Abstract
Germline protein truncating variants (PTVs) in the FANCM gene have been associated with a 2–4-fold increased breast cancer risk in case-control studies conducted in different European populations. However, the distribution and the frequency of FANCM PTVs in Europe have never been investigated. In the present study, we collected the data of 114 European female breast cancer cases with FANCM PTVs ascertained in 20 centers from 13 European countries. We identified 27 different FANCM PTVs. The p.Gln1701* PTV is the most common PTV in Northern Europe with a maximum frequency in Finland and a lower relative frequency in Southern Europe. On the contrary, p.Arg1931* seems to be the most common PTV in Southern Europe. We also showed that p.Arg658*, the third most common PTV, is more frequent in Central Europe, and p.Gln498Thrfs*7 is probably a founder variant from Lithuania. Of the 23 rare or unique FANCM PTVs, 15 have not been previously reported. We provide here the initial spectrum of FANCM PTVs in European breast cancer cases.
1. Introduction
FANCM, the Fanconi anemia (FA) complementation group M gene (OMIM609644), was originally described as one of the members of the FA molecular pathway [1] that is primarily responsible for the repair of the DNA inter-strand crosslinks through homologous recombination. FA is a recessive DNA repair disease characterized by bone marrow failure, congenital malformations, chromosome fragility, and cancer. Although FANCM is part of the core complex in the FA pathway, accumulating evidence indicates that protein truncating variants (PTVs) in this gene are not causative of FA. Recently, eight individuals who were not diagnosed with FA were found to harbor bi-allelic FANCM PTVs. Three individuals developed early-onset cancers including lymphoblastic leukemia and squamous cancers [2]. In addition, five carriers of homozygous FANCM PTVs were identified among females diagnosed with breast cancer, two of which were diagnosed with early onset disease [3]. Hence, it seems that bi-allelic FANCM PTVs cause cancer predisposition with a greater risk for early onset development.
In the last years, results from case-control studies have indicated that mono-allelic FANCM PTVs are breast cancer risk factors. The c.5101C > T (p.Gln1701*, rs147021911) is frequent in the Finnish population with carrier frequency = 0.0162 versus 0.00168 in non-Finnish Europeans (NFE), as reported in gnomAD v2.1.1 (https://gnomad.broadinstitute.org/; [4]). In the Finnish population, p.Gln1701* was found to be associated with breast cancer risk with an odds ratio (OR) of 1.86 and with greater effects in familial cases, and for estrogen receptor-negative (ER-negative) and triple-negative breast cancer (TNBC) subtypes [5]. This variant was later found to be also associated with breast cancer survival and treatment outcome [6]. A second FANCM PTV, the c.5791C > T (p.Gly1906Alafs12*, rs144567652), which is annotated and hereafter referred to as p.Arg1931*, showed association with breast cancer risk in familial cases with OR = 3.93 [7]. This PTV was later found to also be associated with risk for TNBC in Finnish cases with OR = 5.14 [8]. A sequencing analysis of the entire FANCM coding region showed an excess of a third common PTV, the c.1972C > T (p.Arg658*, rs368728266), in German breast cancer cases versus controls, and confirmed that FANCM PTVs have a particularly high risk (OR = 3.75) for TNBC [9]. We recently tested p.Arg658*, p.Gln1701*, and p.Arg1931* in 67,112 European breast cancer cases and 53,766 controls. In these analyses, we observed that p.Arg658* was associated with increased risk of ER-negative disease and TNBC with ORs of 2.44 and 3.79, respectively, and that p.Arg1931* was associated with the risk of ER-negative breast cancer with an OR of 1.96 [10].
BRCA1, BRCA2, and PALB2 are established breast cancer predisposition genes conferring high risk especially for ER-negative disease and TNBC. Similarly to other genes such as BARD1, RAD51D, BRIP1, and RAD51C [11], FANCM is emerging as a breast cancer predisposing factor conferring a greater risk specifically for these breast cancer subtypes. However, the spectrum of FANCM PTVs in Europe has never been investigated. Through a project call addressed to ENIGMA consortium collaborators [12], we collected data from European female breast cancer probands who were subjected to the sequencing of the FANCM coding region, resulting as carriers of a FANCM PTV. This study describes the spectrum of FANCM PTVs in this cohort of European breast cancer cases.
2. Results and Discussion
The 114 European breast cancer probands included in this study carried 27 different FANCM PTVs. Four, namely c.1491dupA (p.Gln498Thrfs*7, rs797045117), p.Arg658*, p.Gln1701*, and p.Arg1931*, were each identified in at least 8 probands and were classified as “common FANCM PTVs”. The remaining 23 variants were unique or were found in not more than three probands and were classified as “rare FANCM PTVs”. Additional data on common and rare FANCM PTVs were derived from previously published studies based on FANCM sequencing of German breast cancer probands [9,13]. Altogether, these and our data allowed us to show the geographic distribution of the four common PTVs and the rare PTVs combined in a cohort of breast cancer probands (Figure 1).
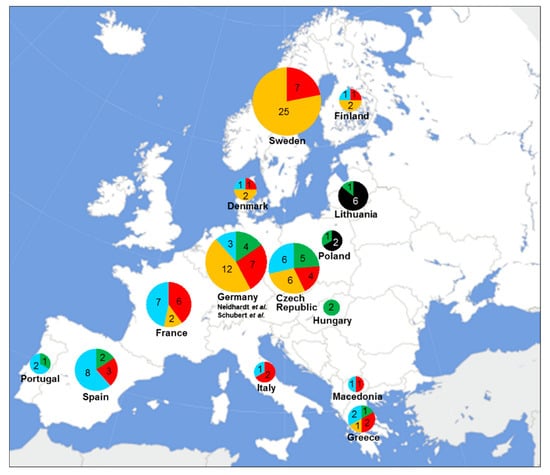
Figure 1.
Distribution of the common and rare FANCM PTVs found in the 114 European female breast cancer probands. The common PTVs p.Gln498Thrfs*7, p.Arg658*, p.Gln1701*, and p.Arg1931* are indicated in black, green, orange, and red, respectively; the rare PTVs are indicated in light blue. Numbers of PTV carriers are reported according to their country of origin. The 26 PTV carriers from Germany are derived from previously published studies (Neidhardt et al., 21 probands [9]; Schubert et al., 5 probands [13]).
Of the 114 breast cancer probands, 86 (75.4%) carried one of the four common FANCM PTVs (Table 1). Of these 86 probands, 8 (9.3%), carried the p.Gln498Thrfs*7, 13 (15.1%) the p.Arg658*, 38 (44.2%) the p.Gln1701*, and 27 (31.4%) carried the p.Arg1931*. We observed that the relative frequency of the two most common PTVs, p.Gln1701* and p.Arg1931*, differ with respect to the country of origin of carriers. The p.Gln1701* is the most common PTV in Northern Europe while it has a lower relative frequency in Southern Europe. An opposite frequency gradient along the North–South axis appeared to exist for p.Arg1931*, which is the most common PTV in Southern Europe but more rare in Northern Europe. Consistently, in Central Europe, p.Gln1701* and p.Arg1931* are both common with similar relative frequencies. The relative frequencies of the FANCM:p.Arg658*, the third most common FANCM PTV, suggest that this variant has the highest frequency in Central Europe, is present in Southern Europe, and seems to be absent or very rare in Scandinavia. The fourth common FANCM PTV p.Gln498Thrfs*7 was identified in 6 of the 7 probands from Lithuania, suggesting it is a founder variant from this specific geographic area (Table 1, Figure 1).

Table 1.
Eighty-six breast cancer probands who carry one of the four common FANCM PTVs.
The high relative frequency of FANCM:p.Gln498Thrfs*7 observed in Lithuania could indicate a founder effect in this relatively homogenous Baltic population. Similarly, haplotype analyses previously conducted in this population found a strong founder effect for several recurrent pathogenic variants in the high-risk predisposition genes BRCA1 and BRCA2 [14,15]. Calculations from haplotype analysis and paleo-demographic data indicate that a bottleneck phenomenon occurring approximately in the 4th–5th century AD in the Lithuanian region could have contributed to decreased genetic diversity and subsequent expansion of specific alleles [14].
We compared the geographic distribution of the four common PTVs observed in our study cohort with that reported in the gnomAD database v2.1.1 [4]. In gnomAD, 630 European individuals carried one of the p.Gln498Thrfs*7, p.Arg658*, p.Gln1701* or p.Arg1931* PTVs with relative proportions of 3.2% (20/630), 3.3% (21/630), 54.0% (340/630), and 39.5% (249/630), respectively. About half of the p.Gln498Thrfs*7 gnomAD carriers (9/20) were from Estonia, confirming that this variant has a specific origin in the Baltic countries. In gnomAD, p.Arg658* was prevalently found in NFE with the higher frequency of the carries (8/21) found in North-western Europeans and only one carrier found among Southern Europeans, which is similar to what we observed. Having being found in 340 gnomAD individuals, FANCM:p.Gln1701* is by far the most common PTV in Europe. As mentioned before, p.Gln1701* was particularly frequent in Finland and was observed more frequently in North-western than Southern Europeans, which is consistent with our data. FANCM:p.Arg1931* was the second most common FANCM PTV in Europe and the most common FANCM PTV in Southern Europeans in the gnomAD dataset. However, this variant was reported to be the most common also in North-western Europeans, not confirming the South–North frequency gradient observed in our cohort.
Of the 114 breast cancer probands, 29 (25.4%) carried one of the 23 rare FANCM PTVs (Table 2). Interestingly, a proband from the Czech Republic was a bi-allelic carrier of FANCM PTVs having inherited the rare p.Arg1030* variant from the mother, and the common p.Arg658* variant from the paternal side of the family (Table 1 and Table 2). This carrier was diagnosed with breast cancer at a young age (28 years), which is consistent with published data showing that females with bi-allelic FANCM PTV may develop early onset disease [3]. Eight of the 23 rare PTVs were annotated in the public ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/), LOVD (https://www.lovd.nl/), or gnomAD databases and 15 were novel. Of the 23 rare PTVs, 19 were unique; two, p.Arg754* and p.Glu774*, were found in three probands; and two, p.Lys1615* and p.Thr1923Profs*2, in two probands. Given these low frequencies, we could not speculate on the origins or the geographic distribution of any of the rare FANCM PTVs. Considering the numbers of carriers of common PTVs and rare PTVs in each country, we observed that in Spain and France, there was an apparent excess of carriers of rare PTVs. On the contrary, in Sweden and Lithuania, no carriers of rare PTVs were found (Figure 1).

Table 2.
Twenty-nine breast cancer probands who carry one of the 23 rare FANCM PTVs.
3. Materials and Methods
The individuals included in our analysis were contributed by ENIGMA consortium collaborators [12] interested in studying the effect/role of FANCM on breast cancer predisposition. They were originally ascertained at several Centers or National Studies and considered eligible for mutation sequencing analysis of breast and/or ovarian cancer predisposing genes in the frame of diagnostic or research purposes. In general, the ascertainment criteria for mutation testing were based on the presence of family history for breast or ovarian cancer, and on the early onset or specific clinical subtype of breast cancer. In the present study, we included 114 probands who received (i) a diagnosis of breast cancer, (ii) tested negative for BRCA1 or BRCA2 pathogenic variants, (iii) underwent complete sequencing of the FANCM coding region, resulting as a carrier of a PTV. Unfortunately, it was not possible to retrieve the number of probands that were originally subjected to mutation sequencing analysis at each participating Center or National Study. The 114 carriers of FANCM PTVs were ascertained in 20 Centers or National Studies from 13 European countries (Table S1) including Czech Republic (20, 17.5%), Denmark (4, 3.5%), Finland (4, 3.5%), France (15, 13.2%), Greece (6, 5.3%), Hungary (2, 1.8%), Italy (3, 2.6%), Lithuania (7, 6.1%), Macedonia (2, 1.8%), Poland (3, 2.6%), Portugal (3, 2.6%), Spain (15, 11.4%), and Sweden (32, 28.1%). Some of these FANCM PTVs carriers were described in studies previously published, as indicated (Table S1). Of the 114 probands, 58 (50.9%) were ER-positive, 27 (23.7%) were ER-negative, and 80 (70.2%) had a positive family history of breast or ovarian cancer. The mean age at breast cancer diagnosis was 46.7 years (min = 21, max = 85). All the 114 probands provided written informed consent for their data to be used for research purposes or specifically agreed to be tested for new variants in potential breast cancer associated genes. The participation of all the Centers or National Studies in the present work (Table S1) was approved by their ethic committees. Specifically, these are the Ethics Committee of the General University Hospital in Prague (Approval code 87/14); Ethics Committee of Carlos III Institute of Health in Madrid (CEI PI 06_2013-v2); by the appropriate Advisory Committee on the Treatment of Health Research Information (Comité Consultatif de Protection des Personnes dans la Recherche Biomédicale (CCPPRB) Ile-de-France III) and by the National Data Protection authority; Ethics Committee of Fondazione IRCCS Istituto Nazionale dei Tumori in Milan (INT 7/15); Ethics Committee of the Capital Region of Denmark (H-4-2010-050); Ethics Committee of Hospital District of Helsinki and Uusimaa (HUS/1597/2016); Ethical Sub-Committee on Medicine, Pharmacy, Veterinary and Dental Medicine, Macedonian Academy of Sciences and Arts (09-1047/1 from 20/04/2016); IPO (Instituto Português de Oncologia) Porto institutional review board (nr. 219-019); Comité Territorial de Ética de la Investigación de Santiago -Lugo, Conselleria de Sanidade, Xunta de Galicia (2018/200); Ethics Committee of the Area Vasta Romagna e I.R.S.T (CEAV, 1727/2012 I.5/43); Provincial Ethics Committee (209/16) and the Modena Hospital (3387); Ethics Committee of University of Rzeszow, Poland (resolution no. 9/06/2015), University of Melbourne Human Research Ethics Committee, Melbourne, Australia; National Scientific and Ethics Committee (ETT-TUKEB), 20998-0/2010-1018EKU (845/PI/010); IDIBELL (Institut d’Investigacio Biomedica de Bellvitge, Barcelona) Ethics Committee PR278/19; Lithuanian Bioethics Committee (LT-HCCA-003 protocol), Vilnius Regional Ethics Committee (Nr. 58200-12-27(1-61); Ethics Committee of Lund, Sweden (Dnr 2011/349 and 2011/652); Bioethics Committee of NSCR (National Centre of Scientific Research) Demokritos (BcncSrD-240/eHΔ/11.3, updated on 29 June 2015).
4. Conclusions
We described the geographic distribution and relative prevalence of 27 different FANCM PTVs detected in 114 female European breast cancer probands with no pathogenic variants in BRCA1 or BRCA2 genes. As far as we know, this is the largest available collection of breast cancer probands carrying FANCM PTVs. In our study-cohort, we found that the FANCM: p.Gln1701* had high relative frequency in Northern Europe and low relative frequency in Southern Europe, whereas p.Arg1931* was the most common PTV in Southern Europe and less common in Northern Europe. The p.Arg658* had higher frequency in Central Europe, while p.Gln498Thrfs*7 is probably a founder variant from Lithuania. Fifteen of the 27 PTVs described are novel, including one found in three probands, and one found in two probands. Further data are warranted to provide an extensive spectrum of FANCM PTVs in Europe.
Supplementary Materials
The following are available online at https://www.mdpi.com/2072-6694/12/2/292/s1, Table S1: Center or Study contributing the 114 FANCM PTV probands. Some of these probands were previously described as indicated; sequencing details are provided.
Author Contributions
Conceptualization, G.F., R.J. and P.P.; Formal analysis, G.F. and P.P; Data curation (providing samples and clinical data), G.F., A.K., E.T., J.S., P.K., T.A.M., N.A., J.A., J.B., A.B., J.B., B.B., A.B., B.B., Å.B., J.B., D.C., M.C., S.C., L.C., E.D., J.D.V., O.D., S.E.M., F.F., F.G., C.H., M.J., J.I.K., I.K., K.K.S., C.L., F.L., S.M., R.M., U.M., V.M., A.M., T.N.-D., F.C.N., R.N., E.O., A.O., L.P., B.P., A.P., D.P.K., T.P., P.R., M.R., V.R., M.S., C.S., S.S., Melissa C Southey, D.S.L., M.T., T.T., A.T., M.U., A.V., Z.V., D.Y., V.Z., Z.K., H.N., H.E., R.J. and P.P.; Writing—original draft preparation, G.F., R.J. and P.P.; Writing—review and editing, G.F., A.K., E.T., J.S., P.K., T.A.M., F.L., D.P.K., P.R., M.R., Z.K., H.N., H.E., R.J. and P.P.; Supervision, P.P. All authors have read and agreed to the published version of the manuscript.
Funding
This research was partially funded by Associazione Italiana Ricerca sul Cancro (AIRC; IG2015 no.16732) to P. Peterlongo, a fellowship from Fondazione Umberto Veronesi to G. Figlioli and by the Italian Ministry of Health with Ricerca Corrente and 5x1000 funds. E. Tham is supported by Region Stockholm (ALF). The Czech study was supported by a grant of the Ministry of Health of the Czech Republic NV16-29959A. CNIO study was partially supported by projects PI16/00440 and PI19/00640, supported by the Instituto de Salud Carlos III, cofunded by European Regional Development Fund (ERDF), the Spanish Network on Rare Diseases (CIBERER) and BRIDGES project H2020. Financial support for GENESIS resource and genotyping was provided by the Ligue Nationale contre le Cancer (grants PRE05/DSL, PRE07/DSL, PRE11/NA), the French National Institute of Cancer (INCa grant No b2008-029/LL-LC) and the comprehensive cancer center SiRIC, (Site de Recherche Intégrée sur le Cancer: Grant INCa-DGOS-4654). HEBCS was funded by Helsinki University Hospital Research Fund, Sigrid Juselius Foundation, The cancer Society of Finland. A.Vega is supported by the Spanish Health Research Foundation, Instituto de Salud Carlos III (ISCIII) through Research Activity Intensification Program (contract grant numbers: INT15/00070, INT16/00154, INT17/00133), and through Centro de Investigación Biomédica en Red de Enferemdades Raras CIBERER (ACCI 2016: ER17P1AC7112/2018); Autonomous Government of Galicia (Consolidation and structuring program: IN607B), and by the Fundación Mutua Madrileña (call 2018). This work was supported by the Australian National Health and Medical Research Council (APP1029974 and APP1074383) and by a Victorian Life Sciences Computation Initiative grant (number VR0182) on its Peak Computing Facility, an initiative of the Victorian Government. T.N-D is a Career Development Fellow of the National Breast Cancer Foundation (Australia, ECF-17-001). M.C.S. is a National Health and Medical Research Council (Australia) Senior Research Fellow. The Hungarian Breast and Ovarian Cancer Study was supported by Hungarian Research Grants KTIA-OTKA CK-80745 and NKFI OTKA K-112228 to E. Olah. Lithuanian study was supported by The Research Council of Lithuania grant SEN18/2015 and P-MIP-20-25 to R. Janavicius. The ICO was supported by the Carlos III National Health Institute funded by FEDER funds—a way to build Europe—[PI16/00563, PI19/00553 and CIBERONC]; the Government of Catalonia [Pla estratègic de recerca i innovació en salut (PERIS) Project MedPerCan, 2017SGR1282 and 2017SGR496]; and CERCA program.
Acknowledgments
The authors are particularly grateful to individuals participating in this study and their families. We would like to thank the personnel of the Genetic Testing for Hereditary Tumors Laboratory – Cogentech, Spiros Tavandzis from the AGEL Laboratories in Novy Jicin for his valuable contribution to the study, and Daniela Zaffaroni (Istituto Nazionale dei Tumori, Milan). GENESIS (GENE SISters) is a French national study sponsored by UNICANCER (Sinilnikova et al. BMC Cancer 2016). We wish to thank the genetic epidemiology platform (the PIGE, Plateforme d’Investigation en Génétique et Epidemiologie: Séverine Eon-Marchais, Marie-Gabrielle Dondon, M. Marcou, D. Le Gal, L. Toulemonde, J. Beauvallet, N. Mebirouk, E. Cavaciuti), the biological resource centre (S. Mazoyer, F. Damiola, L. Barjhoux, C. Verny-Pierre, V. Sornin) and all the GENESIS collaborating cancer clinics.. HEBCS thanks Drs. Kristiina Aittomäki and Carl Blomqvist and research nurse Outi Malkavaara for their help with patient data and samples. FPGMX: members of the Cancer Genetics group (IDIS): Belinda Rodríguez, Olivia Fuentes and Miguel Aguado. ICO wants to thank all the members of the ICO Hereditary Cancer Program team led by Dr. Gabriel Capella. SWE-BRCA (The Swedish BRCA1 & BRCA2 Study Collaborators): Gothenburg, Sahlgrenska University Hospital: Zakaria Einbeigi, Anna Öfverholm. Linköping University Hospital: Marie Stenmark-Askmalm, Ekaterina Kuchinskaya. Lund University Hospital: Hans Ehrencrona, Therese Törngren, Anders Kvist, Åke Borg. Stockholm, Karolinska University Hospital: Brita Arver, Annika Lindblom, Emma Tham. Umeå University Hospital: Beatrice Melin. Uppsala University Hospital: Ylva Paulsson-Karlsson.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Meetei, A.R.; Medhurst, A.L.; Ling, C.; Xue, Y.; Singh, T.R.; Bier, P.; Steltenpool, J.; Stone, S.; Dokal, I.; Mathew, C.G.; et al. A human ortholog of archaeal DNA repair protein Hef is defective in Fanconi anemia complementation group M. Nat. Genet. 2005, 37, 958–963. [Google Scholar] [CrossRef] [PubMed]
- Bogliolo, M.; Bluteau, D.; Lespinasse, J.; Pujol, R.; Vasquez, N.; d’Enghien, C.D.; Stoppa-Lyonnet, D.; Leblanc, T.; Soulier, J.; Surralles, J. Biallelic truncating FANCM mutations cause early-onset cancer but not Fanconi anemia. Genet. Med. 2018, 20, 458–463. [Google Scholar] [CrossRef] [PubMed]
- Catucci, I.; Osorio, A.; Arver, B.; Neidhardt, G.; Bogliolo, M.; Zanardi, F.; Riboni, M.; Minardi, S.; Pujol, R.; Azzollini, J.; et al. Individuals with FANCM biallelic mutations do not develop Fanconi anemia, but show risk for breast cancer, chemotherapy toxicity and may display chromosome fragility. Genet. Med. 2018, 20, 452–457. [Google Scholar] [CrossRef] [PubMed]
- Lek, M.; Karczewski, K.J.; Minikel, E.V.; Samocha, K.E.; Banks, E.; Fennell, T.; O’Donnell-Luria, A.H.; Ware, J.S.; Hill, A.J.; Cummings, B.B.; et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature 2016, 536, 285–291. [Google Scholar] [CrossRef] [PubMed]
- Kiiski, J.I.; Pelttari, L.M.; Khan, S.; Freysteinsdottir, E.S.; Reynisdottir, I.; Hart, S.N.; Shimelis, H.; Vilske, S.; Kallioniemi, A.; Schleutker, J.; et al. Exome sequencing identifies FANCM as a susceptibility gene for triple-negative breast cancer. Proc. Natl. Acad. Sci. USA 2014, 111, 15172–15177. [Google Scholar] [CrossRef] [PubMed]
- Kiiski, J.I.; Fagerholm, R.; Tervasmaki, A.; Pelttari, L.M.; Khan, S.; Jamshidi, M.; Mantere, T.; Pylkas, K.; Bartek, J.; Bartkova, J.; et al. FANCM c.5101C>T mutation associates with breast cancer survival and treatment outcome. Int. J. Cancer 2016, 139, 2760–2770. [Google Scholar] [CrossRef] [PubMed]
- Peterlongo, P.; Catucci, I.; Colombo, M.; Caleca, L.; Mucaki, E.; Bogliolo, M.; Marin, M.; Damiola, F.; Bernard, L.; Pensotti, V.; et al. FANCM c.5791C>T nonsense mutation (rs144567652) induces exon skipping, affects DNA repair activity and is a familial breast cancer risk factor. Hum. Mol. Genet. 2015, 24, 5345–5355. [Google Scholar] [CrossRef] [PubMed]
- Kiiski, J.I.; Tervasmaki, A.; Pelttari, L.M.; Khan, S.; Mantere, T.; Pylkas, K.; Mannermaa, A.; Tengstrom, M.; Kvist, A.; Borg, A.; et al. FANCM mutation c.5791C>T is a risk factor for triple-negative breast cancer in the Finnish population. Breast Cancer Res. Treat. 2017, 166, 217–226. [Google Scholar] [CrossRef] [PubMed]
- Neidhardt, G.; Hauke, J.; Ramser, J.; Gross, E.; Gehrig, A.; Muller, C.R.; Kahlert, A.K.; Hackmann, K.; Honisch, E.; Niederacher, D.; et al. Association Between Loss-of-Function Mutations Within the FANCM Gene and Early-Onset Familial Breast Cancer. JAMA Oncol. 2017, 3, 1245–1248. [Google Scholar] [CrossRef]
- Figlioli, G.; Bogliolo, M.; Catucci, I.; Caleca, L.; Lasheras, S.V.; Pujol, R.; Kiiski, J.I.; Muranen, T.A.; Barnes, D.R.; Dennis, J.; et al. The FANCM:p.Arg658* truncating variant is associated with risk of triple-negative breast cancer. npj Breast Cancer 2019, 5. [Google Scholar] [CrossRef]
- Shimelis, H.; LaDuca, H.; Hu, C.; Hart, S.N.; Na, J.; Thomas, A.; Akinhanmi, M.; Moore, R.M.; Brauch, H.; Cox, A.; et al. Triple-Negative Breast Cancer Risk Genes Identified by Multigene Hereditary Cancer Panel Testing. J. Natl. Cancer Inst. 2018, 110, 855–862. [Google Scholar] [CrossRef]
- Spurdle, A.B.; Healey, S.; Devereau, A.; Hogervorst, F.B.; Monteiro, A.N.; Nathanson, K.L.; Radice, P.; Stoppa-Lyonnet, D.; Tavtigian, S.; Wappenschmidt, B.; et al. ENIGMA—Evidence-based network for the interpretation of germline mutant alleles: An international initiative to evaluate risk and clinical significance associated with sequence variation in BRCA1 and BRCA2 genes. Hum. Mutat. 2012, 33, 2–7. [Google Scholar] [CrossRef]
- Schubert, S.; van Luttikhuizen, J.L.; Auber, B.; Schmidt, G.; Hofmann, W.; Penkert, J.; Davenport, C.F.; Hille-Betz, U.; Wendeburg, L.; Bublitz, J.; et al. The identification of pathogenic variants in BRCA1/2 negative, high risk, hereditary breast and/or ovarian cancer patients: High frequency of FANCM pathogenic variants. Int. J. Cancer 2019, 144, 2683–2694. [Google Scholar] [CrossRef] [PubMed]
- Janavicius, R.; Rudaitis, V.; Feng, B.J.; Ozolina, S.; Griskevicius, L.; Goldgar, D.; Tihomirova, L. Haplotype analysis and ancient origin of the BRCA1 c.4035delA Baltic founder mutation. Eur. J. Med. Genet. 2013, 56, 125–130. [Google Scholar] [CrossRef] [PubMed]
- Janavicius, R.; Rudaitis, V.; Mickys, U.; Elsakov, P.; Griskevicius, L. Comprehensive BRCA1 and BRCA2 mutational profile in Lithuania. Cancer Genet. 2014, 207, 195–205. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).