Cooperation between SS18-SSX1 and miR-214 in Synovial Sarcoma Development and Progression
Abstract
1. Introduction
2. Results
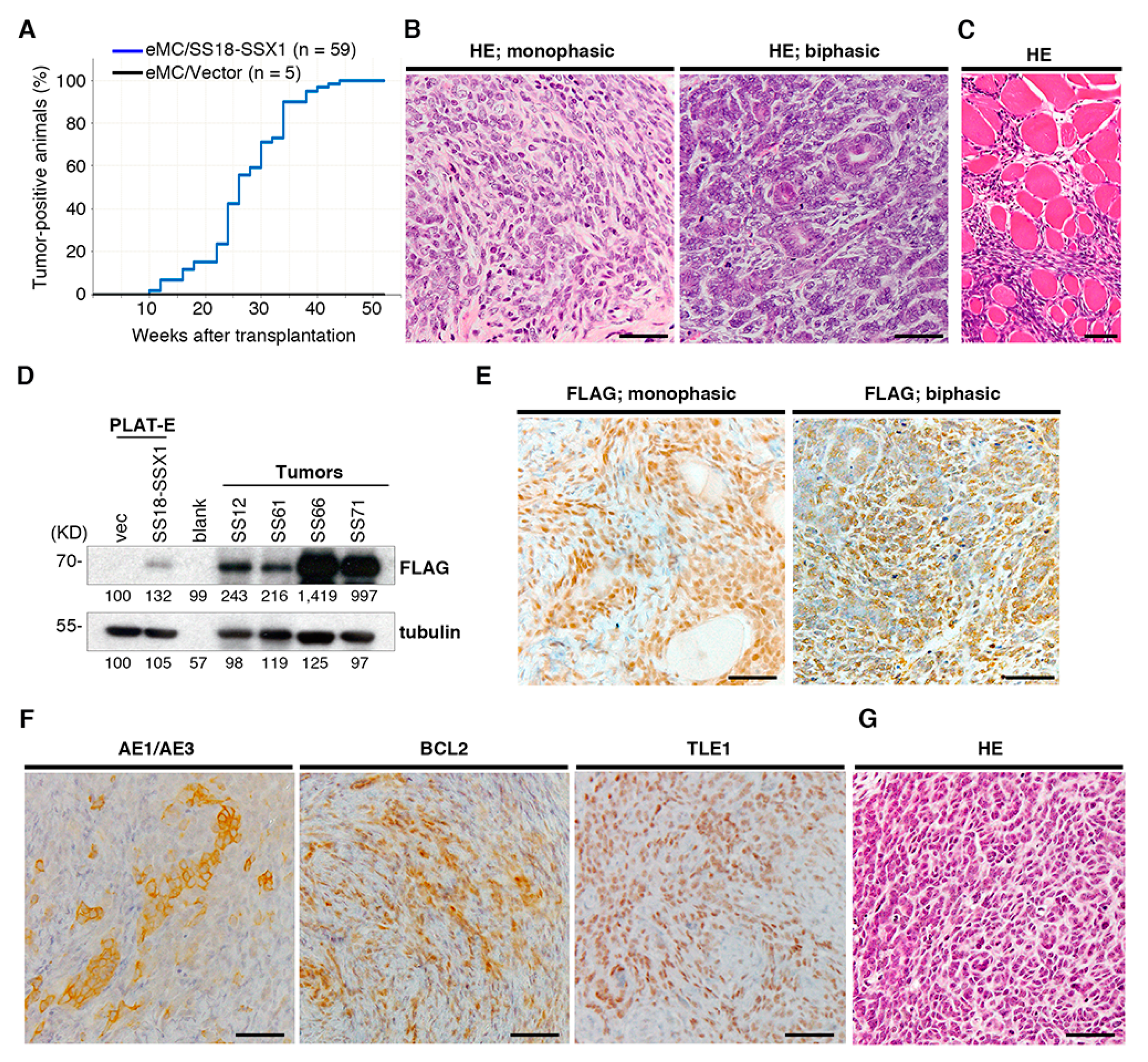
2.1. Generation and Characterization of the Ex Vivo Model of Synovial Sarcoma
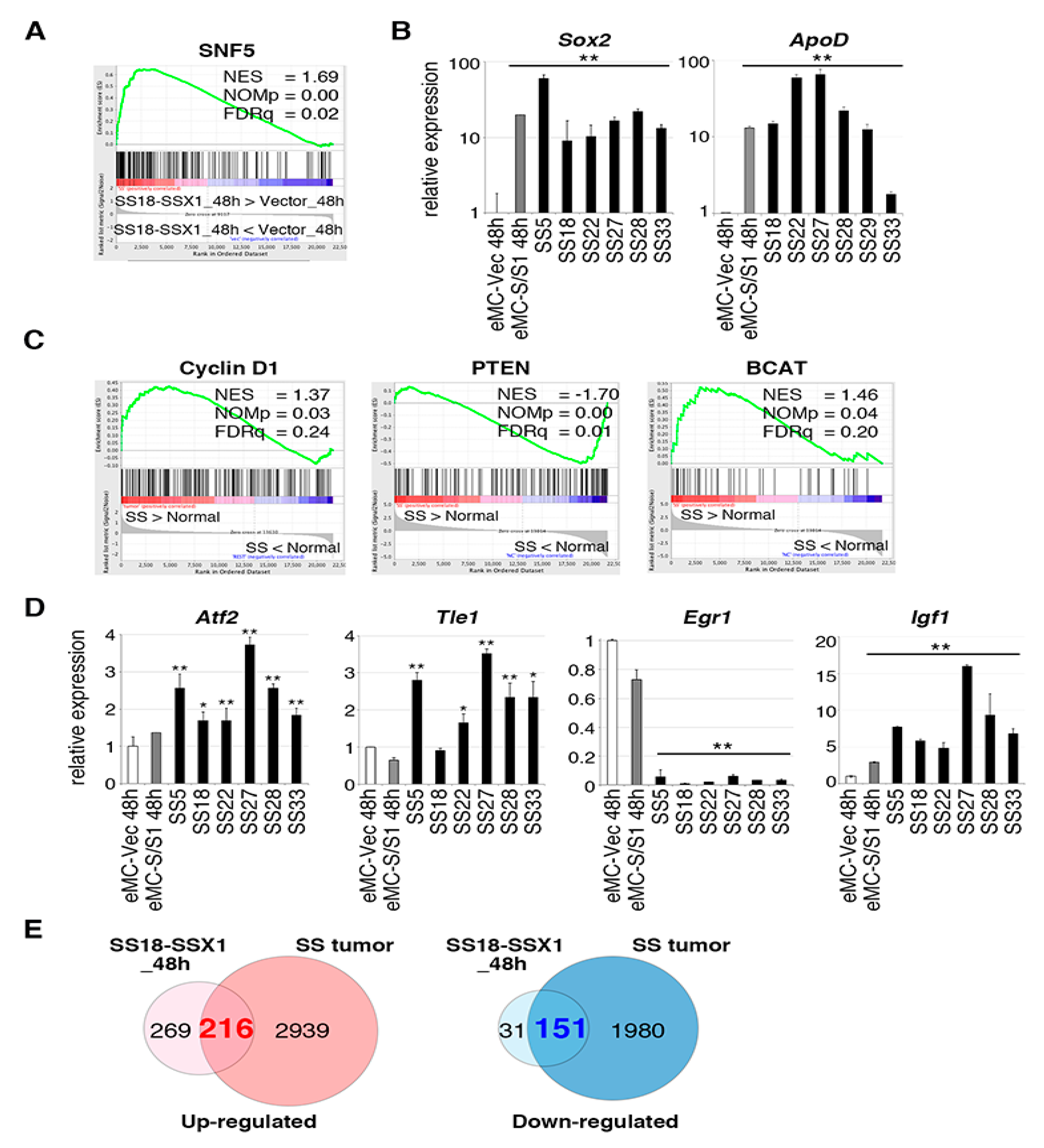
2.2. Gene Expression Profile of Mouse Synovial Sarcoma
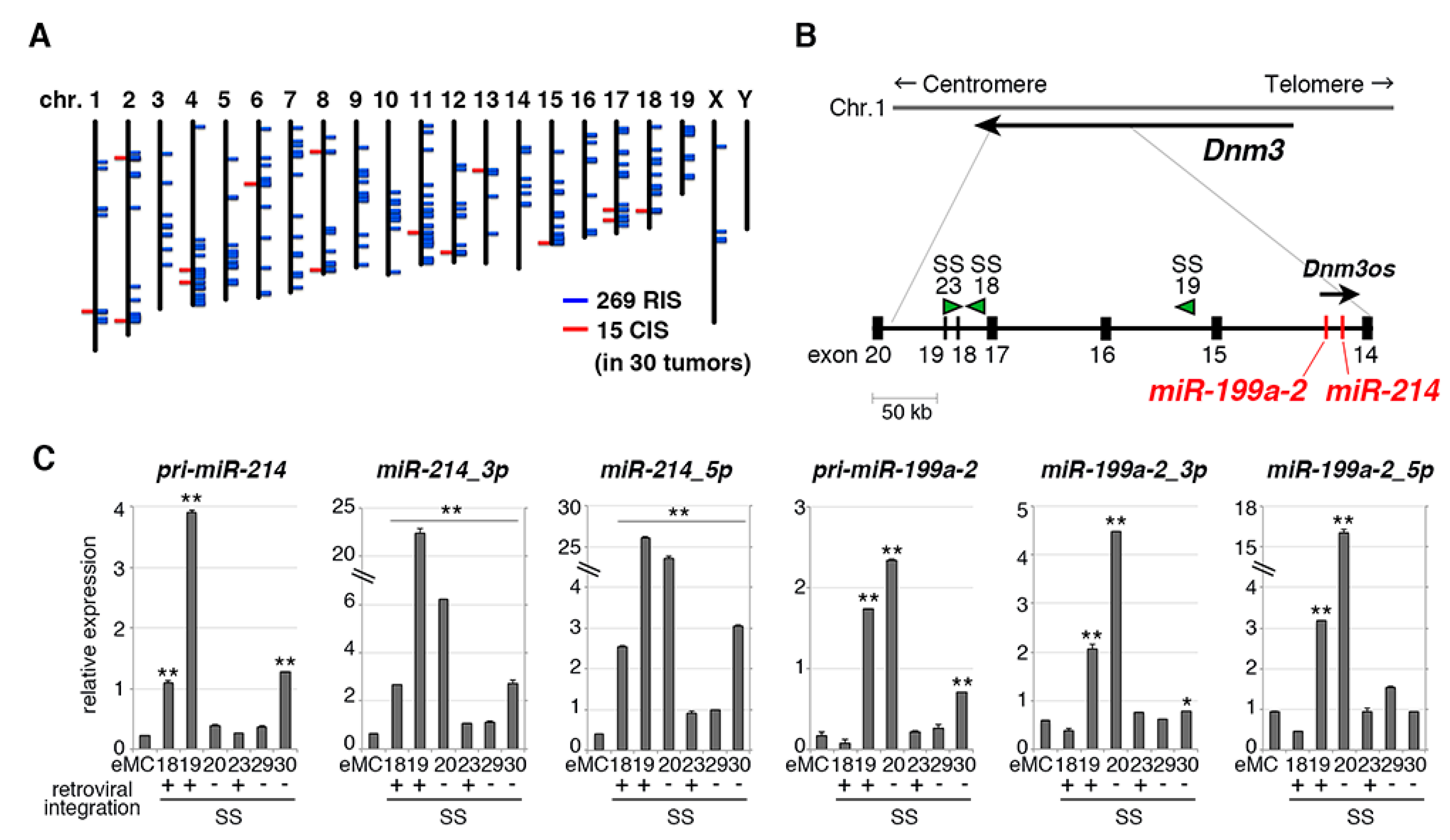
2.3. Retroviral Tagging Identifies Candidate Cooperative Genes for SS18-SSX1
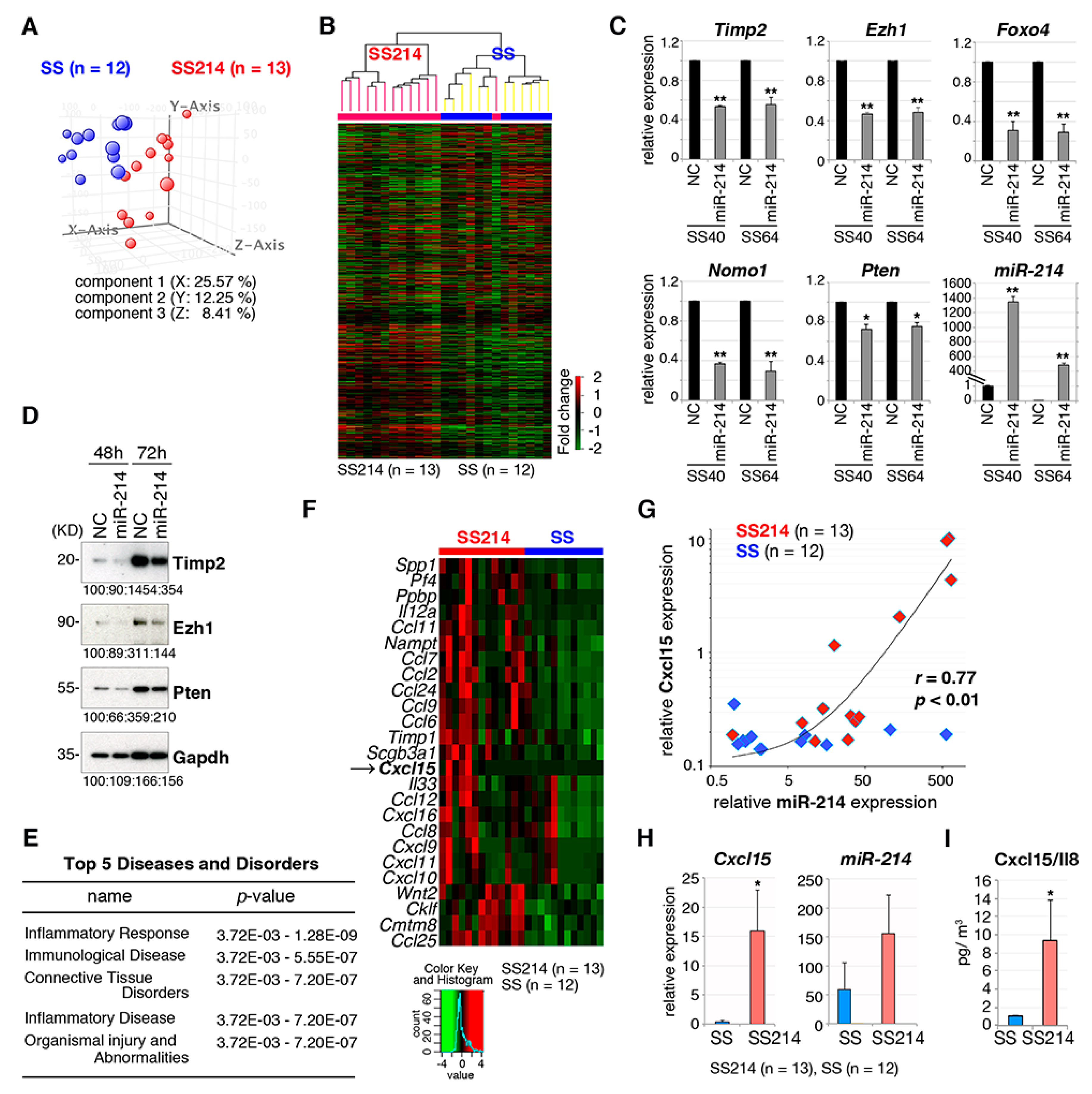
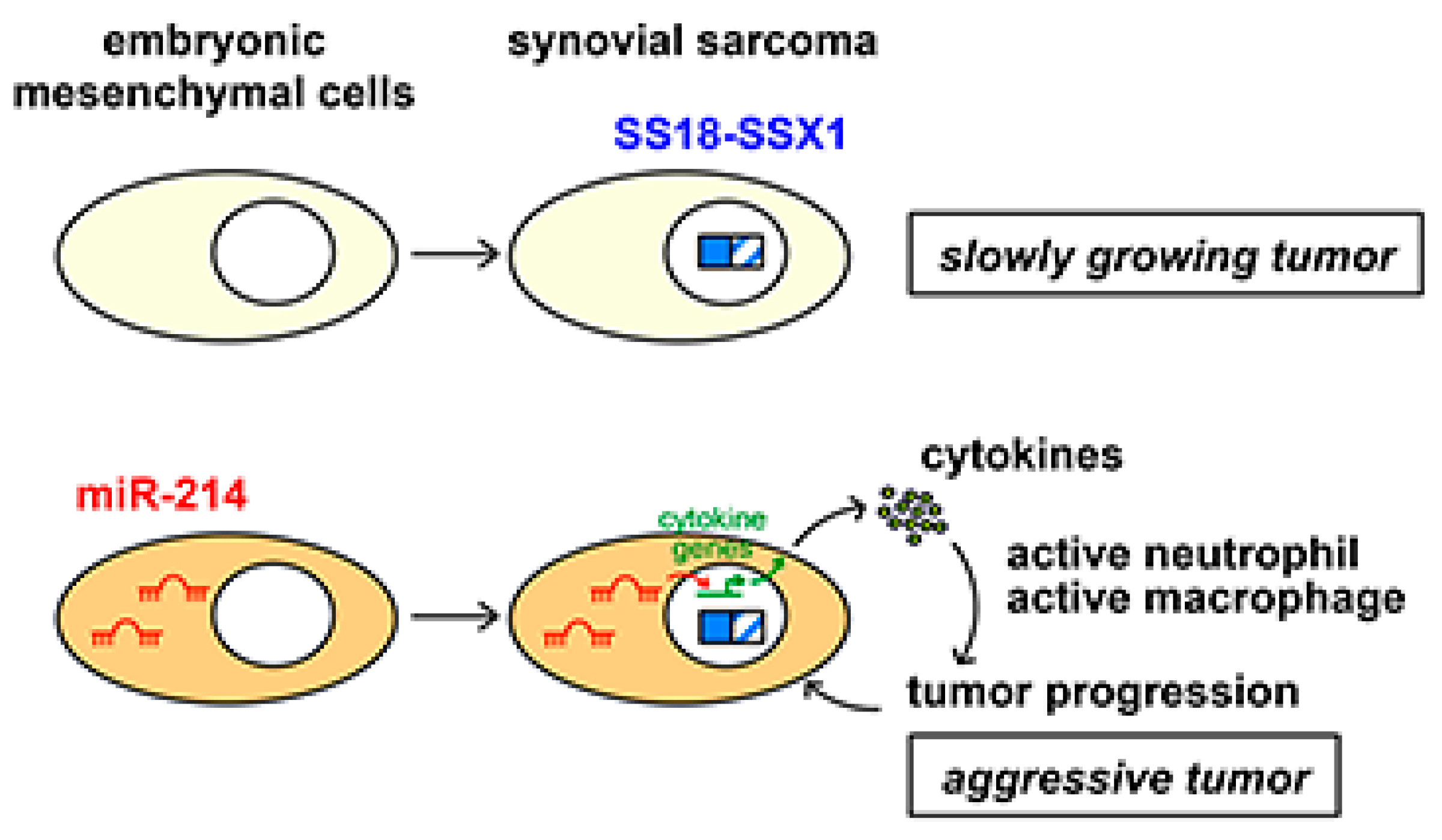
2.4. miR-214 Cooperates with SS18-SSX1 in Synovial Sarcoma Development
2.5. Effects of miR-214 Expression on Synovial Sarcoma
3. Discussion
4. Materials and Methods
4.1. Plasmid Construction
4.2. Generation and Characterization of the Mouse Synovial Sarcoma Model
4.3. Human Sarcoma Specimens
4.4. Histology and Immunohistochemistry
4.5. Immunoblotting
4.6. Microarray Analysis
4.7. Real-Time Quantitative Polymerase Chain Reaction (RT-qPCR)
4.8. Cloning of Retroviral Integration Sites
4.9. In Situ Hybridization
4.10. ELISA
4.11. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Enzinger, F.M.; Weiss, S.W. Synovial sarcoma. In Soft Tissue Tumors, 5th ed.; Weiss, S.W., Goldblum, J.R., Eds.; Elsevier: Philadelphia, PA, USA, 2008; pp. 1161–1182. [Google Scholar]
- Clark, J.; Rocques, P.J.; Crew, A.J.; Gill, S.; Shipley, J.; Chan, A.M.; Gusterson, B.A.; Cooper, C.S. Identification of novel genes, SYT and SSX, involved in the t(X;18)(p11.2;q11.2) translocation found in human synovial sarcoma. Nat. Genet. 1994, 7, 502–508. [Google Scholar] [CrossRef] [PubMed]
- Kawai, A.; Woodruff, J.; Healey, J.H.; Brennan, M.F.; Antonescu, C.R.; Ladanyi, M. SYT-SSX gene fusion as a determinant of morphology and prognosis in synovial sarcoma. N. Engl. J. Med. 1998, 338, 153–160. [Google Scholar] [CrossRef] [PubMed]
- Ladanyi, M. Fusions of the SYT and SSX genes in synovial sarcoma. Oncogene 2001, 20, 5755–5762. [Google Scholar] [CrossRef]
- Nielsen, T.O.; Poulin, N.M.; Ladanyi, M. Synovial sarcoma: Recent discoveries as a roadmap to new avenues for therapy. Cancer Discov. 2015, 4, 123–134. [Google Scholar] [CrossRef]
- Kadoch, C.; Crabtree, G.G. Reversible disruption of mSWI/SNF (BAF) complexes by the SS18-SSX oncogenic fusion in synovial sarcoma. Cell 2013, 153, 71–85. [Google Scholar] [CrossRef]
- McBride, M.J.; Pulice, J.L.; Beird, H.C.; Ingram, D.R.; D’Avino, A.R.; Shern, J.F.; Charville, G.W.; Hornick, J.L.; Nakayama, R.T.; Garcia-Rivera, E.M.; et al. The SS18-SSX fusion oncoprotein hijacks BAF complex targeting and function to drive synovial sarcoma. Cancer Cell 2018, 33, 1128–1141. [Google Scholar] [CrossRef]
- Su, L.; Sampaio, A.V.; Jones, K.B.; Pacheco, M.; Goytain, A.; Lin, S.; Poulin, N.; Yi, L.; Rossi, F.M.; Kast, J.; et al. Deconstruction of the SS18-SSX fusion oncoprotein complex: Insights into disease etiology and therapeutics. Cancer Cell 2012, 21, 333–347. [Google Scholar] [CrossRef]
- Barham, W.; Frump, A.L.; Sherill, T.P.; Garcia, C.B.; Saito-Diaz, K.; VanSaun, M.N.; Fingleton, B.; Gleaves, L.; Orton, D.; Capecchi, M.R.; et al. Targeting the Wnt pathway in synovial sarcoma models. Cancer Discov. 2013, 3, 1286–1301. [Google Scholar] [CrossRef]
- Banito, A.; Li, X.; Laporte, A.N.; Roe, J.S.; Sanchez-Vega, F.; Huang, C.H.; Dancsok, A.R.; Hatzi, K.; Chen, C.C.; Tschaharganeh, D.F.; et al. The SS18-SSX oncoprotein hijacks KDM2B-PRC1.1 to drive synovial sarcoma. Cancer Cell 2018, 33, 527–541. [Google Scholar] [CrossRef] [PubMed]
- Vlenterie, M.; Hillebrandt-Roeffen, M.H.S.; Flucke, U.E.; Gronen, P.J.T.A.; Tops, B.B.J.; Kamping, E.J.; Pfundt, R.; de Bruijn, D.R.H.; van Kessel, A.H.M.G.; van Krieken, H.J.H.J.M.; et al. Next generation sequencing in synovial sarcoma reveals novel gene mutations. Oncotarget 2015, 6, 34680–34690. [Google Scholar] [CrossRef]
- Haldar, M.; Hancock, J.D.; Coffin, C.M.; Lessnick, S.L.; Capecchi, M.R. A conditional mouse model of synovial sarcoma: Insights into a myogenic origin. Cancer Cell 2007, 11, 375–388. [Google Scholar] [CrossRef]
- Tanaka, M.; Yamazaki, Y.; Kanno, Y.; Igarashi, K.; Aisaki, K.; Kanno, J.; Nakamura, T. Ewing’s sarcoma precursors are highly enriched in embryonic osteochondrogenic progenitors. J. Clin. Investig. 2014, 121, 3061–3074. [Google Scholar] [CrossRef]
- Tanaka, M.; Homme, M.; Yamazaki, Y.; Shimizu, R.; Takazawa, Y.; Nakamura, T. Modeling alveolar soft part sarcoma unveils novel mechanisms of metastasis. Cancer Res. 2017, 77, 897–904. [Google Scholar] [CrossRef] [PubMed]
- Yoshimoto, T.; Tanaka, M.; Homme, M.; Yamazaki, Y.; Takazawa, Y.; Antonescu, C.R.; Nakamura, T. CIC-DUX4 induces small round cell sarcoma distinct from Ewing sarcoma. Cancer Res. 2017, 77, 2927–2937. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, S.; Lui, W.O.; Lee, C.H.; Espinosa, I.; Nielsen, T.O.; Heinrich, M.C.; Corless, C.L.; Fire, A.Z.; van de Rijn, M. MicroRNA expression signature of human sarcomas. Oncogene 2008, 27, 2015–2026. [Google Scholar] [CrossRef]
- Terry, J.; Saito, T.; Subramanian, S.; Ruttan, C.; Antonescu, C.R.; Goldblum, J.R.; Downs-Kelly, E.; Corless, C.L.; Rubin, B.P.; van de Rijn, M.; et al. TLE1 as a diagnostic immunohistochemical marker for synovial sarcoma emerging from gene expression profiling studies. Am. J. Surg. Pathol. 2007, 31, 240–246. [Google Scholar] [CrossRef] [PubMed]
- Krskova, L.; Kalinova, M.; Brizova, H.; Mrhalova, M.; Sumerauer, D.; Kodet, R. Molecular and immunohistochemical analyses of BCL2, KI-67, and cyclin D1 expression in synovial sarcoma. Cancer Genet. Cytogenet. 2009, 193, 1–8. [Google Scholar] [CrossRef]
- Isakoff, M.S.; Sansam, C.G.; Tamayo, P.; Subramanian, A.; Evans, J.A.; Fillmore, C.M.; Wang, X.; Biegel, J.A.; Pomeroy, S.L.; Mesirov, J.P.; et al. Inactivation of the Snf5 tumor suppressor stimulates cell cycle progression and cooperates with p53 loss in oncogenic transformation. Proc. Natl. Acad. Sci. USA 2005, 102, 17445–17450. [Google Scholar] [CrossRef] [PubMed]
- Jones, K.B.; Su, L.; Jin, H.; Lenz, C.; Randall, R.L.; Underhill, T.M.; Nielsen, T.O.; Sharma, S.; Capecchi, M.R. SS18-SSX2 and the mitochondrial apoptosis pathway in mouse and human synovial sarcoma. Oncogene 2013, 32, 2365–2371. [Google Scholar] [CrossRef]
- Endo, M.; Su, L.; Nielsen, T.O. Activating transcription factor 2 in mesenchymal tumors. Hum. Pathol. 2014, 45, 276–284. [Google Scholar] [CrossRef]
- Lanzi, C.; Dal Bo, L.; Favini, E.; Tortoreto, M.; Beretta, G.L.; Arrighetti, N.; Zaffaroni, N.; Cassinelli, C. Overactive IGF1/insulin receptors and NRASQ61R mutation drive mechanisms of resistance to Pazopanib and define rational combination strategies to treat synovial sarcoma. Cancers 2019, 11, E408. [Google Scholar] [CrossRef]
- Nakamura, T. Retroviral insertional mutagenesis identifies oncogene cooperation. Cancer Sci. 2005, 96, 7–12. [Google Scholar] [CrossRef]
- Jin, G.; Yamazaki, Y.; Takuwa, M.; Takahara, T.; Kaneko, K.; Kuwata, T.; Miyata, S.; Nakamura, T. Trib1 and Evi1 cooperate with Hoxa and Meis1 in myeloid leukemogenesis. Blood 2007, 109, 3998–4005. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, M.; Jin, G.; Yamazaki, Y.; Takahara, T.; Takuwa, M.; Nakamura, T. Identification of candidate cooperative genes of the Apc mutation in transformation of the colon epithelial cell by retroviral insertional mutagenesis. Cancer Sci. 2008, 99, 979–985. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, T.; Sato, T.; Amano, T.; Kawamura, Y.; Kawamura, N.; Kawaguchi, H.; Yamashita, N.; Kurihara, H.; Nakaoka, T. Dnm3os, a non-coding RNA, is required for normal growth and skeletal development in mice. Dev. Dyn. 2008, 237, 3738–3748. [Google Scholar]
- Desvignes, T.; Contreras, A.; Postlethwair, J.H. Evolution of the miR199-214 cluster and vertebrate skeletal development. RNA Biol. 2014, 11, 281–294. [Google Scholar] [CrossRef]
- Loebel, D.A.; Tsoi, B.; Wong, N.; Tam, P.P. A conserved noncoding intronic transcript at the mouse Dnm3 locus. Genomics 2005, 85, 782–789. [Google Scholar] [CrossRef]
- Roberto, V.P.; Gavaia, P.; Nunes, M.J.; Rodrigues, E.; Cancela, M.L.; Tiago, D.M. Evidences for a new role of miR-214 in chondrogenesis. Sci. Rep. 2018, 8, 3704. [Google Scholar] [CrossRef]
- Yin, G.; Chen, R.; Alvero, A.B.; Fu, H.H.; Holmberg, J.; Glackin, C.; Rutherford, T.; Mor, G. TWISTing stemness, inflammation and proliferation of epithelial ovarian cancer cells through MIR199A2/214. Oncogene 2010, 29, 3545–3553. [Google Scholar] [CrossRef] [PubMed]
- Hirata, M.; Asano, N.; Katayama, K.; Yoshida, A.; Tsuda, Y.; Sekimizu, M.; Mitani, S.; Kobayashi, E.; Komiyama, M.; Fujimoto, H.; et al. Integrated exome and RNA sequencing of dedifferentiated liposarcoma. Nat. Commun. 2020, in press. [Google Scholar] [CrossRef]
- Yang, H.; Kong, W.; He, L.; Zhao, J.J.; O’Donell, J.D.; Wang, J.; Wenham, R.M.; Coppola, D.; Kruk, P.A.; Nicosia, S.V.; et al. MicroRNA expression profiling in human ovarian cancer: miR-214 induces cell survival and cisplatin resistance by targeting PTEN. Cancer Res. 2008, 68, 425–433. [Google Scholar] [CrossRef] [PubMed]
- Barrott, J.J.; Kafchinski, L.A.; Jin, H.; Potter, J.W.; Kannan, S.D.; Kennedy, R.; Mosbruger, T.; Wang, W.L.; Tsai, J.W.; Araujo, D.M.; et al. Modeling synovial sarcoma metastasis in the mouse: PI3’-lipid signaling and inflammation. J. Exp. Med. 2016, 213, 2989–3005. [Google Scholar] [CrossRef] [PubMed]
- Fang, H.Y.; Greten, F.R. Cell autonomous and non-autonomous functions of IKK and NF- B during the pathogenesis of gastrointestinal tumors. Cancers 2011, 3, 2214–2222. [Google Scholar] [CrossRef]
- Bellazzo, A.; Di Minin, G.; Valentino, E.; Sicari, D.; Torre, D.; Marchionni, L.; Serpi, F.; Stadler, M.B.; Taverna, D.; Zuccolotto, G.; et al. Cell-autonomous and cell non-autonomous downregulation of tumor suppressor DAB2IP by microRNA-149-3p promotes aggressiveness of cancer cells. Cell Death Differ. 2018, 25, 1224–1238. [Google Scholar] [CrossRef]
- Baggiolini, M.; Clark-Lewis, I. Interleukin-8, a chemotactic and inflammatory cytokine. FEBS Lett. 1992, 307, 97–101. [Google Scholar] [CrossRef]
- Xie, K. Interleukin-8 and human cancer biology. Cytokine Growth Factor Rev. 2001, 12, 375–391. [Google Scholar] [CrossRef]
- Abdul-Aziz, A.M.; Shafat, M.S.; Mehta, T.K.; Di Palma, F.; Lawes, M.J.; Rushworth, S.A.; Bowles, K.M. MIF-induced stromal PKC/IL8 is essential in human acute myeloid leukemia. Cancer Res. 2017, 77, 303–311. [Google Scholar] [CrossRef] [PubMed]
- Manfroi, B.; McKee, T.; Mayol, J.F.; Tabruyn, S.; Moret, S.; Villiers, C.; Righini, C.; Dyer, M.; Callanan, M.; Schneider, P.; et al. CXCL-8/IL8 produced by diffuse large B-cell lymphomas recruits neutrophils expressing a proliferation-inducing ligand APRIL. Cancer Res. 2017, 77, 1097–1107. [Google Scholar] [CrossRef] [PubMed]
- Kahraman, D.C.; Kahraman, T.; Cetin-Atalay, R. Targeting PI3K/Akt/mTOR pathway identifies differential expression and functional role of IL8 in liver cancer stem cell enrichment. Mol. Cancer Ther. 2019, 18, 2146–2157. [Google Scholar] [PubMed]
- Maynard, J.P.; Ertunc, O.; Kulac, I.; Baena-Del Valle, J.A.; De Marzo, A.M.; Sfanos, K.S. IL8 expression is associated with prostate cancer aggressiveness and androgen receptor loss in primary and metastatic prostate cancer. Mol. Cancer Res. 2019. [Google Scholar] [CrossRef]
- Nie, M.; Yang, L.; Bi, X.; Wang, Y.; Sun, P.; Yang, H.; Liu, P.; Li, Z.; Xia, Y.; Jiang, W. Neutrophil extracellular traps induced by IL8 promote diffuse large B-cell lymphoma progression via the TLR9 signaling. Clin. Cancer Res. 2019, 25, 1867–1879. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Hua, Y.; Jiang, Z.; Yue, J.; Shi, M.; Zhen, X.; Zhang, X.; Yang, L.; Zhou, R.; Wu, S. Cancer-associated fibroblast-promoted lnc RNA DNM3OS confers radioresistance by regulating DNA damage response in esophageal squamous cell carcinoma. Clin. Cancer Res. 2019, 25, 1989–2000. [Google Scholar] [CrossRef]
- Zhao, L.; Liu, Y.W.; Yang, T.; Gan, L.; Yamg, N.; Dai, S.S.; He, F. The mutual regulation between miR-214 and A2AR signaling plays an important role in inflammatory response. Cell Signal. 2015, 27, 2026–2034. [Google Scholar] [CrossRef]
- Kuninty, P.R.; Bojmar, L.; Tjomsland, V.; Larsson, M.; Storm, G.; Ostman, A.; Sandstrom, P.; Prakash, J. MicroRNA-199a and -214 as potential therapeutic targets in pancreatic stellate cells in pancreatic tumor. Oncotarget 2016, 7, 16396–16408. [Google Scholar] [CrossRef]
- Imamura, K.; Imamachi, N.; Akizuki, G.; Mumakura, M.; Kawaguchi, A.; Nagata, K.; Kato, A.; Kawaguchi, Y.; Sato, H.; Yoneda, M.; et al. Long noncoding RNA NEAT1-dependent SFPQ relocation from promoter region to paraspeckle mediates IL8 expression upon immune stimuli. Mol. Cell 2014, 53, 393–406. [Google Scholar] [CrossRef]
- Kawamura-Saito, M.; Yamazaki, Y.; Kaneko, K.; Kawaguchi, N.; Kanda, H.; Mukai, H.; Gotoh, T.; Motoi, T.; Fukayama, M.; Takizawa, T.; et al. Fusion between CIC and DUX4 up-regulates PEA3 family genes in Ewing-like sarcomas with t(4;19)(q35;q13) translocation. Hum. Mol. Genet. 2006, 15, 2125–2137. [Google Scholar] [CrossRef]
- Subramanian, A.; Kuehn, H.; Gould, J.; Tamayo, P.; Mesirov, J.P. GSEA-P: A desktop application for gene set enrichment analysis. Bioinformatics 2007, 23, 3251–3253. [Google Scholar] [CrossRef] [PubMed]
- Nakayama, R.; Mitani, S.; Nakagawa, T.; Hasegawa, T.; Kawai, A.; Morioka, H.; Yabe, H.; Toyama, Y.; Ogose, A.; Toguchida, J.; et al. Gene expression profiling of synovial sarcoma: Distinct signature of poorly differentiated type. Am. J. Surg. Pathol. 2010, 34, 1599–1607. [Google Scholar] [CrossRef]
- Vella, S.; Tavanti, E.; Hattinger, C.M.; Fanelli, M.; Versteeg, R.; Koster, J.; Picci, P.; Serra, M. Targeting CDKs with Roscovitine increases sensitivity to DNA damaging drugs of human osteosarcoma cells. PLoS ONE 2016, 11, e0166233. [Google Scholar] [CrossRef] [PubMed]
- Scotlandi, K.; Remondini, D.; Castellani, G.; Manara, M.C.; Nardi, F.; Cantiani, L.; Francesconi, M.; Mercuri, M.; Caccuri, A.M.; Serra, M.; et al. Overcoming resistance to concventional drugs in Ewing sarcoma and identification of molecular predictors of outcome. J. Clin. Oncol. 2009, 27, 2209–2216. [Google Scholar] [CrossRef]
- Kummar, S.; Allen, D.; Monks, A.; Polley, E.C.; Hose, C.D.; Ivy, S.P.; Turkbey, I.B.; Lawrence, S.; Kinders, R.J.; Choyke, P.; et al. Cediranib for metastatic alveolar soft part sarcoma. J. Clin. Oncol. 2013, 31, 2296–2302. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Chatterjee, B.; Wang, Y.; Stevenson, H.S.; Edelman, D.C.; Meltzer, P.S.; Barr, F.G. Distinct methylation profiles characterize fusion-positive and fusion-negative rhabdomyosarcoma. Mod. Pathol. 2015, 28, 1214–1224. [Google Scholar] [CrossRef] [PubMed]

| Chromosome Locus | Loci | No. of Integrations (n = 30) |
|---|---|---|
| 1qH2.1 | Dnm3 | 3 |
| 2qH3 | Tshz2 | 2 |
| 2qB | Cstad | |
| 4qD2.2 | Stk40 | |
| 4qD3 | Hspg2 | |
| 6qB3 | Creb5 | |
| 8qA2 | Fgfr1 | |
| 8qE2 | Irf2bp2 | |
| 11qD | Col1a1 | |
| 12qF1 | Trmt61a | |
| 13qA4 | Nedd9 | |
| 15qF3 | Itga5 | |
| 17qE2 | Ltbp1 | |
| 17qE4 | 4933433H22Rik | |
| 18qE3 | Zbtb7c |
| Genes | Fold Change | Products |
|---|---|---|
| Cxcl15 | 12.34 | chemokine (C-X-C motif) ligand 15 |
| Ppbp | 5.80 | pro-platelet basic protein |
| Spp1 | 4.31 | secreted phosphoprotein 1 |
| Ccl6 | 3.63 | chemokine (C-C motif) ligand 6 |
| Ccl9 | 3.57 | chemokine (C-C motif) ligand 9 |
| Ccl11 | 2.48 | chemokine (C-C motif) ligand 11 |
| Cxcl9 | 2.21 | chemokine (C-X-C motif) ligand 9 |
| Ccl2 | 2.16 | chemokine (C-C motif) ligand 2 |
| Pf4 | 2.13 | platelet factor 4 |
| Ccl7 | 1.88 | chemokine (C-C motif) ligand 7 |
| Genes | Fold Change | Products |
|---|---|---|
| BMP7 | 6.79 | bone morphogenetic protein 7 |
| BMP5 | 4.48 | bone morphogenetic protein 5 |
| WNT5A | 4.35 | wingless-type MMTV integration site family, member 5A |
| CXCL12 | 3.35 | chemokine (C-X-C motif) ligand 12 |
| TNFRSF19 | 2.75 | tumor necrosis factor receptor superfamily, member 19 |
| BMP2 | 2.46 | bone morphogenetic protein 2 |
| TGFB2 | 2.14 | transforming growth factor beta 2 |
| CXCL8 | 1.86 | chemokine (C-X-C motif) ligand 8 |
| FAM19A1 | 1.63 | family with sequence similarity 19 (chemokine (C-C motif)-like), member A1 |
| CCL28 | 1.60 | chemokine (C-C motif) ligand 28 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tanaka, M.; Homme, M.; Yamazaki, Y.; Ae, K.; Matsumoto, S.; Subramanian, S.; Nakamura, T. Cooperation between SS18-SSX1 and miR-214 in Synovial Sarcoma Development and Progression. Cancers 2020, 12, 324. https://doi.org/10.3390/cancers12020324
Tanaka M, Homme M, Yamazaki Y, Ae K, Matsumoto S, Subramanian S, Nakamura T. Cooperation between SS18-SSX1 and miR-214 in Synovial Sarcoma Development and Progression. Cancers. 2020; 12(2):324. https://doi.org/10.3390/cancers12020324
Chicago/Turabian StyleTanaka, Miwa, Mizuki Homme, Yukari Yamazaki, Keisuke Ae, Seiichi Matsumoto, Subbaya Subramanian, and Takuro Nakamura. 2020. "Cooperation between SS18-SSX1 and miR-214 in Synovial Sarcoma Development and Progression" Cancers 12, no. 2: 324. https://doi.org/10.3390/cancers12020324
APA StyleTanaka, M., Homme, M., Yamazaki, Y., Ae, K., Matsumoto, S., Subramanian, S., & Nakamura, T. (2020). Cooperation between SS18-SSX1 and miR-214 in Synovial Sarcoma Development and Progression. Cancers, 12(2), 324. https://doi.org/10.3390/cancers12020324






