mol. Sci. 2018, 19, 988. [Google Scholar] [CrossRef]
- Li, W.; Cooper, J.; Zhou, L.; Yang, C.; Erdjument-Bromage, H.; Zagzag, D.; Snuderl, M.; Ladanyi, M.; Hanemann, C.O.; Zhou, P.; et al. Merlin/NF2 loss-driven tumorigenesis linked to CRL4(DCAF1)-mediated inhibition of the hippo pathway kinases Lats1 and 2 in the nucleus. Cancer Cell 2014, 26, 48–60. [Google Scholar] [CrossRef]
- Hikasa, H.; Sekido, Y.; Suzuki, A. Merlin/NF2-Lin28B-let-7 is a tumor-suppressive pathway that is cell-density dependent and hippo independent. Cell Rep. 2016, 14, 2950–2961. [Google Scholar] [CrossRef]
- Sekido, Y.; Bader, S.A.; Carbone, D.P.; Johnson, B.E.; Minna, J.D. Molecular analysis of the HuD gene encoding a paraneoplastic encephalomyelitis antigen in human lung cancer cell lines. Cancer Res. 1994, 54, 4988–4992. [Google Scholar]
- Bianchi, A.B.; Mitsunaga, S.I.; Cheng, J.Q.; Klein, W.M.; Jhanwar, S.C.; Seizinger, B.; Kley, N.; Klein-Szanto, A.J.; Testa, J.R. High frequency of inactivating mutations in the neurofibromatosis type 2 gene (NF2) in primary malignant mesotheliomas. Proc. Natl. Acad. Sci. USA 1995, 92, 10854–10858. [Google Scholar] [CrossRef]
- Gendreau, S.B.; Ventura, R.; Keast, P.; Laird, A.D.; Yakes, F.M.; Zhang, W.; Bentzien, F.; Cancilla, B.; Lutman, J.; Chu, F.; et al. Inhibition of the T790M gatekeeper mutant of the epidermal growth factor receptor by EXEL-7647. Clin. Cancer Res. 2007, 13, 3713–3723. [Google Scholar] [CrossRef]
- Ferrara, N.; Gerber, H.P.; LeCouter, J. The biology of VEGF and its receptors. Nat. Med. 2003, 9, 669–676. [Google Scholar] [CrossRef]
- Wu, B.; Rockel, J.S.; Lagares, D.; Kapoor, M. Ephrins and eph receptor signaling in tissue repair and fibrosis. Curr. Rheumatol. Rep. 2019, 21, 23. [Google Scholar] [CrossRef] [PubMed]
- Roskoski, R., Jr. The ErbB/HER family of protein-tyrosine kinases and cancer. Pharmacol. Res. 2014, 79, 34–74. [Google Scholar] [CrossRef] [PubMed]
- Pietanza, M.C.; Lynch, T.J., Jr.; Lara, P.N., Jr.; Cho, J.; Yanagihara, R.H.; Vrindavanam, N.; Chowhan, N.M.; Gadgeel, S.M.; Pennell, N.A.; Funke, R.; et al. XL647--a multitargeted tyrosine kinase inhibitor: Results of a phase II study in subjects with non-small cell lung cancer who have progressed after responding to treatment with either gefitinib or erlotinib. J. Thorac. Oncol. 2012, 7, 219–226. [Google Scholar] [CrossRef] [PubMed]
- Pietanza, M.C.; Gadgeel, S.M.; Dowlati, A.; Lynch, T.J.; Salgia, R.; Rowland, K.M., Jr.; Wertheim, M.S.; Price, K.A.; Riely, G.J.; Azzoli, C.G.; et al. Phase II study of the multitargeted tyrosine kinase inhibitor XL647 in patients with non-small-cell lung cancer. J. Thorac. Oncol. 2012, 7, 856–865. [Google Scholar] [CrossRef] [PubMed]
- Cross, D.; Ashton, S.; Nebhan, C.; Eberlein, C.; Finlay, M.R.V.; Hughes, G.; Jacobs, V.; Mellor, M.; Brewer, M.R.; Meador, C.; et al. AZD9291: An irreversible, potent and selective third generation tyrosine kinase inhibitor (TKI) targeting EGFR activating (EGFRm plus ) and resistance (T790M) mutations in advanced lung adenocarcinoma. Mol. Cancer Ther. 2013, 12. [Google Scholar] [CrossRef]
- Cross, D.A.E.; Ashton, S.E.; Ghiorghiu, S.; Eberlein, C.; Nebhan, C.A.; Spitzler, P.J.; Orme, J.P.; Finlay, M.R.V.; Ward, R.A.; Mellor, M.J.; et al. AZD9291, an irreversible EGFR TKI, overcomes T790M-Mediated resistance to EGFR inhibitors in lung cancer. Cancer Discov. 2014, 4, 1046–1061. [Google Scholar] [CrossRef] [PubMed]
- Chang, L.K.; Chang, Y.L.; Shih, J.Y. Primary resistance to osimertinib despite acquired T790M. Respirol. Case Rep. 2020, 8, e00532. [Google Scholar] [CrossRef]
- Thress, K.S.; Paweletz, C.P.; Felip, E.; Cho, B.C.; Stetsonl, D.; Dougherty, B.; Lai, Z.W.; Markovets, A.; Vivancos, A.; Kuang, Y.; et al. Acquired EGFR C797S mutation mediates resistance to AZD9291 in non-small cell lung cancer harboring EGFR T790M. Nat. Med. 2015, 21, 560–562. [Google Scholar] [CrossRef]
- Zheng, D.; Hu, M.; Bai, Y.; Zhu, X.; Lu, X.; Wu, C.; Wang, J.; Liu, L.; Wang, Z.; Ni, J.; et al. EGFR G796D mutation mediates resistance to osimertinib. Oncotarget 2017, 8, 49671–49679. [Google Scholar] [CrossRef]
- Ou, S.H.I.; Cui, J.; Schrock, A.B.; Goldberg, M.E.; Zhu, V.W.; Albacker, L.; Stephens, P.J.; Miller, V.A.; Ali, S.M. Emergence of novel and dominant acquired EGFR solvent-front mutations at Gly796 (G796S/R) together with C797S/G and L792F/H mutations in one EGFR (L858R/T790M) NSCLC patient who progressed on osimertinib. Lung Cancer 2019, 138, 141-141. [Google Scholar] [CrossRef]
- Bersanelli, M.; Minari, R.; Bordi, P.; Gnetti, L.; Bozzetti, C.; Squadrilli, A.; Lagrasta, C.A.M.; Bottarelli, L.; Osipova, G.; Capelletto, E.; et al. L718Q Mutation as New Mechanism of Acquired Resistance to AZD9291 in EGFR-Mutated NSCLC. J. Thorac. Oncol. 2016, 11, E121–E123. [Google Scholar] [CrossRef] [PubMed]
- Ercan, D.; Choi, H.G.; Yun, C.H.; Capelletti, M.; Xie, T.; Eck, M.J.; Gray, N.S.; Janne, P.A. EGFR mutations and resistance to irreversible pyrimidine-based EGFR inhibitors. Clin. Cancer Res. 2015, 21, 3913–3923. [Google Scholar] [CrossRef] [PubMed]
- Callegari, D.; Ranaghan, K.E.; Woods, C.J.; Minari, R.; Tiseo, M.; Mor, M.; Mulholland, A.J.; Lodola, A. L718Q mutant EGFR escapes covalent inhibition by stabilizing a non-reactive conformation of the lung cancer drug osimertinib. Chem. Sci. 2018, 9, 2740–2749. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Wang, Y.Y.; Zhai, Y.C.; Wang, J. Non-small cell lung cancer harboring a rare EGFR L747P mutation showing intrinsic resistance to both gefitinib and osimertinib (AZD9291): A case report. Thorac. Cancer 2018, 9, 745–749. [Google Scholar] [CrossRef] [PubMed]
- Zhou, T.; Zhou, X.Y.; Li, P.; Qi, C.; Ling, Y. EGFR L747P mutation in one lung adenocarcinoma patient responded to afatinib treatment: A case report. J. Thorac. Dis. 2018, 10, E802–E805. [Google Scholar] [CrossRef] [PubMed]
- Liang, S.K.; Ko, J.C.; Yang, J.C.H.; Shih, J.Y. Afatinib is effective in the treatment of lung adenocarcinoma with uncommon EGFR p.L747P and p.L747S mutations. Lung Cancer 2019, 133, 103–109. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.T.; Ning, W.W.; Li, J.; Huang, J.A. Exon 19 L747P mutation presented as a primary resistance to EGFR-TKI: A case report. J. Thorac. Dis. 2016, 8, E542–E546. [Google Scholar] [CrossRef]
- Yu, G.H.; Xie, X.X.; Sun, D.J.; Geng, J.Z.; Fu, F.H.; Zhang, L.M.; Wang, H.B. EGFR mutation L747P led to gefitinib resistance and accelerated liver metastases in a Chinese patient with lung adenocarcinoma. Int. J. Clin. Exp. Patho. 2015, 8, 8603–8606. [Google Scholar]
- Yamaguchi, F.; Fukuchi, K.; Yamazaki, Y.; Takayasu, H.; Tazawa, S.; Tateno, H.; Kato, E.; Wakabayashi, A.; Fujimori, M.; Iwasaki, T.; et al. Acquired resistance L747S mutation in an epidermal growth factor receptor-tyrosine kinase inhibitor-naive patient: A report of three cases. Oncol. Lett. 2014, 7, 357–360. [Google Scholar] [CrossRef]
- Leonetti, A.; Sharma, S.; Minari, R.; Perego, P.; Giovannetti, E.; Tiseo, M. Resistance mechanisms to osimertinib in EGFR-mutated non-small cell lung cancer. Br. J. Cancer 2019, 121, 725–737. [Google Scholar] [CrossRef]
- Mehlman, C.; Cadranel, J.; Rousseau-Bussac, G.; Lacave, R.; Pujals, A.; Girard, N.; Callens, C.; Gounant, V.; Theou-Anton, N.; Friard, S.; et al. Resistance mechanisms to osimertinib in EGFR-mutated advanced non-small-cell lung cancer: A multicentric retrospective French study. Lung Cancer 2019, 137, 149–156. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.O.; Cha, M.Y.; Kim, M.; Song, J.Y.; Lee, J.H.; Kim, Y.H.; Lee, Y.M.; Suh, K.H.; Son, J. Discovery of HM61713 as an orally available and mutant EGFR selective inhibitor. Cancer Res. 2014, 74. [Google Scholar] [CrossRef]
- Kim, E.S. Olmutinib: First global approval. Drugs 2016, 76, 1153–1157. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.W.; Lee, D.H.; Han, J.Y.; Lee, J.; Cho, B.C.; Kang, J.H.; Lee, K.H.; Cho, E.K.; Kim, J.S.; Min, Y.J.; et al. Safety, tolerability, and anti-tumor activity of olmutinib in non-small cell lung cancer with T790M mutation: A single arm, open label, phase 1/2 trial. Lung Cancer 2019, 135, 66–72. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Guo, X.; To, K.K.W.; Chen, Z.; Fang, X.; Luo, M.; Ma, C.; Xu, J.; Yan, S.; Fu, L. Olmutinib (HM61713) reversed multidrug resistance by inhibiting the activity of ATP-binding cassette subfamily G member 2 in vitro and in vivo. Acta Pharm. Sin. B 2018, 8, 563–574. [Google Scholar] [CrossRef]
- Quintieri, L.; Fantin, M.; Vizler, C. Identification of molecular determinants of tumor sensitivity and resistance to anticancer drugs. Adv. Exp. Med. Biol. 2007, 593, 95–104. [Google Scholar] [CrossRef] [PubMed]
- Song, H.N.; Jung, K.S.; Yoo, K.H.; Cho, J.; Lee, J.Y.; Lim, S.H.; Kim, H.S.; Sun, J.M.; Lee, S.H.; Ahn, J.S.; et al. Acquired C797S mutation upon treatment with a T790M-Specific third-generation EGFR inhibitor (HM61713) in non-small cell lung cancer. J. Thorac. Oncol. 2016, 11, E45–E47. [Google Scholar] [CrossRef]
- Roskoski, R., Jr. Small molecule inhibitors targeting the EGFR/ErbB family of protein-tyrosine kinases in human cancers. Pharmacol. Res. 2019, 139, 395–411. [Google Scholar] [CrossRef]
- Russo, A.; Franchina, T.; Ricciardi, G.R.R.; Smiroldo, V.; Picciotto, M.; Zanghi, M.; Rolfo, C.; Adamo, V. Third generation EGFR TKIs in EGFR-mutated NSCLC: Where are we now and where are we going. Crit Rev. Oncol. Hematol. 2017, 117, 38–47. [Google Scholar] [CrossRef]
- Meador, C.B.; Hata, A.N. Acquired resistance to targeted therapies in NSCLC: Updates and evolving insights. Pharmacol. Ther. 2020. [Google Scholar] [CrossRef]
- Baraibar, I.; Mezquita, L.; Gil-Bazo, I.; Planchard, D. Novel drugs targeting EGFR and HER2 exon 20 mutations in metastatic NSCLC. Crit. Rev. Oncol. Hematol. 2020, 148, 102906. [Google Scholar] [CrossRef] [PubMed]
- Bottaro, D.P.; Rubin, J.S.; Faletto, D.L.; Chan, A.M.L.; Kmiecik, T.E.; Vandewoude, G.F.; Aaronson, S.A. Identification of the hepatocyte growth-factor receptor as the C-Met protooncogene product. Science 1991, 251, 802–804. [Google Scholar] [CrossRef] [PubMed]
- Naldini, L.; Vigna, E.; Narsimhan, R.P.; Gaudino, G.; Zarnegar, R.; Michalopoulos, G.K.; Comoglio, P.M. Hepatocyte growth-factor (Hgf) stimulates the tyrosine kinase-activity of the receptor encoded by the protooncogene C-Met. Oncogene 1991, 6, 501–504. [Google Scholar] [PubMed]
- Graziani, A.; Gramaglia, D.; Cantley, L.C.; Comoglio, P.M. The tyrosine-phosphorylated hepatocyte growth-factor scatter factor receptor associates with phosphatidylinositol 3-Kinase. J. Biol. Chem. 1991, 266, 22087–22090. [Google Scholar]
- Fan, S.J.; Ma, Y.X.; Wang, J.A.; Yuan, R.Q.; Meng, Q.H.; Cao, Y.J.; Laterra, J.J.; Goldberg, I.D.; Rosen, E.M. The cytokine hepatocyte growth factor/scatter factor inhibits apoptosis and enhances DNA repair by a common mechanism involving signaling through phosphatidyl inositol 3 ’ kinase. Oncogene 2000, 19, 2212–2223. [Google Scholar] [CrossRef]
- Salgia, R. MET in lung cancer: Biomarker selection based on scientific rationale. Mol. Cancer Ther. 2017, 16, 555–565. [Google Scholar] [CrossRef]
- Zhang, Y.Z.; Xia, M.F.; Jin, K.; Wang, S.F.; Wei, H.; Fan, C.M.; Wu, Y.F.; Li, X.L.; Li, X.Y.; Li, G.Y.; et al. Function of the c-Met receptor tyrosine kinase in carcinogenesis and associated therapeutic opportunities. Mol. Cancer 2018, 17. [Google Scholar] [CrossRef]
- Zhang, Y.W.; Wang, L.M.; Jove, R.; Vande Woude, G.F. Requirement of Stat3 signaling for HGF/SF-Met mediated tumorigenesis. Oncogene 2002, 21, 217–226. [Google Scholar] [CrossRef]
- Beau-Faller, M.; Ruppert, A.M.; Voegeli, A.C.; Neuville, A.; Meyer, N.; Guerin, E.; Legrain, M.; Mennecier, B.; Wihlm, J.M.; Massard, G.; et al. MET gene copy number in non-small cell lung cancer: Molecular analysis in a targeted tyrosine kinase inhibitor naive cohort. J. Thorac. Oncol. 2008, 3, 331–339. [Google Scholar] [CrossRef]
- Tong, J.H.; Yeung, S.F.; Chan, A.W.; Chung, L.Y.; Chau, S.L.; Lung, R.W.; Tong, C.Y.; Chow, C.; Tin, E.K.; Yu, Y.H.; et al. MET amplification and exon 14 splice site mutation define unique molecular subgroups of non-small cell lung carcinoma with poor prognosis. Clin. Cancer Res. 2016, 22, 3048–3056. [Google Scholar] [CrossRef]
- Cappuzzo, F.; Janne, P.A.; Skokan, M.; Finocchiaro, G.; Rossi, E.; Ligorio, C.; Zucali, P.A.; Terracciano, L.; Toschi, L.; Roncalli, M.; et al. MET increased gene copy number and primary resistance to gefitinib therapy in non-small-cell lung cancer patients. Ann. Oncol. 2009, 20, 298–304. [Google Scholar] [CrossRef] [PubMed]
- Cappuzzo, F.; Marchetti, A.; Skokan, M.; Rossi, E.; Gajapathy, S.; Felicioni, L.; Del Grammastro, M.; Sciarrotta, M.G.; Buttitta, F.; Incarbone, M.; et al. Increased MET gene copy number negatively affects survival of surgically resected non-small-cell lung cancer patients. J. Clin. Oncol. 2009, 27, 1667–1674. [Google Scholar] [CrossRef]
- Tachibana, K.; Minami, Y.; Shiba-Ishii, A.; Kano, J.; Nakazato, Y.; Sato, Y.; Goya, T.; Noguchi, M. Abnormality of the hepatocyte growth factor/MET pathway in pulmonary adenocarcinogenesis. Lung Cancer 2012, 75, 181–188. [Google Scholar] [CrossRef] [PubMed]
- Kubo, T.; Yamamoto, H.; Lockwood, W.W.; Valencia, I.; Soh, J.; Peyton, M.; Jida, M.; Otani, H.; Fujii, T.; Ouchida, M.; et al. MET gene amplification or EGFR mutation activate MET in lung cancers untreated with EGFR tyrosine kinase inhibitors. Int. J. Cancer 2009, 124, 1778–1784. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Song, L.; Ai, T.; Zhang, Y.; Gao, Y.; Cui, J. Prognostic value of MET, cyclin D1 and MET gene copy number in non-small cell lung cancer. J. Biomed. Res. 2013, 27, 220–230. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; An, S.J.; Chen, Z.H.; Su, J.; Yan, H.H.; Wu, Y.L. MET expression plays differing roles in non-small-cell lung cancer patients with or without EGFR mutation. J. Thorac. Oncol. 2014, 9, 725–728. [Google Scholar] [CrossRef]
- Paik, P.K.; Drilon, A.; Fan, P.D.; Yu, H.; Rekhtman, N.; Ginsberg, M.S.; Borsu, L.; Schultz, N.; Berger, M.F.; Rudin, C.M.L.; et al. Response to MET inhibitors in patients with stage IV lung adenocarcinomas harboring MET mutations causing exon 14 skipping. Cancer Discov. 2015, 5, 842–849. [Google Scholar] [CrossRef]
- Ma, P.C. MET receptor juxtamembrane exon 14 alternative spliced variant: Novel cancer genomic predictive biomarker. Cancer Discov. 2015, 5, 802–805. [Google Scholar] [CrossRef]
- Ma, P.C.; Kijima, T.; Maulik, G.; Fox, E.A.; Sattler, M.; Griffin, J.D.; Johnson, B.E.; Salgia, R. c-MET mutational analysis in small cell lung cancer: Novel juxtamembrane domain mutations regulating cytoskeletal functions. Cancer Res. 2003, 63, 6272–6281. [Google Scholar]
- Parikh, P.K.; Ghate, M.D. Recent advances in the discovery of small molecule c-Met Kinase inhibitors. Eur. J. Med. Chem. 2018, 143, 1103–1138. [Google Scholar] [CrossRef]
- Pasquini, G.; Giaccone, G. C-MET inhibitors for advanced non-small cell lung cancer. Expert Opin. Investig. Drugs 2018, 27, 363–375. [Google Scholar] [CrossRef] [PubMed]
- Drilon, A.; Cappuzzo, F.; Ou, S.I.; Camidge, D.R. Targeting MET in lung cancer: Will expectations finally be MET? J. Thorac. Oncol. 2017, 12, 15–26. [Google Scholar] [CrossRef] [PubMed]
- Mo, H.N.; Liu, P. Targeting MET in cancer therapy. Chronic. Dis. Transl. Med. 2017, 3, 148–153. [Google Scholar] [CrossRef] [PubMed]
- Gozdzik-Spychalska, J.; Szyszka-Barth, K.; Spychalski, L.; Ramlau, K.; Wojtowicz, J.; Batura-Gabryel, H.; Ramlau, R. C-MET inhibitors in the treatment of lung cancer. Curr. Treat. Options Oncol. 2014, 15, 670–682. [Google Scholar] [CrossRef]
- Eathiraj, S.; Palma, R.; Volckova, E.; Hirschi, M.; France, D.S.; Ashwell, M.A.; Chan, T.C. Discovery of a novel mode of protein kinase inhibition characterized by the mechanism of inhibition of human mesenchymal-epithelial transition factor (c-Met) protein autophosphorylation by ARQ 197. J. Biol. Chem. 2011, 286, 20666–20676. [Google Scholar] [CrossRef]
- Gherardi, E.; Birchmeier, W.; Birchmeier, C.; Woude, G.V. Targeting MET in cancer: Rationale and progress. Nat. Rev. Cancer 2012, 12, 89–103. [Google Scholar] [CrossRef]
- Dussault, I.; Bellon, S.F. From Concept to Reality: The Long Road to c-Met and RON Receptor Tyrosine Kinase Inhibitors for the Treatment of Cancer. Anti Cancer Agent Med. Chem. 2009, 9, 221–229. [Google Scholar] [CrossRef]
- Zou, H.Y.; Li, Q.H.; Lee, J.H.; Arango, M.E.; McDonnell, S.R.; Yamazaki, S.; Koudriakova, T.B.; Alton, G.; Cui, J.J.; Kung, P.P.; et al. An orally available small-molecule inhibitor of c-met, PF-2341066, exhibits cytoreductive antitumor efficacy through antiproliferative and antiangiogenic mechanisms. Cancer Res. 2007, 67, 4408–4417. [Google Scholar] [CrossRef]
- Cui, J.J.; Tran-Dube, M.; Shen, H.; Nambu, M.; Kung, P.P.; Pairish, M.; Jia, L.; Meng, J.; Funk, L.; Botrous, I.; et al. Structure based drug design of crizotinib (PF-02341066), a potent and selective dual inhibitor of mesenchymal-epithelial transition factor (c-MET) kinase and anaplastic lymphoma kinase (ALK). J. Med. Chem. 2011, 54, 6342–6363. [Google Scholar] [CrossRef]
- Kang, J.; Chen, H.J.; Wang, Z.; Liu, J.; Li, B.; Zhang, T.; Yang, Z.; Wu, Y.L.; Yang, J.J. Osimertinib and Cabozantinib Combinatorial Therapy in an EGFR-Mutant Lung Adenocarcinoma Patient with Multiple MET Secondary-Site Mutations after Resistance to Crizotinib. J. Thorac. Oncol. 2018, 13, e49–e53. [Google Scholar] [CrossRef]
- Heist, R.S.; Sequist, L.V.; Borger, D.; Gainor, J.F.; Arellano, R.S.; Le, L.P.; Dias-Santagata, D.; Clark, J.W.; Engelman, J.A.; Shaw, A.T.; et al. Acquired resistance to crizotinib in NSCLC with MET exon 14 skipping. J. Thorac. Oncol. 2016, 11, 1242–1245. [Google Scholar] [CrossRef] [PubMed]
- Dong, H.J.; Li, P.; Wu, C.L.; Zhou, X.Y.; Lu, H.J.; Zhou, T. Response and acquired resistance to crizotinib in Chinese patients with lung adenocarcinomas harboring MET Exon 14 splicing alternations. Lung Cancer 2016, 102, 118–121. [Google Scholar] [CrossRef] [PubMed]
- Li, A.; Yang, J.J.; Zhang, X.C.; Zhang, Z.; Su, J.; Gou, L.Y.; Bai, Y.; Zhou, Q.; Yang, Z.; Han-Zhang, H.; et al. Acquired MET Y1248H and D1246N mutations mediate resistance to MET inhibitors in non-small cell lung cancer. Clin. Cancer Res. 2017, 23, 4929–4937. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Peled, N.; Greer, J.; Wu, W.; Choi, P.; Berger, A.H.; Wong, S.; Jen, K.Y.; Seo, Y.; Hann, B.; et al. MET exon 14 mutation encodes an actionable therapeutic target in lung adenocarcinoma. Cancer Res. 2017, 77, 4498–4505. [Google Scholar] [CrossRef] [PubMed]
- Ou, S.H.I.; Young, L.; Schrock, A.B.; Johnson, A.; Klempner, S.J.; Zhu, V.W.; Miller, V.A.; Ali, S.M. Emergence of preexisting MET Y1230C mutation as a resistance mechanism to crizotinib in NSCLC with MET exon 14 skipping. J. Thorac. Oncol. 2017, 12, 137–140. [Google Scholar] [CrossRef]
- Miller, M.; Ginalski, K.; Lesyng, B.; Nakaigawa, N.; Schmidt, L.; Zbar, B. Structural basis of oncogenic activation caused by point mutations in the kinase domain of the MET proto-oncogene: Modeling studies. Proteins 2001, 44, 32–43. [Google Scholar] [CrossRef]
- Liu, X.; Wang, Q.; Yang, G.; Marando, C.; Koblish, H.K.; Hall, L.M.; Fridman, J.S.; Behshad, E.; Wynn, R.; Li, Y.; et al. A novel kinase inhibitor, INCB28060, blocks c-MET-dependent signaling, neoplastic activities, and cross-talk with EGFR and HER-3. Clin. Cancer Res. 2011, 17, 7127–7138. [Google Scholar] [CrossRef]
- Baltschukat, S.; Engstler, B.S.; Huang, A.; Hao, H.X.; Tam, A.; Wang, H.Q.; Liang, J.; DiMare, M.T.; Bhang, H.C.; Wang, Y.; et al. Capmatinib (INC280) is active against models of non-small cell lung cancer and other cancer types with defined mechanisms of MET activation. Clin. Cancer Res. 2019, 25, 3164–3175. [Google Scholar] [CrossRef]
- Jia, H.; Dai, G.; Weng, J.; Zhang, Z.; Wang, Q.; Zhou, F.; Jiao, L.; Cui, Y.; Ren, Y.; Fan, S.; et al. Discovery of (S)-1-(1-(Imidazo[1,2-a]pyridin-6-yl)ethyl)-6-(1-methyl-1H-pyrazol-4-yl)-1H-[1,2, 3]triazolo[4,5-b]pyrazine (volitinib) as a highly potent and selective mesenchymal-epithelial transition factor (c-Met) inhibitor in clinical development for treatment of cancer. J. Med. Chem. 2014, 57, 7577–7589. [Google Scholar] [CrossRef]
- Gavine, P.R.; Ren, Y.; Han, L.; Lv, J.; Fan, S.; Zhang, W.; Xu, W.; Liu, Y.J.; Zhang, T.; Fu, H.; et al. Volitinib, a potent and highly selective c-Met inhibitor, effectively blocks c-Met signaling and growth in c-MET amplified gastric cancer patient-derived tumor xenograft models. Mol. Oncol. 2015, 9, 323–333. [Google Scholar] [CrossRef]
- Schuller, A.G.; Barry, E.R.; Jones, R.D.; Henry, R.E.; Frigault, M.M.; Beran, G.; Linsenmayer, D.; Hattersley, M.; Smith, A.; Wilson, J.; et al. The MET inhibitor AZD6094 (Savolitinib, HMPL-504) induces regression in papillary renal cell carcinoma patient-derived xenograft models. Clin. Cancer Res. 2015, 21, 2811–2819. [Google Scholar] [CrossRef] [PubMed]
- Henry, R.E.; Barry, E.R.; Castriotta, L.; Ladd, B.; Markovets, A.; Beran, G.; Ren, Y.; Zhou, F.; Adam, A.; Zinda, M.; et al. Acquired savolitinib resistance in non-small cell lung cancer arises via multiple mechanisms that converge on MET-independent mTOR and MYC activation. Oncotarget 2016, 7, 57651–57670. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Hu, P.; Fang, D.; Ni, L.; Xu, J. Electrostatic explanation of D1228V/H/N-induced c-Met resistance and sensitivity to type I and type II kinase inhibitors in targeted gastric cancer therapy. J. Mol. Modeling 2019, 25, 13. [Google Scholar] [CrossRef] [PubMed]
- Maritano, D.; Accornero, P.; Bonifaci, N.; Ponzetto, C. Two mutations affecting conserved residues in the Met receptor operate via different mechanisms. Oncogene 2000, 19, 1354–1361. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Huang, H. Anaplastic Lymphoma Kinase (ALK) receptor tyrosine kinase: A catalytic receptor with many faces. Int. J. Mol. Sci. 2018, 19, 3448. [Google Scholar] [CrossRef] [PubMed]
- Roskoski, R., Jr. Anaplastic lymphoma kinase (ALK): Structure, oncogenic activation, and pharmacological inhibition. Pharmacol. Res. 2013, 68, 68–94. [Google Scholar] [CrossRef]
- Guan, J.; Umapathy, G.; Yamazaki, Y.; Wolfstetter, G.; Mendoza, P.; Pfeifer, K.; Mohammed, A.; Hugosson, F.; Zhang, H.; Hsu, A.W.; et al. FAM150A and FAM150B are activating ligands for anaplastic lymphoma kinase. eLife 2015, 4, e09811. [Google Scholar] [CrossRef]
- Ducray, S.P.; Natarajan, K.; Garland, G.D.; Turner, S.D.; Egger, G. The transcriptional roles of ALK fusion proteins in tumorigenesis. Cancers 2019, 11, 1074. [Google Scholar] [CrossRef] [PubMed]
- Morris, S.W.; Kirstein, M.N.; Valentine, M.B.; Dittmer, K.G.; Shapiro, D.N.; Saltman, D.L.; Look, A.T. Fusion of a kinase gene, ALK, to a nucleolar protein gene, NPM, in non-Hodgkin’s lymphoma. Science 1994, 263, 1281–1284. [Google Scholar] [CrossRef] [PubMed]
- Box, J.K.; Paquet, N.; Adams, M.N.; Boucher, D.; Bolderson, E.; O’Byrne, K.J.; Richard, D.J. Nucleophosmin: From structure and function to disease development. BMC Mol. Biol. 2016, 17, 19. [Google Scholar] [CrossRef] [PubMed]
- Lamant, L.; Dastugue, N.; Pulford, K.; Delsol, G.; Mariame, B. A new fusion gene TPM3-ALK in anaplastic large cell lymphoma created by a (1;2)(q25;p23) translocation. Blood 1999, 93, 3088–3095. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, K.; Choi, Y.L.; Togashi, Y.; Soda, M.; Hatano, S.; Inamura, K.; Takada, S.; Ueno, T.; Yamashita, Y.; Satoh, Y.; et al. KIF5B-ALK, a novel fusion oncokinase identified by an immunohistochemistry-based diagnostic system for ALK-positive lung cancer. Clin. Cancer Res. 2009, 15, 3143–3149. [Google Scholar] [CrossRef] [PubMed]
- Togashi, Y.; Soda, M.; Sakata, S.; Sugawara, E.; Hatano, S.; Asaka, R.; Nakajima, T.; Mano, H.; Takeuchi, K. KLC1-ALK: A novel fusion in lung cancer identified using a formalin-fixed paraffin-embedded tissue only. PLoS ONE 2012, 7, e31323. [Google Scholar] [CrossRef] [PubMed]
- Kelly, L.M.; Barila, G.; Liu, P.; Evdokimova, V.N.; Trivedi, S.; Panebianco, F.; Gandhi, M.; Carty, S.E.; Hodak, S.P.; Luo, J.; et al. Identification of the transforming STRN-ALK fusion as a potential therapeutic target in the aggressive forms of thyroid cancer. Proc. Natl. Acad. Sci. USA 2014, 111, 4233–4238. [Google Scholar] [CrossRef] [PubMed]
- Jung, Y.; Kim, P.; Jung, Y.; Keum, J.; Kim, S.N.; Choi, Y.S.; Do, I.G.; Lee, J.; Choi, S.J.; Kim, S.; et al. Discovery of ALK-PTPN3 gene fusion from human non-small cell lung carcinoma cell line using next generation RNA sequencing. Genes Chromosomes Cancer 2012, 51, 590–597. [Google Scholar] [CrossRef] [PubMed]
- Iyevleva, A.G.; Raskin, G.A.; Tiurin, V.I.; Sokolenko, A.P.; Mitiushkina, N.V.; Aleksakhina, S.N.; Garifullina, A.R.; Strelkova, T.N.; Merkulov, V.O.; Ivantsov, A.O.; et al. Novel ALK fusion partners in lung cancer. Cancer Lett. 2015, 362, 116–121. [Google Scholar] [CrossRef]
- Jiang, J.; Wu, X.; Tong, X.; Wei, W.; Chen, A.; Wang, X.; Shao, Y.W.; Huang, J. GCC2-ALK as a targetable fusion in lung adenocarcinoma and its enduring clinical responses to ALK inhibitors. Lung Cancer 2018, 115, 5–11. [Google Scholar] [CrossRef]
- Soda, M.; Choi, Y.L.; Enomoto, M.; Takada, S.; Yamashita, Y.; Ishikawa, S.; Fujiwara, S.I.; Watanabe, H.; Kurashina, K.; Hatanaka, H.; et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature 2007, 448, 561–566. [Google Scholar] [CrossRef]
- Horn, L.; Pao, W. EML4-ALK: Honing in on a new target in non-small-cell lung cancer. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2009, 27, 4232–4235. [Google Scholar] [CrossRef]
- Lin, Y.T.; Yu, C.J.; Yang, J.C.H.; Shih, J.Y. Anaplastic Lymphoma Kinase (ALK) kinase domain mutation following ALK Inhibitor(s) failure in advanced ALK positive Non-Small-Cell Lung Cancer: Analysis and literature review. Clin. Lung Cancer 2016, 17, e77–e94. [Google Scholar] [CrossRef]
- Shaw, A.T.; Yeap, B.Y.; Solomon, B.J.; Riely, G.J.; Gainor, J.; Engelman, J.A.; Shapiro, G.I.; Costa, D.B.; Ou, S.H.I.; Butaney, M.; et al. Effect of crizotinib on overall survival in patients with advanced non-small-cell lung cancer harbouring ALK gene rearrangement: A retrospective analysis. Lancet Oncol. 2011, 12, 1004–1012. [Google Scholar] [CrossRef]
- Narayanan, D.; Gani, O.A.B.S.M.; Gruber, F.X.E.; Engh, R.A. Data driven polypharmacological drug design for lung cancer: Analyses for targeting ALK, MET, and EGFR. J. Cheminform 2017, 9, 43. [Google Scholar] [CrossRef] [PubMed]
- Huang, Q.; Johnson, T.W.; Bailey, S.; Brooun, A.; Bunker, K.D.; Burke, B.J.; Collins, M.R.; Cook, A.S.; Cui, J.J.; Dack, K.N.; et al. Design of potent and selective inhibitors to overcome clinical anaplastic lymphoma kinase mutations resistant to crizotinib. J. Med. Chem. 2014, 57, 1170–1187. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.L.; Soda, M.; Yamashita, Y.; Ueno, T.; Takashima, J.; Nakajima, T.; Yatabe, Y.; Takeuchi, K.; Hamada, T.; Haruta, H.; et al. EML4-ALK mutations in lung cancer that confer resistance to ALK inhibitors. N. Engl. J. Med. 2010, 363, 1734–1739. [Google Scholar] [CrossRef] [PubMed]
- Gainor, J.F.; Dardaei, L.; Yoda, S.; Friboulet, L.; Leshchiner, I.; Katayama, R.; Dagogo-Jack, I.; Gadgeel, S.; Schultz, K.; Singh, M.; et al. Molecular mechanisms of resistance to first- and second-generation ALK inhibitors in ALK-rearranged lung cancer. Cancer Discov. 2016, 6, 1118–1133. [Google Scholar] [CrossRef] [PubMed]
- Katayama, R.; Shaw, A.T.; Khan, T.M.; Mino-Kenudson, M.; Solomon, B.J.; Halmos, B.; Jessop, N.A.; Wain, J.C.; Yeo, A.T.; Benes, C.; et al. Mechanisms of acquired crizotinib resistance in ALK-rearranged lung Cancers. Sci. Transl. Med. 2012, 4, 120ra117. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Kim, T.M.; Kim, D.W.; Go, H.; Keam, B.; Lee, S.H.; Ku, J.L.; Chung, D.H.; Heo, D.S. Heterogeneity of genetic changes associated with acquired crizotinib resistance in ALK-rearranged lung cancer. J. Thorac. Oncol. 2013, 8, 415–422. [Google Scholar] [CrossRef]
- Huang, D.; Kim, D.W.; Kotsakis, A.; Deng, S.; Lira, P.; Ho, S.N.; Lee, N.V.; Vizcarra, P.; Cao, J.Q.; Christensen, J.G.; et al. Multiplexed deep sequencing analysis of ALK kinase domain identifies resistance mutations in relapsed patients following crizotinib treatment. Genomics 2013, 102, 157–162. [Google Scholar] [CrossRef]
- Ignatius Ou, S.H.; Azada, M.; Hsiang, D.J.; Herman, J.M.; Kain, T.S.; Siwak-Tapp, C.; Casey, C.; He, J.; Ali, S.M.; Klempner, S.J.; et al. Next-generation sequencing reveals a Novel NSCLC ALK F1174V mutation and confirms ALK G1202R mutation confers high-level resistance to alectinib (CH5424802/RO5424802) in ALK-rearranged NSCLC patients who progressed on crizotinib. J. Thorac. Oncol. 2014, 9, 549–553. [Google Scholar]
- Friboulet, L.; Li, N.; Katayama, R.; Lee, C.C.; Gainor, J.F.; Crystal, A.S.; Michellys, P.Y.; Awad, M.M.; Yanagitani, N.; Kim, S.; et al. The ALK inhibitor ceritinib overcomes crizotinib resistance in non-small cell lung cancer. Cancer Discov. 2014, 4, 662–673. [Google Scholar] [CrossRef]
- Saber, A.; Van der Wekken, A.J.; Kok, K.; Terpstra, M.M.; Bosman, L.J.; Mastik, M.F.; Timens, W.; Schuuring, E.; Hiltermann, T.J.N.; Groen, H.J.M.; et al. Genomic aberrations in crizotinib resistant lung adenocarcinoma samples identified by transcriptome sequencing. PLoS ONE 2016, 11, e0153065. [Google Scholar] [CrossRef] [PubMed]
- Ai, X.; Niu, X.; Chang, L.; Chen, R.; Ou, S.-H.I.; Lu, S. Next generation sequencing reveals a novel ALK G1128A mutation resistant to crizotinib in an ALK-Rearranged NSCLC patient. Lung Cancer 2018, 123, 83–86. [Google Scholar] [CrossRef] [PubMed]
- Bresler, S.C.; Weiser, D.A.; Huwe, P.J.; Park, J.H.; Krytska, K.; Ryles, H.; Laudenslager, M.; Rappaport, E.F.; Wood, A.C.; McGrady, P.W.; et al. ALK mutations confer differential oncogenic activation and sensitivity to ALK inhibition therapy in neuroblastoma. Cancer Cell 2014, 26, 682–694. [Google Scholar] [CrossRef] [PubMed]
- Toyokawa, G.; Hirai, F.; Inamasu, E.; Yoshida, T.; Nosaki, K.; Takenaka, T.; Yamaguchi, M.; Seto, T.; Takenoyama, M.; Ichinose, Y. Secondary mutations at I1171 in the ALK gene confer resistance to both Crizotinib and Alectinib. J. Thorac. Oncol. 2014, 9, 86–87. [Google Scholar] [CrossRef]
- Sakamoto, H.; Tsukaguchi, T.; Hiroshima, S.; Kodama, T.; Kobayashi, T.; Fukami, T.A.; Oikawa, N.; Tsukuda, T.; Ishii, N.; Aoki, Y. CH5424802, a selective ALK inhibitor capable of blocking the resistant gatekeeper mutant. Cancer Cell 2011, 19, 679–690. [Google Scholar] [CrossRef]
- Kodama, T.; Tsukaguchi, T.; Yoshida, M.; Kondoh, O.; Sakamoto, H. Selective ALK inhibitor alectinib with potent antitumor activity in models of crizotinib resistance. Cancer Lett. 2014, 351, 215–221. [Google Scholar] [CrossRef]
- Paik, J.; Dhillon, S. Alectinib: A Review in Advanced, ALK-Positive NSCLC. Drugs 2018, 78, 1247–1257. [Google Scholar] [CrossRef]
- Ou, S.-H.I.; Greenbowe, J.; Khan, Z.U.; Azada, M.C.; Ross, J.S.; Stevens, P.J.; Ali, S.M.; Miller, V.A.; Gitlitz, B. I1171 missense mutation (particularly I1171N) is a common resistance mutation in ALK-positive NSCLC patients who have progressive disease while on alectinib and is sensitive to ceritinib. Lung Cancer 2015, 88, 231–234. [Google Scholar] [CrossRef]
- Ou, S.H.I.; Klempner, S.J.; Greenbowe, J.R.; Azada, M.; Schrock, A.B.; Ali, S.M.; Ross, J.S.; Stephens, P.J.; Miller, V.A. Identification of a novel HIP1-ALK fusion variant in Non-Small-Cell Lung Cancer (NSCLC) and discovery of ALK I1171 (I1171N/S) mutations in two ALK-rearranged NSCLC patients with resistance to Alectinib. J. Thorac. Oncol. 2014, 9, 1821–1825. [Google Scholar] [CrossRef]
- Ou, S.-H.; Milliken, J.C.; Azada, M.C.; Miller, V.A.; Ali, S.M.; Klempner, S.J. ALK F1174V mutation confers sensitivity while ALK I1171 mutation confers resistance to alectinib. The importance of serial biopsy post progression. Lung Cancer 2016, 91, 70–72. [Google Scholar] [CrossRef]
- He, M.; Li, W.; Zheng, Q.; Zhang, H. A molecular dynamics investigation into the mechanisms of alectinib resistance of three ALK mutants. J. Cell. Biochem. 2018, 119, 5332–5342. [Google Scholar] [CrossRef] [PubMed]
- Katayama, R.; Friboulet, L.; Koike, S.; Lockerman, E.L.; Khan, T.M.; Gainor, J.F.; Iafrate, A.J.; Takeuchi, K.; Taiji, M.; Okuno, Y.; et al. Two novel ALK mutations mediate acquired resistance to the next-generation ALK inhibitor alectinib. Clin. Cancer Res. 2014, 20, 5686–5696. [Google Scholar] [CrossRef] [PubMed]
- Soria, J.C.; Tan, D.S.W.; Chiari, R.; Wu, Y.L.; Paz-Ares, L.; Wolf, J.; Geater, S.L.; Orlov, S.; Cortinovis, D.; Yu, C.J.; et al. First-line ceritinib versus platinum-based chemotherapy in advanced ALK-rearranged non-small-cell lung cancer (ASCEND-4): A randomised, open-label, phase 3 study. Lancet 2017, 389, 917–929. [Google Scholar] [CrossRef]
- Toyokawa, G.; Inamasu, E.; Shimamatsu, S.; Yoshida, T.; Nosaki, K.; Hirai, F.; Yamaguchi, M.; Seto, T.; Takenoyama, M.; Ichinose, Y. Identification of a Novel ALK G1123S Mutation in a Patient with ALK-rearranged Non-small-cell Lung Cancer Exhibiting Resistance to Ceritinib. J. Thorac. Oncol. 2015, 10, 55–57. [Google Scholar] [CrossRef] [PubMed]
- He, M.Y.; Li, W.K.; Meiler, J.; Zheng, Q.C.; Zhang, H.X. Insight on mutation-induced resistance to anaplastic lymphoma kinase inhibitor ceritinib from molecular dynamics simulations. Biopolymers 2019, 110, e23257. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.Y.; Ho, C.C.; Shih, J.Y. Multiple acquired resistance mutations of the ALK tyrosine kinase domainafter sequential use of ALK inhibitors. J. Thorac. Oncol. 2017, 12, e49–e51. [Google Scholar] [CrossRef]
- Swerdlow, S.H.; Campo, E.; Pileri, S.A.; Harris, N.L.; Stein, H.; Siebert, R.; Advani, R.; Ghielmini, M.; Salles, G.A.; Zelenetz, A.D.; et al. The 2016 revision of the World Health Organization classification of lymphoid neoplasms. Blood 2016, 127, 2375–2390. [Google Scholar] [CrossRef]
- Cazzola, M. Introduction to a review series on myeloproliferative neoplasms. Blood 2017, 129, 659. [Google Scholar] [CrossRef]
- Houshmand, M.; Simonetti, G.; Circosta, P.; Gaidano, V.; Cignetti, A.; Martinelli, G.; Saglio, G.; Gale, R.P. Chronic myeloid leukemia stem cells. Leukemia 2019, 33, 1543–1556. [Google Scholar] [CrossRef]
- Hallek, M. Chronic lymphocytic leukemia: 2015 Update on diagnosis, risk stratification, and treatment. Am. J. Hematol. 2015, 90, 446–460. [Google Scholar] [CrossRef]
- Austen, B.; Skowronska, A.; Baker, C.; Powell, J.E.; Gardiner, A.; Oscier, D.; Majid, A.; Dyer, M.; Siebert, R.; Taylor, A.M.; et al. Mutation status of the residual ATM allele is an important determinant of the cellular response to chemotherapy and survival in patients with chronic lymphocytic leukemia containing an 11q deletion. J. Clin. Oncol. 2007, 25, 5448–5457. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.L.; Lawrence, M.S.; Wan, Y.Z.; Stojanov, P.; Sougnez, C.; Stevenson, K.; Werner, L.; Sivachenko, A.; DeLuca, D.S.; Zhang, L.; et al. SF3B1 and other novel cancer genes in chronic lymphocytic leukemia. N. Engl. J. Med. 2011, 365, 2497–2506. [Google Scholar] [CrossRef] [PubMed]
- Rose-Zerilli, M.J.J.; Forster, J.; Parker, H.; Parker, A.; Rodriguez, A.E.; Chaplin, T.; Gardiner, A.; Steele, A.J.; Collins, A.; Young, B.D.; et al. ATM mutation rather than BIRC3 deletion and/or mutation predicts reduced survival in 11q-deleted chronic lymphocytic leukemia: Data from the UK LRF CLL4 trial. Haematologica 2014, 99, 736–742. [Google Scholar] [CrossRef] [PubMed]
- Puente, X.S.; Lopez-Otin, C. The evolutionary biography of chronic lymphocytic leukemia. Nat. Genet. 2013, 45, 229–231. [Google Scholar] [CrossRef] [PubMed]
- Juarez-Salcedo, L.M.; Castillo, J.J. Lymphoplasmacytic lymphoma and marginal zone lymphoma. Hematol. Oncol. Clin. N. Am. 2019, 33, 639–656. [Google Scholar] [CrossRef]
- Diamond, B.; Kumar, A. Mantle cell lymphoma: Current and emerging treatment strategies and unanswered questions. Hematol. Oncol. Clin. N. Am. 2019, 33, 613–626. [Google Scholar] [CrossRef]
- Konopka, J.B.; Watanabe, S.M.; Witte, O.N. An alteration of the human c-abl protein in K562 leukemia cells unmasks associated tyrosine kinase activity. Cell 1984, 37, 1035–1042. [Google Scholar] [CrossRef]
- Pui, C.-H.; Roberts, K.G.; Yang, J.J.; Mullighan, C.G. Philadelphia chromosome-like acute lymphoblastic leukemia. Clin. Lymphoma Myeloma Leuk. 2017, 17, 464–470. [Google Scholar] [CrossRef]
- Wang, J.Y.J. The capable ABL: What is its biological function? Mol. Cell. Biol. 2014, 34, 1188–1197. [Google Scholar] [CrossRef]
- Kharbanda, S.; Ren, R.; Pandey, P.; Shafman, T.D.; Feller, S.M.; Weichselbaum, R.R.; Kufe, D.W. Activation of the c-Abl tyrosine kinase in the stress response to DNA-damaging agents. Nature 1995, 376, 785–788. [Google Scholar] [CrossRef]
- Lewis, J.M.; Baskaran, R.; Taagepera, S.; Schwartz, M.A.; Wang, J.Y. Integrin regulation of c-Abl tyrosine kinase activity and cytoplasmic-nuclear transport. Proc. Natl. Acad. Sci. USA 1996, 93, 15174–15179. [Google Scholar] [CrossRef] [PubMed]
- Barila, D.; Superti-Furga, G. An intramolecular SH3-domain interaction regulates c-Abl activity. Nat. Genet. 1998, 18, 280–282. [Google Scholar] [CrossRef] [PubMed]
- Nowell, P.C.; Hungerford, D.A. Chromosome studies on normal and leukemic human leukocytes. J. Natl. Cancer Inst. 1960, 25, 85–109. [Google Scholar] [PubMed]
- Nowell, P.C.; Hungerford, D.A. Minute chromosome in human chronic granulocytic leukemia. Science 1960, 132, 1497-1497. [Google Scholar]
- Rowley, J.D. New consistent chromosomal abnormality in chronic myelogenous leukemia identified by quinacrine fluorescence and giemsa staining. Nature 1973, 243, 290–293. [Google Scholar] [CrossRef] [PubMed]
- Kurzrock, R.; Kantarjian, H.M.; Druker, B.J.; Talpaz, M. Philadelphia chromosome-positive leukemias: From basic mechanisms to molecular therapeutics. Ann. Intern. Med. 2003, 138, 819–830. [Google Scholar] [CrossRef]
- Laurent, E.; Talpaz, M.; Kantarjian, H.; Kurzrock, R. The BCR gene and Philadelphia chromosome-positive leukemogenesis. Cancer Res. 2001, 61, 2343–2355. [Google Scholar]
- Arlinghaus, R.B. The involvement of Bcr in leukemias with the Philadelphia chromosome. Crit. Rev. Oncog. 1998, 9, 1–18. [Google Scholar] [CrossRef]
- Lugo, T.G.; Pendergast, A.M.; Muller, A.J.; Witte, O.N. Tyrosine kinase-activity and transformation potency of Bcr-Abl oncogene products. Science 1990, 247, 1079–1082. [Google Scholar] [CrossRef]
- Bedi, A.; Zehnbauer, B.A.; Barber, J.P.; Sharkis, S.J.; Jones, R.J. Inhibition of apoptosis by Bcr-Abl in chronic myeloid-leukemia. Blood 1994, 83, 2038–2044. [Google Scholar] [CrossRef]
- Sawyers, C.L.; Mclaughlin, J.; Witte, O.N. Genetic requirement for ras in the transformation of fibroblasts and hematopoietic-cells by the Bcr-Abl oncogene. J. Exp. Med. 1995, 181, 307–313. [Google Scholar] [CrossRef]
- Tauchi, T.; Okabe, S.; Miyazawa, K.; Ohyashiki, K. The tetramerization domain-independent Ras activation by BCR-ABL oncoprotein in hematopoietic cells. Int. J. Oncol. 1998, 12, 1269–1276. [Google Scholar] [CrossRef] [PubMed]
- Cheng, K.D.; Kurzrock, R.; Qiu, X.G.; Estrov, Z.; Ku, S.; Dulski, K.M.; Wang, J.Y.J.; Talpaz, M. Reduced focal adhesion kinase and paxillin phosphorylation in BCR-ABL-transfected cells. Cancer 2002, 95, 440–450. [Google Scholar] [CrossRef] [PubMed]
- Gotoh, A.; Miyazawa, K.; Ohyashiki, K.; Tauchi, T.; Boswell, H.S.; Broxmeyer, H.E.; Toyama, K. Tyrosine phosphorylation and activation of Focal Adhesion Kinase (P125(Fak)) by Bcr-Abl oncoprotein. Exp. Hematol. 1995, 23, 1153–1159. [Google Scholar] [PubMed]
- Ilaria, R.L.; VanEtten, R.A. P210 and P190(BCR/ABL) induce the tyrosine phosphorylation and DNA binding activity of multiple specific STAT family members. J. Biol. Chem. 1996, 271, 31704–31710. [Google Scholar] [CrossRef] [PubMed]
- WilsonRawls, J.; Xie, S.H.; Liu, J.X.; Laneuville, P.; Arlinghaus, R.B. P210 Bcr-Abl interacts with the interleukin 3 receptor beta(c) subunit and constitutively induces its tyrosine phosphorylation. Cancer Res. 1996, 56, 4549. [Google Scholar]
- Takeda, N.; Shibuya, M.; Maru, Y. The BCR-ABL oncoprotein potentially interacts with the xeroderma pigmentosum group B protein. Proc. Natl. Acad. Sci. USA 1999, 96, 203–207. [Google Scholar] [CrossRef]
- Slupianek, A.; Schmutte, C.; Tombline, G.; Nieborowska-Skorska, M.; Hoser, G.; Nowicki, M.O.; Pierce, A.J.; Fishel, R.; Skorski, T. BCR/ABL regulates mammalian RecA homologs, resulting in drug resistance. Mol. Cell 2001, 8, 795–806. [Google Scholar] [CrossRef]
- Kalaycio, M.E. Chronic myelogenous leukemia: The news you have and haven’t heard. Cleve. Clin. J. Med. 2001, 68, 913–927. [Google Scholar] [CrossRef]
- Trela, E.; Glowacki, S.; Blasiak, J. Therapy of chronic myeloid leukemia: Twilight of the imatinib era? ISRN Oncol. 2014, 2014, 596483. [Google Scholar] [CrossRef]
- Mauro, M.J.; Druker, B.J. STI571: Targeting BCR-ABL as therapy for CML. Oncologist 2001, 6, 233–238. [Google Scholar] [CrossRef] [PubMed]
- Nagar, B.; Bornmann, W.G.; Pellicena, P.; Schindler, T.; Veach, D.R.; Miller, W.T.; Clarkson, B.; Kuriyan, J. Crystal structures of the kinase domain of c-Abl in complex with the small molecule inhibitors PD173955 and imatinib (STI-571). Cancer Res. 2002, 62, 4236–4243. [Google Scholar] [PubMed]
- Schindler, T.; Bornmann, W.; Pellicena, P.; Miller, W.T.; Clarkson, B.; Kuriyan, J. Structural mechanism for STI-571 inhibition of abelson tyrosine kinase. Science 2000, 289, 1938–1942. [Google Scholar] [CrossRef] [PubMed]
- Gorre, M.E.; Mohammed, M.; Ellwood, K.; Hsu, N.; Paquette, R.; Rao, P.N.; Sawyers, C.L. Clinical resistance to STI-571 cancer therapy caused by BCR-ABL gene mutation or amplification. Science 2001, 293, 876–880. [Google Scholar] [CrossRef]
- Milojkovic, D.; Apperley, J. Mechanisms of resistance to imatinib and second-generation tyrosine inhibitors in chronic myeloid leukemia. Clin. Cancer Res. 2009, 15, 7519–7527. [Google Scholar] [CrossRef]
- Schnittger, S.; Bacher, U.; Dicker, F.; Kern, W.; Alpermann, T.; Haferlach, T.; Haferlach, C. Associations between imatinib resistance conferring mutations and Philadelphia positive clonal cytogenetic evolution in CML. Genes Chromosomes Cancer 2010, 49, 910–918. [Google Scholar] [CrossRef]
- Tanaka, R.; Kimura, S.; Ashihara, E.; Yoshimura, M.; Takahashi, N.; Wakita, H.; Itoh, K.; Nishiwaki, K.; Suzuki, K.; Nagao, R.; et al. Rapid automated detection of ABL kinase domain mutations in imatinib-resistant patients. Cancer Lett. 2011, 312, 228–234. [Google Scholar] [CrossRef]
- Gibbons, D.L.; Pricl, S.; Kantarjian, H.; Cortes, J.; Quintas-Cardama, A. The rise and fall of gatekeeper mutations? The BCR-ABL1 T315I paradigm. Cancer 2012, 118, 293–299. [Google Scholar] [CrossRef]
- Xu, H.L.; Wang, Z.J.; Liang, X.M.; Li, X.; Shi, Z.; Zhou, N.; Bao, J.K. In silico identification of novel kinase inhibitors targeting wild-type and T315I mutant ABL1 from FDA-approved drugs. Mol. Biosyst. 2014, 10, 1524–1537. [Google Scholar] [CrossRef]
- Young, M.A.; Shah, N.P.; Chao, L.H.; Seeliger, M.; Milanov, Z.V.; Biggs, W.H., 3rd; Treiber, D.K.; Patel, H.K.; Zarrinkar, P.P.; Lockhart, D.J.; et al. Structure of the kinase domain of an imatinib-resistant Abl mutant in complex with the Aurora kinase inhibitor VX-680. Cancer Res. 2006, 66, 1007–1014. [Google Scholar] [CrossRef]
- Branford, S.; Rudzki, Z.; Walsh, S.; Parkinson, I.; Grigg, A.; Szer, J.; Taylor, K.; Herrmann, R.; Seymour, J.F.; Arthur, C.; et al. Detection of BCR-ABL mutations in patients with CML treated with imatinib is virtually always accompanied by clinical resistance, and mutations in the ATP phosphate-binding loop (P-loop) are associated with a poor prognosis. Blood 2003, 102, 276–283. [Google Scholar] [CrossRef] [PubMed]
- White, D.; Saunders, V.; Lyons, A.B.; Branford, S.; Grigg, A.; To, L.B.; Hughes, T. In vitro sensitivity to imatinib-induced inhibition of ABL kinase activity is predictive of molecular response in patients with de novo CML. Blood 2005, 106, 2520–2526. [Google Scholar] [CrossRef]
- Jabbour, E.; Kantarjian, H.; Jones, D.; Talpaz, M.; Bekele, N.; O’Brien, S.; Zhou, X.; Luthra, R.; Garcia-Manero, G.; Giles, F.; et al. Frequency and clinical significance of BCR-ABL mutations in patients with chronic myeloid leukemia treated with imatinib mesylate. Leukemia 2006, 20, 1767–1773. [Google Scholar] [CrossRef] [PubMed]
- Cang, S.; Liu, D. P-loop mutations and novel therapeutic approaches for imatinib failures in chronic myeloid leukemia. J. Hematol. Oncol. 2008, 1, 15. [Google Scholar] [CrossRef] [PubMed]
- Roumiantsev, S.; Shah, N.P.; Gorre, M.E.; Nicoll, J.; Brasher, B.B.; Sawyers, C.L.; Van Etten, R.A. Clinical resistance to the kinase inhibitor STI-571 in chronic myeloid leukemia by mutation of Tyr-253 in the Abl kinase domain P-loop. Proc. Natl. Acad. Sci. USA 2002, 99, 10700–10705. [Google Scholar] [CrossRef] [PubMed]
- Nagar, B.; Hantschel, O.; Seeliger, M.; Davies, J.M.; Weiss, W.I.; Superti-Furga, G.; Kuriyan, J. Organization of the SH3-SH2 unit in active and inactive forms of the c-Abl tyrosine kinase. Mol. Cell 2006, 21, 787–798. [Google Scholar] [CrossRef]
- Smith, K.M.; Yacobi, R.; Van Etten, R.A. Autoinhibition of Bcr-Abl through its SH3 domain. Mol. Cell 2003, 12, 27–37. [Google Scholar] [CrossRef]
- Colicelli, J. ABL tyrosine kinases: Evolution of function, regulation, and specificity. Sci. Signal. 2010, 3. [Google Scholar] [CrossRef]
- Von Bubnoff, N.; Schneller, F.; Peschel, C.; Duyster, J. BCR-ABL gene mutations in relation to clinical resistance of Philadelphia-chromosome-positive leukaemia to STI571: A prospective study. Lancet 2002, 359, 487–491. [Google Scholar] [CrossRef]
- Radich, J.P.; Mauro, M.J. Tyrosine kinase inhibitor treatment for newly diagnosed chronic myeloid leukemia. Hematol. Oncol. Clin. N. Am. 2017, 31, 577–587. [Google Scholar] [CrossRef]
- Kennedy, J.A.; Hobbs, G. Tyrosine kinase inhibitors in the treatment of chronic-phase CML: Strategies for frontline decision-making. Curr. Hematol. Malig. Rep. 2018, 13, 202–211. [Google Scholar] [CrossRef] [PubMed]
- Chopade, P.; Akard, L.P. Improving outcomes in chronic myeloid leukemia over time in the era of tyrosine kinase inhibitors. Clin. Lymph. Myelom. Leuk. 2018, 18, 710–723. [Google Scholar] [CrossRef] [PubMed]
- Padmanabhan, S.; Ravella, S.; Curiel, T.; Giles, F. Current status of therapy for chronic myeloid leukemia: A review of drug development. Future Oncol. 2008, 4, 359–377. [Google Scholar] [CrossRef] [PubMed]
- Shah, N.P.; Tran, C.; Lee, F.Y.; Chen, P.; Norris, D.; Sawyers, C.L. Overriding imatinib resistance with a novel ABL kinase inhibitor. Science 2004, 305, 399–401. [Google Scholar] [CrossRef]
- Lombardo, L.J.; Lee, F.Y.; Chen, P.; Norris, D.; Barrish, J.C.; Behnia, K.; Castaneda, S.; Cornelius, L.A.; Das, J.; Doweyko, A.M.; et al. Discovery of N-(2-chloro-6-methyl- phenyl)-2-(6-(4-(2-hydroxyethyl)- piperazin-1-yl)-2-methylpyrimidin-4- ylamino)thiazole-5-carboxamide (BMS-354825), a dual Src/Abl kinase inhibitor with potent antitumor activity in preclinical assays. J. Med. Chem. 2004, 47, 6658–6661. [Google Scholar] [CrossRef] [PubMed]
- Brattas, M.K.; Reikvam, H.; Tvedt, T.H.A.; Bruserud, O. Dasatinib as an investigational drug for the treatment of Philadelphia chromosome-positive acute lymphoblastic leukemia in adults. Expert Opin. Investig. Drugs 2019, 28, 411–420. [Google Scholar] [CrossRef] [PubMed]
- Schittenhelm, M.M.; Shiraga, S.; Schroeder, A.; Corbin, A.S.; Griffith, D.; Lee, F.Y.; Bokemeyer, C.; Deininger, M.W.; Druker, B.J.; Heinrich, M.C. Dasatinib (BMS-354825), a dual SRC/ABL kinase inhibitor, inhibits the kinase activity of wild-type, juxtamembrane, and activation loop mutant KIT isoforms associated with human malignancies. Cancer Res. 2006, 66, 473–481. [Google Scholar] [CrossRef]
- Agrawal, M.; Garg, R.J.; Cortes, J.; Quintas-Cardama, A. Tyrosine kinase inhibitors: The first decade. Curr. Hematol. Malig. Rep. 2010, 5, 70–80. [Google Scholar] [CrossRef]
- Kantarjian, H.; Ottmann, O.; Cortes, J.; Wassmann, B.; Jones, D.; Hochhaus, A.; Alland, L.; Dugan, M.; Albitar, M.; Giles, F. AMN107, a novel aminopyrimidine inhibitor of Bcr-Abl, has significant activity in imatinib-resistant Bcr-Abl positive chronic myeloid leukemia (CML). J. Clin. Oncol. 2005, 23, 195s-195s. [Google Scholar] [CrossRef]
- Golemovic, M.; Giles, F.J.; Beran, M.; Cortes, J.; Manshouri, T.; Kantarjian, H.; Verstovsek, S. AMN107, a novel aminopyrimidine inhibitor of BCR-ABL, has pre-clinical activity against imatinib mesylate-resistant chronic myeloid leukemia (CML). Blood 2004, 104, 547a-547a. [Google Scholar] [CrossRef]
- Giles, F.G.; Ottmann, O.T.; Cortes, J.; Wassmann, B.; Bhalla, K.; Jones, D.; Hochhaus, A.; Rae, P.E.; Tanaka, C.; Alland, L.; et al. AMN107, a novel aminopyrimidine inhibitor of Bcr-Abl, has significant activity in imatinib-resistant Bcr-Abl positive chronic myeloid leukemia (CML). Brit. J. Haematol. 2005, 129, 3. [Google Scholar]
- Weisberg, E.; Manley, P.W.; Breitenstein, W.; Bruggen, J.; Cowan-Jacob, S.W.; Ray, A.; Huntly, B.; Fabbro, D.; Fendrich, G.; Hall-Meyers, E.; et al. Characterization of AMN107, a selective inhibitor of native and mutant Bcr-Abl. Cancer Cell 2005, 7, 129–141. [Google Scholar] [CrossRef]
- Weisberg, E.; Manley, P.; Mestan, J.; Cowan-Jacob, S.; Ray, A.; Griffin, J.D. AMN107 (nilotinib): A novel and selective inhibitor of BCR-ABL (vol 94, pg 1765, 2016). Brit. J. Cancer 2019, 121, 282. [Google Scholar] [CrossRef] [PubMed]
- Von Bubnoff, N.; Gorantla, S.H.P.; Kancha, R.K.; Lordick, F.; Peschel, C.; Duyster, J. The systemic mastocytosis-specific activating cKit mutation D816V can be inhibited by the tyrosine kinase inhibitor AMN107. Leukemia 2005, 19, 1670–1671. [Google Scholar] [CrossRef] [PubMed]
- Gleixner, K.V.; Mayerhofer, M.; Aichberger, K.J.; Derdak, S.; Sonneck, K.; Bohm, A.; Gruze, A.; Samorapoompichit, P.; Manley, P.W.; Fabbro, D.; et al. PKC412 inhibits in vitro growth of neoplastic human mast cells expressing the D816V-mutated variant of KIT: Comparison with AMN107, imatinib, and cladribine (2CdA) and evaluation of cooperative drug effects. Blood 2006, 107, 752–759. [Google Scholar] [CrossRef]
- Montemurro, M.; Schoffski, P.; Reichardt, P.; Gelderblom, H.; Schutte, J.; Hartmann, J.T.; von Moos, R.; Seddon, B.; Joensuu, H.; Wendtner, C.M.; et al. Nilotinib in the treatment of advanced gastrointestinal stromal tumours resistant to both imatinib and sunitinib. Eur. J. Cancer 2009, 45, 2293–2297. [Google Scholar] [CrossRef] [PubMed]
- Sugase, T.; Takahashi, T.; Ishikawa, T.; Ichikawa, H.; Kanda, T.; Hirota, S.; Nakajima, K.; Tanaka, K.; Miyazaki, Y.; Makino, T.; et al. Surgical resection of recurrent gastrointestinal stromal tumor after interruption of long-term nilotinib therapy. Surg. Case Rep. 2016, 2. [Google Scholar] [CrossRef] [PubMed]
- Huang, K.; Chen, Z.; Zhang, X.M. Two new KIT exon 13 mutations in one gastric gastrointestinal stromal tumor (GIST). Int. J. Clin. Exp. Patho. 2017, 10, 8863–8867. [Google Scholar]
- Boschelli, D.H.; Wang, Y.D.; Ye, F.; Wu, B.; Zhang, N.; Dutia, M.; Powell, D.W.; Wissner, A.; Arndt, K.; Weber, J.M.; et al. Synthesis and Src kinase inhibitory activity of a series of 4-phenylamino-3-quinolinecarbonitriles. J. Med. Chem. 2001, 44, 822–833. [Google Scholar] [CrossRef]
- Golas, J.M.; Arndt, K.; Etienne, C.; Lucas, J.; Nardin, D.; Gibbons, J.; Frost, P.; Ye, F.; Boschelli, D.H.; Boschelli, F. SKI-606, a 4-anilino-3-quinolinecarbonitrile dual inhibitor of Src and Abl kinases, is a potent antiproliferative agent against chronic myelogenous leukemia cells in culture and causes regression of K562 xenografts in nude mice. Cancer Res. 2003, 63, 375–381. [Google Scholar]
- Cortes, J.E.; Kantarjian, H.M.; Brummendorf, T.H.; Kim, D.W.; Turkina, A.G.; Shen, Z.X.; Pasquini, R.; Khoury, H.J.; Arkin, S.; Volkert, A.; et al. Safety and efficacy of bosutinib (SKI-606) in chronic phase Philadelphia chromosome-positive chronic myeloid leukemia patients with resistance or intolerance to imatinib. Blood 2011, 118, 4567–4576. [Google Scholar] [CrossRef] [PubMed]
- Kantarjian, H.M.; Cortes, J.E.; Kim, D.-W.; Khoury, H.J.; Brummendorf, T.H.; Porkka, K.; Martinelli, G.; Durrant, S.; Leip, E.; Kelly, V.; et al. Bosutinib safety and management of toxicity in leukemia patients with resistance or intolerance to imatinib and other tyrosine kinase inhibitors. Blood 2014, 123, 1309–1318. [Google Scholar] [CrossRef] [PubMed]
- Gambacorti-Passerini, C.; Kantarjian, H.M.; Kim, D.W.; Khoury, H.J.; Turkina, A.G.; Brummendorf, T.H.; Matczak, E.; Bardy-Bouxin, N.; Shapiro, M.; Turnbull, K.; et al. Long-term efficacy and safety of bosutinib in patients with advanced leukemia following resistance/intolerance to imatinib and other tyrosine kinase inhibitors. Am. J. Hematol. 2015, 90, 755–768. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Gutierrez, V.; Martinez-Trillos, A.; Lopez Lorenzo, J.L.; Bautista, G.; Martin Mateos, M.L.; Alvarez-Larran, A.; Iglesias Perez, A.; Romo Collado, A.; Fernandez, A.; Portero, A.; et al. Bosutinib shows low cross intolerance, in chronic myeloid leukemia patients treated in fourth line. Results of the Spanish compassionate use program. Am. J. Hematol. 2015, 90, 429–433. [Google Scholar] [CrossRef] [PubMed]
- Isfort, S.; Brummendorf, T.H. Bosutinib in chronic myeloid leukemia: Patient selection and perspectives. J. Blood Med. 2018, 9, 43–50. [Google Scholar] [CrossRef]
- Puttini, M.; Coluccia, A.M.L.; Boschelli, F.; Cleris, L.; Marchesi, E.; Donella-Deana, A.; Ahmed, S.; Redaelli, S.; Piazza, R.; Magistroni, V.; et al. In vitro and in vivo activity of SKI-606, a novel Src-Abl inhibitor, against imatinib-resistant Bcr-Abl+ neoplastic cells. Cancer Res. 2006, 66, 11314–11322. [Google Scholar] [CrossRef]
- Levinson, N.M.; Boxer, S.G. Structural and spectroscopic analysis of the kinase inhibitor bosutinib and an isomer of bosutinib binding to the Abl tyrosine kinase domain. PLoS ONE 2012, 7, e29828. [Google Scholar] [CrossRef]
- Abbas, R.; Hsyu, P.H. Clinical Pharmacokinetics and Pharmacodynamics of Bosutinib. Clin. Pharm. 2016, 55, 1191–1204. [Google Scholar] [CrossRef]
- Redaelli, S.; Piazza, R.; Rostagno, R.; Magistroni, V.; Perini, P.; Marega, M.; Gambacorti-Passerini, C.; Boschelli, F. Activity of bosutinib, dasatinib, and nilotinib against 18 imatinib-resistant BCR/ABL mutants. J. Clin. Oncol. 2009, 27, 469–471. [Google Scholar] [CrossRef]
- Bradeen, H.A.; Eide, C.A.; O’Hare, T.; Johnson, K.J.; Willis, S.G.; Lee, F.Y.; Druker, B.J.; Deininger, M.W. Comparison of imatinib mesylate, dasatinib (BMS-354825), and nilotinib (AMN107) in an N-ethyl-N-nitrosourea (ENU)-based mutagenesis screen: High efficacy of drug combinations. Blood 2006, 108, 2332–2338. [Google Scholar] [CrossRef]
- Redaelli, S.; Mologni, L.; Rostagno, R.; Piazza, R.; Magistroni, V.; Ceccon, M.; Viltadi, M.; Flynn, D.; Gambacorti-Passerini, C. Three novel patient-derived BCR/ABL mutants show different sensitivity to second and third generation tyrosine kinase inhibitors. Am. J. Hematol. 2012, 87, 125–128. [Google Scholar] [CrossRef] [PubMed]
- Burger, J.A.; Wiestner, A. Targeting B cell receptor signalling in cancer: Preclinical and clinical advances. Nat. Rev. Cancer 2018, 18, 148–167. [Google Scholar] [CrossRef]
- Pal Singh, S.; Dammeijer, F.; Hendriks, R.W. Role of Bruton’s tyrosine kinase in B cells and malignancies. Mol. Cancer 2018, 17, 57. [Google Scholar] [CrossRef] [PubMed]
- Lam, K.P.; Kuhn, R.; Rajewsky, K. In vivo ablation of surface immunoglobulin on mature B cells by inducible gene targeting results in rapid cell death. Cell 1997, 90, 1073–1083. [Google Scholar] [CrossRef]
- Rolli, V.; Gallwitz, M.; Wossning, T.; Flemming, A.; Schamel, W.W.A.; Zurn, C.; Reth, M. Amplification of B cell antigen receptor signaling by a Syk/ITAM positive feedback loop. Mol. Cell 2002, 10, 1057–1069. [Google Scholar] [CrossRef]
- Inabe, K.; Ishiai, M.; Scharenberg, A.M.; Freshney, N.; Downward, J.; Kurosaki, T. Vav3 modulates B cell receptor responses by regulating phosphoinositide 3-kinase activation. J. Exp. Med. 2002, 195, 189–200. [Google Scholar] [CrossRef]
- Saito, K.; Scharenberg, A.M.; Kinet, J.P. Interaction between the Btk PH domain and phosphatidylinositol-3,4,5-trisphosphate directly regulates Btk. J. Biol. Chem. 2001, 276, 16201–16206. [Google Scholar] [CrossRef]
- Rawlings, D.J.; Scharenberg, A.M.; Park, H.; Wahl, M.I.; Lin, S.; Kato, R.M.; Fluckiger, A.C.; Witte, O.N.; Kinet, J.P. Activation of BTK by a phosphorylation mechanism initiated by SRC family kinases. Science 1996, 271, 822–825. [Google Scholar] [CrossRef] [PubMed]
- Saouaf, S.J.; Mahajan, S.; Rowley, R.B.; Kut, S.A.; Fargnoli, J.; Burkhardt, A.L.; Tsukada, S.; Witte, O.N.; Bolen, J.B. Temporal differences in the activation of three classes of non-transmembrane protein tyrosine kinases following B-cell antigen receptor surface engagement. Proc. Natl. Acad. Sci. USA 1994, 91, 9524–9528. [Google Scholar] [CrossRef]
- Yamanashi, Y.; Kakiuchi, T.; Mizuguchi, J.; Yamamoto, T.; Toyoshima, K. Association of B cell antigen receptor with protein tyrosine kinase Lyn. Science 1991, 251, 192–194. [Google Scholar] [CrossRef]
- Mahajan, S.; Fargnoli, J.; Burkhardt, A.L.; Kut, S.A.; Saouaf, S.J.; Bolen, J.B. Src family protein tyrosine kinases induce autoactivation of Bruton’s tyrosine kinase. Mol. Cell. Biol. 1995, 15, 5304–5311. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Park, H.; Wahl, M.I.; Afar, D.E.; Turck, C.W.; Rawlings, D.J.; Tam, C.; Scharenberg, A.M.; Kinet, J.P.; Witte, O.N. Regulation of Btk function by a major autophosphorylation site within the SH3 domain. Immunity 1996, 4, 515–525. [Google Scholar] [CrossRef]
- Feng, S.; Chen, J.K.; Yu, H.; Simon, J.A.; Schreiber, S.L. Two binding orientations for peptides to the Src SH3 domain: Development of a general model for SH3-ligand interactions. Science 1994, 266, 1241–1247. [Google Scholar] [CrossRef]
- Goudreau, N.; Cornille, F.; Duchesne, M.; Parker, F.; Tocque, B.; Garbay, C.; Roques, B.P. NMR structure of the N-terminal SH3 domain of GRB2 and its complex with a proline-rich peptide from Sos. Nat. Struct. Biol. 1994, 1, 898–907. [Google Scholar] [CrossRef] [PubMed]
- Koyama, S.; Yu, H.; Dalgarno, D.C.; Shin, T.B.; Zydowsky, L.D.; Schreiber, S.L. Structure of the PI3K SH3 domain and analysis of the SH3 family. Cell 1993, 72, 945–952. [Google Scholar] [CrossRef]
- Wahl, M.I.; Fluckiger, A.C.; Kato, R.M.; Park, H.; Witte, O.N.; Rawlings, D.J. Phosphorylation of two regulatory tyrosine residues in the activation of Bruton’s tyrosine kinase via alternative receptors. Proc. Natl. Acad. Sci. USA 1997, 94, 11526–11533. [Google Scholar] [CrossRef]
- Tsukada, S.; Saffran, D.C.; Rawlings, D.J.; Parolini, O.; Allen, R.C.; Klisak, I.; Sparkes, R.S.; Kubagawa, H.; Mohandas, T.; Quan, S. Deficient expression of a B cell cytoplasmic tyrosine kinase in human X-linked agammaglobulinemia. Cell 1993, 72, 279–290. [Google Scholar] [CrossRef]
- Valiaho, J.; Smith, C.I.E.; Vihinen, M. BTKbase: The mutation database for X-linked agammaglobulinemia. Hum. Mutat. 2006, 27, 1209–1217. [Google Scholar] [CrossRef]
- Singh, S.P.; Pillai, S.Y.; de Bruijn, M.J.W.; Stadhouders, R.; Corneth, O.B.J.; van den Ham, H.J.; Muggen, A.; van Ijcken, W.; Slinger, E.; Kuil, A.; et al. Cell lines generated from a chronic lymphocytic leukemia mouse model exhibit constitutive Btk and Akt signaling. Oncotarget 2017, 8, 71981–71995. [Google Scholar] [CrossRef]
- Kil, L.P.; de Bruijn, M.J.; van Hulst, J.A.; Langerak, A.W.; Yuvaraj, S.; Hendriks, R.W. Bruton’s tyrosine kinase mediated signaling enhances leukemogenesis in a mouse model for chronic lymphocytic leukemia. Am. J. Blood Res. 2013, 3, 71–83. [Google Scholar]
- Cinar, M.; Hamedani, F.; Mo, Z.; Cinar, B.; Amin, H.M.; Alkan, S. Bruton tyrosine kinase is commonly overexpressed in mantle cell lymphoma and its attenuation by Ibrutinib induces apoptosis. Leuk. Res. 2013, 37, 1271–1277. [Google Scholar] [CrossRef] [PubMed]
- Chang, B.Y.; Francesco, M.; De Rooij, M.F.M.; Magadala, P.; Steggerda, S.M.; Huang, M.M.; Kuil, A.; Herman, S.E.M.; Chang, S.; Pals, S.T.; et al. Egress of CD19(+)CD5(+) cells into peripheral blood following treatment with the Bruton tyrosine kinase inhibitor ibrutinib in mantle cell lymphoma patients. Blood 2013, 122, 2412–2424. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.; Zhou, Y.; Liu, X.; Xu, L.; Cao, Y.; Manning, R.J.; Patterson, C.J.; Buhrlage, S.J.; Gray, N.; Tai, Y.T.; et al. A mutation in MYD88 (L265P) supports the survival of lymphoplasmacytic cells by activation of Bruton tyrosine kinase in Waldenstrom macroglobulinemia. Blood 2013, 122, 1222–1232. [Google Scholar] [CrossRef] [PubMed]
- Davis, R.E.; Ngo, V.N.; Lenz, G.; Tolar, P.; Young, R.M.; Romesser, P.B.; Kohlhammer, H.; Lamy, L.; Zhao, H.; Yang, Y.; et al. Chronic active B-cell-receptor signalling in diffuse large B-cell lymphoma. Nature 2010, 463, 88–92. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.O. Development of BTK inhibitors for the treatment of B-cell malignancies. Arch. Pharm. Res. 2019, 42, 171–181. [Google Scholar] [CrossRef]
- Pan, Z.Y.; Scheerens, H.; Li, S.J.; Schultz, B.E.; Sprengeler, P.A.; Burrill, L.C.; Mendonca, R.V.; Sweeney, M.D.; Scott, K.C.K.; Grothaus, P.G.; et al. Discovery of selective irreversible inhibitors for Bruton’s tyrosine kinase. Chemmedchem 2007, 2, 58–61. [Google Scholar] [CrossRef]
- Honigberg, L.A.; Smith, A.M.; Sirisawad, M.; Verner, E.; Loury, D.; Chang, B.; Li, S.; Pan, Z.; Thamm, D.H.; Miller, R.A.; et al. The Bruton tyrosine kinase inhibitor PCI-32765 blocks B-cell activation and is efficacious in models of autoimmune disease and B-cell malignancy. Proc. Natl. Acad. Sci. USA 2010, 107, 13075–13080. [Google Scholar] [CrossRef]
- Akinleye, A.; Avvaru, P.; Furqan, M.; Song, Y.; Liu, D. Phosphatidylinositol 3-kinase (PI3K) inhibitors as cancer therapeutics. J. Hematol. Oncol. 2013, 6, 88. [Google Scholar] [CrossRef]
- Parmar, S.; Patel, K.; Pinilla-Ibarz, J. Ibrutinib (imbruvica): A novel targeted therapy for chronic lymphocytic leukemia. Pharm. Ther. 2014, 39, 483–519. [Google Scholar]
- Cheng, S.; Guo, A.; Lu, P.; Ma, J.; Coleman, M.; Wang, Y.L. Functional characterization of BTK(C481S) mutation that confers ibrutinib resistance: Exploration of alternative kinase inhibitors. Leukemia 2015, 29, 895–900. [Google Scholar] [CrossRef] [PubMed]
- Woyach, J.A.; Furman, R.R.; Liu, T.M.; Ozer, H.G.; Zapatka, M.; Ruppert, A.S.; Xue, L.; Li, D.H.; Steggerda, S.M.; Versele, M.; et al. Resistance mechanisms for the Bruton’s tyrosine kinase inhibitor ibrutinib. N. Engl. J. Med. 2014, 370, 2286–2294. [Google Scholar] [CrossRef] [PubMed]
- Furman, R.R.; Cheng, S.; Lu, P.; Setty, M.; Perez, A.R.; Guo, A.; Racchumi, J.; Xu, G.; Wu, H.; Ma, J.; et al. Ibrutinib resistance in chronic lymphocytic leukemia. N. Engl. J. Med. 2014, 370, 2352–2354. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Galanina, N.; Guo, A.; Lee, J.; Kadri, S.; Van Slambrouck, C.; Long, B.; Wang, W.; Ming, M.; Furtado, L.V.; et al. Identification of a structurally novel BTK mutation that drives ibrutinib resistance in CLL. Oncotarget 2016, 7, 68833–68841. [Google Scholar] [CrossRef] [PubMed]
- Kazi, J.U.; Ronnstrand, L. The role of SRC family kinases in FLT3 signaling. Int. J. Biochem. Cell Biol. 2019, 107, 32–37. [Google Scholar] [CrossRef]
- Gilliland, D.G.; Griffin, J.D. The roles of FLT3 in hematopoiesis and leukemia. Blood 2002, 100, 1532–1542. [Google Scholar] [CrossRef]
- Moriki, T.; Maruyama, H.; Maruyama, I.N. Activation of preformed EGF receptor dimers by ligand-induced rotation of the transmembrane domain. J. Mol. Biol. 2001, 311, 1011–1026. [Google Scholar] [CrossRef]
- Griffith, J.; Black, J.; Faerman, C.; Swenson, L.; Wynn, M.; Lu, F.; Lippke, J.; Saxena, K. The structural basis for autoinhibition of FLT3 by the juxtamembrane domain. Mol. Cell 2004, 13, 169–178. [Google Scholar] [CrossRef]
- Lemmon, M.A.; Schlessinger, J. Cell signaling by receptor tyrosine kinases. Cell 2010, 141, 1117–1134. [Google Scholar] [CrossRef]
- Kiyoi, H.; Ohno, R.; Ueda, R.; Saito, H.; Naoe, T. Mechanism of constitutive activation of FLT3 with internal tandem duplication in the juxtamembrane domain. Oncogene 2002, 21, 2555–2563. [Google Scholar] [CrossRef]
- Horiike, S.; Yokota, S.; Nakao, M.; Iwai, T.; Sasai, Y.; Kaneko, H.; Taniwaki, M.; Kashima, K.; Fujii, H.; Abe, T.; et al. Tandem duplications of the FLT3 receptor gene are associated with leukemic transformation of myelodysplasia. Leukemia 1997, 11, 1442–1446. [Google Scholar] [CrossRef]
- Iwai, T.; Yokota, S.; Nakao, M.; Okamoto, T.; Taniwaki, M.; Onodera, N.; Watanabe, A.; Kikuta, A.; Tanaka, A.; Asami, K.; et al. Internal tandem duplication of the FLT3 gene and clinical evaluation in childhood acute myeloid leukemia. The Children’s Cancer and Leukemia Study Group, Japan. Leukemia 1999, 13, 38–43. [Google Scholar] [CrossRef] [PubMed]
- Kiyoi, H.; Naoe, T.; Yokota, S.; Nakao, M.; Minami, S.; Kuriyama, K.; Takeshita, A.; Saito, K.; Hasegawa, S.; Shimodaira, S.; et al. Internal tandem duplication of FLT3 associated with leukocytosis in acute promyelocytic leukemia. Leukemia Study Group of the Ministry of Health and Welfare (Kohseisho). Leukemia 1997, 11, 1447–1452. [Google Scholar] [CrossRef] [PubMed]
- Yokota, S.; Kiyoi, H.; Nakao, M.; Iwai, T.; Misawa, S.; Okuda, T.; Sonoda, Y.; Abe, T.; Kahsima, K.; Matsuo, Y.; et al. Internal tandem duplication of the FLT3 gene is preferentially seen in acute myeloid leukemia and myelodysplastic syndrome among various hematological malignancies. A study on a large series of patients and cell lines. Leukemia 1997, 11, 1605–1609. [Google Scholar] [CrossRef] [PubMed]
- Meshinchi, S.; Woods, W.G.; Stirewalt, D.L.; Sweetser, D.A.; Buckley, J.D.; Tjoa, T.K.; Bernstein, I.D.; Radich, J.P. Prevalence and prognostic significance of Flt3 internal tandem duplication in pediatric acute myeloid leukemia. Blood 2001, 97, 89–94. [Google Scholar] [CrossRef]
- Abu-Duhier, F.M.; Goodeve, A.C.; Wilson, G.A.; Care, R.S.; Peake, I.R.; Reilly, J.T. Identification of novel FLT-3 Asp835 mutations in adult acute myeloid leukaemia. Br. J. Haematol. 2001, 113, 983–988. [Google Scholar] [CrossRef] [PubMed]
- Smith, C.C.; Wang, Q.; Chin, C.S.; Salerno, S.; Damon, L.E.; Levis, M.J.; Perl, A.E.; Travers, K.J.; Wang, S.; Hunt, J.P.; et al. Validation of ITD mutations in FLT3 as a therapeutic target in human acute myeloid leukaemia. Nature 2012, 485, 260–263. [Google Scholar] [CrossRef]
- Yamaguchi, H.; Hanawa, H.; Uchida, N.; Inamai, M.; Sawaguchi, K.; Mitamura, Y.; Shimada, T.; Dan, K.; Inokuchi, K. Multistep pathogenesis of leukemia via the MLL-AF4 chimeric gene/Flt3 gene tyrosine kinase domain (TKD) mutation-related enhancement of S100A6 expression. Exp. Hematol. 2009, 37, 701–714. [Google Scholar] [CrossRef]
- Chen, Y.; Pan, Y.; Guo, Y.; Zhao, W.; Ho, W.T.; Wang, J.; Xu, M.; Yang, F.C.; Zhao, Z.J. Tyrosine kinase inhibitors targeting FLT3 in the treatment of acute myeloid leukemia. Stem. Cell Investig. 2017, 4, 48. [Google Scholar] [CrossRef]
- Leick, M.B.; Levis, M.J. The Future of Targeting FLT3 Activation in AML. Curr. Hematol. Malig. Rep. 2017, 12, 153–167. [Google Scholar] [CrossRef]
- Lowinger, T.B.; Riedl, B.; Dumas, J.; Smith, R.A. Design and discovery of small molecules targeting raf-1 kinase. Curr. Pharm. Des. 2002, 8, 2269–2278. [Google Scholar] [CrossRef]
- Wilhelm, S.; Carter, C.; Lynch, M.; Lowinger, T.; Dumas, J.; Smith, R.A.; Schwartz, B.; Simantov, R.; Kelley, S. Discovery and development of sorafenib: A multikinase inhibitor for treating cancer. Nat. Rev. Drug Discov. 2006, 5, 835–844. [Google Scholar] [CrossRef] [PubMed]
- Adnane, L.; Trail, P.A.; Taylor, I.; Wilhelm, S.M. Sorafenib (BAY 43-9006, Nexavar (R)), a dual-action inhibitor that targets RAF/MEK/ERK pathway in tumor cells and tyrosine kinases VEGFR/PDGFR in tumor vasculature. Methods Enzymol. 2006, 407, 597–612. [Google Scholar] [CrossRef] [PubMed]
- Auclair, D.; Miller, D.; Yatsula, V.; Pickett, W.; Carter, C.; Chang, Y.; Zhang, X.; Wilkie, D.; Burd, A.; Shi, H.; et al. Antitumor activity of sorafenib in FLT3-driven leukemic cells. Leukemia 2007, 21, 439–445. [Google Scholar] [CrossRef] [PubMed]
- Wilhelm, S.M.; Carter, C.; Tang, L.; Wilkie, D.; McNabola, A.; Rong, H.; Chen, C.; Zhang, X.; Vincent, P.; McHugh, M.; et al. BAY 43-9006 exhibits broad spectrum oral antitumor activity and targets the RAF/MEK/ERK pathway and receptor tyrosine kinases involved in tumor progression and angiogenesis. Cancer Res. 2004, 64, 7099–7109. [Google Scholar] [CrossRef]
- Carlomagno, F.; Anaganti, S.; Guida, T.; Salvatore, G.; Troncone, G.; Wilhelm, S.M.; Santoro, M. BAY 43-9006 inhibition of oncogenic RET mutants. J. Natl. Cancer Inst. 2006, 98, 326–334. [Google Scholar] [CrossRef]
- Hasskarl, J. Sorafenib: Targeting multiple tyrosine kinases in cancer. Recent Results Cancer Res. 2014, 201, 145–164. [Google Scholar] [CrossRef]
- Man, C.H.; Fung, T.K.; Ho, C.; Han, H.H.C.; Chow, H.C.H.; Ma, A.C.H.; Choi, W.W.L.; Lok, S.; Cheung, A.M.S.; Eaves, C.; et al. Sorafenib treatment of FLT3-ITD+ acute myeloid leukemia: Favorable initial outcome and mechanisms of subsequent nonresponsiveness associated with the emergence of a D835 mutation. Blood 2012, 119, 5133–5143. [Google Scholar] [CrossRef]
- Baker, S.D.; Zimmerman, E.I.; Wang, Y.D.; Orwick, S.; Zatechka, D.S.; Buaboonnam, J.; Neale, G.A.; Olsen, S.R.; Enemark, E.J.; Shurtleff, S.; et al. Emergence of polyclonal FLT3 tyrosine kinase domain mutations during sequential therapy with sorafenib and sunitinib in FLT3-ITD-Positive acute myeloid leukemia. Clin. Cancer Res. 2013, 19, 5758–5768. [Google Scholar] [CrossRef]
- Smith, C.C.; Lin, K.; Stecula, A.; Sali, A.; Shah, N.P. FLT3 D835 mutations confer differential resistance to type II FLT3 inhibitors. Leukemia 2015, 29, 2390–2392. [Google Scholar] [CrossRef]
- Chao, Q.; Sprankle, K.G.; Grotzfeld, R.M.; Lai, A.G.; Carter, T.A.; Velasco, A.M.; Gunawardane, R.N.; Cramer, M.D.; Gardner, M.F.; James, J.; et al. Identification of N-(5-tert-Butyl-isoxazol-3-yl)-N ’-{4-[7-(2-morpholin-4-yl-ethoxy)imidazo[2,1-b][1,3]benzothiazol-2-yl]phenyl}urea dihydrochloride (AC220), a uniquely potent, selective, and efficacious FMS-Like tyrosine Kinase-3 (FLT3) inhibitor. J. Med. Chem. 2009, 52, 7808–7816. [Google Scholar] [CrossRef]
- Zarrinkar, P.P.; Gunawardane, R.N.; Cramer, M.D.; Gardner, M.F.; Brigham, D.; Belli, B.; Karaman, M.W.; Pratz, K.W.; Pallares, G.; Chao, Q.; et al. AC220 is a uniquely potent and selective inhibitor of FLT3 for the treatment of acute myeloid leukemia (AML). Blood 2009, 114, 2984–2992. [Google Scholar] [CrossRef] [PubMed]
- Naqvi, K.; Ravandi, F. FLT3 inhibitor quizartinib (AC220). Leuk Lymphoma 2019, 60, 1866–1876. [Google Scholar] [CrossRef] [PubMed]
- Zhou, F.; Ge, Z.; Chen, B. Quizartinib (AC220): A promising option for acute myeloid leukemia. Drug Des. Devel Ther. 2019, 13, 1117–1125. [Google Scholar] [CrossRef] [PubMed]
- Zorn, J.A.; Wang, Q.; Fujimura, E.; Barros, T.; Kuriyan, J. Crystal structure of the FLT3 kinase domain bound to the inhibitor Quizartinib (AC220). PLoS ONE 2015, 10, e0121177. [Google Scholar] [CrossRef] [PubMed]
- Albers, C.; Leischner, H.; Verbeek, M.; Yu, C.; Illert, A.L.; Peschel, C.; von Bubnoff, N.; Duyster, J. The secondary FLT3-ITD F691L mutation induces resistance to AC220 in FLT3-ITD+ AML but retains in vitro sensitivity to PKC412 and Sunitinib. Leukemia 2013, 27, 1416–1418. [Google Scholar] [CrossRef] [PubMed]
- Moore, A.S.; Faisal, A.; de Castro, D.G.; Bavetsias, V.; Sun, C.; Atrash, B.; Valenti, M.; Brandon, A.D.; Avery, S.; Mair, D.; et al. Selective FLT3 inhibition of FLT3-ITD+ acute myeloid leukaemia resulting in secondary D835Y mutation: A model for emerging clinical resistance patterns. Leukemia 2012, 26, 1462–1470. [Google Scholar] [CrossRef] [PubMed]
- Chow, L.Q.M.; Eckhardt, S.G. Sunitinib: From rational design to clinical efficacy. J. Clin. Oncol. 2007, 25, 884–896. [Google Scholar] [CrossRef] [PubMed]
- Deeks, E.D.; Keating, G.M. Sunitinib. Drugs 2006, 66, 2255–2266. [Google Scholar] [CrossRef] [PubMed]
- Cheng, P.; Levesque, M.P.; Dummer, R.; Mangana, J. Targeting complex, adaptive responses in melanoma therapy. Cancer Treat. Rev. 2020, 86, 101997. [Google Scholar] [CrossRef]
- Klener, P.; Klanova, M. Drug Resistance in Non-Hodgkin Lymphomas. Int. J. Mol. Sci. 2020, 21, 2081. [Google Scholar] [CrossRef]
- Zhao, X.; Yue, C. Gastrointestinal stromal tumor. J. Gastrointest. Oncol. 2012, 3, 189–208. [Google Scholar] [CrossRef] [PubMed]
- Miettinen, M.; Lasota, J. Gastrointestinal stromal tumors--definition, clinical, histological, immunohistochemical, and molecular genetic features and differential diagnosis. Virchows Arch. 2001, 438, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Khoshnood, A. Gastrointestinal stromal tumor—A review of clinical studies. J. Oncol. Pharm. Pract. 2019, 25, 1473–1485. [Google Scholar] [CrossRef] [PubMed]
- Sanders, K.M.; Koh, S.D.; Ward, S.M. Interstitial cells of Cajal as pacemakers in the gastrointestinal tract. Annu. Rev. Physiol. 2006, 68, 307–343. [Google Scholar] [CrossRef]
- Hirota, S.; Isozaki, K.; Moriyama, Y.; Hashimoto, K.; Nishida, T.; Ishiguro, S.; Kawano, K.; Hanada, M.; Kurata, A.; Takeda, M.; et al. Gain-of-function mutations of c-kit in human gastrointestinal stromal tumors. Science 1998, 279, 577–580. [Google Scholar] [CrossRef]
- Heinrich, M.C.; Corless, C.L.; Duensing, A.; McGreevey, L.; Chen, C.J.; Joseph, N.; Singer, S.; Griffith, D.J.; Haley, A.; Town, A.; et al. PDGFRA activating mutations in gastrointestinal stromal tumors. Science 2003, 299, 708–710. [Google Scholar] [CrossRef]
- Ashman, L.K. The biology of stem cell factor and its receptor C-kit. Int. J. Biochem. Cell Biol. 1999, 31, 1037–1051. [Google Scholar] [CrossRef]
- Wang, H.; Boussouar, A.; Mazelin, L.; Tauszig-Delamasure, S.; Sun, Y.; Goldschneider, D.; Paradisi, A.; Mehlen, P. The Proto-oncogene c-Kit Inhibits Tumor Growth by Behaving as a Dependence Receptor. Mol. Cell 2018, 72, 413–425. [Google Scholar] [CrossRef]
- Abbaspour Babaei, M.; Kamalidehghan, B.; Saleem, M.; Huri, H.Z.; Ahmadipour, F. Receptor tyrosine kinase (c-Kit) inhibitors: A potential therapeutic target in cancer cells. Drug Des. Devel. Ther. 2016, 10, 2443–2459. [Google Scholar] [CrossRef]
- Liang, J.; Wu, Y.L.; Chen, B.J.; Zhang, W.; Tanaka, Y.; Sugiyama, H. The C-kit receptor-mediated signal transduction and tumor-related diseases. Int. J. Biol. Sci. 2013, 9, 435–443. [Google Scholar] [CrossRef]
- Corless, C.L.; McGreevey, L.; Town, A.; Schroeder, A.; Bainbridge, T.; Harrell, P.; Fletcher, J.A.; Heinrich, M.C. KIT gene deletions at the intron 10-exon 11 boundary in GI stromal tumors. J. Mol. Diagn. 2004, 6, 366–370. [Google Scholar] [CrossRef]
- Corless, C.L. Assessing the prognosis of gastrointestinal stromal tumors: A growing role for molecular testing. Am. J. Clin. Pathol. 2004, 122, 11–13. [Google Scholar] [CrossRef] [PubMed]
- Isozaki, K.; Terris, B.; Belghiti, J.; Schiffmann, S.; Hirota, S.; Vanderwinden, J.M. Germline-activating mutation in the kinase domain of KIT gene in familial gastrointestinal stromal tumors. Am. J. Pathol. 2000, 157, 1581–1585. [Google Scholar] [CrossRef]
- Yarden, Y.; Kuang, W.J.; Yang-Feng, T.; Coussens, L.; Munemitsu, S.; Dull, T.J.; Chen, E.; Schlessinger, J.; Francke, U.; Ullrich, A. Human proto-oncogene c-kit: A new cell surface receptor tyrosine kinase for an unidentified ligand. EMBO J. 1987, 6, 3341–3351. [Google Scholar] [CrossRef]
- Schwaab, J.; Schnittger, S.; Sotlar, K.; Walz, C.; Fabarius, A.; Pfirrmann, M.; Kohlmann, A.; Grossmann, V.; Meggendorfer, M.; Horny, H.P.; et al. Comprehensive mutational profiling in advanced systemic mastocytosis. Blood 2013, 122, 2460–2466. [Google Scholar] [CrossRef] [PubMed]
- Heinrich, M.C.; Griffith, D.J.; Druker, B.J.; Wait, C.L.; Ott, K.A.; Zigler, A.J. Inhibition of c-kit receptor tyrosine kinase activity by STI 571, a selective tyrosine kinase inhibitor. Blood 2000, 96, 925–932. [Google Scholar] [CrossRef] [PubMed]
- Craig, J.W.; Hasserjian, R.P.; Kim, A.S.; Aster, J.C.; Pinkus, G.S.; Hornick, J.L.; Steensma, D.P.; Coleman Lindsley, R.; DeAngelo, D.J.; Morgan, E.A. Detection of the KIT(D816V) mutation in myelodysplastic and/or myeloproliferative neoplasms and acute myeloid leukemia with myelodysplasia-related changes predicts concurrent systemic mastocytosis. Mod. Pathol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Kristensen, T.; Preiss, B.; Broesby-Olsen, S.; Vestergaard, H.; Friis, L.; Moller, M.B. Systemic mastocytosis is uncommon in KIT D816V mutation positive core-binding factor acute myeloid leukemia. Leukemia Lymphoma 2012, 53, 1338–1344. [Google Scholar] [CrossRef] [PubMed]
- Niinuma, T.; Suzuki, H.; Sugai, T. Molecular characterization and pathogenesis of gastrointestinal stromal tumor. Transl. Gastroenterol. Hepatol. 2018, 3. [Google Scholar] [CrossRef]
- Wozniak, A.; Rutkowski, P.; Schoffski, P.; Ray-Coquard, I.; Hostein, I.; Schildhaus, H.U.; Le Cesne, A.; Bylina, E.; Limon, J.; Blay, J.Y.; et al. Tumor genotype is an independent prognostic factor in primary gastrointestinal stromal tumors of gastric origin: A european multicenter analysis based on conticaGIST. Clin. Cancer Res. 2014, 20, 6105–6116. [Google Scholar] [CrossRef]
- Croom, K.F.; Perry, C.M. Imatinib mesylate: In the treatment of gastrointestinal stromal tumours. Drugs 2003, 63, 513–522. [Google Scholar] [CrossRef]
- Cools, J.; DeAngelo, D.J.; Gotlib, J.; Stover, E.H.; Legare, R.D.; Cortes, J.; Kutok, J.; Clark, J.; Galinsky, I.; Griffin, J.D.; et al. A tyrosine kinase created by fusion of the PDGFRA and FIP1L1 genes as a therapeutic target of imatinib in idiopathic hypereosinophilic syndrome. N. Engl. J. Med. 2003, 348, 1201–1214. [Google Scholar] [CrossRef] [PubMed]
- Rajasekaran, R.; Sethumadhavan, R. Exploring the cause of drug resistance by the detrimental missense mutations in KIT receptor: Computational approach. Amino Acids 2010, 39, 651–660. [Google Scholar] [CrossRef] [PubMed]
- Corless, C.L.; Schroeder, A.; Griffith, D.; Town, A.; McGreevey, L.; Harrell, P.; Shiraga, S.; Bainbridge, T.; Morich, J.; Heinrich, M.C. PDGFRA mutations in gastrointestinal stromal tumors: Frequency, spectrum and in vitro sensitivity to imatinib. J. Clin. Oncol. 2005, 23, 5357–5364. [Google Scholar] [CrossRef]
- Liang, L.; Yan, X.E.; Yin, Y.; Yun, C.H. Structural and biochemical studies of the PDGFRA kinase domain. Biochem. Bioph. Res. Commun. 2016, 477, 667–672. [Google Scholar] [CrossRef] [PubMed]
- Cho, J.; Kim, S.Y.; Kim, Y.J.; Sim, M.H.; Kim, S.T.; Kim, N.K.D.; Kim, K.; Park, W.; Kim, J.H.; Jang, K.T.; et al. Emergence of CTNNB1 mutation at acquired resistance to KIT inhibitor in metastatic melanoma. Clin. Transl. Oncol. 2017, 19, 1247–1252. [Google Scholar] [CrossRef]
- Noujaim, J.; Gonzalez, D.; Thway, K.; Jones, R.L.; Judson, I. p. (L576P) -KIT mutation in GIST: Favorable prognosis and sensitive to imatinib? Cancer Biol. Ther. 2016, 17, 543–545. [Google Scholar] [CrossRef][Green Version]
- Conca, E.; Negri, T.; Gronchi, A.; Fumagalli, E.; Tamborini, E.; Pavan, G.M.; Fermeglia, M.; Pierotti, M.A.; Pricl, S.; Pilotti, S. Activate and resist: L576P-KIT in GIST. Mol. Cancer Ther. 2009, 8, 2491–2495. [Google Scholar] [CrossRef][Green Version]
- Pai, S.G.; Carneiro, B.A.; Mota, J.M.; Costa, R.; Leite, C.A.; Barroso-Sousa, R.; Kaplan, J.B.; Chae, Y.K.; Giles, F.J. Wnt/beta-catenin pathway: Modulating anticancer immune response. J. Hematol. Oncol. 2017, 10, 101. [Google Scholar] [CrossRef]
- Chiarini, F.; Paganelli, F.; Martelli, A.M.; Evangelisti, C. The role played by wnt/beta-catenin signaling pathway in acute lymphoblastic leukemia. Int. J. Mol. Sci. 2020, 21, 1098. [Google Scholar] [CrossRef]
- Provost, E.; Yamamoto, Y.; Lizardi, I.; Stern, J.; D’Aquila, T.G.; Gaynor, R.B.; Rimm, D.L. Functional correlates of mutations in beta-catenin exon 3 phosphorylation sites. J. Biol. Chem. 2003, 278, 31781–31789. [Google Scholar] [CrossRef] [PubMed]
- Maeda, D.; Shibahara, J.; Sakuma, T.; Isobe, M.; Teshima, S.; Mori, M.; Oda, K.; Nakagawa, S.; Taketani, Y.; Ishikawa, S.; et al. Beta-catenin (CTNNB1) S33C mutation in ovarian microcystic stromal tumors. American J. Surg. Pathol. 2011, 35, 1429–1440. [Google Scholar] [CrossRef] [PubMed]
- Dziadziuszko, R.; Le, A.T.; Wrona, A.; Jassem, J.; Camidge, D.R.; Varella-Garcia, M.; Aisner, D.L.; Doebele, R.C. An activating KIT mutation induces crizotinib resistance in ROS1-Positive lung cancer. J. Thorac. Oncol. 2016, 11, 1273–1281. [Google Scholar] [CrossRef]
- Birchmeier, C.; Sharma, S.; Wigler, M. Expression and rearrangement of the Ros1 gene in human glioblastoma cells. Proc. Natl. Acad. Sci. USA 1987, 84, 9270–9274. [Google Scholar] [CrossRef] [PubMed]
- Arbour, K.C.; Riely, G.J. Systemic therapy for locally advanced and metastatic non-small cell lung cancer a review. J. Am. Med. Assoc. 2019, 322, 764–774. [Google Scholar] [CrossRef]
- Asao, T.; Takahashi, F.; Takahashi, K. Resistance to molecularly targeted therapy in non-small-cell lung cancer. Respir. Investig. 2019, 57, 20–26. [Google Scholar] [CrossRef]
- Skoulidis, F.; Heymach, J.V. Co-occurring genomic alterations in non-small-cell lung cancer biology and therapy. Nat. Rev. Cancer 2019, 19, 495–509. [Google Scholar] [CrossRef]
- Arai, Y.; Totoki, Y.; Takahashi, H.; Nakamura, H.; Hama, N.; Kohno, T.; Tsuta, K.; Yoshida, A.; Asamura, H.; Mutoh, M.; et al. Mouse model for ros1-rearranged lung cancer. PLoS ONE 2013, 8. [Google Scholar] [CrossRef]
- Bergethon, K.; Shaw, A.T.; Ou, S.H.; Katayama, R.; Lovly, C.M.; McDonald, N.T.; Massion, P.P.; Siwak-Tapp, C.; Gonzalez, A.; Fang, R.; et al. ROS1 rearrangements define a unique molecular class of lung cancers. J. Clin. Oncol. 2012, 30, 863–870. [Google Scholar] [CrossRef]
- Davies, K.D.; Le, A.T.; Theodoro, M.F.; Skokan, M.C.; Aisner, D.L.; Berge, E.M.; Terracciano, L.M.; Cappuzzo, F.; Incarbone, M.; Roncalli, M.; et al. Identifying and targeting ROS1 gene fusions in non-small cell lung cancer. Clin. Cancer Res. 2012, 18, 4570–4579. [Google Scholar] [CrossRef]
- Shaw, A.T.; Ou, S.H.I.; Bang, Y.J.; Camidge, D.R.; Solomon, B.J.; Salgia, R.; Riely, G.J.; Varella-Garcia, M.; Shapiro, G.I.; Costa, D.B.; et al. Crizotinib in ROS1-rearranged non-small-cell lung cancer. N. Engl. J. Med. 2014, 371, 1963–1971. [Google Scholar] [CrossRef]
- Shaw, A.T.; Kim, D.W.; Mehra, R.; Tan, D.S.W.; Felip, E.; Chow, L.Q.M.; Camidge, D.R.; Vansteenkiste, J.; Sharma, S.; De Pas, T.; et al. Ceritinib in ALK-rearranged non-small-cell lung cancer. N. Engl. J. Med. 2014, 370, 1189–1197. [Google Scholar] [CrossRef] [PubMed]
- Mazieres, J.; Zalcman, G.; Crino, L.; Biondani, P.; Barlesi, F.; Filleron, T.; Dingemans, A.M.C.; Lena, H.; Monnet, I.; Rothschild, S.I.; et al. Crizotinib therapy for advanced lung adenocarcinoma and a ROS1 rearrangement: Results from the EUROS1 cohort. J. Clin. Oncol. 2015, 33, 992–999. [Google Scholar] [CrossRef]
- Mort, R.L.; Jackson, I.J.; Patton, E.E. The melanocyte lineage in development and disease. Development 2015, 142, 1387. [Google Scholar] [CrossRef] [PubMed]
- Bastian, B.C. The molecular pathology of melanoma: An integrated taxonomy of melanocytic neoplasia. Annu. Rev. Pathol. 2014, 9, 239–271. [Google Scholar] [CrossRef]
- Shain, A.H.; Bastian, B.C. From melanocytes to melanomas. Nat. Rev. Cancer 2016, 16, 345–358. [Google Scholar] [CrossRef] [PubMed]
- Haugh, A.M.; Johnson, D.B. Management of V600E and V600K BRAF-Mutant Melanoma. Curr. Treat. Options Oncol. 2019, 20, 81. [Google Scholar] [CrossRef] [PubMed]
- O’Leary, C.G.; Andelkovic, V.; Ladwa, R.; Pavlakis, N.; Zhou, C.; Hirsch, F.; Richard, D.; O’Byrne, K. Targeting BRAF mutations in non-small cell lung cancer. Transl. Lung Cancer Res. 2019, 8, 1119–1124. [Google Scholar] [CrossRef]
- Curtin, J.A.; Fridlyand, J.; Kageshita, T.; Patel, H.N.; Busam, K.J.; Kutzner, H.; Cho, K.H.; Aiba, S.; Brocker, E.B.; LeBoit, P.E.; et al. Distinct sets of genetic alterations in melanoma. N. Engl. J. Med. 2005, 353, 2135–2147. [Google Scholar] [CrossRef]
- Davies, H.; Bignell, G.R.; Cox, C.; Stephens, P.; Edkins, S.; Clegg, S.; Teague, J.; Woffendin, H.; Garnett, M.J.; Bottomley, W.; et al. Mutations of the BRAF gene in human cancer. Nature 2002, 417, 949–954. [Google Scholar] [CrossRef]
- Cantwell-Dorris, E.R.; O’Leary, J.J.; Sheils, O.M. BRAFV600E: Implications for carcinogenesis and molecular therapy. Mol. Cancer Ther. 2011, 10, 385–394. [Google Scholar] [CrossRef] [PubMed]
- Lassam, N.; Bickford, S. Loss of c-kit expression in cultured melanoma cells. Oncogene 1992, 7, 51–56. [Google Scholar] [PubMed]
- Shain, A.H.; Yeh, I.; Kovalyshyn, I.; Sriharan, A.; Talevich, E.; Gagnon, A.; Dummer, R.; North, J.; Pincus, L.; Ruben, B.; et al. The genetic evolution of melanoma from precursor lesions. N. Engl. J. Med. 2015, 373, 1926–1936. [Google Scholar] [CrossRef] [PubMed]
- Philpott, C.; Tovell, H.; Frayling, I.M.; Cooper, D.N.; Upadhyaya, M. The NF1 somatic mutational landscape in sporadic human cancers. Hum. Genom. 2017, 11, 13. [Google Scholar] [CrossRef] [PubMed]
- Maertens, O.; Johnson, B.; Hollstein, P.; Frederick, D.T.; Cooper, Z.A.; Messiaen, L.; Bronson, R.T.; McMahon, M.; Granter, S.; Flaherty, K.; et al. Elucidating distinct roles for NF1 in melanomagenesis. Cancer Discov. 2013, 3, 338–349. [Google Scholar] [CrossRef]
- Franken, M.G.; Leeneman, B.; Gheorghe, M.; Uyl-de Groot, C.A.; Haanen, J.; van Baal, P.H.M. A systematic literature review and network meta-analysis of effectiveness and safety outcomes in advanced melanoma. Eur. J. Cancer 2019, 123, 58–71. [Google Scholar] [CrossRef]
- Tsai, J.; Lee, J.T.; Wang, W.; Zhang, J.; Cho, H.; Mamo, S.; Bremer, R.; Gillette, S.; Kong, J.; Haass, N.K.; et al. Discovery of a selective inhibitor of oncogenic B-Raf kinase with potent antimelanoma activity. Proc. Natl. Acad. Sci. USA 2008, 105, 3041–3046. [Google Scholar] [CrossRef]
- Bollag, G.; Hirth, P.; Tsai, J.; Zhang, J.; Ibrahim, P.N.; Cho, H.; Spevak, W.; Zhang, C.; Zhang, Y.; Habets, G.; et al. Clinical efficacy of a RAF inhibitor needs broad target blockade in BRAF-mutant melanoma. Nature 2010, 467, 596–599. [Google Scholar] [CrossRef]
- Li, H.F.; Chen, Y.; Rao, S.S.; Chen, X.M.; Liu, H.C.; Qin, J.H.; Tang, W.F.; Yue, W.; Zhou, X.; Lu, T. Recent advances in the research and development of B-Raf inhibitors. Curr. Med. Chem. 2010, 17, 1618–1634. [Google Scholar] [CrossRef]
- Smith, R.A.; Dumas, J.; Adnane, L.; Wilhelm, S.M. Recent advances in the research and development of RAF kinase inhibitors. Curr. Top. Med. Chem. 2006, 6, 1071–1089. [Google Scholar] [CrossRef]
- Yang, H.; Higgins, B.; Kolinsky, K.; Packman, K.; Go, Z.; Iyer, R.; Kolis, S.; Zhao, S.; Lee, R.; Grippo, J.F.; et al. RG7204 (PLX4032), a selective BRAFV600E inhibitor, displays potent antitumor activity in preclinical melanoma models. Cancer Res. 2010, 70, 5518–5527. [Google Scholar] [CrossRef]
- Flaherty, K.T.; Puzanov, I.; Kim, K.B.; Ribas, A.; McArthur, G.A.; Sosman, J.A.; O’Dwyer, P.J.; Lee, R.J.; Grippo, J.F.; Nolop, K.; et al. Inhibition of mutated, activated BRAF in metastatic melanoma. N. Engl. J. Med. 2010, 363, 809–819. [Google Scholar] [CrossRef]
- Chapman, P.B.; Hauschild, A.; Robert, C.; Haanen, J.B.; Ascierto, P.; Larkin, J.; Dummer, R.; Garbe, C.; Testori, A.; Maio, M.; et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N. Engl. J. Med. 2011, 364, 2507–2516. [Google Scholar] [CrossRef] [PubMed]
- Hyman, D.M.; Puzanov, I.; Subbiah, V.; Faris, J.E.; Chau, I.; Blay, J.Y.; Wolf, J.; Raje, N.S.; Diamond, E.L.; Hollebecque, A.; et al. Vemurafenib in multiple nonmelanoma cancers with BRAF V600 mutations. N. Engl. J. Med. 2015, 373, 726–736. [Google Scholar] [CrossRef] [PubMed]
- Mazieres, J.; Cropet, C.; Montane, L.; Barlesi, F.; Souquet, P.J.; Quantin, X.; Dubos-Arvis, C.; Otto, J.; Favier, L.; Avrillon, V.; et al. Vemurafenib in non-small-cell lung cancer patients with BRAF(V600) and BRAF(nonV600) mutations. Ann. Oncol. 2020, 31, 289–294. [Google Scholar] [CrossRef] [PubMed]
- Subbiah, V.; Puzanov, I.; Blay, J.Y.; Chau, I.; Lockhart, A.C.; Raje, N.S.; Wolf, J.; Baselga, J.; Meric-Bernstam, F.; Roszik, J.; et al. Pan-cancer efficacy of vemurafenib in BRAFV600-mutant non-melanoma cancers. Cancer Discov. 2020. [Google Scholar] [CrossRef] [PubMed]
- Roskoski, R., Jr. RAF protein-serine/threonine kinases: Structure and regulation. Biochem. Biophys. Res. Commun. 2010, 399, 313–317. [Google Scholar] [CrossRef]
- Luebker, S.A.; Koepsell, S.A. Diverse mechanisms of BRAF inhibitor resistance in melanoma identified in clinical and preclinical studies. Front. Oncol. 2019, 9. [Google Scholar] [CrossRef]
- Wagle, N.; Emery, C.; Berger, M.F.; Davis, M.J.; Sawyer, A.; Pochanard, P.; Kehoe, S.M.; Johannessen, C.M.; Macconaill, L.E.; Hahn, W.C.; et al. Dissecting therapeutic resistance to RAF inhibition in melanoma by tumor genomic profiling. J. Clin. Oncol. 2011, 29, 3085–3096. [Google Scholar] [CrossRef]
- Van Allen, E.M.; Wagle, N.; Sucker, A.; Treacy, D.J.; Johannessen, C.M.; Goetz, E.M.; Place, C.S.; Taylor-Weiner, A.; Whittaker, S.; Kryukov, G.V.; et al. The genetic landscape of clinical resistance to RAF inhibition in metastatic melanoma. Cancer Discov. 2014, 4, 94–109. [Google Scholar] [CrossRef]
- Rizos, H.; Menzies, A.M.; Pupo, G.M.; Carlino, M.S.; Fung, C.; Hyman, J.; Haydu, L.E.; Mijatov, B.; Becker, T.M.; Boyd, S.C.; et al. BRAF inhibitor resistance mechanisms in metastatic melanoma: Spectrum and clinical impact. Clin. Cancer Res. 2014, 20, 1965–1977. [Google Scholar] [CrossRef] [PubMed]
- Kwong, L.N.; Boland, G.M.; Frederick, D.T.; Helms, T.L.; Akid, A.T.; Miller, J.P.; Jiang, S.; Cooper, Z.A.; Song, X.; Seth, S.; et al. Co-clinical assessment identifies patterns of BRAF inhibitor resistance in melanoma. J. Clin. Investig. 2015, 125, 1459–1470. [Google Scholar] [CrossRef] [PubMed]
- Lim, S.Y.; Menzies, A.M.; Rizos, H. Mechanisms and strategies to overcome resistance to molecularly targeted therapy for melanoma. Cancer 2017, 123, 2118–2129. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.D.; Lee, N.H. Aberrant RNA splicing in cancer and drug resistance. Cancers 2018, 10, 458. [Google Scholar] [CrossRef] [PubMed]
- Poulikakos, P.I.; Persaud, Y.; Janakiraman, M.; Kong, X.; Ng, C.; Moriceau, G.; Shi, H.; Atefi, M.; Titz, B.; Gabay, M.T.; et al. RAF inhibitor resistance is mediated by dimerization of aberrantly spliced BRAF(V600E). Nature 2011, 480, 387–390. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.; Landrette, S.F.; Wang, T.; Evans, P.; Bacchiocchi, A.; Bjornson, R.; Cheng, E.; Stiegler, A.L.; Gathiaka, S.; Acevedo, O.; et al. Identification of PLX4032-resistance mechanisms and implications for novel RAF inhibitors. Pigment Cell Melanoma Res. 2014, 27, 253–262. [Google Scholar] [CrossRef] [PubMed]
- Hoogstraat, M.; Gadellaa-van Hooijdonk, C.G.; Ubink, I.; Besselink, N.J.; Pieterse, M.; Veldhuis, W.; van Stralen, M.; Meijer, E.F.; Willems, S.M.; Hadders, M.A.; et al. Detailed imaging and genetic analysis reveal a secondary BRAF(L505H) resistance mutation and extensive intrapatient heterogeneity in metastatic BRAF mutant melanoma patients treated with vemurafenib. Pigment Cell Melanoma Res. 2015, 28, 318–323. [Google Scholar] [CrossRef]
- Romano, E.; Pradervand, S.; Paillusson, A.; Weber, J.; Harshman, K.; Muehlethaler, K.; Speiser, D.; Peters, S.; Rimoldi, D.; Michielin, O. Identification of multiple mechanisms of resistance to vemurafenib in a patient with BRAFV600E-mutated cutaneous melanoma successfully rechallenged after progression. Clin. Cancer Res. 2013, 19, 5749–5757. [Google Scholar] [CrossRef]
- Mao, M.; Tian, F.; Mariadason, J.M.; Tsao, C.C.; Lemos, R., Jr.; Dayyani, F.; Gopal, Y.N.; Jiang, Z.Q.; Wistuba, II.; Tang, X.M.; et al. Resistance to BRAF inhibition in BRAF-mutant colon cancer can be overcome with PI3K inhibition or demethylating agents. Clin. Cancer Res. 2013, 19, 657–667. [Google Scholar] [CrossRef]
- Menzies, A.M.; Long, G.V.; Murali, R. Dabrafenib and its potential for the treatment of metastatic melanoma. Drug Des. Devel. Ther. 2012, 6, 391–405. [Google Scholar] [CrossRef]
- Zhang, J.; Yang, P.L.; Gray, N.S. Targeting cancer with small molecule kinase inhibitors. Nat. Rev. Cancer 2009, 9, 28–39. [Google Scholar] [CrossRef]
- Roskoski, R., Jr. Targeting oncogenic Raf protein-serine/threonine kinases in human cancers. Pharmacol. Res. 2018, 135, 239–258. [Google Scholar] [CrossRef] [PubMed]
- Falchook, G.S.; Long, G.V.; Kurzrock, R.; Kim, K.B.; Arkenau, T.H.; Brown, M.P.; Hamid, O.; Infante, J.R.; Millward, M.; Pavlick, A.C.; et al. Dabrafenib in patients with melanoma, untreated brain metastases, and other solid tumours: A phase 1 dose-escalation trial. Lancet 2012, 379, 1893–1901. [Google Scholar] [CrossRef]
- Rheault, T.R.; Stellwagen, J.C.; Adjabeng, G.M.; Hornberger, K.R.; Petrov, K.G.; Waterson, A.G.; Dickerson, S.H.; Mook, R.A.; Laquerre, S.G.; King, A.J.; et al. Discovery of dabrafenib: a selective inhibitor of raf kinases with antitumor activity against B-Raf-driven tumors. Acs Med. Chem. Lett. 2013, 4, 358–362. [Google Scholar] [CrossRef] [PubMed]
- Gibney, G.T.; Zager, J.S. Clinical development of dabrafenib in BRAF mutant melanoma and other malignancies. Expert Opin. Drug Metab. Toxicol. 2013, 9, 893–899. [Google Scholar] [CrossRef] [PubMed]
- Laquerre, S.; Amone, M.; Moss, K.; Yang, J.; Fisher, K.; Kane-Carson, L.S.; Smitheman, K.; Ward, J.; Heidrich, B.; Rheault, T.; et al. A selective Raf kinase inhibitor induces cell death and tumor regression of human cancer cell lines encoding B-Raf(V600E) mutation. Mol. Cancer Ther. 2009, 8. [Google Scholar] [CrossRef]
- Roskoski, R., Jr. Targeting ERK1/2 protein-serine/threonine kinases in human cancers. Pharmacol. Res. 2019, 142, 151–168. [Google Scholar] [CrossRef]
- Carlino, M.S.; Gowrishankar, K.; Saunders, C.A.B.; Pupo, G.M.; Snoyman, S.; Zhang, X.D.; Saw, R.; Becker, T.M.; Kefford, R.F.; Long, G.V.; et al. Antiproliferative effects of continued mitogen-activated protein kinase pathway inhibition following acquired resistance to BRAF and/or MEK inhibition in melanoma. Mol. Cancer Ther. 2013, 12, 1332–1342. [Google Scholar] [CrossRef]
- Kakadia, S.; Yarlagadda, N.; Awad, R.; Kundranda, M.; Niu, J.; Naraev, B.; Mina, L.; Dragovich, T.; Gimbel, M.; Mahmoud, F. Mechanisms of resistance to BRAF and MEK inhibitors and clinical update of US Food and Drug Administration-approved targeted therapy in advanced melanoma. Oncotargets Ther. 2018, 11, 7095–7107. [Google Scholar] [CrossRef]
- Long, G.V.; Fung, C.; Menzies, A.M.; Pupo, G.M.; Carlino, M.S.; Hyman, J.; Shahheydari, H.; Tembe, V.; Thompson, J.F.; Saw, R.P.; et al. Increased MAPK reactivation in early resistance to dabrafenib/trametinib combination therapy of BRAF-mutant metastatic melanoma. Nat. Commun. 2014, 5, 5694. [Google Scholar] [CrossRef]
- Lian, T.; Li, C.; Wang, H. Trametinib in the treatment of multiple malignancies harboring MEK1 mutations. Cancer Treat. Rev. 2019, 81, 101907. [Google Scholar] [CrossRef] [PubMed]
- Cocco, E.; Schram, A.M.; Kulick, A.; Misale, S.; Won, H.H.; Yaeger, R.; Razavi, P.; Ptashkin, R.; Hechtman, J.F.; Toska, E.; et al. Resistance to TRK inhibition mediated by convergent MAPK pathway activation. Nat. Med. 2019, 25, 1422–1427. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Rauch, J.; Kolch, W. Targeting MAPK signaling in cancer: Mechanisms of drug resistance and sensitivity. Int. J. Mol. Sci. 2020, 21, 1102. [Google Scholar] [CrossRef] [PubMed]
- Yeh, T.C.; Marsh, V.; Bernat, B.A.; Ballard, J.; Colwell, H.; Evans, R.J.; Parry, J.; Smith, D.; Brandhuber, B.J.; Gross, S.; et al. Biological characterization of ARRY-142886 (AZD6244), a potent, highly selective mitogen-activated protein kinase kinase 1/2 inhibitor. Clin. Cancer Res. 2007, 13, 1576–1583. [Google Scholar] [CrossRef] [PubMed]
- Davies, B.R.; Logie, A.; McKay, J.S.; Martin, P.; Steele, S.; Jenkins, R.; Cockerill, M.; Cartlidge, S.; Smith, P.D. AZD6244 (ARRY-142886), a potent inhibitor of mitogen-activated protein kinase/extracellular signal-regulated kinase kinase 1/2 kinases: Mechanism of action in vivo, pharmacokinetic/pharmacodynamic relationship, and potential for combination in preclinical models. Mol. Cancer Ther. 2007, 6, 2209–2219. [Google Scholar] [CrossRef]
- Kim, D.W.; Patel, S.P. Profile of selumetinib and its potential in the treatment of melanoma. OncoTargets Ther. 2014, 7, 1631–1639. [Google Scholar] [CrossRef][Green Version]
- Kim, S.; Ding, W.; Zhang, L.; Tian, W.; Chen, S. Clinical response to sunitinib as a multitargeted tyrosine-kinase inhibitor (TKI) in solid cancers: A review of clinical trials. OncoTargets Ther. 2014, 7, 719–728. [Google Scholar] [CrossRef]
- Casaluce, F.; Sgambato, A.; Maione, P.; Sacco, P.C.; Santabarbara, G.; Gridelli, C. Selumetinib for the treatment of non-small cell lung cancer. Expert Opin. Investig. Drugs 2017, 26, 973–984. [Google Scholar] [CrossRef]
- Bernabe, R.; Patrao, A.; Carter, L.; Blackhall, F.; Dean, E. Selumetinib in the treatment of non-small-cell lung cancer. Future Oncol. 2016, 12, 2545–2560. [Google Scholar] [CrossRef]
- Engelman, J.A.; Chen, L.; Tan, X.H.; Crosby, K.; Guimaraes, A.R.; Upadhyay, R.; Maira, M.; McNamara, K.; Perera, S.A.; Song, Y.C.; et al. Effective use of PI3K and MEK inhibitors to treat mutant Kras G12D and PIK3CA H1047R murine lung cancers. Nat. Med. 2008, 14, 1351–1356. [Google Scholar] [CrossRef]
- Dreaden, E.C.; Kong, Y.W.; Morton, S.W.; Correa, S.; Choi, K.Y.; Shopsowitz, K.E.; Renggli, K.; Drapkin, R.; Yaffe, M.B.; Hammond, P.T. Tumor-targeted synergistic blockade of MAPK and PI3K from a layer-by-layer nanoparticle. Clin. Cancer Res. 2015, 21, 4410–4419. [Google Scholar] [CrossRef]
- Atefi, M.; von Euw, E.; Attar, N.; Ng, C.; Chu, C.; Guo, D.L.; Nazarian, R.; Chmielowski, B.; Glaspy, J.A.; Comin-Anduix, B.; et al. Reversing melanoma cross-resistance to BRAF and MEK inhibitors by Co-targeting the AKT/mTOR pathway. PLoS ONE 2011, 6. [Google Scholar] [CrossRef]
- Emery, C.M.; Vijayendran, K.G.; Zipser, M.C.; Sawyer, A.M.; Niu, L.; Kim, J.J.; Hatton, C.; Chopra, R.; Oberholzer, P.A.; Karpova, M.B.; et al. MEK1 mutations confer resistance to MEK and B-RAF inhibition. Proc. Natl. Acad. Sci. USA 2009, 106, 20411–20416. [Google Scholar] [CrossRef] [PubMed]
- Poole, R.M. Pembrolizumab: First global approval. Drugs 2014, 74, 1973–1981. [Google Scholar] [CrossRef] [PubMed]
- Flynn, J.P.; Gerriets, V. Pembrolizumab. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2020. [Google Scholar]
- Pardoll, D.M. The blockade of immune checkpoints in cancer immunotherapy. Nat. Rev. Cancer 2012, 12, 252–264. [Google Scholar] [CrossRef] [PubMed]
- Mamalis, A.; Garcha, M.; Jagdeo, J. Targeting the PD-1 pathway: A promising future for the treatment of melanoma. Arch. Dermatol. Res. 2014, 306, 511–519. [Google Scholar] [CrossRef] [PubMed]
- Tang, P.A.; Heng, D.Y.C. Programmed Death 1 Pathway inhibition in Metastatic Renal Cell Cancer and Prostate Cancer. Curr. Oncol. Rep. 2013, 15, 98–104. [Google Scholar] [CrossRef]
- Sahni, S.; Valecha, G.; Sahni, A. Role of Anti-PD-1 Antibodies in Advanced Melanoma: The Era of Immunotherapy. Cureus 2018, 10. [Google Scholar] [CrossRef]
- Hargadon, K.M.; Johnson, C.E.; Williams, C.J. Immune checkpoint blockade therapy for cancer: An overview of FDA-approved immune checkpoint inhibitors. Int. Immunopharmacol. 2018, 62, 29–39. [Google Scholar] [CrossRef]
- Simsek, M.; Tekin, S.B.; Bilici, M. Immunological Agents Used in Cancer Treatment. Eurasian J. Med. 2019, 51, 90–94. [Google Scholar] [CrossRef]
- Hodi, F.S. Improved Survival with Ipilimumab in Patients with Metastatic Melanoma. N. Engl. J. Med. 2010, 363, 1290-1290. [Google Scholar] [CrossRef] [PubMed]
- Peggs, K.S.; Quezada, S.A.; Chambers, C.A.; Korman, A.J.; Allison, J.P. Blockade of CTLA-4 on both effector and regulatory T cell compartments contributes to the antitumor activity of anti-CTLA-4 antibodies. J. Exp. Med. 2009, 206, 1717–1725. [Google Scholar] [CrossRef]
- Wei, S.C.; Levine, J.H.; Cogdill, A.P.; Zhao, Y.; Anang, N.A.A.S.; Andrews, M.C.; Sharma, P.; Wang, J.; Wargo, J.A.; Pe’er, D.; et al. Distinct cellular mechanisms underlie Anti-CTLA-4 and Anti-PD-1 checkpoint blockade. Cell 2017, 170, 1120–1133. [Google Scholar] [CrossRef] [PubMed]
- Zaretsky, J.M.; Garcia-Diaz, A.; Shin, D.S.; Escuin-Ordinas, H.; Hugo, W.; Hu-Lieskovan, S.; Torrejon, D.Y.; Abril-Rodriguez, G.; Sandoval, S.; Barthly, L.; et al. Mutations associated with acquired resistance to PD-1 blockade in melanoma. N. Engl. J. Med. 2016, 375, 819–829. [Google Scholar] [CrossRef] [PubMed]
- Bach, E.A.; Aguet, M.; Schreiber, R.D. The IFN gamma receptor: A paradigm for cytokine receptor signaling. Annu. Rev. Immunol. 1997, 15, 563–591. [Google Scholar] [CrossRef]
- Muller, M.; Briscoe, J.; Laxton, C.; Guschin, D.; Ziemiecki, A.; Silvennoinen, O.; Harpur, A.G.; Barbieri, G.; Witthuhn, B.A.; Schindler, C.; et al. The protein-tyrosine kinase Jak1 complements defects in interferon-alpha/beta and interferon-gamma signal-transduction. Nature 1993, 366, 129–135. [Google Scholar] [CrossRef]
- Zhuang, Y.; Liu, C.; Liu, J.; Li, G. Resistance mechanism of PD-1/PD-L1 blockade in the cancer-immunity cycle. Onco. Targets Ther. 2020, 13, 83–94. [Google Scholar] [CrossRef]
- Robarge, K.D.; Brunton, S.A.; Castanedo, G.M.; Cui, Y.; Dina, M.S.; Goldsmith, R.; Gould, S.E.; Guichert, O.; Gunzner, J.L.; Halladay, J.; et al. GDC-0449-A potent inhibitor of the hedgehog pathway. Bioorganic Med. Chem. Lett. 2009, 19, 5576–5581. [Google Scholar] [CrossRef]
- Huang, P.X.; Zheng, S.D.; Wierbowski, B.M.; Kim, Y.; Nedelcu, D.; Aravena, L.; Liu, J.; Kruse, A.C.; Salic, A. Structural basis of smoothened activation in hedgehog signaling. Cell 2018, 174, 312–324. [Google Scholar] [CrossRef]
- Alcedo, J.; Ayzenzon, M.; VonOhlen, T.; Noll, M.; Hooper, J.E. The Drosophila smoothened gene encodes a seven-pass membrane protein, a putative receptor for the hedgehog signal. Cell 1996, 86, 221–232. [Google Scholar] [CrossRef]
- Van den Heuvel, M.; Ingham, P.W. Smoothened encodes a receptor-like serpentine protein required for hedgehog signalling. Nature 1996, 382, 547–551. [Google Scholar] [CrossRef] [PubMed]
- Carballo, G.B.; Honorato, J.R.; de Lopes, G.P.F.; Spohr, T. A highlight on Sonic hedgehog pathway. Cell Commun. Signal. 2018, 16, 11. [Google Scholar] [CrossRef] [PubMed]
- Skoda, A.M.; Simovic, D.; Karin, V.; Kardum, V.; Vranic, S.; Serman, L. The role of the Hedgehog signaling pathway in cancer: A comprehensive review. Bosn. J. Basic Med. Sci. 2018, 18, 8–20. [Google Scholar] [CrossRef] [PubMed]
- Yauch, R.L.; Gould, S.E.; Scales, S.J.; Tang, T.; Tian, H.; Ahn, C.P.; Marshall, D.; Fu, L.; Januario, T.; Kallop, D.; et al. A paracrine requirement for hedgehog signalling in cancer. Nature 2008, 455, 406–410. [Google Scholar] [CrossRef] [PubMed]
- Fattahi, S.; Langroudi, M.P.; Akhavan-Niaki, H. Hedgehog signaling pathway: Epigenetic regulation and role in disease and cancer development. J. Cell. Physiol. 2018, 233, 5726–5735. [Google Scholar] [CrossRef] [PubMed]
- Kinzler, K.W.; Ruppert, J.M.; Bigner, S.H.; Vogelstein, B. The GLI gene is a member of the Kruppel family of zinc finger proteins. Nature 1988, 332, 371–374. [Google Scholar] [CrossRef] [PubMed]
- Ingham, P.W.; McMahon, A.P. Hedgehog signaling in animal development: Paradigms and principles. Genes Dev. 2001, 15, 3059–3087. [Google Scholar] [CrossRef]
- Rubin, L.L.; de Sauvage, F.J. Targeting the Hedgehog pathway in cancer. Nat. Rev. Drug Discov. 2006, 5, 1026–1033. [Google Scholar] [CrossRef]
- Salaritabar, A.; Berindan-Neagoe, I.; Darvish, B.; Hadjiakhoondi, F.; Manayi, A.; Devi, K.P.; Barreca, D.; Orhan, I.E.; Suntar, I.; Farooqi, A.A.; et al. Targeting Hedgehog signaling pathway: Paving the road for cancer therapy. Pharmacol. Res. 2019, 141, 466–480. [Google Scholar] [CrossRef]
- Girardi, D.; Barrichello, A.; Fernandes, G.; Pereira, A. Targeting the hedgehog pathway in cancer: current evidence and future perspectives. Cells 2019, 8, 153. [Google Scholar] [CrossRef]
- Chahal, K.K.; Parle, M.; Abagyan, R. Hedgehog pathway and smoothened inhibitors in cancer therapies. Anti Cancer Drug 2018, 29, 387–401. [Google Scholar] [CrossRef]
- Lorusso, P.M.; Jimeno, A.; Dy, G.; Adjei, A.; Berlin, J.; Leichman, L.; Low, J.A.; Colburn, D.; Chang, I.; Cheeti, S.; et al. Pharmacokinetic dose-scheduling study of hedgehog pathway inhibitor vismodegib (GDC-0449) in patients with locally advanced or metastatic solid tumors. Clin. Cancer Res. 2011, 17, 5774–5782. [Google Scholar] [CrossRef] [PubMed]
- Keating, G.M. Vismodegib: In locally advanced or metastatic basal cell carcinoma. Drugs 2012, 72, 1535–1541. [Google Scholar] [CrossRef] [PubMed]
- Frampton, J.E.; Basset-Seguin, N. Vismodegib: A review in advanced basal cell carcinoma. Drugs 2018, 78, 1145–1156. [Google Scholar] [CrossRef] [PubMed]
- Fecher, L.A.; Sharfman, W.H. Advanced basal cell carcinoma, the hedgehog pathway, and treatment options—Role of smoothened inhibitors. Biologics 2015, 9, 129–140. [Google Scholar] [CrossRef] [PubMed]
- Sekulic, A.; Migden, M.R.; Oro, A.E.; Dirix, L.; Lewis, K.D.; Hainsworth, J.D.; Solomon, J.A.; Yoo, S.; Arron, S.T.; Friedlander, P.A.; et al. Efficacy and safety of vismodegib in advanced basal-cell carcinoma. N. Engl. J. Med. 2012, 366, 2171–2179. [Google Scholar] [CrossRef]
- Sharpe, H.J.; Pau, G.; Dijkgraaf, G.J.; Basset-Seguin, N.; Modrusan, Z.; Januario, T.; Tsui, V.; Durham, A.B.; Dlugosz, A.A.; Haverty, P.M.; et al. Genomic analysis of smoothened inhibitor resistance in basal cell carcinoma. Cancer Cell 2015, 27, 327–341. [Google Scholar] [CrossRef]
- Atwood, S.X.; Sarin, K.Y.; Whitson, R.J.; Li, J.R.; Kim, G.; Rezaee, M.; Ally, M.S.; Kim, J.; Yao, C.; Chang, A.L.; et al. Smoothened variants explain the majority of drug resistance in basal cell carcinoma. Cancer Cell 2015, 27, 342–353. [Google Scholar] [CrossRef]
- Wang, C.; Wu, H.; Katritch, V.; Han, G.W.; Huang, X.P.; Liu, W.; Siu, F.Y.; Roth, B.L.; Cherezov, V.; Stevens, R.C. Structure of the human smoothened receptor bound to an antitumour agent. Nature 2013, 497, 338–343. [Google Scholar] [CrossRef]
- Pusztai, L.; Mazouni, C.; Anderson, K.; Wu, Y.; Symmans, W.F. Molecular classification of breast cancer: Limitations and potential. Oncologist 2006, 11, 868–877. [Google Scholar] [CrossRef]
- Sorlie, T.; Perou, C.M.; Tibshirani, R.; Aas, T.; Geisler, S.; Johnsen, H.; Hastie, T.; Eisen, M.B.; van de Rijn, M.; Jeffrey, S.S.; et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc. Natl. Acad. Sci. USA 2001, 98, 10869–10874. [Google Scholar] [CrossRef] [PubMed]
- Jia, M.; Dahlman-Wright, K.; Gustafsson, J.A. Estrogen receptor alpha and beta in health and disease. Best Pract. Res. Clin. Endocrinol. Metab. 2015, 29, 557–568. [Google Scholar] [CrossRef] [PubMed]
- Gourdy, P.; Guillaume, M.; Fontaine, C.; Adlanmerini, M.; Montagner, A.; Laurell, H.; Lenfant, F.; Arnal, J.F. Estrogen receptor subcellular localization and cardiometabolism. Mol. Metab. 2018, 15, 56–69. [Google Scholar] [CrossRef] [PubMed]
- Livezey, M.; Kim, J.E.; Shapiro, D.J. A new role for estrogen receptor alpha in cell proliferation and cancer: Activating the anticipatory unfolded protein response. Front. Endocrinol. 2018, 9, 325. [Google Scholar] [CrossRef]
- Klinge, C.M. Estrogen receptor interaction with estrogen response elements. Nucleic Acids Res. 2001, 29, 2905–2919. [Google Scholar] [CrossRef]
- Ayaz, G.; Yasar, P.; Olgun, C.E.; Karakaya, B.; Kars, G.; Razizadeh, N.; Yavuz, K.; Turan, G.; Muyan, M. Dynamic transcriptional events mediated by estrogen receptor alpha. Front. Biosci. 2019, 24, 245–276. [Google Scholar]
- Kladde, M.P.; Xu, M.; Simpson, R.T. Direct study of DNA-protein interactions in repressed and active chromatin in living cells. EMBO J. 1996, 15, 6290–6300. [Google Scholar] [CrossRef]
- Gronemeyer, H. Transcription activation by estrogen and progesterone receptors. Annu. Rev. Genet. 1991, 25, 89–123. [Google Scholar] [CrossRef]
- Weigel, N.L.; Zhang, Y. Ligand-independent activation of steroid hormone receptors. J. Mol. Med. 1998, 76, 469–479. [Google Scholar] [CrossRef]
- Berger, C.E.; Qian, Y.; Liu, G.; Chen, H.; Chen, X. p53, a target of estrogen receptor (ER) alpha, modulates DNA damage-induced growth suppression in ER-positive breast cancer cells. J. Biol. Chem. 2012, 287, 30117–30127. [Google Scholar] [CrossRef]
- Xu, C.F.; Chambers, J.A.; Solomon, E. Complex regulation of the BRCA1 gene. J. Biol. Chem. 1997, 272, 20994–20997. [Google Scholar] [CrossRef] [PubMed]
- Fang, H.; Huang, D.; Yang, F.; Guan, X. Potential biomarkers of CDK4/6 inhibitors in hormone receptor-positive advanced breast cancer. Breast Cancer Res. Treat. 2018, 168, 287–297. [Google Scholar] [CrossRef] [PubMed]
- Osborne, C.K.; Schiff, R. Estrogen-receptor biology: Continuing progress and therapeutic implications. J. Clin. Oncol. 2005, 23, 1616–1622. [Google Scholar] [CrossRef] [PubMed]
- Nagaraj, G.; Ma, C. Revisiting the estrogen receptor pathway and its role in endocrine therapy for postmenopausal women with estrogen receptor-positive metastatic breast cancer. Breast Cancer Res. Treat. 2015, 150, 231–242. [Google Scholar] [CrossRef] [PubMed]
- Tang, S.; Han, H.; Bajic, V.B. ERGDB: Estrogen Responsive Genes Database. Nucleic Acids Res. 2004, 32, 533–536. [Google Scholar] [CrossRef]
- Ikeda, K.; Horie-Inoue, K.; Inoue, S. Identification of estrogen-responsive genes based on the DNA binding properties of estrogen receptors using high-throughput sequencing technology. Acta Pharmacol. Sin. 2015, 36, 24–31. [Google Scholar] [CrossRef] [PubMed]
- Kamalakaran, S.; Radhakrishnan, S.K.; Beck, W.T. Identification of estrogen-responsive genes using a genome-wide analysis of promoter elements for transcription factor binding sites. J. Biol. Chem. 2005, 280, 21491–21497. [Google Scholar] [CrossRef]
- Greenwalt, I.; Zaza, N.; Das, S.; Li, B.D. Precision medicine and targeted therapies in breast cancer. Surg. Oncol. Clin. N. Am. 2020, 29, 51–62. [Google Scholar] [CrossRef]
- Herrada, J.; Iyer, R.B.; Atkinson, E.N.; Sneige, N.; Buzdar, A.U.; Hortobagyi, G.N. Relative value of physical examination, mammography, and breast sonography in evaluating the size of the primary tumor and regional lymph node metastases in women receiving neoadjuvant chemotherapy for locally advanced breast carcinoma. Clin. Cancer Res. 1997, 3, 1565–1569. [Google Scholar]
- Goldhirsch, A.; Gelber, R.D. Endocrine therapies of breast cancer. Semin. Oncol. 1996, 23, 494–505. [Google Scholar]
- Metwally, I.H.; Hamdy, O.; Elbalka, S.S.; Elbadrawy, M.; Elsaid, D.M. Oophorectomy as a hormonal ablation therapy in metastatic and recurrent breast cancer: Current indications and results. India J. Surg. Oncol. 2019, 10, 542–546. [Google Scholar] [CrossRef] [PubMed]
- Abderrahman, B.; Jordan, V.C. Successful targeted therapies for breast cancer: The worcester foundation and future opportunities in women’s health. Endocrinology 2018, 159, 2980–2990. [Google Scholar] [CrossRef] [PubMed]
- Dustin, D.; Gu, G.W.; Fuqua, S.A.W. ESR1 mutations in breast cancer. Cancer 2019, 125, 3714–3728. [Google Scholar] [CrossRef] [PubMed]
- Haque, M.M.; Desai, K.V. Pathways to endocrine therapy resistance in breast cancer. Front. Endocrinol. 2019, 10. [Google Scholar] [CrossRef]
- Robinson, D.R.; Wu, Y.M.; Vats, P.; Su, F.Y.; Lonigro, R.J.; Cao, X.H.; Kalyana-Sundaram, S.; Wang, R.; Ning, Y.; Hodges, L.; et al. Activating ESR1 mutations in hormone-resistant metastatic breast cancer. Nat. Genet. 2013, 45, 1446–1451. [Google Scholar] [CrossRef]
- Jordan, V.C. Tamoxifen (ICI46,474) as a targeted therapy to treat and prevent breast cancer. Brit. J. Pharmacol. 2006, 147, S269–S276. [Google Scholar] [CrossRef]
- Jordan, V.C.; Koerner, S. Tamoxifen (Ici 46,474) and Human Carcinoma 8s Estrogen-Receptor. Eur. J. Cancer 1975, 11, 205–206. [Google Scholar] [CrossRef]
- Grainger, D.J.; Metcalfe, J.C. Tamoxifen: Teaching an old drug new tricks? Nat. Med. 1996, 2, 381–385. [Google Scholar] [CrossRef]
- Shiau, A.K.; Barstad, D.; Loria, P.M.; Cheng, L.; Kushner, P.J.; Agard, D.A.; Greene, G.L. The structural basis of estrogen receptor/coactivator recognition and the antagonism of this interaction by tamoxifen. Cell 1998, 95, 927–937. [Google Scholar] [CrossRef]
- Ascenzi, P.; Bocedi, A.; Marino, M. Structure-function relationship of estrogen receptor alpha and beta: Impact on human health. Mol. Asp. Med. 2006, 27, 299–402. [Google Scholar] [CrossRef]
- Merenbakh-Lamin, K.; Ben-Baruch, N.; Yeheskel, A.; Dvir, A.; Soussan-Gutman, L.; Jeselsohn, R.; Yelensky, R.; Brown, M.; Miller, V.A.; Sarid, D.; et al. D538G mutation in estrogen receptor-alpha: A novel mechanism for acquired endocrine resistance in breast cancer. Cancer Res. 2013, 73, 6856–6864. [Google Scholar] [CrossRef] [PubMed]
- Guttery, D.S.; Page, K.; Hills, A.; Woodley, L.; Marchese, S.D.; Rghebi, B.; Hastings, R.K.; Luo, J.L.; Pringle, J.H.; Stebbing, J.; et al. Noninvasive detection of activating estrogen receptor 1 (ESR1) mutations in estrogen receptor-positive metastatic breast cancer. Clin. Chem. 2015, 61, 974–982. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.S.; Martin, M.; Rugo, H.S.; Jones, S.; Im, S.A.; Gelmon, K.; Harbeck, N.; Lipatov, O.N.; Walshe, J.M.; Moulder, S.; et al. Palbociclib and letrozole in advanced breast cancer. N. Engl. J. Med. 2016, 375, 1925–1936. [Google Scholar] [CrossRef] [PubMed]
- Goetz, M.P.; Toi, M.; Campone, M.; Sohn, J.; Paluch-Shimon, S.; Huober, J.; Park, I.H.; Tredan, O.; Chen, S.C.; Manso, L.; et al. MONARCH 3: Abemaciclib as initial therapy for advanced breast cancer. J. Clin. Oncol. 2017, 35, 3638–3646. [Google Scholar] [CrossRef] [PubMed]
- Rodrik-Outmezguine, V.S.; Okaniwa, M.; Yao, Z.; Novotny, C.J.; McWhirter, C.; Banaji, A.; Won, H.; Wong, W.; Berger, M.; de Stanchina, E.; et al. Overcoming mTOR resistance mutations with a new-generation mTOR inhibitor. Nature 2016, 534, 272–276. [Google Scholar] [CrossRef]
- Murphy, C.G. The Role of CDK4/6 Inhibitors in Breast Cancer. Curr. Treat. Options Oncol. 2019, 20, 52. [Google Scholar] [CrossRef]
- Hamadeh, I.S.; Patel, J.N.; Rusin, S.; Tan, A.R. Personalizing aromatase inhibitor therapy in patients with breast cancer. Cancer Treat. Rev. 2018, 70, 47–55. [Google Scholar] [CrossRef]
- Ariazi, E.A.; Lewis-Wambi, J.S.; Gill, S.D.; Pyle, J.R.; Ariazi, J.L.; Kim, H.R.; Sharma, C.G.; Cordera, F.; Shupp, H.A.; Li, T.; et al. Emerging principles for the development of resistance to antihormonal therapy: implications for the clinical utility of fulvestrant. J. Steroid. Biochem. Mol. Biol. 2006, 102, 128–138. [Google Scholar] [CrossRef][Green Version]
- Rao, R.D.; Cobleigh, M.A. Adjuvant endocrine therapy for breast cancer. Oncology (Williston Park) 2012, 26, 541–547, 550, 552. [Google Scholar]
- Huerta-Reyes, M.; Maya-Nunez, G.; Perez-Solis, M.A.; Lopez-Munoz, E.; Guillen, N.; Olivo-Marin, J.C.; Aguilar-Rojas, A. Treatment of Breast Cancer With Gonadotropin-Releasing Hormone Analogs. Front. Oncol. 2019, 9, 943. [Google Scholar] [CrossRef]
- Fribbens, C.; O’Leary, B.; Kilburn, L.; Hrebien, S.; Garcia-Murillas, I.; Beaney, M.; Cristofanilli, M.; Andre, F.; Loi, S.; Loibl, S.; et al. Plasma ESR1 mutations and the treatment of estrogen receptor-positive advanced breast cancer. J. Clin. Oncol. 2016, 34, 2961–2968. [Google Scholar] [CrossRef] [PubMed]
- Chandarlapaty, S.; Chen, D.; He, W.; Sung, P.; Samoila, A.; You, D.Q.; Bhatt, T.; Patel, P.; Voi, M.; Gnant, M.; et al. Prevalence of ESR1 mutations in Cell-Free DNA and outcomes in metastatic breast cancer a secondary analysis of the BOLERO-2 clinical trial. JAMA Oncol. 2016, 2, 1310–1315. [Google Scholar] [CrossRef] [PubMed]
- Spoerke, J.M.; Gendreau, S.; Walter, K.; Qiu, J.H.; Wilson, T.R.; Savage, H.; Aimi, J.; Derynck, M.K.; Chen, M.; Chan, I.T.; et al. Heterogeneity and clinical significance of ESR1 mutations in ER-positive metastatic breast cancer patients receiving fulvestrant. Nat. Commun. 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Toy, W.; Weir, H.; Razavi, P.; Lawson, M.; Goeppert, A.U.; Mazzola, A.M.; Smith, A.; Wilson, J.; Morrow, C.; Wong, W.L.; et al. Activating ESR1 mutations differentially affect the efficacy of ER antagonists. Cancer Discov. 2017, 7, 277–287. [Google Scholar] [CrossRef]
- Takeshita, T.; Yamamoto, Y.; Yamamoto-Ibusuki, M.; Sueta, A.; Tomiguchi, M.; Murakami, K.; Omoto, Y.; Iwase, H. Prevalence of ESR1 E380Q mutation in tumor tissue and plasma from Japanese breast cancer patients. BMC Cancer 2017, 17. [Google Scholar] [CrossRef]
- Ross, D.S.; Zehir, A.; Brogi, E.; Konno, F.; Krystel-Whittemore, M.; Edelweiss, M.; Berger, M.F.; Toy, W.; Chandarlapaty, S.; Razavi, P.; et al. Immunohistochemical analysis of estrogen receptor in breast cancer with ESR1 mutations detected by hybrid capture-based next-generation sequencing. Mod. Pathol. 2019, 32, 81–87. [Google Scholar] [CrossRef]
- Najim, O.; Huizing, M.; Papadimitriou, K.; Trinh, X.B.; Pauwels, P.; Goethals, S.; Zwaenepoel, K.; Peterson, K.; Weyler, J.; Altintas, S.; et al. The prevalence of estrogen receptor-1 mutation in advanced breast cancer: The estrogen receptor one study (EROS1). Cancer Treat. Res. Commun. 2019, 19, 100123. [Google Scholar] [CrossRef]
- Toy, W.; Weir, H.; Razavi, P.; Berger, M.; Wong, W.L.; De Stanchina, E.; Baselga, J.; Chandarlapaty, S. Differential activity and SERD sensitivity of clinical ESR1 mutations. Cancer Res. 2016, 76. [Google Scholar] [CrossRef]
- Jeannot, E.; Darrigues, L.; Michel, M.; Stern, M.H.; Pierga, J.Y.; Rampanou, A.; Melaabi, S.; Benoist, C.; Bieche, I.; Vincent-Salomon, A.; et al. A single droplet digital PCR for ESR1 activating mutations detection in plasma. Oncogene 2020. [Google Scholar] [CrossRef]
- Khan, A.; Ashfaq Ur, R.; Junaid, M.; Li, C.D.; Saleem, S.; Humayun, F.; Shamas, S.; Ali, S.S.; Babar, Z.; Wei, D.Q. Dynamics insights into the gain of flexibility by Helix-12 in ESR1 as a mechanism of resistance to drugs in breast cancer cell lines. Front. Mol. Biosci. 2019, 6, 159. [Google Scholar] [CrossRef]
- Gucalp, A.; Traina, T.A. The Androgen Receptor: Is It a Promising Target? Ann. Surg. Oncol. 2017, 24, 2876–2880. [Google Scholar] [CrossRef] [PubMed]
- Brinkmann, A.O.; Trapman, J. Prostate cancer schemes for androgen escape. Nat. Med. 2000, 6, 628–629. [Google Scholar] [CrossRef] [PubMed]
- Tan, M.H.; Li, J.; Xu, H.E.; Melcher, K.; Yong, E.L. Androgen receptor: Structure, role in prostate cancer and drug discovery. Acta Pharmacol. Sin. 2015, 36, 3–23. [Google Scholar] [CrossRef] [PubMed]
- Cleutjens, K.B.; van Eekelen, C.C.; van der Korput, H.A.; Brinkmann, A.O.; Trapman, J. Two androgen response regions cooperate in steroid hormone regulated activity of the prostate-specific antigen promoter. J. Biol. Chem. 1996, 271, 6379–6388. [Google Scholar] [CrossRef]
- Pang, S.; Taneja, S.; Dardashti, K.; Cohan, P.; Kaboo, R.; Sokoloff, M.; Tso, C.L.; Dekernion, J.B.; Belldegrun, A.S. Prostate tissue specificity of the prostate-specific antigen promoter isolated from a patient with prostate cancer. Hum. Gene Ther. 1995, 6, 1417–1426. [Google Scholar] [CrossRef]
- Shang, Y.; Myers, M.; Brown, M. Formation of the androgen receptor transcription complex. Mol. Cell 2002, 9, 601–610. [Google Scholar] [CrossRef]
- Maximum androgen blockade in advanced prostate cancer: An overview of 22 randomised trials with 3283 deaths in 5710 patients. Prostate Cancer Trialists’ Collaborative Group. Lancet 1995, 346, 265–269.
- Litvinov, I.V.; De Marzo, A.M.; Isaacs, J.T. Is the Achilles’ heel for prostate cancer therapy a gain of function in androgen receptor signaling? J. Clin. Endocrinol. Metab. 2003, 88, 2972–2982. [Google Scholar] [CrossRef][Green Version]
- Singla, N.; Ghandour, R.A.; Raj, G.V. Investigational luteinizing hormone releasing hormone (LHRH) agonists and other hormonal agents in early stage clinical trials for prostate cancer. Expert Opin. Investig. Drugs 2019, 28, 249–259. [Google Scholar] [CrossRef]
- Feng, Q.; He, B. Androgen receptor signaling in the development of castration-resistant prostate cancer. Front. Oncol. 2019, 9, 858. [Google Scholar] [CrossRef]
- Hobisch, A.; Culig, Z.; Radmayr, C.; Bartsch, G.; Klocker, H.; Hittmair, A. Androgen receptor status of lymph node metastases from prostate cancer. Prostate 1996, 28, 129–135. [Google Scholar] [CrossRef]
- Taplin, M.E.; Bubley, G.J.; Ko, Y.J.; Small, E.J.; Upton, M.; Rajeshkumar, B.; Balk, S.P. Selection for androgen receptor mutations in prostate cancers treated with androgen antagonist. Cancer Res. 1999, 59, 2511–2515. [Google Scholar] [PubMed]
- Linja, M.J.; Savinainen, K.J.; Saramaki, O.R.; Tammela, T.L.; Vessella, R.L.; Visakorpi, T. Amplification and overexpression of androgen receptor gene in hormone-refractory prostate cancer. Cancer Res. 2001, 61, 3550–3555. [Google Scholar] [PubMed]
- Brown, R.S.; Edwards, J.; Dogan, A.; Payne, H.; Harland, S.J.; Bartlett, J.M.; Masters, J.R. Amplification of the androgen receptor gene in bone metastases from hormone-refractory prostate cancer. J. Pathol. 2002, 198, 237–244. [Google Scholar] [CrossRef]
- Culig, Z.; Hobisch, A.; Cronauer, M.V.; Cato, A.C.; Hittmair, A.; Radmayr, C.; Eberle, J.; Bartsch, G.; Klocker, H. Mutant androgen receptor detected in an advanced-stage prostatic carcinoma is activated by adrenal androgens and progesterone. Mol. Endocrinol. 1993, 7, 1541–1550. [Google Scholar] [CrossRef] [PubMed]
- Steinkamp, M.P.; O’Mahony, O.A.; Brogley, M.; Rehman, H.; Lapensee, E.W.; Dhanasekaran, S.; Hofer, M.D.; Kuefer, R.; Chinnaiyan, A.; Rubin, M.A.; et al. Treatment-dependent androgen receptor mutations in prostate cancer exploit multiple mechanisms to evade therapy. Cancer Res. 2009, 69, 4434–4442. [Google Scholar] [CrossRef] [PubMed]
- Rees, I.; Lee, S.; Kim, H.; Tsai, F.T. The E3 ubiquitin ligase CHIP binds the androgen receptor in a phosphorylation-dependent manner. Biochim. Biophys. Acta 2006, 1764, 1073–1079. [Google Scholar] [CrossRef] [PubMed]
- Veldscholte, J.; Ris-Stalpers, C.; Kuiper, G.G.; Jenster, G.; Berrevoets, C.; Claassen, E.; van Rooij, H.C.; Trapman, J.; Brinkmann, A.O.; Mulder, E. A mutation in the ligand binding domain of the androgen receptor of human LNCaP cells affects steroid binding characteristics and response to anti-androgens. Biochem. Biophys. Res. Commun. 1990, 173, 534–540. [Google Scholar] [CrossRef]
- Fenton, M.A.; Shuster, T.D.; Fertig, A.M.; Taplin, M.E.; Kolvenbag, G.; Bubley, G.J.; Balk, S.P. Functional characterization of mutant androgen receptors from androgen-independent prostate cancer. Clin. Cancer Res. 1997, 3, 1383–1388. [Google Scholar]
- Hobisch, A.; Colleselli, K.; Ennemoser, O.; Horninger, W.; Poisel, S.; Janetschek, G.; Bartsch, G. Modified retroperitoneal lymphadenectomy for testicular tumor: Anatomical approach, operative technique and results. Eur. Urol. 1993, 23, 39–43. [Google Scholar] [CrossRef]
- Brown, T.R.; Lubahn, D.B.; Wilson, E.M.; French, F.S.; Migeon, C.J.; Corden, J.L. Functional characterization of naturally occurring mutant androgen receptors from subjects with complete androgen insensitivity. Mol. Endocrinol. 1990, 4, 1759–1772. [Google Scholar] [CrossRef] [PubMed]
- Marcelli, M.; Tilley, W.D.; Zoppi, S.; Griffin, J.E.; Wilson, J.D.; Mcphaul, M.J. Androgen resistance associated with a mutation of the androgen receptor at amino-acid 772 (Arg-]Cys) results from a combination of decreased messenger-ribonucleic-acid levels and impairment of receptor function. J. Clin. Endocr. Metab. 1991, 73, 318–325. [Google Scholar] [CrossRef] [PubMed]
- Reichert, Z.R.; Hussain, M. Androgen receptor and beyond, targeting androgen signaling in castration-resistant prostate cancer. Cancer J. 2016, 22, 326–329. [Google Scholar] [CrossRef] [PubMed]
- Cabeza, M.; Sanchez-Marquez, A.; Garrido, M.; Silva, A.; Bratoeff, E. Recent advances in drug design and drug discovery for androgen-dependent diseases. Curr. Med. Chem. 2016, 23, 792–815. [Google Scholar] [CrossRef]
- Yap, T.A.; Carden, C.P.; Attard, G.; de Bono, J.S. Targeting CYP17: Established and novel approaches in prostate cancer. Curr. Opin. Pharmacol. 2008, 8, 449–457. [Google Scholar] [CrossRef]
- Hall, P.F. Cytochrome P-450 C21scc: One enzyme with two actions: Hydroxylase and lyase. J. Steroid Biochem. Mol. Biol. 1991, 40, 527–532. [Google Scholar] [CrossRef]
- Chen, Y.; Clegg, N.J.; Scher, H.I. Anti-androgens and androgen-depleting therapies in prostate cancer: New agents for an established target. Lancet Oncol. 2009, 10, 981–991. [Google Scholar] [CrossRef]
- Gupta, E.; Guthrie, T.; Tan, W. Changing paradigms in management of metastatic Castration Resistant Prostate Cancer (mCRPC). BMC Urol. 2014, 14, 55. [Google Scholar] [CrossRef]
- Potter, G.A.; Barrie, S.E.; Jarman, M.; Rowlands, M.G. Novel steroidal inhibitors of human cytochrome P45017 alpha (17 alpha-hydroxylase-C17,20-lyase): Potential agents for the treatment of prostatic cancer. J. Med. Chem. 1995, 38, 2463–2471. [Google Scholar] [CrossRef]
- Rowlands, M.G.; Barrie, S.E.; Chan, F.; Houghton, J.; Jarman, M.; Mccague, R.; Potter, G.A. Esters of 3-pyridylacetic acid that combine potent inhibition of 17-Alpha-Hydroxylase C-17,C-20-Lyase (Cytochrome P45017-Alpha) with resistance to esterase hydrolysis. J. Med. Chem. 1995, 38, 4191–4197. [Google Scholar] [CrossRef]
- McCague, R.; Rowlands, M.G.; Barrie, S.E.; Houghton, J. Inhibition of enzymes of estrogen and androgen biosynthesis by esters of 4-pyridylacetic acid. J. Med. Chem. 1990, 33, 3050–3055. [Google Scholar] [CrossRef] [PubMed]
- Attard, G.; Belldegrun, A.S.; de Bono, J.S. Selective blockade of androgenic steroid synthesis by novel lyase inhibitors as a therapeutic strategy for treating metastatic prostate cancer. BJU Int. 2005, 96, 1241–1246. [Google Scholar] [CrossRef] [PubMed]
- Ryan, C.J.; Smith, M.R.; de Bono, J.S.; Molina, A.; Logothetis, C.J.; de Souza, P.; Fizazi, K.; Mainwaring, P.; Piulats, J.M.; Ng, S.; et al. Abiraterone in metastatic prostate cancer without previous chemotherapy. N. Engl. J. Med. 2013, 368, 138–148. [Google Scholar] [CrossRef] [PubMed]
- Scott, L.J. Enzalutamide: A review in castration-resistant prostate cancer. Drugs 2018, 78, 1913–1924. [Google Scholar] [CrossRef] [PubMed]
- Tran, C.; Ouk, S.; Clegg, N.J.; Chen, Y.; Watson, P.A.; Arora, V.; Wongvipat, J.; Smith-Jones, P.M.; Yoo, D.; Kwon, A.; et al. Development of a second-generation antiandrogen for treatment of advanced prostate cancer. Science 2009, 324, 787–790. [Google Scholar] [CrossRef]
- Hejmej, A.; Bilinska, B. The effects of flutamide on cell-cell junctions in the testis, epididymis, and prostate. Reprod. Toxicol. 2018, 81, 1–16. [Google Scholar] [CrossRef]
- Neri, R.; Peets, E.; Watnick, A. Anti-Androgenicity of Flutamide and its Metabolite Sch 16423. Biochem. Soc. T 1979, 7, 565–569. [Google Scholar] [CrossRef]
- Anahara, R.; Toyama, Y.; Mori, C. Review of the histological effects of the anti-androgen, flutamide, on mouse testis. Reprod. Toxicol. 2008, 25, 139–143. [Google Scholar] [CrossRef]
- Romanel, A.; Tandefelt, D.G.; Conteduca, V.; Jayaram, A.; Casiraghi, N.; Wetterskog, D.; Salvi, S.; Amadori, D.; Zafeiriou, Z.; Rescigno, P.; et al. Plasma AR and abiraterone-resistant prostate cancer. Sci. Transl. Med. 2015, 7. [Google Scholar] [CrossRef]
- Chen, E.J.; Sowalsky, A.G.; Gao, S.; Cai, C.M.; Voznesensky, O.; Schaefer, R.; Loda, M.; True, L.D.; Ye, H.H.; Troncoso, P.; et al. Abiraterone treatment in castration-resistant prostate cancer selects for progesterone responsive mutant androgen receptors. Clin. Cancer Res. 2015, 21, 1273–1280. [Google Scholar] [CrossRef]
- Azad, A.A.; Volik, S.V.; Wyatt, A.W.; Haegert, A.; Le Bihan, S.; Bell, R.H.; Anderson, S.A.; McConeghy, B.; Shukin, R.; Bazov, J.; et al. Androgen receptor gene aberrations in circulating cell-free DNA: Biomarkers of therapeutic resistance in castration-resistant prostate cancer. Clin. Cancer Res. 2015, 21, 2315–2324. [Google Scholar] [CrossRef] [PubMed]
- Sperger, J.M.; Strotman, L.N.; Welsh, A.; Casavant, B.P.; Chalmers, Z.; Horn, S.; Heninger, E.; Thiede, S.M.; Tokar, J.; Gibbs, B.K.; et al. Integrated analysis of multiple biomarkers from circulating tumor cells enabled by exclusion-based analyte isolation. Clin. Cancer Res. 2017, 23, 746–756. [Google Scholar] [CrossRef] [PubMed]
- Korpal, M.; Korn, J.M.; Gao, X.; Rakiec, D.P.; Ruddy, D.A.; Doshi, S.; Yuan, J.; Kovats, S.G.; Kim, S.; Cooke, V.G.; et al. An F876L mutation in androgen receptor confers genetic and phenotypic resistance to MDV3100 (enzalutamide). Cancer Discov. 2013, 3, 1030–1043. [Google Scholar] [CrossRef] [PubMed]
- Prekovic, S.; van Royen, M.E.; Voet, A.R.; Geverts, B.; Houtman, R.; Melchers, D.; Zhang, K.Y.; Van den Broeck, T.; Smeets, E.; Spans, L.; et al. The effect of F877L and T878A mutations on androgen receptor response to enzalutamide. Mol. Cancer Ther. 2016, 15, 1702–1712. [Google Scholar] [CrossRef] [PubMed]
- Guagnano, V.; Furet, P.; Spanka, C.; Bordas, V.; Le Douget, M.; Stamm, C.; Brueggen, J.; Jensen, M.R.; Schnell, C.; Schmid, H.; et al. Discovery of 3-(2,6-dichloro-3,5-dimethoxy-phenyl)-1-{6-[4-(4-ethyl-piperazin-1-yl)-phenylamin o]-pyrimidin-4-yl}-1-methyl-urea (NVP-BGJ398), a potent and selective inhibitor of the fibroblast growth factor receptor family of receptor tyrosine kinase. J. Med. Chem. 2011, 54, 7066–7083. [Google Scholar] [CrossRef]
- Nogova, L.; Sequist, L.V.; Perez Garcia, J.M.; Andre, F.; Delord, J.P.; Hidalgo, M.; Schellens, J.H.; Cassier, P.A.; Camidge, D.R.; Schuler, M.; et al. Evaluation of BGJ398, a fibroblast growth factor receptor 1-3 kinase inhibitor, in patients with advanced solid tumors harboring genetic alterations in fibroblast growth factor receptors: Results of a global phase i, dose-escalation and dose-expansion study. J. Clin. Oncol. 2017, 35, 157–165. [Google Scholar] [CrossRef]
- Harimoto, N.; Taguchi, K.; Shirabe, K.; Adachi, E.; Sakaguchi, Y.; Toh, Y.; Okamura, T.; Kayashima, H.; Taketomi, A.; Maehara, Y. The significance of fibroblast growth factor receptor 2 expression in differentiation of hepatocellular carcinoma. Oncology 2010, 78, 361–368. [Google Scholar] [CrossRef]
- Touat, M.; Ileana, E.; Postel-Vinay, S.; Andre, F.; Soria, J.C. Targeting FGFR signaling in cancer. Clin. Cancer Res. 2015, 21, 2684–2694. [Google Scholar] [CrossRef]
- Casadei, C.; Dizman, N.; Schepisi, G.; Cursano, M.C.; Basso, U.; Santini, D.; Pal, S.K.; De Giorgi, U. Targeted therapies for advanced bladder cancer: New strategies with FGFR inhibitors. Ther. Adv. Med. Oncol. 2019, 11, 1758835919890285. [Google Scholar] [CrossRef]
- Huynh, H.; Lee, L.Y.; Goh, K.Y.; Ong, R.; Hao, H.X.; Huang, A.L.; Wang, Y.Z.; Porta, D.G.; Chow, P.; Chung, A. Infigratinib mediates vascular normalization, impairs metastasis, and improves chemotherapy in hepatocellular carcinoma. Hepatology 2019, 69, 943–958. [Google Scholar] [CrossRef]
- Javle, M.; Lowery, M.; Shroff, R.T.; Weiss, K.H.; Springfeld, C.; Borad, M.J.; Ramanathan, R.K.; Goyal, L.; Sadeghi, S.; Macarulla, T.; et al. Phase II study of BGJ398 in patients with FGFR-Altered advanced cholangiocarcinoma. J. Clin. Oncol. 2018, 36, 276. [Google Scholar] [CrossRef] [PubMed]
- Javle, M.; Borbath, I.; Clarke, S.; Hitre, E.; Louvet, C.; Macarulla, T.; Oh, D.; Spratlin, J.; Valle, J.; Weiss, K.; et al. Phase 3 multicenter, open-label, randomized study of infigratinib versus gemcitabine plus cisplatin in the first-line treatment of patients with advanced cholangiocarcinoma with FGFR2 gene fusions/translocations: The PROOF trial. Ann. Oncol. 2019, 30. [Google Scholar] [CrossRef] [PubMed]
- Goyal, L.; Saha, S.K.; Liu, L.Y.; Siravegna, G.; Leshchiner, I.; Ahronian, L.G.; Lennerz, J.K.; Vu, P.; Deshpande, V.; Kambadakone, A.; et al. Polyclonal secondary FGFR2 mutations drive acquired resistance to FGFR inhibition in patients with FGFR2 fusion-positive cholangiocarcinoma. Cancer Discov. 2017, 7, 252–263. [Google Scholar] [CrossRef] [PubMed]
- Hasskarl, J. Everolimus. Recent Results Cancer Res. 2014, 201, 373–392. [Google Scholar] [CrossRef] [PubMed]
- Wagle, N.; Grabiner, B.C.; Van Allen, E.M.; Amin-Mansour, A.; Taylor-Weiner, A.; Rosenberg, M.; Gray, N.; Barletta, J.A.; Guo, Y.; Swanson, S.J.; et al. Response and acquired resistance to everolimus in anaplastic thyroid cancer. N. Engl. J. Med. 2014, 371, 1426–1433. [Google Scholar] [CrossRef] [PubMed]
- Lorenz, M.C.; Heitman, J. Tor mutations confer rapamycin resistance by preventing interaction with Fkbp12-rapamycin. J. Biol. Chem. 1995, 270, 27531–27537. [Google Scholar] [CrossRef]
- Zou, H.Y.; Li, Q.; Lee, J.H.; Arango, M.E.; Burgess, K.; Qiu, M.; Engstrom, L.D.; Yamazaki, S.; Parker, M.; Timofeevski, S.; et al. Sensitivity of selected human tumor models to PF-04217903, a novel selective c-Met kinase inhibitor. Mol. Cancer Ther. 2012, 11, 1036–1047. [Google Scholar] [CrossRef]
- Diamond, J.R.; Salgia, R.; Varella-Garcia, M.; Kanteti, R.; LoRusso, P.M.; Clark, J.W.; Xu, L.G.; Wilner, K.; Eckhardt, S.G.; Ching, K.A.; et al. Initial clinical sensitivity and acquired resistance to MET inhibition in MET-mutated papillary renal cell carcinoma. J. Clin. Oncol. 2013, 31, e254–e258. [Google Scholar] [CrossRef]
- Sebolt-Leopold, J.S. Advances in the development of cancer therapeutics directed against the RAS-mitogen-activated protein kinase pathway. Clin. Cancer Res. 2008, 14, 3651–3656. [Google Scholar] [CrossRef]
- Brown, A.P.; Carlson, T.C.G.; Loi, C.M.; Graziano, M.J. Pharmacodynamic and toxicokinetic evaluation of the novel MEK inhibitor, PD0325901, in the rat following oral and intravenous administration. Cancer Chemoth. Pharmacol. 2007, 59, 671–679. [Google Scholar] [CrossRef]
- Kohno, M.; Pouyssegur, J. Targeting the ERK signaling pathway in cancer therapy. Ann. Med. 2006, 38, 200–211. [Google Scholar] [CrossRef] [PubMed]
- Thompson, N.; Lyons, J. Recent progress in targeting the Raf/MEK/ERK pathway with inhibitors in cancer drug discovery. Curr. Opin. Pharmacol. 2005, 5, 350–356. [Google Scholar] [CrossRef] [PubMed]
- Hatzivassiliou, G.; Liu, B.; O’Brien, C.; Spoerke, J.M.; Hoeflich, K.P.; Haverty, P.M.; Soriano, R.; Forrest, W.F.; Heldens, S.; Chen, H.; et al. ERK inhibition overcomes acquired resistance to MEK inhibitors. Mol. Cancer Ther. 2012, 11, 1143–1154. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Daouti, S.; Li, W.H.; Wen, Y.; Rizzo, C.; Higgins, B.; Packman, K.; Rosen, N.; Boylan, J.F.; Heimbrook, D.; et al. Identification of the MEK1(F129L) activating mutation as a potential mechanism of acquired resistance to MEK inhibition in human cancers carrying the B-RafV600E mutation. Cancer Res. 2011, 71, 5535–5545. [Google Scholar] [CrossRef]
- Jubb, H.C.; Saini, H.K.; Verdonk, M.L.; Forbes, S.A. COSMIC-3D provides structural perspectives on cancer genetics for drug discovery. Nat. Genet. 2018, 50, 1200–1202. [Google Scholar] [CrossRef]
- Bedard, P.L.; Hyman, D.M.; Davids, M.S.; Siu, L.L. Small molecules, big impact: 20 years of targeted therapy in oncology. Lancet 2020, 395, 1078–1088. [Google Scholar] [CrossRef]
- Yi, M.; Xu, L.; Jiao, Y.; Luo, S.; Li, A.; Wu, K. The role of cancer-derived microRNAs in cancer immune escape. J. Hematol. Oncol. 2020, 13, 25. [Google Scholar] [CrossRef]
- Singh, S.P.; Singh, R.; Gupta, O.P.; Gupta, S.; Brahma Bhatt, M.L. Immunotherapy: Newer therapeutic armamentarium against cancer stem cells. J. Oncol. 2020, 2020, 3963561. [Google Scholar] [CrossRef]
- Guzel, E.; Okyay, T.M.; Yalcinkaya, B.; Karacaoglu, S.; Gocmen, M.; Akcakuyu, M.H. Tumor suppressor and oncogenic role of long non-coding RNAs in cancer. North. Clin. Istanb. 2020, 7, 81–86. [Google Scholar] [CrossRef]
- Behl, T.; Kumar, C.; Makkar, R.; Gupta, A.; Sachdeva, M. Intercalating the role of MicroRNAs in cancer: As enemy or protector. Asian Pac. J. Cancer Prev. 2020, 21, 593–598. [Google Scholar] [CrossRef]
- Jiang, W.; Xia, J.; Xie, S.; Zou, R.; Pan, S.; Wang, Z.W.; Assaraf, Y.G.; Zhu, X. Long non-coding RNAs as a determinant of cancer drug resistance: Towards the overcoming of chemoresistance via modulation of lncRNAs. Drug Resist. Updates 2020, 50, 100683. [Google Scholar] [CrossRef] [PubMed]
- Allegra, A.; Musolino, C.; Tonacci, A.; Pioggia, G.; Gangemi, S. Interactions between the MicroRNAs and microbiota in cancer development: Roles and therapeutic opportunities. Cancers 2020, 12, 805. [Google Scholar] [CrossRef] [PubMed]
- Galon, J.; Bruni, D. Tumor immunology and tumor evolution: Intertwined histories. Immunity 2020, 52, 55–81. [Google Scholar] [CrossRef] [PubMed]
- Egen, J.G.; Ouyang, W.; Wu, L.C. Human anti-tumor immunity: Insights from immunotherapy clinical trials. Immunity 2020, 52, 36–54. [Google Scholar] [CrossRef]
- Luo, Y.; Yang, J.; Yu, J.; Liu, X.; Yu, C.; Hu, J.; Shi, H.; Ma, X. Long non-coding RNAs: Emerging roles in the immunosuppressive tumor microenvironment. Front. Oncol. 2020, 10, 48. [Google Scholar] [CrossRef]
- Li, H.; Yang, Y.; Hong, W.; Huang, M.; Wu, M.; Zhao, X. Applications of genome editing technology in the targeted therapy of human diseases: Mechanisms, advances and prospects. Signal. Transduct. Target. Ther. 2020, 5, 1. [Google Scholar] [CrossRef]
- Zatloukalova, P.; Krejcir, R.; Valik, D.; Vojtesek, B. CRISPR-Cas9 as a tool in cancer therapy. Klin. Onkol. 2019, 32, 13–18. [Google Scholar] [CrossRef]
- Datta, P.; Ray, S. Nanoparticulate formulations of radiopharmaceuticals: Strategy to improve targeting and biodistribution properties. J. Label. Comp. Radiopharm. 2020. [Google Scholar] [CrossRef]
- Haider, M.; Abdin, S.M.; Kamal, L.; Orive, G. Nanostructured lipid carriers for delivery of chemotherapeutics: A review. Pharmaceutics 2020, 12, 288. [Google Scholar] [CrossRef]
- Song, W.; Das, M.; Chen, X. Nanotherapeutics for immuno-oncology: A crossroad for new paradigms. Trends Cancer 2020, 6, 288–298. [Google Scholar] [CrossRef]
- Habibi, N.; Quevedo, D.F.; Gregory, J.V.; Lahann, J. Emerging methods in therapeutics using multifunctional nanoparticles. Wiley Interdiscip Rev. Nanomed Nanobiotechnol. 2020, e1625. [Google Scholar] [CrossRef] [PubMed]
- Chivere, V.T.; Kondiah, P.P.D.; Choonara, Y.E.; Pillay, V. Nanotechnology-based biopolymeric oral delivery platforms for advanced cancer treatment. Cancers 2020, 12, 522. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).