Chromosome Instability; Implications in Cancer Development, Progression, and Clinical Outcomes
Abstract
1. Introduction
2. Genome and Chromosome Instability; Definition and Types of CIN
2.1. Critical Distinctions Between Aneuploidy and CIN
2.2. Fundamental Concepts in Assessing CIN: Benefits and Limitations
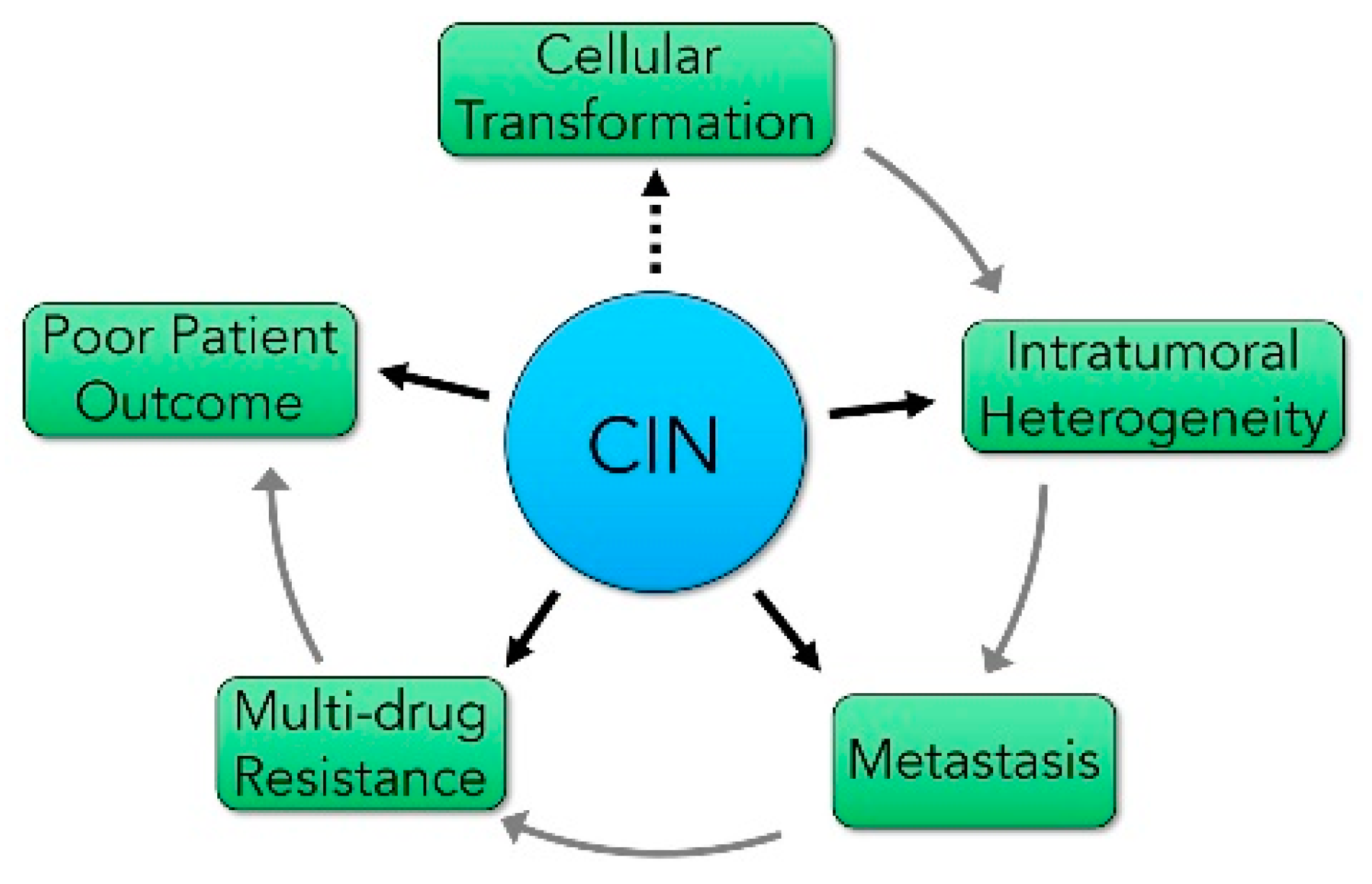
3. The impact of CIN on Cancer Development and Progression
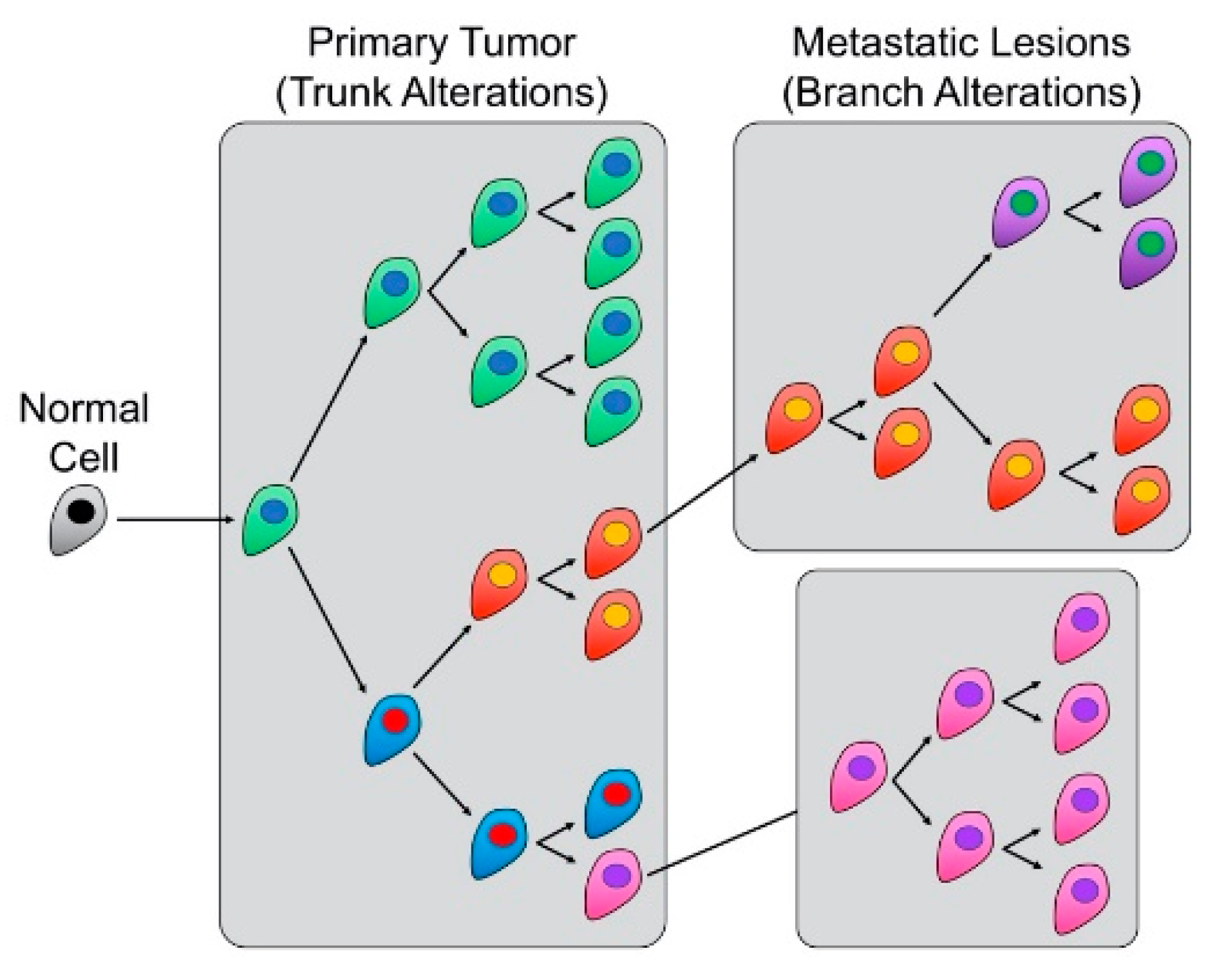
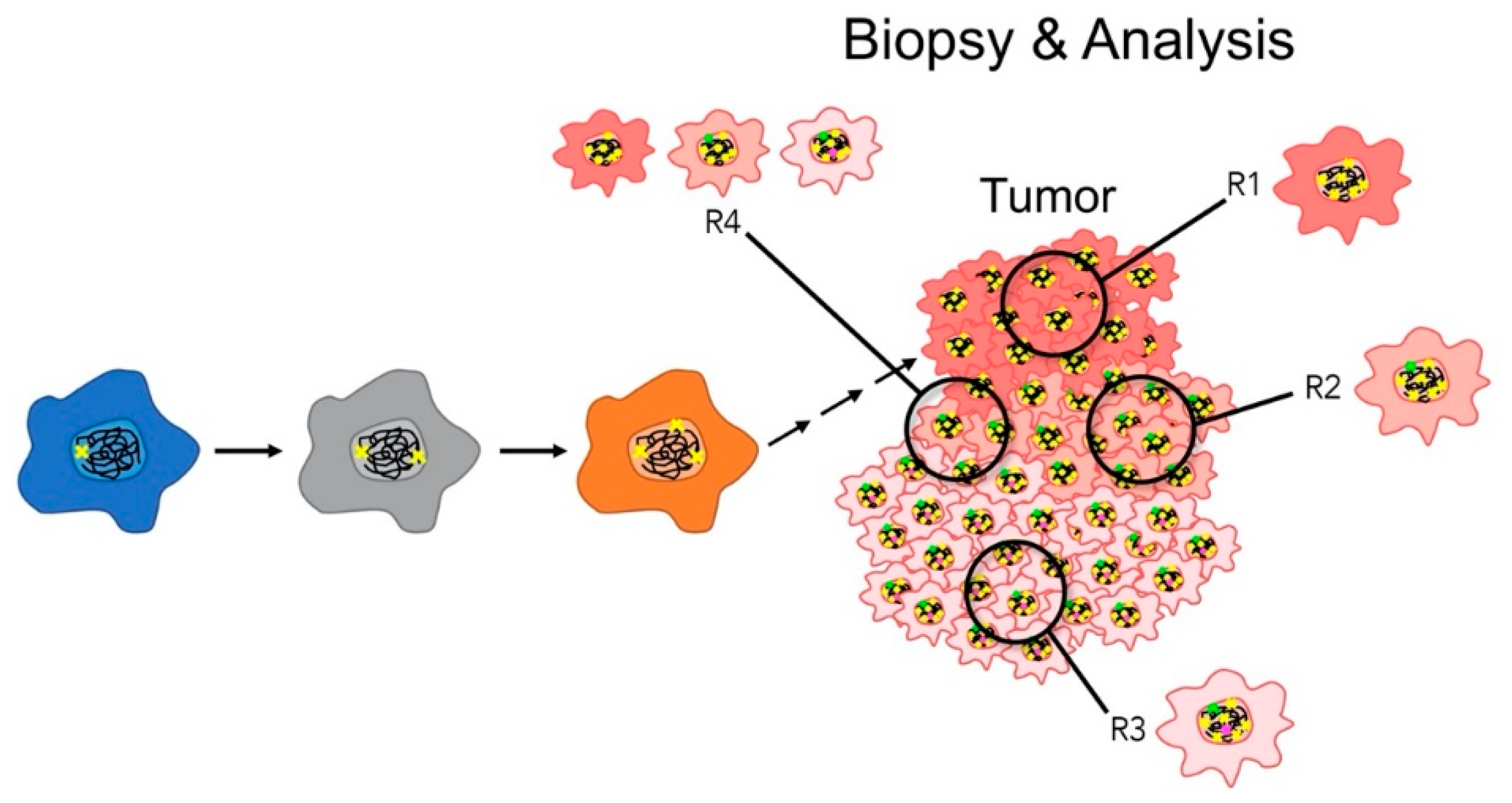
The Relationship between CIN and Intertumoral and Intratumoral Heterogeneity
4. CIN Influences the Metastatic Potential of Many Cancer Types
5. CIN and Cancer Prognosis
6. CIN and its Impact on Precision Medicine Strategies
6.1. The Impact of CIN on Therapeutic Targeting
7. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Nowell, P.C. Discovery of the Philadelphia chromosome: A personal perspective. J. Clin. Investig. 2007, 117, 2033–2035. [Google Scholar] [CrossRef] [PubMed]
- Rowley, J.D. Letter: A new consistent chromosomal abnormality in chronic myelogenous leukaemia identified by quinacrine fluorescence and Giemsa staining. Nature 1973, 243, 290–293. [Google Scholar] [CrossRef] [PubMed]
- Cancer Genome Atlas Research Network; Weinstein, J.N.; Collisson, E.A.; Mills, G.B.; Shaw, K.R.; Ozenberger, B.A.; Ellrott, K.; Shmulevich, I.; Sander, C.; Stuart, J.M. The Cancer Genome Atlas Pan-Cancer analysis project. Nat. Genet. 2013, 45, 1113–1120. [Google Scholar] [CrossRef]
- The ICGC/TCGA Pan-Cancer Analysis of Whole Genomes Consortium. Pan-cancer analysis of whole genomes. Nature 2020, 578, 82–93. [Google Scholar] [CrossRef]
- Comings, D.E. A general theory of carcinogenesis. Proc. Natl. Acad. Sci. USA 1973, 70, 3324–3328. [Google Scholar] [CrossRef]
- Fidler, I.J. Commentary on “Tumor Heterogeneity and the Biology of Cancer Invasion and Metastasis”. Cancer Res. 2016, 76, 3441–3442. [Google Scholar] [CrossRef]
- Lee, J.K.; Choi, Y.L.; Kwon, M.; Park, P.J. Mechanisms and Consequences of Cancer Genome Instability: Lessons from Genome Sequencing Studies. Annu. Rev. Pathol. 2016, 11, 283–312. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Holland, A.J.; Cleveland, D.W. Boveri revisited: Chromosomal instability, aneuploidy and tumorigenesis. Nat. Rev. Mol. Cell Biol. 2009, 10, 478–487. [Google Scholar] [CrossRef]
- Geigl, J.B.; Obenauf, A.C.; Schwarzbraun, T.; Speicher, M.R. Defining ’chromosomal instability’. Trends Genet. 2008, 24, 64–69. [Google Scholar] [CrossRef]
- Lepage, C.C.; Morden, C.R.; Palmer, M.C.L.; Nachtigal, M.W.; McManus, K.J. Detecting Chromosome Instability in Cancer: Approaches to Resolve Cell-to-Cell Heterogeneity. Cancers (Basel) 2019, 11, 226. [Google Scholar] [CrossRef] [PubMed]
- Bakhoum, S.F.; Kabeche, L.; Murnane, J.P.; Zaki, B.I.; Compton, D.A. DNA-damage response during mitosis induces whole-chromosome missegregation. Cancer Discov. 2014, 4, 1281–1289. [Google Scholar] [CrossRef] [PubMed]
- Ganmore, I.; Smooha, G.; Izraeli, S. Constitutional aneuploidy and cancer predisposition. Hum. Mol. Genet. 2009, 18, 84–93. [Google Scholar] [CrossRef] [PubMed]
- Biron-Shental, T.; Liberman, M.; Sharvit, M.; Sukenik-Halevy, R.; Amiel, A. Amniocytes from aneuploidy embryos have enhanced random aneuploidy and signs of senescence-can these findings be related to medical problems? Gene 2015, 562, 232–235. [Google Scholar] [CrossRef] [PubMed]
- Nicholson, J.M.; Macedo, J.C.; Mattingly, A.J.; Wangsa, D.; Camps, J.; Lima, V.; Gomes, A.M.; Doria, S.; Ried, T.; Logarinho, E.; et al. Chromosome mis-segregation and cytokinesis failure in trisomic human cells. Elife 2015, 4. [Google Scholar] [CrossRef]
- Zhu, J.; Tsai, H.J.; Gordon, M.R.; Li, R. Cellular Stress Associated with Aneuploidy. Dev. Cell 2018, 44, 420–431. [Google Scholar] [CrossRef]
- Ben-David, U.; Arad, G.; Weissbein, U.; Mandefro, B.; Maimon, A.; Golan-Lev, T.; Narwani, K.; Clark, A.T.; Andrews, P.W.; Benvenisty, N.; et al. Aneuploidy induces profound changes in gene expression, proliferation and tumorigenicity of human pluripotent stem cells. Nat. Commun. 2014, 5, e4825. [Google Scholar] [CrossRef]
- McGranahan, N.; Swanton, C. Clonal Heterogeneity and Tumor Evolution: Past, Present, and the Future. Cell 2017, 168, 613–628. [Google Scholar] [CrossRef]
- Pampalona, J.; Roscioli, E.; Silkworth, W.T.; Bowden, B.; Genesca, A.; Tusell, L.; Cimini, D. Chromosome Bridges Maintain Kinetochore-Microtubule Attachment throughout Mitosis and Rarely Break during Anaphase. PLoS ONE 2016, 11, e0147420. [Google Scholar] [CrossRef]
- Robertson, E.G.; Baxter, G. Tumour seeding following percutaneous needle biopsy: The real story! Clin. Radiol. 2011, 66, 1007–1014. [Google Scholar] [CrossRef]
- Tyagi, R.; Dey, P. Needle tract seeding: An avoidable complication. Diagn. Cytopathol. 2014, 42, 636–640. [Google Scholar] [CrossRef] [PubMed]
- Penner-Goeke, S.; Lichtensztejn, Z.; Neufeld, M.; Ali, J.L.; Altman, A.D.; Nachtigal, M.W.; McManus, K.J. The temporal dynamics of chromosome instability in ovarian cancer cell lines and primary patient samples. PLoS Genet. 2017, 13, e1006707. [Google Scholar] [CrossRef]
- Bates, S.E. Classical cytogenetics: Karyotyping techniques. Methods Mol. Biol. 2011, 767, 177–190. [Google Scholar] [CrossRef] [PubMed]
- Schrock, E.; du Manoir, S.; Veldman, T.; Schoell, B.; Wienberg, J.; Ferguson-Smith, M.A.; Ning, Y.; Ledbetter, D.H.; Bar-Am, I.; Soenksen, D.; et al. Multicolor spectral karyotyping of human chromosomes. Science 1996, 273, 494–497. [Google Scholar] [CrossRef] [PubMed]
- Klinger, K.; Landes, G.; Shook, D.; Harvey, R.; Lopez, L.; Locke, P.; Lerner, T.; Osathanondh, R.; Leverone, B.; Houseal, T.; et al. Rapid detection of chromosome aneuploidies in uncultured amniocytes by using fluorescence in situ hybridization (FISH). Am. J. Hum. Genet. 1992, 51, 55–65. [Google Scholar] [PubMed]
- Bakker, B.; Taudt, A.; Belderbos, M.E.; Porubsky, D.; Spierings, D.C.; de Jong, T.V.; Halsema, N.; Kazemier, H.G.; Hoekstra-Wakker, K.; Bradley, A.; et al. Single-cell sequencing reveals karyotype heterogeneity in murine and human malignancies. Genome Biol. 2016, 17, e115. [Google Scholar] [CrossRef]
- Navin, N.; Kendall, J.; Troge, J.; Andrews, P.; Rodgers, L.; McIndoo, J.; Cook, K.; Stepansky, A.; Levy, D.; Esposito, D.; et al. Tumour evolution inferred by single-cell sequencing. Nature 2011, 472, 90–94. [Google Scholar] [CrossRef]
- Lepage, C.C.; Thompson, L.L.; Larson, B.; McManus, K.J. An Automated, Single Cell Quantitative Imaging Microscopy Approach to Assess Micronucleus Formation, Genotoxicity and Chromosome Instability. Cells 2020, 9, 344. [Google Scholar] [CrossRef]
- Thompson, L.L.; McManus, K.J. A novel multiplexed, image-based approach to detect phenotypes that underlie chromosome instability in human cells. PLoS ONE 2015, 10, e0123200. [Google Scholar] [CrossRef] [PubMed]
- Kouprina, N.; Liskovykh, M.; Petrov, N.; Larionov, V. Human artificial chromosome (HAC) for measuring chromosome instability (CIN) and identification of genes required for proper chromosome transmission. Exp. Cell Res. 2020, 387, e111805. [Google Scholar] [CrossRef]
- Bhatia, A.; Kumar, Y. Cancer cell micronucleus: An update on clinical and diagnostic applications. APMIS 2013, 121, 569–581. [Google Scholar] [CrossRef] [PubMed]
- Stopper, H.; Muller, S.O. Micronuclei as a biological endpoint for genotoxicity: A minireview. Toxicol. In Vitro 1997, 11, 661–667. [Google Scholar] [CrossRef]
- Ye, C.J.; Sharpe, Z.; Alemara, S.; Mackenzie, S.; Liu, G.; Abdallah, B.; Horne, S.; Regan, S.; Heng, H.H. Micronuclei and Genome Chaos: Changing the System Inheritance. Genes (Basel) 2019, 10, 366. [Google Scholar] [CrossRef] [PubMed]
- Asbaghi, Y.; Thompson, L.L.; Lichtensztejn, Z.; McManus, K.J. KIF11 silencing and inhibition induces chromosome instability that may contribute to cancer. Genes Chromosomes Cancer 2017, 56, 668–680. [Google Scholar] [CrossRef] [PubMed]
- Baergen, A.K.; Jeusset, L.M.; Lichtensztejn, Z.; McManus, K.J. Diminished Condensin Gene Expression Drives Chromosome Instability That May Contribute to Colorectal Cancer Pathogenesis. Cancers (Basel) 2019, 11, 1066. [Google Scholar] [CrossRef] [PubMed]
- Leylek, T.R.; Jeusset, L.M.; Lichtensztejn, Z.; McManus, K.J. Reduced Expression of Genes Regulating Cohesion Induces Chromosome Instability that May Promote Cancer and Impact Patient Outcomes. Sci. Rep. 2020, 10, e592. [Google Scholar] [CrossRef]
- Thompson, L.L.; Baergen, A.K.; Lichtensztejn, Z.; McManus, K.J. Reduced SKP1 Expression Induces Chromosome Instability through Aberrant Cyclin E1 Protein Turnover. Cancers (Basel) 2020, 12, 531. [Google Scholar] [CrossRef]
- Kouprina, N.; Pommier, Y.; Larionov, V. Novel screen for anti-cancer drugs that elevate chromosome instability (CIN) using human artificial chromosome (HAC). Oncotarget 2018, 9, 36833–36835. [Google Scholar] [CrossRef]
- Worrall, J.T.; Tamura, N.; Mazzagatti, A.; Shaikh, N.; van Lingen, T.; Bakker, B.; Spierings, D.C.J.; Vladimirou, E.; Foijer, F.; McClelland, S.E. Non-random Mis-segregation of Human Chromosomes. Cell Rep. 2018, 23, 3366–3380. [Google Scholar] [CrossRef]
- Covo, S.; Puccia, C.M.; Argueso, J.L.; Gordenin, D.A.; Resnick, M.A. The sister chromatid cohesion pathway suppresses multiple chromosome gain and chromosome amplification. Genetics 2014, 196, 373–384. [Google Scholar] [CrossRef][Green Version]
- Barber, T.D.; McManus, K.; Yuen, K.W.; Reis, M.; Parmigiani, G.; Shen, D.; Barrett, I.; Nouhi, Y.; Spencer, F.; Markowitz, S.; et al. Chromatid cohesion defects may underlie chromosome instability in human colorectal cancers. Proc. Natl. Acad. Sci. USA 2008, 105, 3443–3448. [Google Scholar] [CrossRef] [PubMed]
- Smith, L.; Plug, A.; Thayer, M. Delayed replication timing leads to delayed mitotic chromosome condensation and chromosomal instability of chromosome translocations. Proc. Natl. Acad. Sci. USA 2001, 98, 13300–13305. [Google Scholar] [CrossRef] [PubMed]
- McManus, K.J.; Biron, V.L.; Heit, R.; Underhill, D.A.; Hendzel, M.J. Dynamic changes in histone H3 lysine 9 methylations: Identification of a mitosis-specific function for dynamic methylation in chromosome congression and segregation. J. Biol. Chem. 2006, 281, 8888–8897. [Google Scholar] [CrossRef]
- Houston, S.I.; McManus, K.J.; Adams, M.M.; Sims, J.K.; Carpenter, P.B.; Hendzel, M.J.; Rice, J.C. Catalytic function of the PR-Set7 histone H4 lysine 20 monomethyltransferase is essential for mitotic entry and genomic stability. J. Biol. Chem. 2008, 283, 19478–19488. [Google Scholar] [CrossRef] [PubMed]
- Guppy, B.J.; McManus, K.J. Mitotic accumulation of dimethylated lysine 79 of histone H3 is important for maintaining genome integrity during mitosis in human cells. Genetics 2015, 199, 423–433. [Google Scholar] [CrossRef]
- Thompson, L.L.; Guppy, B.J.; Sawchuk, L.; Davie, J.R.; McManus, K.J. Regulation of chromatin structure via histone post-translational modification and the link to carcinogenesis. Cancer Metastasis Rev. 2013, 32, 363–376. [Google Scholar] [CrossRef]
- Green, R.A.; Kaplan, K.B. Chromosome instability in colorectal tumor cells is associated with defects in microtubule plus-end attachments caused by a dominant mutation in APC. J. Cell Biol. 2003, 163, 949–961. [Google Scholar] [CrossRef]
- Marshall, H.; Bhaumik, M.; Aviv, H.; Moore, D.; Yao, M.; Dutta, J.; Rahim, H.; Gounder, M.; Ganesan, S.; Saleem, A.; et al. Deficiency of the dual ubiquitin/SUMO ligase Topors results in genetic instability and an increased rate of malignancy in mice. BMC Mol. Biol. 2010, 11, e31. [Google Scholar] [CrossRef]
- Wu, M.; Tu, H.Q.; Chang, Y.; Tan, B.; Wang, G.; Zhou, J.; Wang, L.; Mu, R.; Zhang, W.N. USP19 deubiquitinates HDAC1/2 to regulate DNA damage repair and control chromosomal stability. Oncotarget 2017, 8, 2197–2208. [Google Scholar] [CrossRef]
- Storchova, Z.; Kuffer, C. The consequences of tetraploidy and aneuploidy. J. Cell Sci. 2008, 121, 3859–3866. [Google Scholar] [CrossRef]
- Swanton, C. Intratumor heterogeneity: Evolution through space and time. Cancer Res. 2012, 72, 4875–4882. [Google Scholar] [CrossRef] [PubMed]
- Janiszewska, M. The microcosmos of intratumor heterogeneity: The space-time of cancer evolution. Oncogene 2020, 39, 2031–2039. [Google Scholar] [CrossRef] [PubMed]
- Oh, B.Y.; Shin, H.T.; Yun, J.W.; Kim, K.T.; Kim, J.; Bae, J.S.; Cho, Y.B.; Lee, W.Y.; Yun, S.H.; Park, Y.A.; et al. Intratumor heterogeneity inferred from targeted deep sequencing as a prognostic indicator. Sci. Rep. 2019, 9, 4542. [Google Scholar] [CrossRef] [PubMed]
- Bakhoum, S.F.; Ngo, B.; Laughney, A.M.; Cavallo, J.A.; Murphy, C.J.; Ly, P.; Shah, P.; Sriram, R.K.; Watkins, T.B.K.; Taunk, N.K.; et al. Chromosomal instability drives metastasis through a cytosolic DNA response. Nature 2018, 553, 467–472. [Google Scholar] [CrossRef] [PubMed]
- Gao, C.; Su, Y.; Koeman, J.; Haak, E.; Dykema, K.; Essenberg, C.; Hudson, E.; Petillo, D.; Khoo, S.K.; Vande Woude, G.F. Chromosome instability drives phenotypic switching to metastasis. Proc. Natl. Acad. Sci. USA 2016, 113, 14793–14798. [Google Scholar] [CrossRef]
- Tijhuis, A.E.; Johnson, S.C.; McClelland, S.E. The emerging links between chromosomal instability (CIN), metastasis, inflammation and tumor immunity. Mol. Cytogenet. 2019, 12, e17. [Google Scholar] [CrossRef]
- Lee, A.J.; Endesfelder, D.; Rowan, A.J.; Walther, A.; Birkbak, N.J.; Futreal, P.A.; Downward, J.; Szallasi, Z.; Tomlinson, I.P.; Howell, M.; et al. Chromosomal instability confers intrinsic multidrug resistance. Cancer Res. 2011, 71, 1858–1870. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Zhang, Y.; Chen, R.; Tian, Z.; Zhai, Y.; Janz, S.; Gu, C.; Yang, Y. Chromosomal instability and acquired drug resistance in multiple myeloma. Oncotarget 2017, 8, 78234–78244. [Google Scholar] [CrossRef]
- Choi, C.M.; Seo, K.W.; Jang, S.J.; Oh, Y.M.; Shim, T.S.; Kim, W.S.; Lee, D.S.; Lee, S.D. Chromosomal instability is a risk factor for poor prognosis of adenocarcinoma of the lung: Fluorescence in situ hybridization analysis of paraffin-embedded tissue from Korean patients. Lung Cancer 2009, 64, 66–70. [Google Scholar] [CrossRef]
- Kikutake, C.; Yoshihara, M.; Sato, T.; Saito, D.; Suyama, M. Pan-cancer analysis of intratumor heterogeneity associated with patient prognosis using multidimensional measures. Oncotarget 2018, 9, 37689–37699. [Google Scholar] [CrossRef][Green Version]
- Sato, H.; Uzawa, N.; Takahashi, K.; Myo, K.; Ohyama, Y.; Amagasa, T. Prognostic utility of chromosomal instability detected by fluorescence in situ hybridization in fine-needle aspirates from oral squamous cell carcinomas. BMC Cancer 2010, 10, e182. [Google Scholar] [CrossRef] [PubMed]
- Walther, A.; Houlston, R.; Tomlinson, I. Association between chromosomal instability and prognosis in colorectal cancer: A meta-analysis. Gut 2008, 57, 941–950. [Google Scholar] [CrossRef] [PubMed]
- Birkbak, N.J.; Eklund, A.C.; Li, Q.; McClelland, S.E.; Endesfelder, D.; Tan, P.; Tan, I.B.; Richardson, A.L.; Szallasi, Z.; Swanton, C. Paradoxical relationship between chromosomal instability and survival outcome in cancer. Cancer Res. 2011, 71, 3447–3452. [Google Scholar] [CrossRef] [PubMed]
- Jamal-Hanjani, M.; A’Hern, R.; Birkbak, N.J.; Gorman, P.; Gronroos, E.; Ngang, S.; Nicola, P.; Rahman, L.; Thanopoulou, E.; Kelly, G.; et al. Extreme chromosomal instability forecasts improved outcome in ER-negative breast cancer: A prospective validation cohort study from the TACT trial. Ann. Oncol. 2015, 26, 1340–1346. [Google Scholar] [CrossRef] [PubMed]
- Roylance, R.; Endesfelder, D.; Gorman, P.; Burrell, R.A.; Sander, J.; Tomlinson, I.; Hanby, A.M.; Speirs, V.; Richardson, A.L.; Birkbak, N.J.; et al. Relationship of extreme chromosomal instability with long-term survival in a retrospective analysis of primary breast cancer. Cancer Epidemiol. Biomark. Prev. 2011, 20, 2183–2194. [Google Scholar] [CrossRef] [PubMed]
- Cunningham, C.E.; MacAuley, M.J.; Yadav, G.; Vizeacoumar, F.S.; Freywald, A.; Vizeacoumar, F.J. Targeting the CINful genome: Strategies to overcome tumor heterogeneity. Prog. Biophys. Mol. Biol. 2019, 147, 77–91. [Google Scholar] [CrossRef]
- Thompson, L.L.; Jeusset, L.M.; Lepage, C.C.; McManus, K.J. Evolving Therapeutic Strategies to Exploit Chromosome Instability in Cancer. Cancers (Basel) 2017, 9, 151. [Google Scholar] [CrossRef]
- Stirling, P.C.; Bloom, M.S.; Solanki-Patil, T.; Smith, S.; Sipahimalani, P.; Li, Z.; Kofoed, M.; Ben-Aroya, S.; Myung, K.; Hieter, P. The complete spectrum of yeast chromosome instability genes identifies candidate CIN cancer genes and functional roles for ASTRA complex components. PLoS Genet. 2011, 7, e1002057. [Google Scholar] [CrossRef]
- Levine, M.S.; Holland, A.J. The impact of mitotic errors on cell proliferation and tumorigenesis. Genes Dev. 2018, 32, 620–638. [Google Scholar] [CrossRef]
- Cahill, D.P.; Lengauer, C.; Yu, J.; Riggins, G.J.; Willson, J.K.; Markowitz, S.D.; Kinzler, K.W.; Vogelstein, B. Mutations of mitotic checkpoint genes in human cancers. Nature 1998, 392, 300–303. [Google Scholar] [CrossRef]
- Jessulat, M.; Malty, R.H.; Nguyen-Tran, D.H.; Deineko, V.; Aoki, H.; Vlasblom, J.; Omidi, K.; Jin, K.; Minic, Z.; Hooshyar, M.; et al. Spindle Checkpoint Factors Bub1 and Bub2 Promote DNA Double-Strand Break Repair by Nonhomologous End Joining. Mol. Cell Biol. 2015, 35, 2448–2463. [Google Scholar] [CrossRef] [PubMed]
- Ryan, S.D.; Britigan, E.M.; Zasadil, L.M.; Witte, K.; Audhya, A.; Roopra, A.; Weaver, B.A. Up-regulation of the mitotic checkpoint component Mad1 causes chromosomal instability and resistance to microtubule poisons. Proc. Natl. Acad. Sci. USA 2012, 109, 2205–2214. [Google Scholar] [CrossRef] [PubMed]
- Chan, G.K.; Jablonski, S.A.; Starr, D.A.; Goldberg, M.L.; Yen, T.J. Human Zw10 and ROD are mitotic checkpoint proteins that bind to kinetochores. Nat. Cell Biol. 2000, 2, 944–947. [Google Scholar] [CrossRef] [PubMed]
- Kops, G.J.; Kim, Y.; Weaver, B.A.; Mao, Y.; McLeod, I.; Yates, J.R., 3rd; Tagaya, M.; Cleveland, D.W. ZW10 links mitotic checkpoint signaling to the structural kinetochore. J. Cell Biol. 2005, 169, 49–60. [Google Scholar] [CrossRef]
- Jelluma, N.; Brenkman, A.B.; McLeod, I.; Yates, J.R., 3rd; Cleveland, D.W.; Medema, R.H.; Kops, G.J. Chromosomal instability by inefficient Mps1 auto-activation due to a weakened mitotic checkpoint and lagging chromosomes. PLoS ONE 2008, 3, e2415. [Google Scholar] [CrossRef]
- Schaar, B.T.; Chan, G.K.; Maddox, P.; Salmon, E.D.; Yen, T.J. CENP-E function at kinetochores is essential for chromosome alignment. J. Cell Biol. 1997, 139, 1373–1382. [Google Scholar] [CrossRef]
- Bakhoum, S.F.; Genovese, G.; Compton, D.A. Deviant kinetochore microtubule dynamics underlie chromosomal instability. Curr. Biol. 2009, 19, 1937–1942. [Google Scholar] [CrossRef]
- Cimini, D.; Fioravanti, D.; Salmon, E.D.; Degrassi, F. Merotelic kinetochore orientation versus chromosome mono-orientation in the origin of lagging chromosomes in human primary cells. J. Cell Sci. 2002, 115, 507–515. [Google Scholar]
- Putkey, F.R.; Cramer, T.; Morphew, M.K.; Silk, A.D.; Johnson, R.S.; McIntosh, J.R.; Cleveland, D.W. Unstable kinetochore-microtubule capture and chromosomal instability following deletion of CENP-E. Dev. Cell 2002, 3, 351–365. [Google Scholar] [CrossRef]
- Cheng, X.; Shen, Z.; Yang, J.; Lu, S.H.; Cui, Y. ECRG2 disruption leads to centrosome amplification and spindle checkpoint defects contributing chromosome instability. J. Biol. Chem. 2008, 283, 5888–5898. [Google Scholar] [CrossRef]
- Kuhn, E.; Wang, T.L.; Doberstein, K.; Bahadirli-Talbott, A.; Ayhan, A.; Sehdev, A.S.; Drapkin, R.; Kurman, R.J.; Shih Ie, M. CCNE1 amplification and centrosome number abnormality in serous tubal intraepithelial carcinoma: Further evidence supporting its role as a precursor of ovarian high-grade serous carcinoma. Mod. Pathol. 2016, 29, 1254–1261. [Google Scholar] [CrossRef] [PubMed]
- Lentini, L.; Amato, A.; Schillaci, T.; Di Leonardo, A. Simultaneous Aurora-A/STK15 overexpression and centrosome amplification induce chromosomal instability in tumor cells with a MIN phenotype. BMC Cancer 2007, 7, e212. [Google Scholar] [CrossRef]
- Boardman, L.A.; Johnson, R.A.; Viker, K.B.; Hafner, K.A.; Jenkins, R.B.; Riegert-Johnson, D.L.; Smyrk, T.C.; Litzelman, K.; Seo, S.; Gangnon, R.E.; et al. Correlation of chromosomal instability, telomere length and telomere maintenance in microsatellite stable rectal cancer: A molecular subclass of rectal cancer. PLoS ONE 2013, 8, e80015. [Google Scholar] [CrossRef] [PubMed]
- Finley, J.C.; Reid, B.J.; Odze, R.D.; Sanchez, C.A.; Galipeau, P.; Li, X.; Self, S.G.; Gollahon, K.A.; Blount, P.L.; Rabinovitch, P.S. Chromosomal instability in Barrett’s esophagus is related to telomere shortening. Cancer Epidemiol. Biomark. Prev. 2006, 15, 1451–1457. [Google Scholar] [CrossRef] [PubMed]
- Murnane, J.P. Telomere dysfunction and chromosome instability. Mutat. Res. 2012, 730, 28–36. [Google Scholar] [CrossRef] [PubMed]
- Burrell, R.A.; McClelland, S.E.; Endesfelder, D.; Groth, P.; Weller, M.C.; Shaikh, N.; Domingo, E.; Kanu, N.; Dewhurst, S.M.; Gronroos, E.; et al. Replication stress links structural and numerical cancer chromosomal instability. Nature 2013, 494, 492–496. [Google Scholar] [CrossRef] [PubMed]
- Wilhelm, T.; Olziersky, A.M.; Harry, D.; De Sousa, F.; Vassal, H.; Eskat, A.; Meraldi, P. Mild replication stress causes chromosome mis-segregation via premature centriole disengagement. Nat. Commun. 2019, 10, e3585. [Google Scholar] [CrossRef]
- Burrell, R.A.; Swanton, C. Tumour heterogeneity and the evolution of polyclonal drug resistance. Mol. Oncol. 2014, 8, 1095–1111. [Google Scholar] [CrossRef]
- Rosai, J.; Ackerman, L.V. The pathology of tumors, part III: Grading, staging & classification. CA Cancer J. Clin. 1979, 29, 66–77. [Google Scholar] [CrossRef]
- Sobin, L.H. The international histological classification of tumours. Bull. World Health Organ. 1981, 59, 813–819. [Google Scholar]
- DeVita, V.T., Jr.; Chu, E. A history of cancer chemotherapy. Cancer Res. 2008, 68, 8643–8653. [Google Scholar] [CrossRef] [PubMed]
- Twelves, C.; Jove, M.; Gombos, A.; Awada, A. Cytotoxic chemotherapy: Still the mainstay of clinical practice for all subtypes metastatic breast cancer. Crit. Rev. Oncol. Hematol. 2016, 100, 74–87. [Google Scholar] [CrossRef] [PubMed]
- Bakhoum, S.F.; Landau, D.A. Chromosomal Instability as a Driver of Tumor Heterogeneity and Evolution. Cold Spring Harb Perspect. Med. 2017, 7. [Google Scholar] [CrossRef] [PubMed]
- McGranahan, N.; Swanton, C. Biological and therapeutic impact of intratumor heterogeneity in cancer evolution. Cancer Cell 2015, 27, 15–26. [Google Scholar] [CrossRef]
- Stanta, G.; Bonin, S. A Practical Approach to Tumor Heterogeneity in Clinical Research and Diagnostics. Pathobiology 2018, 85, 7–17. [Google Scholar] [CrossRef]
- Prasetyanti, P.R.; Medema, J.P. Intra-tumor heterogeneity from a cancer stem cell perspective. Mol. Cancer 2017, 16, e41. [Google Scholar] [CrossRef]
- Gerlinger, M.; Horswell, S.; Larkin, J.; Rowan, A.J.; Salm, M.P.; Varela, I.; Fisher, R.; McGranahan, N.; Matthews, N.; Santos, C.R.; et al. Genomic architecture and evolution of clear cell renal cell carcinomas defined by multiregion sequencing. Nat. Genet. 2014, 46, 225–233. [Google Scholar] [CrossRef]
- Gerlinger, M.; Rowan, A.J.; Horswell, S.; Math, M.; Larkin, J.; Endesfelder, D.; Gronroos, E.; Martinez, P.; Matthews, N.; Stewart, A.; et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N. Engl. J. Med. 2012, 366, 883–892. [Google Scholar] [CrossRef]
- Alix-Panabieres, C.; Pantel, K. Clinical Applications of Circulating Tumor Cells and Circulating Tumor DNA as Liquid Biopsy. Cancer Discov. 2016, 6, 479–491. [Google Scholar] [CrossRef]
- Torres, L.; Ribeiro, F.R.; Pandis, N.; Andersen, J.A.; Heim, S.; Teixeira, M.R. Intratumor genomic heterogeneity in breast cancer with clonal divergence between primary carcinomas and lymph node metastases. Breast Cancer Res. Treat. 2007, 102, 143–155. [Google Scholar] [CrossRef]
- Wei, Q.; Ye, Z.; Zhong, X.; Li, L.; Wang, C.; Myers, R.E.; Palazzo, J.P.; Fortuna, D.; Yan, A.; Waldman, S.A.; et al. Multiregion whole-exome sequencing of matched primary and metastatic tumors revealed genomic heterogeneity and suggested polyclonal seeding in colorectal cancer metastasis. Ann. Oncol. 2017, 28, 2135–2141. [Google Scholar] [CrossRef] [PubMed]
- Grzywa, T.M.; Paskal, W.; Wlodarski, P.K. Intratumor and Intertumor Heterogeneity in Melanoma. Transl. Oncol. 2017, 10, 956–975. [Google Scholar] [CrossRef]
- Sutherland, K.D.; Visvader, J.E. Cellular Mechanisms Underlying Intertumoral Heterogeneity. Trends Cancer 2015, 1, 15–23. [Google Scholar] [CrossRef]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef]
- Dillekas, H.; Rogers, M.S.; Straume, O. Are 90% of deaths from cancer caused by metastases? Cancer Med. 2019, 8, 5574–5576. [Google Scholar] [CrossRef]
- Amin, M.B.; Greene, F.L.; Edge, S.B.; Compton, C.C.; Gershenwald, J.E.; Brookland, R.K.; Meyer, L.; Gress, D.M.; Byrd, D.R.; Winchester, D.P. The Eighth Edition AJCC Cancer Staging Manual: Continuing to build a bridge from a population-based to a more "personalized" approach to cancer staging. CA Cancer J. Clin. 2017, 67, 93–99. [Google Scholar] [CrossRef]
- Xiao, W.; Zheng, S.; Yang, A.; Zhang, X.; Zou, Y.; Tang, H.; Xie, X. Breast cancer subtypes and the risk of distant metastasis at initial diagnosis: A population-based study. Cancer Manag. Res. 2018, 10, 5329–5338. [Google Scholar] [CrossRef]
- Salvador, S.; Rempel, A.; Soslow, R.A.; Gilks, B.; Huntsman, D.; Miller, D. Chromosomal instability in fallopian tube precursor lesions of serous carcinoma and frequent monoclonality of synchronous ovarian and fallopian tube mucosal serous carcinoma. Gynecol. Oncol. 2008, 110, 408–417. [Google Scholar] [CrossRef]
- Turajlic, S.; Xu, H.; Litchfield, K.; Rowan, A.; Chambers, T.; Lopez, J.I.; Nicol, D.; O’Brien, T.; Larkin, J.; Horswell, S.; et al. Tracking Cancer Evolution Reveals Constrained Routes to Metastases: TRACERx Renal. Cell 2018, 173, 581–594. [Google Scholar] [CrossRef]
- Pastushenko, I.; Blanpain, C. EMT Transition States during Tumor Progression and Metastasis. Trends Cell Biol. 2019, 29, 212–226. [Google Scholar] [CrossRef]
- Naxerova, K.; Jain, R.K. Using tumor phylogenetics to identify the roots of metastasis in humans. Nat. Rev. Clin. Oncol. 2015, 12, 258–272. [Google Scholar] [CrossRef]
- Turajlic, S.; Swanton, C. Metastasis as an evolutionary process. Science 2016, 352, 169–175. [Google Scholar] [CrossRef]
- Macintyre, G.; Van Loo, P.; Corcoran, N.M.; Wedge, D.C.; Markowetz, F.; Hovens, C.M. How Subclonal Modeling Is Changing the Metastatic Paradigm. Clin. Cancer Res. 2017, 23, 630–635. [Google Scholar] [CrossRef]
- Talmadge, J.E. Clonal selection of metastasis within the life history of a tumor. Cancer Res. 2007, 67, 11471–11475. [Google Scholar] [CrossRef]
- Nguyen, D.X.; Massague, J. Genetic determinants of cancer metastasis. Nat. Rev. Genet. 2007, 8, 341–352. [Google Scholar] [CrossRef]
- Onder, T.T.; Gupta, P.B.; Mani, S.A.; Yang, J.; Lander, E.S.; Weinberg, R.A. Loss of E-cadherin promotes metastasis via multiple downstream transcriptional pathways. Cancer Res. 2008, 68, 3645–3654. [Google Scholar] [CrossRef]
- Petrova, Y.I.; Schecterson, L.; Gumbiner, B.M. Roles for E-cadherin cell surface regulation in cancer. Mol. Biol. Cell 2016, 27, 3233–3244. [Google Scholar] [CrossRef]
- Carter, S.L.; Eklund, A.C.; Kohane, I.S.; Harris, L.N.; Szallasi, Z. A signature of chromosomal instability inferred from gene expression profiles predicts clinical outcome in multiple human cancers. Nat. Genet. 2006, 38, 1043–1048. [Google Scholar] [CrossRef]
- Lengauer, C.; Kinzler, K.W.; Vogelstein, B. Genetic instability in colorectal cancers. Nature 1997, 386, 623–627. [Google Scholar] [CrossRef]
- Orsetti, B.; Selves, J.; Bascoul-Mollevi, C.; Lasorsa, L.; Gordien, K.; Bibeau, F.; Massemin, B.; Paraf, F.; Soubeyran, I.; Hostein, I.; et al. Impact of chromosomal instability on colorectal cancer progression and outcome. BMC Cancer 2014, 14, e121. [Google Scholar] [CrossRef]
- Bartlett, J.M.; Munro, A.F.; Dunn, J.A.; McConkey, C.; Jordan, S.; Twelves, C.J.; Cameron, D.A.; Thomas, J.; Campbell, F.M.; Rea, D.W.; et al. Predictive markers of anthracycline benefit: A prospectively planned analysis of the UK National Epirubicin Adjuvant Trial (NEAT/BR9601). Lancet Oncol. 2010, 11, 266–274. [Google Scholar] [CrossRef]
- Munro, A.F.; Twelves, C.; Thomas, J.S.; Cameron, D.A.; Bartlett, J.M. Chromosome instability and benefit from adjuvant anthracyclines in breast cancer. Br. J. Cancer 2012, 107, 71–74. [Google Scholar] [CrossRef][Green Version]
- Swanton, C.; Nicke, B.; Schuett, M.; Eklund, A.C.; Ng, C.; Li, Q.; Hardcastle, T.; Lee, A.; Roy, R.; East, P.; et al. Chromosomal instability determines taxane response. Proc. Natl. Acad. Sci. USA 2009, 106, 8671–8676. [Google Scholar] [CrossRef]
- Spears, M.; Yousif, F.; Lyttle, N.; Boutros, P.C.; Munro, A.F.; Twelves, C.; Pritchard, K.I.; Levine, M.N.; Shepherd, L.; Bartlett, J.M. A four gene signature predicts benefit from anthracyclines: Evidence from the BR9601 and MA.5 clinical trials. Oncotarget 2015, 6, 31693–31701. [Google Scholar] [CrossRef]
- Smeets, D.; Miller, I.S.; O’Connor, D.P.; Das, S.; Moran, B.; Boeckx, B.; Gaiser, T.; Betge, J.; Barat, A.; Klinger, R.; et al. Copy number load predicts outcome of metastatic colorectal cancer patients receiving bevacizumab combination therapy. Nat. Commun. 2018, 9, e4112. [Google Scholar] [CrossRef]
- Lagarde, P.; Perot, G.; Kauffmann, A.; Brulard, C.; Dapremont, V.; Hostein, I.; Neuville, A.; Wozniak, A.; Sciot, R.; Schoffski, P.; et al. Mitotic checkpoints and chromosome instability are strong predictors of clinical outcome in gastrointestinal stromal tumors. Clin. Cancer Res. 2012, 18, 826–838. [Google Scholar] [CrossRef]
- Tang, R.; Ho, Y.S.; You, Y.T.; Hsu, K.C.; Chen, J.S.; Changchien, C.R.; Wang, J.Y. Prognostic evaluation of DNA flow cytometric and histopathologic parameters of colorectal cancer. Cancer 1995, 76, 1724–1730. [Google Scholar] [CrossRef]
- Habermann, J.K.; Doering, J.; Hautaniemi, S.; Roblick, U.J.; Bundgen, N.K.; Nicorici, D.; Kronenwett, U.; Rathnagiriswaran, S.; Mettu, R.K.; Ma, Y.; et al. The gene expression signature of genomic instability in breast cancer is an independent predictor of clinical outcome. Int. J. Cancer 2009, 124, 1552–1564. [Google Scholar] [CrossRef]
- How, C.; Bruce, J.; So, J.; Pintilie, M.; Haibe-Kains, B.; Hui, A.; Clarke, B.A.; Hedley, D.W.; Hill, R.P.; Milosevic, M.; et al. Chromosomal instability as a prognostic marker in cervical cancer. BMC Cancer 2015, 15, 361. [Google Scholar] [CrossRef]
- Kasprzyk, M.; Dyszkiewicz, W.; Piwkowski, C.; Gasiorowski, L.; Kaczmarek, E. Prognostic value of DNA ploidy: 5-year follow-up of patients with resectable squamous cell carcinoma (SCC) of the lung. Lung Cancer 2006, 51, 201–206. [Google Scholar] [CrossRef]
- Lykkesfeldt, A.E.; Balslev, I.; Christensen, I.J.; Larsen, J.K.; Molgaard, H.; Rasmussen, B.B.; Thorpe, S.; Rose, C. DNA ploidy and S-phase fraction in primary breast carcinomas in relation to prognostic factors and survival for premenopausal patients at high risk for recurrent disease. Acta. Oncol. 1988, 27, 749–756. [Google Scholar] [CrossRef] [PubMed]
- Nakazawa, M. The prognostic significance of DNA ploidy for neuroblastoma. Surg. Today 1993, 23, 215–219. [Google Scholar] [CrossRef] [PubMed]
- Pradhan, M.; Abeler, V.M.; Danielsen, H.E.; Sandstad, B.; Trope, C.G.; Kristensen, G.B.; Risberg, B.A. Prognostic importance of DNA ploidy and DNA index in stage I and II endometrioid adenocarcinoma of the endometrium. Ann. Oncol. 2012, 23, 1178–1184. [Google Scholar] [CrossRef]
- Rodenburg, C.J.; Cornelisse, C.J.; Heintz, P.A.; Hermans, J.; Fleuren, G.J. Tumor ploidy as a major prognostic factor in advanced ovarian cancer. Cancer 1987, 59, 317–323. [Google Scholar] [CrossRef]
- Song, T.; Lee, J.W.; Kim, H.J.; Kim, M.K.; Choi, C.H.; Kim, T.J.; Bae, D.S.; Kim, B.G. Prognostic significance of DNA ploidy in stage I endometrial cancer. Gynecol. Oncol. 2011, 122, 79–82. [Google Scholar] [CrossRef]
- Tsavaris, N.; Kavantzas, N.; Tsigritis, K.; Xynos, I.D.; Papadoniou, N.; Lazaris, A.; Kosmas, C.; Agrogiannis, G.; Dokou, A.; Felekouras, E.; et al. Evaluation of DNA ploidy in relation with established prognostic factors in patients with locally advanced (unresectable) or metastatic pancreatic adenocarcinoma: A retrospective analysis. BMC Cancer 2009, 9, 264. [Google Scholar] [CrossRef] [PubMed]
- Janssen, A.; Kops, G.J.; Medema, R.H. Elevating the frequency of chromosome mis-segregation as a strategy to kill tumor cells. Proc. Natl. Acad. Sci. USA 2009, 106, 19108–19113. [Google Scholar] [CrossRef]
- Bryant, H.E.; Schultz, N.; Thomas, H.D.; Parker, K.M.; Flower, D.; Lopez, E.; Kyle, S.; Meuth, M.; Curtin, N.J.; Helleday, T. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature 2005, 434, 913–917. [Google Scholar] [CrossRef]
- Farmer, H.; McCabe, N.; Lord, C.J.; Tutt, A.N.; Johnson, D.A.; Richardson, T.B.; Santarosa, M.; Dillon, K.J.; Hickson, I.; Knights, C.; et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 2005, 434, 917–921. [Google Scholar] [CrossRef]
- McManus, K.J.; Barrett, I.J.; Nouhi, Y.; Hieter, P. Specific synthetic lethal killing of RAD54B-deficient human colorectal cancer cells by FEN1 silencing. Proc. Natl. Acad. Sci. USA 2009, 106, 3276–3281. [Google Scholar] [CrossRef]
- Sajesh, B.V.; Guppy, B.J.; McManus, K.J. Synthetic genetic targeting of genome instability in cancer. Cancers (Basel) 2013, 5, 739–761. [Google Scholar] [CrossRef] [PubMed]
- Sajesh, B.V.; McManus, K.J. Targeting SOD1 induces synthetic lethal killing in BLM- and CHEK2-deficient colorectal cancer cells. Oncotarget 2015, 6, 27907–27922. [Google Scholar] [CrossRef] [PubMed]
- McAndrew, E.N.; Lepage, C.C.; McManus, K.J. The synthetic lethal killing of RAD54B-deficient colorectal cancer cells by PARP1 inhibition is enhanced with SOD1 inhibition. Oncotarget 2016, 7, 87417–87430. [Google Scholar] [CrossRef] [PubMed]
- Friedl, P.; Alexander, S. Cancer invasion and the microenvironment: Plasticity and reciprocity. Cell 2011, 147, 992–1009. [Google Scholar] [CrossRef] [PubMed]
- Stanta, G.; Bonin, S. Overview on Clinical Relevance of Intra-Tumor Heterogeneity. Front. Med. (Lausanne) 2018, 5, 85. [Google Scholar] [CrossRef]
- Witsch, E.; Sela, M.; Yarden, Y. Roles for growth factors in cancer progression. Physiology (Bethesda) 2010, 25, 85–101. [Google Scholar] [CrossRef]
- Gupta, R.G.; Somer, R.A. Intratumor Heterogeneity: Novel Approaches for Resolving Genomic Architecture and Clonal Evolution. Mol. Cancer Res. 2017, 15, 1127–1137. [Google Scholar] [CrossRef]
- Turajlic, S.; McGranahan, N.; Swanton, C. Inferring mutational timing and reconstructing tumour evolutionary histories. Biochim. Biophys. Acta. 2015, 1855, 264–275. [Google Scholar] [CrossRef]
- Russo, M.; Lamba, S.; Lorenzato, A.; Sogari, A.; Corti, G.; Rospo, G.; Mussolin, B.; Montone, M.; Lazzari, L.; Arena, S.; et al. Reliance upon ancestral mutations is maintained in colorectal cancers that heterogeneously evolve during targeted therapies. Nat. Commun. 2018, 9, e2287. [Google Scholar] [CrossRef]
- Misale, S.; Arena, S.; Lamba, S.; Siravegna, G.; Lallo, A.; Hobor, S.; Russo, M.; Buscarino, M.; Lazzari, L.; Sartore-Bianchi, A.; et al. Blockade of EGFR and MEK intercepts heterogeneous mechanisms of acquired resistance to anti-EGFR therapies in colorectal cancer. Sci. Transl. Med. 2014, 6, 224ra226. [Google Scholar] [CrossRef]
- Chen, G.; Mulla, W.A.; Kucharavy, A.; Tsai, H.J.; Rubinstein, B.; Conkright, J.; McCroskey, S.; Bradford, W.D.; Weems, L.; Haug, J.S.; et al. Targeting the adaptability of heterogeneous aneuploids. Cell 2015, 160, 771–784. [Google Scholar] [CrossRef] [PubMed]
- Bergshoeff, V.E.; Van der Heijden, S.J.; Haesevoets, A.; Litjens, S.G.; Bot, F.J.; Voogd, A.C.; Chenault, M.N.; Hopman, A.H.; Schuuring, E.; Van der Wal, J.M.; et al. Chromosome instability predicts progression of premalignant lesions of the larynx. Pathology 2014, 46, 216–224. [Google Scholar] [CrossRef] [PubMed]
- Giaretti, W.; Monteghirfo, S.; Pentenero, M.; Gandolfo, S.; Malacarne, D.; Castagnola, P. Chromosomal instability, DNA index, dysplasia, and subsite in oral premalignancy as intermediate endpoints of risk of cancer. Cancer Epidemiol. Biomark. Prev. 2013, 22, 1133–1141. [Google Scholar] [CrossRef] [PubMed]
- Wanders, L.K.; Cordes, M.; Voorham, Q.; Sie, D.; de Vries, S.D.; d’Haens, G.; de Boer, N.K.H.; Ylstra, B.; van Grieken, N.C.T.; Meijer, G.A.; et al. IBD-Associated Dysplastic Lesions Show More Chromosomal Instability Than Sporadic Adenomas. Inflamm. Bowel Dis. 2020, 26, 167–180. [Google Scholar] [CrossRef]
- Hiley, C.T.; Swanton, C. Spatial and temporal cancer evolution: Causes and consequences of tumour diversity. Clin. Med. (Lond) 2014, 14, 33–37. [Google Scholar] [CrossRef]
- Salk, J.J.; Schmitt, M.W.; Loeb, L.A. Enhancing the accuracy of next-generation sequencing for detecting rare and subclonal mutations. Nat. Rev. Genet. 2018, 19, 269–285. [Google Scholar] [CrossRef]
- McGranahan, N.; Burrell, R.A.; Endesfelder, D.; Novelli, M.R.; Swanton, C. Cancer chromosomal instability: Therapeutic and diagnostic challenges. EMBO Rep. 2012, 13, 528–538. [Google Scholar] [CrossRef]
- Kapeleris, J.; Kulasinghe, A.; Warkiani, M.E.; Vela, I.; Kenny, L.; O’Byrne, K.; Punyadeera, C. The Prognostic Role of Circulating Tumor Cells (CTCs) in Lung Cancer. Front. Oncol. 2018, 8, e311. [Google Scholar] [CrossRef]
- Wang, J.; Chang, S.; Li, G.; Sun, Y. Application of liquid biopsy in precision medicine: Opportunities and challenges. Front. Med. 2017, 11, 522–527. [Google Scholar] [CrossRef]
- Balakrishnan, A.; Koppaka, D.; Anand, A.; Deb, B.; Grenci, G.; Viasnoff, V.; Thompson, E.W.; Gowda, H.; Bhat, R.; Rangarajan, A.; et al. Circulating Tumor Cell cluster phenotype allows monitoring response to treatment and predicts survival. Sci. Rep. 2019, 9, 7933. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vishwakarma, R.; McManus, K.J. Chromosome Instability; Implications in Cancer Development, Progression, and Clinical Outcomes. Cancers 2020, 12, 824. https://doi.org/10.3390/cancers12040824
Vishwakarma R, McManus KJ. Chromosome Instability; Implications in Cancer Development, Progression, and Clinical Outcomes. Cancers. 2020; 12(4):824. https://doi.org/10.3390/cancers12040824
Chicago/Turabian StyleVishwakarma, Raghvendra, and Kirk J. McManus. 2020. "Chromosome Instability; Implications in Cancer Development, Progression, and Clinical Outcomes" Cancers 12, no. 4: 824. https://doi.org/10.3390/cancers12040824
APA StyleVishwakarma, R., & McManus, K. J. (2020). Chromosome Instability; Implications in Cancer Development, Progression, and Clinical Outcomes. Cancers, 12(4), 824. https://doi.org/10.3390/cancers12040824




