Commercialized Blood-, Urinary- and Tissue-Based Biomarker Tests for Prostate Cancer Diagnosis and Prognosis
Abstract
:Simple Summary
Abstract
1. Introduction
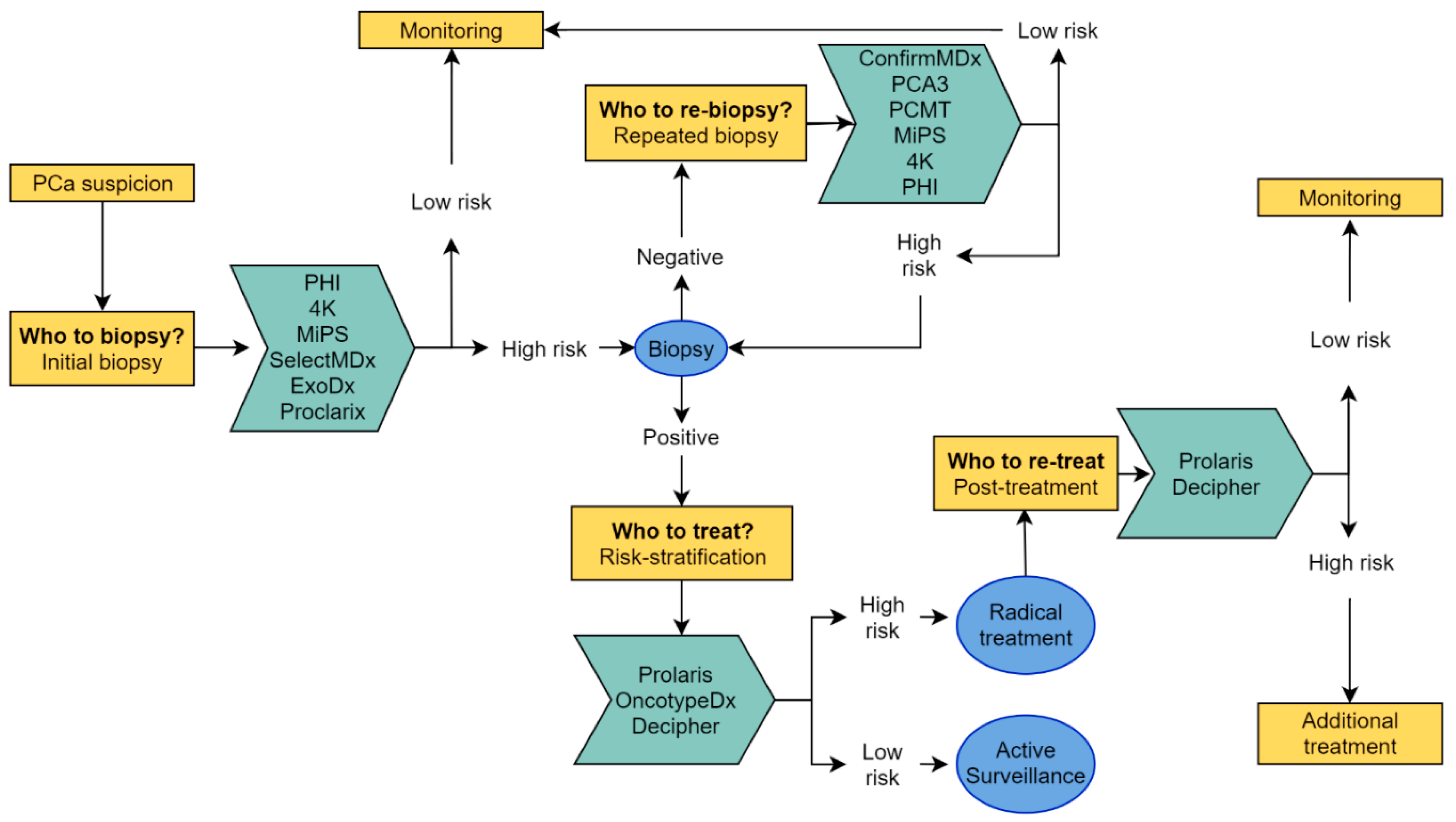
2. Biomarker Tests
2.1. Pre-Biopsy: Who to Biopsy?
2.1.1. PHI
2.1.2. 4K Score
2.1.3. Michigan Prostate Score
2.1.4. ExoDx Prostate Test (IntelliScore)
2.1.5. Proclarix
2.1.6. SelectMDx
2.2. Post-Negative Biopsy: Who to re-Biopsy?
2.2.1. Progensa PCA3-Test
2.2.2. ConfirmMDx
2.2.3. Prostate Core Mitomic Test (PCMT)
2.3. Post-Positive Biopsy and Post-Definitive Treatment: Who to (Re)-Treat?
2.3.1. OncotypeDx
2.3.2. Decipher
2.3.3. Prolaris
3. Comparing Biomarker Tests
4. MRI and Biomarkers
5. Discussion
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lilja, H.; Ulmert, D.; Vickers, A.J. Prostate-specific antigen and prostate cancer: Prediction, detection and monitoring. Nat. Rev. Cancer 2008, 8, 268–278. [Google Scholar] [CrossRef] [PubMed]
- Loeb, S.; Sanda, M.G.; Broyles, D.L.; Shin, S.S.; Bangma, C.H.; Wei, J.T.; Partin, A.W.; Klee, G.G.; Slawin, K.M.; Marks, L.S.; et al. The prostate health index selectively identifies clinically significant prostate cancer. J. Urol. 2015, 193, 1163–1169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Catalona, W.J.; Partin, A.W.; Sanda, M.G.; Wei, J.T.; Klee, G.G.; Bangma, C.H.; Slawin, K.M.; Marks, L.S.; Loeb, S.; Broyles, D.L.; et al. A multicenter study of [-2]pro-prostate specific antigen combined with prostate specific antigen and free prostate specific antigen for prostate cancer detection in the 2.0 to 10.0 ng/mL prostate specific antigen range. J. Urol. 2011, 185, 1650–1655. [Google Scholar] [CrossRef] [Green Version]
- Stephan, C.; Vincendeau, S.; Houlgatte, A.; Cammann, H.; Jung, K.; Semjonow, A. Multicenter evaluation of [-2]proprostate-specific antigen and the prostate health index for detecting prostate cancer. Clin. Chem. 2013, 59, 306–314. [Google Scholar] [CrossRef] [Green Version]
- De La Calle, C.; Patil, D.; Wei, J.T.; Scherr, D.S.; Sokoll, L.; Chan, D.W.; Siddiqui, J.; Mosquera, J.M.; Rubin, M.A.; Sanda, M.G. Multicenter evaluation of the prostate health index to detect aggressive prostate cancer in biopsy Naïve men. J. Urol. 2015, 194, 65–72. [Google Scholar] [CrossRef] [Green Version]
- Wang, W.; Wang, M.; Wang, L.; Adams, T.S.; Tian, Y.; Xu, J. Diagnostic ability of %p2PSA and prostate health index for aggressive prostate cancer: A meta-analysis. Sci. Rep. 2014, 4, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Park, H.; Lee, S.W.; Song, G.; Kang, T.W.; Jung, J.H.; Chung, H.C.; Kim, S.J.; Park, J.Y.; Shin, T.Y.; Kim, J.H. Preoperative prostate health index and %p2PSA as the significant biomarkers of postoperative pathological outcomes of prostate cancer in Korean males: A prospective multi-institutional study. Investig. Clin. Urol. 2020, 61, 42–50. [Google Scholar] [CrossRef]
- Guazzoni, G.; Lazzeri, M.; Nava, L.; Lughezzani, G.; Larcher, A.; Scattoni, V.; Gadda, G.M.; Bini, V.; Cestari, A.; Buffi, N.M.; et al. Preoperative prostate-specific antigen isoform p2PSA and its derivatives, %p2PSA and prostate health index, predict pathologic outcomes in patients undergoing radical prostatectomy for prostate cancer. Eur. Urol. 2012, 61, 455–466. [Google Scholar] [CrossRef]
- Fossati, N.; Buffi, N.M.; Haese, A.; Stephan, C.; Larcher, A.; McNicholas, T.; De La Taille, A.; Freschi, M.; Lughezzani, G.; Abrate, A.; et al. Preoperative prostate-specific antigen isoform p2PSA and its derivatives, %p2PSA and prostate health index, predict pathologic outcomes in patients undergoing radical prostatectomy for prostate cancer: Results from a multicentric european prospective study. Eur. Urol. 2015, 68, 132–138. [Google Scholar] [CrossRef]
- Lazzeri, M.; Haese, A.; De La Taille, A.; Palou Redorta, J.; McNicholas, T.; Lughezzani, G.; Scattoni, V.; Bini, V.; Freschi, M.; Sussman, A.; et al. Serum isoform [-2]proPSA derivatives significantly improve prediction of prostate cancer at initial biopsy in a total PSA range of 2–10 ng/mL: A multicentric european study. Eur. Urol. 2013, 63, 986–994. [Google Scholar] [CrossRef] [PubMed]
- Filella, X.; Foj, L.; Augé, J.M.; Molina, R.; Alcover, J. Clinical utility of %p2PSA and prostate health index in the detection of prostate cancer. Clin. Chem. Lab. Med. 2014, 52, 1347–1355. [Google Scholar] [CrossRef]
- Parekh, D.J.; Punnen, S.; Sjoberg, D.D.; Asroff, S.W.; Bailen, J.L.; Cochran, J.S.; Concepcion, R.; David, R.D.; Deck, K.B.; Dumbadze, I.; et al. A Multi-institutional Prospective Trial in the USA Confirms that the 4Kscore Accurately Identifies Men with High-grade Prostate Cancer. Eur. Urol. 2015, 68, 464–470. [Google Scholar] [CrossRef] [PubMed]
- Punnen, S.; Freedland, S.J.; Polascik, T.J.; Loeb, S.; Risk, M.C.; Savage, S.; Mathur, S.C.; Uchio, E.; Dong, Y.; Silberstein, J.L. A Multi-Institutional Prospective Trial Confirms Noninvasive Blood Test Maintains Predictive Value in African American Men. J. Urol. 2018, 199, 1459–1463. [Google Scholar] [CrossRef] [PubMed]
- Van Neste, L.; Hendriks, R.J.; Dijkstra, S.; Trooskens, G.; Cornel, E.B.; Jannink, S.A.; de Jong, H.; Hessels, D.; Smit, F.P.; Melchers, W.J.G.; et al. Detection of High-grade Prostate Cancer Using a Urinary Molecular Biomarker–Based Risk Score. Eur. Urol. 2016, 70, 740–748. [Google Scholar] [CrossRef] [PubMed]
- Haese, A.; Trooskens, G.; Steyaert, S.; Hessels, D.; Brawer, M.; Vlaeminck-Guillem, V.; Ruffion, A.; Tilki, D.; Schalken, J.; Groskopf, J.; et al. Multicenter Optimization and Validation of a 2-Gene mRNA Urine Test for Detection of Clinically Significant Prostate Cancer before Initial Prostate Biopsy. J. Urol. 2019, 202, 256–262. [Google Scholar] [CrossRef] [PubMed]
- McKiernan, J.; Donovan, M.J.; O’Neill, V.; Bentink, S.; Noerholm, M.; Belzer, S.; Skog, J.; Kattan, M.W.; Partin, A.; Andriole, G.; et al. A novel urine exosome gene expression assay to predict high-grade prostate cancer at initial biopsy. JAMA Oncol. 2016, 2, 882–889. [Google Scholar] [CrossRef] [Green Version]
- McKiernan, J.; Donovan, M.J.; Margolis, E.; Partin, A.; Carter, B.; Brown, G.; Torkler, P.; Noerholm, M.; Skog, J.; Shore, N.; et al. A Prospective Adaptive Utility Trial to Validate Performance of a Novel Urine Exosome Gene Expression Assay to Predict High-grade Prostate Cancer in Patients with Prostate-specific Antigen 2–10ng/mL at Initial Biopsy. Eur. Urol. 2018, 74, 731–738. [Google Scholar] [CrossRef] [Green Version]
- Sanda, M.G.; Feng, Z.; Howard, D.H.; Tomlins, S.A.; Sokoll, L.J.; Chan, D.W.; Regan, M.M.; Groskopf, J.; Chipman, J.; Patil, D.H.; et al. Association Between Combined TMPRSS2:ERG and PCA3 RNA Urinary Testing and Detection of Aggressive Prostate Cancer. JAMA Oncol. 2017, 3, 1085–1093. [Google Scholar] [CrossRef]
- Tomlins, S.A.; Day, J.R.; Lonigro, R.J.; Hovelson, D.H.; Siddiqui, J.; Kunju, L.P.; Dunn, R.L.; Meyer, S.; Hodge, P.; Groskopf, J.; et al. Urine TMPRSS2:ERG Plus PCA3 for Individualized Prostate Cancer Risk Assessment. Eur. Urol. 2016, 70, 45–53. [Google Scholar] [CrossRef] [Green Version]
- Klocker, H.; Golding, B.; Weber, S.; Steiner, E.; Tennstedt, P.; Keller, T.; Schiess, R.; Gillessen, S.; Horninger, W.; Steuber, T. Development and validation of a novel multivariate risk score to guide biopsy decision for the diagnosis of clinically significant prostate cancer. BJUI Compass 2020, 1, 15–20. [Google Scholar] [CrossRef]
- Lebastchi, A.H.; Russell, C.M.; Niknafs, Y.S.; Eyrich, N.W.; Chopra, Z.; Botbyl, R.; Kabeer, R.; Osawa, T.; Siddiqui, J.; Siddiqui, R.; et al. Impact of the MyProstateScore (MPS) test on the clinical decision to undergo prostate biopsy: Results from a contemporary academic practice. medRxiv 2020. [Google Scholar] [CrossRef] [PubMed]
- Boukouris, S.; Mathivanan, S. Exosomes in bodily fluids are a highly stable resource of disease biomarkers. Proteomics. Clin. Appl. 2015, 9, 358–367. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Donovan, M.J.; Noerholm, M.; Bentink, S.; Belzer, S.; Skog, J.; O’Neill, V.; Cochran, J.S.; Brown, G.A. A molecular signature of PCA3 and ERG exosomal RNA from non-DRE urine is predictive of initial prostate biopsy result. Prostate Cancer Prostatic Dis. 2015, 18, 370–375. [Google Scholar] [CrossRef]
- Tutrone, R.; Donovan, M.J.; Torkler, P.; Tadigotla, V.; McLain, T.; Noerholm, M.; Skog, J.; McKiernan, J. Clinical utility of the exosome based ExoDx Prostate(IntelliScore) EPI test in men presenting for initial Biopsy with a PSA 2-10 ng/mL. Prostate Cancer Prostatic Dis. 2020. [Google Scholar] [CrossRef]
- McKiernan, J.; Noerholm, M.; Tadigotla, V.; Kumar, S.; Torkler, P.; Sant, G.; Alter, J.; Donovan, M.J.; Skog, J. A urine-based Exosomal gene expression test stratifies risk of high-grade prostate Cancer in men with prior negative prostate biopsy undergoing repeat biopsy. BMC Urol. 2020, 20, 138. [Google Scholar] [CrossRef]
- Robinson, K.; Creed, J.; Reguly, B.; Powell, C.; Wittock, R.; Klein, D.; Maggrah, A.; Klotz, L.; Parr, R.L.; Dakubo, G.D. Accurate prediction of repeat prostate biopsy outcomes by a mitochondrial DNA deletion assay. Prostate Cancer Prostatic Dis. 2010, 13, 126–131. [Google Scholar] [CrossRef] [Green Version]
- Van Neste, L.; Partin, A.W.; Stewart, G.D.; Epstein, J.I.; Harrison, D.J.; Van Criekinge, W. Risk score predicts high-grade prostate cancer in DNA-methylation positive, histopathologically negative biopsies. Prostate 2016, 76, 1078–1087. [Google Scholar] [CrossRef] [Green Version]
- Waterhouse, R.L.J.; Van Neste, L.; Moses, K.A.; Barnswell, C.; Silberstein, J.L.; Jalkut, M.; Tutrone, R.; Sylora, J.; Anglade, R.; Murdock, M.; et al. Evaluation of an Epigenetic Assay for Predicting Repeat Prostate Biopsy Outcome in African American Men. Urology 2019, 128, 62–65. [Google Scholar] [CrossRef] [Green Version]
- Ibrahim, N.H.; Abdellateif, M.S.; Thabet, G.; Kassem, S.H.; El-Salam, M.A.; El-Leithy, A.A.; Selim, M.M. Combining PHI and miRNAs as Biomarkers in Prostate Cancer Diagnosis and Prognosis. Clin. Lab. 2019, 65. [Google Scholar] [CrossRef]
- Foj, L.; Filella, X. Development and internal validation of a novel PHI-nomogram to identify aggressive prostate cancer. Clin. Chim. Acta 2020, 501, 174–178. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.-S.; Fu, X.-J.; Na, R.; Ye, D.-W.; Qi, J.; Lin, X.-L.; Liu, F.; Gong, J.; Zhang, N.; Jiang, G.-L.; et al. Phi-based risk calculators performed better in the prediction of prostate cancer in the Chinese population. Asian J. Androl. 2019, 21, 592–597. [Google Scholar] [CrossRef] [PubMed]
- Vickers, A.J.; Cronin, A.M.; Aus, G.; Pihl, C.-G.; Becker, C.; Pettersson, K.; Scardino, P.T.; Hugosson, J.; Lilja, H. Impact of recent screening on predicting the outcome of prostate cancer biopsy in men with elevated prostate-specific antigen: Data from the European Randomized Study of Prostate Cancer Screening in Gothenburg, Sweden. Cancer 2010, 116, 2612–2620. [Google Scholar] [CrossRef] [PubMed]
- Vickers, A.J.; Cronin, A.M.; Roobol, M.J.; Savage, C.J.; Peltola, M.; Pettersson, K.; Scardino, P.T.; Schröder, F.H.; Lilja, H. A four-kallikrein panel predicts prostate cancer in men with recent screening: Data from the european randomized study of screening for prostate cancer, Rotterdam. Clin. Cancer Res. 2010, 16, 3232–3239. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gupta, A.; Roobol, M.J.; Savage, C.J.; Peltola, M.; Pettersson, K.; Scardino, P.T.; Vickers, A.J.; Schröder, F.H.; Lilja, H. A four-kallikrein panel for the prediction of repeat prostate biopsy: Data from the European Randomized Study of Prostate Cancer Screening in Rotterdam, Netherlands. Br. J. Cancer 2010, 103, 708–714. [Google Scholar] [CrossRef] [Green Version]
- Punnen, S.; Pavan, N.; Parekh, D.J. Finding the Wolf in Sheep’s Clothing: The 4Kscore Is a Novel Blood Test That Can Accurately Identify the Risk of Aggressive Prostate Cancer. Rev. Urol. 2015, 17, 3–13. [Google Scholar] [CrossRef]
- Stattin, P.; Vickers, A.J.; Sjoberg, D.D.; Johansson, R.; Granfors, T.; Johansson, M.; Pettersson, K.; Scardino, P.T.; Hallmans, G.; Lilja, H. Improving the Specificity of Screening for Lethal Prostate Cancer Using Prostate-specific Antigen and a Panel of Kallikrein Markers: A Nested Case-Control Study. Eur. Urol. 2015, 68, 207–213. [Google Scholar] [CrossRef] [Green Version]
- Punnen, S.; Nahar, B.; Prakash, N.S.; Sjoberg, D.D.; Zappala, S.M.; Parekh, D.J. The 4Kscore Predicts the Grade and Stage of Prostate Cancer in the Radical Prostatectomy Specimen: Results from a Multi-institutional Prospective Trial. Eur. Urol. Focus 2017, 3, 94–99. [Google Scholar] [CrossRef]
- Haese, A.; Tin, A.L.; Carlsson, S.V.; Sjoberg, D.D.; Pehrke, D.; Steuber, T.; Huland, H.; Graefen, M.; Scardino, P.T.; Schlomm, T.; et al. A pre-specified model based on four kallikrein markers in blood improves predictions of adverse pathology and biochemical recurrence after radical prostatectomy. Br. J. Cancer 2020, 123, 604–609. [Google Scholar] [CrossRef]
- Hessels, D.; Klein Gunnewiek, J.M.; Oort, I. DD3(PCA3)-based molecular urine analysis for the diagnosis of prostate cancer. Eur. Urol. 2003, 44, 8–15. [Google Scholar] [CrossRef]
- Magi-Galluzzi, C.; Tsusuki, T.; Elson, P.; Simmerman, K.; LaFargue, C.; Esgueva, R.; Klein, E.; Rubin, M.A.; Zhou, M. TMPRSS2-ERG gene fusion prevalence and class are significantly different in prostate cancer of Caucasian, African-American and Japanese patients. Prostate 2011, 71, 489–497. [Google Scholar] [CrossRef] [PubMed]
- Laxman, B.; Tomlins, S.A.; Mehra, R.; Morris, D.S.; Wang, L.; Helgeson, B.E.; Shah, R.B.; Rubin, M.A.; Wei, J.T.; Chinnaiyan, A.M. Noninvasive detection of TMPRSS2:ERG fusion transcripts in the urine of men with prostate cancer. Neoplasia 2006, 8, 885–888. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miyata, Y.; Sakai, H. Thrombospondin-1 in urological cancer: Pathological role, clinical significance, and therapeutic prospects. Int. J. Mol. Sci. 2013, 14, 12249–12272. [Google Scholar] [CrossRef] [Green Version]
- Vetvicka, V. Procathepsin D in cancer development. J. Cancer Ther. Res. 2012, 1. [Google Scholar] [CrossRef] [Green Version]
- Endt, K.; Goepfert, J.; Omlin, A.; Athanasiou, A.; Tennstedt, P.; Guenther, A.; Rainisio, M.; Engeler, D.S.; Steuber, T.; Gillessen, S. Development and clinical testing of individual immunoassays for the quantification of serum glycoproteins to diagnose prostate cancer. PLoS ONE 2017, 12, e0181557. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Steuber, T.; Tennstedt, P.; Macagno, A.; Athanasiou, A.; Wittig, A.; Huber, R.; Golding, B.; Schiess, R.; Gillessen, S. Thrombospondin 1 and cathepsin D improve prostate cancer diagnosis by avoiding potentially unnecessary prostate biopsies. BJU Int. 2019, 123, 826–833. [Google Scholar] [CrossRef] [PubMed]
- Leyten, G.H.J.M.; Hessels, D.; Smit, F.P.; Jannink, S.A.; De Jong, H.; Melchers, W.J.G.; Cornel, E.B.; De Reijke, T.M.; Vergunst, H.; Kil, P.; et al. Identification of a candidate gene panel for the early diagnosis of prostate cancer. Clin. Cancer Res. 2015, 21, 3061–3070. [Google Scholar] [CrossRef] [Green Version]
- Groskopf, J.; Aubin, S.M.; Deras, I.L.; Blase, A.; Bodrug, S.; Clark, C.; Cass, M. APTIMA PCA3 molecular urine test: Development of a method to aid in the diagnosis of prostate cancer. Clin. Chem. 2006, 52, 1089–1095. [Google Scholar] [CrossRef] [Green Version]
- Progensa PCA3 Assay. Available online: https://www.hologic.com/sites/default/files/2019-05/502083-IFU-PI_003_01.pdf (accessed on 28 August 2020).
- Marks, L.S.; Fradet, Y.; Lim Deras, I.; Blase, A.; Mathis, J.; Aubin, S.M.J.; Cancio, A.T.; Desaulniers, M.; Ellis, W.J.; Rittenhouse, H.; et al. PCA3 Molecular Urine Assay for Prostate Cancer in Men Undergoing Repeat Biopsy. Urology 2007, 69, 532–535. [Google Scholar] [CrossRef]
- Rodríguez, S.V.M.; García-Perdomo, H.A. Diagnostic accuracy of prostate cancer antigen 3 (PCA3) prior to first prostate biopsy: A systematic review and meta-analysis. Can. Urol. Assoc. J. 2020, 14, E214–E219. [Google Scholar] [CrossRef]
- Ploussard, G.; de la Taille, A. The role of prostate cancer antigen 3 (PCA3) in prostate cancer detection. Expert Rev. Anticancer Ther. 2018, 18, 1013–1020. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.; Shim, S.R.; Ahn, S.T.; Oh, M.M.; Moon, D.G.; Park, H.S.; Cheon, J.; Kim, J.W. Diagnostic Performance of the Prostate Cancer Antigen 3 Test in Prostate Cancer: Systematic Review and Meta-analysis. Clin. Genitourin. Cancer 2020. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Cao, W.; Li, Q.; Shen, H.; Liu, C.; Deng, J.; Xu, J.; Shao, Q. Evaluation of prostate cancer antigen 3 for detecting prostate cancer: A systematic review and meta-analysis. Sci. Rep. 2016, 6, 2–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haese, A.; de la Taille, A.; van Poppel, H.; Marberger, M.; Stenzl, A.; Mulders, P.F.A.; Huland, H.; Abbou, C.C.; Remzi, M.; Tinzl, M.; et al. Clinical Utility of the PCA3 Urine Assay in European Men Scheduled for Repeat Biopsy. Eur. Urol. 2008, 54, 1081–1088. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deras, I.L.; Aubin, S.M.; Blase, A.; Day, J.R.; Koo, S.; Partin, A.W.; Groskopf, J. PCA3: A molecular urine assay for predicting prostate biopsy outcome. J. Urol. 2008, 179, 1587–1592. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Gou, X.; Huang, P.; Mou, C. The PCA3 test for guiding repeat biopsy of prostate cancer and its cut-off score: A systematic review and meta-analysis. Asian J. Androl. 2014, 16, 487–492. [Google Scholar] [CrossRef] [PubMed]
- Partin, A.W.; Van Neste, L.; Klein, E.A.; Marks, L.S.; Gee, J.R.; Troyer, D.A.; Rieger-Christ, K.; Jones, J.S.; Magi-Galluzzi, C.; Mangold, L.A.; et al. Clinical validation of an epigenetic assay to predict negative histopathological results in repeat prostate biopsies. J. Urol. 2014, 192, 1081–1087. [Google Scholar] [CrossRef]
- Stewart, G.D.; Van Neste, L.; Delvenne, P.; Delrée, P.; Delga, A.; McNeill, S.A.; O’Donnell, M.; Clark, J.; Van Criekinge, W.; Bigley, J.; et al. Clinical utility of an epigenetic assay to detect occult prostate cancer in histopathologically negative biopsies: Results of the MATLOC study. J. Urol. 2013, 189, 1110–1116. [Google Scholar] [CrossRef]
- Dakubo, G.D.; Jakupciak, J.P.; Birch-Machin, M.A.; Parr, R.L. Clinical implications and utility of field cancerization. Cancer Cell Int. 2007, 7, 2. [Google Scholar] [CrossRef] [Green Version]
- Cullen, J.; Rosner, I.L.; Brand, T.C.; Zhang, N.; Tsiatis, A.C.; Moncur, J.; Ali, A.; Chen, Y.; Knezevic, D.; Maddala, T.; et al. A biopsy-based 17-gene genomic prostate score predicts recurrence after radical prostatectomy and adverse surgical pathology in a racially diverse population of men with clinically low- and intermediate-risk prostate cancer. Eur. Urol. 2015, 68, 123–131. [Google Scholar] [CrossRef] [Green Version]
- Van Den Eeden, S.K.; Lu, R.; Zhang, N.; Quesenberry, C.P.J.; Shan, J.; Han, J.S.; Tsiatis, A.C.; Leimpeter, A.D.; Lawrence, H.J.; Febbo, P.G.; et al. A Biopsy-based 17-gene Genomic Prostate Score as a Predictor of Metastases and Prostate Cancer Death in Surgically Treated Men with Clinically Localized Disease. Eur. Urol. 2018, 73, 129–138. [Google Scholar] [CrossRef] [PubMed]
- Klein, E.A.; Yousefi, K.; Haddad, Z.; Choeurng, V.; Buerki, C.; Stephenson, A.J.; Li, J.; Kattan, M.W.; Magi-Galluzzi, C.; Davicioni, E. A genomic classifier improves prediction of metastatic disease within 5 years after surgery in node-negative high-risk prostate cancer patients managed by radical prostatectomy without adjuvant therapy. Eur. Urol. 2015, 67, 778–786. [Google Scholar] [CrossRef] [PubMed]
- Karnes, R.J.; Choeurng, V.; Ross, A.E.; Schaeffer, E.M.; Klein, E.A.; Freedland, S.J.; Erho, N.; Yousefi, K.; Takhar, M.; Davicioni, E.; et al. Validation of a Genomic Risk Classifier to Predict Prostate Cancer-specific Mortality in Men with Adverse Pathologic Features. Eur. Urol. 2018, 73, 168–175. [Google Scholar] [CrossRef] [PubMed]
- Erho, N.; Crisan, A.; Vergara, I.A.; Mitra, A.P.; Ghadessi, M.; Buerki, C.; Bergstralh, E.J.; Kollmeyer, T.; Fink, S.; Haddad, Z.; et al. Discovery and Validation of a Prostate Cancer Genomic Classifier that Predicts Early Metastasis Following Radical Prostatectomy. PLoS ONE 2013, 8, e66855. [Google Scholar] [CrossRef] [Green Version]
- Ross, A.E.; Feng, F.Y.; Ghadessi, M.; Erho, N.; Crisan, A.; Buerki, C.; Sundi, D.; Mitra, A.P.; Vergara, I.A.; Thompson, D.J.S.; et al. A genomic classifier predicting metastatic disease progression in men with biochemical recurrence after prostatectomy. Prostate Cancer Prostatic Dis. 2014, 17, 64–69. [Google Scholar] [CrossRef] [Green Version]
- Klein, E.A.; Haddad, Z.; Yousefi, K.; Lam, L.L.C.; Wang, Q.; Choeurng, V.; Palmer-Aronsten, B.; Buerki, C.; Davicioni, E.; Li, J.; et al. Decipher Genomic Classifier Measured on Prostate Biopsy Predicts Metastasis Risk. Urology 2016, 90, 148–152. [Google Scholar] [CrossRef] [Green Version]
- Freedland, S.J.; Choeurng, V.; Howard, L.; De Hoedt, A.; du Plessis, M.; Yousefi, K.; Lam, L.L.; Buerki, C.; Ra, S.; Robbins, B.; et al. Utilization of a Genomic Classifier for Prediction of Metastasis Following Salvage Radiation Therapy after Radical Prostatectomy. Eur. Urol. 2016, 70, 588–596. [Google Scholar] [CrossRef] [Green Version]
- Bishoff, J.T.; Freedland, S.J.; Gerber, L.; Tennstedt, P.; Reid, J.; Welbourn, W.; Graefen, M.; Sangale, Z.; Tikishvili, E.; Park, J.; et al. Prognostic utility of the cell cycle progression score generated from biopsy in men treated with prostatectomy. J. Urol. 2014, 192, 409–414. [Google Scholar] [CrossRef]
- Cooperberg, M.R.; Davicioni, E.; Crisan, A.; Jenkins, R.B.; Ghadessi, M.; Karnes, R.J. Combined value of validated clinical and genomic risk stratification tools for predicting prostate cancer mortality in a high-risk prostatectomy cohort. Eur. Urol. 2015, 67, 326–333. [Google Scholar] [CrossRef] [Green Version]
- Cuzick, J.; Swanson, G.P.; Fisher, G.; Brothman, A.R.; Berney, D.M.; Reid, J.E.; Mesher, D.; Speights, V.O.; Stankiewicz, E.; Foster, C.S.; et al. Prognostic value of an RNA expression signature derived from cell cycle proliferation genes in patients with prostate cancer: A retrospective study. Lancet Oncol. 2011, 12, 245–255. [Google Scholar] [CrossRef] [Green Version]
- Canter, D.J.; Freedland, S.; Rajamani, S.; Latsis, M.; Variano, M.; Halat, S.; Tward, J.; Cohen, T.; Stone, S.; Schlomm, T.; et al. Analysis of the prognostic utility of the cell cycle progression (CCP) score generated from needle biopsy in men treated with definitive therapy. Prostate Cancer Prostatic Dis. 2020, 23, 102–107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klein, E.A.; Cooperberg, M.R.; Magi-Galluzzi, C.; Simko, J.P.; Falzarano, S.M.; Maddala, T.; Chan, J.M.; Li, J.; Cowan, J.E.; Tsiatis, A.C.; et al. A 17-gene assay to predict prostate cancer aggressiveness in the context of gleason grade heterogeneity, tumor multifocality, and biopsy undersampling. Eur. Urol. 2014, 66, 550–560. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brand, T.C.; Zhang, N.; Crager, M.R.; Maddala, T.; Dee, A.; Sesterhenn, I.A.; Simko, J.P.; Cooperberg, M.R.; Srivastava, S.; Rosner, I.L.; et al. Patient-specific Meta-analysis of 2 Clinical Validation Studies to Predict Pathologic Outcomes in Prostate Cancer Using the 17-Gene Genomic Prostate Score. Urology 2016, 89, 69–75. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jeffrey Karnes, R.; Bergstralh, E.J.; Davicioni, E.; Ghadessi, M.; Buerki, C.; Mitra, A.P.; Crisan, A.; Erho, N.; Vergara, I.A.; Lam, L.L.; et al. Validation of a genomic classifier that predicts metastasis following radical prostatectomy in an at risk Patient population. J. Urol. 2013, 190, 2047–2053. [Google Scholar] [CrossRef] [PubMed]
- Knudsen, B.S.; Kim, H.L.; Erho, N.; Shin, H.; Alshalalfa, M.; Lam, L.L.C.; Tenggara, I.; Chadwich, K.; Van Der Kwast, T.; Fleshner, N.; et al. Application of a Clinical Whole-Transcriptome Assay for Staging and Prognosis of Prostate Cancer Diagnosed in Needle Core Biopsy Specimens. J. Mol. Diagn. 2016, 18, 395–406. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, H.J.; Yousefi, K.; Haddad, Z.; Abdollah, F.; Lam, L.L.; Shin, H.; Alshalalfa, M.; Godebu, E.; Wang, S.; Shabaik, A.; et al. Evaluation of a genomic classifier in radical prostatectomy patients with lymph node metastasis. Res. Rep. Urol. 2016, 8, 77–84. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cuzick, J.; Berney, D.M.; Fisher, G.; Mesher, D.; Møller, H.; Reid, J.E.; Perry, M.; Park, J.; Younus, A.; Gutin, A.; et al. Prognostic value of a cell cycle progression signature for prostate cancer death in a conservatively managed needle biopsy cohort. Br. J. Cancer 2012, 106, 1095–1099. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sommariva, S.; Tarricone, R.; Lazzeri, M.; Ricciardi, W.; Montorsi, F. Prognostic Value of the Cell Cycle Progression Score in Patients with Prostate Cancer: A Systematic Review and Meta-analysis. Eur. Urol. 2016, 69, 107–115. [Google Scholar] [CrossRef]
- Moul, J.W. Secondary screening tests for prostate cancer: Is more information better? Which test is best? Can. J. Urol. 2020, 27, 10086. [Google Scholar]
- Nordström, T.; Vickers, A.; Assel, M.; Lilja, H.; Grönberg, H.; Eklund, M. Comparison between the four-kallikrein panel and prostate health index for predicting prostate cancer. Eur. Urol. 2015, 68, 139–146. [Google Scholar] [CrossRef] [Green Version]
- Vedder, M.M.; de Bekker-Grob, E.W.; Lilja, H.G.; Vickers, A.J.; van Leenders, G.J.L.H.; Steyerberg, E.W.; Roobol, M.J. The added value of percentage of free to total prostate-specific antigen, PCA3, and a kallikrein panel to the ERSPC risk calculator for prostate cancer in prescreened men. Eur. Urol. 2014, 66, 1109–1115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stephan, C.; Jung, K.; Semjonow, A.; Schulze-Forster, K.; Cammann, H.; Hu, X.; Meyer, H.A.; Bögemann, M.; Miller, K.; Friedersdorff, F. Comparative assessment of urinary prostate cancer antigen 3 and TMPRSS2:ERG gene fusion with the serum [-2]proprostate-specific antigen-based prostate health index for detection of prostate cancer. Clin. Chem. 2013, 59, 280–288. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scattoni, V.; Lazzeri, M.; Lughezzani, G.; De Luca, S.; Passera, R.; Bollito, E.; Randone, D.; Abdollah, F.; Capitanio, U.; Larcher, A.; et al. Head-to-head comparison of prostate health index and urinary PCA3 for predicting cancer at initial or repeat biopsy. J. Urol. 2013, 190, 496–501. [Google Scholar] [CrossRef] [PubMed]
- Perdonà, S.; Bruzzese, D.; Ferro, M.; Autorino, R.; Marino, A.; Mazzarella, C.; Perruolo, G.; Longo, M.; Spinelli, R.; Di Lorenzo, G.; et al. Prostate health index (phi) and prostate cancer antigen 3 (PCA3) significantly improve diagnostic accuracy in patients undergoing prostate biopsy. Prostate 2013, 73, 227–235. [Google Scholar] [CrossRef]
- Seisen, T.; Rouprêt, M.; Brault, D.; Léon, P.; Cancel-Tassin, G.; Compérat, E.; Renard-Penna, R.; Mozer, P.; Guechot, J.; Cussenot, O. Accuracy of the prostate health index versus the urinary prostate cancer antigen 3 score to predict overall and significant prostate cancer at initial biopsy. Prostate 2015, 75, 103–111. [Google Scholar] [CrossRef]
- Ferro, M.; Bruzzese, D.; Perdonà, S.; Mazzarella, C.; Marino, A.; Sorrentino, A.; Di Carlo, A.; Autorino, R.; Di Lorenzo, G.; Buonerba, C.; et al. Predicting prostate biopsy outcome: Prostate health index (phi) and prostate cancer antigen 3 (PCA3) are useful biomarkers. Clin. Chim. Acta 2012, 413, 1274–1278. [Google Scholar] [CrossRef]
- Ferro, M.; Bruzzese, D.; Perdonà, S.; Marino, A.; Mazzarella, C.; Perruolo, G.; D’Esposito, V.; Cosimato, V.; Buonerba, C.; Di Lorenzo, G.; et al. Prostate Health Index (Phi) and Prostate Cancer Antigen 3 (PCA3) Significantly Improve Prostate Cancer Detection at Initial Biopsy in a Total PSA Range of 2–10 ng/mL. PLoS ONE 2013, 8, e67687. [Google Scholar] [CrossRef]
- Hendriks, R.J.; Van Oort, I.M.; Schalken, J.A. Blood-based and urinary prostate cancer biomarkers: A review and comparison of novel biomarkers for detection and treatment decisions. Prostate Cancer Prostatic Dis. 2017, 20, 12–19. [Google Scholar] [CrossRef]
- Alam, S.; Tortora, J.; Staff, I.; McLaughlin, T.; Wagner, J. Prostate cancer genomics: Comparing results from three molecular assays. Can. J. Urol. 2019, 26, 9758–9762. [Google Scholar]
- Kasivisvanathan, V.; Rannikko, A.S.; Borghi, M.; Panebianco, V.; Mynderse, L.A.; Vaarala, M.H.; Briganti, A.; Budäus, L.; Hellawell, G.; Hindley, R.G.; et al. MRI-Targeted or Standard Biopsy for Prostate-Cancer Diagnosis. N. Engl. J. Med. 2018, 378, 1767–1777. [Google Scholar] [CrossRef]
- Abdi, H.; Pourmalek, F.; Zargar, H.; Walshe, T.; Harris, A.C.; Chang, S.D.; Eddy, C.; So, A.I.; Gleave, M.E.; Machan, L.; et al. Multiparametric magnetic resonance imaging enhances detection of significant tumor in patients on active surveillance for prostate cancer. Urology 2015, 85, 423–428. [Google Scholar] [CrossRef] [PubMed]
- Gaur, S.; Turkbey, B. Prostate MR Imaging for Posttreatment Evaluation and Recurrence. Urol. Clin. North Am. 2018, 45, 467–479. [Google Scholar] [CrossRef] [PubMed]
- Mottet, N.; Bellmunt, J.; Briers, E.; Bolla, M.; Bourke, L.; Cornford, P.; De Santis, M.; Henry, A.; Joniau, S.; Lam, T.; et al. EAU–ESTRO–ESUR–SIOG Guidelines on Prostate Cancer; Edn. Presented at the EAU Annual Congress Amsterdam 2020; EAU Guidelines Office: Arnhem, The Netherlands, 2020; ISBN 978-94-92671-07-3. [Google Scholar]
- Drost, F.H.; Osses, D.F.; Nieboer, D.; Steyerberg, E.W.; Bangma, C.H.; Roobol, M.J.; Schoots, I.G. Prostate MRI, with or without MRI--targeted biopsy, and systematic biopsy for detecting prostate cancer. Cochrane Database Syst. Rev. 2019, 77, 78–94. [Google Scholar] [CrossRef] [PubMed]
- Hendriks, R.J.; van der Leest, M.M.G.; Dijkstra, S.; Barentsz, J.O.; Van Criekinge, W.; Hulsbergen-van de Kaa, C.A.; Schalken, J.A.; Mulders, P.F.A.; van Oort, I.M. A urinary biomarker-based risk score correlates with multiparametric MRI for prostate cancer detection. Prostate 2017, 77, 1401–1407. [Google Scholar] [CrossRef] [PubMed]
- Falagario, U.G.; Martini, A.; Wajswol, E.; Treacy, P.-J.; Ratnani, P.; Jambor, I.; Anastos, H.; Lewis, S.; Haines, K.; Cormio, L.; et al. Avoiding Unnecessary Magnetic Resonance Imaging (MRI) and Biopsies: Negative and Positive Predictive Value of MRI According to Prostate-specific Antigen Density, 4Kscore and Risk Calculators. Eur. Urol. Oncol. 2019. [Google Scholar] [CrossRef] [Green Version]
- Punnen, S.; Nahar, B.; Soodana-Prakash, N.; Koru-Sengul, T.; Stoyanova, R.; Pollack, A.; Kava, B.; Gonzalgo, M.L.; Ritch, C.R.; Parekh, D.J. Optimizing patient’s selection for prostate biopsy: A single institution experience with multi-parametric MRI and the 4Kscore test for the detection of aggressive prostate cancer. PLoS ONE 2018, 13, e0201384. [Google Scholar] [CrossRef]
- Marzouk, K.; Ehdaie, B.; Vertosick, E.; Zappala, S.; Vickers, A. Developing an effective strategy to improve the detection of significant prostate cancer by combining the 4Kscore and multiparametric MRI. Urol. Oncol. 2019, 37, 672–677. [Google Scholar] [CrossRef]
- Thompson, I.M.; Ankerst, D.P.; Chi, C.; Goodman, P.J.; Tangen, C.M.; Lucia, M.S.; Feng, Z.; Parnes, H.L.; Coltman, C.A., Jr. Assessing Prostate Cancer Risk: Results from the Prostate Cancer Prevention Trial. J. Natl. Cancer Inst. 2006, 98, 529–534. [Google Scholar] [CrossRef] [Green Version]
- Van Den Bergh, R.C.N.; Roobol, M.J.; Wolters, T.; Van Leeuwen, P.J.; Schröder, F.H. The Prostate Cancer Prevention Trial and European Randomized Study of Screening for Prostate Cancer risk calculators indicating a positive prostate biopsy: A comparison. BJU Int. 2008, 102, 1068–1073. [Google Scholar] [CrossRef]
- Your Prostate Cancer Risk Calculator. Available online: http://www.prostatecancer-riskcalculator.com/ (accessed on 25 August 2020).
- Borque-Fernando, A.; Esteban-Escano, L.M.; Rubio-Briones, J.; Lou-Mercade, A.C.; Garcia-Ruiz, R.; Tejero-Sanchez, A.; Munoz-Rivero, M.V.; Cabanuz-Plo, T.; Alfaro-Torres, J.; Marquina-Ibanez, I.M.; et al. A Preliminary Study of the Ability of the 4Kscore test, the Prostate Cancer Prevention Trial-Risk Calculator and the European Research Screening Prostate-Risk Calculator for Predicting High-Grade Prostate Cancer. Actas Urol. Esp. 2016, 40, 155–163. [Google Scholar] [CrossRef] [Green Version]
- Verbeek, J.F.M.; Roobol, M.J. Reply by authors: Reducing unnecessary biopsies while detecting clinically significant prostate cancer including cribriform growth with the ERSPC Rotterdam risk calculator and 4Kscore. Urol. Oncol. 2019, 37, 341–342. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Li, Y.; Yang, D.; Yang, C.; Mao, L. Current state of biomarkers for the diagnosis and assessment of treatment efficacy of prostate cancer. Discov. Med. 2019, 27, 235–243. [Google Scholar] [PubMed]
- Wang, J.; Ni, J.; Beretov, J.; Thompson, J.; Graham, P.; Li, Y. Exosomal microRNAs as liquid biopsy biomarkers in prostate cancer. Crit. Rev. Oncol. Hematol. 2020, 145, 102860. [Google Scholar] [CrossRef] [PubMed]
- Urabe, F.; Matsuzaki, J.; Yamamoto, Y.; Kimura, T.; Hara, T.; Ichikawa, M.; Takizawa, S.; Aoki, Y.; Niida, S.; Sakamoto, H.; et al. Large-scale Circulating microRNA Profiling for the Liquid Biopsy of Prostate Cancer. Clin. Cancer Res. 2019, 25, 3016–3025. [Google Scholar] [CrossRef] [Green Version]
| Test | Specimen | Setting (Pre-) | Biomarkers | Clinical Variables | Clinical Endpoint | AUC hg-PCa | Cutoff | Se | Sp | NPV | PPV | Ref |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PHI | Blood | IBx, RBx | p2PSA, fPSA, tPSA | - | PCa | 0.70–0.73 * | 24.1–31.94 | 90–90.3 | 23–31.1 | - | - | [3,4,11,12] |
| 4K | Blood | IBx, RBx | tPSA, fPSA, iPSA, hK2 | Age, DRE result, prior Bx- | hg-PCa, 10-y metastasis | 0.81–0.82 | - | - | - | - | - | [13,14] |
| SelectMDx | Post-DRE urine | IBx | DLX1, HOXC6, KLK3 mRNA, PSAd | Age, DRE result | PCa, hg-PCa | 0.82–0.86 | −2.8 | 89 | 53 | 95 | 34 | [15,16] |
| ExoDx | Urine | IBx | PCA3, ERG, SPDEF ex-mRNA | - | hg-PCa | 0.70–0.71 | 15.6 | 91.89–93 | 26.1–34 | 89.1–91.3 | 35.7–36.6 | [17,18] |
| MiPS | Post-DRE urine | IBx, RBx | tPSA, PCA3, T2-ERG mRNA | - | PCa, hg-PCa | 0.77 | Not provided | 92.6 | 33.4 | 92.6 | 33.2 | [19,20] |
| Proclarix | Blood | IBx | THBS1, CTSD, tPSA, %fPSA | Age | hg-PCa | - | Not provided | 90 | 43 | 95 | 25 | [21] |
| PCA3 | Post-DRE urine | RBx | PCA3, PSA mRNA | - | PCa | 0.59–0.69 * | 35 | 42–75.2 | 41.8–72 | 68–90.5 | 15.7–38.9 | [17,18,22,23,24,25,26] |
| PCMT | PBx-tissue | RBx | mtDNA del | - | Presence mtDNA del | 0.75 | Ct: 31 | 84 | 54 | 91 | - | [27] |
| ConfirmMDx | PBx-tissue | RBx | GSTP1, APC, RASSF1-hypm., PSA | Age, DRE result, histopath. IBx | PCa, hg-PCa, hyperm. site | 0.76 | Not provided | 78 | 94.2 | 96 | - | [28,29] |
| Test | Specimen | Setting | Biomarkers | Clinical Variables | Clinical Endpoint (Risk on) | Performance | Ref. |
|---|---|---|---|---|---|---|---|
| OncotypeDx | FFPE tissue (from Bx) | Post-positive Bx: NCCN-low risk | 17 genes | GS, PSA, Clinical Stage, PV, PSA Density | hg-PCa, high-stage PCa (≥pT3) upon RP, 10-y metastasis, 10-y PCa specific mortality | hg-PCa upon RP: OR/20 units GPS: 2.3 High-stage PCa upon RP: OR/20 units GPS: 1.9 10-y Metastasis: HR/20 units GPS: 3.8 HR/20 units GPS: 2.75 10-y PCa Mortality: HR/20 GPS units = 3.23 | [61,62,63] |
| Decipher | FFPE tissue (from Bx or RP) | Post-positive Bx and post-RP | 22 genes | No clinical parameters | Post-positive Bx
| 5-y Metastasis: Bx tissue: HR/0.1 point: 1.72 RP tissue: AUC range: 0.75–0.82 10-y PCa Mortality: RP tissue: AUC:0.73–0.78 | [64,65,66,67] |
| Prolaris | FFPE tissue (from Bx or RP) | Post-positive Bx and post-RP | 46 genes | Age at Bx, PSA, clinical T stage, percentage positive cores, GS, NCCN-risk category | Post-positive Bx
| BCR: HR: 1.3–2.53 (multivar. model) 10-y PCa Mortality: HR: 1.7–2.6 (multivar. model) 10-y Metastasis: HR: 2.21–4.19 (multivar model) | [68,69,70,71,72] |
| Compared Tests | Clinical Setting | Endpoint | AUC (95% CI) | Ref. |
|---|---|---|---|---|
| 4K and PHI | IBx | PCa and hg-PCa | PCa 4K: 0.69 (0.65–0.73), PHI: 0.70 (0.66–0.75) hg-PCa 4K: 0.72 (0.67–0.77), PHI: 0.71 (0.66–0.76) | [81] |
| PCA3 and 4K | IBx | PCa | 4K: 0.78 (0.69–0.85), PCA3: 0.62 (0.52–0.73) | [82] |
| PCA3 and PHI | IBx and RBx | PCa | PHI: 0.68 (0.62–0.74), PCA3: 0.74 (0.68 –0.79) | [83] |
| PCA3 and PHI | IBx and RBx | PCa | PHI: 0.70, PCA3: 0.59 | [84] |
| PCA3 and PHI | IBx | PCa | PHI: 0.77 (0.72–0.83), PCA3: 0.73 (0.68–0.79) | [65] |
| PCA3 and PHI | IBx | PCa | PHI: 0.71 (0.61–0.80), PCA3: 0.66 (0.57–0.75) | [85] |
| PCA3 and PHI | IBx | PCa and hg-PCa | PCa PHI: 0.65, PCA3: 0.71 hg-PCa PHI: 0.80, PCA3: 0.55 | [86] |
| PCA3 and PHI | IBx | PCa | PHI: 0.77, PCA3: 0.71 | [87] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Visser, W.C.H.; de Jong, H.; Melchers, W.J.G.; Mulders, P.F.A.; Schalken, J.A. Commercialized Blood-, Urinary- and Tissue-Based Biomarker Tests for Prostate Cancer Diagnosis and Prognosis. Cancers 2020, 12, 3790. https://doi.org/10.3390/cancers12123790
Visser WCH, de Jong H, Melchers WJG, Mulders PFA, Schalken JA. Commercialized Blood-, Urinary- and Tissue-Based Biomarker Tests for Prostate Cancer Diagnosis and Prognosis. Cancers. 2020; 12(12):3790. https://doi.org/10.3390/cancers12123790
Chicago/Turabian StyleVisser, Wieke C. H., Hans de Jong, Willem J. G. Melchers, Peter F. A. Mulders, and Jack A. Schalken. 2020. "Commercialized Blood-, Urinary- and Tissue-Based Biomarker Tests for Prostate Cancer Diagnosis and Prognosis" Cancers 12, no. 12: 3790. https://doi.org/10.3390/cancers12123790
APA StyleVisser, W. C. H., de Jong, H., Melchers, W. J. G., Mulders, P. F. A., & Schalken, J. A. (2020). Commercialized Blood-, Urinary- and Tissue-Based Biomarker Tests for Prostate Cancer Diagnosis and Prognosis. Cancers, 12(12), 3790. https://doi.org/10.3390/cancers12123790





