The Crosstalk between Tumor Cells and the Microenvironment in Hepatocellular Carcinoma: The Role of Exosomal microRNAs and Their Clinical Implications
Abstract
1. Introduction
1.1. The Tumor Microenvironment Consists of a Mass of Heterogeneous Cell Types
1.1.1. Lymphoid Cells
1.1.2. Myeloid Cells
1.1.3. Other Cells
1.2. Characteristics of Extracellular Vesicles
1.3. Exosomal miRNAs as Regulators of the Tumor Microenvironment
1.3.1. The Transfer of HCC-Derived Exosomal miRNA Induces Neighbor Cancer Cells to Proliferate in the TME
1.3.2. Cancer-Derived Exosomal miRNA Promote Angiogenesis by Targeting Endothelial Cells
1.3.3. CAFs Foster HCC Growth through Exosomal miRNA Exchange
1.3.4. The Interplay between Hepatocellular Carcinoma Cells and Adipocytes Creates a Favorable Microenvironment for Tumor Growth
1.3.5. Exosomes Play a Critical Role in Modulating Cancer Immune Escape
1.4. The Cellular Crosstalk in the Tumor Microenvironment Promotes Metastasis
1.5. Targeting Tumor Microenvironment Cells Using Exosomal miRNAs as a Future Antitumor Strategy
1.5.1. Exosomal miRNAs in HCC and Their Implication for Current Therapies
1.5.2. Exosomal miRNAs’ Future Implications for Immunotherapy in HCC
1.6. Exosomal miRNAs as Potential Biomarkers in HCC
2. Conclusions
Author Contributions
Funding
Conflicts of Interest
Appendix A
References
- Poltavets, V.; Kochetkova, M.; Pitson, S.M.; Samuel, M.S. The Role of the Extracellular Matrix and Its Molecular and Cellular Regulators in Cancer Cell Plasticity. Front. Oncol. 2018, 8. [Google Scholar] [CrossRef] [PubMed]
- Maia, J.; Caja, S.; Strano Moraes, M.C.; Couto, N.; Costa-Silva, B. Exosome-Based Cell-Cell Communication in the Tumor Microenvironment. Front. Cell Dev. Biol. 2018, 6. [Google Scholar] [CrossRef] [PubMed]
- Doyle, L.M.; Wang, M.Z. Overview of Extracellular Vesicles, Their Origin, Composition, Purpose, and Methods for Exosome Isolation and Analysis. Cells 2019, 8, 727. [Google Scholar] [CrossRef] [PubMed]
- Minciacchi, V.R.; Freeman, M.R.; Di Vizio, D. Extracellular Vesicles in Cancer: Exosomes, Microvesicles and the Emerging Role of Large Oncosomes. Semin. Cell Dev. Biol. 2015, 40, 41–51. [Google Scholar] [CrossRef] [PubMed]
- Romero-Cordoba, S.L.; Salido-Guadarrama, I.; Rodriguez-Dorantes, M.; Hidalgo-Miranda, A. miRNA biogenesis: Biological impact in the development of cancer. Cancer Biol. Ther. 2014, 15, 1444–1455. [Google Scholar] [CrossRef]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA. Cancer J. Clin. 2018, 68, 394–4240. [Google Scholar] [CrossRef]
- Balkwill, F.R.; Capasso, M.; Hagemann, T. The tumor microenvironment at a glance. J. Cell. Sci. 2012, 125, 5591–5596. [Google Scholar] [CrossRef]
- Parker, T.M.; Henriques, V.; Beltran, A.; Nakshatri, H.; Gogna, R. Cell competition and tumor heterogeneity. Semin. Cancer Biol. 2019, 9. pii: S1044-579X(18)30173-1. [Google Scholar] [CrossRef]
- Fu, Y.; Liu, S.; Zeng, S.; Shen, H. From bench to bed: The tumor immune microenvironment and current immunotherapeutic strategies for hepatocellular carcinoma. J. Exp. Clin. Cancer Res. 2019, 38, 396. [Google Scholar] [CrossRef]
- Delhem, N.; Carpentier, A.; Moralès, O.; Miroux, C.; Groux, H.; Auriault, C.; Pancré, V. [Regulatory T-cells and hepatocellular carcinoma: Implication of the regulatory T lymphocytes in the control of the immune response]. Bull. Cancer 2008, 95, 1219–1225. [Google Scholar]
- Bates, G.J.; Fox, S.B.; Han, C.; Leek, R.D.; Garcia, J.F.; Harris, A.L.; Banham, A.H. Quantification of regulatory T cells enables the identification of high-risk breast cancer patients and those at risk of late relapse. J. Clin. Oncol. 2006, 24, 5373–5380. [Google Scholar] [CrossRef] [PubMed]
- Curiel, T.J.; Coukos, G.; Zou, L.; Alvarez, X.; Cheng, P.; Mottram, P.; Evdemon-Hogan, M.; Conejo-Garcia, J.R.; Zhang, L.; Burow, M.; et al. Specific recruitment of regulatory T cells in ovarian carcinoma fosters immune privilege and predicts reduced survival. Nat. Med. 2004, 10, 942–949. [Google Scholar] [CrossRef] [PubMed]
- Imai, H.; Saio, M.; Nonaka, K.; Suwa, T.; Umemura, N.; Ouyang, G.-F.; Nakagawa, J.; Tomita, H.; Osada, S.; Sugiyama, Y.; et al. Depletion of CD4+CD25+ regulatory T cells enhances interleukin-2-induced antitumor immunity in a mouse model of colon adenocarcinoma. Cancer Sci. 2007, 98, 416–423. [Google Scholar] [CrossRef] [PubMed]
- Hannani, D.; Ma, Y.; Yamazaki, T.; Déchanet-Merville, J.; Kroemer, G.; Zitvogel, L. Harnessing γδ T cells in anticancer immunotherapy. Trends Immunol. 2012, 33, 199–206. [Google Scholar] [CrossRef]
- Flores-Borja, F.; Bosma, A.; Ng, D.; Reddy, V.; Ehrenstein, M.R.; Isenberg, D.A.; Mauri, C. CD19+CD24hiCD38hi B cells maintain regulatory T cells while limiting TH1 and TH17 differentiation. Sci. Transl. Med. 2013, 5, 173ra23. [Google Scholar] [CrossRef]
- Wang, W.; Yuan, X.; Chen, H.; Xie, G.; Ma, Y.; Zheng, Y.; Zhou, Y.; Shen, L. CD19+CD24hiCD38hiBregs involved in downregulate helper T cells and upregulate regulatory T cells in gastric cancer. Oncotarget 2015, 6, 33486–33499. [Google Scholar]
- Terrén, I.; Orrantia, A.; Vitallé, J.; Zenarruzabeitia, O.; Borrego, F. NK Cell Metabolism and Tumor Microenvironment. Front. Immunol. 2019, 10, 2278. [Google Scholar] [CrossRef]
- Chew, V.; Chen, J.; Lee, D.; Loh, E.; Lee, J.; Lim, K.H.; Weber, A.; Slankamenac, K.; Poon, R.T.P.; Yang, H.; et al. Chemokine-driven lymphocyte infiltration: An early intratumoural event determining long-term survival in resectable hepatocellular carcinoma. Gut 2012, 61, 427–438. [Google Scholar] [CrossRef]
- Cai, L.; Zhang, Z.; Zhou, L.; Wang, H.; Fu, J.; Zhang, S.; Shi, M.; Zhang, H.; Yang, Y.; Wu, H.; et al. Functional impairment in circulating and intrahepatic NK cells and relative mechanism in hepatocellular carcinoma patients. Clin. Immunol. 2008, 129, 428–437. [Google Scholar] [CrossRef]
- Sun, H.; Liu, L.; Huang, Q.; Liu, H.; Huang, M.; Wang, J.; Wen, H.; Lin, R.; Qu, K.; Li, K.; et al. Accumulation of Tumor-Infiltrating CD49a+ NK Cells Correlates with Poor Prognosis for Human Hepatocellular Carcinoma. Cancer Immunol. Res. 2019, 7, 1535–1546. [Google Scholar] [CrossRef]
- Gabrilovich, D.I.; Ostrand-Rosenberg, S.; Bronte, V. Coordinated regulation of myeloid cells by tumours. Nat. Rev. Immunol. 2012, 12, 253–268. [Google Scholar] [CrossRef] [PubMed]
- Mantovani, A.; Sozzani, S.; Locati, M.; Allavena, P.; Sica, A. Macrophage polarization: Tumor-associated macrophages as a paradigm for polarized M2 mononuclear phagocytes. Trends Immunol. 2002, 23, 549–555. [Google Scholar] [CrossRef]
- Minami, K.; Hiwatashi, K.; Ueno, S.; Sakoda, M.; Iino, S.; Okumura, H.; Hashiguchi, M.; Kawasaki, Y.; Kurahara, H.; Mataki, Y.; et al. Prognostic significance of CD68, CD163 and Folate receptor-β positive macrophages in hepatocellular carcinoma. Exp. Ther. Med. 2018, 15, 4465–4476. [Google Scholar] [CrossRef] [PubMed]
- Tian, Z.; Hou, X.; Liu, W.; Han, Z.; Wei, L. Macrophages and hepatocellular carcinoma. Cell Biosci. 2019, 9, 79. [Google Scholar] [CrossRef]
- Yeung, O.W.H.; Lo, C.-M.; Ling, C.-C.; Qi, X.; Geng, W.; Li, C.-X.; Ng, K.T.P.; Forbes, S.J.; Guan, X.-Y.; Poon, R.T.P.; et al. Alternatively activated (M2) macrophages promote tumour growth and invasiveness in hepatocellular carcinoma. J. Hepatol. 2015, 62, 607–616. [Google Scholar] [CrossRef]
- De Larco, J.E.; Wuertz, B.R.K.; Furcht, L.T. The potential role of neutrophils in promoting the metastatic phenotype of tumors releasing interleukin-8. Clin. Cancer Res. 2004, 10, 4895–4900. [Google Scholar] [CrossRef]
- Granot, Z.; Henke, E.; Comen, E.A.; King, T.A.; Norton, L.; Benezra, R. Tumor entrained neutrophils inhibit seeding in the premetastatic lung. Cancer Cell 2011, 20, 300–314. [Google Scholar] [CrossRef]
- Bronte, V.; Serafini, P.; Mazzoni, A.; Segal, D.M.; Zanovello, P. L-arginine metabolism in myeloid cells controls T-lymphocyte functions. Trends Immunol. 2003, 24, 302–306. [Google Scholar] [CrossRef]
- Yin, Z.; Dong, C.; Jiang, K.; Xu, Z.; Li, R.; Guo, K.; Shao, S.; Wang, L. Heterogeneity of cancer-associated fibroblasts and roles in the progression, prognosis, and therapy of hepatocellular carcinoma. J. Hematol. Oncol. 2019, 12, 1–9. [Google Scholar] [CrossRef]
- Hida, K.; Maishi, N.; Annan, D.A.; Hida, Y. Contribution of Tumor Endothelial Cells in Cancer Progression. Int. J. Mol. Sci. 2018, 19, 1272. [Google Scholar] [CrossRef]
- Nieman, K.M.; Kenny, H.A.; Penicka, C.V.; Ladanyi, A.; Buell-Gutbrod, R.; Zillhardt, M.R.; Romero, I.L.; Carey, M.S.; Mills, G.B.; Hotamisligil, G.S.; et al. Adipocytes promote ovarian cancer metastasis and provide energy for rapid tumor growth. Nat. Med. 2011, 17, 1498–1503. [Google Scholar] [CrossRef] [PubMed]
- Yáñez-Mó, M.; Siljander, P.R.-M.; Andreu, Z.; Zavec, A.B.; Borràs, F.E.; Buzas, E.I.; Buzas, K.; Casal, E.; Cappello, F.; Carvalho, J.; et al. Biological properties of extracellular vesicles and their physiological functions. J. Extracell Vesicles 2015, 4, 27066. [Google Scholar] [CrossRef] [PubMed]
- Al-Nedawi, K.; Meehan, B.; Micallef, J.; Lhotak, V.; May, L.; Guha, A.; Rak, J. Intercellular transfer of the oncogenic receptor EGFRvIII by microvesicles derived from tumour cells. Nat. Cell Biol. 2008, 10, 619–624. [Google Scholar] [CrossRef]
- Muralidharan-Chari, V.; Clancy, J.W.; Sedgwick, A.; D’Souza-Schorey, C. Microvesicles: Mediators of extracellular communication during cancer progression. J. Cell. Sci. 2010, 123, 1603–1611. [Google Scholar] [CrossRef] [PubMed]
- Ratajczak, J.; Wysoczynski, M.; Hayek, F.; Janowska-Wieczorek, A.; Ratajczak, M.Z. Membrane-derived microvesicles: Important and underappreciated mediators of cell-to-cell communication. Leukemia 2006, 20, 1487–1495. [Google Scholar] [CrossRef]
- Clancy, J.W.; Zhang, Y.; Sheehan, C.; D’Souza-Schorey, C. An ARF6-Exportin-5 axis delivers pre-miRNA cargo to tumour microvesicles. Nat. Cell Biol. 2019, 21, 856–866. [Google Scholar] [CrossRef]
- Tricarico, C.J.; Clancy, J.W.; D’Souza-Schorey, C. Biology and biogenesis of shed microvesicles. Small GTPases 2017, 8, 220–232. [Google Scholar] [CrossRef]
- Halicka, H.D.; Bedner, E.; Darzynkiewicz, Z. Segregation of RNA and Separate Packaging of DNA and RNA in Apoptotic Bodies during Apoptosis. Exp. Cell Res. 2000, 260, 248–256. [Google Scholar] [CrossRef]
- Morvan, J.; Rinaldi, B.; Friant, S. Pkh1/2-dependent phosphorylation of Vps27 regulates ESCRT-I recruitment to endosomes. Mol. Biol. Cell 2012, 23, 4054–4064. [Google Scholar] [CrossRef]
- Adell, M.A.Y.; Vogel, G.F.; Pakdel, M.; Müller, M.; Lindner, H.; Hess, M.W.; Teis, D. Coordinated binding of Vps4 to ESCRT-III drives membrane neck constriction during MVB vesicle formation. J. Cell Biol. 2014, 205, 33–49. [Google Scholar] [CrossRef]
- van Balkom, B.W.M.; Eisele, A.S.; Pegtel, D.M.; Bervoets, S.; Verhaar, M.C. Quantitative and qualitative analysis of small RNAs in human endothelial cells and exosomes provides insights into localized RNA processing, degradation and sorting. J. Extracell Vesicles 2015, 4, 26760. [Google Scholar] [CrossRef] [PubMed]
- Mathivanan, S.; Fahner, C.J.; Reid, G.E.; Simpson, R.J. ExoCarta 2012: Database of exosomal proteins, RNA and lipids. Nucleic Acids Res. 2012, 40, D1241–D1244. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.-K.; Kang, B.; Kim, O.Y.; Choi, D.-S.; Lee, J.; Kim, S.R.; Go, G.; Yoon, Y.J.; Kim, J.H.; Jang, S.C.; et al. EVpedia: An integrated database of high-throughput data for systemic analyses of extracellular vesicles. J. Extracell Vesicles 2013, 19, 2. [Google Scholar] [CrossRef] [PubMed]
- Hessvik, N.P.; Llorente, A. Current knowledge on exosome biogenesis and release. Cell. Mol. Life Sci. CMLS 2018, 75, 193–208. [Google Scholar] [CrossRef] [PubMed]
- Valadi, H.; Ekström, K.; Bossios, A.; Sjöstrand, M.; Lee, J.J.; Lötvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef] [PubMed]
- Kogure, T.; Lin, W.-L.; Yan, I.K.; Braconi, C.; Patel, T. Inter-cellular nanovesicle mediated microRNA transfer: A mechanism of environmental modulation of hepatocellular cancer cell growth. Hepatology 2011, 54, 1237–1248. [Google Scholar] [CrossRef]
- Villarroya-Beltri, C.; Gutiérrez-Vázquez, C.; Sánchez-Cabo, F.; Pérez-Hernández, D.; Vázquez, J.; Martin-Cofreces, N.; Martinez-Herrera, D.J.; Pascual-Montano, A.; Mittelbrunn, M.; Sánchez-Madrid, F. Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat. Commun. 2013, 4, 2980. [Google Scholar] [CrossRef]
- Kosaka, N.; Iguchi, H.; Hagiwara, K.; Yoshioka, Y.; Takeshita, F.; Ochiya, T. Neutral Sphingomyelinase 2 (nSMase2)-dependent Exosomal Transfer of Angiogenic MicroRNAs Regulate Cancer Cell Metastasis. J. Biol. Chem. 2013, 288, 10849–10859. [Google Scholar] [CrossRef]
- Chiche, J.; Brahimi-Horn, M.C.; Pouysségur, J. Tumour hypoxia induces a metabolic shift causing acidosis: A common feature in cancer. J. Cell. Mol. Med. 2010, 14, 771–794. [Google Scholar] [CrossRef]
- Lee, S.H.; Seo, G.S.; Park, Y.N.; Yoo, T.M.; Sohn, D.H. Effects and regulation of osteopontin in rat hepatic stellate cells. Biochem. Pharmacol. 2004, 68, 2367–2378. [Google Scholar] [CrossRef]
- Gatenby, R.A.; Gillies, R.J. Why do cancers have high aerobic glycolysis? Nat. Rev. Cancer 2004, 4, 891–899. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Ge, Z.; Yang, X.; Luo, Q.; Wang, C.; You, H.; Ge, T.; Deng, Y.; Lin, H.; Cui, Y.; et al. Hepatic stellate cells activated by acidic tumor microenvironment promote the metastasis of hepatocellular carcinoma via osteopontin. Cancer Lett. 2015, 356, 713–720. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Fukumura, D.; Jain, R.K. Acidic extracellular pH induces vascular endothelial growth factor (VEGF) in human glioblastoma cells via ERK1/2 MAPK signaling pathway: Mechanism of low pH-induced VEGF. J. Biol. Chem. 2002, 277, 11368–11374. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Chen, C.-H.; Tung, P.-Y.; Huang, S.-H.; Wang, S.-M. An acidic extracellular pH disrupts adherens junctions in HepG2 cells by Src kinases-dependent modification of E-cadherin. J. Cell. Biochem. 2009, 108, 851–859. [Google Scholar] [CrossRef] [PubMed]
- Gatenby, R.A.; Gawlinski, E.T.; Gmitro, A.F.; Kaylor, B.; Gillies, R.J. Acid-mediated tumor invasion: A multidisciplinary study. Cancer Res. 2006, 66, 5216–5223. [Google Scholar] [CrossRef] [PubMed]
- Tian, X.-P.; Wang, C.-Y.; Jin, X.-H.; Li, M.; Wang, F.-W.; Huang, W.-J.; Yun, J.-P.; Xu, R.-H.; Cai, Q.-Q.; Xie, D. Acidic Microenvironment Up-Regulates Exosomal miR-21 and miR-10b in Early-Stage Hepatocellular Carcinoma to Promote Cancer Cell Proliferation and Metastasis. Theranostics 2019, 9, 1965–1979. [Google Scholar] [CrossRef]
- Hujie, G.; Zhou, S.-H.; Zhang, H.; Qu, J.; Xiong, X.-W.; Hujie, O.; Liao, C.-G.; Yang, S.-E. MicroRNA-10b regulates epithelial-mesenchymal transition by modulating KLF4/KLF11/Smads in hepatocellular carcinoma. Cancer Cell Int. 2018, 18, 10. [Google Scholar] [CrossRef]
- Cao, L.-Q.; Yang, X.-W.; Chen, Y.-B.; Zhang, D.-W.; Jiang, X.-F.; Xue, P. Exosomal miR-21 regulates the TETs/PTENp1/PTEN pathway to promote hepatocellular carcinoma growth. Mol. Cancer 2019, 18, 148. [Google Scholar] [CrossRef]
- Gao, L.; Ren, W.; Zhang, L.; Li, S.; Kong, X.; Zhang, H.; Dong, J.; Cai, G.; Jin, C.; Zheng, D.; et al. PTENp1, a natural sponge of miR-21, mediates PTEN expression to inhibit the proliferation of oral squamous cell carcinoma. Mol. Carcinog. 2017, 56, 1322–1334. [Google Scholar] [CrossRef]
- Guo, X.; Lv, X.; Lv, X.; Ma, Y.; Chen, L.; Chen, Y. Circulating miR-21 serves as a serum biomarker for hepatocellular carcinoma and correlated with distant metastasis. Oncotarget 2017, 8, 44050–44058. [Google Scholar] [CrossRef]
- Wan, X.; Wang, H.; Jiang, M.; He, Y.; Liu, S.; Cao, H.; Qiu, X.; Tang, L.; Wu, M. [PTEN expression and its significance in human primary hepatocellular carcinoma]. Zhonghua Gan Zang Bing Za Zhi 2003, 11, 490–492. [Google Scholar] [PubMed]
- Sun, J.-F.; Zhang, D.; Gao, C.-J.; Zhang, Y.-W.; Dai, Q.-S. Exosome-Mediated MiR-155 Transfer Contributes to Hepatocellular Carcinoma Cell Proliferation by Targeting PTEN. Med. Sci. Monit. Basic Res. 2019, 25, 218–228. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Xu, H.-F.; Liu, M.-Y.; Xu, Y.-J.; He, J.-C.; Zhou, Y.; Cang, S.-D. Mechanism of exosomal microRNA-224 in development of hepatocellular carcinoma and its diagnostic and prognostic value. World J. Gastroenterol. 2019, 25, 1890–1898. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Tan, J.; Ou, S.; Chen, J.; Chen, L. Adipose-derived exosomes deliver miR-23a/b to regulate tumor growth in hepatocellular cancer by targeting the VHL/HIF axis. J. Physiol. Biochem. 2019, 75, 391–401. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Ren, H.; Dai, B.; Li, J.; Shang, L.; Huang, J.; Shi, X. Hepatocellular carcinoma-derived exosomal miRNA-21 contributes to tumor progression by converting hepatocyte stellate cells to cancer-associated fibroblasts. J. Exp. Clin. Cancer Res. 2018, 37, 324. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Li, X.; Sun, W.; Yue, S.; Yang, J.; Li, J.; Ma, B.; Wang, J.; Yang, X.; Pu, M.; et al. Loss of exosomal miR-320a from cancer-associated fibroblasts contributes to HCC proliferation and metastasis. Cancer Lett. 2017, 397, 33–42. [Google Scholar] [CrossRef]
- Lin, X.-J.; Fang, J.-H.; Yang, X.-J.; Zhang, C.; Yuan, Y.; Zheng, L.; Zhuang, S.-M. Hepatocellular Carcinoma Cell-Secreted Exosomal MicroRNA-210 Promotes Angiogenesis In Vitro and In Vivo. Mol. Ther. Nucl. Acids 2018, 11, 243–252. [Google Scholar] [CrossRef]
- Matsuura, Y.; Wada, H.; Eguchi, H.; Gotoh, K.; Kobayashi, S.; Kinoshita, M.; Kubo, M.; Hayashi, K.; Iwagami, Y.; Yamada, D.; et al. Exosomal miR-155 Derived from Hepatocellular Carcinoma Cells Under Hypoxia Promotes Angiogenesis in Endothelial Cells. Dig. Dis. Sci. 2019, 64, 792–802. [Google Scholar] [CrossRef]
- Zhao, S.; Li, J.; Zhang, G.; Wang, Q.; Wu, C.; Zhang, Q.; Wang, H.; Sun, P.; Xiang, R.; Yang, S. Exosomal miR-451a Functions as a Tumor Suppressor in Hepatocellular Carcinoma by Targeting LPIN1. Cell. Physiol. Biochem. 2019, 53, 19–35. [Google Scholar]
- Liu, J.; Fan, L.; Yu, H.; Zhang, J.; He, Y.; Feng, D.; Wang, F.; Li, X.; Liu, Q.; Li, Y.; et al. Endoplasmic Reticulum Stress Causes Liver Cancer Cells to Release Exosomal miR-23a-3p and Up-regulate Programmed Death Ligand 1 Expression in Macrophages. Hepatology 2019, 70, 241–258. [Google Scholar] [CrossRef]
- Yin, C.; Han, Q.; Xu, D.; Zheng, B.; Zhao, X.; Zhang, J. SALL4-mediated upregulation of exosomal miR-146a-5p drives T-cell exhaustion by M2 tumor-associated macrophages in HCC. Oncoimmunology 2019, 8, 1601479. [Google Scholar] [CrossRef] [PubMed]
- Nakano, T.; Chen, I.-H.; Wang, C.-C.; Chen, P.-J.; Tseng, H.-P.; Huang, K.-T.; Hu, T.-H.; Li, L.-C.; Goto, S.; Cheng, Y.-F.; et al. Circulating exosomal miR-92b: Its role for cancer immunoediting and clinical value for prediction of posttransplant hepatocellular carcinoma recurrence. Am. J. Transplant. 2019, 19, 3250–3262. [Google Scholar] [CrossRef] [PubMed]
- Yen, C.-H.; Lin, Y.-T.; Chen, H.-L.; Chen, S.-Y.; Chen, Y.-M.A. The multi-functional roles of GNMT in toxicology and cancer. Toxicol. Appl. Pharmacol. 2013, 266, 67–75. [Google Scholar] [CrossRef] [PubMed]
- Fernández, M.; Semela, D.; Bruix, J.; Colle, I.; Pinzani, M.; Bosch, J. Angiogenesis in liver disease. J. Hepatol. 2009, 50, 604–620. [Google Scholar] [CrossRef] [PubMed]
- Yasuda, S.; Arii, S.; Mori, A.; Isobe, N.; Yang, W.; Oe, H.; Fujimoto, A.; Yonenaga, Y.; Sakashita, H.; Imamura, M. Hexokinase II and VEGF expression in liver tumors: Correlation with hypoxia-inducible factor 1 alpha and its significance. J. Hepatol. 2004, 40, 117–123. [Google Scholar] [CrossRef]
- Mise, M.; Arii, S.; Higashituji, H.; Furutani, M.; Niwano, M.; Harada, T.; Ishigami, S.; Toda, Y.; Nakayama, H.; Fukumoto, M.; et al. Clinical significance of vascular endothelial growth factor and basic fibroblast growth factor gene expression in liver tumor. Hepatology 1996, 23, 455–464. [Google Scholar] [CrossRef]
- Gerber, H.P.; McMurtrey, A.; Kowalski, J.; Yan, M.; Keyt, B.A.; Dixit, V.; Ferrara, N. Vascular endothelial growth factor regulates endothelial cell survival through the phosphatidylinositol 3’-kinase/Akt signal transduction pathway. Requirement for Flk-1/KDR activation. J. Biol. Chem. 1998, 273, 30336–30343. [Google Scholar] [CrossRef]
- Vogel, C.; Bauer, A.; Wiesnet, M.; Preissner, K.T.; Schaper, W.; Marti, H.H.; Fischer, S. Flt-1, but not Flk-1 mediates hyperpermeability through activation of the PI3-K/Akt pathway. J. Cell. Physiol. 2007, 212, 236–243. [Google Scholar] [CrossRef]
- Li, X.; Li, X.; Lv, X.; Xiao, J.; Liu, B.; Zhang, Y. Smad4 Inhibits VEGF-A and VEGF-C Expressions via Enhancing Smad3 Phosphorylation in Colon Cancer. Anat. Rec. 2017, 300, 1560–1569. [Google Scholar] [CrossRef]
- Majmundar, A.J.; Wong, W.J.; Simon, M.C. Hypoxia-Inducible Factors and the Response to Hypoxic Stress. Mol. Cell 2010, 40, 294–309. [Google Scholar] [CrossRef]
- von Marschall, Z.; Cramer, T.; Höcker, M.; Finkenzeller, G.; Wiedenmann, B.; Rosewicz, S. Dual mechanism of vascular endothelial growth factor upregulation by hypoxia in human hepatocellular carcinoma. Gut 2001, 48, 87–96. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Wang, J.; Xiao, M.; Zhou, T.; Shi, X. Role of Mir-155 in Controlling HIF-1α Level and Promoting Endothelial Cell Maturation. Sci. Rep. 2016, 6, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Zhang, A.; Xiang, J.; Lv, Y.; Zhang, X. miR-451 acts as a suppressor of angiogenesis in hepatocellular carcinoma by targeting the IL-6R-STAT3 pathway. Oncol. Rep. 2016, 36, 1385–1392. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Bu, Z.; Zhao, F.; Xiao, D. Increased T-helper 17 cell differentiation mediated by exosome-mediated microRNA-451 redistribution in gastric cancer infiltrated T cells. Cancer Sci. 2018, 109, 65–73. [Google Scholar] [CrossRef] [PubMed]
- Baglieri, J.; Brenner, D.A.; Kisseleva, T. The Role of Fibrosis and Liver-Associated Fibroblasts in the Pathogenesis of Hepatocellular Carcinoma. Int. J. Mol. Sci. 2019, 20, 1723. [Google Scholar] [CrossRef] [PubMed]
- Balaban, S.; Shearer, R.F.; Lee, L.S.; van Geldermalsen, M.; Schreuder, M.; Shtein, H.C.; Cairns, R.; Thomas, K.C.; Fazakerley, D.J.; Grewal, T.; et al. Adipocyte lipolysis links obesity to breast cancer growth: Adipocyte-derived fatty acids drive breast cancer cell proliferation and migration. Cancer Metab. 2017, 13, 1. [Google Scholar] [CrossRef]
- Dirat, B.; Bochet, L.; Dabek, M.; Daviaud, D.; Dauvillier, S.; Majed, B.; Wang, Y.Y.; Meulle, A.; Salles, B.; Gonidec, S.L.; et al. Cancer-Associated Adipocytes Exhibit an Activated Phenotype and Contribute to Breast Cancer Invasion. Cancer Res. 2011, 71, 2455–2465. [Google Scholar] [CrossRef]
- Lazar, I.; Clement, E.; Dauvillier, S.; Milhas, D.; Ducoux-Petit, M.; LeGonidec, S.; Moro, C.; Soldan, V.; Dalle, S.; Balor, S.; et al. Adipocyte Exosomes Promote Melanoma Aggressiveness through Fatty Acid Oxidation: A Novel Mechanism Linking Obesity and Cancer. Cancer Res. 2016, 76, 4051–4057. [Google Scholar] [CrossRef]
- Estes, C.; Razavi, H.; Loomba, R.; Younossi, Z.; Sanyal, A.J. Modeling the epidemic of nonalcoholic fatty liver disease demonstrates an exponential increase in burden of disease. Hepatology 2018, 67, 123–133. [Google Scholar] [CrossRef]
- Golabi, P.; Rhea, L.; Henry, L.; Younossi, Z.M. Hepatocellular carcinoma and non-alcoholic fatty liver disease. Hepatol. Int. 2019, 13, 688–694. [Google Scholar] [CrossRef]
- Kaelin, W.G., Jr. The von Hippel-Lindau tumour suppressor protein: O2 sensing and cancer. Nat. Rev. Cancer 2008, 8, 865–873. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Xu, M.; Li, X.; Su, X.; Xiao, X.; Keating, A.; Zhao, R.C. Exosomes released by hepatocarcinoma cells endow adipocytes with tumor-promoting properties. J. Hematol. Oncol. 2018, 11, 82. [Google Scholar] [CrossRef]
- Han, Q.; Zhao, H.; Jiang, Y.; Yin, C.; Zhang, J. HCC-Derived Exosomes: Critical Player and Target for Cancer Immune Escape. Cells 2019, 8, 558. [Google Scholar] [CrossRef]
- Zhang, K.; Kaufman, R.J. From endoplasmic-reticulum stress to the inflammatory response. Nature 2008, 454, 455–462. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, J.; Yang, J.; Yu, X.; Chen, Z.; Chen, Y.; Kuang, M.; Zhu, Y.; Zhuang, S. Lnc-UCID Promotes G1/S Transition and Hepatoma Growth by Preventing DHX9-Mediated CDK6 Down-regulation. Hepatology 2019, 70, 259–275. [Google Scholar] [CrossRef] [PubMed]
- Doherty, D.G.; Norris, S.; Madrigal-Estebas, L.; McEntee, G.; Traynor, O.; Hegarty, J.E.; O’Farrelly, C. The human liver contains multiple populations of NK cells, T cells, and CD3+CD56+ natural T cells with distinct cytotoxic activities and Th1, Th2, and Th0 cytokine secretion patterns. J. Immunol. 1999, 163, 2314–2321. [Google Scholar] [PubMed]
- Wu, Y.; Kuang, D.-M.; Pan, W.-D.; Wan, Y.-L.; Lao, X.-M.; Wang, D.; Li, X.-F.; Zheng, L. Monocyte/macrophage-elicited natural killer cell dysfunction in hepatocellular carcinoma is mediated by CD48/2B4 interactions. Hepatology 2013, 57, 1107–1116. [Google Scholar] [CrossRef]
- Borrego, F.; Robertson, M.J.; Ritz, J.; Peña, J.; Solana, R. CD69 is a stimulatory receptor for natural killer cell and its cytotoxic effect is blocked by CD94 inhibitory receptor. Immunology 1999, 97, 159–165. [Google Scholar] [CrossRef]
- Zhuang, L.K.; Yang, Y.T.; Ma, X.; Han, B.; Wang, Z.S.; Zhao, Q.Y.; Wu, L.Q.; Qu, Z.Q. MicroRNA-92b promotes hepatocellular carcinoma progression by targeting Smad7 and is mediated by long non-coding RNA XIST. Cell Death Dis. 2016, 7, e2203. [Google Scholar] [CrossRef]
- Guo, S.; Deng, C.-X. Effect of Stromal Cells in Tumor Microenvironment on Metastasis Initiation. Int. J. Biol. Sci. 2018, 14, 2083–2093. [Google Scholar] [CrossRef]
- Kubo, N.; Araki, K.; Kuwano, H.; Shirabe, K. Cancer-associated fibroblasts in hepatocellular carcinoma. World J. Gastroenterol. 2016, 22, 6841–6850. [Google Scholar] [CrossRef] [PubMed]
- Fang, J.-H.; Zhang, Z.-J.; Shang, L.-R.; Luo, Y.-W.; Lin, Y.-F.; Yuan, Y.; Zhuang, S.-M. Hepatoma cell-secreted exosomal microRNA-103 increases vascular permeability and promotes metastasis by targeting junction proteins. Hepatology 2018, 68, 1459–1475. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Chen, W.; Zhi, X.; Chen, E.-J.; Wei, T.; Zhang, J.; Shen, J.; Hu, L.-Q.; Zhao, B.; Feng, X.-H.; et al. Tumor-derived exosomes promote tumor self-seeding in hepatocellular carcinoma by transferring miRNA-25-5p to enhance cell motility. Oncogene 2018, 37, 4964–4978. [Google Scholar] [CrossRef] [PubMed]
- Fang, T.; Lv, H.; Lv, G.; Li, T.; Wang, C.; Han, Q.; Yu, L.; Su, B.; Guo, L.; Huang, S.; et al. Tumor-derived exosomal miR-1247-3p induces cancer-associated fibroblast activation to foster lung metastasis of liver cancer. Nat. Commun. 2018, 9, 191. [Google Scholar] [CrossRef] [PubMed]
- Aucher, A.; Rudnicka, D.; Davis, D.M. MicroRNAs transfer from human macrophages to hepato-carcinoma cells and inhibit proliferation. J. Immunol. 2013, 191, 6250–6260. [Google Scholar] [CrossRef]
- Wang, F.; Li, L.; Piontek, K.; Sakaguchi, M.; Selaru, F.M. Exosome miR-335 as a novel therapeutic strategy in hepatocellular carcinoma. Hepatology 2018, 67, 940–954. [Google Scholar] [CrossRef]
- Liang, G.; Kan, S.; Zhu, Y.; Feng, S.; Feng, W.; Gao, S. Engineered exosome-mediated delivery of functionally active miR-26a and its enhanced suppression effect in HepG2 cells. Int. J. Nanomed. 2018, 13, 585–599. [Google Scholar] [CrossRef]
- Li, I.; Nabet, B.Y. Exosomes in the tumor microenvironment as mediators of cancer therapy resistance. Mol. Cancer 2019, 18, 32. [Google Scholar] [CrossRef]
- Le Grazie, M.; Biagini, M.R.; Tarocchi, M.; Polvani, S.; Galli, A. Chemotherapy for hepatocellular carcinoma: The present and the future. World J. Hepatol. 2017, 9, 907–920. [Google Scholar] [CrossRef]
- Chiou, J.-F.; Tai, C.-J.; Huang, M.-T.; Wei, P.-L.; Wang, Y.-H.; An, J.; Wu, C.-H.; Liu, T.-Z.; Chang, Y.-J. Glucose-regulated protein 78 is a novel contributor to acquisition of resistance to sorafenib in hepatocellular carcinoma. Ann. Surg. Oncol. 2010, 17, 603–612. [Google Scholar] [CrossRef]
- Chen, K.-F.; Chen, H.-L.; Tai, W.-T.; Feng, W.-C.; Hsu, C.-H.; Chen, P.-J.; Cheng, A.-L. Activation of phosphatidylinositol 3-kinase/Akt signaling pathway mediates acquired resistance to sorafenib in hepatocellular carcinoma cells. J. Pharmacol. Exp. Ther. 2011, 337, 155–161. [Google Scholar] [CrossRef] [PubMed]
- Pratama, M.Y.; Pascut, D.; Massi, M.N.; Tiribelli, C. The role of microRNA in the resistance to treatment of hepatocellular carcinoma. Ann. Transl. Med. 2019, 7, 577. [Google Scholar] [CrossRef] [PubMed]
- Fu, X.; Liu, M.; Qu, S.; Ma, J.; Zhang, Y.; Shi, T.; Wen, H.; Yang, Y.; Wang, S.; Wang, J.; et al. Exosomal microRNA-32-5p induces multidrug resistance in hepatocellular carcinoma via the PI3K/Akt pathway. J. Exp. Clin. Cancer Res. 2018, 37, 52. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Zhao, W.; Wang, H.; Qiu, G.; Jiang, Z.; Wei, G.; Li, X. Exosomal MiR-744 Inhibits Proliferation and Sorafenib Chemoresistance in Hepatocellular Carcinoma by Targeting PAX2. Med. Sci. Monit. 2019, 25, 7209–7217. [Google Scholar] [CrossRef]
- Lou, G.; Song, X.; Yang, F.; Wu, S.; Wang, J.; Chen, Z.; Liu, Y. Exosomes derived from miR-122-modified adipose tissue-derived MSCs increase chemosensitivity of hepatocellular carcinoma. J. Hematol. Oncol. 2015, 8, 122. [Google Scholar] [CrossRef]
- Nishida, N.; Arizumi, T.; Hagiwara, S.; Ida, H.; Sakurai, T.; Kudo, M. MicroRNAs for the Prediction of Early Response to Sorafenib Treatment in Human Hepatocellular Carcinoma. Liver Cancer 2017, 6, 113. [Google Scholar] [CrossRef]
- Aguilar, E.J.; Ricciuti, B.; Gainor, J.F.; Kehl, K.L.; Kravets, S.; Dahlberg, S.; Nishino, M.; Sholl, L.M.; Adeni, A.; Subegdjo, S.; et al. Outcomes to first-line pembrolizumab in patients with non-small-cell lung cancer and very high PD-L1 expression. Ann. Oncol. 2019, 30, 1653–1659. [Google Scholar] [CrossRef]
- Jazirehi, A.R.; Lim, A.; Dinh, T. PD-1 inhibition and treatment of advanced melanoma-role of pembrolizumab. Am. J. Cancer Res. 2016, 6, 2117–2128. [Google Scholar]
- Liu, X.; Qin, S. Immune Checkpoint Inhibitors in Hepatocellular Carcinoma: Opportunities and Challenges. Oncologist 2019, 24, S3–S10. [Google Scholar] [CrossRef]
- Finn, R.S.; Ryoo, B.-Y.; Merle, P.; Kudo, M.; Bouattour, M.; Lim, H.Y.; Breder, V.; Edeline, J.; Chao, Y.; Ogasawara, S.; et al. Pembrolizumab As Second-Line Therapy in Patients With Advanced Hepatocellular Carcinoma in KEYNOTE-240: A Randomized, Double-Blind, Phase III Trial. J. Clin. Oncol. 2020, 38, 193–202. [Google Scholar] [CrossRef]
- Yau, T.; Park, J.W.; Finn, R.S.; Cheng, A.-L.; Mathurin, P.; Edeline, J.; Kudo, M.; Han, K.-H.; Harding, J.J.; Merle, P.; et al. LBA38_PR - CheckMate 459: A randomized, multi-center phase III study of nivolumab (NIVO) vs. sorafenib (SOR) as first-line (1L) treatment in patients (pts) with advanced hepatocellular carcinoma (aHCC). Ann. Oncol. 2019, 30, v874–v875. [Google Scholar] [CrossRef]
- Georganaki, M.; van Hooren, L.; Dimberg, A. Vascular Targeting to Increase the Efficiency of Immune Checkpoint Blockade in Cancer. Front. Immunol. 2018, 9, 3081. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Patel, S.P.; Roszik, J.; Qin, Y. Hypoxia-Driven Immunosuppressive Metabolites in the Tumor Microenvironment: New Approaches for Combinational Immunotherapy. Front. Immunol. 2018, 9, 1591. [Google Scholar] [CrossRef]
- Pinato, D.J.; Black, J.R.; Trousil, S.; Dina, R.E.; Trivedi, P.; Mauri, F.A.; Sharma, R. Programmed cell death ligands expression in phaeochromocytomas and paragangliomas: Relationship with the hypoxic response, immune evasion and malignant behavior. Oncoimmunology 2017, 6, e1358332. [Google Scholar] [CrossRef] [PubMed]
- Faivre, S.; Rimassa, L.; Finn, R.S. Molecular therapies for HCC: Looking outside the box. J. Hepatol. 2020, 72, 342–352. [Google Scholar] [CrossRef]
- D’Souza-Schorey, C.; Clancy, J.W. Tumor-derived microvesicles: Shedding light on novel microenvironment modulators and prospective cancer biomarkers. Genes Dev. 2012, 26, 1287–1299. [Google Scholar] [CrossRef]
- Wang, H.; Hou, L.; Li, A.; Duan, Y.; Gao, H.; Song, X. Expression of Serum Exosomal MicroRNA-21 in Human Hepatocellular Carcinoma. Biomed Res. Int. 2014, 2014, 864894. [Google Scholar] [CrossRef]
- Gao, D.; Zhou, Z.; Huang, H. miR-30b-3p Inhibits Proliferation and Invasion of Hepatocellular Carcinoma Cells via Suppressing PI3K/Akt Pathway. Front. Genet. 2019, 10, 1274. [Google Scholar] [CrossRef]
- Morishita, A.; Fujita, K.; Iwama, H.; Chiyo, T.; Fujihara, S.; Oura, K.; Tadokoro, T.; Mimura, S.; Nomura, T.; Tani, J.; et al. Role of microRNA-210-3p in hepatitis B virus-related hepatocellular carcinoma. Am. J. Physiol. Gastrointest. Liver Physiol. 2020, 318, G401–G409. [Google Scholar] [CrossRef]
- Sugimachi, K.; Matsumura, T.; Hirata, H.; Uchi, R.; Ueda, M.; Ueo, H.; Shinden, Y.; Iguchi, T.; Eguchi, H.; Shirabe, K.; et al. Identification of a bona fide microRNA biomarker in serum exosomes that predicts hepatocellular carcinoma recurrence after liver transplantation. Br. J. Cancer 2015, 112, 532–538. [Google Scholar] [CrossRef]
- Sohn, W.; Kim, J.; Kang, S.H.; Yang, S.R.; Cho, J.-Y.; Cho, H.C.; Shim, S.G.; Paik, Y.-H. Serum exosomal microRNAs as novel biomarkers for hepatocellular carcinoma. Exp. Mol. Med. 2015, 47, e184. [Google Scholar] [CrossRef] [PubMed]
- Cho, H.J.; Eun, J.W.; Baek, G.O.; Seo, C.W.; Ahn, H.R.; Kim, S.S.; Cho, S.W.; Cheong, J.Y. Serum Exosomal MicroRNA, miR-10b-5p, as a Potential Diagnostic Biomarker for Early-Stage Hepatocellular Carcinoma. J. Clin. Med. 2020, 9, 281. [Google Scholar] [CrossRef] [PubMed]
- Hassan, A.S.; Elgendy, N.A.; Tawfik, N.A.; Elnasser, A.M. Serum miR-483-5p and miR-133a as Biomarkers for Diagnosis of Hepatocellular Carcinoma Post-Hepatitis C Infection in Egyptian Patients. Egypt J. Immunol. 2019, 26, 31–40. [Google Scholar] [PubMed]
- Lin, H.; Zhang, Z. Diagnostic value of a microRNA signature panel in exosomes for patients with hepatocellular carcinoma. Int. J. Clin. Exp. Pathol. 2019, 12, 1478–1487. [Google Scholar]
- Wang, Y.; Zhang, C.; Zhang, P.; Guo, G.; Jiang, T.; Zhao, X.; Jiang, J.; Huang, X.; Tong, H.; Tian, Y. Serum exosomal microRNAs combined with alpha-fetoprotein as diagnostic markers of hepatocellular carcinoma. Cancer Med. 2018, 7, 1670–1679. [Google Scholar] [CrossRef]
- Liu, W.; Hu, J.; Zhou, K.; Chen, F.; Wang, Z.; Liao, B.; Dai, Z.; Cao, Y.; Fan, J.; Zhou, J. Serum exosomal miR-125b is a novel prognostic marker for hepatocellular carcinoma. Oncol. Targets Ther. 2017, 10, 3843–3851. [Google Scholar] [CrossRef]
- Shi, M.; Jiang, Y.; Yang, L.; Yan, S.; Wang, Y.-G.; Lu, X.-J. Decreased levels of serum exosomal miR-638 predict poor prognosis in hepatocellular carcinoma. J. Cell. Biochem. 2018, 119, 4711–4716. [Google Scholar] [CrossRef]
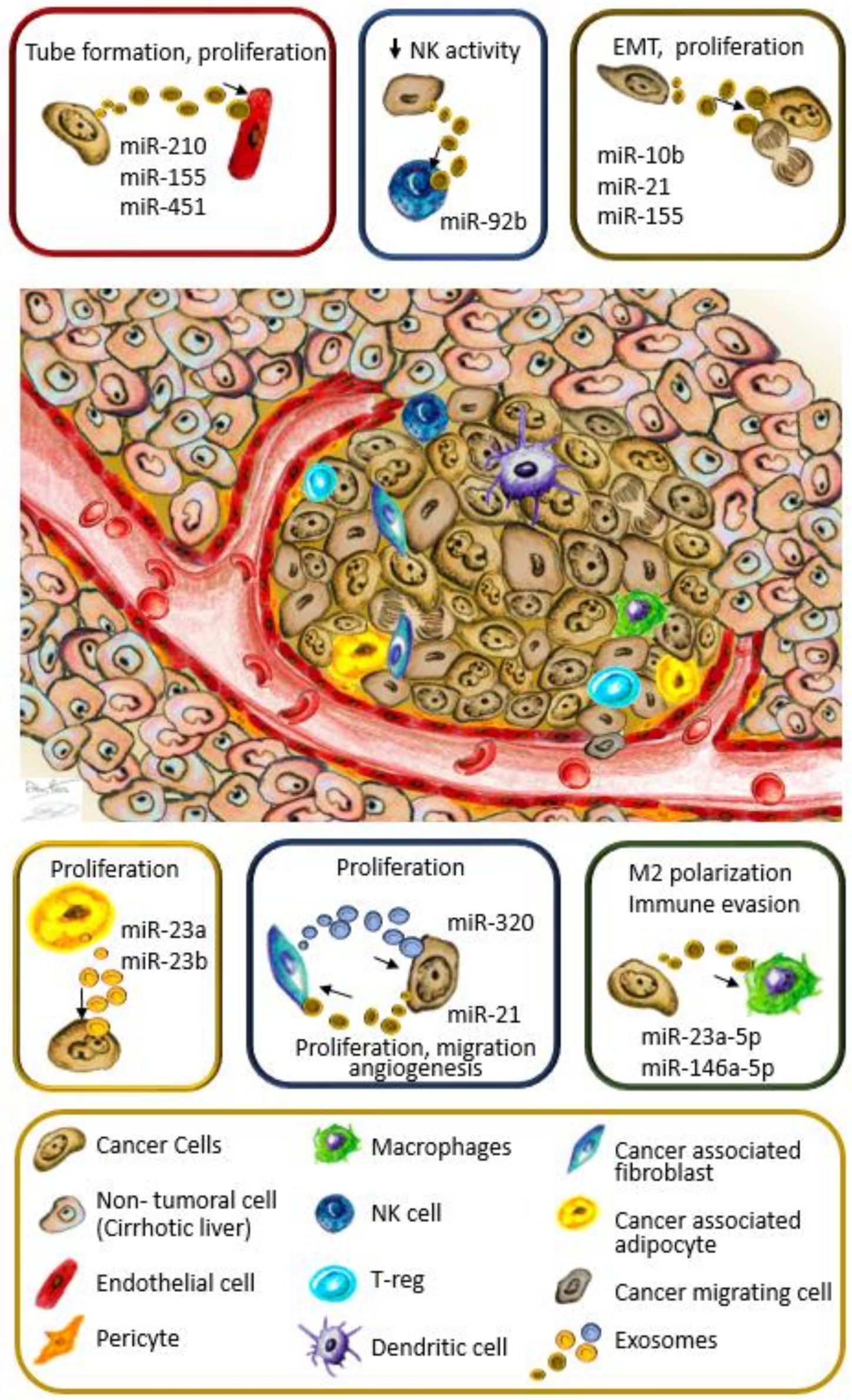
| Exo-Releasing Cells | miRNA | Recipient Cells | Target | Effect | Validated in Mouse Model | Reference |
|---|---|---|---|---|---|---|
| Hep-3B, SMMC-7721 | miR-21, miR-10b | Hep-3B, SMMC-7721 | n.d. | EMT proliferation | Yes | [56] |
| Hep-3B | miR-155 | HepG2 | PTEN | Proliferation | Yes | [62] |
| SNU-449 | miR-21 | HepG2, Hep-3B | TETs/PTEN/PTENp1 | Proliferation | Yes | [58] |
| HepG2 | miR-224 | SKHEP1 (adenocarcinoma) | GNMT | Cell proliferation | No | [63] |
| Mature adipocytes | miR-23a, miR-23b | BEL-7402 | VHL | Proliferation | Yes | [64] |
| LM3, 97H liver cancer cells | miR-21 | LX2 | PTEN/PDK1/AKT pathway | Proliferation, migration, angiogenesis | Yes | [65] |
| Primary CAFs | miR-320a downregulation | MHCC97-H | PBX3 | Proliferation, metastasis | [66] | |
| QGY-7703, Hep-G2, SK-Hep-1, Huh7, EXOs from HCC patients | miR-210 | HUVEC | SMAD4, STAT6 | Tube formation, high MVD and larger tumors | Yes | [67] |
| PLC/PRF/5, Huh7 | miR-155 | HUVEC | n.d. | Tube formation | No | [68] |
| SMMC-7721 | miR-451 | HUVEC | LPN1 (in cancer cells) | Increased cell death, decreased cell migration | [69] | |
| Hep-G2, Hep-3B | miR-23a-3p | THP-1 | PTEN | Immune evasion by PD-L1 overexpression | Yes, partially | [70] |
| Hep-G2, H7402 | miR-146a-5p | THP-1 | n.d. | M2 polarization, T cell dysfunction | Yes | [71] |
| Hep-3B | miR-92b | NK cells | CD69 | Reduced NK activity | No | [72] |
| miRNAs | Patients | Clinical Significance | Reference | |
|---|---|---|---|---|
| Diagnosis | ||||
| miR-21 | ↑ | 30 HCC, 30 CHB, 30 healthy | Discrimination between HCC and CHB or LC | [127] |
| miR-30b-3p | ↓ | 50 paired HCC tissues (non-tumor tissues) | Biomarker diagnosis and treatment HCC | [128] |
| miR-210-3p | ↑ | 29 HCC | Biomarker for the risk of HBV-related HCC | [129] |
| miRNA-224 | ↑ | 9 HCC and 50 normal serum samples | Biomarker of diagnosis and prognosis of HCC patient | [63] |
| miR-718 | ↓ | 59 HCC | Predicting biomarker for recurrence after LT | [130] |
| miR-18a miR-221 miR-222 miR-224 miR-101 miR-106b miR-122 miR-195 | ↑ ↑ ↑ ↑ ↓ ↓ ↓ ↓ | 20 HCC vs. 20 CHB vs. 20 LC | Discrimination between HCC and CHB or LC | [131] |
| miR-10b-5p miR-18a-5p miR-215-5p miR-940 | ↑ ↑ ↑ ↑ | 28 healthy, 60 CLD, 90 HCC | miR-10b-5p biomarker for early stage HCC | [132] |
| miR-483-5p miR-133a | ↑ ↑ | 20 HCC, 20 CHB, 20 healthy | Noninvasive diagnostic biomarkers for HCC | [133] |
| miRNA-26a miRNA-29c miRNA-21 | ↓ ↓ ↓ | 72 HCC, 72 LC, and 72 HBV | Diagnostic biomarkers for patients with HCC | [134] |
| miR-122↑ miR-148a↑ miR-1246↑ | 5 HCC vs. 5 LC | Diagnostic biomarker discriminating HCC from LC | [135] |
| miRNAs | Patients | Clinical Significance | Reference | |
|---|---|---|---|---|
| Prognosis | ||||
| miR-125b | ↑ | 158 HCC vs. 30 CHB vs. 30 LC | Predicting biomarker for recurrence and survival | [136] |
| miR-638 | ↓ | 126 HCC | Poor prognosis marker for patients with HCC | [137] |
| miR-10b-5p miR-18a-5p miR-215-5p miR-940 | ↑ ↑ ↑ ↑ | 28 healthy, 60 CLD, 90 HCC | miR-215-5p: prognostic biomarker for HCC | [132] |
| miR-744 | ↓ | 68 HCC and 52 normal liver tissue samples | Proliferation and chemoresistance | [114] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pascut, D.; Pratama, M.Y.; Vo, N.V.T.; Masadah, R.; Tiribelli, C. The Crosstalk between Tumor Cells and the Microenvironment in Hepatocellular Carcinoma: The Role of Exosomal microRNAs and Their Clinical Implications. Cancers 2020, 12, 823. https://doi.org/10.3390/cancers12040823
Pascut D, Pratama MY, Vo NVT, Masadah R, Tiribelli C. The Crosstalk between Tumor Cells and the Microenvironment in Hepatocellular Carcinoma: The Role of Exosomal microRNAs and Their Clinical Implications. Cancers. 2020; 12(4):823. https://doi.org/10.3390/cancers12040823
Chicago/Turabian StylePascut, Devis, Muhammad Yogi Pratama, Niem V.T. Vo, Rina Masadah, and Claudio Tiribelli. 2020. "The Crosstalk between Tumor Cells and the Microenvironment in Hepatocellular Carcinoma: The Role of Exosomal microRNAs and Their Clinical Implications" Cancers 12, no. 4: 823. https://doi.org/10.3390/cancers12040823
APA StylePascut, D., Pratama, M. Y., Vo, N. V. T., Masadah, R., & Tiribelli, C. (2020). The Crosstalk between Tumor Cells and the Microenvironment in Hepatocellular Carcinoma: The Role of Exosomal microRNAs and Their Clinical Implications. Cancers, 12(4), 823. https://doi.org/10.3390/cancers12040823





