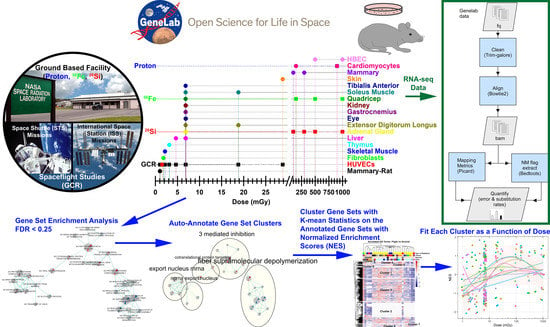
NASA GeneLab Platform Utilized for Biological Response to Space Radiation in Animal Models
Abstract
1. Introduction
2. Results
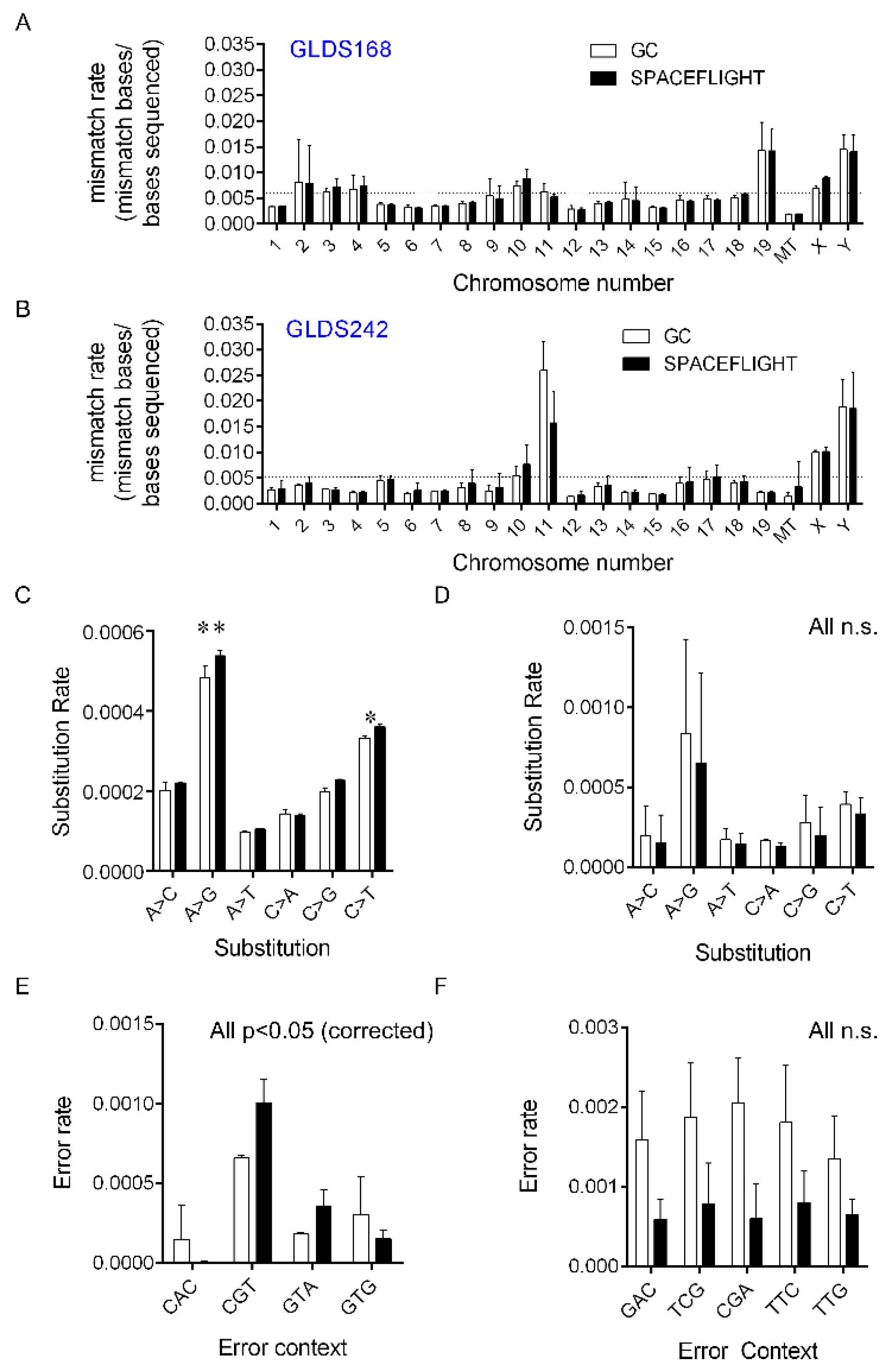
2.1. Changes in RNA-Specific Error Rates Comparing Spaceflight Versus Ground Control Samples
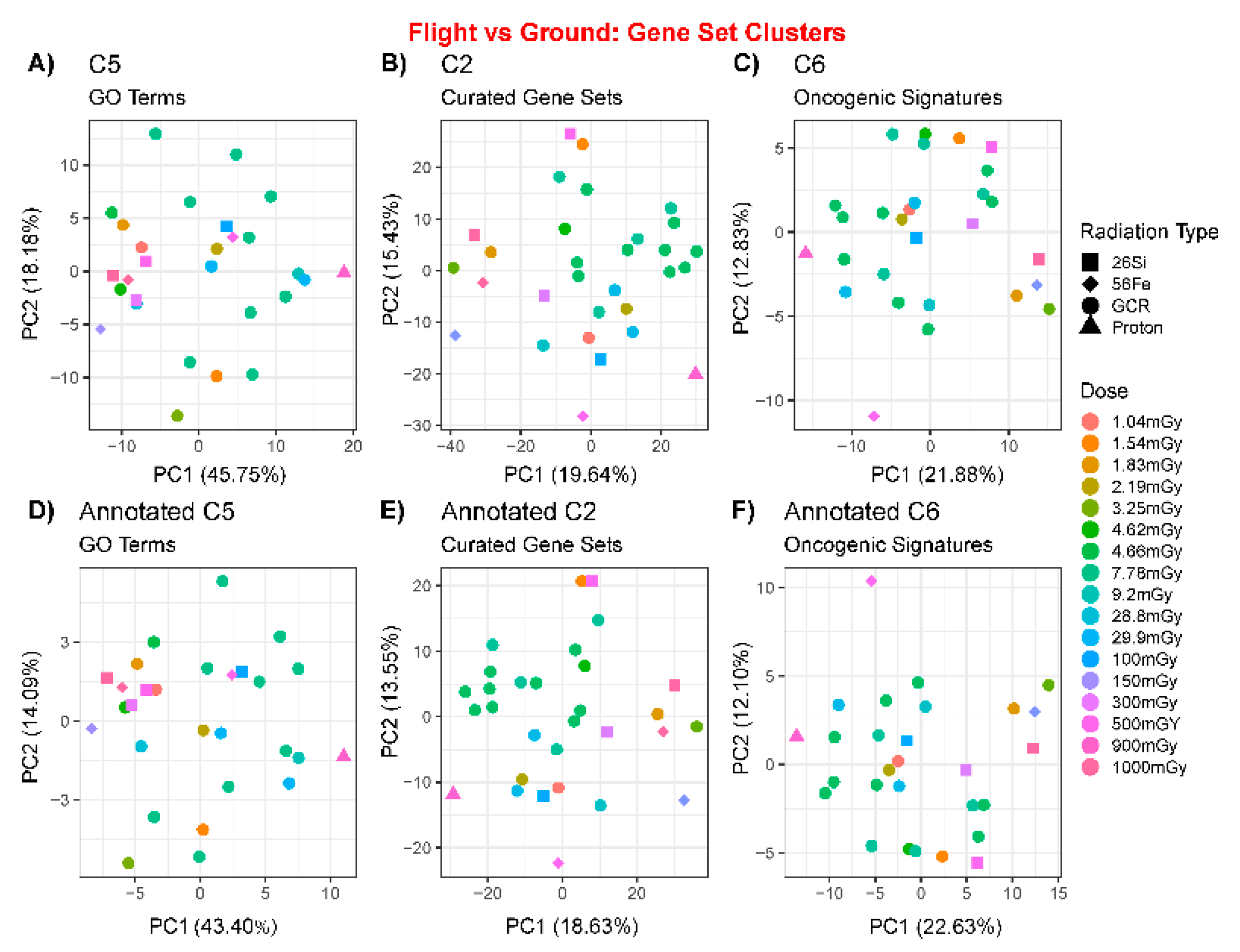
2.2. Comparison of Multiple Genelab Datasets Using Principal Component Analysis
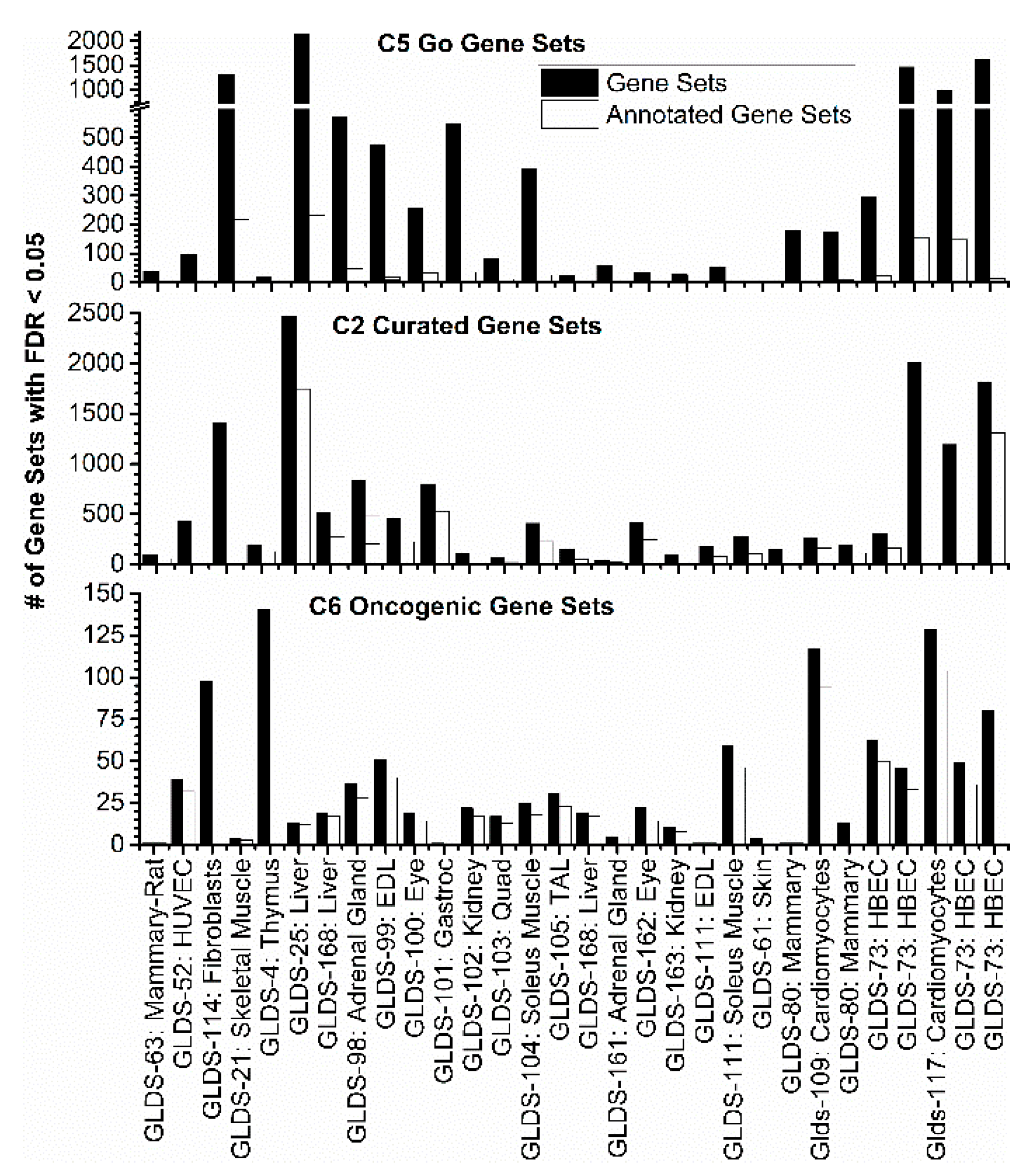
2.3. Transcriptome Evaluation of Biological Functions Impacted by Spaceflight Exposure in Animal Models
2.3.1. GSEA C5: Gene Ontology Results
2.3.2. GSEA C2: Curated Dataset Results
2.3.3. GSEA C6: Oncogenic Signatures
2.4. Common Genes as a Function of Dose and Association with Biological Pathways
3. Discussion
4. Materials and Methods
4.1. Chromosomal and Subsitution Error Rate Mapping
4.2. Bioinformatics, Gene Set Enrichment Analysis, and Visulization Using Cytoscape
4.3. Principal Component Analysis, t-Distributed Stochastic Neighbor Embedding, k-Means Clustering, Heatmaps, and Generalized Additive Model Fits
4.4. Analysis on Common Individual Genes with Fold-Change Values and Fits as a Function of Dose
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Chancellor, J.C.; Scott, G.B.; Sutton, J.P. Space Radiation: The Number One Risk to Astronaut Health beyond Low Earth Orbit. Life (Basel) 2014, 4, 491–510. [Google Scholar] [CrossRef]
- Blaber, E.A.; Dvorochkin, N.; Lee, C.; Alwood, J.S.; Yousuf, R.; Pianetta, P.; Globus, R.K.; Burns, B.P.; Almeida, E.A. Microgravity induces pelvic bone loss through osteoclastic activity, osteocytic osteolysis, and osteoblastic cell cycle inhibition by CDKN1a/p21. PLoS ONE 2013, 8, e61372. [Google Scholar] [CrossRef]
- Riley, D.A. Review of primary spaceflight-induced and secondary reloading-induced changes in slow antigravity muscles of rats. Adv. Space Res. 1998, 21, 1073–1075. [Google Scholar] [CrossRef]
- Baldwin, K.M. Effect of spaceflight on the functional, biochemical, and metabolic properties of skeletal muscle. Med. Sci. Sports Exerc. 1996, 28, 983–987. [Google Scholar] [CrossRef] [PubMed]
- Gambara, G.; Salanova, M.; Ciciliot, S.; Furlan, S.; Gutsmann, M.; Schiffl, G.; Ungethuem, U.; Volpe, P.; Gunga, H.C.; Blottner, D. Gene Expression Profiling in Slow-Type Calf Soleus Muscle of 30 Days Space-Flown Mice. PLoS ONE 2017, 12, e0169314. [Google Scholar] [CrossRef] [PubMed]
- Mishra, B.; Luderer, U. Reproductive hazards of space travel in women and men. Nat. Rev. Endocrinol. 2019, 15, 713–730. [Google Scholar] [CrossRef]
- Cucinotta, F.A.; Cacao, E. Risks of cognitive detriments after low dose heavy ion and proton exposures. Int. J. Radiat. Biol. 2019, 95, 985–998. [Google Scholar] [CrossRef]
- Rabin, B.M.; Shukitt-Hale, B.; Joseph, J.; Todd, P. Diet as a factor in behavioral radiation protection following exposure to heavy particles. Gravit. Space Biol. Bull. 2005, 18, 71–77. [Google Scholar]
- Dutta, S.M.; Hadley, M.M.; Peterman, S.; Jewell, J.S.; Duncan, V.D.; Britten, R.A. Quantitative Proteomic Analysis of the Hippocampus of Rats with GCR-Induced Spatial Memory Impairment. Radiat. Res. 2018, 189, 136–145. [Google Scholar] [CrossRef]
- Shen, M.; Frishman, W.H. Effects of Spaceflight on Cardiovascular Physiology and Health. Cardiol. Rev. 2019, 27, 122–126. [Google Scholar] [CrossRef]
- Boerma, M.; Nelson, G.A.; Sridharan, V.; Mao, X.W.; Koturbash, I.; Hauer-Jensen, M. Space radiation and cardiovascular disease risk. World J. Cardiol. 2015, 7, 882–888. [Google Scholar] [CrossRef] [PubMed]
- Raghunathan, D.; Khilji, M.I.; Hassan, S.A.; Yusuf, S.W. Radiation-Induced Cardiovascular Disease. Curr. Atheroscler. Rep. 2017, 19, 22. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: the next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Barcellos-Hoff, M.H.; Blakely, E.A.; Burma, S.; Fornace, A.J.; Gerson, S.; Hlatky, L.; Kirsch, D.G.; Luderer, U.; Shay, J.; Wang, Y.; et al. Concepts and challenges in cancer risk prediction for the space radiation environment. Life Sci. Space Res. (Amst) 2015, 6, 92–103. [Google Scholar] [CrossRef] [PubMed]
- Vidyasekar, P.; Shyamsunder, P.; Arun, R.; Santhakumar, R.; Kapadia, N.K.; Kumar, R.; Verma, R.S. Genome Wide Expression Profiling of Cancer Cell Lines Cultured in Microgravity Reveals Significant Dysregulation of Cell Cycle and MicroRNA Gene Networks. PLoS ONE 2015, 10, e0135958. [Google Scholar] [CrossRef] [PubMed]
- Yatagai, F.; Honma, M.; Dohmae, N.; Ishioka, N. Biological effects of space environmental factors: A possible interaction between space radiation and microgravity. Life Sci. Space Res. (Amst) 2019, 20, 113–123. [Google Scholar] [CrossRef]
- Maxwell, C.A.; Fleisch, M.C.; Costes, S.V.; Erickson, A.C.; Boissiere, A.; Gupta, R.; Ravani, S.A.; Parvin, B.; Barcellos-Hoff, M.H. Targeted and nontargeted effects of ionizing radiation that impact genomic instability. Cancer Res. 2008, 68, 8304–8311. [Google Scholar] [CrossRef]
- Crucian, B.E.; Chouker, A.; Simpson, R.J.; Mehta, S.; Marshall, G.; Smith, S.M.; Zwart, S.R.; Heer, M.; Ponomarev, S.; Whitmire, A.; et al. Immune System Dysregulation During Spaceflight: Potential Countermeasures for Deep Space Exploration Missions. Front Immunol. 2018, 9, 1437. [Google Scholar] [CrossRef]
- Michaletti, A.; Gioia, M.; Tarantino, U.; Zolla, L. Effects of microgravity on osteoblast mitochondria: A proteomic and metabolomics profile. Sci. Rep. 2017, 7, 15376. [Google Scholar] [CrossRef]
- Andreazzoli, A.; Fossati, C.; Spaccamiglio, A.; Salvo, R.; Quaranta, F.; Minganti, C.; Di Luigi, L.; Borrione, P. Assessment of pN-GAL as a marker of renal function in elite cyclists during professional competitions. J. Biol. Regul. Homeost Agents 2017, 31, 829–835. [Google Scholar]
- Garrett-Bakelman, F.E.; Darshi, M.; Green, S.J.; Gur, R.C.; Lin, L.; Macias, B.R.; McKenna, M.J.; Meydan, C.; Mishra, T.; Nasrini, J.; et al. The NASA Twins Study: A multidimensional analysis of a year-long human spaceflight. Science 2019, 364. [Google Scholar] [CrossRef]
- Gao, Y.; Xu, D.; Zhao, L.; Zhang, M.; Sun, Y. Effects of microgravity on DNA damage response in Caenorhabditis elegans during Shenzhou-8 spaceflight. Int. J. Radiat. Biol. 2015, 91, 531–539. [Google Scholar] [CrossRef] [PubMed]
- Ray, S.; Gebre, S.; Fogle, H.; Berrios, D.C.; Tran, P.B.; Galazka, J.M.; Costes, S.V. GeneLab: Omics database for spaceflight experiments. Bioinformatics 2019, 35, 1753–1759. [Google Scholar] [CrossRef] [PubMed]
- La Tessa, C.; Sivertz, M.; Chiang, I.H.; Lowenstein, D.; Rusek, A. Overview of the NASA space radiation laboratory. Life Sci. Space Res. (Amst) 2016, 11, 18–23. [Google Scholar] [CrossRef]
- Casey, T.; Patel, O.V.; Plaut, K. Transcriptomes reveal alterations in gravity impact circadian clocks and activate mechanotransduction pathways with adaptation through epigenetic change. Physiol. Genom. 2015, 47, 113–128. [Google Scholar] [CrossRef]
- Patel, O.V.; Casey, T.; Dover, H.; Plaut, K. Homeorhetic adaptation to lactation: comparative transcriptome analysis of mammary, liver, and adipose tissue during the transition from pregnancy to lactation in rats. Funct. Integr. Genom. 2011, 11, 193–202. [Google Scholar] [CrossRef]
- Casey, T.; Patel, O.; Dykema, K.; Dover, H.; Furge, K.; Plaut, K. Molecular signatures reveal circadian clocks may orchestrate the homeorhetic response to lactation. PLoS ONE 2009, 4, e7395. [Google Scholar] [CrossRef]
- Versari, S.; Longinotti, G.; Barenghi, L.; Maier, J.A.; Bradamante, S. The challenging environment on board the International Space Station affects endothelial cell function by triggering oxidative stress through thioredoxin interacting protein overexpression: the ESA-SPHINX experiment. FASEB J. 2013, 27, 4466–4475. [Google Scholar] [CrossRef]
- Beheshti, A.; McDonald, J.T.; Miller, J.; Grabham, P.; Costes, S.V. GeneLab Database Analyses Suggest Long-Term Impact of Space Radiation on the Cardiovascular System by the Activation of FYN Through Reactive Oxygen Species. Int. J. Mol. Sci. 2019, 20, 661. [Google Scholar] [CrossRef]
- Lu, T.; Zhang, Y.; Kidane, Y.; Feiveson, A.; Stodieck, L.; Karouia, F.; Ramesh, G.; Rohde, L.; Wu, H. Cellular responses and gene expression profile changes due to bleomycin-induced DNA damage in human fibroblasts in space. PLoS ONE 2017, 12, e0170358. [Google Scholar] [CrossRef]
- Allen, D.L.; Bandstra, E.R.; Harrison, B.C.; Thorng, S.; Stodieck, L.S.; Kostenuik, P.J.; Morony, S.; Lacey, D.L.; Hammond, T.G.; Leinwand, L.L.; et al. Effects of spaceflight on murine skeletal muscle gene expression. J. Appl. Physiol. (1985) 2009, 106, 582–595. [Google Scholar] [CrossRef] [PubMed]
- Beheshti, A.; Ray, S.; Fogle, H.; Berrios, D.; Costes, S.V. A microRNA signature and TGF-beta1 response were identified as the key master regulators for spaceflight response. PLoS ONE 2018, 13, e0199621. [Google Scholar] [CrossRef] [PubMed]
- Beheshti, A.; Cekanaviciute, E.; Smith, D.J.; Costes, S.V. Global transcriptomic analysis suggests carbon dioxide as an environmental stressor in spaceflight: A systems biology GeneLab case study. Sci. Rep. 2018, 8, 4191. [Google Scholar] [CrossRef] [PubMed]
- Lebsack, T.W.; Fa, V.; Woods, C.C.; Gruener, R.; Manziello, A.M.; Pecaut, M.J.; Gridley, D.S.; Stodieck, L.S.; Ferguson, V.L.; Deluca, D. Microarray analysis of spaceflown murine thymus tissue reveals changes in gene expression regulating stress and glucocorticoid receptors. J. Cell Biochem. 2010, 110, 372–381. [Google Scholar] [CrossRef]
- Globus, R.; Cadena, S. Rodent Research-1 (RR1) National Lab Validation Flight: Mouse Liver Transcriptomic, Proteomic, and Epigenomic Data. Available online: https://genelab-data.ndc.nasa.gov/genelab/accession/GLDS-47/ (accessed on 5 February 2020).
- Gridley, D.S.; Mao, X.W.; Stodieck, L.S.; Ferguson, V.L.; Bateman, T.A.; Moldovan, M.; Cunningham, C.E.; Jones, T.A.; Slater, J.M.; Pecaut, M.J. Changes in mouse thymus and spleen after return from the STS-135 mission in space. PLoS ONE 2013, 8, e75097. [Google Scholar] [CrossRef]
- Mao, X.W.; Pecaut, M.J.; Stodieck, L.S.; Ferguson, V.L.; Bateman, T.A.; Bouxsein, M.; Jones, T.A.; Moldovan, M.; Cunningham, C.E.; Chieu, J.; et al. Spaceflight environment induces mitochondrial oxidative damage in ocular tissue. Radiat. Res. 2013, 180, 340–350. [Google Scholar] [CrossRef]
- Jonscher, K.R.; Alfonso-Garcia, A.; Suhalim, J.L.; Orlicky, D.J.; Potma, E.O.; Ferguson, V.L.; Bouxsein, M.L.; Bateman, T.A.; Stodieck, L.S.; Levi, M.; et al. Spaceflight Activates Lipotoxic Pathways in Mouse Liver. PLoS ONE 2016, 11, e0152877. [Google Scholar] [CrossRef]
- Pecaut, M.J.; Mao, X.W.; Bellinger, D.L.; Jonscher, K.R.; Stodieck, L.S.; Ferguson, V.L.; Bateman, T.A.; Mohney, R.P.; Gridley, D.S. Is spaceflight-induced immune dysfunction linked to systemic changes in metabolism? PLoS ONE 2017, 12, e0174174. [Google Scholar] [CrossRef]
- Blaber, E.A.; Pecaut, M.J.; Jonscher, K.R. Spaceflight Activates Autophagy Programs and the Proteasome in Mouse Liver. Int. J. Mol. Sci. 2017, 18, 2062. [Google Scholar] [CrossRef]
- Mao, X.W.; Pecaut, M.J.; Stodieck, L.S.; Ferguson, V.L.; Bateman, T.A.; Bouxsein, M.L.; Gridley, D.S. Biological and metabolic response in STS-135 space-flown mouse skin. Free Radic. Res. 2014, 48, 890–897. [Google Scholar] [CrossRef]
- Beheshti, A.; Chakravarty, K.; Fogle, H.; Fazelinia, H.; Silveira, W.A.D.; Boyko, V.; Polo, S.L.; Saravia-Butler, A.M.; Hardiman, G.; Taylor, D.; et al. Multi-omics analysis of multiple missions to space reveal a theme of lipid dysregulation in mouse liver. Sci. Rep. 2019, 9, 19195. [Google Scholar] [CrossRef] [PubMed]
- Globus R, G.N. Rodent Research-1 (RR1) NASA Validation Flight: Mouse Eye Transcriptomic and Epigenomic Data. Available online: https://genelab-data.ndc.nasa.gov/genelab/accession/GLDS-100/ (accessed on 5 February 2020).
- Jiang, P.; Green, S.J.; Chlipala, G.E.; Turek, F.W.; Vitaterna, M.H. Reproducible changes in the gut microbiome suggest a shift in microbial and host metabolism during spaceflight. Microbiome 2019, 7, 113. [Google Scholar] [CrossRef] [PubMed]
- Galazka, J. Effect of Spaceflight on Liver from Mice Flown on the ISS for 33 Days: Transcriptional Analysis. NASA GENELAB: 2019. Available online: https://genelab-data.ndc.nasa.gov/genelab/accession/GLDS-242/. (accessed on 5 February 2020).
- Smith R, C.M.G.R.G.N. Rodent Research-3-CASIS: Mouse Adrenal Gland Transcriptomic, Proteomic, and Epigenomic Data. Available online: https://genelab-data.ndc.nasa.gov/genelab/accession/GLDS-161/ (accessed on 5 February 2020).
- Smith R, C.M.G.R.G.N. Rodent Research-3-CASIS: Mouse Eye Transcriptomic and Proteomic Data. Available online: https://genelab-data.ndc.nasa.gov/genelab/accession/GLDS-162/ (accessed on 5 February 2020).
- Smith R, C.M.G.R.G.N. Rodent Research-3-CASIS: Mouse Kidney Transcriptomic, Proteomic, and Epigenomic Data. Available online: https://genelab-data.ndc.nasa.gov/genelab/accession/GLDS-163/ (accessed on 5 February 2020).
- Neutelings, T.; Nusgens, B.V.; Liu, Y.; Tavella, S.; Ruggiu, A.; Cancedda, R.; Gabriel, M.; Colige, A.; Lambert, C. Skin physiology in microgravity: A 3-month stay aboard ISS induces dermal atrophy and affects cutaneous muscle and hair follicles cycling in mice. NPJ Microgravity 2015, 1, 15002. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Fernandez-Garcia, I.; Vijayakumar, S.; Martinez-Ruis, H.; Illa-Bochaca, I.; Nguyen, D.H.; Mao, J.H.; Costes, S.V.; Barcellos-Hoff, M.H. Irradiation of juvenile, but not adult, mammary gland increases stem cell self-renewal and estrogen receptor negative tumors. Stem Cells 2014, 32, 649–661. [Google Scholar] [CrossRef] [PubMed]
- Coleman, M.A.; Sasi, S.P.; Onufrak, J.; Natarajan, M.; Manickam, K.; Schwab, J.; Muralidharan, S.; Peterson, L.E.; Alekseyev, Y.O.; Yan, X.; et al. Low-dose radiation affects cardiac physiology: gene networks and molecular signaling in cardiomyocytes. Am. J. Physiol. Heart Circ. Physiol. 2015, 309, H1947–H1963. [Google Scholar] [CrossRef]
- Ding, L.H.; Park, S.; Peyton, M.; Girard, L.; Xie, Y.; Minna, J.D.; Story, M.D. Distinct transcriptome profiles identified in normal human bronchial epithelial cells after exposure to gamma-rays and different elemental particles of high Z and energy. BMC Genom. 2013, 14, 372. [Google Scholar] [CrossRef]
- Alexeyev, M.; Shokolenko, I.; Wilson, G.; LeDoux, S. The maintenance of mitochondrial DNA integrity--critical analysis and update. Cold Spring Harb. Perspect. Biol. 2013, 5, a012641. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Hastie, T.; Tibshirani, R. Generalized Additive Models. Stat. Sci. 1986, 1, 297–310. [Google Scholar] [CrossRef]
- Wood, S.N.; Pya, N.; Säfken, B. Smoothing Parameter and Model Selection for General Smooth Models. J. Am. Stat. Assoc. 2015, 111, 1548–1563. [Google Scholar] [CrossRef]
- Moreno-Villanueva, M.; Wong, M.; Lu, T.; Zhang, Y.; Wu, H. Interplay of space radiation and microgravity in DNA damage and DNA damage response. NPJ Microgravity 2017, 3, 14. [Google Scholar] [CrossRef] [PubMed]
- Raber, J.; Allen, A.R.; Sharma, S.; Allen, B.; Rosi, S.; Olsen, R.H.; Davis, M.J.; Eiwaz, M.; Fike, J.R.; Nelson, G.A. Effects of Proton and Combined Proton and (56)Fe Radiation on the Hippocampus. Radiat. Res. 2016, 185, 20–30. [Google Scholar] [CrossRef] [PubMed]
- Martinez, E.M.; Yoshida, M.C.; Candelario, T.L.; Hughes-Fulford, M. Spaceflight and simulated microgravity cause a significant reduction of key gene expression in early T-cell activation. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2015, 308, R480–R488. [Google Scholar] [CrossRef] [PubMed]
- Beheshti, A.; Peluso, M.; Lamont, C.; Hahnfeldt, P.; Hlatky, L. Proton irradiation augments the suppression of tumor progression observed with advanced age. Radiat. Res. 2014, 181, 272–283. [Google Scholar] [CrossRef]
- Beheshti, A.; Sachs, R.K.; Peluso, M.; Rietman, E.; Hahnfeldt, P.; Hlatky, L. Age and space irradiation modulate tumor progression: implications for carcinogenesis risk. Radiat. Res. 2013, 179, 208–220. [Google Scholar] [CrossRef] [PubMed]
- Wage, J.; Ma, L.; Peluso, M.; Lamont, C.; Evens, A.M.; Hahnfeldt, P.; Hlatky, L.; Beheshti, A. Proton irradiation impacts age-driven modulations of cancer progression influenced by immune system transcriptome modifications from splenic tissue. J. Radiat. Res. 2015, 56, 792–803. [Google Scholar] [CrossRef]
- Balbo, B.E.; Amaral, A.G.; Fonseca, J.M.; de Castro, I.; Salemi, V.M.; Souza, L.E.; Dos Santos, F.; Irigoyen, M.C.; Qian, F.; Chammas, R.; et al. Cardiac dysfunction in Pkd1-deficient mice with phenotype rescue by galectin-3 knockout. Kidney Int. 2016, 90, 580–597. [Google Scholar] [CrossRef]
- Galant, D.; Gaborit, B.; Desgrouas, C.; Abdesselam, I.; Bernard, M.; Levy, N.; Merono, F.; Coirault, C.; Roll, P.; Lagarde, A.; et al. A Heterozygous ZMPSTE24 Mutation Associated with Severe Metabolic Syndrome, Ectopic Fat Accumulation, and Dilated Cardiomyopathy. Cells 2016, 5, 21. [Google Scholar] [CrossRef]
- Olabi, A.A.; Lawless, H.T.; Hunter, J.B.; Levitsky, D.A.; Halpern, B.P. The effect of microgravity and space flight on the chemical senses. J. Food Sci. 2002, 67, 468–478. [Google Scholar] [CrossRef]
- Mukhopadhyay, S.; Saha, R.; Palanisamy, A.; Ghosh, M.; Biswas, A.; Roy, S.; Pal, A.; Sarkar, K.; Bagh, S. A systems biology pipeline identifies new immune and disease related molecular signatures and networks in human cells during microgravity exposure. Sci. Rep. 2016, 6, 25975. [Google Scholar] [CrossRef]
- Vickers, Z.M.; Rice, B.L.; Rose, M.S.; Lane, H.W. Simulated microgravity [bed rest] has little influence on taste, odor or trigeminal sensitivity. J. Sens. Stud. 2001, 16, 23–32. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.N.; Su, X.X.; Cheng, S.Q.; Sun, Z.Y.; Huang, Z.S.; Ou, T.M. MYC modulators in cancer: A patent review. Expert Opin. Ther. Pat. 2019, 29, 353–367. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Gutierrez, L.; Delgado, M.D.; Leon, J. MYC Oncogene Contributions to Release of Cell Cycle Brakes. Genes (Basel) 2019, 10, 244. [Google Scholar] [CrossRef] [PubMed]
- Xu, C. A review of somatic single nucleotide variant calling algorithms for next-generation sequencing data. Comput. Struct. Biotechnol. J. 2018, 16, 15–24. [Google Scholar] [CrossRef]
- Beheshti, A.; Miller, J.; Kidane, Y.; Berrios, D.; Gebre, S.G.; Costes, S.V. NASA GeneLab Project: Bridging Space Radiation Omics with Ground Studies. Radiat Res. 2018, 189, 553–559. [Google Scholar] [CrossRef]
- Asaithamby, A.; Chen, D.J. Mechanism of cluster DNA damage repair in response to high-atomic number and energy particles radiation. Mutat. Res. 2011, 711, 87–99. [Google Scholar] [CrossRef]
- Lanza, V.; Pretazzoli, V.; Olivieri, G.; Pascarella, G.; Panconesi, A.; Negri, R. Transcriptional response of human umbilical vein endothelial cells to low doses of ionizing radiation. J. Radiat. Res. 2005, 46, 265–276. [Google Scholar] [CrossRef][Green Version]
- Eberhard, R.; Stergiou, L.; Hofmann, E.R.; Hofmann, J.; Haenni, S.; Teo, Y.; Furger, A.; Hengartner, M.O. Ribosome synthesis and MAPK activity modulate ionizing radiation-induced germ cell apoptosis in Caenorhabditis elegans. PLoS Genet. 2013, 9, e1003943. [Google Scholar] [CrossRef]
- Pucci, M.; Masi, A.; Marsilii, G. Ribosome changes induced by irradiation of isolated rat liver in vitro. Acta Radiol. Oncol. 1982, 21, 449–453. [Google Scholar] [CrossRef][Green Version]
- Kulkarni, R.; Marples, B.; Balasubramaniam, M.; Thomas, R.A.; Tucker, J.D. Mitochondrial gene expression changes in normal and mitochondrial mutant cells after exposure to ionizing radiation. Radiat. Res. 2010, 173, 635–644. [Google Scholar] [CrossRef]
- Barjaktarovic, Z.; Schmaltz, D.; Shyla, A.; Azimzadeh, O.; Schulz, S.; Haagen, J.; Dorr, W.; Sarioglu, H.; Schafer, A.; Atkinson, M.J.; et al. Radiation-induced signaling results in mitochondrial impairment in mouse heart at 4 weeks after exposure to X-rays. PLoS ONE 2011, 6, e27811. [Google Scholar] [CrossRef] [PubMed]
- Pearce, L.L.; Epperly, M.W.; Greenberger, J.S.; Pitt, B.R.; Peterson, J. Identification of Respiratory Complexes I and III as Mitochondrial Sites of Damage Following Exposure to Ionizing Radiation and Nitric Oxide. Nitric. Oxide 2001, 5, 128–136. [Google Scholar] [CrossRef] [PubMed]
- Yin, Z.; Yang, G.; Deng, S.; Wang, Q. Oxidative stress levels and dynamic changes in mitochondrial gene expression in a radiation-induced lung injury model. J. Radiat. Res. 2018, 60, 204–214. [Google Scholar] [CrossRef] [PubMed]
- Brush, J.; Lipnick, S.L.; Phillips, T.; Sitko, J.; McDonald, J.T.; McBride, W.H. Molecular mechanisms of late normal tissue injury. Semin. Radiat. Oncol. 2007, 17, 121–130. [Google Scholar] [CrossRef]
- Schaue, D.; Kachikwu, E.L.; McBride, W.H. Cytokines in radiobiological responses: A review. Radiat. Res. 2012, 178, 505–523. [Google Scholar] [CrossRef]
- McBride, W.H.; Ganapathy, E.; Lee, M.H.; Nesseler, J.P.; Nguyen, C.; Schaue, D. A perspective on the impact of radiation therapy on the immune rheostat. Br. J. Radiol. 2017, 90, 20170272. [Google Scholar] [CrossRef]
- Smith, J.K. IL-6 and the dysregulation of immune, bone, muscle, and metabolic homeostasis during spaceflight. NPJ Microgravity 2018, 4, 24. [Google Scholar] [CrossRef]
- Crucian, B.E.; Zwart, S.R.; Mehta, S.; Uchakin, P.; Quiriarte, H.D.; Pierson, D.; Sams, C.F.; Smith, S.M. Plasma cytokine concentrations indicate that in vivo hormonal regulation of immunity is altered during long-duration spaceflight. J. Interferon Cytokine Res. 2014, 34, 778–786. [Google Scholar] [CrossRef]
- Shibuya, H.; Yoshitomi, H.; Murata, K.; Kobayashi, S.; Furu, M.; Ishikawa, M.; Fujii, T.; Ito, H.; Matsuda, S. TNFalpha, PDGF, and TGFbeta synergistically induce synovial lining hyperplasia via inducible PI3Kdelta. Mod. Rheumatol. 2015, 25, 72–78. [Google Scholar] [CrossRef]
- Aula, H.; Skytta, T.; Tuohinen, S.; Luukkaala, T.; Hamalainen, M.; Virtanen, V.; Raatikainen, P.; Moilanen, E.; Kellokumpu-Lehtinen, P.L. Decreases in TGF-beta1 and PDGF levels are associated with echocardiographic changes during adjuvant radiotherapy for breast cancer. Radiat. Oncol. 2018, 13, 201. [Google Scholar] [CrossRef]
- Tsuchida, N.; Murugan, A.K.; Grieco, M. Kirsten Ras* oncogene: significance of its discovery in human cancer research. Oncotarget 2016, 7, 46717–46733. [Google Scholar] [CrossRef]
- Grabham, P.; Sharma, P. The effects of radiation on angiogenesis. Vascular Cell 2013, 5, 19. [Google Scholar] [CrossRef] [PubMed]
- Hailesellasse Sene, K.; Porter, C.J.; Palidwor, G.; Perez-Iratxeta, C.; Muro, E.M.; Campbell, P.A.; Rudnicki, M.A.; Andrade-Navarro, M.A. Gene function in early mouse embryonic stem cell differentiation. BMC Genom. 2007, 8, 85. [Google Scholar] [CrossRef]
- Mieloch, A.A.; Suchorska, W.M. The concept of radiation-enhanced stem cell differentiation. Radiol. Oncol. 2015, 49, 209–216. [Google Scholar] [CrossRef][Green Version]
- Chang, H.Y.; Sneddon, J.B.; Alizadeh, A.A.; Sood, R.; West, R.B.; Montgomery, K.; Chi, J.T.; van de Rijn, M.; Botstein, D.; Brown, P.O. Gene expression signature of fibroblast serum response predicts human cancer progression: similarities between tumors and wounds. PLoS Biol. 2004, 2, E7. [Google Scholar] [CrossRef] [PubMed]
- Demontis, G.C.; Germani, M.M.; Caiani, E.G.; Barravecchia, I.; Passino, C.; Angeloni, D. Human Pathophysiological Adaptations to the Space Environment. Front. Physiol. 2017, 8, 547. [Google Scholar] [CrossRef] [PubMed]
- Chancellor, J.C.; Blue, R.S.; Cengel, K.A.; Aunon-Chancellor, S.M.; Rubins, K.H.; Katzgraber, H.G.; Kennedy, A.R. Limitations in predicting the space radiation health risk for exploration astronauts. NPJ Microgravity 2018, 4, 8. [Google Scholar] [CrossRef]
- Reisz, J.A.; Bansal, N.; Qian, J.; Zhao, W.; Furdui, C.M. Effects of ionizing radiation on biological molecules--mechanisms of damage and emerging methods of detection. Antioxid. Redox Signal. 2014, 21, 260–292. [Google Scholar] [CrossRef]
- Farin, A.M.; Manzo, N.D.; Kirsch, D.G.; Stripp, B.R. Low- and high-LET radiation drives clonal expansion of lung progenitor cells in vivo. Radiat. Res. 2015, 183, 124–132. [Google Scholar] [CrossRef]
- Merico, D.; Isserlin, R.; Stueker, O.; Emili, A.; Bader, G.D. Enrichment map: A network-based method for gene-set enrichment visualization and interpretation. PLoS ONE 2010, 5, e13984. [Google Scholar] [CrossRef]
- Weiner, J., 3rd; Parida, S.K.; Maertzdorf, J.; Black, G.F.; Repsilber, D.; Telaar, A.; Mohney, R.P.; Arndt-Sullivan, C.; Ganoza, C.A.; Fae, K.C.; et al. Biomarkers of inflammation, immunosuppression and stress with active disease are revealed by metabolomic profiling of tuberculosis patients. PLoS ONE 2012, 7, e40221. [Google Scholar] [CrossRef]
- Brun, R.; Rademakers, F. ROOT — An object oriented data analysis framework. Nucl. Instrum. Meth. A. 1997, 389, 81–86. [Google Scholar] [CrossRef]
- Kaimal, V.; Bardes, E.E.; Tabar, S.C.; Jegga, A.G.; Aronow, B.J. ToppCluster: A multiple gene list feature analyzer for comparative enrichment clustering and network-based dissection of biological systems. Nucleic Acids Res. 2010, 38, W96–W102. [Google Scholar] [CrossRef] [PubMed]


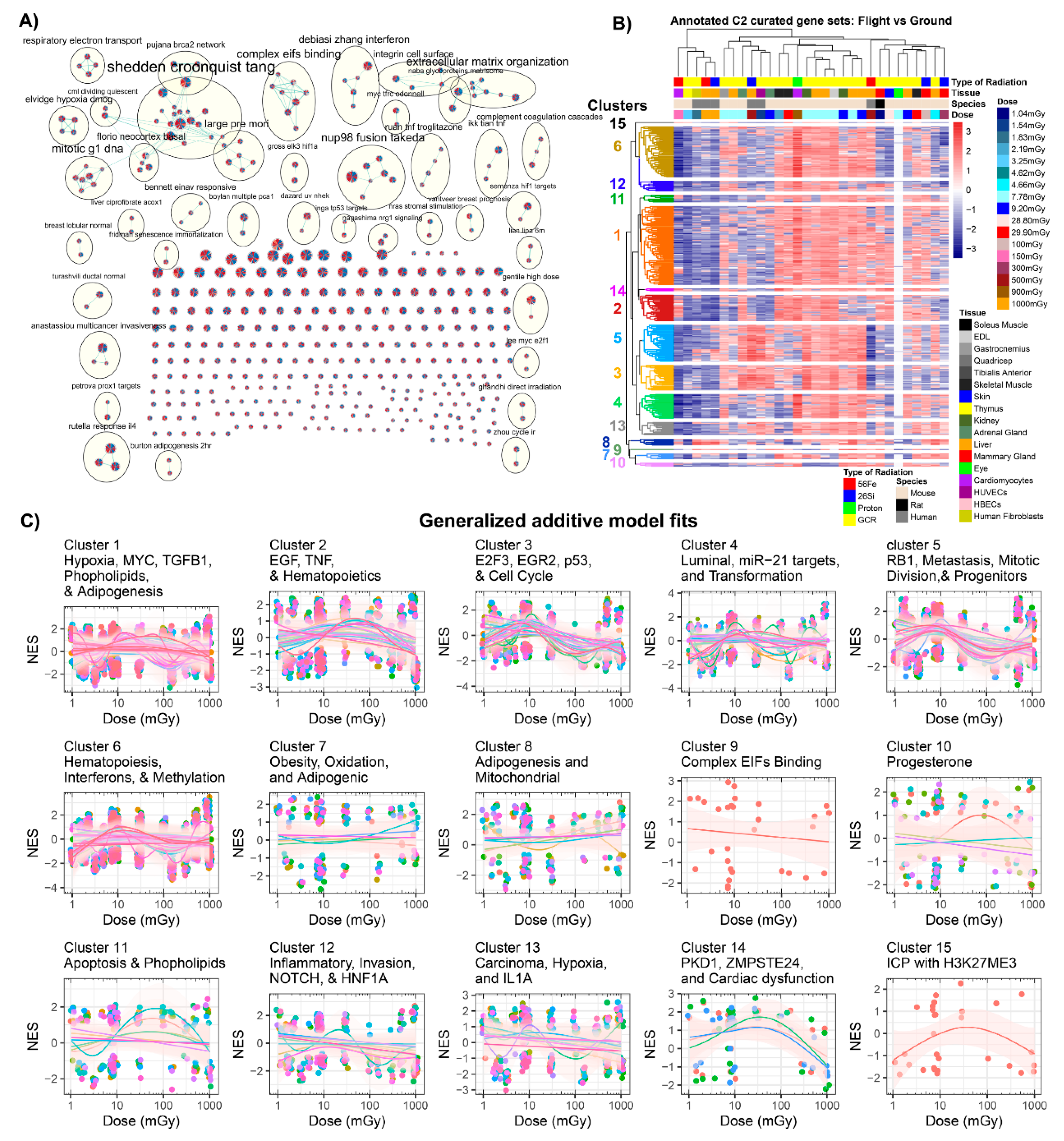
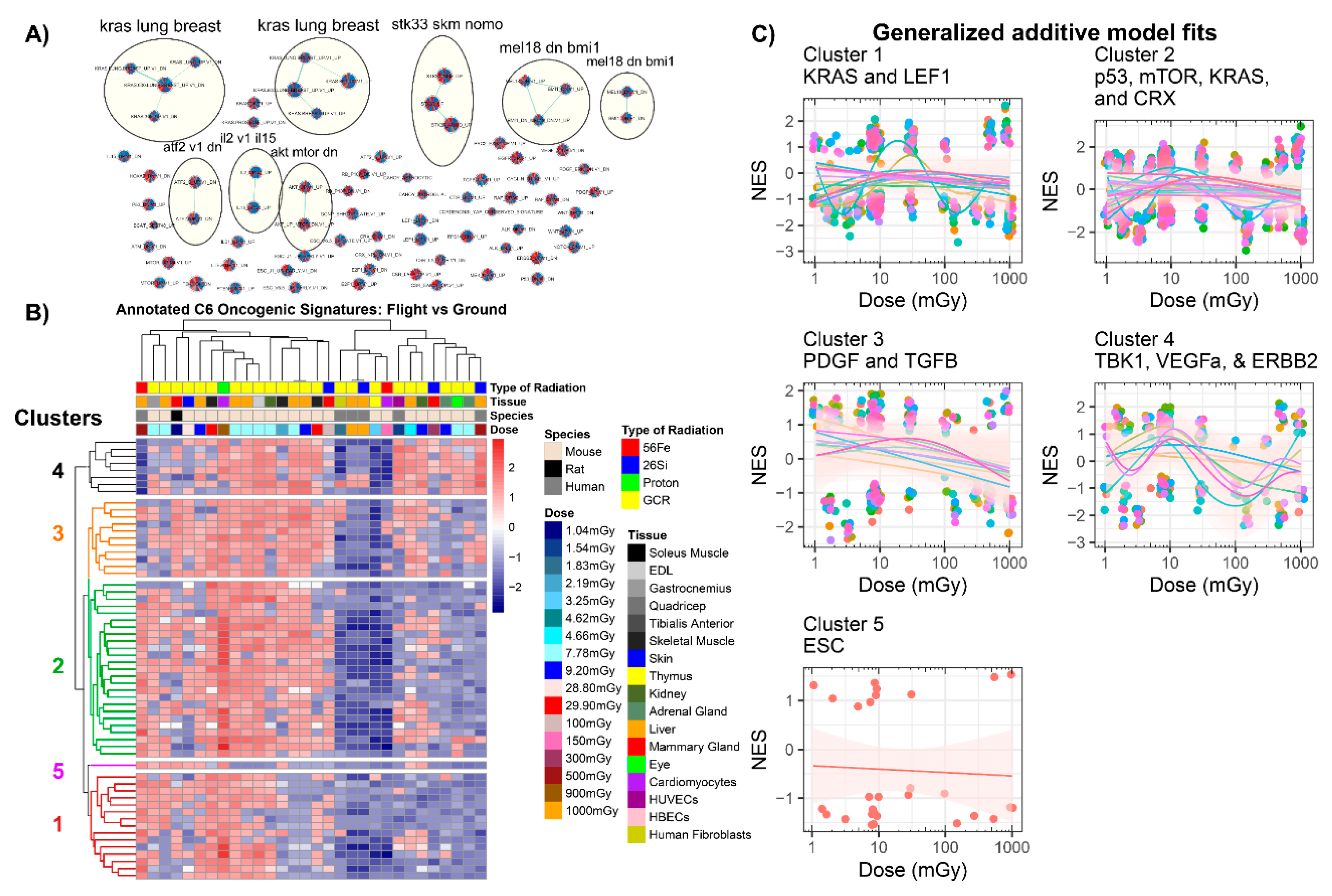
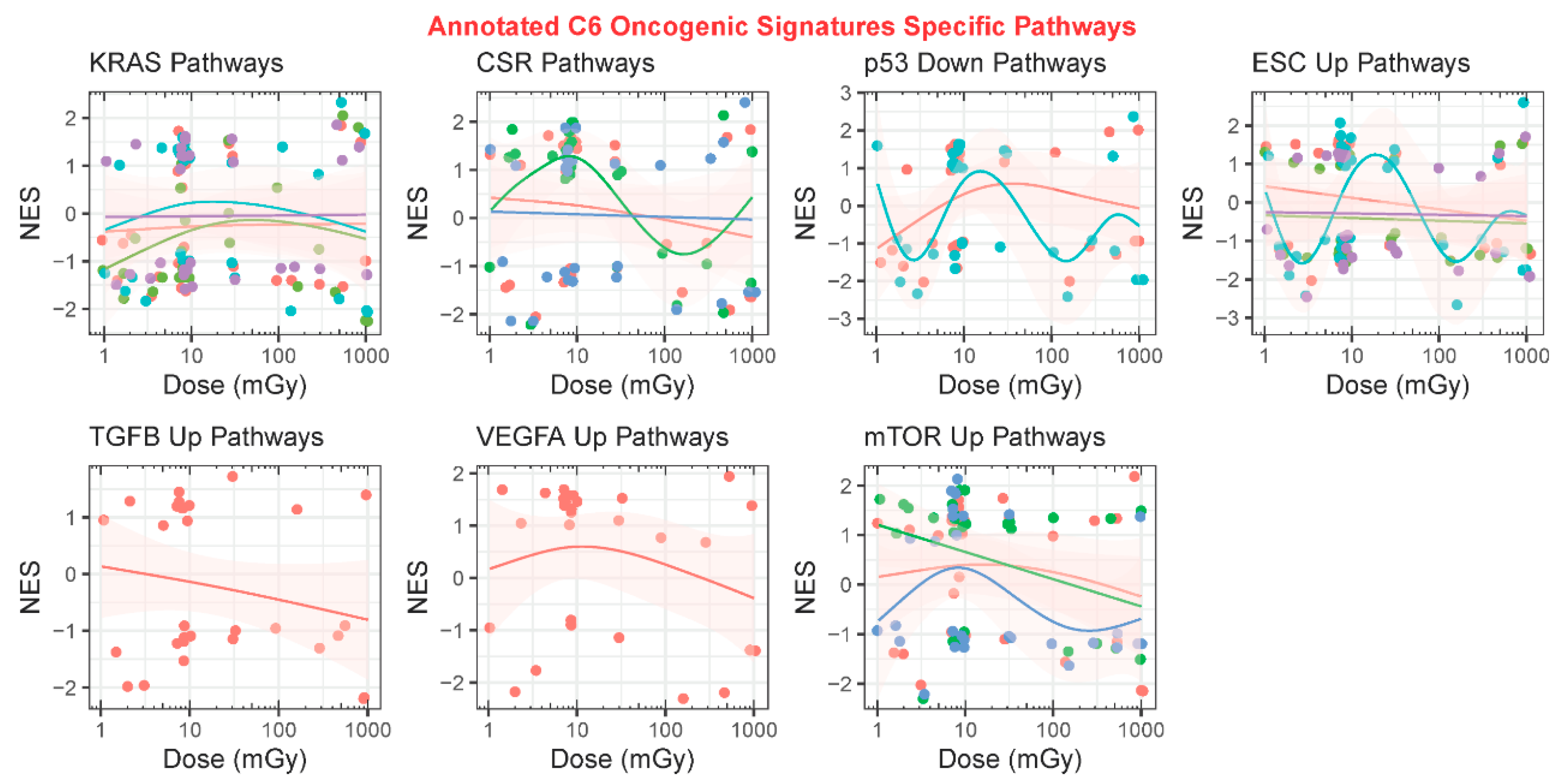
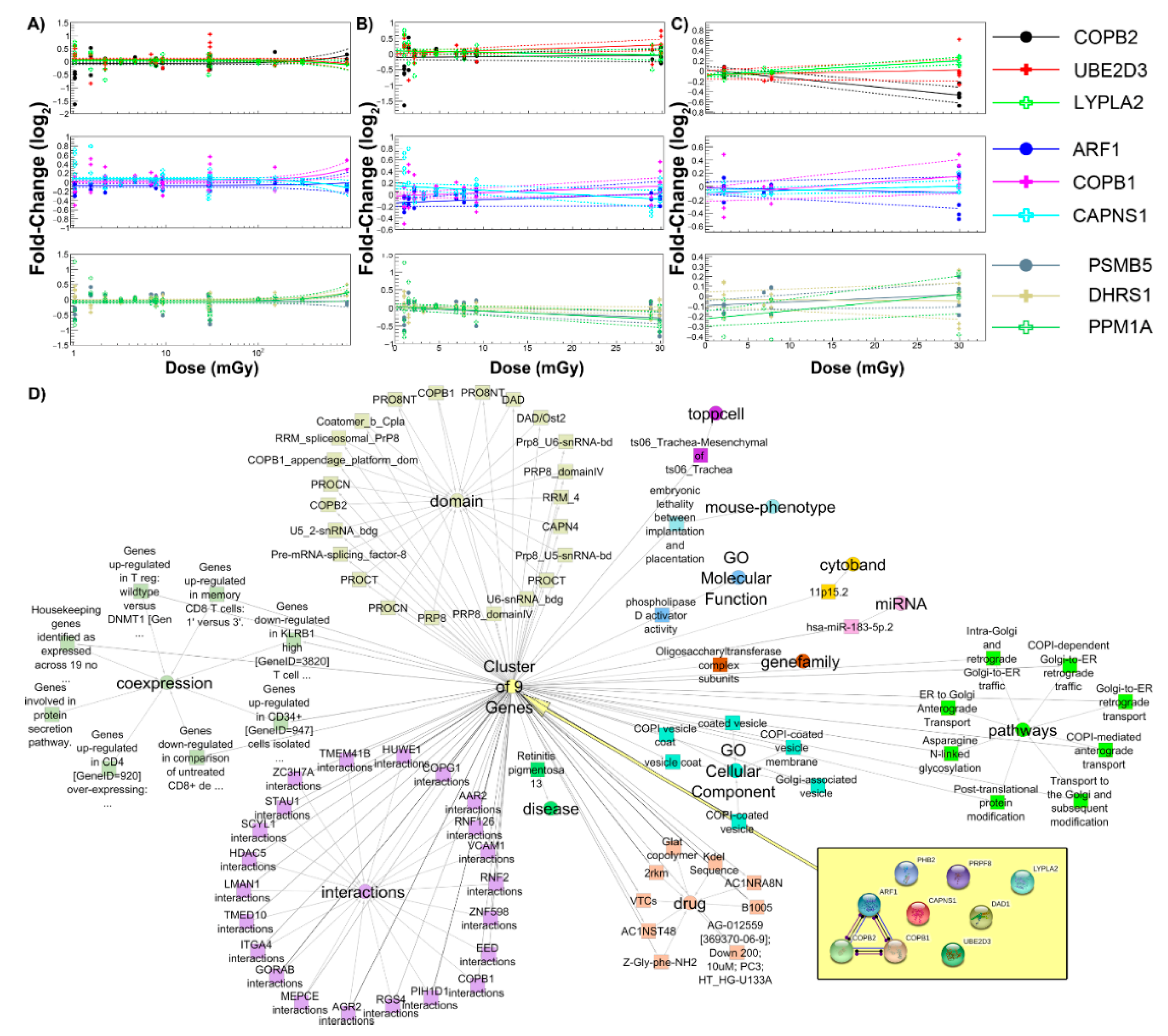
| GLDS | Condition | Dose (mGy) | Radiation Type * | Species | Tissue/Cell Culture | Ref. |
|---|---|---|---|---|---|---|
| 63 | Spaceflight | 1.04 | LEO | Rat | Mammary | [25,26,27] |
| 52 | Spaceflight | 1.54 | LEO | Human | Endothelial cells (HUVEC) | [28,29] |
| 114 | Spaceflight | 1.83 | LEO | Human | Fibroblast cells (AG01522) | [30] |
| 21 | Spaceflight | 2.19 | LEO | Mouse (C57BL/6) | Skeletal muscle | [31,32,33] |
| 4 | Spaceflight | 3.25 | LEO | Mouse (C57BL/6) | Thymus | [32,34] |
| 47 | Spaceflight | 4.62 | LEO | Mouse (C57BL/6) | Liver | [35] |
| 25 | Spaceflight | 4.66 | LEO | Mouse (C57BL/6) | Liver | [32,33,36,37,38,39,40,41,42] |
| 98 | Spaceflight | 7.78 | LEO | Mouse (C57BL/6) | Adrenal gland | [32] |
| 99 | Spaceflight | 7.78 | LEO | Mouse (C57BL/6) | Muscle (extensor digitorum longus) | [32] |
| 100 | Spaceflight | 7.78 | LEO | Mouse (C57BL/6) | Eye | [43] |
| 101 | Spaceflight | 7.78 | LEO | Mouse (C57BL/6) | Muscle (gastrocnemius) | [32] |
| 102 | Spaceflight | 7.78 | LEO | Mouse (C57BL/6) | Kidney | [32] |
| 103 | Spaceflight | 7.78 | LEO | Mouse (C57BL/6) | Muscle (quadriceps) | [32] |
| 104 | Spaceflight | 7.78 | LEO | Mouse (C57BL/6) | Muscle (soleus) | [32] |
| 105 | Spaceflight | 7.78 | LEO | Mouse (C57BL/6) | Muscle (tibialis anterior) | [32] |
| 168 | Spaceflight | 7.78 | LEO | Mouse (C57BL/6) | Liver | [42,44] |
| 242 | Spaceflight | 8.03 | LEO | Mouse (C57BL/6) | Liver | [45] |
| 161 | Spaceflight | 9.20 | LEO | Mouse (BALB/C) | Adrenal gland | [46] |
| 162 | Spaceflight | 9.20 | LEO | Mouse (BALB/C) | Eye | [47] |
| 163 | Spaceflight | 9.20 | LEO | Mouse (BALB/C) | Kidney | [48] |
| 168 | Spaceflight | 9.20 | LEO | Mouse (BALB/C) | Liver | [42,44] |
| 61 | Spaceflight | 28.80 | LEO | Mouse (C57BL/6) | Skin | [32,49] |
| 111 | Spaceflight | 29.90 | LEO | Mouse (C57BL/6) | Muscle (extensor digitorum longus) | [5,32,33] |
| 111 | Spaceflight | 29.90 | LEO | Mouse (C57BL/6) | Muscle (soleus) | [5,32,33] |
| 80 | Ground | 100 | Silicon | Mouse (BALB/C) | Mammary tissue | [50] |
| 109 | Ground | 150 | Iron | Mouse (C57BL/6) | Cardiomyocytes | [29,51] |
| 80 | Ground | 300 | Silicon | Mouse (BALB/C) | Mammary tissue | [50] |
| 73 | Ground | 500 | Iron | Human | Bronchial epithelial cells | [52] |
| 73 | Ground | 500 | Silicon | Human | Bronchial epithelial cells | [52] |
| 117 | Ground | 900 | Proton | Mouse (C57BL/6) | Cardiomyocytes | [29,51] |
| 73 | Ground | 1000 | Iron | Human | Bronchial epithelial cells | [52] |
| 73 | Ground | 1000 | Silicon | Human | Bronchial epithelial cells | [52] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
McDonald, J.T.; Stainforth, R.; Miller, J.; Cahill, T.; Silveira, W.A.d.; Rathi, K.S.; Hardiman, G.; Taylor, D.; Costes, S.V.; Chauhan, V.; et al. NASA GeneLab Platform Utilized for Biological Response to Space Radiation in Animal Models. Cancers 2020, 12, 381. https://doi.org/10.3390/cancers12020381
McDonald JT, Stainforth R, Miller J, Cahill T, Silveira WAd, Rathi KS, Hardiman G, Taylor D, Costes SV, Chauhan V, et al. NASA GeneLab Platform Utilized for Biological Response to Space Radiation in Animal Models. Cancers. 2020; 12(2):381. https://doi.org/10.3390/cancers12020381
Chicago/Turabian StyleMcDonald, J. Tyson, Robert Stainforth, Jack Miller, Thomas Cahill, Willian A. da Silveira, Komal S. Rathi, Gary Hardiman, Deanne Taylor, Sylvain V. Costes, Vinita Chauhan, and et al. 2020. "NASA GeneLab Platform Utilized for Biological Response to Space Radiation in Animal Models" Cancers 12, no. 2: 381. https://doi.org/10.3390/cancers12020381
APA StyleMcDonald, J. T., Stainforth, R., Miller, J., Cahill, T., Silveira, W. A. d., Rathi, K. S., Hardiman, G., Taylor, D., Costes, S. V., Chauhan, V., Meller, R., & Beheshti, A. (2020). NASA GeneLab Platform Utilized for Biological Response to Space Radiation in Animal Models. Cancers, 12(2), 381. https://doi.org/10.3390/cancers12020381