Differential Dependency of Human Pancreatic Cancer Cells on Targeting PTEN via PLK 1 Expression
Abstract
1. Introduction
2. Results
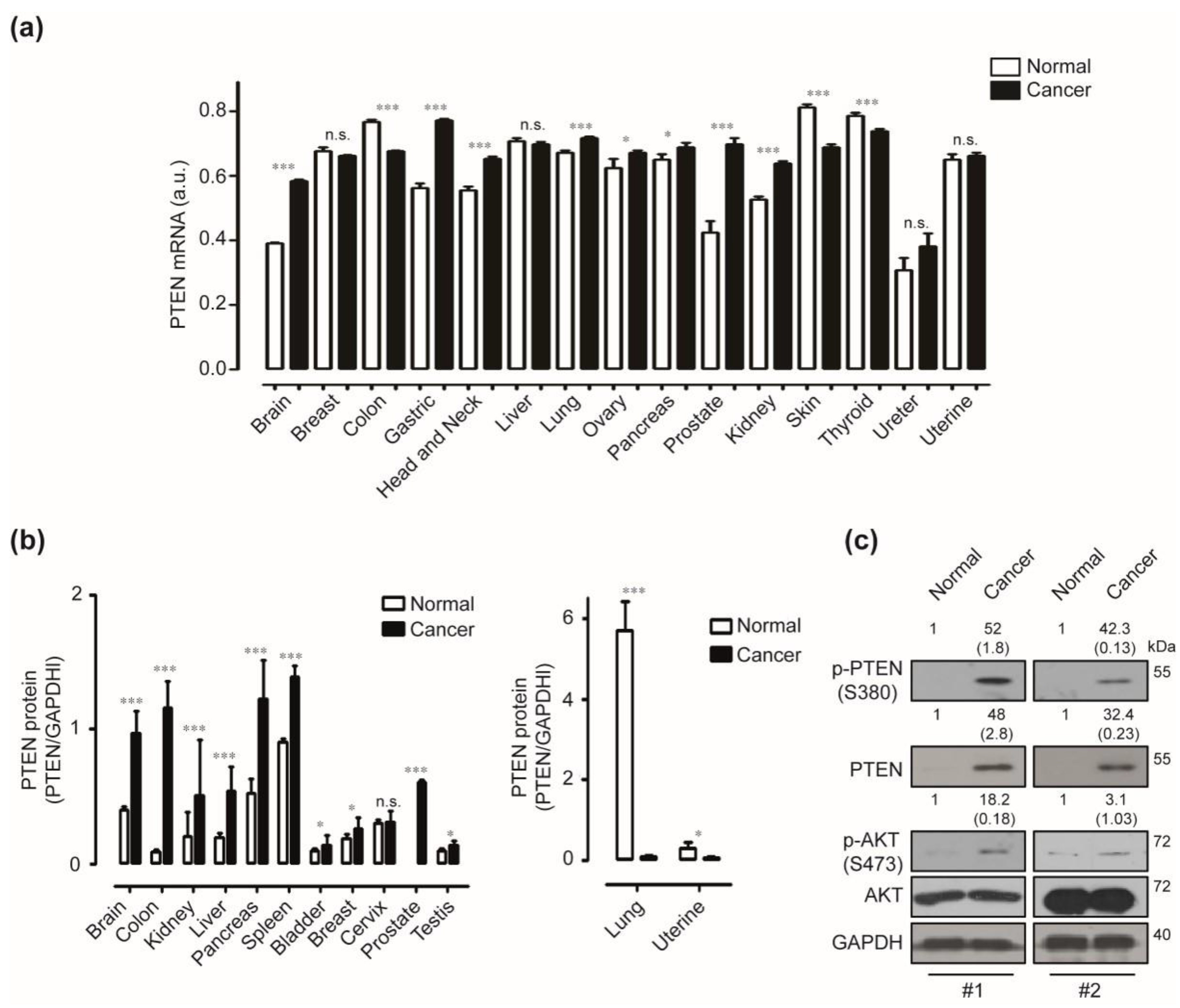
2.1. Expression of Phosphatase and Tensin Homologue (PTEN) in Human Pancreatic Cancer
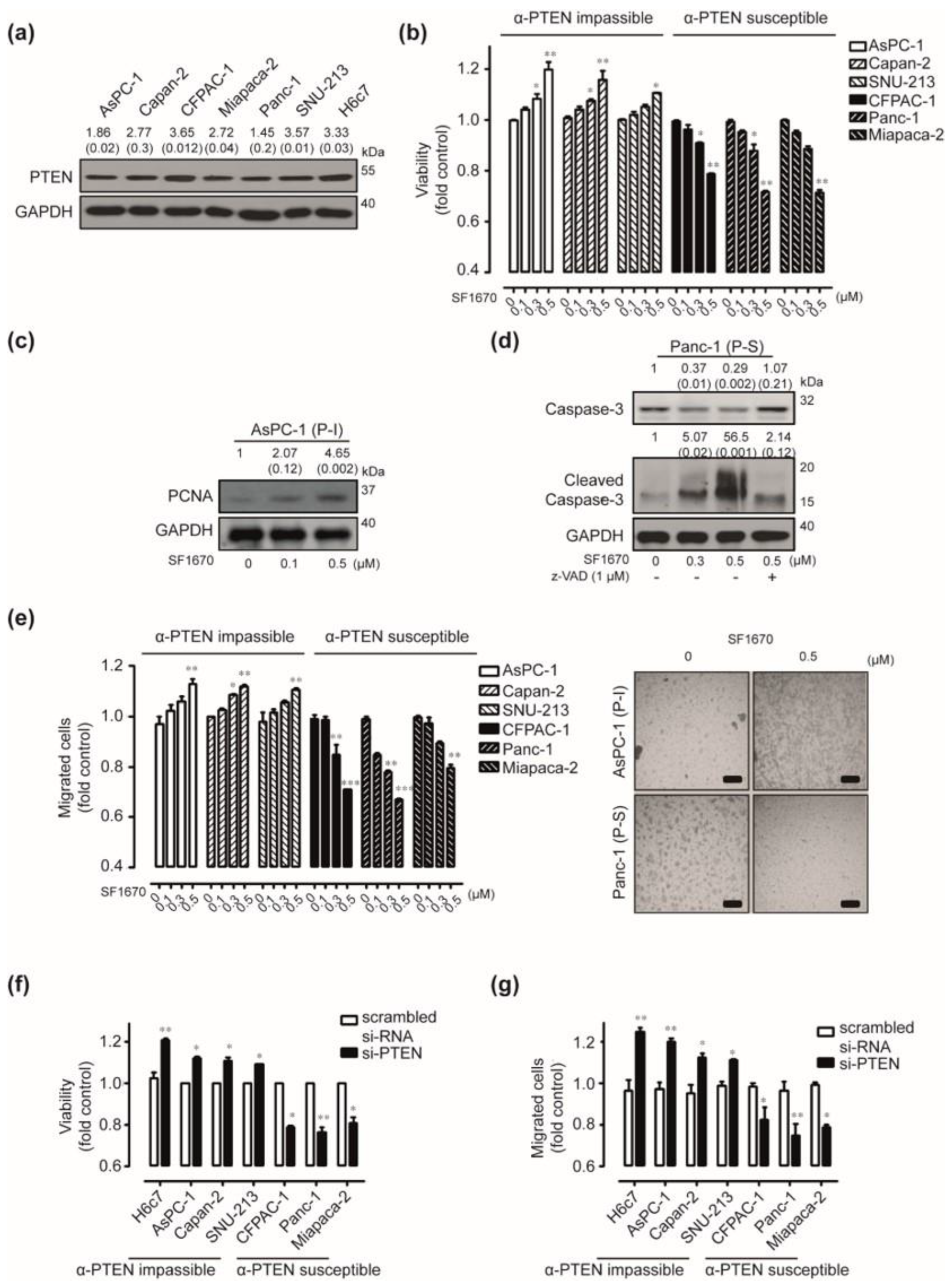
2.2. In Vitro Effects of PTEN Inhibition
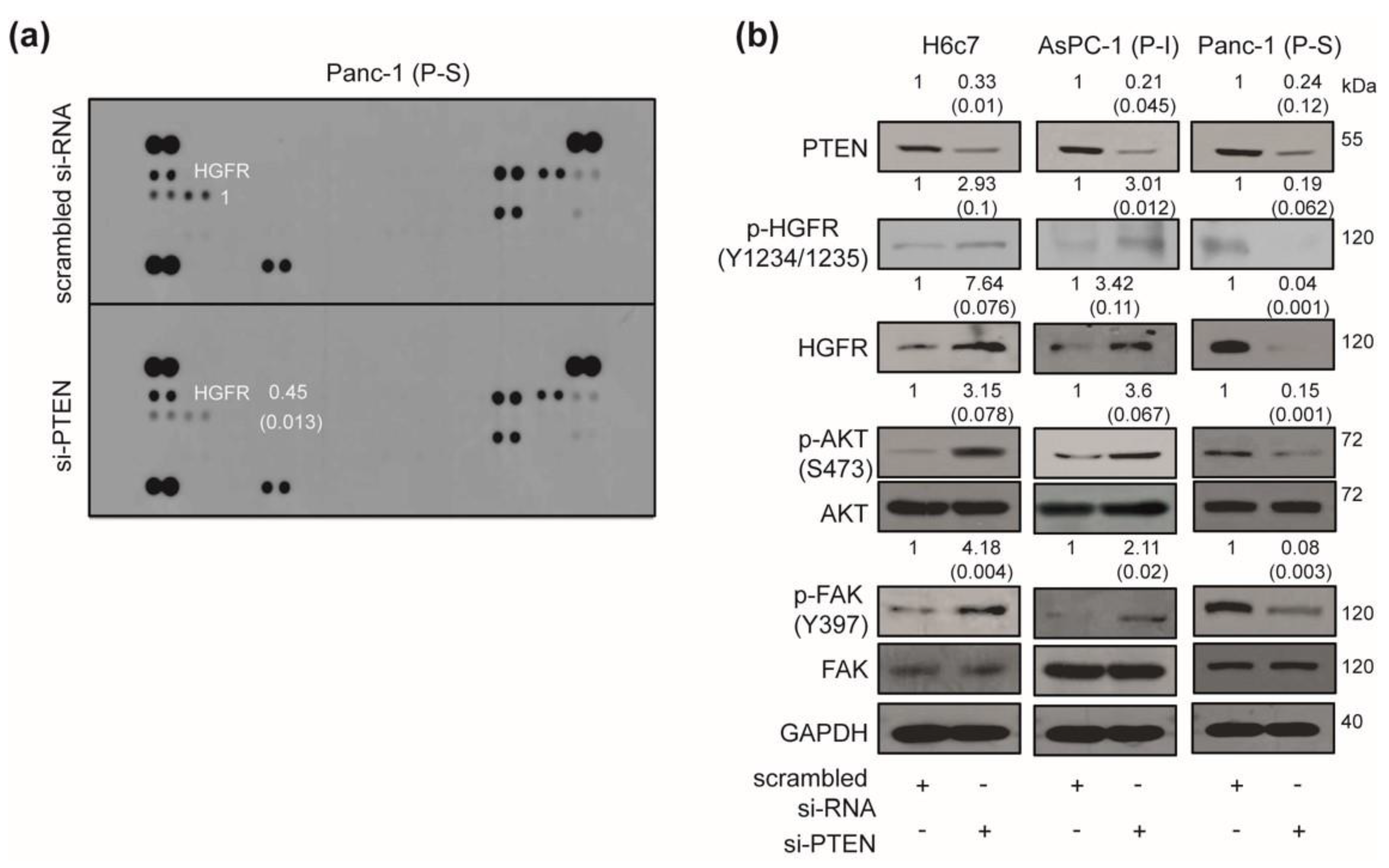
2.3. Effect of PTEN Inhibition on the Intracellular Signaling Pathway
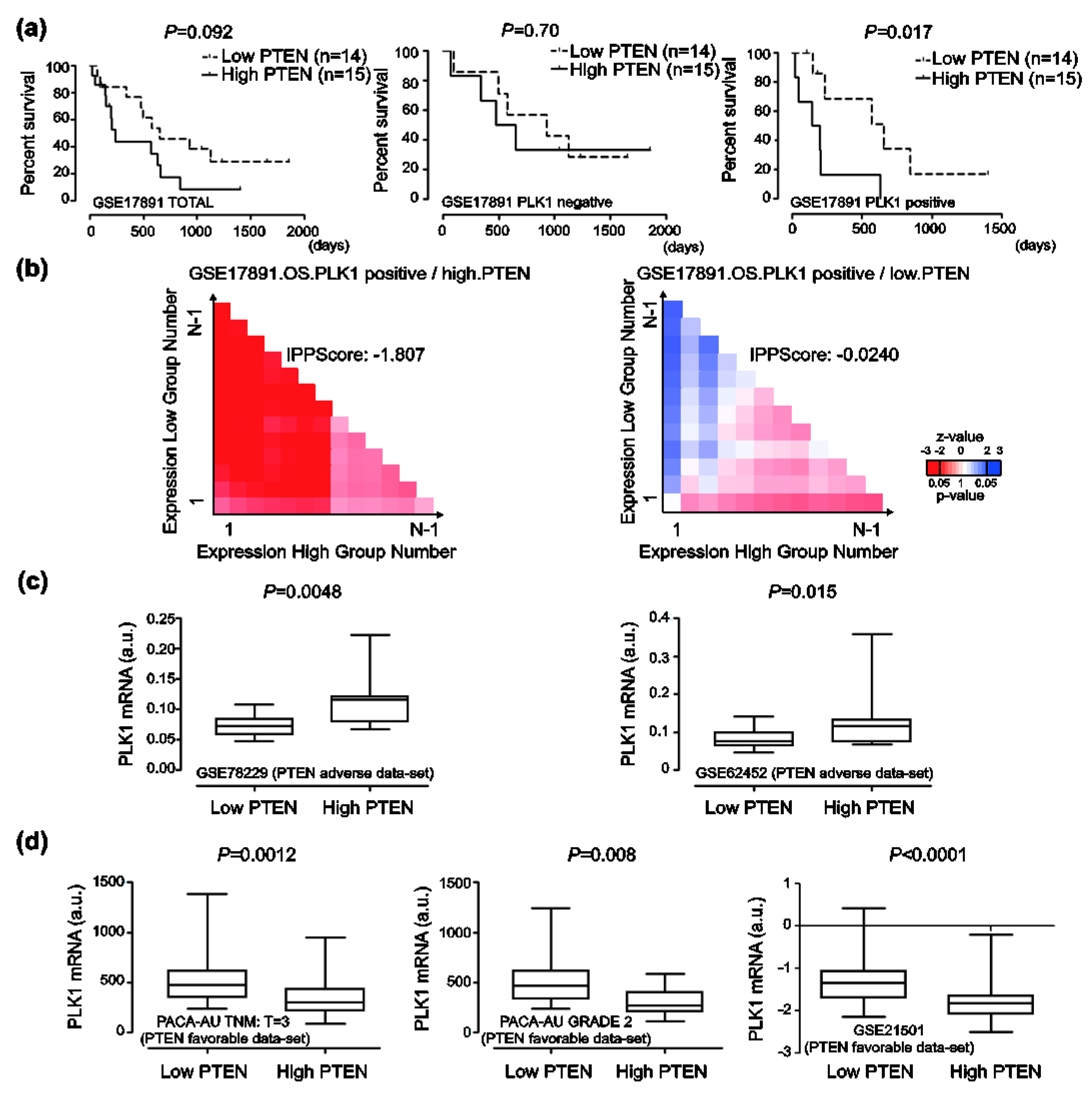
2.4. Correlation Between Polo Like Kinase1 (PLK1) Expressions and Responses to PTEN Regulation in Human Pancreatic Cancer
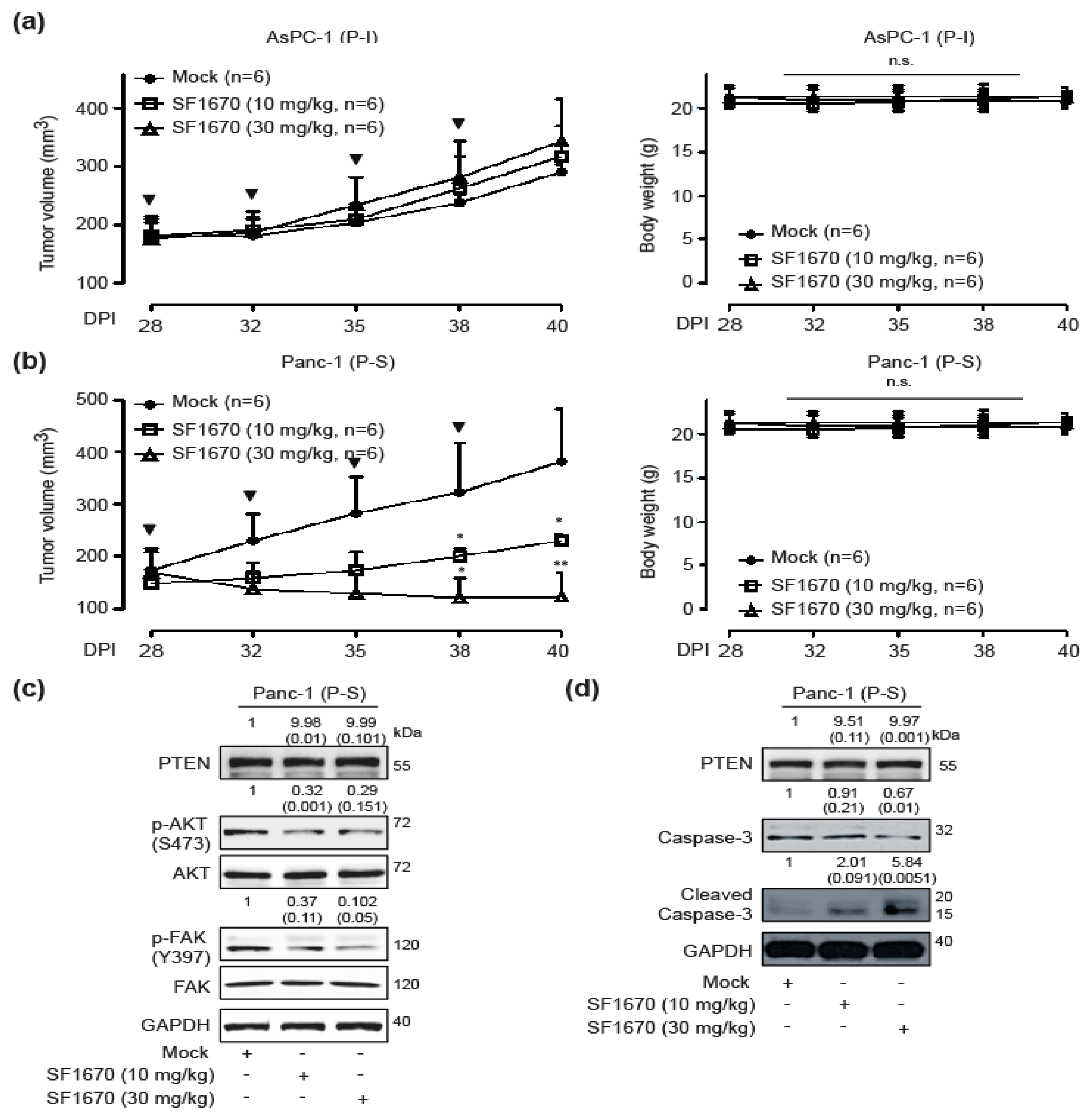
2.5. In Vivo Effects of PTEN Inhibition
2.6. Prognosis Associated with PLK1 Expression in Human Pancreatic Cancer
3. Discussion
4. Materials and Methods
4.1. Gene Expression Analysis
4.2. Cell Culture and Reagents
4.3. Transfection with Small Interfering RNA (siRNA) and Overexpression Construct
4.4. Phosphor-RTK Array
4.5. Measurement of Cell Viability
4.6. Trans-Well Migration Assay
4.7. Xenograft Tumour Model
4.8. GSE Data-Set Analysis
4.9. Iterative Patient Partitioning (IPP) Method
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Abubakar, I.I.; Tillmann, T.; Banerjee, A. Global, regional, and national age-sex specific all-cause and cause-specific mortality for 240 causes of death, 1990–2013: A systematic analysis for the Global Burden of Disease Study 2013. Lancet 2015, 385, 117–171. [Google Scholar] [CrossRef]
- Ryan, D.P.; Hong, T.S.; Bardeesy, N. Pancreatic adenocarcinoma. N. Engl. J. Med. 2014, 371, 2140–2141. [Google Scholar] [CrossRef]
- Blanco-Aparicio, C.; Renner, O.; Leal, J.F.; Carnero, A. PTEN, more than the AKT pathway. Carcinogenesis 2007, 28, 1379–1386. [Google Scholar] [CrossRef] [PubMed]
- Song, M.S.; Salmena, L.; Pandolfi, P.P. The functions and regulation of the PTEN tumour suppressor. Nat. Rev. Mol. Cell Biol. 2012, 13, 283–296. [Google Scholar] [CrossRef] [PubMed]
- Freeman, D.J.; Li, A.G.; Wei, G.; Li, H.H.; Kertesz, N.; Lesche, R.; Whale, A.D.; Martinez-Diaz, H.; Rozengurt, N.; Cardiff, R.D.; et al. PTEN tumor suppressor regulates p53 protein levels and activity through phosphatase-dependent and -independent mechanisms. Cancer Cell 2003, 3, 117–130. [Google Scholar] [CrossRef]
- Gu, J.; Tamura, M.; Yamada, K.M. Tumor suppressor PTEN inhibits integrin- and growth factor-mediated mitogen-activated protein (MAP) kinase signaling pathways. J. Cell Biol. 1998, 143, 1375–1383. [Google Scholar] [CrossRef]
- Gu, J.; Tamura, M.; Pankov, R.; Danen, E.H.; Takino, T.; Matsumoto, K.; Yamada, K.M. Shc and FAK differentially regulate cell motility and directionality modulated by PTEN. J. Cell Biol. 1999, 146, 389–403. [Google Scholar] [CrossRef]
- Tamura, M.; Gu, J.; Takino, T.; Yamada, K.M. Tumor suppressor PTEN inhibition of cell invasion, migration, and growth: Differential involvement of focal adhesion kinase and p130Cas. Cancer Res. 1999, 59, 442–449. [Google Scholar]
- Li, Z.; Li, J.; Bi, P.; Lu, Y.; Burcham, G.; Elzey, B.D.; Ratliff, T.; Konieczny, S.F.; Ahmad, N.; Kuang, S.; et al. Plk1 phosphorylation of PTEN causes a tumor-promoting metabolic state. Mol. Cell. Biol. 2014, 34, 3642–3661. [Google Scholar] [CrossRef]
- Gutteridge, R.E.; Ndiaye, M.A.; Liu, X.; Ahmad, N. Plk1 Inhibitors in Cancer Therapy: From Laboratory to Clinics. Mol. Cancer Ther. 2016, 15, 1427–1435. [Google Scholar] [CrossRef]
- Liu, Z.; Sun, Q.; Wang, X. PLK1, A Potential Target for Cancer Therapy. Transl. Oncol. 2017, 10, 22–32. [Google Scholar] [CrossRef] [PubMed]
- Lin, P.; Xiong, D.D.; Dang, Y.W.; Yang, H.; He, Y.; Wen, D.Y.; Qin, X.G.; Chen, G. The anticipating value of PLK1 for diagnosis, progress and prognosis and its prospective mechanism in gastric cancer: A comprehensive investigation based on high-throughput data and immunohistochemical validation. Oncotarget 2017, 8, 92497–92521. [Google Scholar] [CrossRef]
- Degenhardt, Y.; Lampkin, T. Targeting Polo-like kinase in cancer therapy. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2010, 16, 384–389. [Google Scholar] [CrossRef] [PubMed]
- Khleif, S.N.; Doroshow, J.H.; Hait, W.N.; Collaborative, A.F.N.C.B. AACR-FDA-NCI Cancer Biomarkers Collaborative consensus report: Advancing the use of biomarkers in cancer drug development. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2010, 16, 3299–3318. [Google Scholar] [CrossRef]
- Duffy, M.J.; Crown, J. Companion biomarkers: Paving the pathway to personalized treatment for cancer. Clin. Chem. 2013, 59, 1447–1456. [Google Scholar] [CrossRef] [PubMed]
- Jorgensen, J.T.; Hersom, M. Companion diagnostics-a tool to improve pharmacotherapy. Ann. Transl. Med. 2016, 4, 482. [Google Scholar] [CrossRef] [PubMed]
- Bhardwaj, V.; Cascone, T.; Cortez, M.A.; Amini, A.; Evans, J.; Komaki, R.U.; Heymach, J.V.; Welsh, J.W. Modulation of c-Met signaling and cellular sensitivity to radiation: Potential implications for therapy. Cancer 2013, 119, 1768–1775. [Google Scholar] [CrossRef]
- Zhao, S.; Cao, L.; Freeman, J.W. Knockdown of RON receptor kinase delays but does not prevent tumor progression while enhancing HGF/MET signaling in pancreatic cancer cell lines. Oncogenesis 2013, 2, e76. [Google Scholar] [CrossRef]
- Li, C.; Zhao, Y.; Yang, D.; Yu, Y.; Guo, H.; Zhao, Z.; Zhang, B.; Yin, X. Inhibitory effects of kaempferol on the invasion of human breast carcinoma cells by downregulating the expression and activity of matrix metalloproteinase-9. Biochem. Cell Biol. 2015, 93, 16–27. [Google Scholar] [CrossRef]
- Schmidt, J.; Abel, U.; Debus, J.; Harig, S.; Hoffmann, K.; Herrmann, T.; Bartsch, D.; Klein, J.; Mansmann, U.; Jager, D.; et al. Open-label, multicenter, randomized phase III trial of adjuvant chemoradiation plus interferon Alfa-2b versus fluorouracil and folinic acid for patients with resected pancreatic adenocarcinoma. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2012, 30, 4077–4083. [Google Scholar] [CrossRef]
- Yamada, K.M.; Araki, M. Tumor suppressor PTEN: Modulator of cell signaling, growth, migration and apoptosis. J. Cell Sci. 2001, 114, 2375–2382. [Google Scholar] [PubMed]
- Burris, H.A., 3rd; Moore, M.J.; Andersen, J.; Green, M.R.; Rothenberg, M.L.; Modiano, M.R.; Cripps, M.C.; Portenoy, R.K.; Storniolo, A.M.; Tarassoff, P.; et al. Improvements in survival and clinical benefit with gemcitabine as first-line therapy for patients with advanced pancreas cancer: A randomized trial. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 1997, 15, 2403–2413. [Google Scholar] [CrossRef] [PubMed]
- Heinemann, V.; Boeck, S.; Hinke, A.; Labianca, R.; Louvet, C. Meta-analysis of randomized trials: Evaluation of benefit from gemcitabine-based combination chemotherapy applied in advanced pancreatic cancer. BMC Cancer 2008, 8, 82. [Google Scholar] [CrossRef] [PubMed]
- Ychou, M.; Conroy, T.; Seitz, J.F.; Gourgou, S.; Hua, A.; Mery-Mignard, D.; Kramar, A. An open phase I study assessing the feasibility of the triple combination: Oxaliplatin plus irinotecan plus leucovorin/ 5-fluorouracil every 2 weeks in patients with advanced solid tumors. Ann. Oncol. Off. J. Eur. Soc. Med Oncol. 2003, 14, 481–489. [Google Scholar] [CrossRef]
- Conroy, T.; Paillot, B.; Francois, E.; Bugat, R.; Jacob, J.H.; Stein, U.; Nasca, S.; Metges, J.P.; Rixe, O.; Michel, P.; et al. Irinotecan plus oxaliplatin and leucovorin-modulated fluorouracil in advanced pancreatic cancer—A Groupe Tumeurs Digestives of the Federation Nationale des Centres de Lutte Contre le Cancer study. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2005, 23, 1228–1236. [Google Scholar] [CrossRef]
- Chatelut, E.; Delord, J.P.; Canal, P. Toxicity patterns of cytotoxic drugs. Investig. New Drugs 2003, 21, 141–148. [Google Scholar] [CrossRef]
- Borsoi, C.; Leonard, F.; Lee, Y.; Zaid, M.; Elganainy, D.; Alexander, J.F.; Kai, M.; Liu, Y.T.; Kang, Y.; Liu, X.; et al. Gemcitabine enhances the transport of nanovector-albumin-bound paclitaxel in gemcitabine-resistant pancreatic ductal adenocarcinoma. Cancer Lett. 2017, 403, 296–304. [Google Scholar] [CrossRef]
- Ito, T.; Deguchi, K.; Yoshii, K.; Kashii, M. A Case of Cystoid Macular Edema Secondary to Albumin-Bound Paclitaxel Therapy. Gan Kagaku Ryoho. Cancer Chemother. 2017, 44, 599–602. [Google Scholar]
- Giordano, G.; Pancione, M.; Olivieri, N.; Parcesepe, P.; Velocci, M.; Di Raimo, T.; Coppola, L.; Toffoli, G.; D’Andrea, M.R. Nano albumin bound-paclitaxel in pancreatic cancer: Current evidences and future directions. World J. Gastroenterol. 2017, 23, 5875–5886. [Google Scholar] [CrossRef]
- Patel, M.B.; Pothula, S.P.; Xu, Z.; Lee, A.K.; Goldstein, D.; Pirola, R.C.; Apte, M.V.; Wilson, J.S. The role of the hepatocyte growth factor/c-MET pathway in pancreatic stellate cell-endothelial cell interactions: Antiangiogenic implications in pancreatic cancer. Carcinogenesis 2014, 35, 1891–1900. [Google Scholar] [CrossRef]
- Li, C.; Wu, J.J.; Hynes, M.; Dosch, J.; Sarkar, B.; Welling, T.H.; Pasca di Magliano, M.; Simeone, D.M. c-Met Is a Marker of Pancreatic Cancer Stem Cells and Therapeutic Target. Gastroenterology 2011, 141, 2218–2227. [Google Scholar] [CrossRef] [PubMed]
- Abounader, R.; Reznik, T.; Colantuoni, C.; Martinez-Murillo, F.; Rosen, E.M.; Laterra, J. Regulation of c-Met-dependent gene expression by PTEN. Oncogene 2004, 23, 9173–9182. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.H.; Zhang, Y.P. Maximizing the commercial value of personalized therapeutics and companion diagnostics. Nat. Biotechnol. 2013, 31, 803–805. [Google Scholar] [CrossRef] [PubMed]
- Milne, C.P.; Cohen, J.P.; Chakravarthy, R. Market watch: Where is personalized medicine in industry heading? Nat. Rev. Drug Discov. 2015, 14, 812–813. [Google Scholar] [CrossRef] [PubMed]
- Havel, J.J.; Chowell, D.; Chan, T.A. The evolving landscape of biomarkers for checkpoint inhibitor immunotherapy. Nat. Rev. Cancer 2019, 19, 133–150. [Google Scholar] [CrossRef]
- MacConaill, L.E.; Van Hummelen, P.; Meyerson, M.; Hahn, W.C. Clinical implementation of comprehensive strategies to characterize cancer genomes: Opportunities and challenges. Cancer Discov. 2011, 1, 297–311. [Google Scholar] [CrossRef]
- Lee, J.; Choi, C. Oncopression: Gene expression compendium for cancer with matched normal tissues. Bioinformatics 2017, 33, 2068–2070. [Google Scholar] [CrossRef]
- Lee, J.; Hun Yun, J.; Lee, J.; Choi, C.; Hoon Kim, J. Blockade of dual-specificity phosphatase 28 decreases chemo-resistance and migration in human pancreatic cancer cells. Sci. Rep. 2015, 5, 12296. [Google Scholar] [CrossRef]
- Radulovich, N.; Qian, J.Y.; Tsao, M.S. Human pancreatic duct epithelial cell model for KRAS transformation. Methods Enzymol. 2008, 439, 1–13. [Google Scholar] [CrossRef]
- Lee, J.; Kim, J.H. Kaempferol Inhibits Pancreatic Cancer Cell Growth and Migration through the Blockade of EGFR-Related Pathway In Vitro. PLoS ONE 2016, 11, e0155264. [Google Scholar] [CrossRef]
- Lee, J.; Lee, J.; Yun, J.H.; Choi, C.; Cho, S.; Kim, S.J.; Kim, J.H. Autocrine DUSP28 signaling mediates pancreatic cancer malignancy via regulation of PDGF-A. Sci. Rep. 2017, 7, 12760. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Ku, T.; Yu, H.; Chong, K.; Ryu, S.W.; Choi, K.; Choi, C. Blockade of VEGF-A suppresses tumor growth via inhibition of autocrine signaling through FAK and AKT. Cancer Lett. 2012, 318, 221–225. [Google Scholar] [CrossRef] [PubMed]
- Zaykin, D.V. Optimally weighted Z-test is a powerful method for combining probabilities in meta-analysis. J. Evol. Biol. 2011, 24, 1836–1841. [Google Scholar] [CrossRef] [PubMed]


| Data-Set | SQRT Low Group (N) | SQRT High Group (N) | PLK1 Low Group IPPScore | PLK1 High Group IPPScore |
|---|---|---|---|---|
| E-MEXP-2780 | 4.3588 (19) | 3.3166 (11) | −1.02099 | −1.1888 |
| GSE17891 | 3.6055 (13) | 3.7416 (14) | −0.024019 | −1.8071 |
| GSE62452 | 7 (49) | 4.1231 (17) | 0.502234 | 0.594234 |
| GSE21501 | 7.6157 (58) | 6.6332 (44) | 0.533184 | −0.247134 |
| GSE57495 | 5.6568 (32) | 5.5677 (31) | −0.790192 | −0.710717 |
| GSE71729 | 8.3066 (69) | 7.4833 (56) | −0.899932 | −0.188088 |
| GSE84219 | 4.1231 (17) | 3.6055 (13) | 1.29252 | 0.809448 |
| PAAD-US | 9.2195 (85) | 7.2801 (53) | −1.09615 | −1.23564 |
| PACA-AU | 6.9282 (48) | 5.5677 (31) | 0.85567 | −1.15497 |
| Liptak’s z-value | −0.392674332 | −1.68977768 | ||
| Liptak’s p-value | 0.347280015 | 0.045535248 | ||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, J.; Lee, J.; Sim, W.; Kim, J.-H. Differential Dependency of Human Pancreatic Cancer Cells on Targeting PTEN via PLK 1 Expression. Cancers 2020, 12, 277. https://doi.org/10.3390/cancers12020277
Lee J, Lee J, Sim W, Kim J-H. Differential Dependency of Human Pancreatic Cancer Cells on Targeting PTEN via PLK 1 Expression. Cancers. 2020; 12(2):277. https://doi.org/10.3390/cancers12020277
Chicago/Turabian StyleLee, Jungwhoi, Jungsul Lee, Woogwang Sim, and Jae-Hoon Kim. 2020. "Differential Dependency of Human Pancreatic Cancer Cells on Targeting PTEN via PLK 1 Expression" Cancers 12, no. 2: 277. https://doi.org/10.3390/cancers12020277
APA StyleLee, J., Lee, J., Sim, W., & Kim, J.-H. (2020). Differential Dependency of Human Pancreatic Cancer Cells on Targeting PTEN via PLK 1 Expression. Cancers, 12(2), 277. https://doi.org/10.3390/cancers12020277





