Identification of Potential Serum Protein Biomarkers and Pathways for Pancreatic Cancer Cachexia Using an Aptamer-Based Discovery Platform
Abstract
:Simple Summary
Abstract
1. Introduction
2. Results
2.1. Patient Demographics/Clinical Characteristics and SOMAscan Quality Control Results
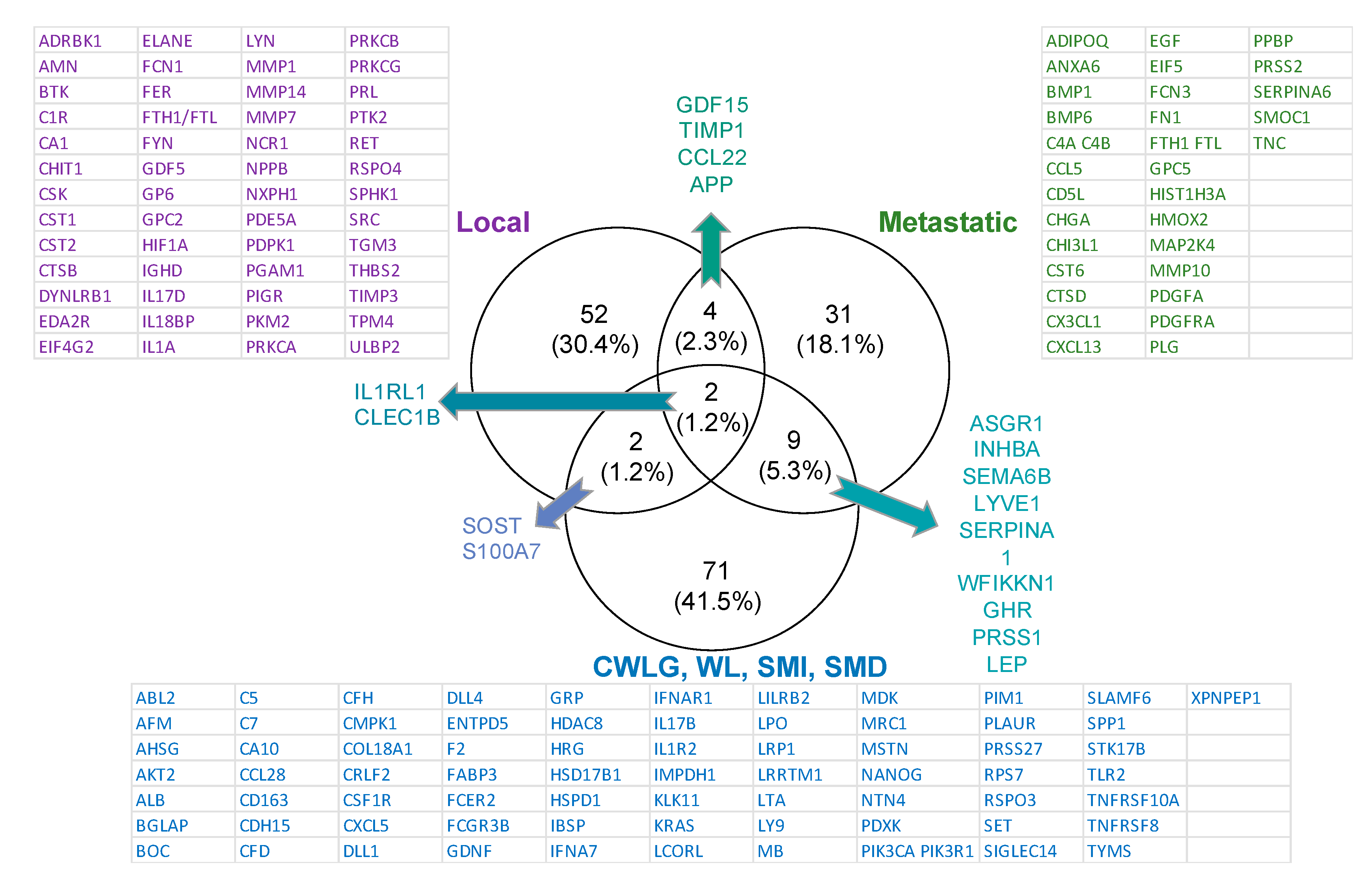
2.2. SOMAscan Quality Control and Results by Cancer Stage
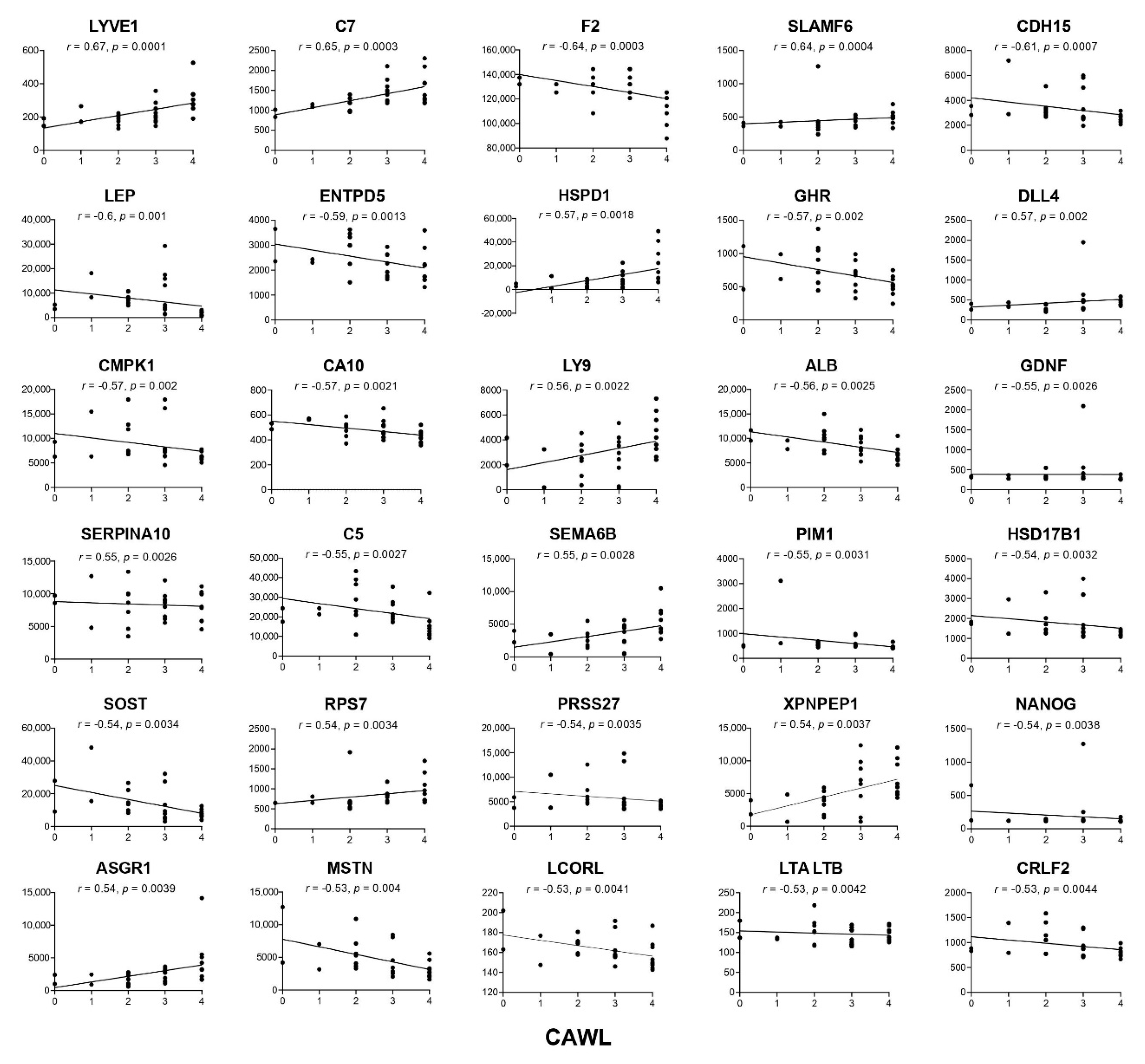
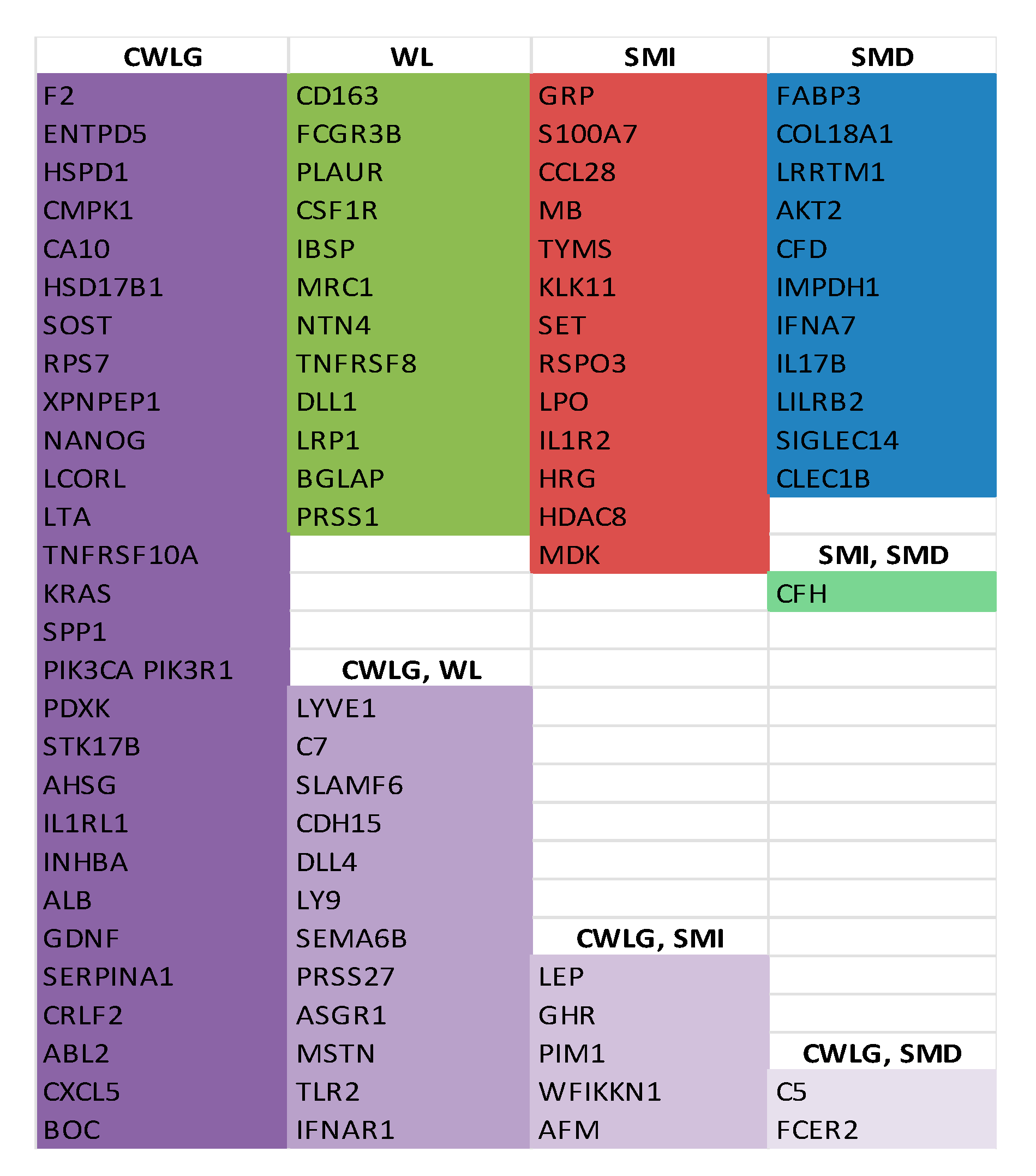
Proteins Associated with Clinical Variables of Cachexia
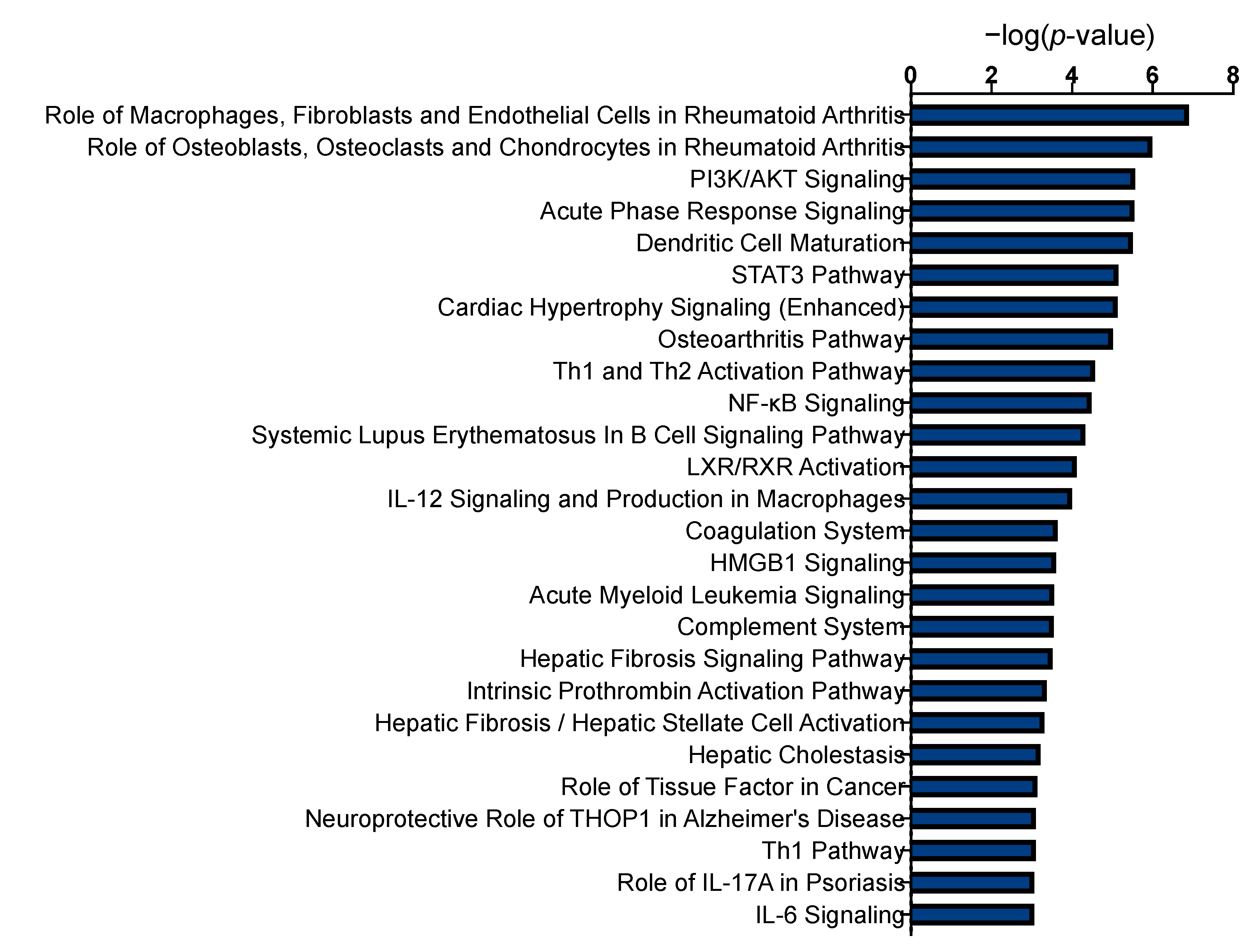
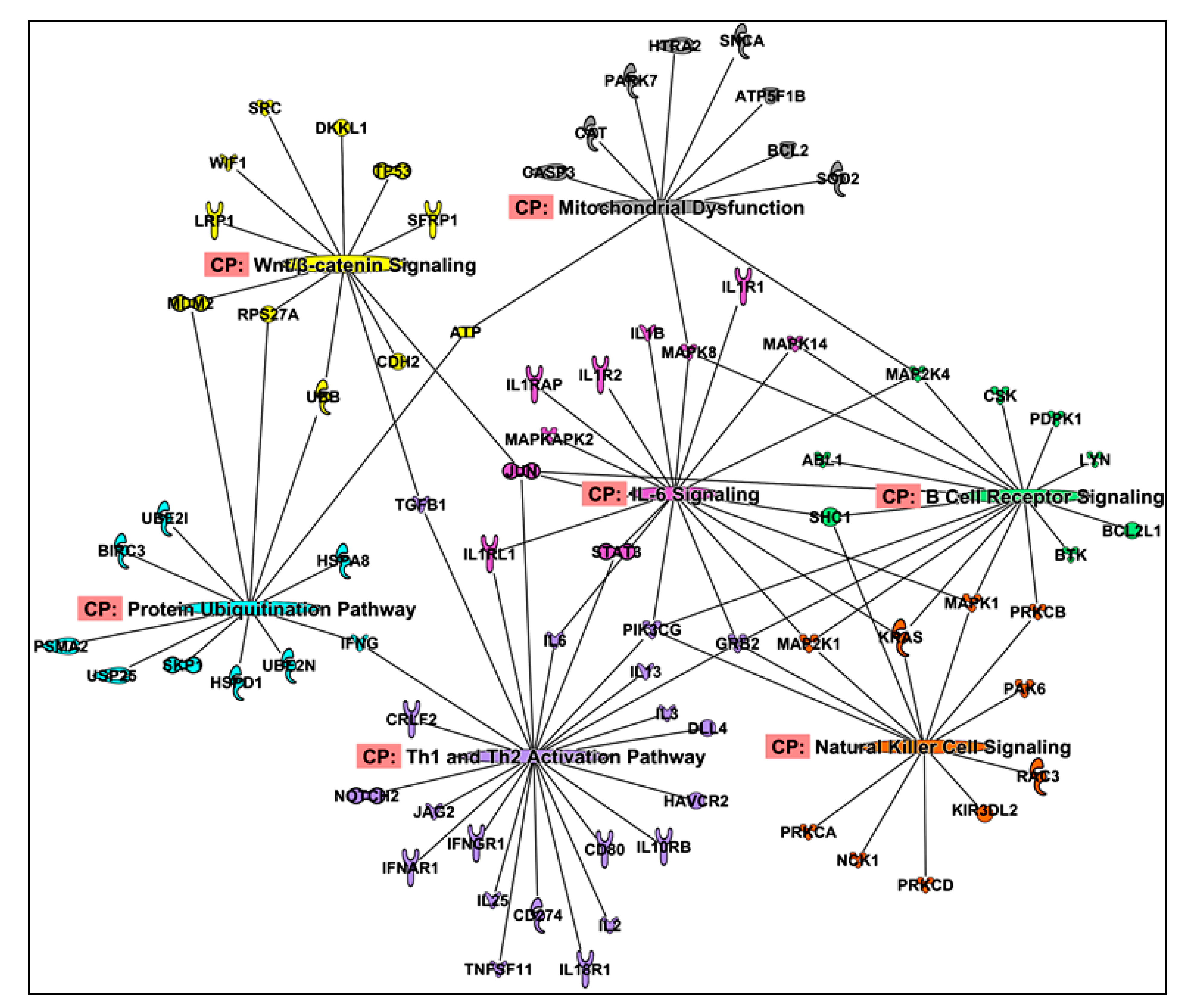
2.3. Functional Enrichment of Proteins
2.4. Upstream Regulators
2.5. Protein Co-Expression Analysis
3. Discussion
4. Materials and Methods
4.1. Recruitment of Study Participants
4.2. Assessment of Clinical Variables for Cachexia
4.3. Protein Measurements Using SOMAscan
4.4. Statistical Analyses
4.5. Identification of Upstream Regulators of Proteins Associated with Cachexia
4.6. Protein Co-Expression Analysis to Identify Novel Pathways Associated with Cachexia
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Fearon, K.; Strasser, F.; Anker, S.D.; Bosaeus, I.; Bruera, E.; Fainsinger, R.L.; Jatoi, A.; Loprinzi, C.; MacDonald, N.; Mantovani, G.; et al. Definition and classification of cancer cachexia: An international consensus. Lancet Oncol. 2011, 12, 489–495. [Google Scholar] [CrossRef]
- Baracos, V.E.; Martin, L.; Korc, M.; Guttridge, D.C.; Fearon, K.C.H. Cancer-associated cachexia. Nat. Rev. Dis. Primers 2018, 4, 17105. [Google Scholar] [CrossRef] [PubMed]
- Martin, L.; Senesse, P.; Gioulbasanis, I.; Antoun, S.; Bozzetti, F.; Deans, C.; Strasser, F.; Thoresen, L.; Jagoe, R.T.; Chasen, M.; et al. Diagnostic criteria for the classification of cancer-associated weight loss. J. Clin. Oncol. 2015, 33, 90–99. [Google Scholar] [CrossRef] [PubMed]
- Nemer, L.; Krishna, S.G.; Shah, Z.K.; Conwell, D.L.; Cruz-Monserrate, Z.; Dillhoff, M.; Guttridge, D.C.; Hinton, A.; Manilchuk, A.; Pawlik, T.M.; et al. Predictors of Pancreatic Cancer-Associated Weight Loss and Nutritional Interventions. Pancreas 2017, 46, 1152–1157. [Google Scholar] [CrossRef] [PubMed]
- Hendifar, A.E.; Chang, J.I.; Huang, B.Z.; Tuli, R.; Wu, B.U. Cachexia, and not obesity, prior to pancreatic cancer diagnosis worsens survival and is negated by chemotherapy. J. Gastrointest. Oncol. 2018, 9, 17–23. [Google Scholar] [CrossRef] [Green Version]
- Bieliuniene, E.; Brondum Frokjaer, J.; Pockevicius, A.; Kemesiene, J.; Lukosevicius, S.; Basevicius, A.; Atstupenaite, V.; Barauskas, G.; Ignatavicius, P.; Gulbinas, A.; et al. CT- and MRI-Based Assessment of Body Composition and Pancreatic Fibrosis Reveals High Incidence of Clinically Significant Metabolic Changes That Affect the Quality of Life and Treatment Outcomes of Patients with Chronic Pancreatitis and Pancreatic Cancer. Medicina 2019, 55, 649. [Google Scholar] [CrossRef] [Green Version]
- Naumann, P.; Eberlein, J.; Farnia, B.; Hackert, T.; Debus, J.; Combs, S.E. Continued Weight Loss and Sarcopenia Predict Poor Outcomes in Locally Advanced Pancreatic Cancer Treated with Chemoradiation. Cancers 2019, 11, 709. [Google Scholar] [CrossRef] [Green Version]
- Griffin, O.M.; Duggan, S.N.; Ryan, R.; McDermott, R.; Geoghegan, J.; Conlon, K.C. Characterising the impact of body composition change during neoadjuvant chemotherapy for pancreatic cancer. Pancreatology 2019, 19, 850–857. [Google Scholar] [CrossRef]
- Kays, J.K.; Shahda, S.; Stanley, M.; Bell, T.M.; O’Neill, B.H.; Kohli, M.D.; Couch, M.E.; Koniaris, L.G.; Zimmers, T.A. Three cachexia phenotypes and the impact of fat-only loss on survival in FOLFIRINOX therapy for pancreatic cancer. J. Cachexia Sarcopenia Muscle 2018, 9, 673–684. [Google Scholar] [CrossRef]
- Naumann, P.; Eberlein, J.; Farnia, B.; Liermann, J.; Hackert, T.; Debus, J.; Combs, S.E. Cachectic Body Composition and Inflammatory Markers Portend a Poor Prognosis in Patients with Locally Advanced Pancreatic Cancer Treated with Chemoradiation. Cancers 2019, 11, 1655. [Google Scholar] [CrossRef] [Green Version]
- Yuen, E.Y.N.; Zaleta, A.K.; McManus, S.; Buzaglo, J.S.; LeBlanc, T.W.; Hamilton, K.; Stein, K. Unintentional weight loss, its associated burden, and perceived weight status in people with cancer. Support. Care Cancer 2020, 28, 329–339. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, R.R.; Fero, K.E.; Seible, D.M.; Panjwani, N.; Matsuno, R.K.; Murphy, J.D. A Population-Based Study of Morbidity After Pancreatic Cancer Diagnosis. J. Natl. Compr. Canc. Netw. 2019, 17, 432–440. [Google Scholar] [CrossRef] [PubMed]
- Zimmers, T.A.; Fishel, M.L.; Bonetto, A. STAT3 in the systemic inflammation of cancer cachexia. Semin. Cell Dev. Biol. 2016, 54, 28–41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roeland, E.J.; Bohlke, K.; Baracos, V.E.; Bruera, E.; Del Fabbro, E.; Dixon, S.; Fallon, M.; Herrstedt, J.; Lau, H.; Platek, M.; et al. Management of Cancer Cachexia: ASCO Guideline. J. Clin. Oncol. 2020, 38, 2438–2453. [Google Scholar] [CrossRef] [PubMed]
- Bonetto, A.; Rupert, J.E.; Barreto, R.; Zimmers, T.A. The Colon-26 Carcinoma Tumor-bearing Mouse as a Model for the Study of Cancer Cachexia. J. Vis. Exp. 2016. [Google Scholar] [CrossRef] [Green Version]
- Zhong, X.; Pons, M.; Poirier, C.; Jiang, Y.; Liu, J.; Sandusky, G.E.; Shahda, S.; Nakeeb, A.; Schmidt, C.M.; House, M.G.; et al. The systemic activin response to pancreatic cancer: Implications for effective cancer cachexia therapy. J. Cachexia Sarcopenia Muscle 2019, 10, 1083–1101. [Google Scholar] [CrossRef] [Green Version]
- Michaelis, K.A.; Norgard, M.A.; Zhu, X.; Levasseur, P.R.; Sivagnanam, S.; Liudahl, S.M.; Burfeind, K.G.; Olson, B.; Pelz, K.R.; Angeles Ramos, D.M.; et al. The TLR7/8 agonist R848 remodels tumor and host responses to promote survival in pancreatic cancer. Nat. Commun. 2019, 10, 4682. [Google Scholar] [CrossRef] [Green Version]
- Shukla, S.K.; Markov, S.D.; Attri, K.S.; Vernucci, E.; King, R.J.; Dasgupta, A.; Grandgenett, P.M.; Hollingsworth, M.A.; Singh, P.K.; Yu, F.; et al. Macrophages potentiate STAT3 signaling in skeletal muscles and regulate pancreatic cancer cachexia. Cancer Lett. 2020, 484, 29–39. [Google Scholar] [CrossRef]
- Burfeind, K.G.; Zhu, X.; Norgard, M.A.; Levasseur, P.R.; Huisman, C.; Buenafe, A.C.; Olson, B.; Michaelis, K.A.; Torres, E.R.; Jeng, S.; et al. Circulating myeloid cells invade the central nervous system to mediate cachexia during pancreatic cancer. eLife 2020, 9. [Google Scholar] [CrossRef]
- Talbert, E.E.; Cuitino, M.C.; Ladner, K.J.; Rajasekerea, P.V.; Siebert, M.; Shakya, R.; Leone, G.W.; Ostrowski, M.C.; Paleo, B.; Weisleder, N.; et al. Modeling Human Cancer-induced Cachexia. Cell Rep. 2019, 28, 1612–1622. [Google Scholar] [CrossRef] [Green Version]
- Lerner, L.; Tao, J.; Liu, Q.; Nicoletti, R.; Feng, B.; Krieger, B.; Mazsa, E.; Siddiquee, Z.; Wang, R.; Huang, L.; et al. MAP3K11/GDF15 axis is a critical driver of cancer cachexia. J. Cachexia Sarcopenia Muscle 2016, 7, 467–482. [Google Scholar] [CrossRef] [PubMed]
- Au, E.D.; Desai, A.P.; Koniaris, L.G.; Zimmers, T.A. The MEK-Inhibitor Selumetinib Attenuates Tumor Growth and Reduces IL-6 Expression but Does Not Protect against Muscle Wasting in Lewis Lung Cancer Cachexia. Front. Physiol. 2016, 7, 682. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Benny Klimek, M.E.; Aydogdu, T.; Link, M.J.; Pons, M.; Koniaris, L.G.; Zimmers, T.A. Acute inhibition of myostatin-family proteins preserves skeletal muscle in mouse models of cancer cachexia. Biochem. Biophys. Res. Commun. 2010, 391, 1548–1554. [Google Scholar] [CrossRef] [PubMed]
- Huot, J.R.; Pin, F.; Narasimhan, A.; Novinger, L.J.; Keith, A.S.; Zimmers, T.A.; Willis, M.S.; Bonetto, A. ACVR2B antagonism as a countermeasure to multi-organ perturbations in metastatic colorectal cancer cachexia. J. Cachexia Sarcopenia Muscle 2020. [Google Scholar] [CrossRef] [PubMed]
- Suriben, R.; Chen, M.; Higbee, J.; Oeffinger, J.; Ventura, R.; Li, B.; Mondal, K.; Gao, Z.; Ayupova, D.; Taskar, P.; et al. Antibody-mediated inhibition of GDF15-GFRAL activity reverses cancer cachexia in mice. Nat. Med. 2020, 26, 1264–1270. [Google Scholar] [CrossRef]
- Miller, J.; Skipworth, R.J.E. Novel molecular targets of muscle wasting in cancer patients. Curr. Opin. Clin. Nutr. Metab. Care 2019, 22, 196–204. [Google Scholar] [CrossRef]
- Martin, L.; Birdsell, L.; Macdonald, N.; Reiman, T.; Clandinin, M.T.; McCargar, L.J.; Murphy, R.; Ghosh, S.; Sawyer, M.B.; Baracos, V.E. Cancer cachexia in the age of obesity: Skeletal muscle depletion is a powerful prognostic factor, independent of body mass index. J. Clin. Oncol. 2013, 31, 1539–1547. [Google Scholar] [CrossRef]
- Rohloff, J.C.; Gelinas, A.D.; Jarvis, T.C.; Ochsner, U.A.; Schneider, D.J.; Gold, L.; Janjic, N. Nucleic Acid Ligands With Protein-like Side Chains: Modified Aptamers and Their Use as Diagnostic and Therapeutic Agents. Mol. Ther. Nucleic Acids 2014, 3, e201. [Google Scholar] [CrossRef]
- Sullivan, G.M.; Feinn, R. Using Effect Size—Or Why the P Value Is Not Enough. J. Grad. Med. Educ. 2012, 4, 279–282. [Google Scholar] [CrossRef] [Green Version]
- Bonetto, A.; Kays, J.K.; Parker, V.A.; Matthews, R.R.; Barreto, R.; Puppa, M.J.; Kang, K.S.; Carson, J.A.; Guise, T.A.; Mohammad, K.S.; et al. Differential Bone Loss in Mouse Models of Colon Cancer Cachexia. Front. Physiol. 2016, 7, 679. [Google Scholar] [CrossRef] [Green Version]
- Park, H.K.; Ahima, R.S. Physiology of leptin: Energy homeostasis, neuroendocrine function and metabolism. Metabolism 2015, 64, 24–34. [Google Scholar] [CrossRef] [Green Version]
- Argiles, J.M.; Busquets, S.; Stemmler, B.; Lopez-Soriano, F.J. Cancer cachexia: Understanding the molecular basis. Nat. Rev. Cancer 2014, 14, 754–762. [Google Scholar] [CrossRef] [PubMed]
- Costamagna, D.; Duelen, R.; Penna, F.; Neumann, D.; Costelli, P.; Sampaolesi, M. Interleukin-4 administration improves muscle function, adult myogenesis, and lifespan of colon carcinoma-bearing mice. J. Cachexia Sarcopenia Muscle 2020, 11, 783–801. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kandarian, S.C.; Nosacka, R.L.; Delitto, A.E.; Judge, A.R.; Judge, S.M.; Ganey, J.D.; Moreira, J.D.; Jackman, R.W. Tumour-derived leukaemia inhibitory factor is a major driver of cancer cachexia and morbidity in C26 tumour-bearing mice. J. Cachexia Sarcopenia Muscle 2018, 9, 1109–1120. [Google Scholar] [CrossRef] [PubMed]
- Takenaga, K.; Akimoto, M.; Koshikawa, N.; Nagase, H. Cancer cell-derived interleukin-33 decoy receptor sST2 enhances orthotopic tumor growth in a murine pancreatic cancer model. PLoS ONE 2020, 15, e0232230. [Google Scholar] [CrossRef] [PubMed]
- Kieler, M.; Unseld, M.; Wojta, J.; Kaider, A.; Bianconi, D.; Demyanets, S.; Prager, G.W. Plasma levels of interleukin-33 and soluble suppression of tumorigenicity 2 in patients with advanced pancreatic ductal adenocarcinoma undergoing systemic chemotherapy. Med. Oncol. 2018, 36, 1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaur, H.; Dhall, A.; Kumar, R.; Raghava, G.P.S. Identification of Platform-Independent Diagnostic Biomarker Panel for Hepatocellular Carcinoma Using Large-Scale Transcriptomics Data. Front. Genet. 2019, 10, 1306. [Google Scholar] [CrossRef]
- Hu, K.; Wang, Z.M.; Li, J.N.; Zhang, S.; Xiao, Z.F.; Tao, Y.M. CLEC1B Expression and PD-L1 Expression Predict Clinical Outcome in Hepatocellular Carcinoma with Tumor Hemorrhage. Transl. Oncol. 2018, 11, 552–558. [Google Scholar] [CrossRef]
- Li, C.; Wang, J.; Kong, J.; Tang, J.; Wu, Y.; Xu, E.; Zhang, H.; Lai, M. GDF15 promotes EMT and metastasis in colorectal cancer. Oncotarget 2016, 7, 860–872. [Google Scholar] [CrossRef]
- Peng, H.; Pan, S.; Yan, Y.; Brand, R.E.; Petersen, G.M.; Chari, S.T.; Lai, L.A.; Eng, J.K.; Brentnall, T.A.; Chen, R. Systemic Proteome Alterations Linked to Early Stage Pancreatic Cancer in Diabetic Patients. Cancers 2020, 12, 1534. [Google Scholar] [CrossRef]
- Prokopchuk, O.; Grunwald, B.; Nitsche, U.; Jager, C.; Prokopchuk, O.L.; Schubert, E.C.; Friess, H.; Martignoni, M.E.; Kruger, A. Elevated systemic levels of the matrix metalloproteinase inhibitor TIMP-1 correlate with clinical markers of cachexia in patients with chronic pancreatitis and pancreatic cancer. BMC Cancer 2018, 18, 128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qi, Q.; Zhuang, L.; Shen, Y.; Geng, Y.; Yu, S.; Chen, H.; Liu, L.; Meng, Z.; Wang, P.; Chen, Z. A novel systemic inflammation response index (SIRI) for predicting the survival of patients with pancreatic cancer after chemotherapy. Cancer 2016, 122, 2158–2167. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hansel, D.E.; Rahman, A.; Wehner, S.; Herzog, V.; Yeo, C.J.; Maitra, A. Increased expression and processing of the Alzheimer amyloid precursor protein in pancreatic cancer may influence cellular proliferation. Cancer Res. 2003, 63, 7032–7037. [Google Scholar] [PubMed]
- Tanaka, T.; Biancotto, A.; Moaddel, R.; Moore, A.Z.; Gonzalez-Freire, M.; Aon, M.A.; Candia, J.; Zhang, P.; Cheung, F.; Fantoni, G.; et al. Plasma proteomic signature of age in healthy humans. Aging Cell 2018, 17, e12799. [Google Scholar] [CrossRef] [Green Version]
- Lehallier, B.; Gate, D.; Schaum, N.; Nanasi, T.; Lee, S.E.; Yousef, H.; Moran Losada, P.; Berdnik, D.; Keller, A.; Verghese, J.; et al. Undulating changes in human plasma proteome profiles across the lifespan. Nat. Med. 2019, 25, 1843–1850. [Google Scholar] [CrossRef]
- Loumaye, A.; de Barsy, M.; Nachit, M.; Lause, P.; Frateur, L.; van Maanen, A.; Trefois, P.; Gruson, D.; Thissen, J.P. Role of Activin A and myostatin in human cancer cachexia. J. Clin. Endocrinol. Metab. 2015, 100, 2030–2038. [Google Scholar] [CrossRef] [Green Version]
- Parente, A.; Boukredine, A.; Baraige, F.; Duprat, N.; Gondran-Tellier, V.; Magnol, L.; Blanquet, V. GASP-2 overexpressing mice exhibit a hypermuscular phenotype with contrasting molecular effects compared to GASP-1 transgenics. FASEB J. 2020, 34, 4026–4040. [Google Scholar] [CrossRef] [Green Version]
- Blyuss, O.; Zaikin, A.; Cherepanova, V.; Munblit, D.; Kiseleva, E.M.; Prytomanova, O.M.; Duffy, S.W.; Crnogorac-Jurcevic, T. Development of PancRISK, a urine biomarker-based risk score for stratified screening of pancreatic cancer patients. Br. J. Cancer 2020, 122, 692–696. [Google Scholar] [CrossRef]
- Sikdar, N.; Saha, G.; Dutta, A.; Ghosh, S.; Shrikhande, S.V.; Banerjee, S. Genetic Alterations of Periampullary and Pancreatic Ductal Adenocarcinoma: An Overview. Curr. Genom. 2018, 19, 444–463. [Google Scholar] [CrossRef]
- Soreide, K.; Roalso, M.; Aunan, J.R. Is There a Trojan Horse to Aggressive Pancreatic Cancer Biology? A Review of the Trypsin-PAR2 Axis to Proliferation, Early Invasion, and Metastasis. J. Pancreat. Cancer 2020, 6, 12–20. [Google Scholar] [CrossRef] [Green Version]
- Yi, Q.; Dong, F.; Lin, L.; Liu, Q.; Chen, S.; Gao, F.; He, Q. PRSS1 mutations and the proteinase/antiproteinase imbalance in the pathogenesis of pancreatic cancer. Tumor Biol. 2016, 37, 5805–5810. [Google Scholar] [CrossRef] [PubMed]
- Nakanishi, T.; Forgetta, V.; Handa, T.; Hirai, T.; Mooser, V.; Lathrop, M.G.; Cookson, W.; Richards, J.B. The Undiagnosed Disease Burden Associated with Alpha-1 Antitrypsin Deficiency Genotypes. Eur. Respir. J. 2020. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, S.; Prokopchuk, O.; Esefeld, K.; Groschel, S.; Bachmann, J.; Lorenzen, S.; Friess, H.; Halle, M.; Martignoni, M.E. The clinical picture of cachexia: A mosaic of different parameters (experience of 503 patients). BMC Cancer 2017, 17, 130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, E.Y.; Kim, Y.S.; Seo, J.Y.; Park, I.; Ahn, H.K.; Jeong, Y.M.; Kim, J.H.; Kim, N. The Relationship between Sarcopenia and Systemic Inflammatory Response for Cancer Cachexia in Small Cell Lung Cancer. PLoS ONE 2016, 11, e0161125. [Google Scholar] [CrossRef]
- Golan, T.; Geva, R.; Richards, D.; Madhusudan, S.; Lin, B.K.; Wang, H.T.; Walgren, R.A.; Stemmer, S.M. LY2495655, an antimyostatin antibody, in pancreatic cancer: A randomized, phase 2 trial. J. Cachexia Sarcopenia Muscle 2018, 9, 871–879. [Google Scholar] [CrossRef] [Green Version]
- Fujiwara, Y.; Kobayashi, T.; Chayahara, N.; Imamura, Y.; Toyoda, M.; Kiyota, N.; Mukohara, T.; Nishiumi, S.; Azuma, T.; Yoshida, M.; et al. Metabolomics evaluation of serum markers for cachexia and their intra-day variation in patients with advanced pancreatic cancer. PLoS ONE 2014, 9, e113259. [Google Scholar] [CrossRef] [Green Version]
- Kuroda, K.; Nakashima, J.; Kanao, K.; Kikuchi, E.; Miyajima, A.; Horiguchi, Y.; Nakagawa, K.; Oya, M.; Ohigashi, T.; Murai, M. Interleukin 6 is associated with cachexia in patients with prostate cancer. Urology 2007, 69, 113–117. [Google Scholar] [CrossRef]
- Gonzalez-Freire, M.; Semba, R.D.; Ubaida-Mohien, C.; Fabbri, E.; Scalzo, P.; Hojlund, K.; Dufresne, C.; Lyashkov, A.; Ferrucci, L. The Human Skeletal Muscle Proteome Project: A reappraisal of the current literature. J. Cachexia Sarcopenia Muscle 2017, 8, 5–18. [Google Scholar] [CrossRef]
- Shum, A.M.Y.; Poljak, A.; Bentley, N.L.; Turner, N.; Tan, T.C.; Polly, P. Proteomic profiling of skeletal and cardiac muscle in cancer cachexia: Alterations in sarcomeric and mitochondrial protein expression. Oncotarget 2018, 9, 22001–22022. [Google Scholar] [CrossRef] [Green Version]
- Miller, A.; McLeod, L.; Alhayyani, S.; Szczepny, A.; Watkins, D.N.; Chen, W.; Enriori, P.; Ferlin, W.; Ruwanpura, S.; Jenkins, B.J. Blockade of the IL-6 trans-signalling/STAT3 axis suppresses cachexia in Kras-induced lung adenocarcinoma. Oncogene 2017, 36, 3059–3066. [Google Scholar] [CrossRef]
- Bonetto, A.; Aydogdu, T.; Jin, X.; Zhang, Z.; Zhan, R.; Puzis, L.; Koniaris, L.G.; Zimmers, T.A. JAK/STAT3 pathway inhibition blocks skeletal muscle wasting downstream of IL-6 and in experimental cancer cachexia. Am. J. Physiol. Endocrinol. Metab. 2012, 303, E410–E421. [Google Scholar] [CrossRef] [Green Version]
- Attaix, D.; Ventadour, S.; Codran, A.; Bechet, D.; Taillandier, D.; Combaret, L. The ubiquitin-proteasome system and skeletal muscle wasting. Essays Biochem. 2005, 41, 173–186. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.L.; Rosa-Caldwell, M.E.; Lee, D.E.; Blackwell, T.A.; Brown, L.A.; Perry, R.A.; Haynie, W.S.; Hardee, J.P.; Carson, J.A.; Wiggs, M.P.; et al. Mitochondrial degeneration precedes the development of muscle atrophy in progression of cancer cachexia in tumour-bearing mice. J. Cachexia Sarcopenia Muscle 2017, 8, 926–938. [Google Scholar] [CrossRef] [PubMed]
- Skipworth, R.J.; Stewart, G.D.; Dejong, C.H.; Preston, T.; Fearon, K.C. Pathophysiology of cancer cachexia: Much more than host-tumour interaction? Clin. Nutr. 2007, 26, 667–676. [Google Scholar] [CrossRef] [PubMed]
- Tidball, J.G. Regulation of muscle growth and regeneration by the immune system. Nat. Rev. Immunol. 2017, 17, 165–178. [Google Scholar] [CrossRef] [PubMed]
- Wehling, M.; Spencer, M.J.; Tidball, J.G. A nitric oxide synthase transgene ameliorates muscular dystrophy in mdx mice. J. Cell. Biol. 2001, 155, 123–131. [Google Scholar] [CrossRef]
- Castiglioni, A.; Corna, G.; Rigamonti, E.; Basso, V.; Vezzoli, M.; Monno, A.; Almada, A.E.; Mondino, A.; Wagers, A.J.; Manfredi, A.A.; et al. FOXP3+ T Cells Recruited to Sites of Sterile Skeletal Muscle Injury Regulate the Fate of Satellite Cells and Guide Effective Tissue Regeneration. PLoS ONE 2015, 10, e0128094. [Google Scholar] [CrossRef]
- Sciorati, C.; Rigamonti, E.; Manfredi, A.A.; Rovere-Querini, P. Cell death, clearance and immunity in the skeletal muscle. Cell Death Differ. 2016, 23, 927–937. [Google Scholar] [CrossRef] [Green Version]
- Argiles, J.M.; Busquets, S.; Toledo, M.; Lopez-Soriano, F.J. The role of cytokines in cancer cachexia. Curr. Opin. Support. Palliat. Care 2009, 3, 263–268. [Google Scholar] [CrossRef]
- Mattila, N.; Seppanen, H.; Mustonen, H.; Przybyla, B.; Haglund, C.; Lassila, R. Preoperative Biomarker Panel, Including Fibrinogen and FVIII, Improves Diagnostic Accuracy for Pancreatic Ductal Adenocarcinoma. Clin. Appl. Thromb. Hemost. 2018, 24, 1267–1275. [Google Scholar] [CrossRef] [Green Version]
- Reddel, C.J.; Allen, J.D.; Ehteda, A.; Taylor, R.; Chen, V.M.; Curnow, J.L.; Kritharides, L.; Robertson, G. Increased thrombin generation in a mouse model of cancer cachexia is partially interleukin-6 dependent. J. Thromb. Haemost. 2017, 15, 477–486. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lerner, L.; Hayes, T.G.; Tao, N.; Krieger, B.; Feng, B.; Wu, Z.; Nicoletti, R.; Chiu, M.I.; Gyuris, J.; Garcia, J.M. Plasma growth differentiation factor 15 is associated with weight loss and mortality in cancer patients. J. Cachexia Sarcopenia Muscle 2015, 6, 317–324. [Google Scholar] [CrossRef] [PubMed]
- Penafuerte, C.A.; Gagnon, B.; Sirois, J.; Murphy, J.; MacDonald, N.; Tremblay, M.L. Identification of neutrophil-derived proteases and angiotensin II as biomarkers of cancer cachexia. Br. J. Cancer 2016, 114, 680–687. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Narsale, A.A.; Carson, J.A. Role of interleukin-6 in cachexia: Therapeutic implications. Curr. Opin. Support. Palliat. Care 2014, 8, 321–327. [Google Scholar] [CrossRef] [Green Version]
- Diehl, S.; Rincon, M. The two faces of IL-6 on Th1/Th2 differentiation. Mol. Immunol. 2002, 39, 531–536. [Google Scholar] [CrossRef]
- Carson, J.A.; Baltgalvis, K.A. Interleukin 6 as a key regulator of muscle mass during cachexia. Exerc. Sport Sci. Rev. 2010, 38, 168–176. [Google Scholar] [CrossRef]
- Esfandiari, N.; Ghosh, S.; Prado, C.M.; Martin, L.; Mazurak, V.; Baracos, V.E. Age, Obesity, Sarcopenia, and Proximity to Death Explain Reduced Mean Muscle Attenuation in Patients with Advanced Cancer. J. Frailty Aging 2014, 3, 3–8. [Google Scholar] [CrossRef]
- Kemik, O.; Sumer, A.; Kemik, A.S.; Hasirci, I.; Purisa, S.; Dulger, A.C.; Demiriz, B.; Tuzun, S. The relationship among acute-phase response proteins, cytokines and hormones in cachectic patients with colon cancer. World J. Surg. Oncol. 2010, 8, 85. [Google Scholar] [CrossRef] [Green Version]
- Sheen, Y.Y.; Kim, M.J.; Park, S.A.; Park, S.Y.; Nam, J.S. Targeting the Transforming Growth Factor-beta Signaling in Cancer Therapy. Biomol. Ther. 2013, 21, 323–331. [Google Scholar] [CrossRef] [Green Version]
- Okada, K.; Naito, A.T.; Higo, T.; Nakagawa, A.; Shibamoto, M.; Sakai, T.; Hashimoto, A.; Kuramoto, Y.; Sumida, T.; Nomura, S.; et al. Wnt/beta-Catenin Signaling Contributes to Skeletal Myopathy in Heart Failure via Direct Interaction with Forkhead Box O. Circ. Heart Fail. 2015, 8, 799–808. [Google Scholar] [CrossRef] [Green Version]
- Yablonka-Reuveni, Z.; Seger, R.; Rivera, A.J. Fibroblast growth factor promotes recruitment of skeletal muscle satellite cells in young and old rats. J. Histochem. Cytochem. 1999, 47, 23–42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moran, A.L.; Nelson, S.A.; Landisch, R.M.; Warren, G.L.; Lowe, D.A. Estradiol replacement reverses ovariectomy-induced muscle contractile and myosin dysfunction in mature female mice. J. Appl. Physiol. 2007, 102, 1387–1393. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sierzega, M.; Kaczor, M.; Kolodziejczyk, P.; Kulig, J.; Sanak, M.; Richter, P. Evaluation of serum microRNA biomarkers for gastric cancer based on blood and tissue pools profiling: The importance of miR-21 and miR-331. Br. J. Cancer 2017, 117, 266–273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Keller, A.; Leidinger, P.; Gislefoss, R.; Haugen, A.; Langseth, H.; Staehler, P.; Lenhof, H.P.; Meese, E. Stable serum miRNA profiles as potential tool for non-invasive lung cancer diagnosis. RNA Biol. 2011, 8, 506–516. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prado, C.M.; Lieffers, J.R.; McCargar, L.J.; Reiman, T.; Sawyer, M.B.; Martin, L.; Baracos, V.E. Prevalence and clinical implications of sarcopenic obesity in patients with solid tumours of the respiratory and gastrointestinal tracts: A population-based study. Lancet Oncol. 2008, 9, 629–635. [Google Scholar] [CrossRef]



| Characteristics | Control (n = 11) | PDAC (n = 30) | p-Value |
|---|---|---|---|
| Age a | 49.2 ± 14.7 | 67.1 ± 11.4 | 0.001 |
| Female | 54.6 ± 12.7 | 70.5 ± 11.7 | 0.015 |
| Male | 34.7 ± 8.5 | 64.2 ± 10.3 | 0.002 |
| Gender b | 0.138 | ||
| Female | 8 | 14 | |
| Male | 3 | 16 | |
| Cancer Stage | |||
| Local/locally advanced | 19 | ||
| Metastatic | 11 | ||
| BMI (kg/m2) † a | 33.2 ± 7.8 | 28.0 ± 7.1 | 0.023 |
| Female | 33.4 ± 8.8 | 26.1 ± 4.5 | 0.042 |
| Male | 32.7 ± 3.8 | 29.7 ± 8.5 | 0.301 |
| Change in weight (% mean) ‡ a | −0.9 ± 9.1 | 11.9 ± 8.1 | 0.001 |
| Female | 0.2 ± 10.4 | −10.8 ± 8.3 | 0.013 |
| Male | −4.0 ± 0.7 | −12.9 ± 7.8 | 0.129 |
| Weight Loss Grade c | 0.02 | ||
| Grade 0 | 3 | 3 | |
| Grade 1 | 4 | 2 | |
| Grade 2 | 3 | 7 | |
| Grade 3 | 1 | 8 | |
| Grade 4 | 0 | 9 | |
| Skeletal muscle index (cm2/m2) * a | |||
| Female | 51.5 ± 9.3 | 39.6 ± 4.2 | 0.013 |
| Male | 59.9 ± 4.4 | 46.1 ± 12.2 | 0.1 |
| Skeletal muscle density (HU) a | |||
| Female | 30.8 ± 7.7 | 27.9 ± 9.5 | 0.99 |
| Male | 40.3 ± 0.9 | 33.2 ± 9.8 | 0.427 |
| Total Adipose Index (cm2/m2) * a | |||
| Female | 183.8 ± 82.1 | 111.1 ± 52.2 | 0.140 |
| Male | 177.4 ± 54.8 | 143.7 ± 83.2 | 0.301 |
| Sarcopenia Status (yes/no) § c | 2/9 | 20/8 | 0.0040 |
| Female | 2/6 | 6/7 | 0.399 |
| Male | 0/3 | 12/6 | 0.0632 |
| Information not available | - | 2 |
| Protein Name | r-Value | p-Value | Protein Name | r-Value | p-Value | Protein Name | r-Value | p-Value |
|---|---|---|---|---|---|---|---|---|
| LYVE1 | 0.67 | 0.0001 | C5 | −0.55 | 0.0027 | TNFRSF10A | −0.52 | 0.0056 |
| C7 | 0.65 | 0.0003 | SEMA6B | 0.55 | 0.0028 | KRAS | −0.52 | 0.0056 |
| F2 | −0.64 | 0.0003 | PIM1 | −0.55 | 0.0031 | SPP1 | 0.52 | 0.0059 |
| SLAMF6 | 0.64 | 0.0004 | HSD17B1 | −0.54 | 0.0032 | PIK3CA PIK3R1 | 0.52 | 0.0059 |
| CDH15 | −0.61 | 0.0007 | SOST | −0.54 | 0.0034 | PDXK | 0.51 | 0.0063 |
| LEP | −0.6 | 0.0010 | RPS7 | 0.54 | 0.0034 | TLR2 | 0.51 | 0.0064 |
| ENTPD5 | −0.59 | 0.0013 | PRSS27 | −0.54 | 0.0035 | IFNAR1 | 0.51 | 0.0065 |
| HSPD1 | 0.57 | 0.0018 | XPNPEP1 | 0.54 | 0.0037 | STK17B | −0.51 | 0.0065 |
| GHR | −0.57 | 0.0020 | NANOG | −0.54 | 0.0038 | WFIKKN1 | −0.52 | 0.0068 |
| DLL4 | 0.57 | 0.0020 | ASGR1 | 0.54 | 0.0039 | FCER2 | −0.50 | 0.0073 |
| CMPK1 | −0.57 | 0.0020 | MSTN | −0.53 | 0.0040 | AHSG | −0.50 | 0.0074 |
| CA10 | −0.57 | 0.0021 | LCORL | −0.53 | 0.0041 | IL1RL1 | 0.50 | 0.0075 |
| LY9 | 0.56 | 0.0022 | LTA | −0.53 | 0.0042 | INHBA | 0.50 | 0.0077 |
| ALB | −0.56 | 0.0025 | CRLF2 | −0.53 | 0.0044 | AFM | −0.50 | 0.0077 |
| GDNF | −0.55 | 0.0026 | ABL2 | −0.53 | 0.0047 | BOC | 0.50 | 0.0078 |
| SERPINA1 | 0.56 | 0.0026 | CXCL5 | −0.53 | 0.0048 |
| Protein Name | r-Value | p-Value | Protein Name | r-Value | p-Value |
|---|---|---|---|---|---|
| GRP | 0.70 | 0.0001 | SET | 0.56 | 0.0028 |
| S100A7 | 0.62 | 0.0007 | LEP | 0.56 | 0.0028 |
| CCL28 | −0.61 | 0.0009 | RSPO3 | −0.56 | 0.0031 |
| CFH | 0.60 | 0.0012 | WFIKKN1 | 0.54 | 0.0041 |
| AFM | 0.59 | 0.0012 | LPO | −0.53 | 0.0050 |
| MB | 0.59 | 0.0015 | IL1R2 | 0.53 | 0.0054 |
| TYMS | 0.57 | 0.0023 | HRG | 0.52 | 0.0066 |
| GHR | 0.57 | 0.0025 | HDAC8 | 0.51 | 0.0076 |
| PIM1 | 0.57 | 0.0026 | MDK | −0.50 | 0.0086 |
| KLK11 | −0.56 | 0.0027 |
| Protein Name | r-Value | p-Value |
|---|---|---|
| FABP3 | −0.77 | 3.7000 × 10−6 |
| COL18A1 | −0.63 | 0.0006 |
| LRRTM1 | −0.61 | 0.0008 |
| CFH | −0.58 | 0.0018 |
| AKT2 | 0.57 | 0.0025 |
| CFD | −0.57 | 0.0026 |
| C5 | −0.56 | 0.0031 |
| IMPDH1 | 0.56 | 0.0032 |
| IFNA7 | −0.55 | 0.0034 |
| IL17B | 0.53 | 0.0051 |
| LILRB2 | −0.53 | 0.0052 |
| FCER2 | −0.51 | 0.0073 |
| SIGLEC14 | −0.51 | 0.0074 |
| CLEC1B | 0.50 | 0.0086 |
| Protein Name | r-Value | p-Value | Protein Name | r-Value | p-Value |
|---|---|---|---|---|---|
| LEP | 0.85 | 3.13 × 10−8 | Human-virus | 0.56 | 0.003 |
| C5 | 0.69 | 0.0001 | CFH | 0.54 | 0.004 |
| CCL28 | −0.68 | 0.0002 | TP53 | 0.54 | 0.004 |
| FABP3 | 0.67 | 0.0002 | DKKL1 | 0.55 | 0.004 |
| BIRC5 | 0.65 | 0.0003 | AFM | 0.55 | 0.004 |
| C1S | 0.65 | 0.0003 | PLG | 0.54 | 0.005 |
| KLRF1 | 0.65 | 0.0003 | SFRP1 | −0.53 | 0.006 |
| PIM1 | 0.65 | 0.0003 | UBE2G2 | 0.52 | 0.006 |
| KLK11 | −0.61 | 0.001 | F9 | 0.53 | 0.006 |
| MDK | −0.61 | 0.001 | KLKB1 | 0.552 | 0.006 |
| PRKAA1 | 0.59 | 0.001 | PTN | −0.52 | 0.007 |
| HRG | 0.6 | 0.001 | LTBR | 0.51 | 0.007 |
| RSPO3 | −0.58 | 0.002 | KEAP1 | −0.51 | 0.008 |
| HMGN1 | 0.58 | 0.002 | APOE | 0.5 | 0.009 |
| LMAN2 | 0.59 | 0.002 | CCL16 | 0.5 | 0.01 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Narasimhan, A.; Shahda, S.; Kays, J.K.; Perkins, S.M.; Cheng, L.; Schloss, K.N.H.; Schloss, D.E.I.; Koniaris, L.G.; Zimmers, T.A. Identification of Potential Serum Protein Biomarkers and Pathways for Pancreatic Cancer Cachexia Using an Aptamer-Based Discovery Platform. Cancers 2020, 12, 3787. https://doi.org/10.3390/cancers12123787
Narasimhan A, Shahda S, Kays JK, Perkins SM, Cheng L, Schloss KNH, Schloss DEI, Koniaris LG, Zimmers TA. Identification of Potential Serum Protein Biomarkers and Pathways for Pancreatic Cancer Cachexia Using an Aptamer-Based Discovery Platform. Cancers. 2020; 12(12):3787. https://doi.org/10.3390/cancers12123787
Chicago/Turabian StyleNarasimhan, Ashok, Safi Shahda, Joshua K. Kays, Susan M. Perkins, Lijun Cheng, Katheryn N. H. Schloss, Daniel E. I. Schloss, Leonidas G. Koniaris, and Teresa A. Zimmers. 2020. "Identification of Potential Serum Protein Biomarkers and Pathways for Pancreatic Cancer Cachexia Using an Aptamer-Based Discovery Platform" Cancers 12, no. 12: 3787. https://doi.org/10.3390/cancers12123787
APA StyleNarasimhan, A., Shahda, S., Kays, J. K., Perkins, S. M., Cheng, L., Schloss, K. N. H., Schloss, D. E. I., Koniaris, L. G., & Zimmers, T. A. (2020). Identification of Potential Serum Protein Biomarkers and Pathways for Pancreatic Cancer Cachexia Using an Aptamer-Based Discovery Platform. Cancers, 12(12), 3787. https://doi.org/10.3390/cancers12123787






