Unique Role of Histone Methyltransferase PRDM8 in the Tumorigenesis of Virus-Negative Merkel Cell Carcinoma
Abstract
1. Introduction
2. Methods
2.1. Patient Samples and MCC Cell Lines
2.2. Immunohistochemistry (IHC), Immunocytochemistry, and Western Blot
2.3. mRNA and miRNA Expression Analyses
2.4. CRISPR-Cas9-Mediated Gene Silencing
2.5. Chromatin Immunoprecipitation Sequencing (ChIP-seq) and ChIP-qPCR
2.6. ChIP-seq Visualization Using IGV Browser
2.7. Quantitative PCR
2.8. Colony Formation Assay
2.9. miRNA Overexpression
2.10. Analysis of ChIP-seq Data from ENCODE Database
3. Results
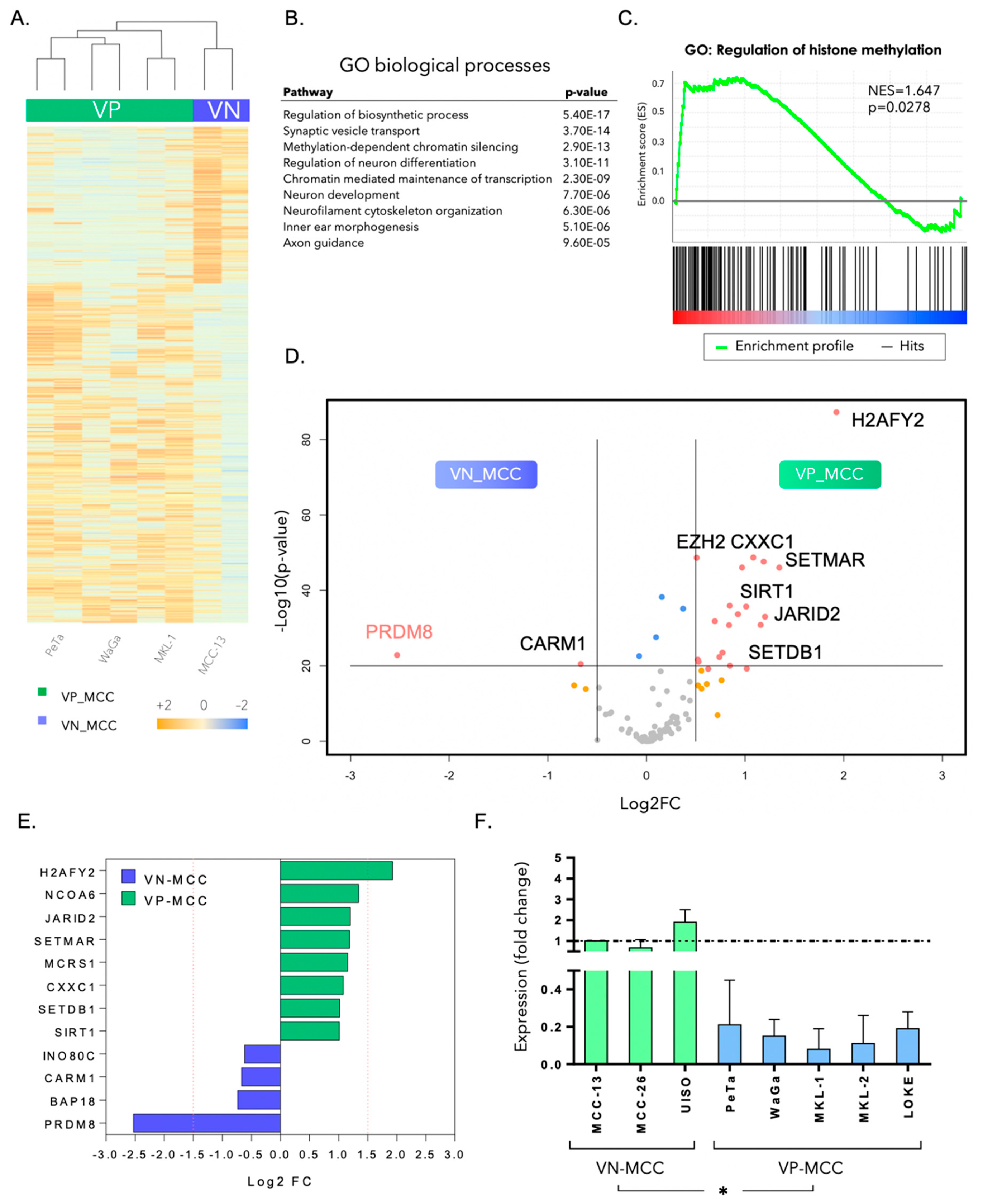
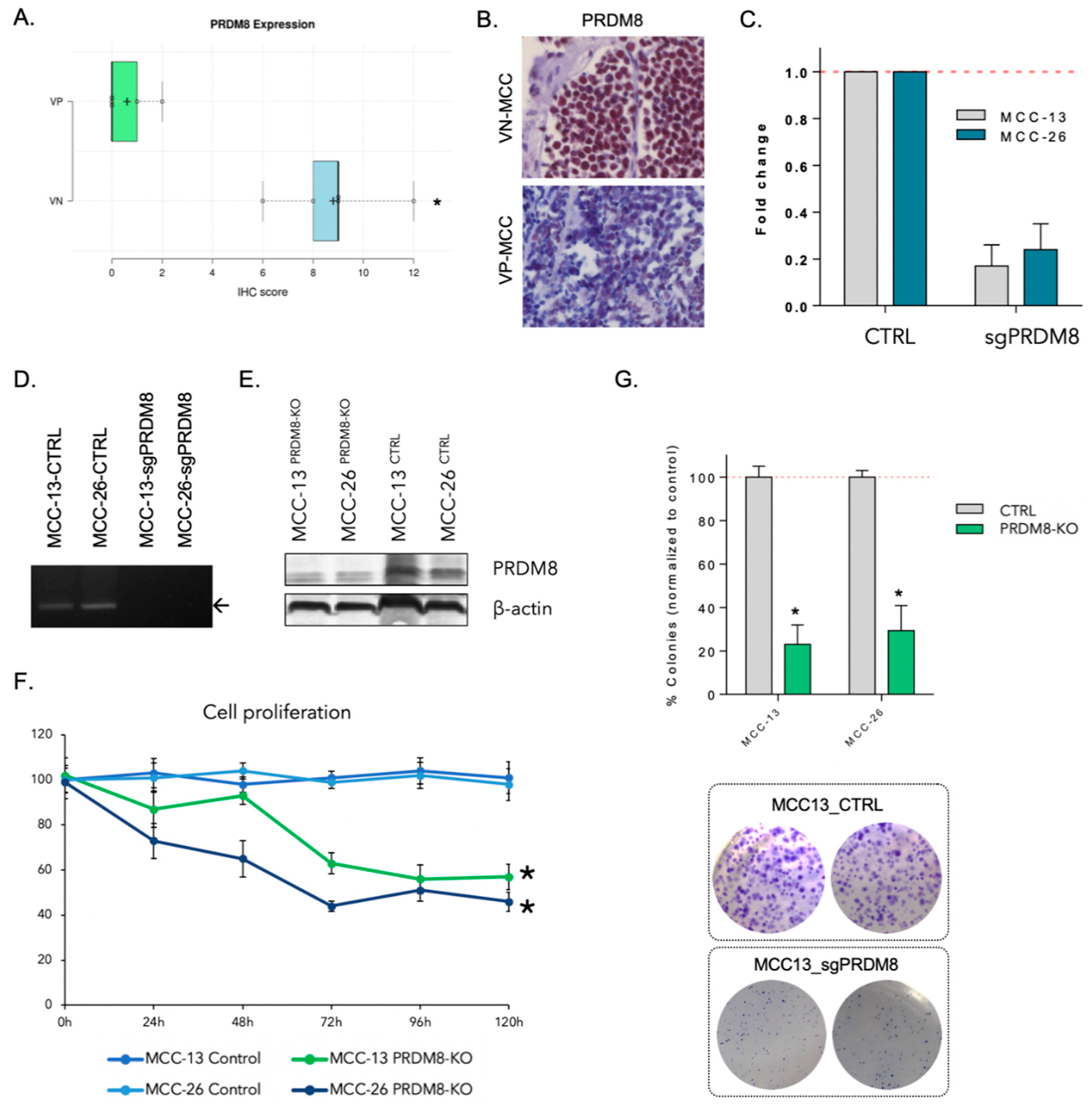
3.1. PRDM8 Is Highly Expressed in Virus-Negative Merkel Cell Carcinoma Cells and Is Correlated with Tumor Proliferation and Clonogenicity
3.2. PRDM8 Expression Was Correlated with Global H3K9me3 Level in VN-MCC
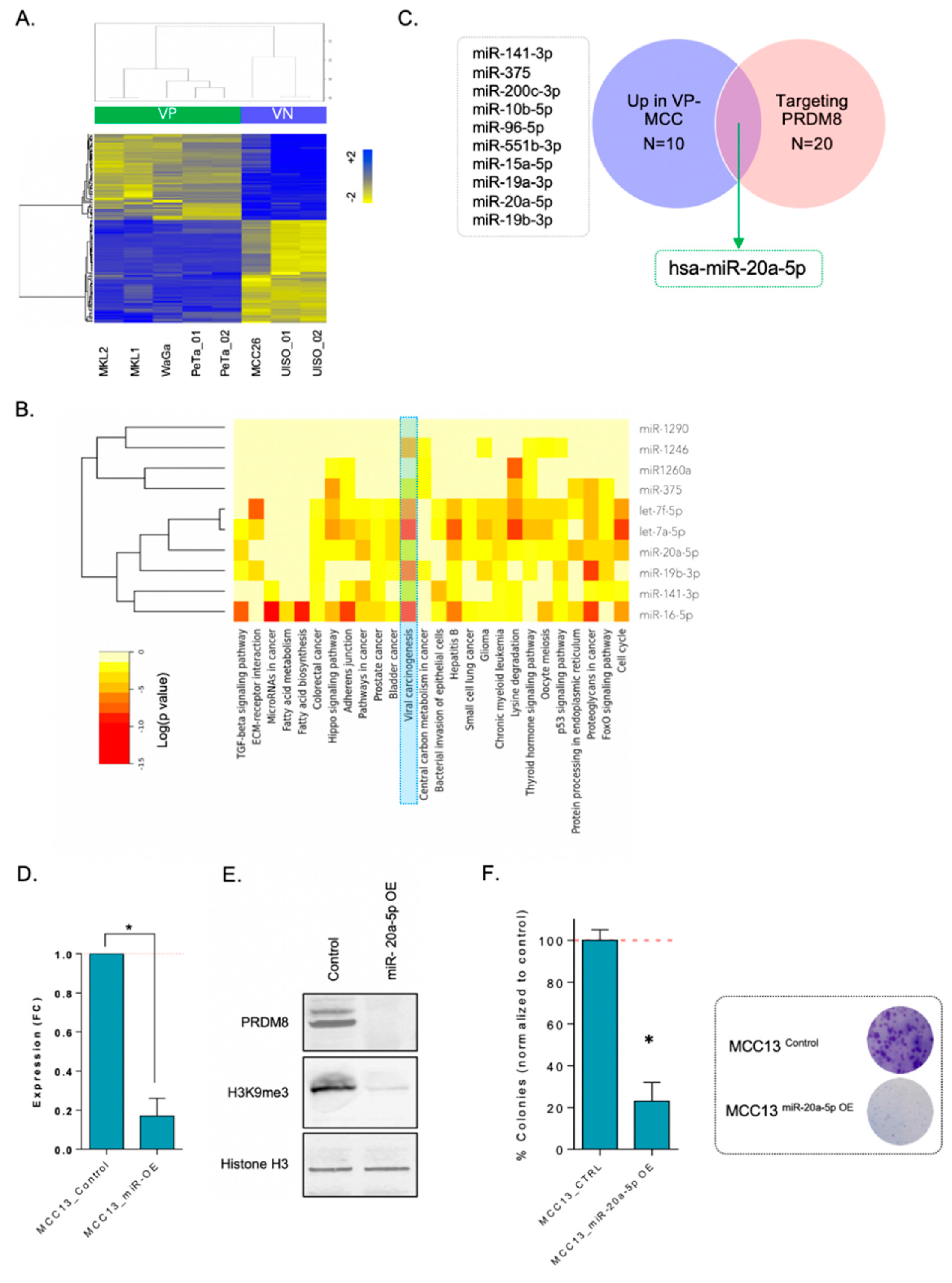
3.3. miR-20a-5p Regulated PRDM8 in VN-MCC
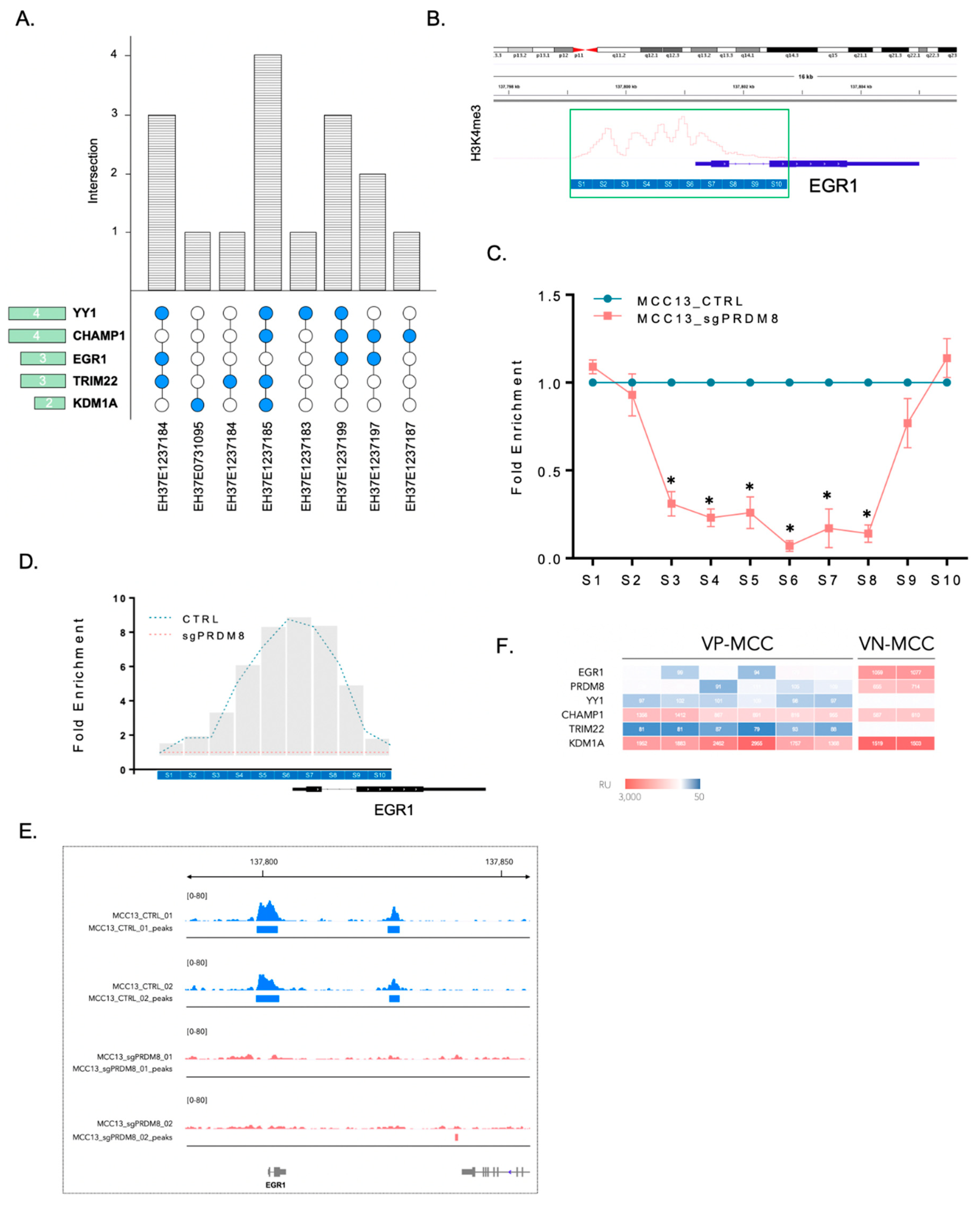
3.4. PRDM8 Acted through Its Partnership with EGR1
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| ChIP | Chromatin immunoprecipitation |
| DEG | Differentially expressed gene |
| ENCODE | Encyclopedia of DNA Elements |
| FDR | False discovery rate |
| FFPE | Formalin-fixed paraffin-embedded |
| GO | Gene Ontology |
| GSEA | Gene set enrichment analysis |
| IGV | Integrative Genomics Viewer |
| IHC | Immunohistochemistry |
| MCC | Merkel cell carcinoma |
| MCPyV | Merkel cell polyomavirus |
| MCV | Merkel cell polyomavirus |
| NES | Normalized enrichment score |
| TMA | Tissue microarray |
| VP-MCC | Virus-positive Merkel cell carcinoma |
| VN-MCC | Virus-negative Merkel cell carcinoma |
References
- Feng, H.; Shuda, M.; Chang, Y.; Moore, P.S. Clonal integration of a polyomavirus in human Merkel cell carcinoma. Science 2008, 319, 1096–1100. [Google Scholar] [CrossRef] [PubMed]
- Schadendorf, D.; Lebbe, C.; Zur Hausen, A.; Avril, M.F.; Hariharan, S.; Bharmal, M.; Becker, J.C. Merkel cell carcinoma: Epidemiology, prognosis, therapy and unmet medical needs. Eur. J. Cancer 2017, 71, 53–69. [Google Scholar] [CrossRef]
- Wong, S.Q.; Waldeck, K.; Vergara, I.A.; Schroder, J.; Madore, J.; Wilmott, J.S.; Colebatch, A.J.; De Paoli-Iseppi, R.; Li, J.; Lupat, R.; et al. UV-Associated Mutations Underlie the Etiology of MCV-Negative Merkel Cell Carcinomas. Cancer Res. 2015, 75, 5228–5234. [Google Scholar] [CrossRef]
- Goh, G.; Walradt, T.; Markarov, V.; Blom, A.; Riaz, N.; Doumani, R.; Stafstrom, K.; Moshiri, A.; Yelistratova, L.; Levinsohn, J.; et al. Mutational landscape of MCPyV-positive and MCPyV-negative Merkel cell carcinomas with implications for immunotherapy. Oncotarget 2016, 7, 3403–3415. [Google Scholar] [CrossRef] [PubMed]
- Harms, P.W.; Vats, P.; Verhaegen, M.E.; Robinson, D.R.; Wu, Y.M.; Dhanasekaran, S.M.; Palanisamy, N.; Siddiqui, J.; Cao, X.; Su, F.; et al. The Distinctive Mutational Spectra of Polyomavirus-Negative Merkel Cell Carcinoma. Cancer Res. 2015, 75, 3720–3727. [Google Scholar] [CrossRef] [PubMed]
- Sunshine, J.C.; Jahchan, N.S.; Sage, J.; Choi, J. Are there multiple cells of origin of Merkel cell carcinoma? Oncogene 2018, 37, 1409–1416. [Google Scholar] [CrossRef] [PubMed]
- Mertz, K.D.; Junt, T.; Schmid, M.; Pfaltz, M.; Kempf, W. Inflammatory monocytes are a reservoir for Merkel cell polyomavirus. J. Investig. Derm. 2010, 130, 1146–1151. [Google Scholar] [CrossRef]
- Liu, W.; Yang, R.; Payne, A.S.; Schowalter, R.M.; Spurgeon, M.E.; Lambert, P.F.; Xu, X.; Buck, C.B.; You, J. Identifying the Target Cells and Mechanisms of Merkel Cell Polyomavirus Infection. Cell Host Microbe 2016, 19, 775–787. [Google Scholar] [CrossRef]
- Becker, J.C.; Stang, A.; DeCaprio, J.A.; Cerroni, L.; Lebbe, C.; Veness, M.; Nghiem, P. Merkel cell carcinoma. Nat. Rev. Dis. Primers 2017, 3, 17077. [Google Scholar] [CrossRef]
- Tilling, T.; Wladykowski, E.; Failla, A.V.; Houdek, P.; Brandner, J.M.; Moll, I. Immunohistochemical analyses point to epidermal origin of human Merkel cells. Histochem. Cell Biol. 2014, 141, 407–421. [Google Scholar] [CrossRef]
- Zur Hausen, A.; Rennspiess, D.; Winnepenninckx, V.; Speel, E.J.; Kurz, A.K. Early B-cell differentiation in Merkel cell carcinomas: Clues to cellular ancestry. Cancer Res. 2013, 73, 4982–4987. [Google Scholar] [CrossRef] [PubMed]
- Cimino, P.J.; Robirds, D.H.; Tripp, S.R.; Pfeifer, J.D.; Abel, H.J.; Duncavage, E.J. Retinoblastoma gene mutations detected by whole exome sequencing of Merkel cell carcinoma. Mod. Pathol. 2014, 27, 1073–1087. [Google Scholar] [CrossRef] [PubMed]
- Orouji, E.; Utikal, J. Tackling malignant melanoma epigenetically: Histone lysine methylation. Clin. Epigenet. 2018, 10, 145. [Google Scholar] [CrossRef] [PubMed]
- Eom, G.H.; Kim, K.; Kim, S.M.; Kee, H.J.; Kim, J.Y.; Jin, H.M.; Kim, J.R.; Kim, J.H.; Choe, N.; Kim, K.B.; et al. Histone methyltransferase PRDM8 regulates mouse testis steroidogenesis. Biochem. Biophys. Res. Commun. 2009, 388, 131–136. [Google Scholar] [CrossRef]
- Houben, R.; Grimm, J.; Willmes, C.; Weinkam, R.; Becker, J.C.; Schrama, D. Merkel cell carcinoma and Merkel cell polyomavirus: Evidence for hit-and-run oncogenesis. J. Investig. Dermatol. 2012, 132, 254–256. [Google Scholar] [CrossRef]
- Houben, R.; Shuda, M.; Weinkam, R.; Schrama, D.; Feng, H.; Chang, Y.; Moore, P.S.; Becker, J.C. Merkel cell polyomavirus-infected Merkel cell carcinoma cells require expression of viral T antigens. J. Virol. 2010, 84, 7064–7072. [Google Scholar] [CrossRef]
- Shuda, M.; Feng, H.; Kwun, H.J.; Rosen, S.T.; Gjoerup, O.; Moore, P.S.; Chang, Y. T antigen mutations are a human tumor-specific signature for Merkel cell polyomavirus. Proc. Natl. Acad. Sci. USA 2008, 105, 16272–16277. [Google Scholar] [CrossRef]
- Van Gele, M.; Kaghad, M.; Leonard, J.H.; Van Roy, N.; Naeyaert, J.M.; Geerts, M.L.; Van Belle, S.; Cocquyt, V.; Bridge, J.; Sciot, R.; et al. Mutation analysis of P73 and TP53 in Merkel cell carcinoma. Br. J. Cancer 2000, 82, 823–826. [Google Scholar] [CrossRef]
- Leonard, J.H.; Dash, P.; Holland, P.; Kearsley, J.H.; Bell, J.R. Characterisation of four Merkel cell carcinoma adherent cell lines. Int. J. Cancer 1995, 60, 100–107. [Google Scholar] [CrossRef]
- Ronan, S.G.; Green, A.D.; Shilkaitis, A.; Huang, T.S.; Das Gupta, T.K. Merkel cell carcinoma: In vitro and in vivo characteristics of a new cell line. J. Am. Acad. Dermatol. 1993, 29, 715–722. [Google Scholar] [CrossRef]
- Houben, R.; Dreher, C.; Angermeyer, S.; Borst, A.; Utikal, J.; Haferkamp, S.; Peitsch, W.K.; Schrama, D.; Hesbacher, S. Mechanisms of p53 restriction in Merkel cell carcinoma cells are independent of the Merkel cell polyoma virus T antigens. J. Investig. Dermatol. 2013, 133, 2453–2460. [Google Scholar] [CrossRef] [PubMed]
- Orouji, E.; Orouji, A.; Gaiser, T.; Larribère, L.; Gebhardt, C.; Utikal, J. MAP kinase pathway gene copy alterations in NRAS/BRAF wild-type advanced melanoma. Int. J. Cancer 2016, 138, 2257–2262. [Google Scholar] [CrossRef] [PubMed]
- Terranova, C.; Tang, M.; Orouji, E.; Maitituoheti, M.; Raman, A.; Amin, S.; Liu, Z.; Rai, K. An Integrated Platform for Genome-wide Mapping of Chromatin States Using High-throughput ChIP-sequencing in Tumor Tissues. J. Vis. Exp. 2018, 134, 56972. [Google Scholar] [CrossRef] [PubMed]
- Vlachos, I.S.; Zagganas, K.; Paraskevopoulou, M.D.; Georgakilas, G.; Karagkouni, D.; Vergoulis, T.; Dalamagas, T.; Hatzigeorgiou, A.G. DIANA-miRPath v3.0: Deciphering microRNA function with experimental support. Nucleic Acids Res. 2015, 43, W460–W466. [Google Scholar] [CrossRef]
- Dweep, H.; Gretz, N. miRWalk2.0: A comprehensive atlas of microRNA-target interactions. Nat. Methods 2015, 12, 697. [Google Scholar] [CrossRef] [PubMed]
- Moshiri, A.S.; Doumani, R.; Yelistratova, L.; Blom, A.; Lachance, K.; Shinohara, M.M.; Delaney, M.; Chang, O.; McArdle, S.; Thomas, H.; et al. Polyomavirus-Negative Merkel Cell Carcinoma: A More Aggressive Subtype Based on Analysis of 282 Cases Using Multimodal Tumor Virus Detection. J. Investig. Derm. 2017, 137, 819–827. [Google Scholar] [CrossRef]
- Starrett, G.J.; Thakuria, M.; Chen, T.; Marcelus, C.; Cheng, J.; Nomburg, J.; Thorner, A.R.; Slevin, M.K.; Powers, W.; Burns, R.T.; et al. Clinical and molecular characterization of virus-positive and virus-negative Merkel cell carcinoma. Genome Med. 2020, 12, 1–22. [Google Scholar] [CrossRef]
- Sahi, H.; Savola, S.; Sihto, H.; Koljonen, V.; Bohling, T.; Knuutila, S. RB1 gene in Merkel cell carcinoma: Hypermethylation in all tumors and concurrent heterozygous deletions in the polyomavirus-negative subgroup. Apmis 2014, 122, 1157–1166. [Google Scholar] [CrossRef]
- Kumar, S.; Xie, H.; Scicluna, P.; Lee, L.; Bjornhagen, V.; Hoog, A.; Larsson, C.; Lui, W.O. MiR-375 Regulation of LDHB Plays Distinct Roles in Polyomavirus-Positive and -Negative Merkel Cell Carcinoma. Cancers 2018, 10, 443. [Google Scholar] [CrossRef]
- Kumar, S.; Xie, H.; Shi, H.; Gao, J.; Juhlin, C.C.; Bjornhagen, V.; Hoog, A.; Lee, L.; Larsson, C.; Lui, W.O. Merkel cell polyomavirus oncoproteins induce microRNAs that suppress multiple autophagy genes. Int. J. Cancer 2019, 146, 1652–1666. [Google Scholar] [CrossRef]
- Lee, S.; Paulson, K.G.; Murchison, E.P.; Afanasiev, O.K.; Alkan, C.; Leonard, J.H.; Byrd, D.R.; Hannon, G.J.; Nghiem, P. Identification and validation of a novel mature microRNA encoded by the Merkel cell polyomavirus in human Merkel cell carcinomas. J. Clin. Virol. 2011, 52, 272–275. [Google Scholar] [CrossRef] [PubMed]
- Chteinberg, E.; Sauer, C.M.; Rennspiess, D.; Beumers, L.; Schiffelers, L.; Eben, J.; Haugg, A.; Winnepenninckx, V.; Kurz, A.K.; Speel, E.J.; et al. Neuroendocrine Key Regulator Gene Expression in Merkel Cell Carcinoma. Neoplasia 2018, 20, 1227–1235. [Google Scholar] [CrossRef] [PubMed]
- Abernathy, D.G.; Kim, W.K.; McCoy, M.J.; Lake, A.M.; Ouwenga, R.; Lee, S.W.; Xing, X.; Li, D.; Lee, H.J.; Heuckeroth, R.O.; et al. MicroRNAs Induce a Permissive Chromatin Environment that Enables Neuronal Subtype-Specific Reprogramming of Adult Human Fibroblasts. Cell Stem Cell 2017, 21, 332–348.e9. [Google Scholar] [CrossRef] [PubMed]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Orouji, E.; Peitsch, W.K.; Orouji, A.; Houben, R.; Utikal, J. Unique Role of Histone Methyltransferase PRDM8 in the Tumorigenesis of Virus-Negative Merkel Cell Carcinoma. Cancers 2020, 12, 1057. https://doi.org/10.3390/cancers12041057
Orouji E, Peitsch WK, Orouji A, Houben R, Utikal J. Unique Role of Histone Methyltransferase PRDM8 in the Tumorigenesis of Virus-Negative Merkel Cell Carcinoma. Cancers. 2020; 12(4):1057. https://doi.org/10.3390/cancers12041057
Chicago/Turabian StyleOrouji, Elias, Wiebke K. Peitsch, Azadeh Orouji, Roland Houben, and Jochen Utikal. 2020. "Unique Role of Histone Methyltransferase PRDM8 in the Tumorigenesis of Virus-Negative Merkel Cell Carcinoma" Cancers 12, no. 4: 1057. https://doi.org/10.3390/cancers12041057
APA StyleOrouji, E., Peitsch, W. K., Orouji, A., Houben, R., & Utikal, J. (2020). Unique Role of Histone Methyltransferase PRDM8 in the Tumorigenesis of Virus-Negative Merkel Cell Carcinoma. Cancers, 12(4), 1057. https://doi.org/10.3390/cancers12041057





