Cancer Glycolytic Dependence as a New Target of Olive Leaf Extract
Abstract
1. Introduction
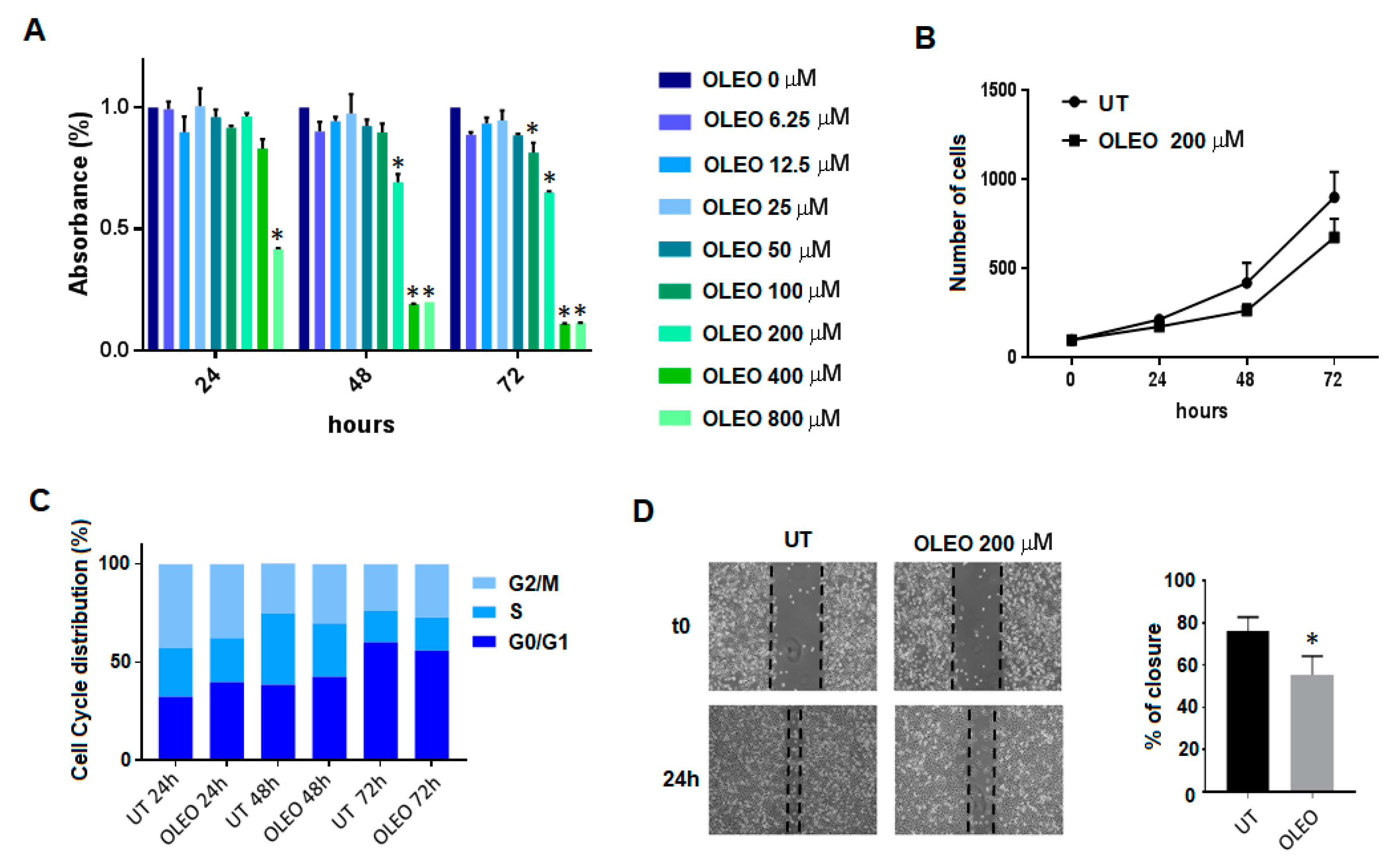
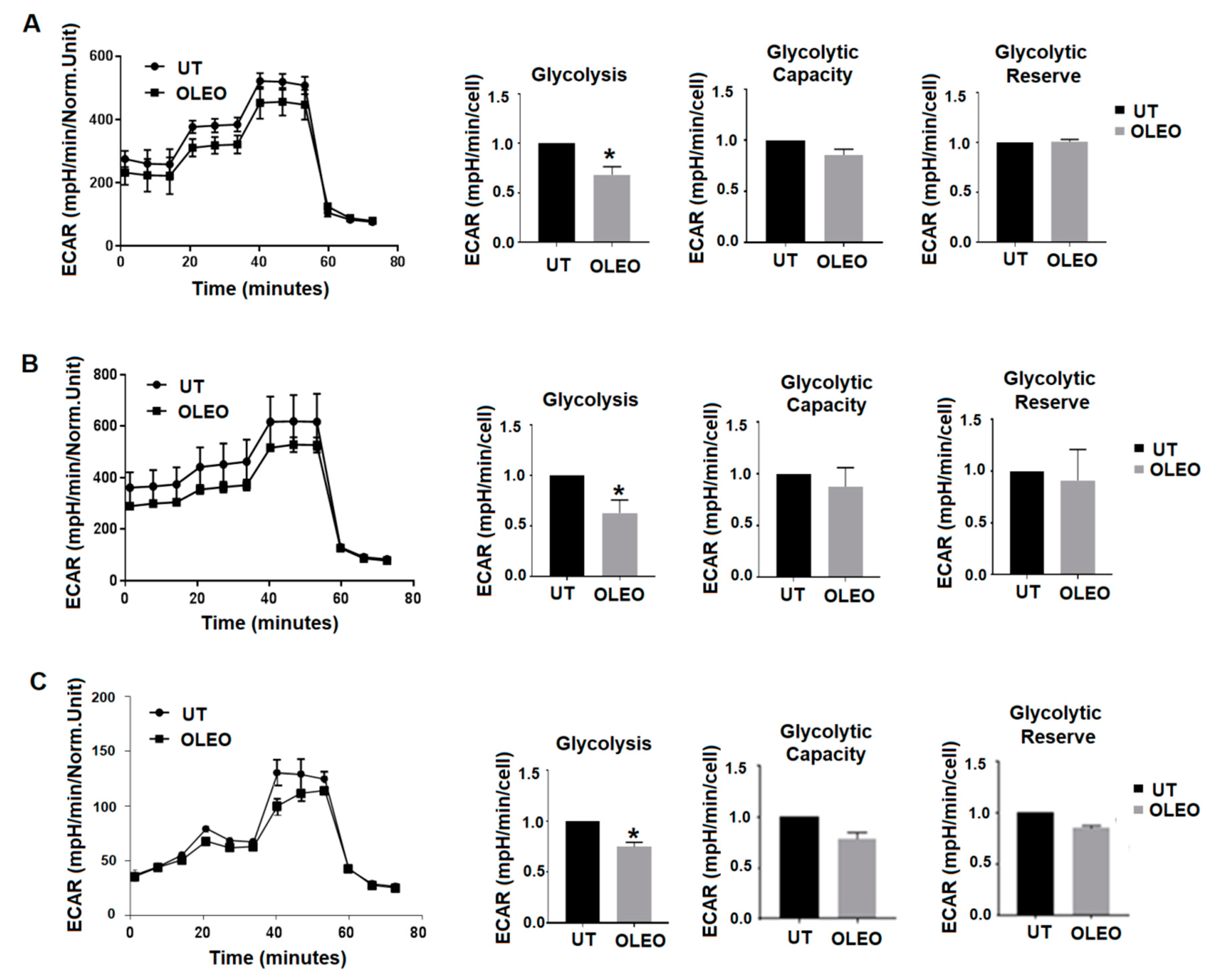
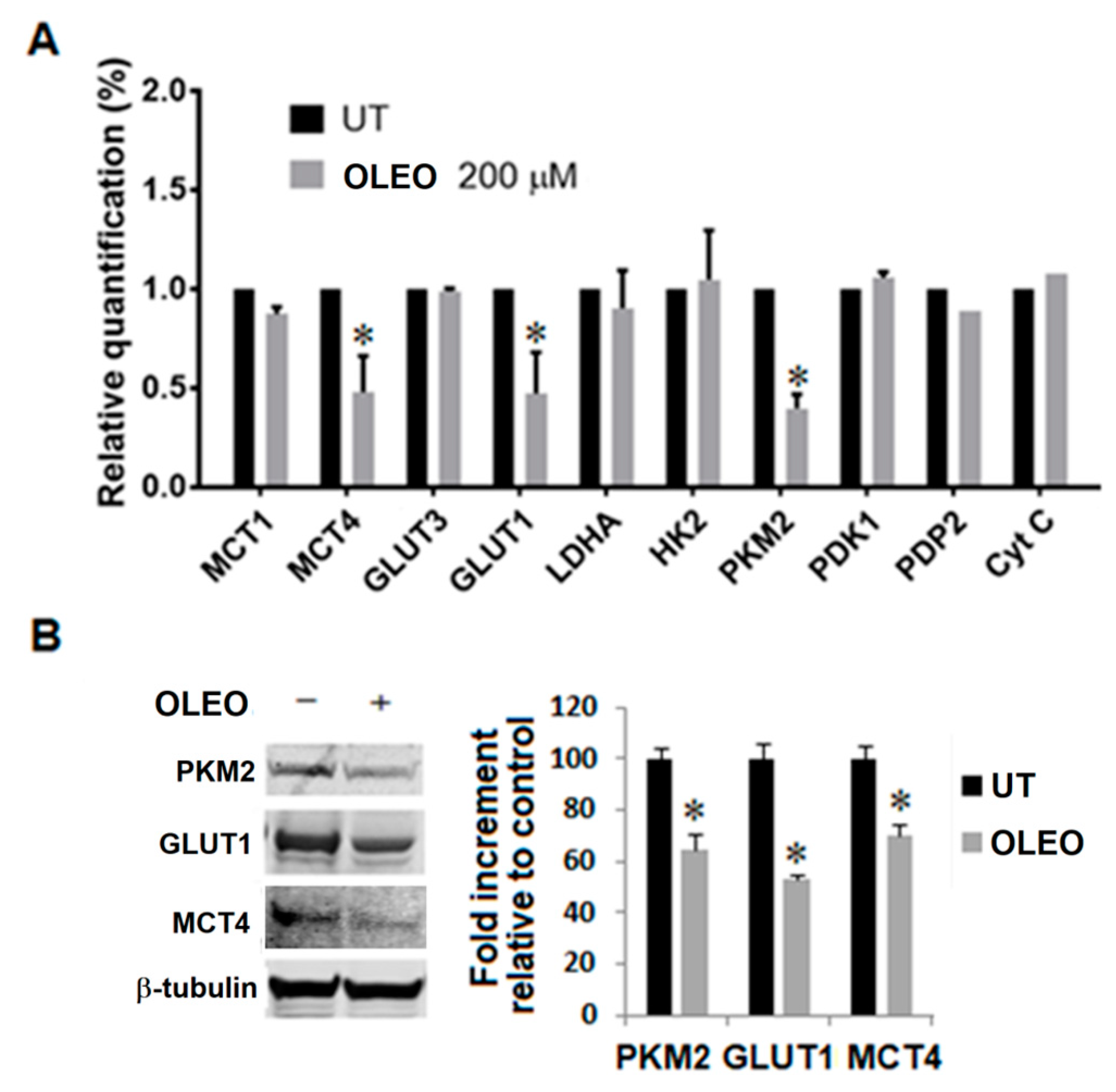
2. Results
3. Discussion
4. Materials and Methods
4.1. Olive Leaf Extract’s Preparation and Toxicity
4.2. Cell Lines and Culture Conditions
4.3. MTT Assay
4.4. Cell Cycle Analysis
4.5. Wound Healing Assay
4.6. Seahorse Analysis
4.7. RNA Isolation and Quantitative PCR (qPCR)
4.8. Western Blotting Analysis
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Barker, J.; Khan, M.A.; Solomos, T. Mechanism of the Pasteur effect. Nature 1964, 201, 1126–1127. [Google Scholar] [CrossRef]
- Warburg, O. On respiratory impairment in cancer cells. Science 1956, 124, 269–270. [Google Scholar] [PubMed]
- Vander Heiden, M.G.; Cantley, L.C.; Thompson, C.B. Understanding the Warburg effect: The metabolic requirements of cell proliferation. Science 2009, 324, 1029–1033. [Google Scholar] [CrossRef] [PubMed]
- Peppicelli, S.; Bianchini, F.; Torre, E.; Calorini, L. Contribution of acidic melanoma cells undergoing epithelial-to-mesenchymal transition to aggressiveness of non-acidic melanoma cells. Clin. Exp. Metastasis 2014, 31, 423–433. [Google Scholar] [CrossRef] [PubMed]
- Ruzzolini, J.; Peppicelli, S.; Andreucci, E.; Bianchini, F.; Margheri, F.; Laurenzana, A.; Fibbi, G.; Pimpinelli, N.; Calorini, L. Everolimus selectively targets vemurafenib resistant BRAFV600E melanoma cells adapted to low pH. Cancer Lett. 2017, 408, 43–54. [Google Scholar] [CrossRef] [PubMed]
- Peppicelli, S.; Ruzzolini, J.; Bianchini, F.; Andreucci, E.; Nediani, C.; Laurenzana, A.; Margheri, F.; Fibbi, G.; Calorini, L. Anoikis Resistance as a Further Trait of Acidic-Adapted Melanoma Cells. J. Oncol. 2019, 2019, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Peppicelli, S.; Bianchini, F.; Calorini, L. Extracellular acidity, a “reappreciated” trait of tumor environment driving malignancy: Perspectives in diagnosis and therapy. Cancer Metastasis Rev. 2014, 33, 823–832. [Google Scholar] [CrossRef] [PubMed]
- Shim, H.; Dolde, C.; Lewis, B.C.; Wu, C.S.; Dang, G.; Jungmann, R.A.; Dalla-Favera, R.; Dang, C.V. c-Myc transactivation of LDH-A: Implications for tumor metabolism and growth. Proc. Natl. Acad. Sci. USA 1997, 94, 6658–6663. [Google Scholar] [CrossRef]
- Dong, G.; Mao, Q.; Xia, W.; Xu, Y.; Wang, J.; Xu, L.; Jiang, F. PKM2 and cancer: The function of PKM2 beyond glycolysis. Oncol. Lett. 2016, 11, 1980–1986. [Google Scholar] [CrossRef]
- Sun, Q.; Chen, X.; Ma, J.; Peng, H.; Wang, F.; Zha, X.; Wang, Y.; Jing, Y.; Yang, H.; Chen, R.; et al. Mammalian target of rapamycin up-regulation of pyruvate kinase isoenzyme type M2 is critical for aerobic glycolysis and tumor growth. Proc. Natl. Acad. Sci. USA 2011, 108, 4129–4134. [Google Scholar] [CrossRef]
- Del Rio, D.; Rodriguez-Mateos, A.; Spencer, J.P.; Tognolini, M.; Borges, G.; Crozier, A. Dietary (poly)phenolics in human health: Structures, bioavailability, and evidence of protective effects against chronic diseases. Antioxid. Redox Signal. 2013, 18, 1818–1892. [Google Scholar] [CrossRef] [PubMed]
- Romani, A.; Ieri, F.; Urciuoli, S.; Noce, A.; Marrone, G.; Nediani, C.; Bernini, R. Health Effects of Phenolic Compounds Found in Extra-Virgin Olive Oil, By-Products, and Leaf of Olea europaea L. Nutrients 2019, 11, 1776. [Google Scholar] [CrossRef] [PubMed]
- Castelli, V.; Grassi, D.; Bocale, R.; d’Angelo, M.; Antonosante, A.; Cimini, A.; Ferri, C.; Desideri, G. Diet and Brain Health: Which Role for Polyphenols? Curr. Pharm. Des. 2018, 24, 227–238. [Google Scholar] [CrossRef] [PubMed]
- Boss, A.; Bishop, K.S.; Marlow, G.; Barnett, M.P.; Ferguson, LR. Evidence to Support the Anti-Cancer Effect of Olive Leaf Extract and Future Directions. Nutrients 2016, 8, 513. [Google Scholar] [CrossRef]
- Milanizadeh, S.; Reza Bigdeli, M. Pro-Apoptotic and Anti-Angiogenesis Effects of Olive Leaf Extract on Spontaneous Mouse Mammary Tumor Model by Balancing Vascular Endothelial Growth Factor and Endostatin Levels. Nutr. Cancer 2019, 71, 1374–1381. [Google Scholar] [CrossRef] [PubMed]
- Butler, M.S.; Robertson, A.A.B.; Cooper, M.A. Natural product and natural product derived drugs in clinical trials. Nat. Prod. Rep. 2014, 31, 1612–1661. [Google Scholar] [CrossRef]
- Seca, A.; Pinto, D. Plant Secondary Metabolites as Anticancer Agents: Successes in Clinical Trials and Therapeutic Application. Int. J. Mol. Sci. 2018, 19, 263. [Google Scholar] [CrossRef]
- Goldsmith, C.D.; Bond, D.R.; Jankowski, H.; Weidenhofer, J.; Stathopoulos, C.E.; Roach, P.D.; Scarlett, C.J. The Olive Biophenols Oleuropein and Hydroxytyrosol Selectively Reduce Proliferation, Influence the Cell Cycle, and Induce Apoptosis in Pancreatic Cancer Cells. Int. J. Mol. Sci. 2018, 19, 1937. [Google Scholar] [CrossRef]
- Siddique, A.B.; Ayoub, N.M.; Tajmim, A.; Meyer, S.A.; Hill, R.A.; El Sayed, K.A. (−)-Oleocanthal Prevents Breast Cancer Locoregional Recurrence After Primary Tumor Surgical Excision and Neoadjuvant Targeted Therapy in Orthotopic Nude Mouse Models. Cancers 2019, 11, 637. [Google Scholar] [CrossRef]
- Zhou, Q.; Pan, H.; Li, J. Molecular Insights into Potential Contributions of Natural Polyphenols to Lung Cancer Treatment. Cancers 2019, 11, 1565. [Google Scholar] [CrossRef]
- Juli, G.; Oliverio, M.; Bellizzi, D.; Gallo Cantafio, M.E.; Grillone, K.; Passarino, G.; Colica, C.; Nardi, M.; Rossi, M.; Procopio, A.; et al. Anti-tumor Activity and Epigenetic Impact of the Polyphenol Oleacein in Multiple Myeloma. Cancers 2019, 11, 990. [Google Scholar] [CrossRef] [PubMed]
- De Stefanis, D.; Scimè, S.; Accomazzo, S.; Catti, A.; Occhipinti, A.; Bertea, C.M.; Costelli, P. Anti-Proliferative Effects of an Extra-Virgin Olive Oil Extract Enriched in Ligstroside Aglycone and Oleocanthal on Human Liver Cancer Cell Lines. Cancers 2019, 11, 1640. [Google Scholar] [CrossRef] [PubMed]
- Nediani, C.; Ruzzolini, J.; Romani, A.; Calorini, L. Oleuropein, a Bioactive Compound from Olea europaea L., as a Potential Preventive and Therapeutic Agent in Non-Communicable Diseases. Antioxidants 2019, 8, 578. [Google Scholar] [CrossRef] [PubMed]
- Shamshoum, H.; Vlavcheski, F.; Tsiani, E. Anticancer effects of oleuropein. BioFactors 2017, 43, 517–528. [Google Scholar] [CrossRef]
- Margheri, F.; Menicacci, B.; Laurenzana, A.; Del Rosso, M.; Fibbi, G.; Cipolleschi, M.G.; Ruzzolini, J.; Nediani, C.; Mocali, A.; Giovannelli, L. Oleuropein aglycone attenuates the pro-angiogenic phenotype of senescent fibroblasts: A functional study in endothelial cells. J. Funct. Foods 2019, 53, 219–226. [Google Scholar] [CrossRef]
- Ruzzolini, J.; Peppicelli, S.; Andreucci, E.; Bianchini, F.; Scardigli, A.; Romani, A.; la Marca, G.; Nediani, C.; Calorini, L. Oleuropein, the Main Polyphenol of Olea europaea Leaf Extract, Has an Anti-Cancer Effect on Human BRAF Melanoma Cells and Potentiates the Cytotoxicity of Current Chemotherapies. Nutrients 2018, 10, 1950. [Google Scholar] [CrossRef] [PubMed]
- Guerra, A.R.; Duarte, M.F.; Duarte, I.F. Targeting Tumor Metabolism with Plant-Derived Natural Products: Emerging Trends in Cancer Therapy. J. Agric. Food Chem. 2018, 66, 10663–10685. [Google Scholar] [CrossRef]
- Gerhäuser, C. Cancer cell metabolism, epigenetics and the potential influence of dietary components-A perspective. Biomed. Res. 2012, 23, 69–89. [Google Scholar]
- Gao, J.L.; Chen, Y.G. Natural compounds regulate glycolysis in hypoxic tumor microenvironment. Biomed. Res. Int. 2015, 2015, 354143. [Google Scholar] [CrossRef]
- Salehi, B.; Zucca, P.; Sharifi-Rad, M.; Pezzani, R.; Rajabi, S.; Setzer, W.N.; Varoni, E.M.; Iriti, M.; Kobarfard, F.; Sharifi-Rad, J. Phytotherapeutics in cancer invasion and metastasis. Phytother. Res. 2018, 32, 1425–1449. [Google Scholar] [CrossRef]
- Sharma, S.H.; Thulasingam, S.; Chellappan, D.R.; Chinnaswamy, P.; Nagarajan, S. Morin and Esculetin supplementation modulates c-myc induced energy metabolism and attenuates neoplastic changes in rats challenged with the procarcinogen 1,2-dimethylhydrazine. Eur. J. Pharmacol. 2017, 796, 20–31. [Google Scholar] [CrossRef] [PubMed]
- Gómez de Cedrón, M.; Vargas, T.; Madrona, A.; Jiménez, A.; Pérez-Pérez, M.J.; Quintela, J.C.; Reglero, G.; San-Félix, A.; Ramírez de Molina, A. Novel Polyphenols That Inhibit Colon Cancer Cell Growth Affecting Cancer Cell Metabolism. J. Pharmacol. Exp. Ther. 2018, 366, 377–389. [Google Scholar] [CrossRef] [PubMed]
- Simmons, R.A. Cell Glucose Transport and Glucose Handling During Fetal and Neonatal Development. In Fetal and Neonatal Physiology, 5th ed.; Elsevier: Amsterdam, The Netherlands, 2017; pp. 428–435. [Google Scholar]
- Barbosa, A.M.; Martel, F. Targeting Glucose Transporters for Breast Cancer Therapy: The Effect of Natural and Synthetic Compounds. Cancers 2020, 12, 154. [Google Scholar] [CrossRef] [PubMed]
- Zambrano, A.; Molt, M.; Uribe, E.; Salas, M. Glut 1 in Cancer Cells and the Inhibitory Action of Resveratrol as A Potential Therapeutic Strategy. Int. J. Mol. Sci. 2019. [Google Scholar] [CrossRef]
- Wiese, E.K.; Hitosugi, T. Tyrosine Kinase Signaling in Cancer Metabolism: PKM2 Paradox in the Warburg Effect. Front. Cell Dev. Biol. 2018, 6, 79. [Google Scholar] [CrossRef]
- Nájera, L.; Alonso-Juarranz, M.; Garrido, M.; Ballestín, C.; Moya, L.; Martínez-Díaz, M.; Carrillo, R.; Juarranz, A.; Rojo, F.; Cuezva, J.M.; et al. Prognostic implications of markers of the metabolic phenotype in human cutaneous melanoma. Br. J. Dermatol. 2019, 181, 114–127. [Google Scholar] [CrossRef]
- Zhang, H.; Feng, C.; Zhang, M.; Zeng, A.; Si, L.; Yu, N.; Bai, M. miR-625-5p/PKM2 negatively regulates melanoma glycolysis state. J. Cell Biochem. 2019, 120, 2964–2972. [Google Scholar] [CrossRef]
- Iqbal, M.A.; Bamezai, R.N. Resveratrol inhibits cancer cell metabolism by down regulating pyruvate kinase M2 via inhibition of mammalian target of rapamycin. PLoS ONE 2012, 7, e36764. [Google Scholar] [CrossRef]
- Li, W.; Ma, X.; Li, N.; Liu, H.; Dong, Q.; Zhang, J.; Yang, C.; Liu, Y.; Liang, Q.; Zhang, S.; et al. Resveratrol inhibits Hexokinases II mediated glycolysis in non-small cell lung cancer via targeting Akt signaling pathway. Exp. Cell Res. 2016, 349, 320–327. [Google Scholar] [CrossRef]
- Saunier, E.; Antonio, S.; Regazzetti, A.; Auzeil, N.; Laprévote, O.; Shay, J.W.; Coumoul, X.; Barouki, R.; Benelli, C.; Huc, L.; et al. Resveratrol reverses the Warburg effect by targeting the pyruvate dehydrogenase complex in colon cancer cells. Sci. Rep. 2017, 7, 6945. [Google Scholar] [CrossRef]
- Siddiqui, F.A.; Prakasam, G.; Chattopadhyay, S.; Rehman, A.U.; Padder, R.A.; Ansari, M.A.; Irshad, R.; Mangalhara, K.; Bamezai, R.N.K.; Husain, M.; et al. Curcumin decreases Warburg effect in cancer cells by down-regulating pyruvate kinase M2 via mTOR-HIF1α inhibition. Sci. Rep. 2018, 8, 8323. [Google Scholar] [CrossRef]
- Pinheiro, C.; Miranda-Gonçalves, V.; Longatto-Filho, A.; Vicente, A.L.; Berardinelli, G.N.; Scapulatempo-Neto, C.; Costa, R.F.; Viana, C.R.; Reis, R.M.; Baltazar, F.; et al. The metabolic microenvironment of melanomas: Prognostic value of MCT1 and MCT4. Cell Cycle 2016, 15, 1462–1470. [Google Scholar] [CrossRef]
- Romani, A.; Mulas, S.; Heimler, D. Polyphenols and secoiridoids in raw material (Olea europaea L. leaves) and commercial food supplements. Eur. Food Res. Technol. 2017, 243, 429–435. [Google Scholar] [CrossRef]
- Romani, A.; Scardigli, A.; Pinelli, P. An environmentally friendly process for the production of extracts rich in phenolic antioxidants from Olea europaea L. and Cynara scolymus L. matrices. Eur. Food Res. Technol. 2017, 243, 1229–1238. [Google Scholar] [CrossRef]
- Guex, C.G.; Reginato, F.Z.; Figueredo, K.C.; da Silva, A.R.H.D.; Pires, F.B.; Jesus, R.D.S.; Lhamas, C.L.; Lopes, G.H.H.; Bauermann, L.F. Safety assessment of ethanolic extract of Olea europaea L. leaves after acute and subacute administration to Wistar rats. Regul. Toxicol. Pharmacol. 2018, 95, 395–399. [Google Scholar] [CrossRef]
- Sepporta, M.V.; Fuccelli, R.; Rosignoli, P.; Ricci, G.; Servili, M.; Fabiani, R. Oleuropein Prevents Azoxymethane-Induced Colon Crypt Dysplasia and Leukocytes DNA Damage in A/J Mice. J. Med. Food 2016, 19, 983–989. [Google Scholar] [CrossRef]
- Giner, E.; Recio, M.C.; Ríos, J.L.; Cerdá-Nicolás, J.M.; Giner, R.M. Chemopreventive effect of oleuropein in colitis-associated colorectal cancer in c57bl/6 mice. Mol. Nutr. Food Res. 2016, 60, 242–255. [Google Scholar] [CrossRef]
- Park, S.; Choi, Y.; Um, S.J.; Yoon, S.K.; Park, T. Oleuropein attenuates hepatic steatosis induced by high-fat diet in mice. J. Hepatol. 2011, 54, 984–993. [Google Scholar] [CrossRef]
- Peppicelli, S.; Bianchini, F.; Toti, A.; Laurenzana, A.; Fibbi, G.; Calorini, L. Extracellular acidity strengthens mesenchymal stem cells to promote melanoma progression. Cell Cycle 2015, 14, 3088–3100. [Google Scholar] [CrossRef]
- Peppicelli, S.; Ruzzolini, J.; Andreucci, E.; Bianchini, F.; Kontos, F.; Yamada, T.; Ferrone, S.; Calorini, L. Potential Role of HLA Class I Antigens in the Glycolytic Metabolism and Motility of Melanoma Cells. Cancers 2019, 11, 1249. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Acquaviva, R.; Di Giacomo, C.; Sorrenti, V.; Galvano, F.; Santangelo, R.; Cardile, V.; Gangia, S.; D’Orazio, N.; Abraham, N.G.; Vanella, L. Antiproliferative effect of oleuropein in prostate cell lines. Int. J. Oncol. 2012, 41, 31–38. [Google Scholar] [CrossRef]
- Samara, P.; Christoforidou, N.; Lemus, C.; Argyropoulou, A.; Ioannou, K.; Vougogiannopoulou, K.; Aligiannis, N.; Paronis, E.; Gaboriaud-Kolar, N.; Tsitsilonis, O.; et al. New semi-synthetic analogs of oleuropein show improved anticancer activity in vitro and in vivo. Eur. J. Med. Chem. 2017, 137, 11–29. [Google Scholar] [CrossRef]

| Gene | FW | RV |
|---|---|---|
| MCT1 | 5’-GTGGCTCAGCTCCGTATTGT-3’ | 5’-GAGCCGACCTAAAAGTGGTG-3’ |
| MCT4 | 5’-CAGTTCGAGGTGCTCATGG-3’ | 5’-ATGTAGAGGTGGGTCGCATC-3’ |
| GLUT1 | 5’-CGGGCCAAGAGTGTGCTAAA-3’ | 5’-TGACGATACCGGAGCCAATG-3’ |
| GLUT3 | 5’-CGAACTTCCTAGTCGGATTG-3’ | 5’-AGGAGGCACGACTTAGACAT-3’ |
| LDHA | 5’-AGCCCGATTCCGTTACCT-3’ | 5′-CACCAGCAACATTCATTCCA-3′ |
| PKM2 | 5’-CAGAGGCTGCCATCTACCAC-3’ | 5’-CCAGACTTGGTGAGGACGAT-3’ |
| PDK1 | 5’-CCAAGACCTCGTGTTGAGACC-3’ | 5’-AATACAGCTTCAGGTCTCCTTGG-3’ |
| HK2 | 5’- CAAAGTGACAGTGGGTGTGG-3’ | 5’- GCCAGGTCCTTCACTGTCTC-3’ |
| 18s | 5’-CGCCGCTAGAGGTGAAATTCT-3’ | 5’-CGAACCTCCGA CTTTCGTTCT-3’ |
| PDP2 | 5’-ACCACCTCCGTGTCTATTGG-3’ | 5’-CCAGCGAGATGTCAGAATCC-3’ |
| CytC | 5’-TTGCACTTACACCGGTACTTAAGC-3’ | 5’-ACGTCCCCACTCTCTAAGTCCAA-3 |
| GLS1 | 5’-TGCTACCTGTCTCCATGGCTT-3’ | 5’-CTTAGATGGCACCTCCTTTGG-3’ |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ruzzolini, J.; Peppicelli, S.; Bianchini, F.; Andreucci, E.; Urciuoli, S.; Romani, A.; Tortora, K.; Caderni, G.; Nediani, C.; Calorini, L. Cancer Glycolytic Dependence as a New Target of Olive Leaf Extract. Cancers 2020, 12, 317. https://doi.org/10.3390/cancers12020317
Ruzzolini J, Peppicelli S, Bianchini F, Andreucci E, Urciuoli S, Romani A, Tortora K, Caderni G, Nediani C, Calorini L. Cancer Glycolytic Dependence as a New Target of Olive Leaf Extract. Cancers. 2020; 12(2):317. https://doi.org/10.3390/cancers12020317
Chicago/Turabian StyleRuzzolini, Jessica, Silvia Peppicelli, Francesca Bianchini, Elena Andreucci, Silvia Urciuoli, Annalisa Romani, Katia Tortora, Giovanna Caderni, Chiara Nediani, and Lido Calorini. 2020. "Cancer Glycolytic Dependence as a New Target of Olive Leaf Extract" Cancers 12, no. 2: 317. https://doi.org/10.3390/cancers12020317
APA StyleRuzzolini, J., Peppicelli, S., Bianchini, F., Andreucci, E., Urciuoli, S., Romani, A., Tortora, K., Caderni, G., Nediani, C., & Calorini, L. (2020). Cancer Glycolytic Dependence as a New Target of Olive Leaf Extract. Cancers, 12(2), 317. https://doi.org/10.3390/cancers12020317











