Comparative Molecular Analysis of Cancer Behavior Cultured In Vitro, In Vivo, and Ex Vivo
Abstract
1. Introduction
2. Results
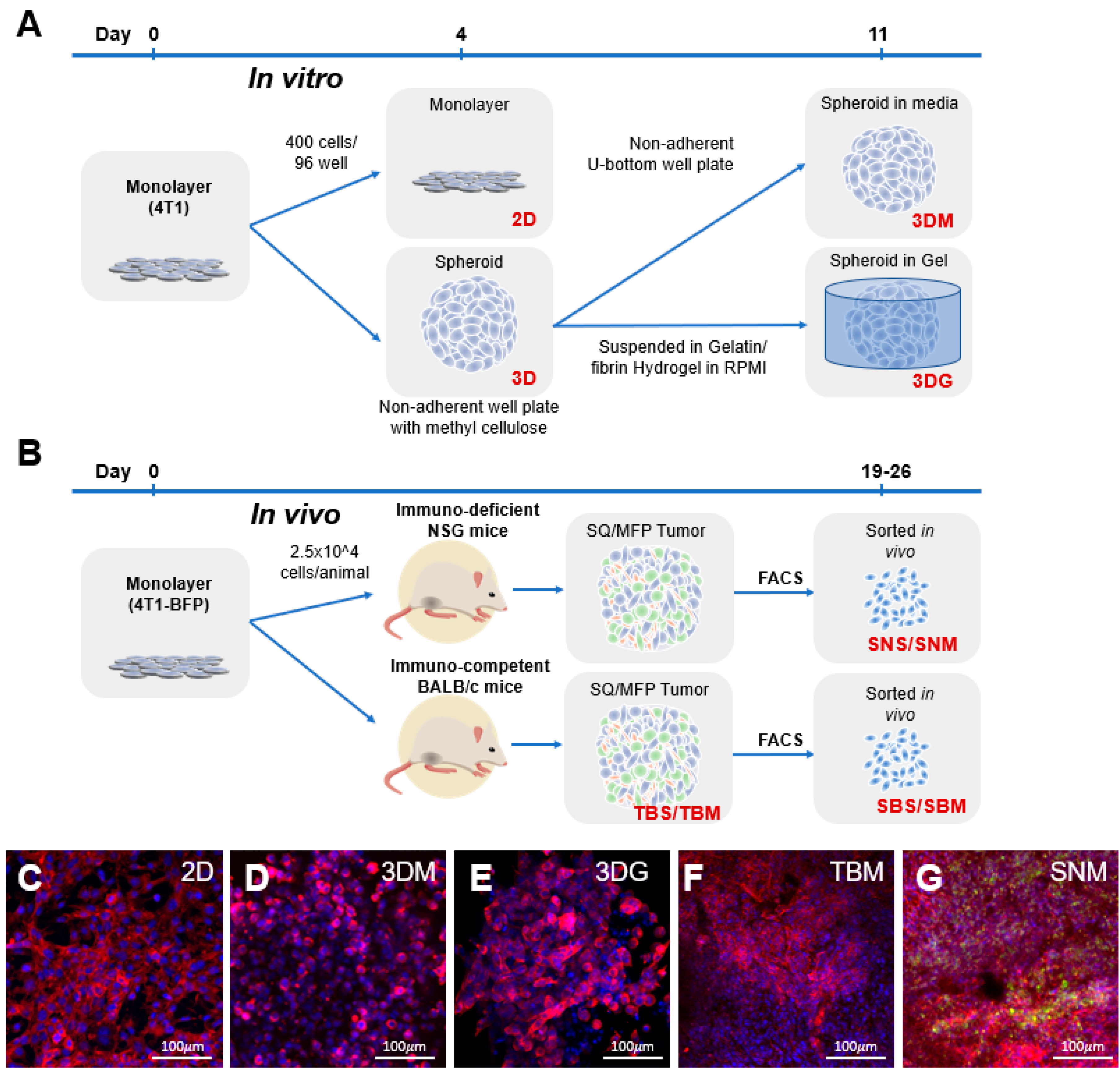
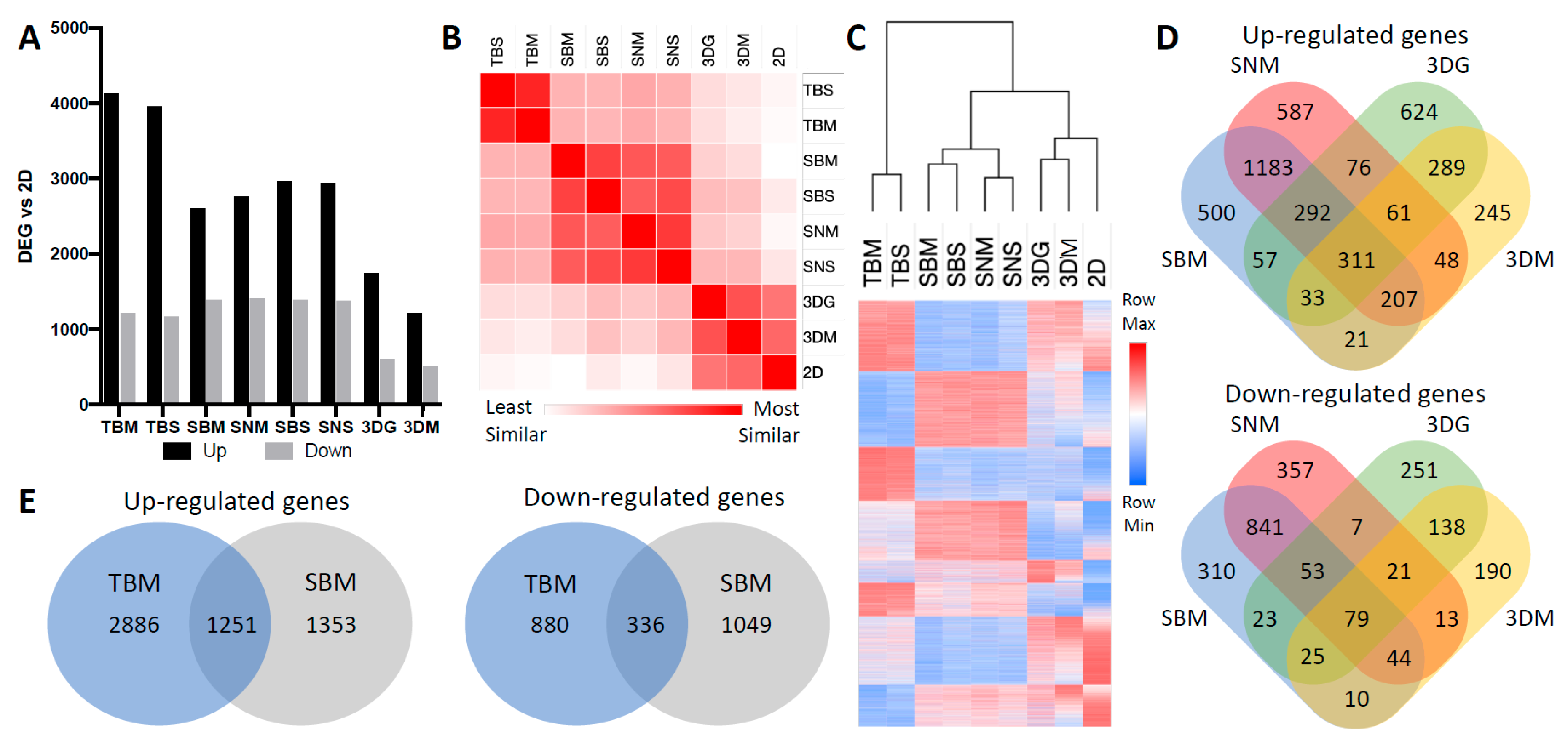
2.1. Cancer Cell Transcriptome is Dictated by Culture Conditions
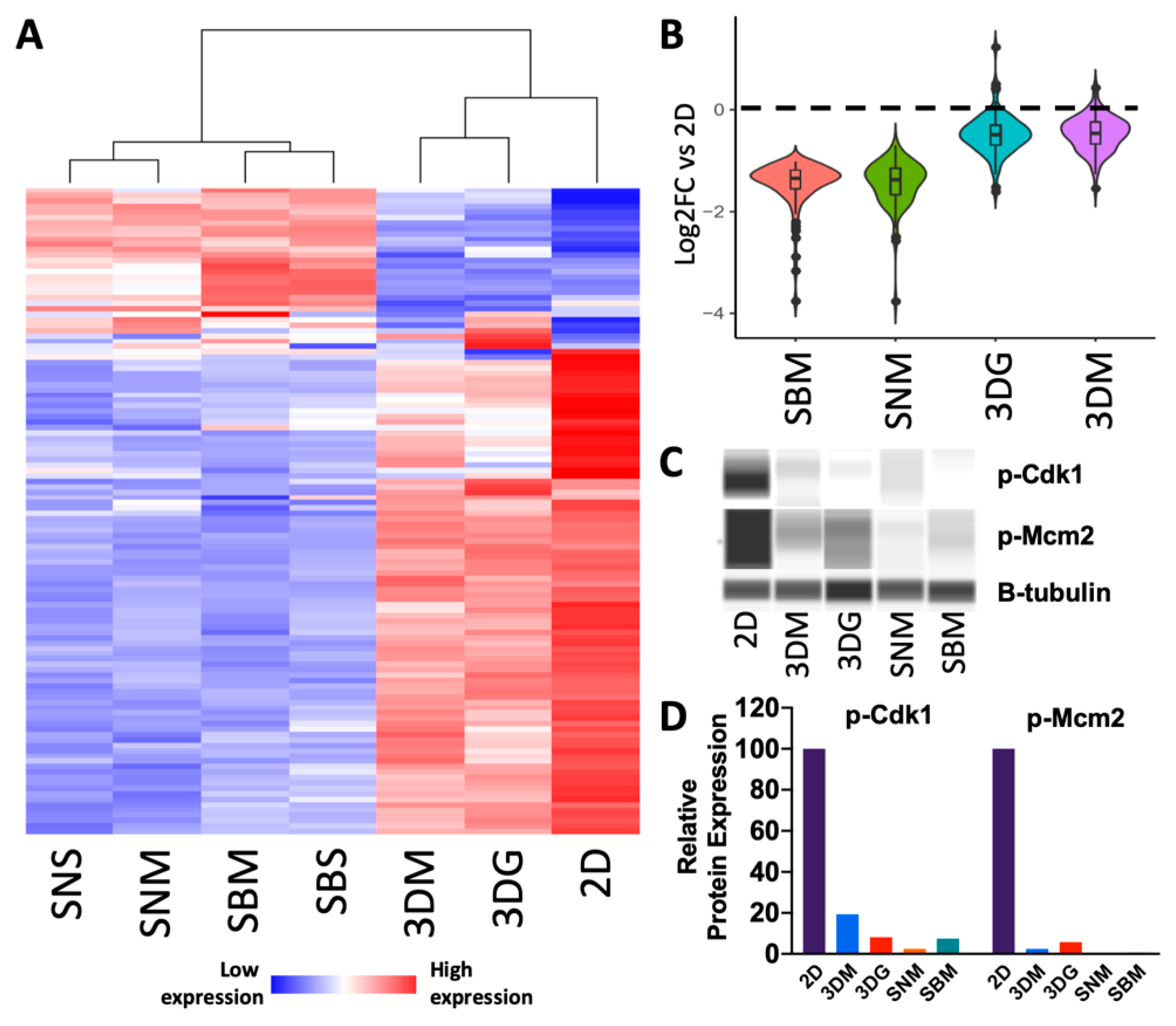
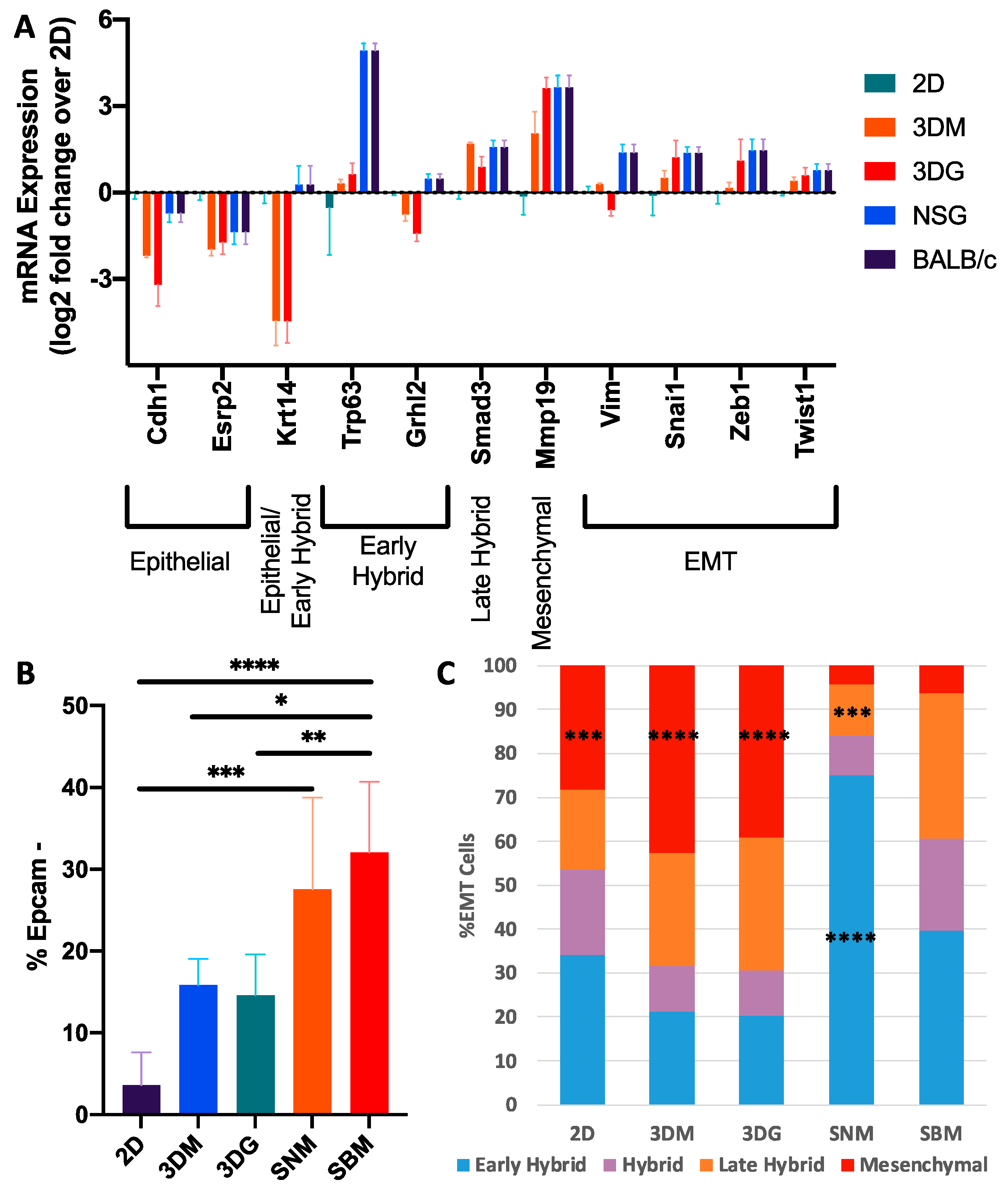
2.2. Culturing Condition Affects Cancer Cell Behavior Critical to Cancer Progression
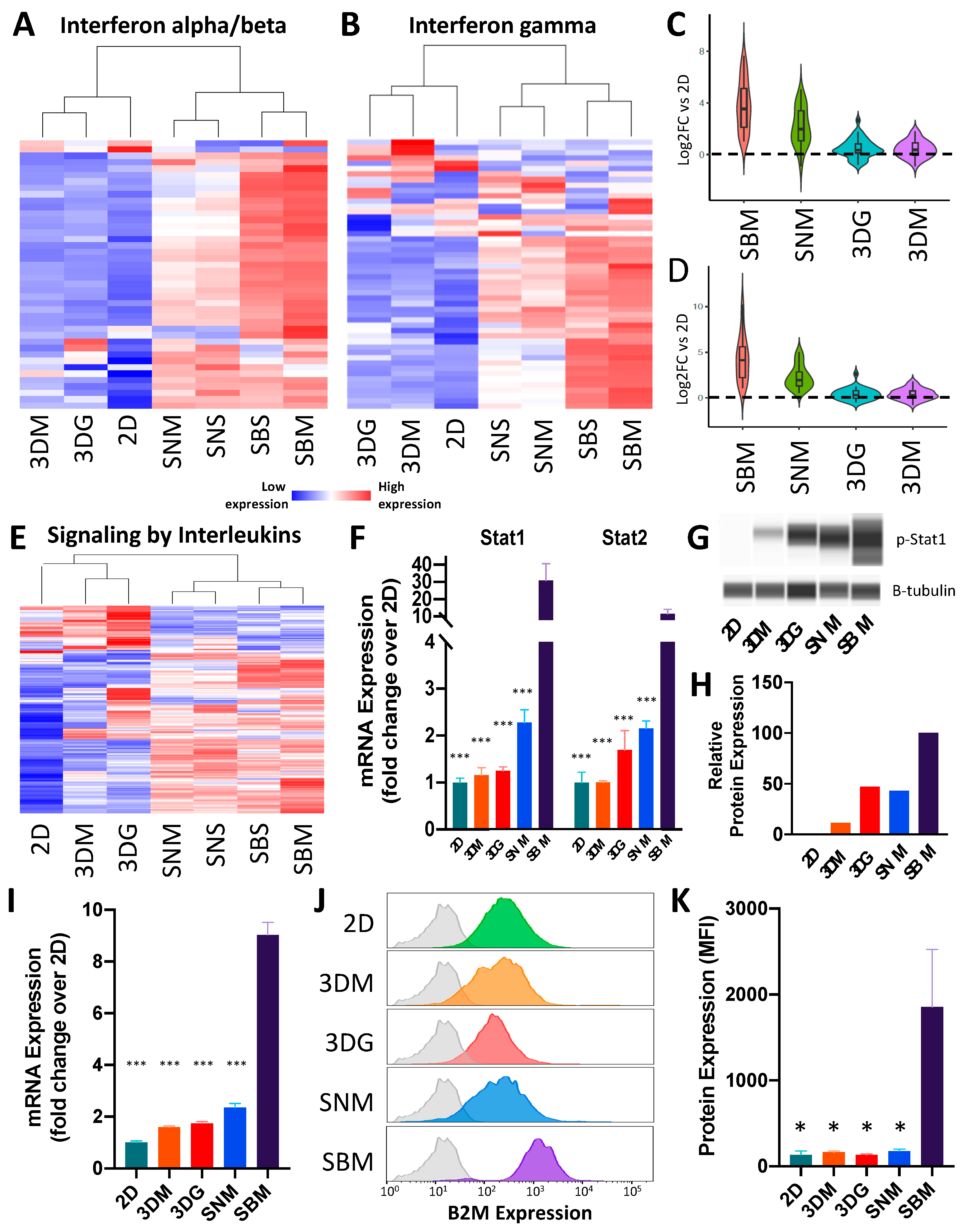
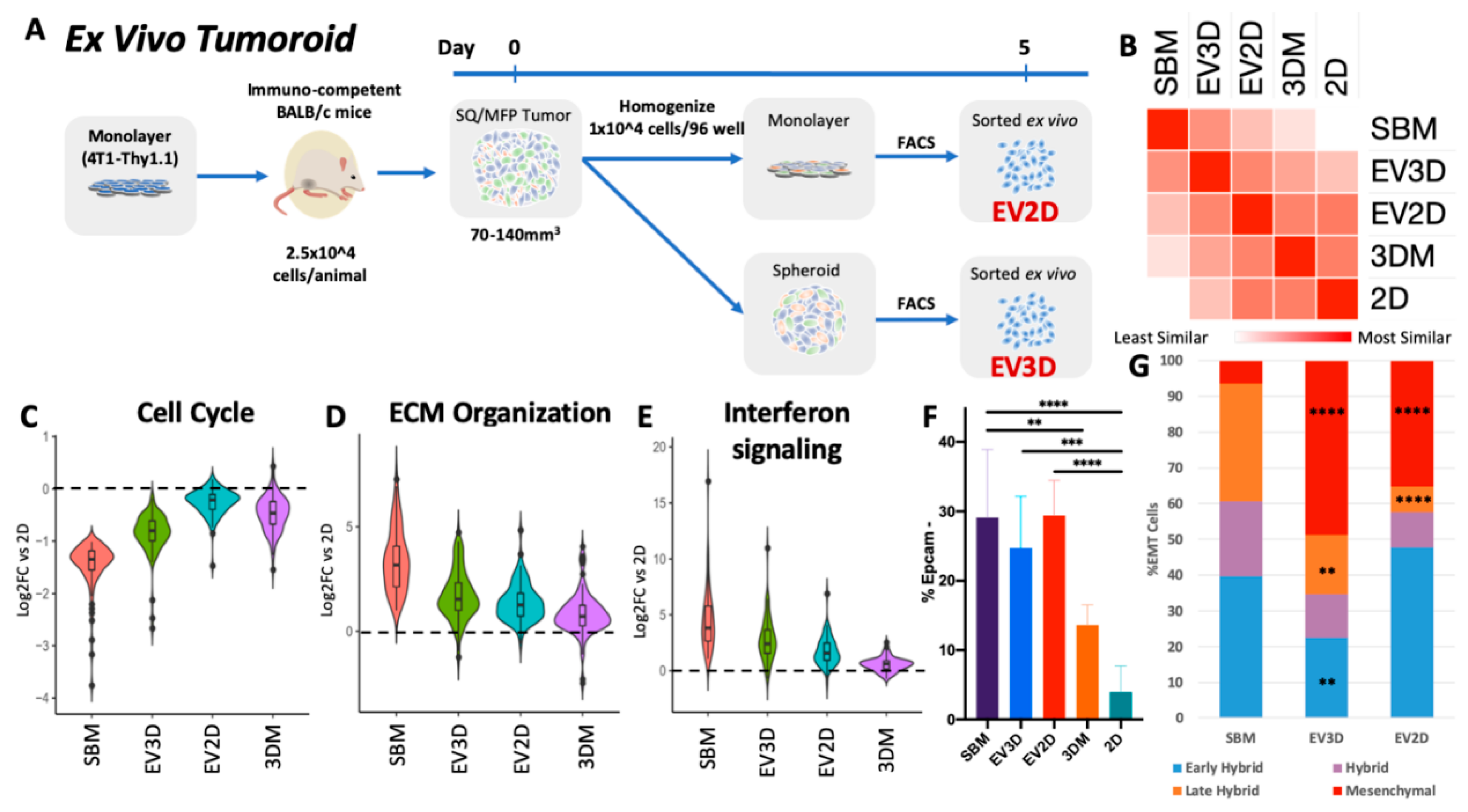
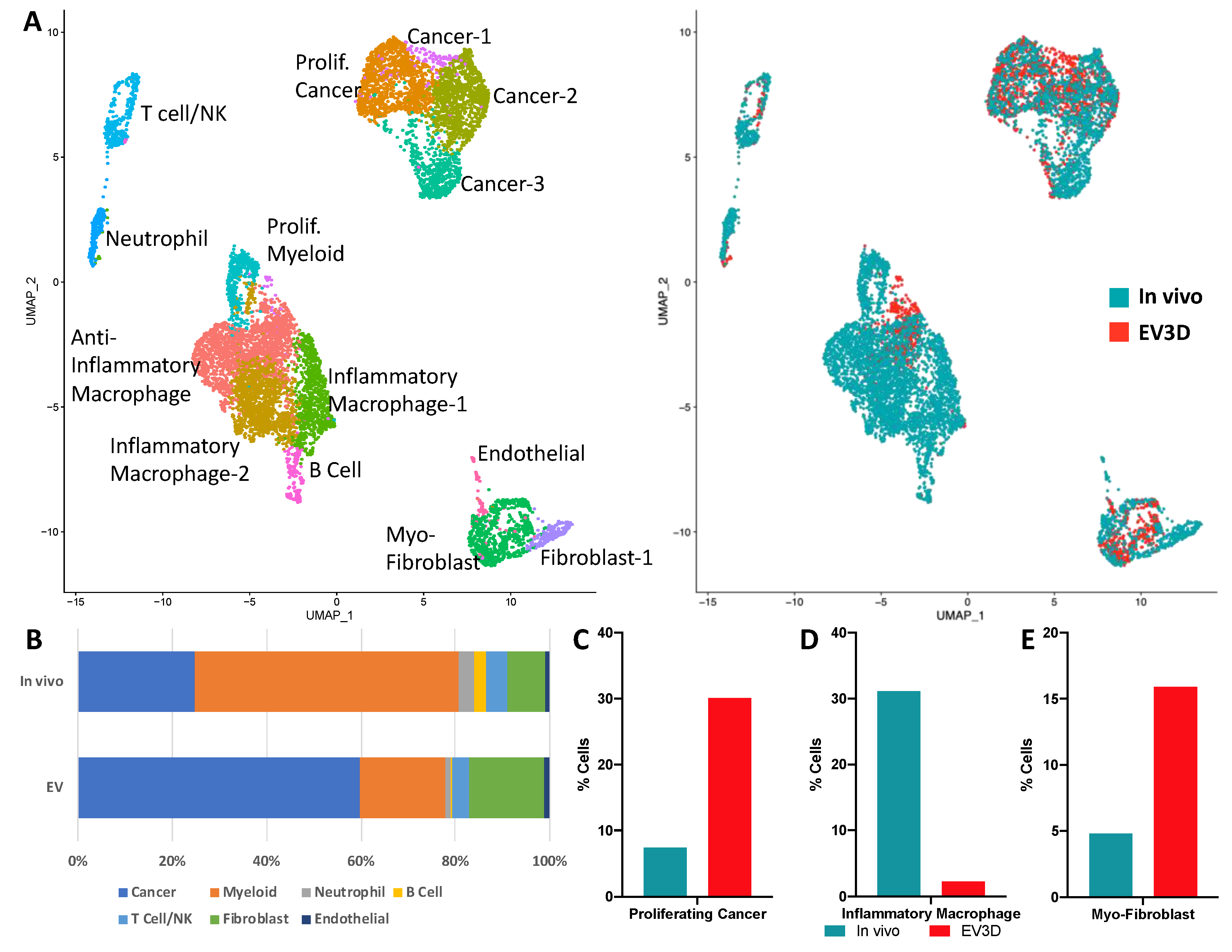
2.3. Ex Vivo Tumoroids Inclusive of Stromal Cells Preserve In Vivo Behavior
3. Discussion
4. Materials and Methods
4.1. The 2D and 3D Cell Culture
4.2. The 4T1-BFP Generation
4.3. Allograft Generation and Tumor Digests
4.4. Histological Sectioning/Staining
4.5. Flow Cytometry and Fluorescent Activated Cell Sorting (FACS)
4.6. RNA Sequencing and Analysis
4.7. Western Blot
4.8. Ex Vivo Tumoroid Culture
4.9. Single-Cell Sequencing and Data Analysis
4.10. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- WHO Global Health Reports 2013. Available online: https://www.who.int/gho/publications/world_health_statistics/EN_WHS2013_Full.pdf (accessed on 15 January 2020).
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2018. CA Cancer J. Clin. 2018, 68, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Pampaloni, F.; Reynaud, E.G.; Stelzer, E.H.K. The third dimension bridges the gap between cell culture and live tissue. Nat. Rev. Mol. Cell Biol. 2007, 8, 839–845. [Google Scholar] [CrossRef] [PubMed]
- Cagan, R.; Meyer, P. Rethinking cancer: Current challenges and opportunities in cancer research. Dis. Model Mech. 2017, 10, 349–352. [Google Scholar] [CrossRef] [PubMed]
- Day, C.-P.; Merlino, G.; Van Dyke, T. Preclinical mouse cancer models: A maze of opportunities and challenges. Cell 2015, 163, 39–53. [Google Scholar] [CrossRef]
- Miura, S.; Gomez, K.; Murillo, O.; Huuki, L.A.; Vu, T.; Buturla, T.; Kumar, S. Predicting clone genotypes from tumor bulk sequencing of multiple samples. Bioinformatics 2018, 34, 4017–4026. [Google Scholar] [CrossRef]
- Zanoni, M.; Piccinini, F.; Arienti, C.; Zamagni, A.; Santi, S.; Polico, R.; Bevilacqua, A.; Tesei, A. 3D tumor spheroid models for in vitro therapeutic screening: A systematic approach to enhance the biological relevance of data obtained. Nat. Publ. Group 2016, 6, 19103. [Google Scholar] [CrossRef]
- Dobrolecki, L.E.; Airhart, S.D.; Alferez, D.G.; Aparicio, S.; Behbod, F.; Bentires-Alj, M.; Brisken, C.; Bult, C.J.; Cai, S.; Clarke, R.B.; et al. Patient-derived xenograft (PDX) models in basic and translational breast cancer research. Cancer Metastasis Rev. 2016, 35, 547–573. [Google Scholar] [CrossRef]
- Ben-David, U.; Ha, G.; Tseng, Y.-Y.; Greenwald, N.F.; Oh, C.; Shih, J.; McFarland, J.M.; Wong, B.; Boehm, J.S.; Beroukhim, R.; et al. Patient-derived xenografts undergo mouse-specific tumor evolution. Nat. Genet. 2017, 49, 1567–1575. [Google Scholar] [CrossRef]
- Mak, I.W.; Evaniew, N.; Ghert, M. Lost in translation: Animal models and clinical trials in cancer treatment. Am. J. Transl. Res. 2014, 6, 114–118. [Google Scholar]
- Dumont, S.; Jan, Z.; Heremans, R.; Van Gorp, T.; Vergote, I.; Timmerman, D. Organoids of epithelial ovarian cancer as an emerging preclinical in vitro tool: A review. J. Ovarian Res. 2019, 12, 105–111. [Google Scholar] [CrossRef]
- Engel, R.M.; Chan, W.H.; Nickless, D.; Hlavca, S.; Richards, E.; Kerr, G.; Oliva, K.; McMurrick, P.J.; Jardé, T.; Abud, H.E. Patient-Derived Colorectal Cancer Organoids Upregulate Revival Stem Cell Marker Genes following Chemotherapeutic Treatment. J. Clin. Med. 2020, 9, 128. [Google Scholar] [CrossRef] [PubMed]
- Boj, S.F.; Hwang, C.-I.; Baker, L.A.; Chio, I.I.C.; Engle, D.D.; Corbo, V.; Jager, M.; Ponz-Sarvise, M.; Tiriac, H.; Spector, M.S.; et al. Organoid Models of Human and Mouse Ductal Pancreatic Cancer. Cell 2015, 160, 324–338. [Google Scholar] [CrossRef] [PubMed]
- Saito, Y. Establishment of an organoid bank of biliary tract and pancreatic cancers and its application for personalized therapy and future treatment. J. Gastroenterol. Hepatol. 2019, 34, 1906–1910. [Google Scholar] [CrossRef] [PubMed]
- Mousavi, N.; Truelsen, S.L.B.; Hagel, G.; Jorgensen, L.N.; Harling, H.; Timmermans, V.; Melchior, L.C.; Thysen, A.H.; Heegaard, S.; Thastrup, J. KRAS mutations in the parental tumour accelerate in vitro growth of tumoroids established from colorectal adenocarcinoma. Int. J. Exp. Pathol. 2019, 100, 12–18. [Google Scholar] [CrossRef] [PubMed]
- Pulaski, B.A.; Ostrand-Rosenberg, S. Mouse 4T1 breast tumor model. Curr. Protoc. Immunol. 2001, 39, 20.2.1–20.2.16. [Google Scholar] [CrossRef]
- Feitelson, M.A.; Arzumanyan, A.; Kulathinal, R.J.; Blain, S.W.; Holcombe, R.F.; Mahajna, J.; Marino, M.; Martinez-Chantar, M.L.; Nawroth, R.; Sanchez-Garcia, I.; et al. Sustained proliferation in cancer: Mechanisms and novel therapeutic targets. Semin. Cancer Biol. 2015, 35, S25–S54. [Google Scholar] [CrossRef]
- Walker, C.; Mojares, E.; Del Río Hernández, A. Role of Extracellular Matrix in Development and Cancer Progression. Int. J. Mol. Sci. 2018, 19, 3028. [Google Scholar] [CrossRef]
- Sever, R.; Brugge, J.S. Signal transduction in cancer. Cold Spring Harb Perspect Med. 2015, 5, a006098. [Google Scholar] [CrossRef]
- Parker, B.S.; Rautela, J.; Hertzog, P.J. Antitumour actions of interferons: Implications for cancer therapy. Nat. Rev. Cancer 2016, 16, 131–144. [Google Scholar] [CrossRef]
- Brabletz, T.; Kalluri, R.; Nieto, M.A.; Weinberg, R.A. EMT in cancer. Nat. Rev. Cancer 2018, 18, 128–134. [Google Scholar] [CrossRef]
- Atkinson, S.P.; Collin, J.; Irina, N.; Anyfantis, G.; Kyung, B.K.; Lako, M.; Armstrong, L. A putative role for the immunoproteasome in the maintenance of pluripotency in human embryonic stem cells. Stem Cells 2012, 30, 1373–1384. [Google Scholar] [CrossRef] [PubMed]
- Au-Yeung, N.; Mandhana, R.; Horvath, C.M. Transcriptional regulation by STAT1 and STAT2 in the interferon JAK-STAT pathway. JAKSTAT 2013, 2, e23931. [Google Scholar] [CrossRef] [PubMed]
- Pastushenko, I.; Brisebarre, A.; Sifrim, A.; Fioramonti, M.; Revenco, T.; Boumahdi, S.; Van Keymeulen, A.; Brown, D.; Moers, V.; Lemaire, S.; et al. Identification of the tumour transition states occurring during EMT. Nature 2018, 556, 463–468. [Google Scholar] [CrossRef] [PubMed]
- Tsubakihara, Y.; Moustakas, A. Epithelial-Mesenchymal Transition and Metastasis under the Control of Transforming Growth Factor β. Int. J. Mol. Sci. 2018, 19, 3672. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.-L.; Huang, H.-C.; Kao, S.-H.; Hsu, Y.-C.; Wang, Y.-T.; Li, K.-C.; Chen, Y.-J.; Yu, S.-L.; Wang, S.-P.; Hsiao, T.-H.; et al. Slug is temporally regulated by cyclin E in cell cycle and controls genome stability. Oncogene 2015, 34, 1116–1125. [Google Scholar] [CrossRef] [PubMed]
- Jolly, M.K.; Tripathi, S.C.; Jia, D.; Mooney, S.M.; Celiktas, M.; Hanash, S.M.; Mani, S.A.; Pienta, K.J.; Ben-Jacob, E.; Levine, H. Stability of the hybrid epithelial/mesenchymal phenotype. Oncotarget 2016, 7, 27067–27084. [Google Scholar] [CrossRef]
- Hong, T.; Watanabe, K.; Ta, C.H.; Villarreal-Ponce, A.; Nie, Q.; Dai, X. An Ovol2-Zeb1 Mutual Inhibitory Circuit Governs Bidirectional and Multi-step Transition between Epithelial and Mesenchymal States. PLoS Comput. Biol. 2015, 11, e1004569. [Google Scholar] [CrossRef]
- Jordan, N.V.; Johnson, G.L.; Abell, A.N. Tracking the intermediate stages of epithelial-mesenchymal transition in epithelial stem cells and cancer. Cell Cycle 2011, 10, 2865–2873. [Google Scholar] [CrossRef]
- Nieto, M.A.; Huang, R.Y.-J.; Jackson, R.A.; Thiery, J.P. EMT: 2016. Cell 2016, 166, 21–45. [Google Scholar] [CrossRef]
- McKeown, S.R. Defining normoxia, physoxia and hypoxia in tumours—implications for treatment response. Br. J. Radiol. 2014, 87, 20130676. [Google Scholar] [CrossRef]
- Reuter, S.; Gupta, S.C.; Chaturvedi, M.M.; Aggarwal, B.B. Oxidative stress, inflammation, and cancer: How are they linked? Free Radic. Biol. Med. 2010, 49, 1603–1616. [Google Scholar] [CrossRef] [PubMed]
- Pompili, L.; Porru, M.; Caruso, C.; Biroccio, A.; Leonetti, C. Patient-derived xenografts: A relevant preclinical model for drug development. J. Exp. Clin. Cancer Res. 2016, 35, 189. [Google Scholar] [CrossRef] [PubMed]
- Dobin, A.; Gingeras, T.R. Mapping RNA-seq Reads with STAR. Curr. Protoc. Bioinform. 2015, 51, 11.14.1–11.14.19. [Google Scholar] [CrossRef] [PubMed]
- Risso, D.; Schwartz, K.; Sherlock, G.; Dudoit, S. GC-content normalization for RNA-Seq data. BMC Bioinform. 2011, 12, 480. [Google Scholar] [CrossRef]
- Risso, D.; Ngai, J.; Speed, T.P.; Dudoit, S. Normalization of RNA-seq data using factor analysis of control genes or samples. Nat. Biotechnol. 2014, 32, 896–902. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Chen, J.; Bardes, E.E.; Aronow, B.J.; Jegga, A.G. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 2009, 37, W305–W311. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Fabregat, A.; Jupe, S.; Matthews, L.; Sidiropoulos, K.; Gillespie, M.; Garapati, P.; Haw, R.; Jassal, B.; Korninger, F.; May, B.; et al. The Reactome Pathway Knowledgebase. Nucleic Acids Res. 2018, 46, D649–D655. [Google Scholar] [CrossRef]
- Sobral-Filho, R.G.; DeVorkin, L.; Macpherson, S.; Jirasek, A.; Lum, J.J.; Brolo, A.G. Ex Vivo Detection of Circulating Tumor Cells from Whole Blood by Direct Nanoparticle Visualization. ACS Nano 2018, 12, 1902–1909. [Google Scholar] [CrossRef]
- Stuart, T.; Butler, A.; Hoffman, P.; Hafemeister, C.; Papalexi, E.; Mauck, W.M., III; Hao, Y.; Stoeckius, M.; Smibert, P.; Satija, R. Comprehensive Integration of Single-Cell Data. Cell 2019, 177, 1888–1902. [Google Scholar] [CrossRef] [PubMed]

| Key Ontology Terms Associated with Genes Highly Expressed in 2D Compared to SBM | ||
| GO ID | Term | No. of Genes |
| GO:1901605 | alpha-amino acid metabolic process | 34 |
| GO:0007049 | cell cycle | 197 |
| GO:0044770 | cell cycle phase transition | 67 |
| GO:0051301 | cell division | 88 |
| GO:0045333 | cellular respiration | 26 |
| GO:0007059 | chromosome segregation | 60 |
| GO:0006259 | DNA metabolic process | 110 |
| GO:0006281 | DNA repair | 56 |
| GO:0006260 | DNA replication | 54 |
| GO:0032543 | mitochondrial translation | 30 |
| GO:0007005 | mitochondrion organization | 101 |
| GO:0034660 | ncRNA metabolic process | 128 |
| GO:0000280 | nuclear division | 88 |
| GO:0048285 | organelle fission | 91 |
| GO:0009126 | purine nucleoside monophosphate metabolic process | 32 |
| GO:0006220 | pyrimidine nucleotide metabolic process | 11 |
| GO:0042254 | ribosome biogenesis | 97 |
| GO:0016072 | rRNA metabolic process | 77 |
| GO:0006360 | transcription by RNA polymerase I | 14 |
| GO:0006412 | translation | 74 |
| GO:0006399 | tRNA metabolic process | 45 |
| Key ontology terms associated with genes highly expressed in SBM compared to 2D | ||
| GO ID | Term | No. of genes |
| GO:0001525 | angiogenesis | 141 |
| GO:0001775 | cell activation | 208 |
| GO:0007155 | cell adhesion | 341 |
| GO:0016477 | cell migration | 307 |
| GO:0032963 | collagen metabolic process | 49 |
| GO:0060429 | epithelium development | 243 |
| GO:0030198 | extracellular matrix organization | 120 |
| GO:0006955 | immune response | 349 |
| GO:0000165 | MAPK cascade | 178 |
| GO:0023056 | positive regulation of signaling | 321 |
| GO:0012501 | programmed cell death | 357 |
| GO:0045595 | regulation of cell differentiation | 358 |
| GO:0042127 | regulation of cell proliferation | 348 |
| GO:0034097 | response to cytokine | 210 |
| GO:0070848 | response to growth factor | 154 |
| GO:0034341 | response to interferon-gamma | 60 |
| GO:0070482 | response to oxygen levels | 83 |
| GO:1901700 | response to oxygen-containing compound | 314 |
| GO:0048771 | tissue remodeling | 56 |
| GO:0001944 | vasculature development | 191 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hum, N.R.; Sebastian, A.; Gilmore, S.F.; He, W.; Martin, K.A.; Hinckley, A.; Dubbin, K.R.; Moya, M.L.; Wheeler, E.K.; Coleman, M.A.; et al. Comparative Molecular Analysis of Cancer Behavior Cultured In Vitro, In Vivo, and Ex Vivo. Cancers 2020, 12, 690. https://doi.org/10.3390/cancers12030690
Hum NR, Sebastian A, Gilmore SF, He W, Martin KA, Hinckley A, Dubbin KR, Moya ML, Wheeler EK, Coleman MA, et al. Comparative Molecular Analysis of Cancer Behavior Cultured In Vitro, In Vivo, and Ex Vivo. Cancers. 2020; 12(3):690. https://doi.org/10.3390/cancers12030690
Chicago/Turabian StyleHum, Nicholas R., Aimy Sebastian, Sean F. Gilmore, Wei He, Kelly A. Martin, Aubree Hinckley, Karen R. Dubbin, Monica L. Moya, Elizabeth K. Wheeler, Matthew A. Coleman, and et al. 2020. "Comparative Molecular Analysis of Cancer Behavior Cultured In Vitro, In Vivo, and Ex Vivo" Cancers 12, no. 3: 690. https://doi.org/10.3390/cancers12030690
APA StyleHum, N. R., Sebastian, A., Gilmore, S. F., He, W., Martin, K. A., Hinckley, A., Dubbin, K. R., Moya, M. L., Wheeler, E. K., Coleman, M. A., & Loots, G. G. (2020). Comparative Molecular Analysis of Cancer Behavior Cultured In Vitro, In Vivo, and Ex Vivo. Cancers, 12(3), 690. https://doi.org/10.3390/cancers12030690







