An Integrated Meta-Analysis of Secretome and Proteome Identify Potential Biomarkers of Pancreatic Ductal Adenocarcinoma
Abstract
1. Introduction
2. Results
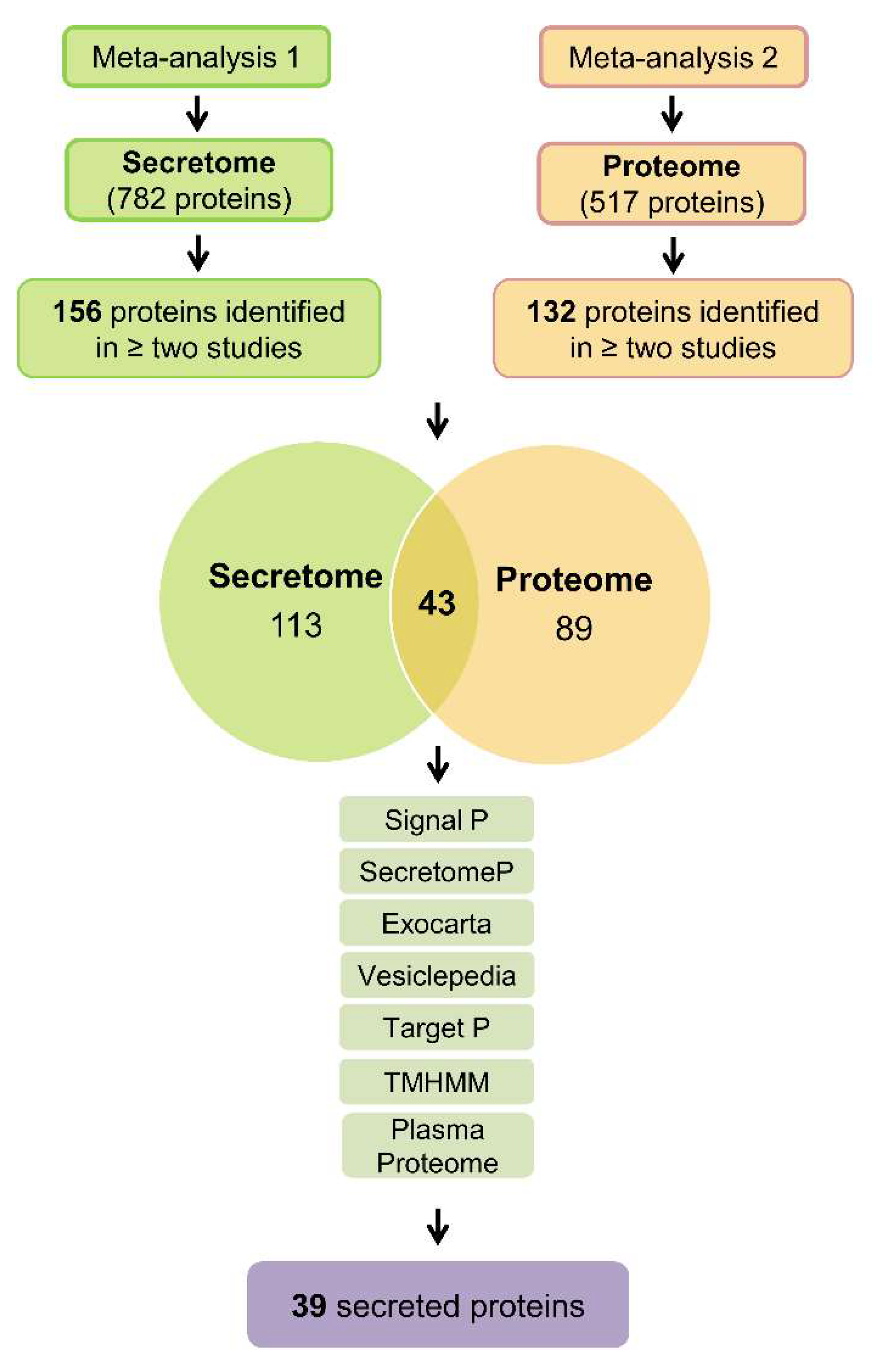
2.1. Integration of Secretome and Proteome Meta-Analysis Identifies 39 Secreted Proteins in Pancreatic Ductal Adenocarcinoma
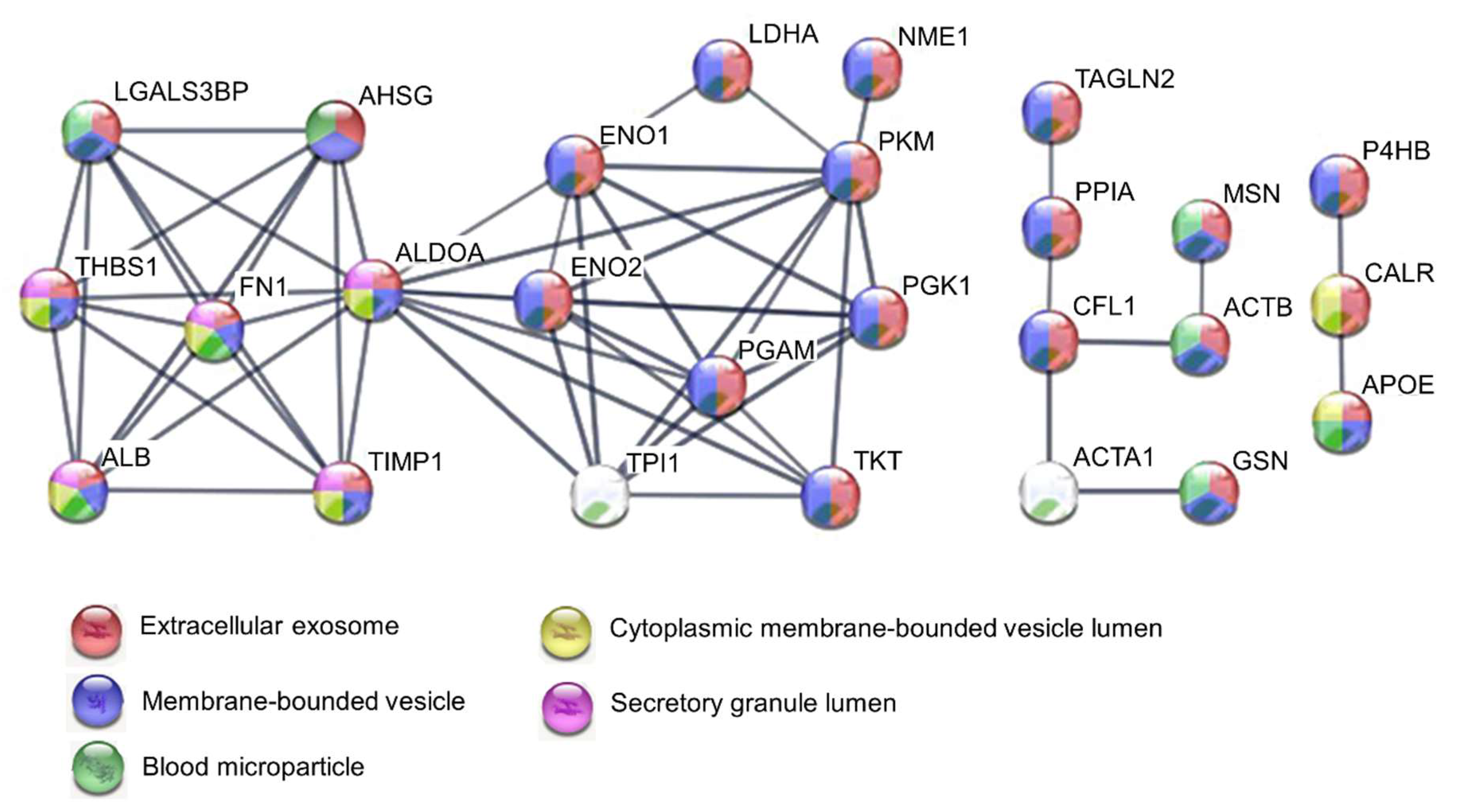
2.2. Protein-Protein Interaction (PPI) Network of 39 Secreted Proteins Enriched in Pancreatic Ductal Adenocarcinoma
2.3. Secretome-Related Gene Expression is Enriched in Pancreatic Ductal Adenocarcinoma
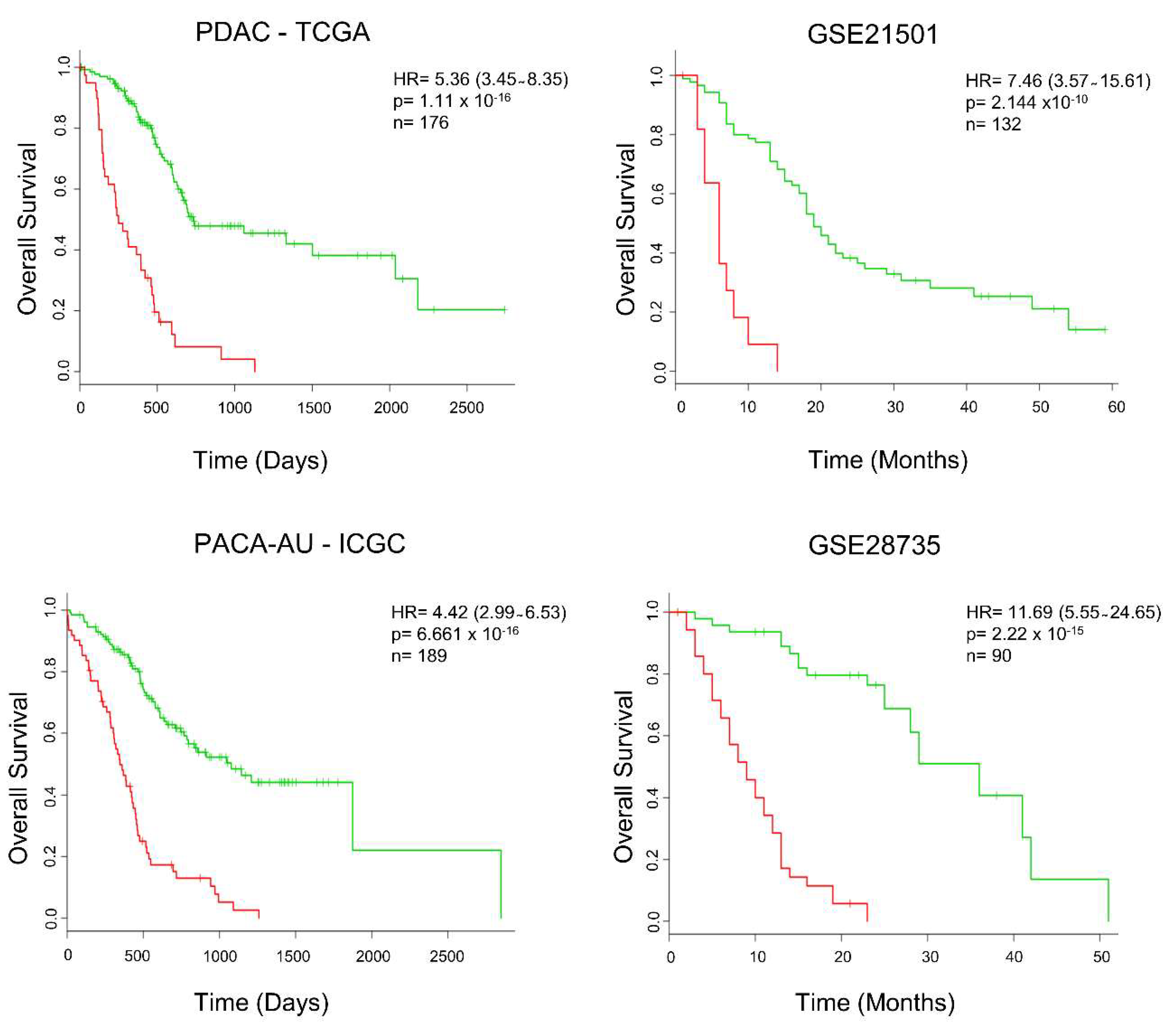
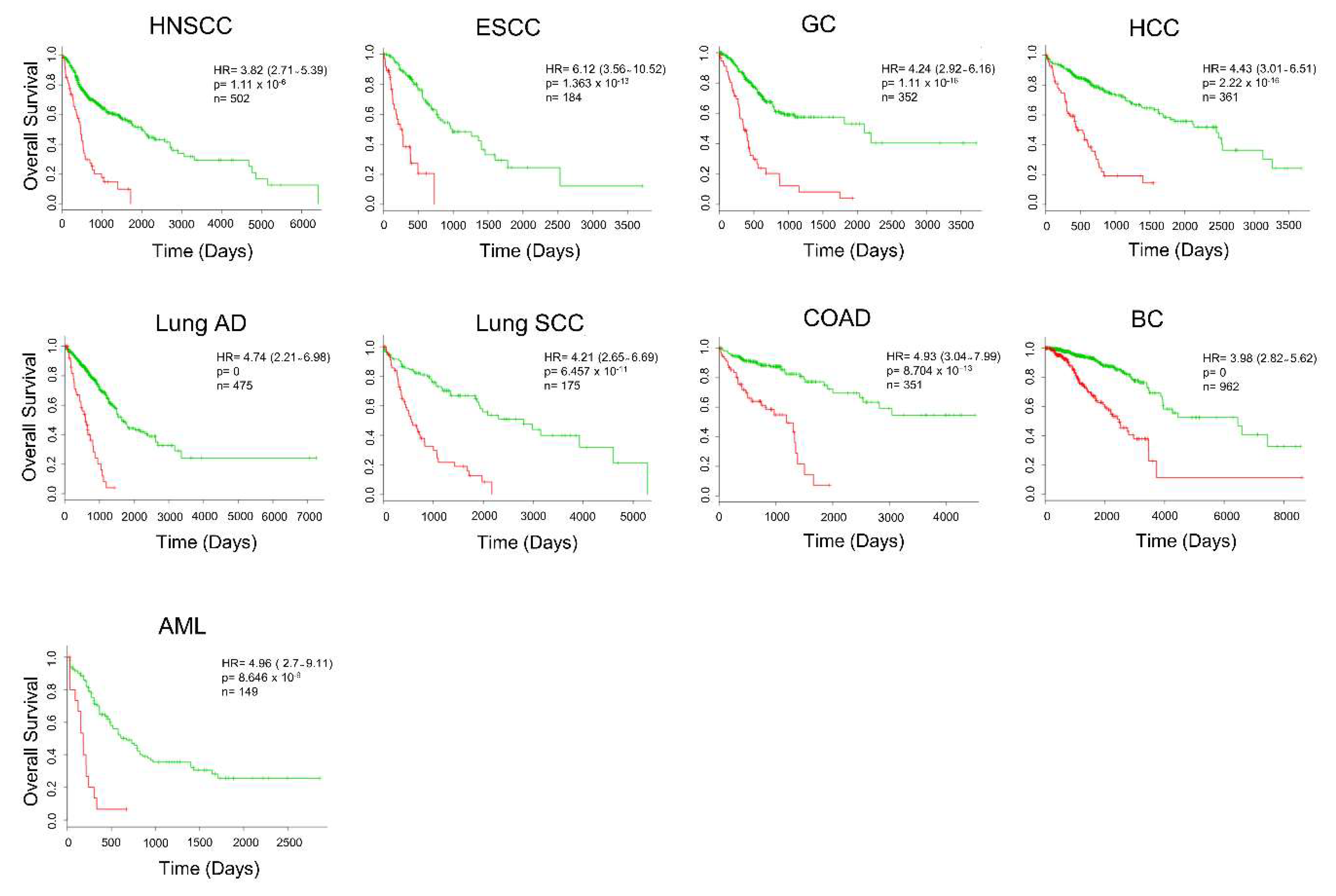
2.4. Secretome-Related Gene Expression Profile of 39 Proteins Predict Shorter Survival in Patients with Pancreatic Ductal Adenocarcinoma
2.5. Secretome Gene Expression Predicts Cancer Outcomes in Different Cancer Studies
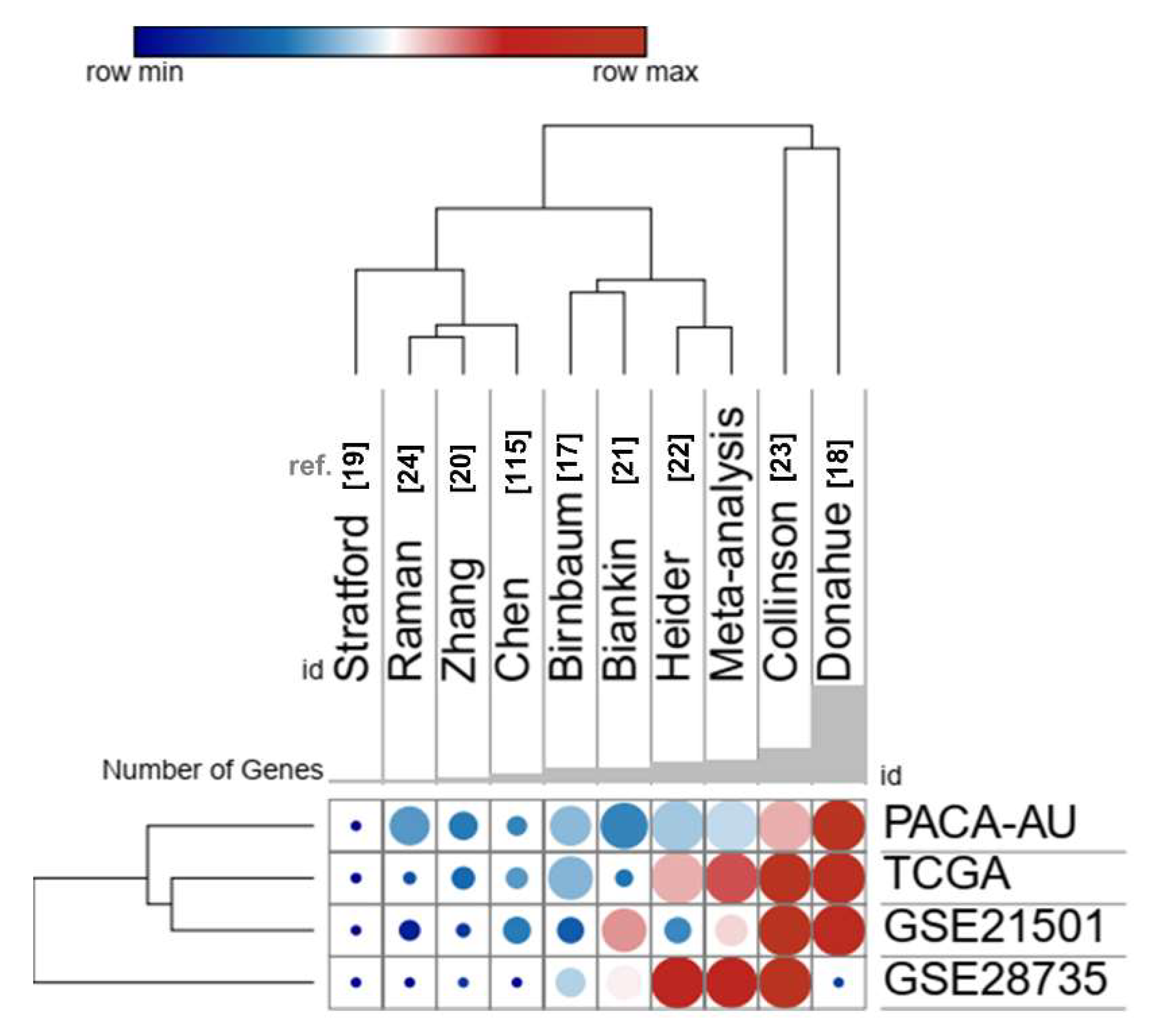
2.6. Comparison with Prognostic Gene Signatures of Pancreatic Ductal Adenocarcinoma
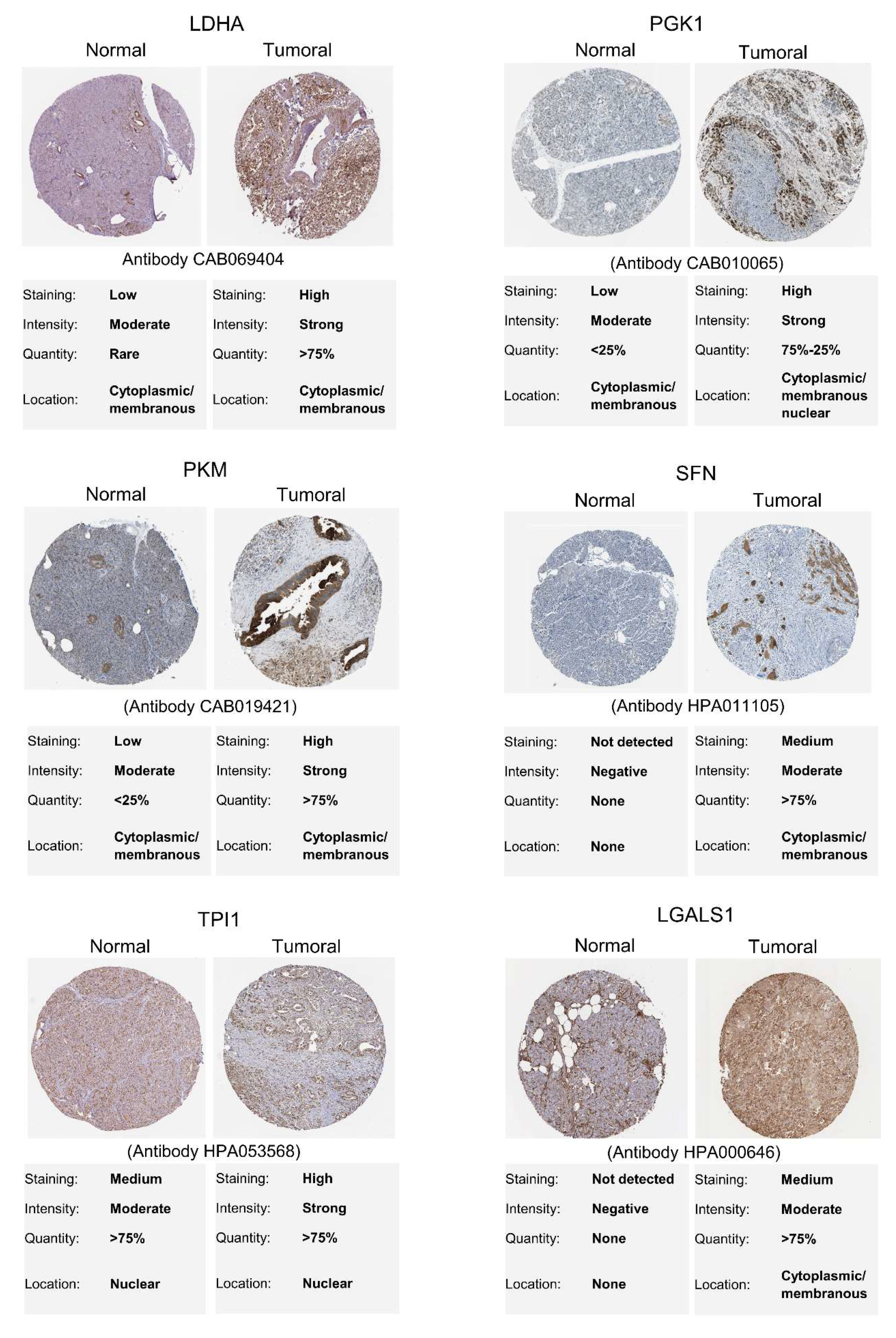
2.7. In Silico Validation of Secreted Proteins
3. Discussion
4. Materials and Methods
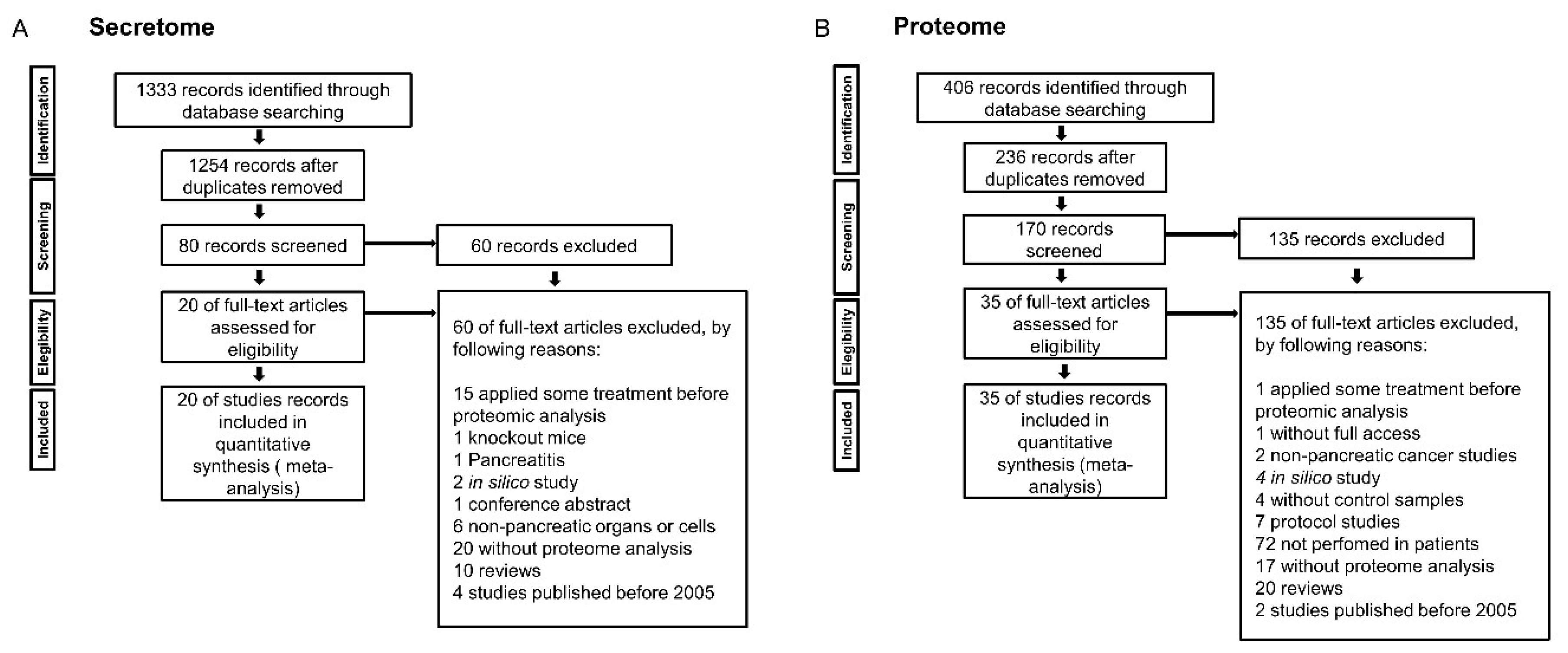
4.1. Integration of Secretome and Proteome Meta-Analysis to Identify Pancreatic Cancer Biomarkers
4.1.1. Pancreatic Cancer Secretome Meta-Analysis
4.1.2. Pancreatic Ductal Adenocarcinoma Proteome Meta-Analysis
4.2. Extraction of Meta-Analysis Data and in Silico Confirmation of Secreted Proteins
4.3. Protein-Protein Interaction Network and Gene Ontology Analysis
4.4. Gene Expression Profile in Pancreatic Ductal Adenocarcinoma
4.5. Prognostic Value of Secreted Protein Translated Transcripts in the Predicting Pancreatic Ductal Adenocarcinoma Outcome
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Rahib, L.; Smith, B.D.; Aizenberg, R.; Rosenzweig, A.B.; Fleshman, J.M.; Matrisian, L.M. Projecting Cancer Incidence and Deaths to 2030: The Unexpected Burden of Thyroid, Liver, and Pancreas Cancers in the United States. Cancer Res. 2014, 74, 2913–2921. [Google Scholar] [CrossRef]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef]
- Makawita, S.; Smith, C.; Batruch, I.; Zheng, Y.; Rückert, F.; Grützmann, R.; Pilarsky, C.; Gallinger, S.; Diamandis, E.P. Integrated Proteomic Profiling of Cell Line Conditioned Media and Pancreatic Juice for the Identification of Pancreatic Cancer Biomarkers. Mol. Cell. Proteom. 2011, 10, M111.008599. [Google Scholar] [CrossRef]
- Zhang, M.; Wang, Z.; Obazee, O.; Jia, J.; Childs, E.J.; Hoskins, J.; Figlioli, G.; Mocci, E.; Collins, I.; Chung, C.C.; et al. Three new pancreatic cancer susceptibility signals identified on chromosomes 1q32.1, 5p15.33 and 8q24.21. Oncotarget 2016, 7, 66328–66343. [Google Scholar] [CrossRef]
- Jemal, A.; Siegel, R.; Ward, E.; Hao, Y.; Xu, J.; Thun, M.J. Cancer Statistics, 2009. CA Cancer J. Clin. 2009, 59, 225–249. [Google Scholar] [CrossRef] [PubMed]
- Sitek, B.; Sipos, B.; Alkatout, I.; Poschmann, G.; Stephan, C.; Schulenborg, T.; Marcus, K.; Lu, J.; Dittert, D.; Baretton, G.; et al. Analysis of the Pancreatic Tumor Progression by a Quantitative Proteomic Approach and Immunhistochemical Validation research articles. J. Proteome Res. 2009, 1647–1656. [Google Scholar] [CrossRef] [PubMed]
- Kamisawa, T.; Wood, L.D.; Itoi, T.; Takaori, K. Pancreatic cancer. Lancet 2016, 388, 73–85. [Google Scholar] [CrossRef]
- Yachida, S.; Jones, S.; Bozic, I.; Antal, T.; Leary, R.; Fu, B.; Kamiyama, M.; Hruban, R.H.; Eshleman, J.R.; Nowak, M.A.; et al. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature 2010, 467, 1114–1117. [Google Scholar] [CrossRef] [PubMed]
- Liang, C.; Shi, S.; Meng, Q.; Liang, D.; Ji, S.; Zhang, B.; Qin, Y.; Xu, J.; Ni, Q.; Yu, X. Complex roles of the stroma in the intrinsic resistance to gemcitabine in pancreatic cancer: Where we are and where we are going. Exp. Mol. Med. 2017, 49, e406. [Google Scholar] [CrossRef] [PubMed]
- Chand, S.; O’Hayer, K.; Blanco, F.F.; Winter, J.M.; Brody, J.R. The landscape of pancreatic cancer therapeutic resistance mechanisms. Int. J. Biol. Sci. 2016, 12, 273–283. [Google Scholar] [CrossRef]
- Lewis, R.; Drebin, J.A.; Callery, M.P.; Fraker, D.; Kent, T.S.; Gates, J.; Vollmer, C.M., Jr. A contemporary analysis of survival for resected pancreatic ductal adenocarcinoma. HPB 2013, 15, 49–60. [Google Scholar] [CrossRef] [PubMed]
- Ducreux, M.; Cuhna, A.S.; Caramella, C.; Hollebecque, A.; Burtin, P.; Goéré, D.; Seufferlein, T.; Haustermans, K.; Van Laethem, J.L.; Conroy, T.; et al. Cancer of the pancreas: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2015, 26, v56–v68. [Google Scholar] [CrossRef] [PubMed]
- Collisson, E.A.; Bailey, P.; Chang, D.K.; Biankin, A.V. Molecular subtypes of pancreatic cancer. Nat. Rev. Gastroenterol. Hepatol. 2019, 531, 47–52. [Google Scholar] [CrossRef] [PubMed]
- Kern, S.E.; Shi, C.; Hruban, R.H. The complexity of pancreatic ductal cancers and multidimensional strategies for therapeutic targeting. J. Pathol. 2011, 223, 296–307. [Google Scholar] [CrossRef]
- Kleeff, J.; Korc, M.; Apte, M.; La Vecchia, C.; Johnson, C.D.; Biankin, A.V.; Neale, R.E.; Tempero, M.; Tuveson, D.A.; Hruban, R.H.; et al. Pancreatic cancer. Nat. Rev. Dis. Prim. 2016, 2, 16022. [Google Scholar] [CrossRef]
- Fong, Z.V.; Winter, J.M. Biomarkers in Pancreatic Cancer. Cancer J. 2012, 18, 530–538. [Google Scholar] [CrossRef]
- Birnbaum, D.J.; Finetti, P.; Lopresti, A.; Gilabert, M.; Poizat, F.; Raoul, J.L.; Delpero, J.R.; Moutardier, V.; Birnbaum, D.; Mamessier, E.; et al. A 25-gene classifier predicts overall survival in resectable pancreatic cancer. BMC Med. 2017, 15, 170. [Google Scholar] [CrossRef]
- Donahue, T.R.; Tran, L.M.; Hill, R.; Li, Y.; Kovochich, A.; Calvopina, J.H.; Patel, S.G.; Wu, N.; Hindoyan, A.; Farrell, J.J.; et al. Integrative Survival-Based Molecular Profiling of Human Pancreatic Cancer. Clin. Cancer Res. 2012, 18, 1352–1363. [Google Scholar] [CrossRef]
- Stratford, J.K.; Bentrem, D.J.; Anderson, J.M.; Fan, C.; Volmar, K.A.; Marron, J.S.; Routh, E.D.; Caskey, L.S.; Samuel, J.C.; Der, C.J.; et al. A Six-Gene Signature Predicts Survival of Patients with Localized Pancreatic Ductal Adenocarcinoma. PLoS Med. 2010, 7, e1000307. [Google Scholar] [CrossRef]
- Zhang, G.; Schetter, A.; He, P.; Funamizu, N.; Gaedcke, J.; Ghadimi, B.M.; Ried, T.; Hassan, R.; Yfantis, H.G.; Lee, D.H.; et al. DPEP1 Inhibits Tumor Cell Invasiveness, Enhances Chemosensitivity and Predicts Clinical Outcome in Pancreatic Ductal Adenocarcinoma. PLoS ONE 2012, 7, e31507. [Google Scholar] [CrossRef]
- Biankin, A.V.; Waddell, N.; Kassahn, K.S.; Gingras, M.-C.; Muthuswamy, L.B.; Johns, A.L.; Miller, D.K.; Wilson, P.J.; Patch, A.-M.; Wu, J.; et al. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature 2012, 491, 399–405. [Google Scholar] [CrossRef] [PubMed]
- Haider, S.; Wang, J.; Nagano, A.; Desai, A.; Arumugam, P.; Dumartin, L.; Fitzgibbon, J.; Hagemann, T.; Marshall, J.F.; Kocher, H.M.; et al. A multi-gene signature predicts outcome in patients with pancreatic ductal adenocarcinoma. Genome Med. 2014, 6, 105. [Google Scholar] [CrossRef] [PubMed]
- Collisson, E.A.; Sadanandam, A.; Olson, P.; Gibb, W.J.; Truitt, M.; Gu, S.; Cooc, J.; Weinkle, J.; Kim, G.E.; Jakkula, L.; et al. Subtypes of pancreatic ductal adenocarcinoma and their differing responses to therapy. Nat. Med. 2011, 17, 500–503. [Google Scholar] [CrossRef] [PubMed]
- Raman, P.; Maddipati, R.; Lim, K.H.; Tozeren, A. Pancreatic cancer survival analysis defines a signature that predicts outcome. PLoS ONE 2018, 13, e0201751. [Google Scholar] [CrossRef] [PubMed]
- Pradet-Balade, B.; Boulmé, F.; Beug, H.; Müllner, E.W.; Garcia-Sanz, J.A. Translation control: Bridging the gap between genomics and proteomics? Trends Biochem. Sci. 2001, 26, 225–229. [Google Scholar] [CrossRef]
- Liu, Y.; Beyer, A.; Aebersold, R. On the Dependency of Cellular Protein Levels on mRNA Abundance. Cell 2016, 165, 535–550. [Google Scholar] [CrossRef]
- Hanash, S.; Taguchi, A. The grand challenge to decipher the cancer proteome. Nat. Rev. Cancer 2010, 10, 652–660. [Google Scholar] [CrossRef]
- Domon, B.; Aebersold, R. Challenges and Opportunities in Proteomics Data Analysis. Mol. Cell. Proteom. 2006, 5, 1921–1926. [Google Scholar] [CrossRef]
- Iuga, C.; Seicean, A.; Iancu, C.; Buiga, R.; Sappa, P.K.; Völker, U.; Hammer, E. Proteomic identification of potential prognostic biomarkers in resectable pancreatic ductal adenocarcinoma. Proteomics 2014, 14, 945–955. [Google Scholar] [CrossRef]
- Jimenez-Luna, C.; Torres, C.; Ortiz, R.; Dieguez, C.; Martinez-Galan, J.; Melguizo, C.; Prados, J.C.; Caba, O. Proteomic biomarkers in body fluids associated with pancreatic cancer. Oncotarget 2018, 9, 16573–16587. [Google Scholar] [CrossRef]
- Brandi, J.; Manfredi, M.; Speziali, G.; Gosetti, F.; Marengo, E.; Cecconi, D. Proteomic approaches to decipher cancer cell secretome. Semin. Cell Dev. Biol. 2018, 78, 93–101. [Google Scholar] [CrossRef] [PubMed]
- Donadelli, M. The cancer secretome and secreted biomarkers. Semin. Cell Dev. Biol. 2018, 78, 1–2. [Google Scholar] [CrossRef] [PubMed]
- Houg, D.S.; Bijlsma, M.F. The hepatic pre-metastatic niche in pancreatic ductal adenocarcinoma. Mol. Cancer 2018, 17, 95. [Google Scholar] [CrossRef] [PubMed]
- Costa-Silva, B.; Aiello, N.M.; Ocean, A.J.; Singh, S.; Zhang, H.; Thakur, B.K.; Becker, A.; Hoshino, A.; Mark, M.T.; Molina, H.; et al. Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. Nat. Cell Biol. 2015, 17, 816–826. [Google Scholar] [CrossRef] [PubMed]
- Melo, S.; Luecke, L.B.; Kahlert, C.; Fernandez, A.F.; Gammon, S.T.; Kaye, J.; Lebleu, V.S.; Mittendorf, E.A.; Weitz, J.; Rahbari, N.; et al. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature 2015, 523, 177–182. [Google Scholar] [CrossRef] [PubMed]
- Lobb, R.J.; Lima, L.G.; Möller, A. Exosomes: Key mediators of metastasis and pre-metastatic niche formation. Semin. Cell Dev. Biol. 2017, 67, 3–10. [Google Scholar] [CrossRef]
- Grønborg, M.; Kristiansen, T.Z.; Iwahori, A.; Chang, R.; Reddy, R.; Sato, N.; Molina, H.; Jensen, O.N.; Hruban, R.H.; Goggins, M.G.; et al. Biomarker Discovery from Pancreatic Cancer Secretome Using a Differential Proteomic Approach. Mol. Cell. Proteom. 2006, 5, 157–171. [Google Scholar] [CrossRef]
- Belczacka, I.; Latosinska, A.; Metzger, J.; Marx, D.; Vlahou, A.; Mischak, H.; Frantzi, M. Proteomics biomarkers for solid tumors: Current status and future prospects. Mass Spectrom. Rev. 2018, 47–78. [Google Scholar] [CrossRef]
- Makridakis, M.; Vlahou, A. Secretome proteomics for discovery of cancer biomarkers. J. Proteom. 2010, 73, 2291–2305. [Google Scholar] [CrossRef]
- Pavlou, M.P.; Diamandis, E.P. The cancer cell secretome: A good source for discovering biomarkers? J. Proteom. 2010, 73, 1896–1906. [Google Scholar] [CrossRef]
- Brandi, J.; Pozza, E.D.; Dando, I.; Biondani, G.; Robotti, E.; Jenkins, R.; Elliott, V.; Park, K.; Marengo, E.; Costello, E.; et al. Secretome protein signature of human pancreatic cancer stem-like cells. J. Proteom. 2016, 136, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Ray, P.; Rialon-Guevara, K.L.; Veras, E.; Sullenger, B.A.; White, R.R. Comparing human pancreatic cell secretomes by in vitro aptamer selection identifies cyclophilin B as a candidate pancreatic cancer biomarker. J. Clin. Investig. 2012, 122, 1734–1741. [Google Scholar] [CrossRef] [PubMed]
- Schiarea, S.; Solinas, G.; Allavena, P.; Scigliuolo, G.M.; Bagnati, R.; Fanelli, R.; Chiabrando, C. Secretome analysis of multiple pancreatic cancer cell lines reveals perturbations of key functional networks. J. Proteome Res. 2010, 9, 4376–4392. [Google Scholar] [CrossRef] [PubMed]
- Xue, H.; Lu, B.; Lai, M. The cancer secretome: A reservoir of biomarkers. J. Transl. Med. 2008, 6, 52. [Google Scholar] [CrossRef] [PubMed]
- Klein-Scory, S.; Tehrani, M.M.; Eilert-Micus, C.; Adamczyk, K.A.; Wojtalewicz, N.; Schnölzer, M.; Hahn, S.A.; Schmiegel, W.; Schwarte-Waldhoff, I. New insights in the composition of extracellular vesicles from pancreatic cancer cells: Implications for biomarkers and functions. Proteome Sci. 2014, 12, 50. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.T.; Wu, C.C.; Shyr, Y.M.; Chen, T.C.; Hwang, T.L.; Yeh, T.S.; Chang, K.P.; Liu, H.P.; Liu, Y.L.; Tsai, M.H.; et al. Secretome-based identification of ULBP2 as a novel serum marker for pancreatic cancer detection. PLoS ONE 2011, 6, e20029. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Weng, Y.; Sui, Z.; Wu, Y.; Meng, X.; Wu, M.; Jin, H.; Tan, X.; Zhang, L.; Zhang, Y. Quantitative secretomic analysis of pancreatic cancer cells in serum-containing conditioned medium. Sci. Rep. 2016, 6, 37606. [Google Scholar] [CrossRef]
- Borrebaeck, C.A.K. Precision diagnostics: Moving towards protein biomarker signatures of clinical utility in cancer. Nat. Rev. Cancer 2017, 17, 199–204. [Google Scholar] [CrossRef]
- Adamczyk, K.A.; Klein-Scory, S.; Tehrani, M.M.; Warnken, U.; Schmiegel, W.; Schnölzer, M.; Schwarte-Waldhoff, I. Characterization of soluble and exosomal forms of the EGFR released from pancreatic cancer cells. Life Sci. 2011, 89, 304–312. [Google Scholar] [CrossRef]
- Ristorcelli, E.; Beraud, E.; Verrando, P.; Villard, C.; Lafitte, D.; Sbarra, V.; Lombardo, D.; Verine, A. Human tumor nanoparticles induce apoptosis of pancreatic cancer cells. FASEB J. 2008, 22, 3358–3369. [Google Scholar] [CrossRef]
- Hyo, S.L.; Jeong, J.; Lee, K.J. Characterization of vesicles secreted from insulinoma NIT-1 cells. J. Proteome Res. 2009, 8, 2851–2862. [Google Scholar]
- Que, R.; Lin, C.; Ding, G.-P.; Wu, Z.-R.; Cao, L.-P. Increasing the immune activity of exosomes: The effect of miRNA-depleted exosome proteins on activating dendritic cell/cytokine-induced killer cells against pancreatic cancer. J. Zhejiang Univ. B 2016, 17, 352–360. [Google Scholar] [CrossRef] [PubMed]
- Walsh, N.; Dowling, P.; O’Donovan, N.; Henry, M.; Meleady, P.; Clynes, M. Aldehyde dehydrogenase 1A1 and gelsolin identified as novel invasion-modulating factors in conditioned medium of pancreatic cancer cells. J. Proteom. 2008, 71, 561–571. [Google Scholar] [CrossRef] [PubMed]
- McKinney, K.Q.; Lee, Y.Y.; Choi, H.S.; Groseclose, G.; Iannitti, D.A.; Martinie, J.B.; Russo, M.W.; Lundgren, D.H.; Han, D.K.; Bonkovsky, H.L.; et al. Discovery of putative pancreatic cancer biomarkers using subcellular proteomics. J. Proteom. 2011, 74, 79–88. [Google Scholar] [CrossRef]
- Srirajaskanthan, R.; Caplin, M.E.; Waugh, M.G.; Watkins, J.; Meyer, T.; Hsuan, J.J.; Beaumont, N.J. Identification of Mac-2-binding protein as a putative marker of neuroendocrine tumors from the analysis of cell line secretomes. Mol. Cell. Proteom. 2010, 9, 656–666. [Google Scholar] [CrossRef]
- Baron, B.; Kitagawa, T.; Nakamura, K.; Kuramitsu, Y. Isolation of a growth factor stress-induced pancreatic cancer sub-population: Insight into changes due to micro-environment. Cancer Genom. Proteom. 2015, 12, 49–55. [Google Scholar]
- Zhang, H.; Lv, L.; Liu, H.; Cui, L.; Chen, G.; Bi, P.; Li, Z. Profiling the potential biomarkers for cell differentiation of pancreatic cancer using iTRAQ and 2-D LC-MS/MS. Proteom. Clin. Appl. 2009, 3, 862–871. [Google Scholar] [CrossRef]
- Xiao, J.; Lee, W.; Zhao, Y.; Cao, R.; Go, V. Profiling Pancreatic Cancer–Secreted Proteome Using 15N Amino Acids and Serum-Free Media. Pancreas 2010, 39, e17–e23. [Google Scholar] [CrossRef]
- Yu, K.H.; Barry, C.G.; Austin, D.; Busch, C.M.; Sangar, V.; Rustgi, A.K.; Blair, I.A. Stable isotope dilution multidimensional liquid chromatography-tandem mass spectrometry for pancreatic cancer serum biomarker discovery. J. Proteome Res. 2009, 8, 1565–1576. [Google Scholar] [CrossRef]
- Sitek, B.; Luttges, J.; Marcus, K.; Kloppel, G.; Schmiegel, W.; Meyer, H.E.; Hahn, S.A.; Stuhler, K. Application of fluorescence difference gel electrophoresis saturation labelling for the analysis of microdissected precursor lesions of pancreatic ductal adenocarcinoma. Proteomics 2005, 5, 2665–2679. [Google Scholar] [CrossRef]
- Qi, T.; Han, J.; Cui, Y.; Zong, M.; Liu, X.; Zhu, B. Comparative proteomic analysis for the detection of biomarkers in pancreatic ductal adenocarcinomas. J. Clin. Pathol. 2008, 61, 49–58. [Google Scholar] [CrossRef] [PubMed]
- Turtoi, A.; Musmeci, D.; Wang, Y.H.; Dumont, B.; Somja, J.; Bevilacqua, G.; De Pauw, E.; Delvenne, P.; Castronovo, V. Identification of Novel Accessible Proteins Bearing Diagnostic and Therapeutic Potential in Human Pancreatic Ductal Adenocarcinoma. J. Proteome Res. 2011, 10, 4302–4313. [Google Scholar] [CrossRef] [PubMed]
- Satoh, M.; Takano, S.; Sogawa, K.; Noda, K.; Yoshitomi, H.; Ishibashi, M.; Mogushi, K.; Takizawa, H.; Otsuka, M.; Shimizu, H.; et al. Immune-complex level of cofilin-1 in sera is associated with cancer progression and poor prognosis in pancreatic cancer. Cancer Sci. 2017, 108, 795–803. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.; Wu, W.-C.; Zhao, G.-C.; Wang, D.-S.; Lou, W.-H.; Jin, D.-Y. ITRAQ-based quantitative proteomics reveals apolipoprotein A-I and transferrin as potential serum markers in CA19-9 negative pancreatic ductal adenocarcinoma. Medicine 2016, 95, e4527. [Google Scholar] [CrossRef]
- Kosanam, H.; Prassas, I.; Chrystoja, C.C.; Soleas, I.; Chan, A.; Dimitromanolakis, A.; Blasutig, I.M.; Rückert, F.; Gruetzmann, R.; Pilarsky, C.; et al. Laminin, gamma 2 (LAMC2): A Promising New Putative Pancreatic Cancer Biomarker Identified by Proteomic Analysis of Pancreatic Adenocarcinoma Tissues. Mol. Cell. Proteom. 2013, 12, 2820–2832. [Google Scholar] [CrossRef]
- Mayerle, J.; Kalthoff, H.; Reszka, R.; Kamlage, B.; Peter, E.; Schniewind, B.; Maldonado, S.G.; Pilarsky, C.; Heidecke, C.D.; Schatz, P.; et al. Metabolic biomarker signature to differentiate pancreatic ductal adenocarcinoma from chronic pancreatitis. Gut 2017, 61, 128–137. [Google Scholar] [CrossRef]
- Takadate, T.; Onogawa, T.; Fukuda, T.; Motoi, F.; Suzuki, T.; Fujii, K.; Kihara, M.; Mikami, S.; Bando, Y.; Maeda, S.; et al. Novel prognostic protein markers of resectable pancreatic cancer identified by coupled shotgun and targeted proteomics using formalin-fixed paraffin-embedded tissues. Int. J. Cancer 2013, 132, 1368–1382. [Google Scholar] [CrossRef]
- Hwang, T.-L.; Liang, Y.; Chien, K.-Y.; Yu, J.-S. Overexpression and elevated serum levels of phosphoglycerate kinase 1 in pancreatic ductal adenocarcinoma. Proteomics 2006, 6, 2259–2272. [Google Scholar] [CrossRef]
- Kuwae, Y.; Kakehashi, A.; Wakasa, K.; Wei, M. Paraneoplastic Ma Antigen—Like 1 as a Potential Prognostic. Pancreas 2014, 44, 106–115. [Google Scholar] [CrossRef]
- Tian, R.; Wei, L.-M.; Qin, R.-Y.; Li, Y.; Du, Z.-Y.; Xia, W.; Shi, C.-J.; Jin, H. Proteome analysis of human pancreatic ductal adenocarcinoma tissue using two-dimensional gel electrophoresis and tandem mass spectrometry for identification of disease-related proteins. Dig. Dis. Sci. 2008, 53, 65–72. [Google Scholar] [CrossRef]
- Cui, Y.; Tian, M.; Zong, M.; Teng, M.; Chen, Y.; Lu, J.; Jiang, J.; Liu, X.; Han, J. Proteomic analysis of pancreatic ductal adenocarcinoma compared with normal adjacent pancreatic tissue and pancreatic benign cystadenoma. Pancreatology 2009, 9, 89–98. [Google Scholar] [CrossRef] [PubMed]
- Chung, J.C.; Oh, M.J.; Choi, S.H.; Bae, C.D. Proteomic analysis to identify biomarker proteins in pancreatic ductal adenocarcinoma. ANZ J. Surg. 2008, 78, 245–251. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Zhang, D.; Jia, Q.; Li, T.; Zhang, W.; Han, J. Proteomic and tissue array profiling identifies elevated hypoxia-regulated proteins in pancreatic ductal adenocarcinoma. Cancer Investig. 2009, 27, 747–755. [Google Scholar] [CrossRef] [PubMed]
- Britton, D.; Zen, Y.; Quaglia, A.; Selzer, S.; Mitra, V.; Löbner, C.; Jung, S.; Böhm, G.; Schmid, P.; Prefot, P.; et al. Quantification of pancreatic cancer proteome and phosphorylome: Indicates molecular events likely contributing to cancer and activity of drug targets. PLoS ONE 2014, 9. [Google Scholar] [CrossRef]
- Pan, S.; Chen, R.; Tamura, Y.; Crispin, D.A.; Lai, L.A.; May, D.H.; McIntosh, M.W.; Goodlett, D.R.; Brentnall, T. A Quantitative Glycoproteomics Analysis Reveals Changes in N-Glycosylation Level Associated with Pancreatic Ductal Adenocarcinoma. J. Proteome Res. 2014, 13, 1293–1306. [Google Scholar] [CrossRef]
- Kawahara, T.; Hotta, N.; Ozawa, Y.; Kato, S.; Kano, K.; Yokoyama, Y.; Nagino, M.; Takahashi, T.; Yanagisawa, K. Quantitative proteomic profiling identifies DPYSL3 as pancreatic ductal adenocarcinoma-associated molecule that regulates cell adhesion and migration by stabilization of focal adhesion complex. PLoS ONE 2013, 8, e79654. [Google Scholar] [CrossRef]
- Chen, R.; Pan, S.; Ottenhof, N.A.; de Wilde, R.F.; Wolfgang, C.L.; Lane, Z.; Post, J.; Bronner, M.P.; Willmann, J.K.; Maitra, A.; et al. Stromal galectin-1 expression is associated with long-term survival in resectable pancreatic ductal adenocarcinoma. Cancer Biol. Ther. 2012, 13, 899–907. [Google Scholar] [CrossRef]
- Kojima, K.; Bowersock, G.J.; Kojima, C.; Klug, C.A.; Grizzle, W.E.; Mobley, J.A. Validation of a robust proteomic analysis carried out on formalin-fixed paraffin-embedded tissues of the pancreas obtained from mouse and human. Proteomics 2012, 12, 3393–3402. [Google Scholar] [CrossRef]
- Zhang, G.; He, P.; Tan, H.; Budhu, A.; Gaedcke, J.; Michael Ghadimi, B.; Ried, T.; Yfantis, H.G.; Lee, D.H.; Maitra, A.; et al. Integration of metabolomics and transcriptomics revealed a fatty acid network exerting growth inhibitory effects in human pancreatic cancer. Clin. Cancer Res. 2013, 19, 4983–4993. [Google Scholar] [CrossRef]
- Weeks, M.E.; Hariharan, D.; Petronijevic, L.; Radon, T.P.; Whiteman, H.J.; Kocher, H.M.; Timms, J.F.; Lemoine, N.R.; Crnogorac-Jurcevic, T. Analysis of the urine proteome in patients with pancreatic ductal adenocarcinoma. Proteom. Clin. Appl. 2008, 2, 1047–1057. [Google Scholar] [CrossRef]
- Chen, J.; Anderson, M.; Misek, D.E.; Simeone, D.M.; Lubman, D.M. Characterization of apolipoprotein and apolipoprotein precursors in pancreatic cancer serum samples via two-dimensional liquid chromatography and mass spectrometry. J. Chromatogr. A 2007, 1162, 117–125. [Google Scholar] [CrossRef] [PubMed]
- Jenkinson, C.; Elliott, V.L.; Evans, A.; Oldfield, L.; Jenkins, R.E.; O’brien, D.P.; Apostolidou, S.; Gentry-Maharaj, A.; Fourkala, E.O.; Jacobs, I.J.; et al. Decreased serum thrombospondin-1 levels in pancreatic cancer patients up to 24 months prior to clinical diagnosis: Association with diabetes mellitus. Clin. Cancer Res. 2016, 22, 1734–1743. [Google Scholar] [CrossRef] [PubMed]
- Hocker, J.R.; Postier, R.G.; Li, M.; Lerner, M.R.; Lightfoot, S.A.; Peyton, M.D.; Deb, S.J.; Baker, C.M.; Williams, T.L.; Hanas, R.J.; et al. Discriminating patients with early-stage pancreatic cancer or chronic pancreatitis using serum electrospray mass profiling. Cancer Lett. 2015, 359, 314–324. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Lv, X.; Fang, C.; Lv, X.; Wang, F.; Wang, D.; Zhao, J.; Ma, Y.; Xue, Y.; Bai, Q.; et al. Dysbindin as a novel biomarker for pancreatic ductal adenocarcinoma identified by proteomic profiling. Int. J. Cancer 2016, 139, 1821–1829. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Chen, L.J.; Xia, Y.L.; Zhou, H.C.; Yang, R.B.; Wu, W.; Lu, Y.; Hu, L.W.; Zhao, Y. Identification and verification of transthyretin as a potential biomarker for pancreatic ductal adenocarcinoma. J. Cancer Res. Clin. Oncol. 2013, 139, 1117–1127. [Google Scholar] [CrossRef]
- Sogawa, K.; Takano, S.; Iida, F.; Satoh, M.; Tsuchida, S.; Kawashima, Y.; Yoshitomi, H.; Sanda, A.; Kodera, Y.; Takizawa, H.; et al. Identification of a novel serum biomarker for pancreatic cancer, C4b-binding protein α-chain (C4BPA) by quantitative proteomic analysis using tandem mass tags. Br. J. Cancer 2016, 115, 949–956. [Google Scholar] [CrossRef]
- Radon, T.P.; Massat, N.J.; Jones, R.; Alrawashdeh, W.; Dumartin, L.; Ennis, D.; Duffy, S.W.; Kocher, H.M.; Pereira, S.P.; Guarner, L.; et al. Identification of a three-biomarker panel in urine for early detection of pancreatic adenocarcinoma. Clin. Cancer Res. 2015, 21, 3512–3521. [Google Scholar] [CrossRef]
- Lee, M.J.; Na, K.; Jeong, S.-K.; Lim, J.-S.; Kim, S.A.; Lee, M.-J.; Song, S.Y.; Kim, H.; Hancock, W.S.; Paik, Y.-K. Identification of Human Complement Factor B as a Novel Biomarker Candidate for Pancreatic Ductal Adenocarcinoma. J. Proteome Res. 2014, 13, 4878–4888. [Google Scholar] [CrossRef]
- Chen, K.T.; Kim, P.D.; Jones, K.A.; Devarajan, K.; Patel, B.B.; Hoffman, J.P.; Ehya, H.; Huang, M.; Watson, J.C.; Tokar, J.L.; et al. Potential Prognostic Biomarkers of Pancreatic Cancer. Pancreas 2014, 43, 22–27. [Google Scholar] [CrossRef]
- Chen, J.; Wu, W.; Chen, L.; Zhou, H.; Yang, R.; Hu, L.; Zhao, Y. Profiling the potential tumor markers of pancreatic ductal adenocarcinoma using 2D-DIGE and MALDI-TOF-MS: Up-regulation of Complement C3 and alpha-2-HS-glycoprotein. Pancreatology 2013, 13, 290–297. [Google Scholar] [CrossRef]
- Tian, M.; Cui, Y.-Z.; Song, G.-H.; Zong, M.-J.; Zhou, X.-Y.; Chen, Y.; Han, J.-X. Proteomic analysis identifies MMP-9, DJ-1 and A1BG as overexpressed proteins in pancreatic juice from pancreatic ductal adenocarcinoma patients. BMC Cancer 2008, 8, 241. [Google Scholar] [CrossRef] [PubMed]
- Tomaino, B.; Cappello, P.; Capello, M.; Fredolini, C.; Ponzetto, A.; Novarino, A.; Ciuffreda, L.; Bertetto, O.; De Angelis, C.; Gaia, E.; et al. Autoantibody signiture in human ductal pancreatic adenocarcinoma. J. Proteome Res. 2007, 10, 4025–4031. [Google Scholar] [CrossRef] [PubMed]
- Wehr, A.Y.; Hwang, W.T.; Blair, I.A.; Yu, K.H. Relative quantification of serum proteins from pancreatic ductal adenocarcinoma patients by stable isotope dilution liquid chromatography-mass spectrometry. J. Proteome Res. 2012, 11, 1749–1758. [Google Scholar] [CrossRef] [PubMed]
- Petersen, T.N.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nat. Methods 2011, 8, 785–786. [Google Scholar] [CrossRef]
- Bendtsen, J.D.; Jensen, L.J.; Blom, N.; Von Heijne, G.; Brunak, S. Feature-based prediction of non-classical and leaderless protein secretion. Protein Eng. Des. Sel. 2004, 17, 349–356. [Google Scholar] [CrossRef]
- Kalra, H.; Simpson, R.J.; Ji, H.; Aikawa, E.; Altevogt, P.; Askenase, P.; Bond, V.C.; Borràs, F.E.; Breakefield, X.; Budnik, V.; et al. Vesiclepedia: A Compendium for Extracellular Vesicles with Continuous Community Annotation. PLoS Biol. 2012, 10, e1001450. [Google Scholar] [CrossRef]
- Mathivanan, S.; Fahner, C.J.; Reid, G.E.; Simpson, R.J. ExoCarta 2012: Database of exosomal proteins, RNA and lipids. Nucleic Acids Res. 2012, 40, D1241–D1244. [Google Scholar] [CrossRef]
- Nanjappa, V.; Thomas, J.K.; Marimuthu, A.; Muthusamy, B.; Radhakrishnan, A.; Sharma, R.; Ahmad Khan, A.; Balakrishnan, L.; Sahasrabuddhe, N.A.; Kumar, S.; et al. Plasma Proteome Database as a resource for proteomics research: 2014 update. Nucleic Acids Res. 2014, 42, D959–D965. [Google Scholar] [CrossRef]
- Emanuelsson, O.; Nielsen, H.; Brunak, S.; von Heijne, G. Predicting Subcellular Localization of Proteins Based on their N-terminal Amino Acid Sequence. J. Mol. Biol. 2000, 300, 1005–1016. [Google Scholar] [CrossRef]
- Krogh, A.; Larsson, B.; von Heijne, G.; Sonnhammer, E.L. Predicting transmembrane protein topology with a hidden markov model: Application to complete genomes. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Morris, J.H.; Cook, H.; Kuhn, M.; Wyder, S.; Simonovic, M.; Santos, A.; Doncheva, N.T.; Roth, A.; Bork, P.; et al. The STRING database in 2017: Quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017, 45, D362–D368. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.; Li, C.; Kang, B.; Gao, G.; Li, C.; Zhang, Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017, 45, W98–W102. [Google Scholar] [CrossRef] [PubMed]
- Vivian, J.; Rao, A.A.; Nothaft, F.A.; Ketchum, C.; Armstrong, J.; Novak, A.; Pfeil, J.; Narkizian, J.; Deran, A.D.; Musselman-Brown, A.; et al. Toil enables reproducible, open source, big biomedical data analyses. Nat. Biotechnol. 2017, 35, 314–316. [Google Scholar] [CrossRef] [PubMed]
- Lonsdale, J.; Thomas, J.; Salvatore, M.; Phillips, R.; Lo, E.; Shad, S.; Hasz, R.; Walters, G.; Garcia, F.; Young, N.; et al. The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 2013, 45, 580–585. [Google Scholar] [CrossRef]
- Lowenfels, A.B.; Maisonneuve, P.; DiMagno, E.P.; Elitsur, Y.; Gates, L.K.; Perrault, J.; Whitcomb, D.C. Hereditary pancreatitis and the risk of pancreatic cancer. International Hereditary Pancreatitis Study Group. J. Natl. Cancer Inst. 1997, 89, 442–446. [Google Scholar] [CrossRef]
- Lowenfels, A.B.; Maisonneuve, P.; Cavallini, G.; Ammann, R.W.; Lankisch, P.G.; Andersen, J.R.; Dimagno, E.P.; Andren-Sandberg, A.; Domellof, L. Pancreatitis and the Risk of Pancreatic Cancer. N. Engl. J. Med. 1993, 328, 1433–1437. [Google Scholar] [CrossRef]
- Aguirre-Gamboa, R.; Gomez-Rueda, H.; Martínez-Ledesma, E.; Martínez-Torteya, A.; Chacolla-Huaringa, R.; Rodriguez-Barrientos, A.; Tamez-Peña, J.G.; Treviño, V. SurvExpress: An online biomarker validation tool and database for cancer gene expression data using survival analysis. PLoS ONE 2013, 8, e74250. [Google Scholar] [CrossRef]
- Tomczak, K.; Czerwińska, P.; Wiznerowicz, M. The Cancer Genome Atlas (TCGA ): An immeasurable source of knowledge. Contemp. Oncol. 2014, 19, A68–A77. [Google Scholar] [CrossRef]
- International Cancer Genome Consortium, T.I.C.G.; Hudson, T.J.; Anderson, W.; Artez, A.; Barker, A.D.; Bell, C.; Bernabé, R.R.; Bhan, M.K.; Calvo, F.; Eerola, I.; et al. International network of cancer genome projects. Nature 2010, 464, 993–998. [Google Scholar] [CrossRef]
- Chen, D.T.; Davis-Yadley, A.H.; Huang, P.Y.; Husain, K.; Centeno, B.A.; Permuth-Wey, J.; Pimiento, J.M.; Malafa, M. Prognostic fifteen-gene signature for early stage pancreatic ductal adenocarcinoma. PLoS ONE 2015, 10, e0133562. [Google Scholar] [CrossRef]
- Uhlen, M.; Oksvold, P.; Fagerberg, L.; Lundberg, E.; Jonasson, K.; Forsberg, M.; Zwahlen, M.; Kampf, C.; Wester, K.; Hober, S.; et al. Towards a knowledge-based Human Protein Atlas. Nat. Biotechnol. 2010, 28, 1248–1250. [Google Scholar] [CrossRef] [PubMed]
- Garbis, S.; Lubec, G.; Fountoulakis, M. Limitations of current proteomics technologies. J. Chromatogr. A 2005, 1077, 1–18. [Google Scholar] [CrossRef]
- Reymond, M.A.; Schlegel, W. Proteomics in cancer. Adv. Clin. Chem. 2007, 44, 103–142. [Google Scholar] [PubMed]
- Lim, L.C.; Lim, Y.M. Proteome Heterogeneity in Colorectal Cancer. Proteomics 2018, 18, 1700169. [Google Scholar] [CrossRef] [PubMed]
- Bateman, N.W.; Conrads, T.P. Recent Advances and Opportunities in Proteomic Analyses of Tumor Heterogeneity. J. Pathol. 2018, 244, 628–637. [Google Scholar] [CrossRef]
- Anderson, L.; Seilhamer, J. A comparison of selected mRNA and protein abundances in human liver. Electrophoresis 1997, 18, 533–537. [Google Scholar] [CrossRef]
- Gygi, S.P.; Rochon, Y.; Franza, B.R.; Aebersold, R. Correlation between Protein and mRNA Abundance in Yeast. Mol. Cell. Biol. 1999, 19, 1720–1730. [Google Scholar] [CrossRef]
- Hsiao, Y.C.; Chu, L.J.; Chen, J.T.; Yeh, T.S.; Yu, J.S. Proteomic profiling of the cancer cell secretome: Informing clinical research. Expert Rev. Proteom. 2017, 14, 737–756. [Google Scholar] [CrossRef]
- Konigsbrugge, O.; Posch, F.; Riedl, J.; Reitter, E.-M.; Zielinski, C.; Pabinger, I.; Ay, C. Association Between Decreased Serum Albumin With Risk of Venous Thromboembolism and Mortality in Cancer Patients. Oncologist 2016, 21, 252–257. [Google Scholar] [CrossRef]
- Deng, Q.L.; Dong, S.; Wang, L.; Zhang, C.Y.; Ying, H.F.; Li, Z.S.; Shen, X.H.; Guo, Y.B.; Meng, Z.Q.; Yu, J.M.; et al. Development and Validation of a Nomogram for Predicting Survival in Patients with Advanced Pancreatic Ductal Adenocarcinoma. Sci. Rep. 2017, 7, 11524. [Google Scholar] [CrossRef]
- Menapace, L.A.; Peterson, D.R.; Berry, A.; Sousou, T.; Khorana, A.A. Symptomatic and incidental thromboembolism are both associated with mortality in pancreatic cancer. Thromb. Haemost. 2011, 106, 371–378. [Google Scholar] [CrossRef] [PubMed]
- Kondo, S.; Sasaki, M.; Hosoi, H.; Sakamoto, Y.; Morizane, C.; Ueno, H.; Okusaka, T. Incidence and risk factors for venous thromboembolism in patients with pretreated advanced pancreatic carcinoma. Oncotarget 2018, 9, 16883–16890. [Google Scholar] [CrossRef] [PubMed]
- Follia, L.; Ferrero, G.; Mandili, G.; Beccuti, M.; Giordano, D.; Spadi, R.; Satolli, M.A.; Evangelista, A.; Katayama, H.; Hong, W.; et al. Integrative Analysis of Novel Metabolic Subtypes in Pancreatic Cancer Fosters New Prognostic Biomarkers. Front. Oncol. 2019, 9, 115. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Huang, Z.; Tian, Y.; Lin, B.; He, R.; Wang, H.; Ouyang, P.; Chen, H.; Wu, L. Clinical significance and prognostic value of Triosephosphate isomerase expression in gastric cancer. Medicine 2017, 96, e6865. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Luo, Q.; Long, H.; Hu, Z.; Que, T.; Zhang, X.; Li, Z.; Wang, G.; Yi, L.; Liu, Z.; et al. Alpha-enolase as a potential cancer prognostic marker promotes cell growth, migration, and invasion in glioma. Mol. Cancer 2014, 13, 65. [Google Scholar] [CrossRef]
- Hsiao, K.C.; Shih, N.Y.; Fang, H.L.; Huang, T.S.; Kuo, C.C.; Chu, P.Y.; Hung, Y.M.; Chou, S.W.; Yang, Y.Y.; Chang, G.C.; et al. Surface α-Enolase Promotes Extracellular Matrix Degradation and Tumor Metastasis and Represents a New Therapeutic Target. PLoS ONE 2013, 8, e69354. [Google Scholar] [CrossRef]
- Principe, M.; Ceruti, P.; Shih, N.Y.; Chattaragada, M.S.; Rolla, S.; Conti, L.; Bestagno, M.; Zentilin, L.; Yang, S.H.; Migliorini, P.; et al. Targeting of surface alpha-enolase inhibits the invasiveness of pancreatic cancer cells. Oncotarget 2015, 6, 11098–11113. [Google Scholar] [CrossRef]
- Principe, M.; Borgoni, S.; Cascione, M.; Chattaragada, M.S.; Ferri-Borgogno, S.; Capello, M.; Bulfamante, S.; Chapelle, J.; Di Modugno, F.; Defilippi, P.; et al. Alpha-enolase (ENO1) controls alpha v/beta 3 integrin expression and regulates pancreatic cancer adhesion, invasion, and metastasis. J. Hematol. Oncol. 2017, 10, 16. [Google Scholar] [CrossRef]
- Miles, L.A.; Dahlberg, C.M.; Plescia, J.; Felez, J.; Kato, K.; Plow, E.F. Role of cell-surface lysines in plasminogen binding to cells: Identification of alpha-enolase as a candidate plasminogen receptor. Biochemistry 1991, 30, 1682–1691. [Google Scholar] [CrossRef]
- López-Alemany, R.; Longstaff, C.; Hawley, S.; Mirshahi, M.; Fábregas, P.; Jardí, M.; Merton, E.; Miles, L.A.; Félez, J. Inhibition of cell surface mediated plasminogen activation by a monoclonal antibody against α-Enolase. Am. J. Hematol. 2003, 72, 234–242. [Google Scholar] [CrossRef]
- Legler, D.F.; Johnson-Léger, C.; Wiedle, G.; Bron, C.; Imhof, B.A. The αvβ3 integrin as a tumor homing ligand for lymphocytes. Eur. J. Immunol. 2004, 34, 1608–1616. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Wang, F.; Chen, X. Integrin α v β 3-targeted cancer therapy. Drug Dev. Res. 2008, 69, 329–339. [Google Scholar] [CrossRef] [PubMed]
- Yin, H.; Wang, L.; Liu, H.-L. ENO1 Overexpression in Pancreatic Cancer Patients and Its Clinical and Diagnostic Significance. Gastroenterol. Res. Pract. 2018, 2018, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Ziegler, Y.S.; Moresco, J.J.; Yates, J.R.; Nardulli, A.M. Integration of Breast Cancer Secretomes with Clinical Data Elucidates Potential Serum Markers for Disease Detection, Diagnosis, and Prognosis. PLoS ONE 2016, 11, e0158296. [Google Scholar] [CrossRef] [PubMed]
- Tsai, S.-T.; Chien, I.-H.; Shen, W.-H.; Kuo, Y.-Z.; Jin, Y.-T.; Wong, T.-Y.; Hsiao, J.-R.; Wang, H.-P.; Shih, N.-Y.; Wu, L.-W. ENO1, a potential prognostic head and neck cancer marker, promotes transformation partly via chemokine CCL20 induction. Eur. J. Cancer 2010, 46, 1712–1723. [Google Scholar] [CrossRef] [PubMed]
- Liberti, M.V.; Locasale, J.W.; Biology, C.; Biology, C. Review- The Warburg Effect: How Does it Benefit Cancer Cells ? Trends Biochem. Sci. 2017, 41, 211–218. [Google Scholar] [CrossRef]
- Zhu, W.; Ma, L.; Qian, J.; Xu, J.; Xu, T.; Pang, L.; Zhou, H.; Shu, Y.; Zhou, J. The molecular mechanism and clinical significance of LDHA in HER2-mediated progression of gastric cancer. Am. J. Transl. Res. 2018, 10, 2055–2067. [Google Scholar]
- Rong, Y.; Wu, W.; Ni, X.; Kuang, T.; Jin, D.; Wang, D.; Lou, W. Lactate dehydrogenase A is overexpressed in pancreatic cancer and promotes the growth of pancreatic cancer cells. Tumor Biol. 2013, 34, 1523–1530. [Google Scholar] [CrossRef]
- Liu, Y.; Guo, J.Z.; Liu, Y.; Wang, K.; Ding, W.; Wang, H.; Liu, X.; Zhou, S.; Lu, X.C.; Yang, H.B.; et al. Nuclear lactate dehydrogenase A senses ROS to produce α-hydroxybutyrate for HPV-induced cervical tumor growth. Nat. Commun. 2018, 9, 4429. [Google Scholar] [CrossRef]
- Feng, Y.; Xiong, Y.; Qiao, T.; Li, X.; Jia, L.; Han, Y. Lactate dehydrogenase A: A key player in carcinogenesis and potential target in cancer therapy. Cancer Med. 2018, 7, 6124–6136. [Google Scholar] [CrossRef]
- Yu, X.; Li, S. Non-metabolic functions of glycolytic enzymes in tumorigenesis. Oncogene 2017, 36, 2629–2636. [Google Scholar] [CrossRef] [PubMed]
- Steeg, P.S.; Kopper, L.; Thorgeirsson, U.P.; Talmadge, E.; Liotta, L.A.; Sobep, M.E. Evidence for a Novel Gene Associated With Low Tumor Metastatic Potential. J. Natl. Cancer Inst. 1988, 80, 200–204. [Google Scholar] [CrossRef] [PubMed]
- Ohshio, G.; Imamura, T.; Okada, N.; Suwa, H.; Yamaki, K.; Imamura, M.; Ogasahara, K.; Tsukayama, C.; Yamabe, H. Immunohistochemical expression of nm23 gene product, nucleotide diphosphate kinase, in pancreatic neoplasms. Int. J. Gastrointest. Cancer 2008, 22, 59–66. [Google Scholar] [CrossRef] [PubMed]
- Nakamori, S.; Ishikawa, O.; Ohigashi, H.; Imaoka, S.; Sasaki, Y.; Kameyama, M.; Kabuto, T.; Furukawa, H.; Iwanakga, T.; Kimura, N. Clinicopathological features and prognostic significance of nucleoside diphosphate kinase/nm23 gene product in human pancreatic exocrine neoplasms. Int. J. Pancreatol. 1993, 14, 125–133. [Google Scholar]
- Takadate, T.; Onogawa, T.; Fujii, K.; Motoi, F.; Mikami, S.; Fukuda, T.; Kihara, M.; Suzuki, T.; Takemura, T.; Minowa, T.; et al. NM23/nucleoside diphosphate kinase-A as a potent prognostic marker in invasive pancreatic ductal carcinoma identified by proteomic analysis of laser micro-dissected formalin-fixed paraffin-embedded tissue. Clin. Proteom. 2012, 9, 8. [Google Scholar] [CrossRef]
- Liu, L.; Li, M.; Zhang, C.; Zhang, J.; Li, G.; Zhang, Z.; He, X.; Fan, M. Prognostic value and clinicopathologic significance of nm23 in various cancers: A systematic review and meta-analysis. Int. J. Surg. 2018, 60, 257–265. [Google Scholar] [CrossRef]
- Royds, J.A.; Cross, S.S.; Silcocks, P.B.; Scholefield, J.H.; Rees, R.C.; Stephenson, T.J. Nm23 ‘anti-metastatic’ gene product expression in colorectal carcinoma. J. Pathol. 1994, 172, 261–266. [Google Scholar] [CrossRef]
- Orozco, C.A.; Martinez-Bosch, N.; Guerrero, P.E.; Vinaixa, J.; Dalotto-Moreno, T.; Iglesias, M.; Moreno, M.; Djurec, M.; Poirier, F.; Gabius, H.-J.; et al. Targeting galectin-1 inhibits pancreatic cancer progression by modulating tumor–stroma crosstalk. Proc. Natl. Acad. Sci. USA 2018, 115, E3769–E3778. [Google Scholar] [CrossRef]
- Zhang, P.F.; Li, K.S.; Shen, Y.H.; Gao, P.T.; Dong, Z.R.; Cai, J.B.; Zhang, C.; Huang, X.Y.; Tian, M.X.; Hu, Z.Q.; et al. Galectin-1 induces hepatocellular carcinoma EMT and sorafenib resistance by activating FAK/PI3K/AKT signaling. Cell Death Dis. 2016, 7, e2201. [Google Scholar] [CrossRef]
- Van Woensel, M.; Mathivet, T.; Wauthoz, N.; Rosière, R.; Garg, A.D.; Agostinis, P.; Mathieu, V.; Kiss, R.; Lefranc, F.; Boon, L.; et al. Sensitization of glioblastoma tumor micro-environment to chemo- and immunotherapy by Galectin-1 intranasal knock-down strategy. Sci. Rep. 2017, 7, 1217. [Google Scholar] [CrossRef]
- Yeh, C.-C.; Hsu, C.-H.; Shao, Y.-Y.; Ho, W.-C.; Tsai, M.-H.; Feng, W.-C.; Chow, L.-P. Integrated Stable Isotope Labeling by Amino Acids in Cell Culture (SILAC) and Isobaric Tags for Relative and Absolute Quantitation (iTRAQ) Quantitative Proteomic Analysis Identifies Galectin-1 as a Potential Biomarker for Predicting Sorafenib Resistance i. Mol. Cell. Proteom. 2015, 14, 1527–1545. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.C.; Davuluri, G.V.N.; Chen, C.H.; Shiau, D.C.; Chen, C.C.; Chen, C.L.; Lin, Y.S.; Chang, C.P. Galectin-1-induced autophagy facilitates cisplatin resistance of hepatocellular carcinoma. PLoS ONE 2016, 11, e0148408. [Google Scholar] [CrossRef] [PubMed]
- Chung, L.Y.; Tang, S.J.; Sun, G.H.; Chou, T.Y.; Yeh, T.S.; Yu, S.L.; Sun, K.H. Galectin-1 promotes lung cancer progression and chemoresistance by upregulating p38 MAPK, ERK, and cyclooxygenase-2. Clin. Cancer Res. 2012, 18, 4037–4047. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Zhang, P.; Shi, B.; Zhou, M.; Jiang, H.; Zhang, H.; Pan, X.; Gao, H.; Sun, H.; Li, Z. Galectin-1 overexpression promotes progression and chemoresistance to cisplatin in epithelial ovarian cancer. Cell Death Dis. 2014, 5, e991. [Google Scholar] [CrossRef]
- Mathieu, V.; Le Mercier, M.; De Neve, N.; Sauvage, S.; Gras, T.; Roland, I.; Lefranc, F.; Kiss, R. Galectin-1 knockdown increases sensitivity to temozolomide in a B16F10 mouse metastatic melanoma model. J. Investig. Dermatol. 2007, 127, 2399–2410. [Google Scholar] [CrossRef]
- Lykken, J.M.; Horikawa, M.; Minard-Colin, V.; Kamata, M.; Miyagaki, T.; Poe, J.C.; Tedder, T.F. Galectin-1 drives lymphoma CD20 immunotherapy resistance: Validation of a preclinical system to identify resistance mechanisms. Blood 2016, 127, 1886–1895. [Google Scholar] [CrossRef]
- Cui, G.; Cui, M.; Li, Y.; Liang, Y.; Li, W.; Guo, H.; Zhao, S. Galectin-3 knockdown increases gefitinib sensitivity to the inhibition of EGFR endocytosis in gefitinib-insensitive esophageal squamous cancer cells. Med. Oncol. 2015, 32, 124. [Google Scholar] [CrossRef]
- Mirandola, L.; Yu, Y.; Cannon, M.J.; Jenkins, M.R.; Rahman, R.L.; Nguyen, D.D.; Grizzi, F.; Cobos, E.; Figueroa, J.A.; Chiriva-Internati, M. Galectin-3 inhibition suppresses drug resistance, motility, invasion and angiogenic potential in ovarian cancer. Gynecol. Oncol. 2014, 135, 573–579. [Google Scholar] [CrossRef]
- Streetly, M.J.; Maharaj, L.; Joel, S.; Schey, S.A.; Gribben, J.G.; Cotter, F.E. GCS-100, a novel galectin-3 antagonist, modulates MCL-1, NOXA, and cell cycle to induce myeloma cell death. Blood 2010, 115, 3939–3948. [Google Scholar] [CrossRef]
- Mazurek, N.; Byrd, J.C.; Sun, Y.; Hafley, M.; Ramirez, K.; Burks, J.; Bresalier, R.S. Cell-surface galectin-3 confers resistance to TRAIL by impeding trafficking of death receptors in metastatic colon adenocarcinoma cells. Cell Death Differ. 2012, 19, 523–533. [Google Scholar] [CrossRef]
- Kyu, J.C.; Yu, J.P.; Min, J.L.; Jin, H.K.; Ha, J.; Choe, W.; Sung, S.K. Overexpressed cyclophilin A in cancer cells renders resistance to hypoxia- and cisplatin-induced cell death. Cancer Res. 2007, 67, 3654–3662. [Google Scholar]
- Zhu, X.; Miao, X.; Wu, Y.; Li, C.; Guo, Y.; Liu, Y.; Chen, Y.; Lu, X.; Wang, Y.; He, S. ENO1 promotes tumor proliferation and cell adhesion mediated drug resistance (CAM-DR) in Non-Hodgkin’s Lymphomas. Exp. Cell Res. 2015, 335, 216–223. [Google Scholar] [CrossRef]
- Maiso, P.; Huynh, D.; Moschetta, M.; Sacco, A.; Aljawai, Y.; Mishima, Y.; Asara, J.M.; Roccaro, A.M.; Kimmelman, A.C.; Ghobrial, I.M. Metabolic Signature Identifies Novel Targets for Drug Resistance in Multiple Myeloma. Cancer Res. 2015, 75, 2071–2082. [Google Scholar] [CrossRef] [PubMed]
- Zeng, H.Z.; Qu, Y.Q.; Zhang, W.J.; Xiu, B.; Deng, A.M.; Liang, A. Bin Proteomic analysis identified DJ-1 as a cisplatin resistant marker in non-small cell lung cancer. Int. J. Mol. Sci. 2011, 12, 3489–3499. [Google Scholar] [CrossRef] [PubMed]
- Sagulenko, V.; Muth, D.; Sagulenko, E.; Paffhausen, T.; Schwab, M.; Westermann, F. Cathepsin D protects human neuroblastoma cells from doxorubicin-induced cell death. Carcinogenesis 2008, 29, 1869–1877. [Google Scholar] [CrossRef]
- Bai, D.; Ueno, L.; Vogt, P. Akt-mediated regulation of NFkB for the oncogenicity of PI3K and Akt. Int. J. Cancer 2009, 125, 2863–2870. [Google Scholar] [CrossRef]
- Fresno Vara, J.Á.; Casado, E.; de Castro, J.; Cejas, P.; Belda-Iniesta, C.; González-Barón, M. P13K/Akt signalling pathway and cancer. Cancer Treat. Rev. 2004, 30, 193–204. [Google Scholar] [CrossRef]
- Gry, M.; Rimini, R.; Strömberg, S.; Asplund, A.; Pontén, F.; Uhlén, M.; Nilsson, P. Correlations between RNA and protein expression profiles in 23 human cell lines. BMC Genom. 2009, 10, 365. [Google Scholar] [CrossRef]
- Pillai, R.S.; Bhattacharyya, S.N.; Filipowicz, W. Repression of protein synthesis by miRNAs: How many mechanisms? Trends Cell Biol. 2007, 17, 118–126. [Google Scholar] [CrossRef]
- Hausser, J.; Zavolan, M. Identification and consequences of miRNA-target interactions-beyond repression of gene expression. Nat. Rev. Genet. 2014, 15, 599–612. [Google Scholar] [CrossRef]
- Raphael, B.J.; Hruban, R.H.; Aguirre, A.J.; Moffitt, R.A.; Yeh, J.J.; Stewart, C.; Robertson, A.G.; Cherniack, A.D.; Gupta, M.; Getz, G.; et al. Integrated Genomic Characterization of Pancreatic Ductal Adenocarcinoma. Cancer Cell 2017, 32, 185–203. [Google Scholar] [CrossRef] [PubMed]
- Felix, T.F.; Lopez Lapa, R.M.; De Carvalho, M.; Bertoni, N.; Tokar, T.; Oliveira, R.A.; Rodrigues, M.A.M.; Hasimoto, C.N.; Oliveira, W.K.; Pelafsky, L.; et al. MicroRNA modulated networks of adaptive and innate immune response in pancreatic ductal adenocarcinoma. PLoS ONE 2019, 14, e0217421. [Google Scholar] [CrossRef] [PubMed]
- Slater, E.P.; Fendrich, V.; Strauch, K.; Rospleszcz, S.; Ramaswamy, A.; Matthäi, E.; Chaloupka, B.; Gress, T.M.; Langer, P.; Bartsch, D.K. LCN2 and TIMP1 as potential serum markers for the early detection of familial pancreatic cancer. Transl. Oncol. 2013, 6, 99–103. [Google Scholar] [CrossRef] [PubMed]
- Bartsch, D.; Gercke, N.; Strauch, K.; Wieboldt, R.; Matthäi, E.; Wagner, V.; Rospleszcz, S.; Schäfer, A.; Franke, F.; Mintziras, I.; et al. The Combination of MiRNA-196b, LCN2, and TIMP1 is a Potential Set of Circulating Biomarkers for Screening Individuals at Risk for Familial Pancreatic Cancer. J. Clin. Med. 2018, 7, 295. [Google Scholar] [CrossRef]
- Yu, S.; Li, Y.; Liao, Z.; Wang, Z.; Wang, Z.; Li, Y.; Qian, L.; Zhao, J.; Zong, H.; Kang, B.; et al. Plasma extracellular vesicle long RNA profiling identifies a diagnostic signature for the detection of pancreatic ductal adenocarcinoma. Gut 2020, 69, 540–550. [Google Scholar] [CrossRef]
- Xu, Q.; Li, P.; Chen, X.; Zong, L.; Jiang, Z.; Nan, L.; Lei, J.; Duan, W.; Zhang, D.; Li, X.; et al. MiR-221/222 induces pancreatic cancer progression through the regulation of matrix metalloproteinases. Oncotarget 2015, 6, 14153–14164. [Google Scholar] [CrossRef]
- Moriyama, T.; Ohuchida, K.; Mizumoto, K.; Yu, J.; Sato, N.; Nabae, T.; Takahata, S.; Toma, H.; Nagai, E.; Tanaka, M. MicroRNA-21 modulates biological functions of pancreatic cancer cells including their proliferation, invasion, and chemoresistance. Mol. Cancer Ther. 2009, 8, 1067–1074. [Google Scholar] [CrossRef]
- Freire, P.P.; Fernandez, G.J.; Moraes, D.D.; Cury, S.S.; Pai-Silva, M.D.; Pintor, P.; Rogatto, S.R.; Carvalho, R.F. The expression landscape of cachexia-inducing factors in human cancers. J. Cachexia Sarcopenia Muscle 2020. [Google Scholar] [CrossRef]
- Robinson, J.L.; Feizi, A.; Uhlén, M.; Nielsen, J. A Systematic Investigation of the Malignant Functions and Diagnostic Potential of the Cancer Secretome. Cell Rep. 2019, 2622–2635. [Google Scholar] [CrossRef]
- Yuan, F.; Zhang, Y.H.; Wan, S.; Wang, S.; Kong, X.Y. Mining for candidate genes related to pancreatic cancer using protein-protein interactions and a shortest path approach. Biomed Res. Int. 2015, 2015, 623121. [Google Scholar] [CrossRef]
- Vareed, S.K.; Bhat, V.B.; Thompson, C.; Vasu, V.T.; Fermin, D.; Choi, H.; Creighton, C.J.; Gayatri, S.; Lan, L.; Putluri, N.; et al. Metabolites of purine nucleoside Phosphorylase (NP) in serum have the potential to delineate Pancreatic Adenocarcinoma. PLoS ONE 2011, 6, e17177. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Zheng, Z.; Li, J.; Ben, Q.; Liu, J.; Zhang, J.; Ji, J.; Yu, B.; Chen, X.; Su, L.; et al. DJ-1 promotes invasion and metastasis of pancreatic cancer cells by activating SRC/ERK/uPA. Carcinogenesis 2012, 33, 555–562. [Google Scholar] [CrossRef] [PubMed]
- Maurer, H.C.; Holmstrom, S.R.; He, J.; Laise, P.; Su, T.; Ahmed, A.; Hibshoosh, H.; Chabot, J.A.; Oberstein, P.E.; Sepulveda, A.R.; et al. Experimental microdissection enables functional harmonisation of pancreatic cancer subtypes. Gut 2019, 68, 1034–1043. [Google Scholar] [CrossRef] [PubMed]
- Liberati, A.; Altman, D.G.; Tetzlaff, J.; Mulrow, C.; Gøtzsche, P.C.; Ioannidis, J.P.A.; Clarke, M.; Devereaux, P.J.; Kleijnen, J.; Moher, D. The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate health care interventions: Explanation and elaboration. Ann. Intern. Med. 2009, 151, W65–W94. [Google Scholar] [CrossRef]
- Metsalu, T.; Vilo, J. ClustVis: A web tool for visualizing clustering of multivariate data using Principal Component Analysis and heatmap. Nucleic Acids Res. 2015, 43, W566–W570. [Google Scholar] [CrossRef]
- Starruß, J.; De Back, W.; Brusch, L.; Deutsch, A. Morpheus: A user-friendly modeling environment for multiscale and multicellular systems biology. Bioinformatics 2014, 30, 1331–1332. [Google Scholar] [CrossRef]


| Ref. | Cell Line /Tumor Tissue | Labeling | Technique | Validation Method |
|---|---|---|---|---|
| [37] | Panc1 | Silac | LC-MS/MS | WB, IHC, MA |
| [43] | AsPC1, MiaPaCa2, Panc1 | Free | LC-MS/MS | - |
| [41] | Panc1 | iTRAQ | LC-MS/MS | WB, ELISA |
| [45] | Paca44, Panc1, BxPc3, MiaPaca2, HPSC, A818-4 | Free | LC-MS/MS | - |
| [46] | BxPC-3, MIA PaCa-2, Panc1, AsPC-1 | Free | LC-MS/MS | WB, IHC, ELISA |
| [93] | CAPAN-2, RLT-PSC | Silac | LC-MRM/MS | - |
| [47] | PC-1.0, PC-1 (Hamster) | Silac | Nano-RPLC-MS/MS | WB |
| [49] | BxPC3, MiaPaca2, Panc1 | Free | ESI-MS/MS | WB |
| [50] | SOJ-6, BxPC-3, MiaPaCa-2, Panc-1 | Free | MALDI-TOF MS | WB |
| [34] | PAN02 (Mouse) | Free | MS/MS | ELISA |
| [51] | NIT-1 | Free | MS/MS | WB |
| [52] | Panc-1 | Free | LC-MS/MS | - |
| [53] | MiaPaCa-2, BxPc-3, Panc-1, AsPc-1 | Free | MALDI–TOF MS | WB |
| [54] | Adenocarcinoma tissue | Free | LC-MS/MS | WB |
| [55] | BON-1, NCI-H727, SHP-77 | Free | LC-MS/MS | WB |
| [3] | BxPc3, MIA-PaCa2, Panc1, CAPAN1, CFPAC1, SU.86.86, HPDE, PJ | Free | LC-MS/MS | ELISA |
| [56] | KLM, PK-59, MIAPaCa2 | Free | MS/MS | WB |
| [57] | Panc-1, SW1990 | iTRAQ | LC-MS/MS | - |
| [58] | MIA PaCa-2 | labeling | MALDI-TOF MS | WB |
| [59] | CAPAN-2 | Silac | LC-MS/MS | IHC |
| Ref. | Sample | Labeling | Technique | Validation Method |
|---|---|---|---|---|
| [6] | PTT | CD | nanoLC-ESI-MS/MS | IHC |
| [60] | PTT | CD | MALDI-TOF MS, nanoLC-ESI MS/MS | - |
| [61] | PTT | Free | MALDI-TOF/TOF MS, MS/MS | IHC |
| [62] | PTT | Free | MS/MS | WB |
| [63] | PTT | Free | MS/MS | WB, IHC |
| [64] | PTT | Free | LC-MS/MS | ELISA |
| [65] | PTT | Free | LC-MS/MS | ELISA |
| [66] | PTT | Free | MS/MS | - |
| [67] | PTT | Free | LC-MS/MS | IHC |
| [68] | PTT | Free | MALDI-TOF MS | ELISA |
| [69] | PTT | Free | LC-MS/MS | IHC |
| [70] | PTT | Free | MALDI-TOF MS | IHC, WB |
| [71] | PTT | CD | MALDI-TOF/TOF-MS | WB, IHC |
| [72] | PTT | Free | MALDI-TOF MS | WB |
| [73] | PTT | Free | MALDI-TOF/TOF-MS | - |
| [29] | PTT | Free | LC-MS/MS | IHC, WB |
| [74] | PTT | Free | LC-MS/MS | - |
| [75] | PTT | SI | LC-MS/MS | - |
| [76] | PTT | Free | LC-MS/MS | IHC, WB |
| [77] | PTT | LP | MS/MS | - |
| [78] | PTT | Free | LC-MS(MS)2 | - |
| [79] | PTT | Free | UHPLC/MS/MS2 | - |
| [80] | Urine | CD | MALDI-TOF MS | IB |
| [81] | Serum | Free | MALDI-QIT-TOF-MS | |
| [82] | Serum | Free | (MRM) | WB |
| [83] | Serum | Free | MS/MS | - |
| [84] | Blood | Free | MALDI-MS | ELISA |
| [85] | Serum | CD | MALDI-TOF/ TOF–MS | ELISA, WB |
| [86] | Serum | IL | LC-MS / MS | - |
| [87] | Urine | Free | LC/MS /MS | - |
| [88] | Plasma | Free | LC−MS/MS | ELISA, WB, IHC, RT-qPCR |
| [89] | Free | GeLC-MS/MS | - | |
| [90] | Serum | Free | MALDI-TOF MS | PCR, WB, IHC |
| [91] | PJ | Free | MALDI-TOF MS MS/MS | WB, IHC, ELISA |
| [92] | Serum | Free | MALDI-TOF | WB |
| Uniprot Access | Gene Symbol | Description |
|---|---|---|
| ALBU_HUMAN | ALB | Serum albumin |
| ENOA_HUMAN | ENO1 | Alpha-enolase |
| FINC_HUMAN | FN1 | Fibronectin |
| TRFE_HUMAN | TF | Serotransferrin |
| LEG1_HUMAN | LGALS1 | Galectin-1 |
| APOE_HUMAN | APOE | Apolipoprotein E |
| CATD_HUMAN | CTSD | Cathepsin D |
| TPIS_HUMAN | TPI1 | Triosephosphate isomerase |
| GSTP1_HUMAN | GSTP1 | Glutathione S-transferase P |
| PARK7_HUMAN | PARK7 | Protein/nucleic acid deglycase DJ-1 |
| TRY1_HUMAN | PRSS1 | Trypsin-1 |
| MOES_HUMAN | MSN | Moesin |
| PGK1_HUMAN | PGK1 | Phosphoglycerate kinase 1 |
| ANXA5_HUMAN | ANXA5 | Annexin A5 |
| PPIA_HUMAN | PPIA | Peptidyl-prolyl cis-trans isomerase A |
| KPYM_HUMAN | PKM | Pyruvate kinase PKM |
| EF1A1_HUMAN | EEF1A1 | Elongation factor 1-alpha 1 |
| TSP1_HUMAN | THBS1 | Thrombospondin-1 |
| GELS_HUMAN | GSN | Gelsolin |
| LEG3_HUMAN | LGALS3 | Galectin-3 |
| TIMP1_HUMAN | TIMP1 | Metalloproteinase inhibitor 1 |
| COF1_HUMAN | CFL1 | Cofilin-1 |
| FLNA_HUMAN | FLNA | Filamin-A |
| LG3BP_HUMAN | LGALS3BP | Galectin-3-binding protein |
| CALR_HUMAN | CALR | Calreticulin |
| CLIC1_HUMAN | CLIC1 | Nuclear chloride ion channel protein |
| TAGL2_HUMAN | TAGLN2 | Transgelin-2 |
| LDHA_HUMAN | LDHA | L-lactate dehydrogenase A chain |
| NDKA_HUMAN | NME1 | Nucleoside diphosphate kinase A |
| TKT_HUMAN | TKT | Transketolase |
| 1433S_HUMAN | SFN | 14-3-3 protein sigma |
| ALDOA_HUMAN | ALDOA | Fructose-bisphosphate aldolase A |
| ENOG_HUMAN | ENO2 | Gamma-enolase |
| PGAM1_HUMAN | PGAM1 | Phosphoglycerate mutase 1 |
| GDIR1_HUMAN | ARHGDIA | Rho GDP-dissociation inhibitor 1 |
| ACTB_HUMAN | ACTB | Actin, cytoplasmic 1 |
| PDIA1_HUMAN | P4HB | Protein disulfide-isomerase |
| ACTS_HUMAN | ACTA1 | Actin, alpha 1, skeletal muscle |
| FETUA_HUMAN | AHSG | Alpha-2-HS-glycoprotein |
| Cellular Component (GO) | |||
| Pathway ID | Pathway Description | Gene Count | FDR |
| GO.0070062 | Extracellular exosome | 35 | 1.29 × 1024 |
| GO.0031988 | Membrane-bounded vesicle | 34 | 4.57 × 1020 |
| GO.0005615 | Extracellular space | 25 | 1.79 × 1019 |
| GO.0044421 | Extracellular region part | 31 | 3.35 × 1015 |
| GO.0072562 | Blood microparticle | 11 | 1.15 × 1014 |
| GO.0005576 | Extracellular region | 31 | 4.02 × 1013 |
| GO.0060205 | Cytoplasmic membrane-bounded vesicle lumen | 8 | 4.29 × 1010 |
| GO.0034774 | Secretory granule lumen | 6 | 2.35 × 107 |
| GO.0005925 | Focal adhesion | 9 | 1.88 × 106 |
| GO.0031093 | Platelet alpha granule lumen | 5 | 2.83 × 106 |
| GO.0031091 | Platelet alpha granule | 5 | 7.61 × 106 |
| GO.0044433 | Cytoplasmic vesicle part | 9 | 2.46 × 105 |
| GO.0005829 | Cytosol | 19 | 3.4 × 105 |
| GO.0030141 | Secretory granule | 7 | 7.44 × 105 |
| Biological Process (GO) | |||
| Pathway ID | Pathway Description | Gene Count | FDR |
| GO.0006165 | Nucleoside diphosphate phosphorylation | 9 | 1.99 × 1011 |
| GO.0006096 | Glycolytic process | 8 | 3.4 × 1011 |
| GO.0002576 | Platelet degranulation | 9 | 5.25 × 1011 |
| GO.0061621 | Canonical glycolysis | 7 | 5.25 × 1011 |
| GO.0019674 | NAD metabolic process | 8 | 8.75 × 1011 |
| GO.0046496 | Nicotinamide nucleotide metabolic process | 8 | 1.47 × 109 |
| GO.0030168 | Platelet activation | 10 | 5.73 × 109 |
| GO.0046364 | Monosaccharide biosynthetic process | 7 | 5.73 × 109 |
| GO.0009611 | Response to wounding | 14 | 9.81 × 109 |
| GO.0006090 | Pyruvate metabolic process | 7 | 1.09 × 108 |
| GO.0042981 | Regulation of apoptotic process | 17 | 1.51 × 108 |
| GO.0042060 | Wound healing | 13 | 3.84 × 108 |
| GO.0006094 | Gluconeogenesis | 6 | 8.46 × 108 |
| GO.0016192 | Vesicle-mediated transport | 15 | 1.23 × 107 |
| GO.0006950 | Response to stress | 23 | 1.39 × 107 |
| Molecular Function (GO) | |||
| Pathway ID | Pathway Description | Gene Count | FDR |
| GO.0005515 | Protein binding | 24 | 0.000171 |
| GO.0019899 | Enzyme binding | 12 | 0.00461 |
| GO.0044822 | Poly(A) RNA binding | 11 | 0.0069 |
| GO.0003723 | RNA binding | 12 | 0.0117 |
| GO.0004634 | Phosphopyruvate hydratase activity | 2 | 0.0174 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
de Oliveira, G.; Paccielli Freire, P.; Santiloni Cury, S.; de Moraes, D.; Santos Oliveira, J.; Dal-Pai-Silva, M.; Reis, P.P.d.; Francisco Carvalho, R. An Integrated Meta-Analysis of Secretome and Proteome Identify Potential Biomarkers of Pancreatic Ductal Adenocarcinoma. Cancers 2020, 12, 716. https://doi.org/10.3390/cancers12030716
de Oliveira G, Paccielli Freire P, Santiloni Cury S, de Moraes D, Santos Oliveira J, Dal-Pai-Silva M, Reis PPd, Francisco Carvalho R. An Integrated Meta-Analysis of Secretome and Proteome Identify Potential Biomarkers of Pancreatic Ductal Adenocarcinoma. Cancers. 2020; 12(3):716. https://doi.org/10.3390/cancers12030716
Chicago/Turabian Stylede Oliveira, Grasieli, Paula Paccielli Freire, Sarah Santiloni Cury, Diogo de Moraes, Jakeline Santos Oliveira, Maeli Dal-Pai-Silva, Patrícia Pintor do Reis, and Robson Francisco Carvalho. 2020. "An Integrated Meta-Analysis of Secretome and Proteome Identify Potential Biomarkers of Pancreatic Ductal Adenocarcinoma" Cancers 12, no. 3: 716. https://doi.org/10.3390/cancers12030716
APA Stylede Oliveira, G., Paccielli Freire, P., Santiloni Cury, S., de Moraes, D., Santos Oliveira, J., Dal-Pai-Silva, M., Reis, P. P. d., & Francisco Carvalho, R. (2020). An Integrated Meta-Analysis of Secretome and Proteome Identify Potential Biomarkers of Pancreatic Ductal Adenocarcinoma. Cancers, 12(3), 716. https://doi.org/10.3390/cancers12030716








