Stroma Transcriptomic and Proteomic Profile of Prostate Cancer Metastasis Xenograft Models Reveals Prognostic Value of Stroma Signatures
Abstract
:Simple Summary
Abstract
1. Introduction
2. Results
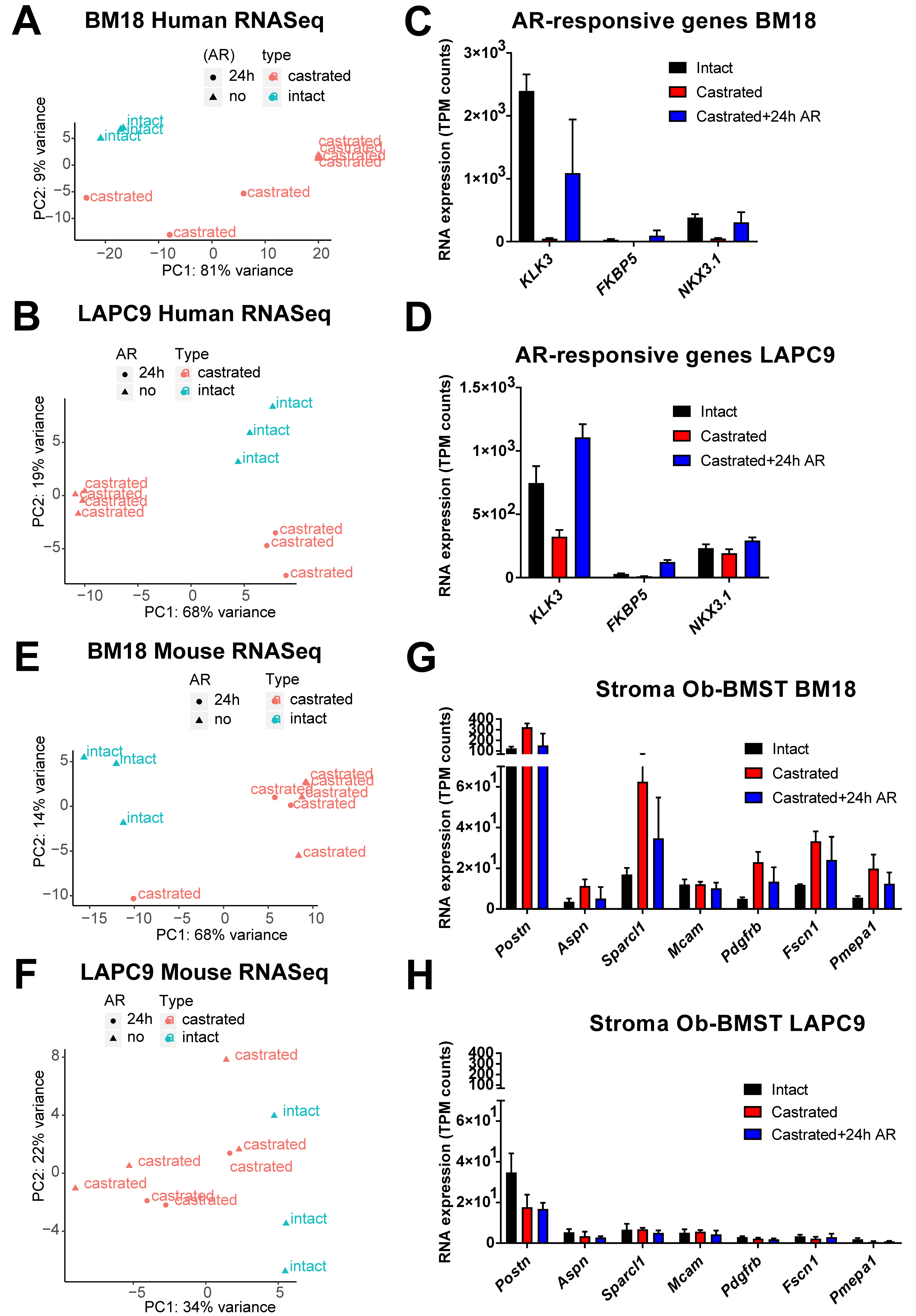
2.1. Simultaneous Transcriptome Analysis of Human and Murine Signatures in PDXs Can Distinguish Androgen-Dependent Expression Changes in Tumor and Host-Derived Stroma
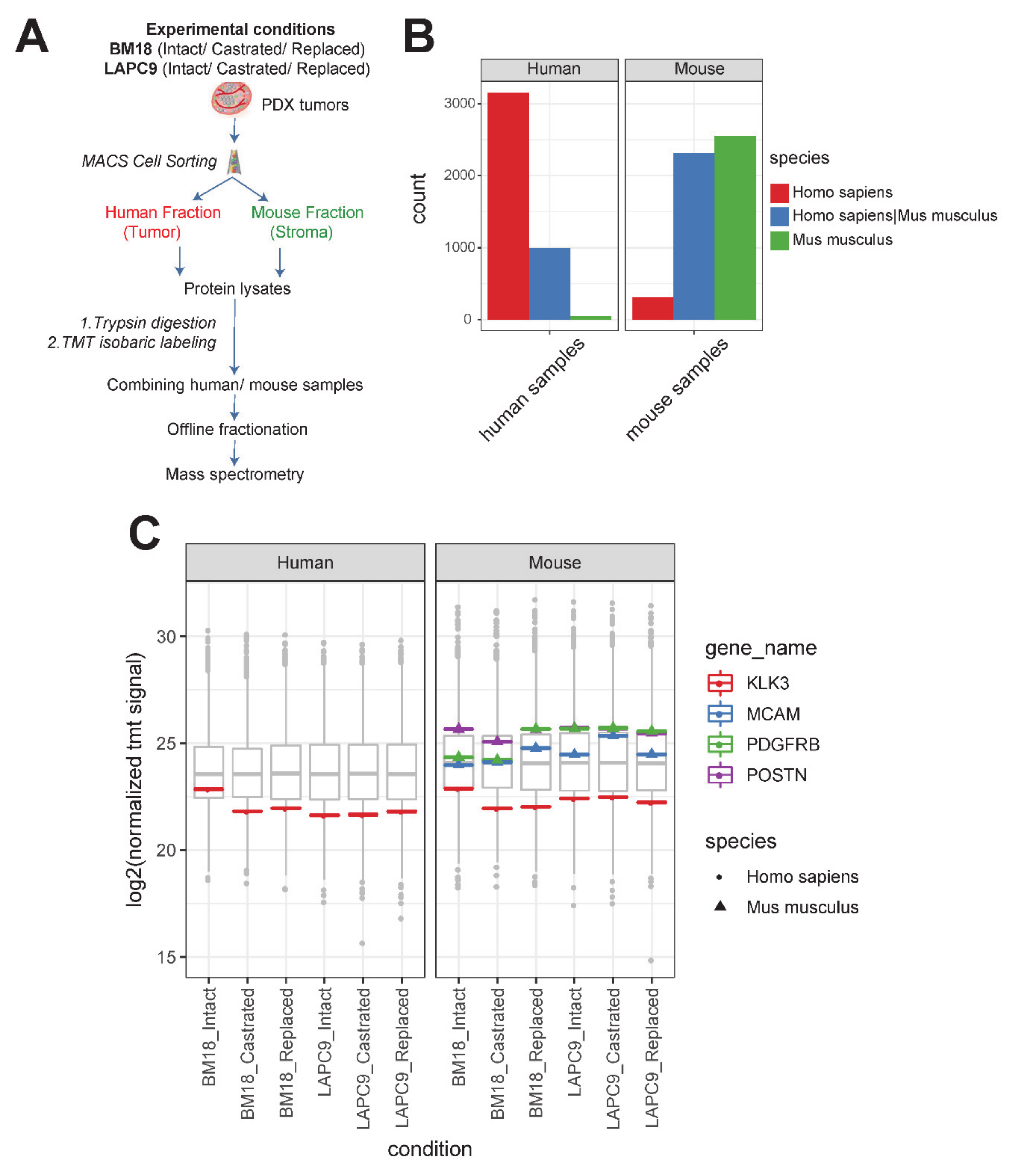
2.2. Proteomic Analysis Provides Functional Information over the Identified Human/Mouse-Specific Transcriptome
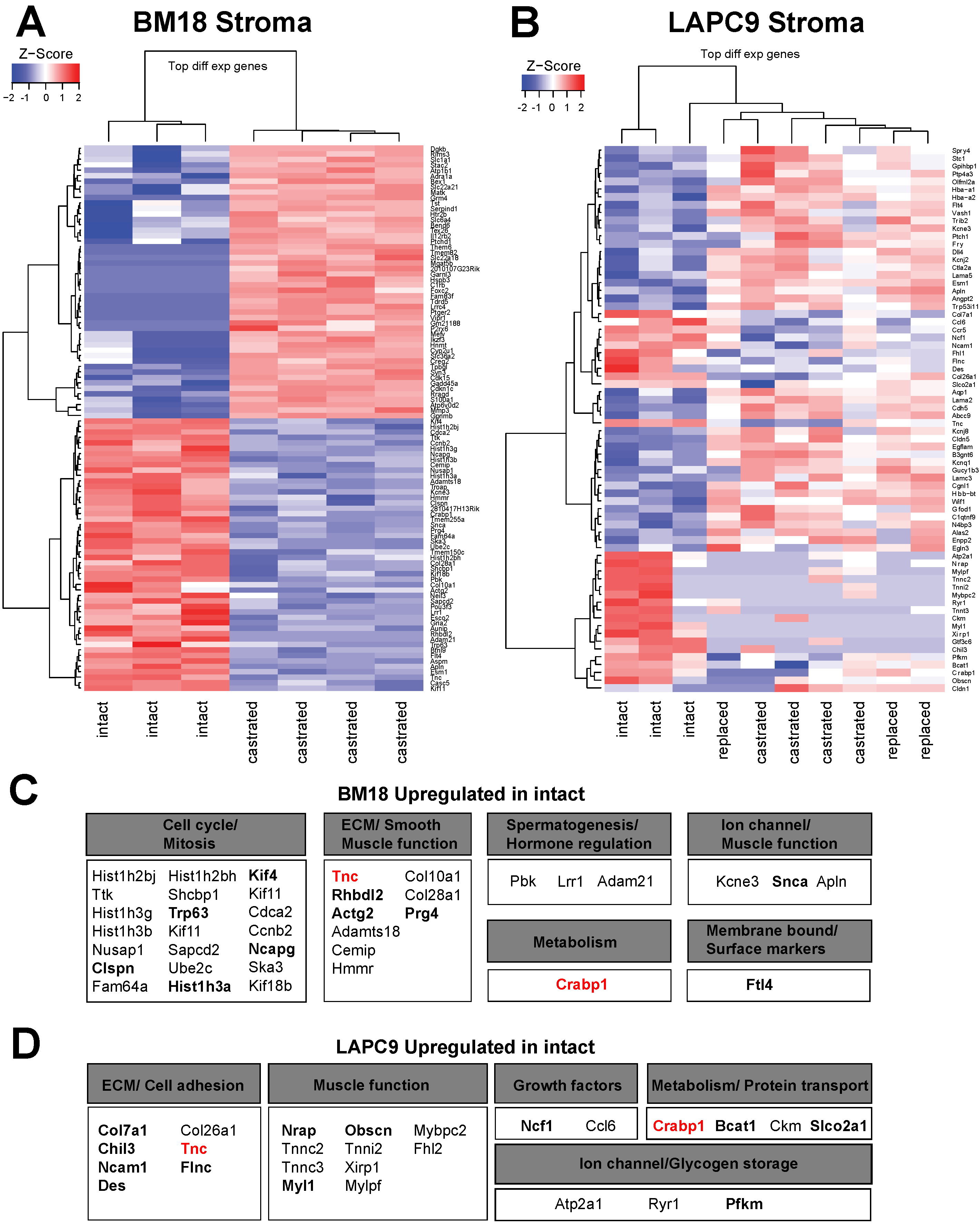
2.3. Differential Expression Analysis Reveals Androgen-Dependent Stromal Gene Modulation in Androgen-Independent PDX Model
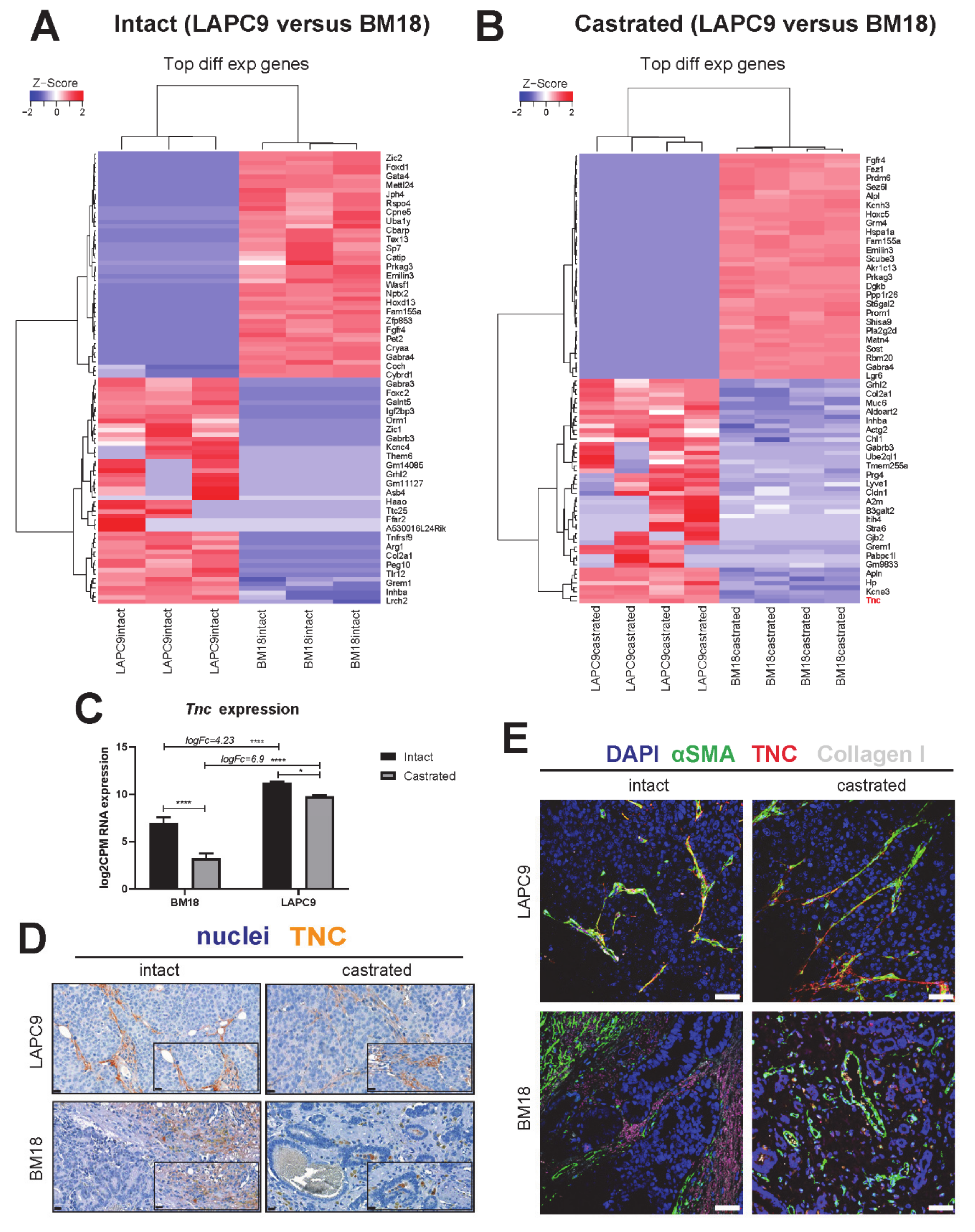
2.4. Cross Comparison of Stromal Transcriptome among Different PDXs Identifies ECM and Cell Adhesion Pathways in the LAPC9 Androgen-Independent Model
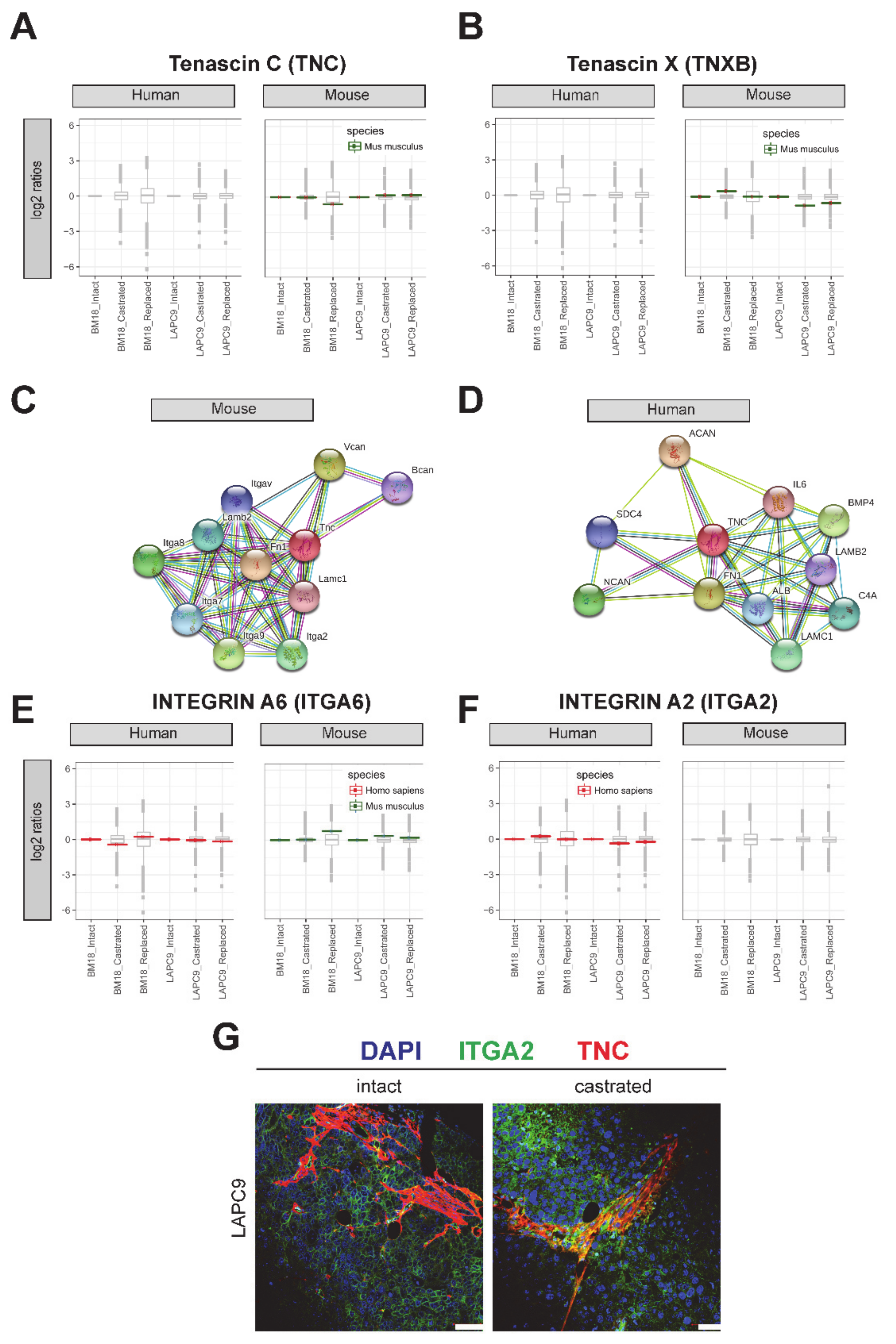
2.5. Protein Expression of Tenascin and Its Interaction Partners
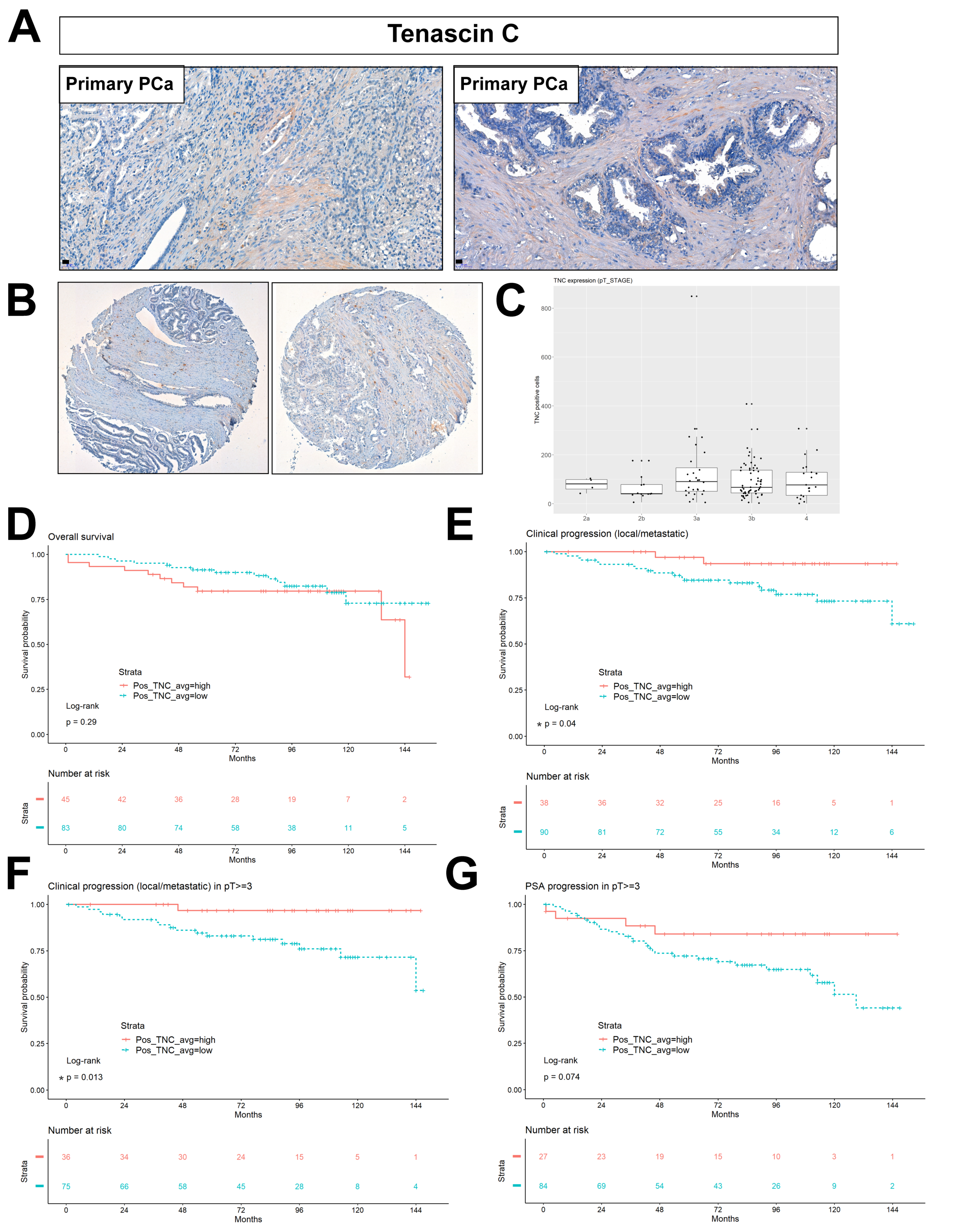
2.6. Stromal Tenascin Expression as a Prognostic Factor of Disease Progression in High-Risk PCa
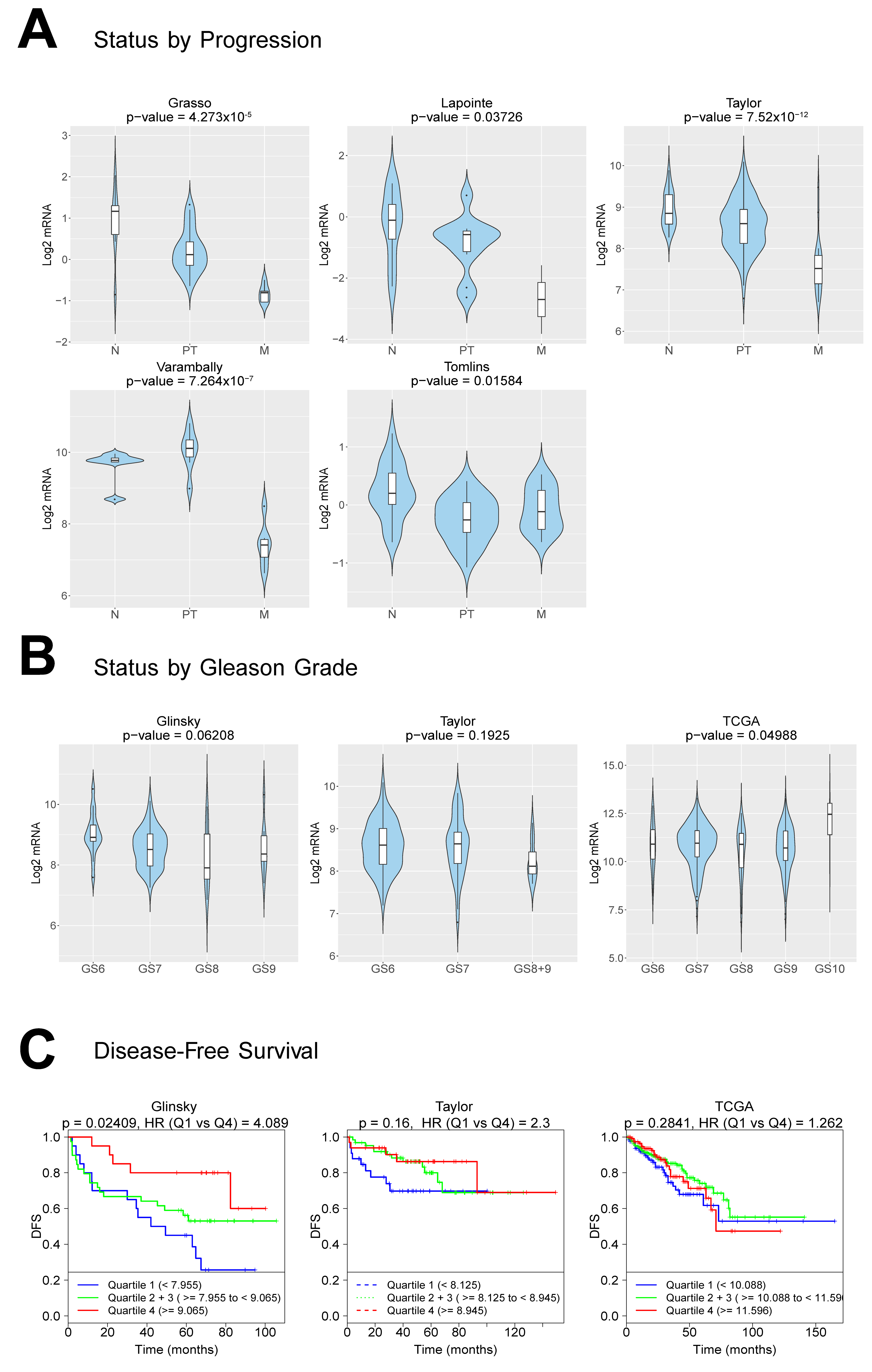
2.7. Stroma Signatures from Androgen-Dependent and -Independent States Correlate with Disease Progression
3. Discussion
4. Materials and Methods
4.1. Tumor Sample Preparation and Xenograft Surgery Procedure
4.2. RNA Isolation from Tissue Samples
4.3. RNA Sequencing
4.4. Signature Validation on TCGA and Other Publically Available Datasets
4.5. Tissue Dissociation and MACS
4.6. Proteomics
4.6.1. Sample Preparation
4.6.2. Mass Spectrometric Analysis
4.6.3. Raw MS Data Analysis
4.6.4. MS Data Analysis
4.7. Tissue Microarray
4.8. Immunohistochemistry
4.9. Immunofluorescence
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Data Availability
References
- Heidenreich, A.; Bastian, P.J.; Bellmunt, J.; Bolla, M.; Joniau, S.; van der Kwast, T.; Mason, M.; Matveev, V.; Wiegel, T.; Zattoni, F.; et al. EAU guidelines on prostate cancer. Part II: Treatment of advanced, relapsing, and castration-resistant prostate cancer. Eur. Urol. 2014, 65, 467–479. [Google Scholar] [CrossRef] [PubMed]
- Malanchi, I.; Santamaria-Martinez, A.; Susanto, E.; Peng, H.; Lehr, H.A.; Delaloye, J.F.; Huelsken, J. Interactions between cancer stem cells and their niche govern metastatic colonization. Nature 2011, 481, 85–89. [Google Scholar] [CrossRef] [PubMed]
- Shiozawa, Y.; Pedersen, E.A.; Havens, A.M.; Jung, Y.; Mishra, A.; Joseph, J.; Kim, J.K.; Patel, L.R.; Ying, C.; Ziegler, A.M.; et al. Human prostate cancer metastases target the hematopoietic stem cell niche to establish footholds in mouse bone marrow. J. Clin. Invest. 2011, 121, 1298–1312. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hensel, J.; Wetterwald, A.; Temanni, R.; Keller, I.; Riether, C.; van der Pluijm, G.; Cecchini, M.G.; Thalmann, G.N. Osteolytic cancer cells induce vascular/axon guidance processes in the bone/bone marrow stroma. Oncotarget 2018, 9, 28877–28896. [Google Scholar] [CrossRef] [PubMed]
- Ozdemir, B.C.; Hensel, J.; Secondini, C.; Wetterwald, A.; Schwaninger, R.; Fleischmann, A.; Raffelsberger, W.; Poch, O.; Delorenzi, M.; Temanni, R.; et al. The molecular signature of the stroma response in prostate cancer-induced osteoblastic bone metastasis highlights expansion of hematopoietic and prostate epithelial stem cell niches. PLoS ONE 2014, 9, e114530. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rucci, N.; Teti, A. Osteomimicry: How the seed grows in the soil. Calcif. Tissue Int. 2018, 102, 131–140. [Google Scholar] [CrossRef]
- Tyekucheva, S.; Bowden, M.; Bango, C.; Giunchi, F.; Huang, Y.; Zhou, C.; Bondi, A.; Lis, R.; Van Hemelrijck, M.; Andrén, O.; et al. Stromal and epithelial transcriptional map of initiation progression and metastatic potential of human prostate cancer. Nat. Commun. 2017, 8, 420. [Google Scholar] [CrossRef] [Green Version]
- Setlur, S.R.; Rubin, M.A. Current thoughts on the role of the androgen receptor and prostate cancer progression. Adv. Anat. Pathol. 2005, 12, 265–270. [Google Scholar] [CrossRef]
- Leach, D.A.; Need, E.F.; Toivanen, R.; Trotta, A.P.; Palenthorpe, H.M.; Tamblyn, D.J.; Kopsaftis, T.; England, G.M.; Smith, E.; Drew, P.A.; et al. Stromal androgen receptor regulates the composition of the microenvironment to influence prostate cancer outcome. Oncotarget 2015, 6, 16135–16150. [Google Scholar] [CrossRef] [Green Version]
- Leach, D.A.; Panagopoulos, V.; Nash, C.; Bevan, C.; Thomson, A.A.; Selth, L.A.; Buchanan, G. Cell-lineage specificity and role of AP-1 in the prostate fibroblast androgen receptor cistrome. Mol. Cell. Endocrinol. 2017, 439, 261–272. [Google Scholar] [CrossRef] [Green Version]
- Nash, C.; Boufaied, N.; Mills, I.G.; Franco, O.E.; Hayward, S.W.; Thomson, A.A. Genome-wide analysis of AR binding and comparison with transcript expression in primary human fetal prostate fibroblasts and cancer associated fibroblasts. Mol. Cell. Endocrinol. 2018, 471, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thalmann, G.N.; Rhee, H.; Sikes, R.A.; Pathak, S.; Multani, A.; Zhau, H.E.; Marshall, F.F.; Chung, L.W.K. Human prostate fibroblasts induce growth and confer castration resistance and metastatic potential in LNCaP Cells. Eur. Urol. 2010, 58, 162–171. [Google Scholar] [CrossRef] [Green Version]
- Thalmann, G.N.; Anezinis, P.E.; Chang, S.M.; Zhau, H.E.; Kim, E.E.; Hopwood, V.L.; Pathak, S.; von Eschenbach, A.C.; Chung, L.W. Androgen-independent cancer progression and bone metastasis in the LNCaP model of human prostate cancer. Cancer Res. 1994, 54, 2577–2581. [Google Scholar] [PubMed]
- Briganti, A.; Spahn, M.; Joniau, S.; Gontero, P.; Bianchi, M.; Kneitz, B.; Chun, F.K.; Sun, M.; Graefen, M.; Abdollah, F.; et al. Impact of age and comorbidities on long-term survival of patients with high-risk prostate cancer treated with radical prostatectomy: A multi-institutional competing-risks analysis. Eur. Urol. 2013, 63, 693–701. [Google Scholar] [CrossRef] [PubMed]
- Tosco, L.; Laenen, A.; Briganti, A.; Gontero, P.; Karnes, R.J.; Bastian, P.J.; Chlosta, P.; Claessens, F.; Chun, F.K.; Everaerts, W.; et al. The EMPaCT classifier: A validated tool to predict postoperative prostate cancer-related death using competing-risk analysis. Eur. Urol. Focus. 2018, 4, 369–375. [Google Scholar] [CrossRef] [PubMed]
- Chys, B.; Devos, G.; Everaerts, W.; Albersen, M.; Moris, L.; Claessens, F.; De Meerleer, G.; Haustermans, K.; Briganti, A.; Chlosta, P.; et al. Preoperative risk-stratification of high-risk prostate cancer: A multicenter analysis. Front. Oncol. 2020, 10, 246. [Google Scholar] [CrossRef] [PubMed]
- D’Amico, A.V.; Whittington, R.; Malkowicz, S.B.; Schultz, D.; Blank, K.; Broderick, G.A.; Tomaszewski, J.E.; Renshaw, A.A.; Kaplan, I.; Beard, C.J.; et al. Biochemical outcome after radical prostatectomy, external beam radiation therapy, or interstitial radiation therapy for clinically localized prostate cancer. JAMA 1998, 280, 969–974. [Google Scholar]
- Kassambara, A. Survminer: Drawing Survival Curves Using ‘ggplot2’. 2018. Available online: http://www.sthda.com/english/rpkgs/survminer/ (accessed on 1 January 2016).
- Cortazar, A.R.; Torrano, V.; Martín-Martín, N.; Caro-Maldonado, A.; Camacho, L.; Hermanova, I.; Guruceaga, E.; Lorenzo-Martín, L.F.; Caloto, R.; Gomis, R.R.; et al. CANCERTOOL: A visualization and representation interface to exploit cancer datasets. Cancer Res. 2018, 78, 6320–6328. [Google Scholar] [CrossRef] [Green Version]
- Glinsky, G.V.; Glinskii, A.B.; Stephenson, A.J.; Hoffman, R.M.; Gerald, W.L. Gene expression profiling predicts clinical outcome of prostate cancer. J. Clin. Invest. 2004. [Google Scholar] [CrossRef]
- Mo, F.; Lin, D.; Takhar, M.; Ramnarine, V.R.; Dong, X.; Bell, R.H.; Volik, S.V.; Wang, K.; Xue, H.; Wang, Y.; et al. Stromal gene expression is predictive for metastatic primary prostate cancer. Eur. Urol. 2018, 73, 524–532. [Google Scholar] [CrossRef]
- Grasso, C.S.; Wu, Y.M.; Robinson, D.R.; Cao, X.; Dhanasekaran, S.M.; Khan, A.P.; Quist, M.J.; Jing, X.; Lonigro, R.J.; Brenner, J.C.; et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature 2012, 487, 239–243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taylor, B.S.; Schultz, N.; Hieronymus, H.; Gopalan, A.; Xiao, Y.; Carver, B.S.; Arora, V.K.; Kaushik, P.; Cerami, E.; Reva, B.; et al. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010, 18, 11–22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sahai, E.; Astsaturov, I.; Cukierman, E.; DeNardo, D.G.; Egeblad, M.; Evans, R.M.; Fearon, D.; Greten, F.R.; Hingorani, S.R.; Hunter, T.; et al. A framework for advancing our understanding of cancer-associated fibroblasts. Nat. Rev. Cancer 2020, 20, 174–186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Corn, P.G. The tumor microenvironment in prostate cancer: Elucidating molecular pathways for therapy development. Cancer Manag. Res. 2012, 4, 183–193. [Google Scholar] [CrossRef] [Green Version]
- Bissell, M.J.; Hines, W.C. Why don’t we get more cancer? A proposed role of the microenvironment in restraining cancer progression. Nat. Med. 2011, 17, 320–329. [Google Scholar] [CrossRef] [Green Version]
- Petersen, O.W.; Rønnov-Jessen, L.; Howlett, A.R.; Bissell, M.J. Interaction with basement membrane serves to rapidly distinguish growth and differentiation pattern of normal and malignant human breast epithelial cells. Proc. Natl. Acad. Sci. USA. 1992, 89, 9064–9068. [Google Scholar] [CrossRef] [Green Version]
- Weaver, V.M.; Petersen, O.W.; Wang, F.; Larabell, C.A.; Briand, P.; Damsky, C.; Bissell, M.J. Reversion of the malignant phenotype of human breast cells in three-dimensional culture and in vivo by integrin blocking antibodies. J. Cell. Biol. 1997, 137, 231–245. [Google Scholar] [CrossRef] [Green Version]
- Sung, S.-Y.; Hsieh, C.-L.; Law, A.; Zhau, H.E.; Pathak, S.; Multani, A.S.; Lim, S.; Coleman, I.M.; Wu, L.-C.; Figg, W.D.; et al. Coevolution of prostate cancer and bone stroma in three-dimensional coculture: Implications for cancer growth and metastasis. Cancer Res. 2008, 68, 9996–10003. [Google Scholar] [CrossRef] [Green Version]
- Craft, N.; Chhor, C.; Tran, C.; Belldegrun, A.; DeKernion, J.; Witte, O.N.; Said, J.; Reiter, R.E.; Sawyers, C.L. Evidence for clonal outgrowth of androgen-independent prostate cancer cells from androgen-dependent tumors through a two-step process. Cancer Res. 1999, 59, 5030–5036. [Google Scholar]
- Germann, M.; Wetterwald, A.; Guzman-Ramirez, N.; van der Pluijm, G.; Culig, Z.; Cecchini, M.G.; Williams, E.D.; Thalmann, G.N. Stem-like cells with luminal progenitor phenotype survive castration in human prostate cancer. Stem Cells 2012, 30, 1076–1086. [Google Scholar] [CrossRef]
- McCulloch, D.R.; Opeskin, K.; Thompson, E.W.; Williams, E.D. BM18: A novel androgen-dependent human prostate cancer xenograft model derived from a bone metastasis. Prostate 2005, 65, 35–43. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Deng, Q.; Chao, H.-P.; Liu, X.; Lu, Y.; Lin, K.; Liu, B.; Tang, G.W.; Zhang, D.; Tracz, A.; et al. Linking prostate cancer cell AR heterogeneity to distinct castration and enzalutamide responses. Nat. Commun. 2018, 9, 3600. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, X.; Li, Q.; Liu, X.; Liu, C.; Liu, R.; Rycaj, K.; Zhang, D.; Liu, B.; Jeter, C.; Calhoun-Davis, T.; et al. Defining a population of stem-like human prostate cancer cells that can generate and propagate castration-resistant prostate cancer. Clin. Cancer Res. 2016, 22, 4505–4516. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cutz, J.-C.; Guan, J.; Bayani, J.; Yoshimoto, M.; Xue, H.; Sutcliffe, M.; English, J.; Flint, J.; LeRiche, J.; Yee, J.; et al. Establishment in severe combined immunodeficiency mice of subrenal capsule xenografts and transplantable tumor lines from a variety of primary human lung cancers: Potential models for studying tumor progression–related changes. Clin. Cancer Res. 2006, 12, 4043–4054. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hidalgo, M.; Amant, F.; Biankin, A.V.; Budinská, E.; Byrne, A.T.; Caldas, C.; Clarke, R.B.; de Jong, S.; Jonkers, J.; Mælandsmo, G.M.; et al. Patient-derived xenograft models: An emerging platform for translational cancer research. Cancer Discov. 2014, 4, 998–1013. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tuxhorn, J.A.; Ayala, G.E.; Smith, M.J.; Smith, V.C.; Dang, T.D.; Rowley, D.R. Reactive stroma in human prostate cancer. Induction of myofibroblast phenotype and extracellular matrix remodeling. Clin. Cancer Res. 2002, 8, 2912–2923. [Google Scholar] [PubMed]
- Ibrahim, S.N.; Lightner, V.A.; Ventimiglia, J.B.; Ibrahim, G.K.; Walther, P.J.; Bigner, D.D.; Humphrey, P.A. Tenascin expression in prostatic hyperplasia, intraepithelial neoplasia, and carcinoma. Hum. Pathol. 1993, 24, 982–989. [Google Scholar] [CrossRef]
- Xue, Y.; Smedts, F.; Latijnhouwers, M.A.; Ruijter, E.T.; Aalders, T.W.; de la Rosette, J.J.; Debruyne, F.M.; Schalken, J.A. Tenascin-C expression in prostatic intraepithelial neoplasia (PIN): A marker of progression? Anticancer Res. 1998, 18, 2679–2684. [Google Scholar]
- Ni, W.-D.; Yang, Z.-T.; Cui, C.-A.; Cui, Y.; Fang, L.-Y.; Xuan, Y.-H. Tenascin-C is a potential cancer-associated fibroblasts marker and predicts poor prognosis in prostate cancer. Biochem. Biophys. Res. Commun. 2017, 486, 607–612. [Google Scholar] [CrossRef]
- Cai, M.; Onoda, K.; Takao, M.; Kyoko, I.-Y.; Shimpo, H.; Yoshida, T.; Yada, I. Degradation of tenascin-C and activity of matrix metalloproteinase-2 are associated with tumor recurrence in early stage non-small cell lung cancer. Clin. Cancer Res. 2002, 8, 1152–1156. [Google Scholar]
- Kusagawa, H.; Onoda, K.; Namikawa, S.; Yada, I.; Okada, A.; Yoshida, T.; Sakakura, T. Expression and degeneration of tenascin-C in human lung cancers. Br. J. Cancer 1998, 77, 98–102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- San Martin, R.; Pathak, R.; Jain, A.; Jung, S.Y.; Hilsenbeck, S.G.; Piña-Barba, M.C.; Sikora, A.G.; Pienta, K.J.; Rowley, D.R. Tenascin-C and integrin α9 mediate interactions of prostate cancer with the bone microenvironment. Cancer Res. 2017, 77, 5977–5988. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Colombel, M.; Eaton, C.L.; Hamdy, F.; Ricci, E.; van der Pluijm, G.; Cecchini, M.; Mege-Lechevallier, F.; Clezardin, P.; Thalmann, G. Increased expression of putative cancer stem cell markers in primary prostate cancer is associated with progression of bone metastases. Prostate 2012, 72, 713–720. [Google Scholar] [CrossRef] [PubMed]
- Lowy, C.M.; Oskarsson, T. Tenascin C in metastasis: A view from the invasive front. Cell Adh. Migr. 2015, 9, 112–124. [Google Scholar] [CrossRef] [Green Version]
- Mishra, P.; Kiebish, M.A.; Cullen, J.; Srinivasan, A.; Patterson, A.; Sarangarajan, R.; Narain, N.R.; Dobi, A. Genomic alterations of Tenascin C in highly aggressive prostate cancer: A meta-analysis. Genes Cancer 2019, 10, 150–159. [Google Scholar] [CrossRef] [Green Version]
- Kiebish, M.A.; Cullen, J.; Mishra, P.; Ali, A.; Milliman, E.; Rodrigues, L.O.; Chen, E.Y.; Tolstikov, V.; Zhang, L.; Panagopoulos, K.; et al. Multi-omic serum biomarkers for prognosis of disease progression in prostate cancer. J. Transl. Med. 2020, 18, 1–10. [Google Scholar] [CrossRef]
- Jachetti, E.; Caputo, S.; Mazzoleni, S.; Brambillasca, C.S.; Parigi, S.M.; Grioni, M.; Piras, I.S.; Restuccia, U.; Calcinotto, A.; Freschi, M.; et al. Tenascin-C protects cancer stem–like cells from immune surveillance by arresting T-cell activation. Cancer Res. 2015, 75, 2095–2108. [Google Scholar] [CrossRef] [Green Version]
- Lapointe, J.; Li, C.; Higgins, J.P.; van de Rijn, M.; Bair, E.; Montgomery, K.; Ferrari, M.; Egevad, L.; Rayford, W.; Bergerheim, U.; et al. Gene expression profiling identifies clinically relevant subtypes of prostate cancer. Proc. Natl. Acad. Sci. USA 2004. [Google Scholar] [CrossRef] [Green Version]
- Varambally, S.; Yu, J.; Laxman, B.; Rhodes, D.R.; Mehra, R.; Tomlins, S.A.; Shah, R.B.; Chandran, U.; Monzon, F.A.; Becich, M.J.; et al. Integrative genomic and proteomic analysis of prostate cancer reveals signatures of metastatic progression. Cancer Cell. 2005. [Google Scholar] [CrossRef] [Green Version]
- Beltran, H.; Eng, K.; Mosquera, J.M.; Sigaras, A.; Romanel, A.; Rennert, H.; Kossai, M.; Pauli, C.; Faltas, B.; Fontugne, J.; et al. Whole-exome sequencing of metastatic cancer and biomarkers of treatment response. JAMA Oncol. 2015, 1, 466–474. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Nikolayeva, O.; Robinson, M.D. Edger for differential RNA-seq and ChIP-seq analysis: An application to stem cell biology. Methods Mol. Biol. 2014, 1150, 45–79. [Google Scholar]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS: J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Hart, T.; Komori, H.K.; LaMere, S.; Podshivalova, K.; Salomon, D.R. Finding the active genes in deep RNA-seq gene expression studies. BMC Genom. 2013, 14, 778. [Google Scholar] [CrossRef] [Green Version]
- Cancer Genome Atlas Research, N. The molecular taxonomy of primary prostate cancer. Cell 2015, 163, 1011–1025. [Google Scholar]
- Hoadley, K.A.; Yau, C.; Hinoue, T.; Wolf, D.M.; Lazar, A.J.; Drill, E.; Shen, R.; Taylor, A.M.; Cherniack, A.D.; Thorsson, V.; et al. Cell-of-origin patterns dominate the molecular classification of 10,000 tumors from 33 types of cancer. Cell 2018, 173, 291–304.e6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hänzelmann, S.; Castelo, R.; Guinney, J. GSVA: Gene set variation analysis for microarray and RNA-seq data. BMC Bioinform. 2013, 14, 7. [Google Scholar] [CrossRef] [Green Version]
- Hughes, C.S.; Foehr, S.; Garfield, D.A.; Furlong, E.E.; Steinmetz, L.M.; Krijgsveld, J. Ultrasensitive proteome analysis using paramagnetic bead technology. Mol. Syst. Biol. 2014, 10, 757. [Google Scholar] [CrossRef]
- Moggridge, S.; Sorensen, P.H.; Morin, G.B.; Hughes, C.S. Extending the compatibility of the SP3 paramagnetic bead processing approach for proteomics. J. Proteome. Res. 2018, 17, 1730–1740. [Google Scholar] [CrossRef]
- Werner, T.; Sweetman, G.; Savitski, M.F.; Mathieson, T.; Bantscheff, M.; Savitski, M.M. Ion coalescence of neutron encoded TMT 10-plex reporter ions. Anal. Chem. 2014, 86, 3594–3601. [Google Scholar] [CrossRef]
- Becher, I.; Andres-Pons, A.; Romanov, N.; Stein, F.; Schramm, M.; Baudin, F.; Helm, D.; Kurzawa, N.; Mateus, A.; Mackmull, M.T.; et al. Pervasive protein thermal stability variation during the cell cycle. Cell 2018, 173, 1495–1507.e18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Franken, H.; Mathieson, T.; Childs, D.; Sweetman, G.M.; Werner, T.; Togel, I.; Doce, C.; Gade, S.; Bantscheff, M.; Drewes, G.; et al. Thermal proteome profiling for unbiased identification of direct and indirect drug targets using multiplexed quantitative mass spectrometry. Nat. Protoc. 2015, 10, 1567–1593. [Google Scholar] [CrossRef] [PubMed]
- Savitski, M.M.; Wilhelm, M.; Hahne, H.; Kuster, B.; Bantscheff, M. A scalable approach for protein false discovery rate estimation in large proteomic data sets. Mol. Cell. Proteomics. 2015, 14, 2394–2404. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huber, W.; von Heydebreck, A.; Sultmann, H.; Poustka, A.; Vingron, M. Variance stabilization applied to microarray data calibration and to the quantification of differential expression. Bioinformatics 2002, 18 (Suppl. S1), S96–S104. [Google Scholar] [CrossRef]
- Efron, B. Large-scale simultaneous hypothesis testing. J. Am. Statist. Assoc. 2004, 99, 96–104. [Google Scholar] [CrossRef]
- Strimmer, K. fdrtool: A versatile R package for estimating local and tail area-based false discovery rates. Bioinformatics 2008, 24, 1461–1462. [Google Scholar] [CrossRef] [Green Version]
- Bankhead, P.; Loughrey, M.B.; Fernández, J.A.; Dombrowski, Y.; McArt, D.G.; Dunne, P.D.; McQuaid, S.; Gray, R.T.; Murray, L.J.; Coleman, H.G.; et al. QuPath: Open source software for digital pathology image analysis. Sci. Rep. 2017, 7, 16878. [Google Scholar] [CrossRef] [Green Version]
- Therneau, T. A Package for Survival Analysis in S. 2015. Available online: https://www.mayo.edu/research/documents/tr53pdf/doc-10027379 (accessed on 28 September 2020).
- Therneau, T. PMG: Modeling Survival Data: Extending the Cox Model; Springer: New York, NY, USA, 2000. [Google Scholar]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016. [Google Scholar]
- RStudio Team. RStudio: Integrated Development for R.; RStudio: Boston, MI, USA, 2016. [Google Scholar]
- R Core Team. R: A language and Environment for Statistical Computing; ARFfSC: Vienna, Austria, 2019. [Google Scholar]


| Descriptive Statistics | Age at Surgery | PSA at Surgery | PSA Progression Time (Months) | Clinical Progression Time (Months) |
|---|---|---|---|---|
| Min | 43 | 20 | 1 | 1 |
| 1st quartile | 62 | 25.33 | 29.5 | 40.5 |
| Median quartile | 67 | 36.99 | 63.5 | 75.5 |
| Mean quartile | 66.18 | 50.56 | 63.47 | 70.89 |
| 3rd quartile | 71 | 61.9 | 90 | 95.75 |
| Max quartile | 81 | 597 | 151 | 153 |
| PSA Progression | Clinical Progression | Pathological Staging (No. of Patient Cases) | ||||
|---|---|---|---|---|---|---|
| 2a | 2b | 3a | 3b | 4 | ||
| no | no | 6 | 15 | 37 | 63 | 26 |
| yes | 0 | 0 | 0 | 1 | 0 | |
| yes | no | 0 | 5 | 7 | 11 | 7 |
| yes | 1 | 7 | 9 | 9 | 6 | |
| Dilution | Antibody | Company | Catalog No. |
|---|---|---|---|
| 1 to 500 | Ki67 | Gene Tex | GTX16667 |
| 1 to 100 | Tnc | R&D | MAB2138 |
| Dilution | Antibody | Company | Catalog No. |
|---|---|---|---|
| 1 to 500 | αSMA | Sigma | A2547 |
| 1 to 500 | ITGA2 | Abcam | ab181548 |
| 1 to 500 | Collagen type I | Southern Biotech | 1310-01 |
| 1 to 50 | Tnc | R&D | MAB2138 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Karkampouna, S.; De Filippo, M.R.; Ng, C.K.Y.; Klima, I.; Zoni, E.; Spahn, M.; Stein, F.; Haberkant, P.; Thalmann, G.N.; Kruithof-de Julio, M. Stroma Transcriptomic and Proteomic Profile of Prostate Cancer Metastasis Xenograft Models Reveals Prognostic Value of Stroma Signatures. Cancers 2020, 12, 3786. https://doi.org/10.3390/cancers12123786
Karkampouna S, De Filippo MR, Ng CKY, Klima I, Zoni E, Spahn M, Stein F, Haberkant P, Thalmann GN, Kruithof-de Julio M. Stroma Transcriptomic and Proteomic Profile of Prostate Cancer Metastasis Xenograft Models Reveals Prognostic Value of Stroma Signatures. Cancers. 2020; 12(12):3786. https://doi.org/10.3390/cancers12123786
Chicago/Turabian StyleKarkampouna, Sofia, Maria R. De Filippo, Charlotte K. Y. Ng, Irena Klima, Eugenio Zoni, Martin Spahn, Frank Stein, Per Haberkant, George N. Thalmann, and Marianna Kruithof-de Julio. 2020. "Stroma Transcriptomic and Proteomic Profile of Prostate Cancer Metastasis Xenograft Models Reveals Prognostic Value of Stroma Signatures" Cancers 12, no. 12: 3786. https://doi.org/10.3390/cancers12123786
APA StyleKarkampouna, S., De Filippo, M. R., Ng, C. K. Y., Klima, I., Zoni, E., Spahn, M., Stein, F., Haberkant, P., Thalmann, G. N., & Kruithof-de Julio, M. (2020). Stroma Transcriptomic and Proteomic Profile of Prostate Cancer Metastasis Xenograft Models Reveals Prognostic Value of Stroma Signatures. Cancers, 12(12), 3786. https://doi.org/10.3390/cancers12123786





