Breast Cancer Stem Cells: Biomarkers, Identification and Isolation Methods, Regulating Mechanisms, Cellular Origin, and Beyond
Abstract
Simple Summary
Abstract
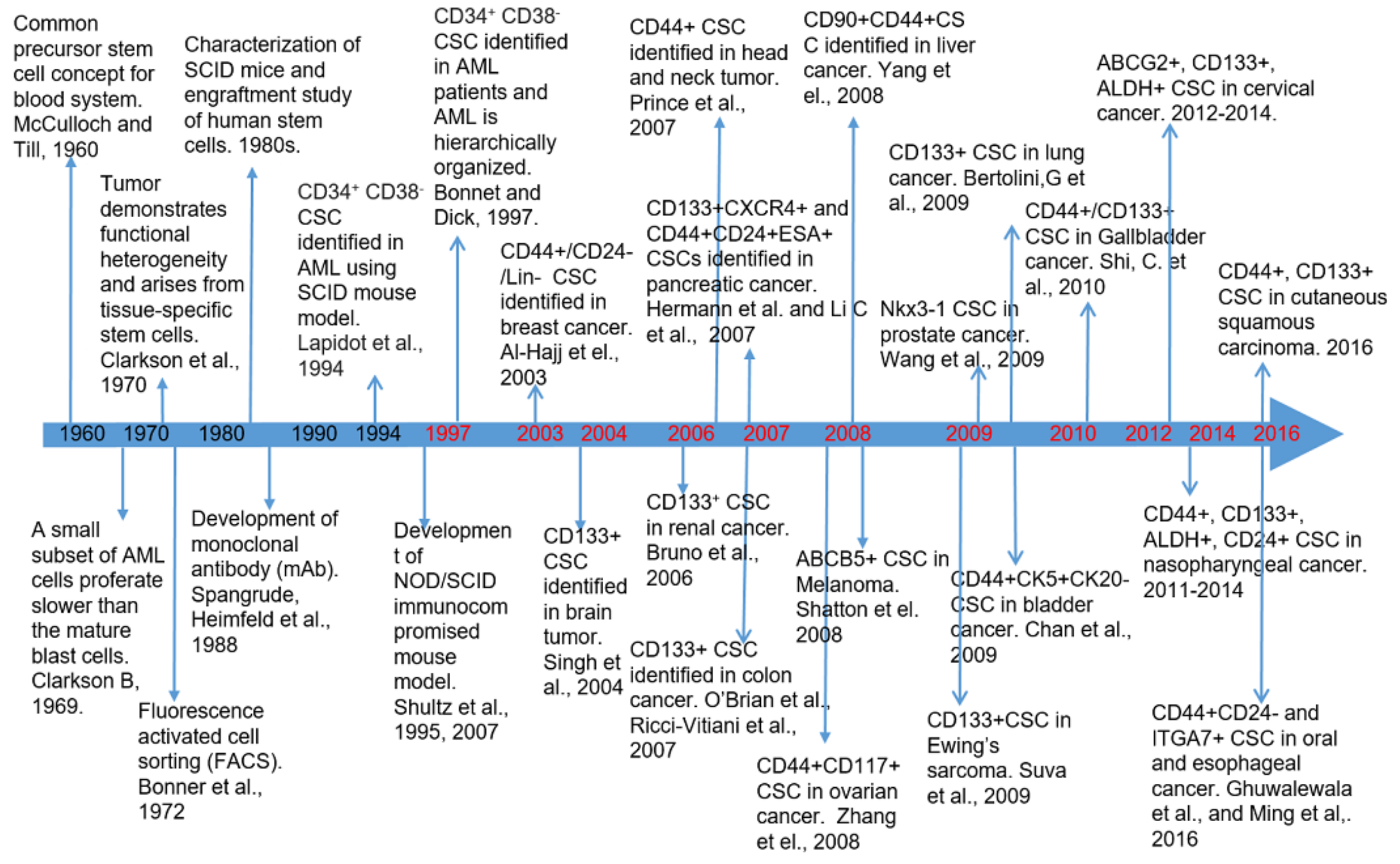
1. Introduction
2. Biomarkers of Breast Cancer Stem Cells and Their Clinical Relevance
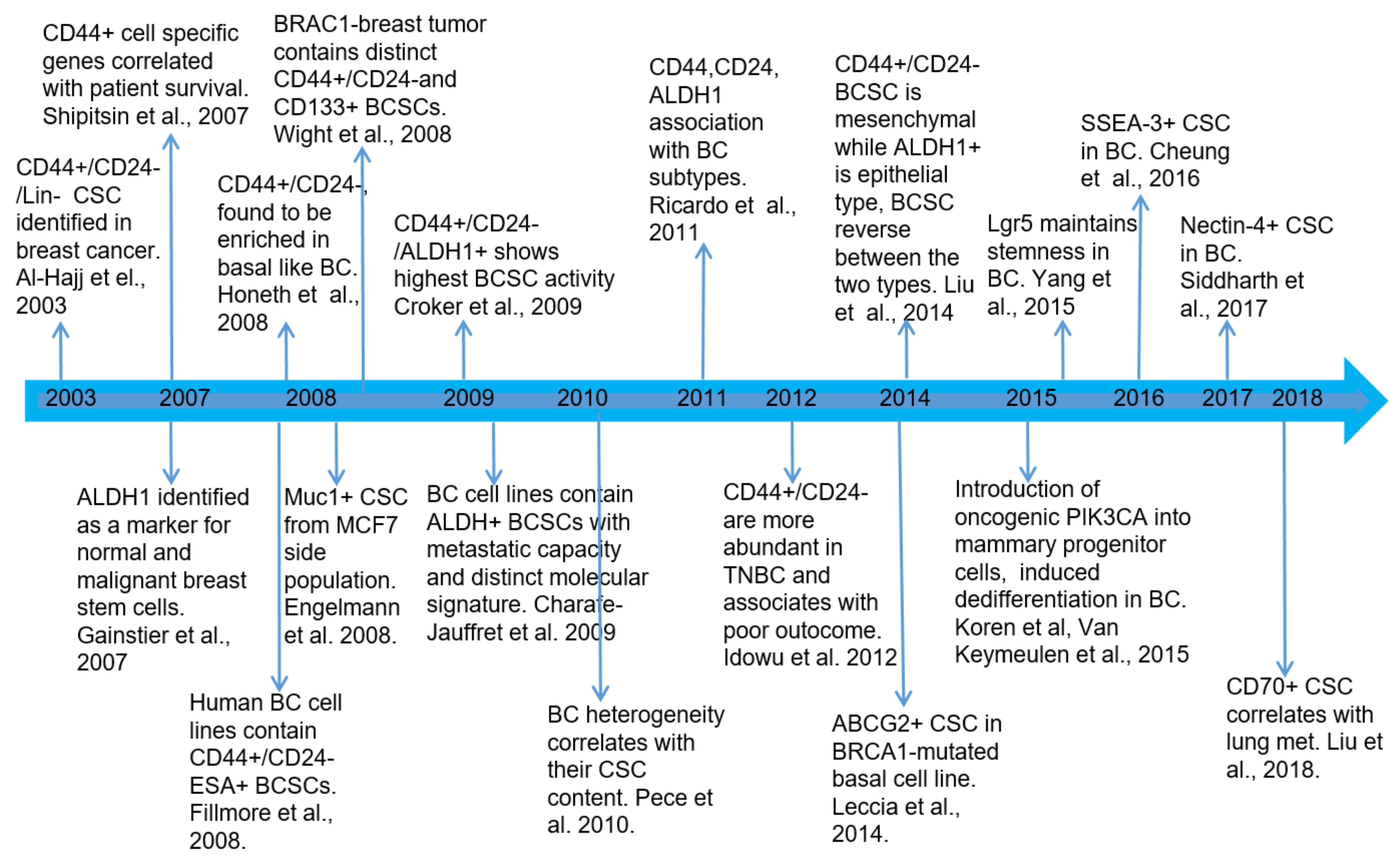
2.1. Breast Cancer Stem Cell Biomarkers
2.2. BCSC Subpopulations Are Heterogeneous with Different Subtypes
2.3. Relative Breast Cancer Stem Cell Abundance in Different Breast Cancer Subtypes and Their Clinical Relevance
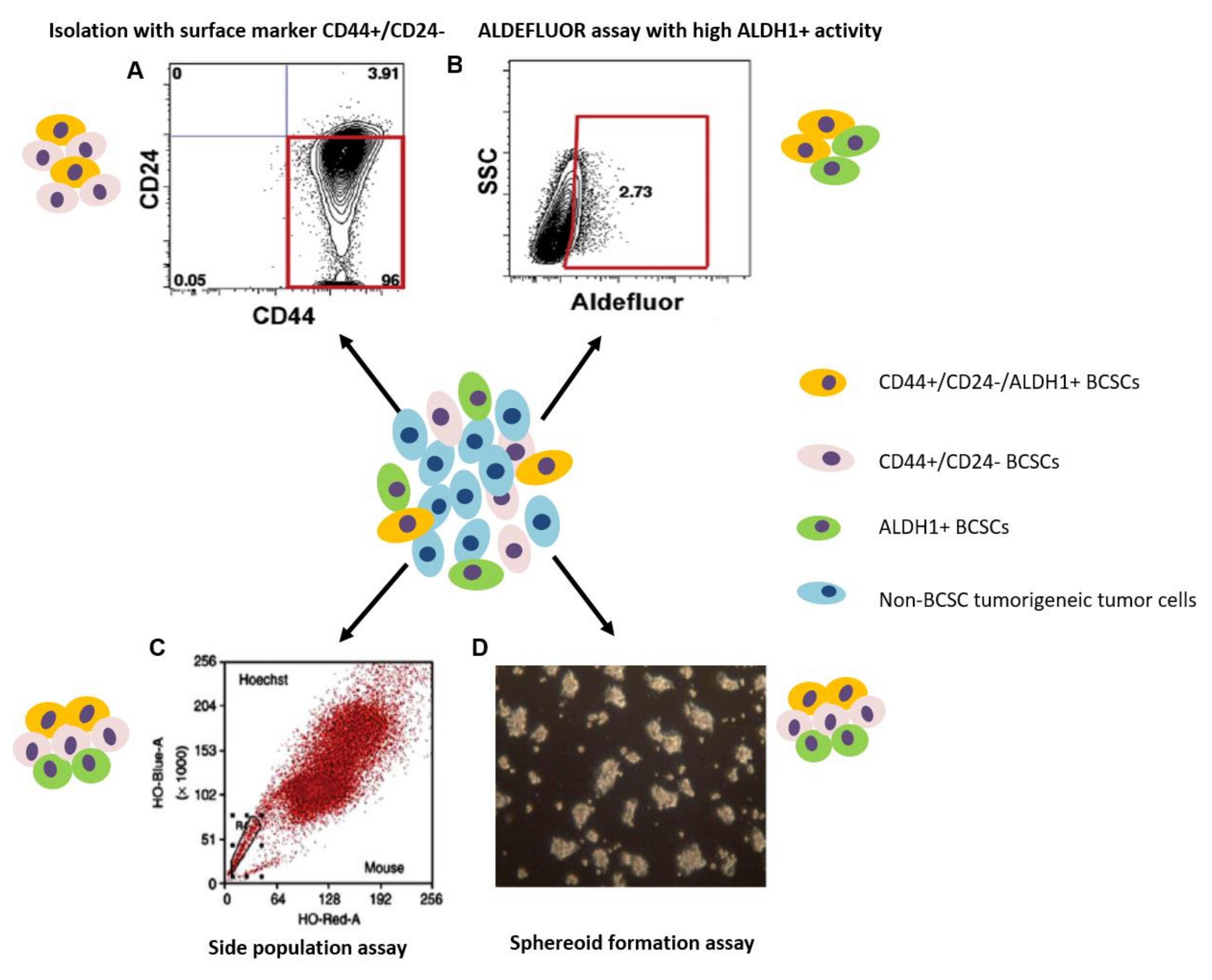
3. Methods for the Identification and Isolation of Breast Cancer Stem Cells
3.1. Isolation with Cell Surface Markers
3.2. Side Population Assay
3.3. Aldehyde Dehydrogenase Assay
3.4. Spheroid Formation Assay
3.5. Isolation and Identification Based on the Combination of Different Methods
4. Signaling Pathways and Molecules in the Regulation of BCSCs
4.1. Major Signaling Pathways That Regulate BCSCs
4.2. The Role of Non-Coding RNAs in BCSCs
4.3. Therapeutic Drugs Targeting BCSC Subpopulations
5. Origin of BCSCs and Methods for Studying BC Origin and Lineage Development
5.1. Origin of BCSCs and Traditional Methods for BCSC Lineage Tracing
5.2. Traditional Methods for BCSC Lineage Tracing
5.3. Single-Cell Transcriptomics in the Study of BCSC Lineages
5.4. Combination of Single-Cell RNA-seq and Single-Cell Genetic Barcode Tracing in the Study of the BCSC Origin
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2016. CA Cancer J. Clin. 2016, 66, 7–30. [Google Scholar] [CrossRef]
- Sin, W.C.; Lim, C.L. Breast cancer stem cells—From origins to targeted therapy. Stem Cell Investig. 2017, 4, 96. [Google Scholar] [CrossRef]
- Fragomeni, S.M.; Sciallis, A.; Jeruss, J.S. Molecular subtypes and local-regional control of breast cancer. Surg. Oncol. Clin. N. Am. 2018, 27, 95–120. [Google Scholar] [CrossRef]
- Toss, A.; Cristofanilli, M. Molecular characterization and targeted therapeutic approaches in breast cancer. Breast Cancer Res. 2015, 17, 1–11. [Google Scholar] [CrossRef]
- Nielsen, N.T.O.; Hsu, F.D.; Jensen, K.; Cheang, M.; Karaca, G.; Hu, Z.; Hernandez-Boussard, T.; Livasy, C.; Cowan, D.; Dressler, L.; et al. Immunohistochemical and clinical characterization of the basal-like subtype of invasive breast carcinoma. Clin. Cancer Res. 2004, 10, 5367–5374. [Google Scholar] [CrossRef]
- Perou, C.M.; Sorlie, T.; Eisen, M.B.; van de Rijn, M.; Jeffrey, S.S.; Rees, C.A. Molecular portraits of human breast tumours. Nature 2000, 406, 747–752. [Google Scholar] [CrossRef]
- Dawson, S.; Provenzano, E.; Caldas, C. Triple negative breast cancers: Clinical and prognostic implications. Eur. J. Cancer 2009, 45, 27–40. [Google Scholar] [CrossRef]
- Kreike, B.; Van Kouwenhove, M.; Horlings, H.M.; Weigelt, B.; Peterse, H.; Bartelink, H.; Van De Vijver, M.J. Gene expression profiling and histopathological characterization of triple-negative/basal-like breast carcinomas. Breast Cancer Res. 2007, 9, 1–14. [Google Scholar] [CrossRef]
- Sotiriou, C.; Pusztai, L. Gene-Expression Signatures in Breast Cancer. N. Engl. J. Med. 2009, 360, 790–800. [Google Scholar] [CrossRef]
- Lin, N.U.; Vanderplas, A.; Hughes, M.E.; Theriault, R.L.; Edge, S.B.; Wong, Y.-N.; Blayney, D.W.; Niland, J.C.; Winer, E.P.; Weeks, J.C. Clinicopathologic features, patterns of recurrence, and survival among women with triple-negative breast cancer in the National Comprehensive Cancer Network. Cancer 2012, 118, 5463–5472. [Google Scholar] [CrossRef]
- Sorlie, T.; Perou, C.M.; Tibshirani, R.; Aas, T.; Geisler, S.; Johnsen, H. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc. Natl. Acad. Sci. USA 2001, 98, 10869–10874. [Google Scholar] [CrossRef]
- Sørlie, T.; Tibshirani, R.; Parker, J.; Hastie, T.; Marron, J.S.; Nobel, A.; Deng, S.; Johnsen, H.; Pesich, R.; Geisler, S.; et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc. Natl. Acad. Sci. USA 2003, 100, 8418–8423. [Google Scholar] [CrossRef] [PubMed]
- Sotiriou, C.; Neo, S.Y.; McShane, L.M.; Korn, E.L.; Long, P.M.; Jazaeri, A. Breast cancer classification and prognosis based on gene expression profiles from a population-based study. Proc. Natl. Acad. Sci. USA 2003, 100, 10393–10398. [Google Scholar] [CrossRef] [PubMed]
- Hubalek, M.; Czech, T.; Müller, H. Biological subtypes of triple-negative breast cancer. Breast Care 2017, 12, 8–14. [Google Scholar] [CrossRef] [PubMed]
- Ricardo, S.; Vieira, A.F.; Gerhard, R.; Leitão, D.; Pinto, R.; Cameselle-Teijeiro, J.F.; Milanezi, F.; Schmitt, F.; Paredes, J. Breast cancer stem cell markers CD44, CD24 and ALDH1: Expression distribution within intrinsic molecular subtype. J. Clin. Pathol. 2011, 64, 937–946. [Google Scholar] [CrossRef]
- Ginestier, C.; Hur, M.H.; Charafe-Jauffret, E.; Monville, F.; Dutcher, J.; Brown, M.; Jacquemier, J.; Viens, P.; Kleer, C.G.; Liu, S.; et al. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell 2007, 1, 555–567. [Google Scholar] [CrossRef]
- Mylona, E.; Giannopoulou, I.; Fasomytakis, E.; Nomikos, A.; Magkou, C.; Bakarakos, P.; Nakopoulou, L. The clinicopathologic and prognostic significance of CD44+/CD24−/low and CD44−/CD24+ tumor cells in invasive breast carcinomas. Hum. Pathol. 2008, 39, 1096–1102. [Google Scholar] [CrossRef]
- Honeth, G.; Bendahl, P.-O.; Ringnér, M.; Saal, L.H.; Gruvberger-Saal, S.K.; Lövgren, K.; Grabau, D.; Fernö, M.; Borg, Å.; Hegardt, C. The CD44+/CD24-phenotype is enriched in basal-like breast tumors. Breast Cancer Res. 2008, 10, 1–12. [Google Scholar] [CrossRef]
- Rabinovich, I.; Sebastião, A.P.M.; Lima, R.S.; Urban, C.D.A.; Schunemann, E., Jr.; Anselmi, K.F.; Elifio-Esposito, S.; De Noronha, L.; Moreno-Amaral, A.N. Cancer stem cell markers ALDH1 and CD44+/CD24– phenotype and their prognosis impact in invasive ductal carcinoma. Eur. J. Histochem. 2018, 62. [Google Scholar] [CrossRef]
- Albini, A.; Bruno, A.; Gallo, C.; Pajardi, G.E.; Noonan, D.M.; Dallaglio, K. Cancer stem cells and the tumor microenvironment: Interplay in tumor heterogeneity. Connect. Tissue Res. 2015, 56, 414–425. [Google Scholar] [CrossRef]
- Clevers, H. The cancer stem cell: Premises, promises and challenges. Nat. Med. 2011, 17, 313–319. [Google Scholar] [CrossRef]
- Lin, C.-Y.; Barry-Holson, K.Q.; Allison, K.H. Breast cancer stem cells: Are we ready to go from bench to bedside? Histopathology 2015, 68, 119–137. [Google Scholar] [CrossRef]
- Tanei, T.; Morimoto, K.; Shimazu, K.; Kim, S.J.; Tanji, Y.; Taguchi, T.; Tamaki, Y.; Noguchi, S. Association of breast cancer stem cells identified by Aldehyde Dehydrogenase 1 expression with resistance to sequential paclitaxel and epirubicin-based chemotherapy for breast cancers. Clin. Cancer Res. 2009, 15, 4234–4241. [Google Scholar] [CrossRef]
- Brabletz, T.; Jung, A.; Spaderna, S.; Hlubek, F.; Kirchner, T. Opinion: Migrating cancer stem cells—An integrated concept of malignant tumour progression. Nat. Rev. Cancer 2005, 5, 744–749. [Google Scholar] [CrossRef]
- Kreso, A.; Dick, J.E. Evolution of the cancer stem cell model. Cell Stem Cell 2014, 14, 275–291. [Google Scholar] [CrossRef]
- Kreso, A.; O’Brien, C.A.; Van Galen, P.; Gan, O.I.; Notta, F.; Brown, A.M.K.; Ng, K.; Ma, J.; Wienholds, E.; Dunant, C.; et al. Variable clonal repopulation dynamics influence chemotherapy response in colorectal cancer. Science 2012, 339, 543–548. [Google Scholar] [CrossRef]
- Pece, S.; Tosoni, D.; Confalonieri, S.; Mazzarol, G.; Vecchi, M.; Ronzoni, S.; Bernard, L.; Viale, G.; Pelicci, P.G.; Di Fiore, P.P. Biological and molecular heterogeneity of breast cancers correlates with their cancer stem cell content. Cell 2010, 140, 62–73. [Google Scholar] [CrossRef]
- Furth, J.K.M.; Breedis, C. The transmission of leukemia of mice with a single cell. Cancer Res. 1937, 31, 62–73. [Google Scholar]
- McDermott, S.P.; Wicha, M.S. Targeting breast cancer stem cells. Mol. Oncol. 2010, 4, 404–419. [Google Scholar] [CrossRef]
- Till, J.E. The radiation sensitivity of normal mouse bone marrow cells, determined by quantitative marrow transplantation into irradiated mice. Radiat. Res. 1960, 13, 115. [Google Scholar] [CrossRef]
- Clarkson, B.; Fried, J.; Strife, A.; Sakai, Y.; Ota, K.; Ohkita, T. Studies of cellular proliferation in human leukemia.III. Behavior of leukemic cells in three adults with acute leukemia given continuous infusions of3H-thymidine for 8 or 10 days. Cancer 1970, 25, 1237–1260. [Google Scholar] [CrossRef]
- Clarkson, B.D. Review of recent studies of cellular proliferation in acute leukemia. Natl. Cancer Inst. Monogr. 1969, 30, 81–120. [Google Scholar] [PubMed]
- Spangrude, G.J.; Heimfeld, S.; Weissman, I.L. Purification and characterization of mouse hematopoietic stem cells. Science 1988, 241, 58–62. [Google Scholar] [CrossRef] [PubMed]
- Wallberg, F.; Tenev, T.; Meier, P. Analysis of apoptosis and necroptosis by fluorescence-activated cell sorting. Cold Spring Harb. Protoc. 2016, 2016. [Google Scholar] [CrossRef]
- Lapidot, T.; Sirard, C.; Vormoor, J.; Murdoch, B.; Hoang, T.; Caceres-Cortes, J.; Minden, M.; Paterson, B.; Caligiuri, M.A.; Dick, J.E. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nat. Cell Biol. 1994, 367, 645–648. [Google Scholar] [CrossRef]
- Shultz, L.D.; Schweitzer, P.A.; Christianson, S.W.; Gott, B.; Schweitzer, I.B.; Tennent, B. Multiple defects in innate and adaptive immunologic function in NOD/LtSz-scid mice. J. Immunol. 1995, 154, 180–191. [Google Scholar]
- Bonnet, D.; Dick, J.E. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat. Med. 1997, 3, 730–737. [Google Scholar] [CrossRef]
- Al-Hajj, M.; Wicha, M.S.; Benito-Hernandez, A.; Morrison, S.J.; Clarke, M.F. Prospective identification of tumorigenic breast cancer cells. Proc. Natl. Acad. Sci. USA 2003, 100, 3983–3988. [Google Scholar] [CrossRef]
- Batlle, E.; Clevers, H. Cancer stem cells revisited. Nat. Med. 2017, 23, 1124–1134. [Google Scholar] [CrossRef]
- Baccelli, I.; Trumpp, A. The evolving concept of cancer and metastasis stem cells. J. Cell Biol. 2012, 198, 281–293. [Google Scholar] [CrossRef]
- Singh, S.K.; Hawkins, C.; Clarke, I.D.; Squire, J.A.; Bayani, J.; Hide, T.; Henkelman, R.M.; Cusimano, M.D.; Dirks, P.B. Identification of human brain tumour initiating cells. Nat. Cell Biol. 2004, 432, 396–401. [Google Scholar] [CrossRef]
- Dalerba, P.; Dylla, S.J.; Park, I.-K.; Liu, R.; Wang, X.; Cho, R.W.; Hoey, T.; Gurney, A.; Huang, E.H.; Simeone, D.M.; et al. Phenotypic characterization of human colorectal cancer stem cells. Proc. Natl. Acad. Sci. USA 2007, 104, 10158–10163. [Google Scholar] [CrossRef]
- Huang, E.H.; Hynes, M.J.; Zhang, T.; Ginestier, C.; Dontu, G.; Appelman, H.; Fields, J.Z.; Wicha, M.S.; Boman, B.M. Aldehyde Dehydrogenase 1 is a marker for normal and malignant human colonic stem cells (SC) and tracks SC overpopulation during colon tumorigenesis. Cancer Res. 2009, 69, 3382–3389. [Google Scholar] [CrossRef]
- O’Brien, C.A.; Pollett, A.; Gallinger, S.; Dick, J.E. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature 2007, 445, 106–110. [Google Scholar] [CrossRef] [PubMed]
- Prince, M.E.; Sivanandan, R.; Kaczorowski, A.; Wolf, G.T.; Kaplan, M.J.; Dalerba, P.; Weissman, I.L.; Clarke, M.F.; Ailles, L.E. Identification of a subpopulation of cells with cancer stem cell properties in head and neck squamous cell carcinoma. Proc. Natl. Acad. Sci. USA 2007, 104, 973–978. [Google Scholar] [CrossRef]
- Hermann, P.C.; Huber, S.L.; Herrler, T.; Aicher, A.; Ellwart, J.W.; Guba, M. Distinct populations of cancer stem cells determine tumor growth and metastatic activity in human pancreatic cancer. Cell Stem Cell 2007, 1, 313–323. [Google Scholar] [CrossRef]
- Li, C.; Heidt, D.G.; Dalerba, P.; Burant, C.F.; Zhang, L.; Adsay, V.; Wicha, M.S.; Clarke, M.F.; Simeone, D.M. Identification of pancreatic cancer stem cells. Cancer Res. 2007, 67, 1030–1037. [Google Scholar] [CrossRef]
- Schatton, T.; Murphy, G.F.; Frank, N.Y.; Yamaura, K.; Waaga-Gasser, A.M.; Gasser, M. Identification of cells initiating human melanomas. Nature 2008, 451, 345–349. [Google Scholar] [CrossRef]
- Yang, Z.F.; Ho, D.W.; Ng, M.N.; Lau, C.K.; Yu, W.C.; Ngai, P.; Chu, P.W.; Lam, C.T.; Poon, R.T.; Fan, S.T. Significance of CD90+ cancer stem cells in human liver cancer. Cancer Cell 2008, 13, 153–166. [Google Scholar] [CrossRef]
- Stack, M.S. Faculty Opinions recommendation of identification and characterization of ovarian cancer-initiating cells from primary human tumors. Fac. Opin. Post-Publ. Peer Rev. Biomed. Lit. 2008, 68, 4311–4320. [Google Scholar] [CrossRef]
- Bertolini, G.; Roz, L.; Perego, P.; Tortoreto, M.; Fontanella, E.; Gatti, L.; Pratesi, G.; Fabbri, A.; Andriani, F.; Tinelli, S.; et al. Highly tumorigenic lung cancer CD133+ cells display stem-like features and are spared by cisplatin treatment. Proc. Natl. Acad. Sci. USA 2009, 106, 16281–16286. [Google Scholar] [CrossRef]
- Wang, X.; Julio, M.K.-D.; Economides, K.D.; Walker, D.; Yu, H.; Halili, M.V.; Hu, Y.-P.; Price, S.M.; Abate-Shen, C.; Shen, M.M. A luminal epithelial stem cell that is a cell of origin for prostate cancer. Nat. Cell Biol. 2009, 461, 495–500. [Google Scholar] [CrossRef] [PubMed]
- Chan, K.S.; Espinosa, I.; Chao, M.; Wong, D.; Ailles, L.; Diehn, M.; Gill, H.; Presti, J.; Chang, H.Y.; Van De Rijn, M.; et al. Identification, molecular characterization, clinical prognosis, and therapeutic targeting of human bladder tumor-initiating cells. Proc. Natl. Acad. Sci. USA 2009, 106, 14016–14021. [Google Scholar] [CrossRef]
- Suva, M.L.; Riggi, N.; Stehle, J.C.; Baumer, K.; Tercier, S.; Joseph, J.M. Identification of cancer stem cells in Ewing’s sarcoma. Cancer Res. 2009, 69, 1776–1781. [Google Scholar] [CrossRef]
- Deonarain, M.P.; Kousparou, C.A.; Epenetos, A.A. Antibodies targeting cancer stem cells: A new paradigm in immunotherapy? MAbs 2009, 1, 12–25. [Google Scholar] [CrossRef]
- Akbarzadeh, M.; Maroufi, N.F.; Tazehkand, A.P.; Akbarzadeh, M.; Bastani, S.; Safdari, R.; Farzane, A.; Fattahi, A.; Nejabati, H.R.; Nouri, M.; et al. Current approaches in identification and isolation of cancer stem cells. J. Cell. Physiol. 2019, 234, 14759–14772. [Google Scholar] [CrossRef]
- Ponti, D.; Costa, A.; Zaffaroni, N.; Pratesi, G.; Petrangolini, G.; Coradini, D.; Pilotti, S.; Pierotti, M.A.; Daidone, M.G. Isolation and in vitro propagation of tumorigenic breast cancer cells with stem/progenitor cell properties. Cancer Res. 2005, 65, 5506–5511. [Google Scholar] [CrossRef]
- Fillmore, C.M.; Kuperwasser, C. Human breast cancer cell lines contain stem-like cells that self-renew, give rise to phenotypically diverse progeny and survive chemotherapy. Breast Cancer Res. 2008, 10, 1–13. [Google Scholar] [CrossRef]
- Yoshida, G.J.; Saya, H. Therapeutic strategies targeting cancer stem cells. Cancer Sci. 2016, 107, 5–11. [Google Scholar] [CrossRef]
- Shipitsin, M.; Campbell, L.L.; Argani, P.; Weremowicz, S.; Bloushtain-Qimron, N.; Yao, J. Molecular definition of breast tumor heterogeneity. Cancer Cell 2007, 11, 259–273. [Google Scholar] [CrossRef]
- Wright, M.H.; Calcagno, A.M.; Salcido, C.D.; Carlson, M.D.; Ambudkar, S.V.; Varticovski, L. Brca1 breast tumors contain distinct CD44+/CD24− and CD133+ cells with cancer stem cell characteristics. Breast Cancer Res. 2008, 10, R10. [Google Scholar] [CrossRef]
- Wang, D.; Cai, C.; Dong, X.; Yu, Q.C.; Zhang, X.O.; Yang, L. Identification of multipotent mammary stem cells by protein C receptor expression. Nature 2015, 517, 81–84. [Google Scholar] [CrossRef] [PubMed]
- Leccia, F.; Del Del Vecchio, L.; Mariotti, E.; Di Noto, R.; Morel, A.-P.; Puisieux, A.; Salvatore, F.; Ansieau, S. ABCG2, a novel antigen to sort luminal progenitors of BRCA1- breast cancer cells. Mol. Cancer 2014, 13, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Tang, H.; Kong, Y.; Xie, X.; Chen, J.; Song, C. LGR5 promotes breast cancer progression and maintains stem-like cells through activation of Wnt/beta-catenin signaling. Stem Cells 2015, 33, 2913–2924. [Google Scholar] [CrossRef] [PubMed]
- Cheung, S.K.; Chuang, P.K.; Huang, H.W.; Hwang-Verslues, W.W.; Cho, C.H.; Yang, W.B. Stage-specific embryonic antigen-3 (SSEA-3) and beta3GalT5 are cancer specific and significant markers for breast cancer stem cells. Proc. Natl. Acad. Sci. USA 2016, 113, 960–965. [Google Scholar] [CrossRef]
- Liu, L.; Yin, B.; Yi, Z.; Liu, X.; Hu, Z.; Gao, W.; Yu, H.; Li, Q. Breast cancer stem cells characterized by CD70 expression preferentially metastasize to the lungs. Breast Cancer 2018, 25, 706–716. [Google Scholar] [CrossRef]
- Charafe-Jauffret, E.; Ginestier, C.; Iovino, F.; Wicinski, J.; Cervera, N.; Finetti, P.; Hur, M.-H.; Diebel, M.E.; Monville, F.; Dutcher, J.; et al. Breast cancer cell lines contain functional cancer stem cells with metastatic capacity and a distinct molecular signature. Cancer Res. 2009, 69, 1302–1313. [Google Scholar] [CrossRef]
- De Beça, F.F.; Caetano, P.; Gerhard, R.; Alvarenga, C.A.; Gomes, M.; Paredes, J.; Schmitt, F. Cancer stem cells markers CD44, CD24 and ALDH1 in breast cancer special histological types. J. Clin. Pathol. 2013, 66, 187–191. [Google Scholar] [CrossRef]
- Bartucci, M.; Dattilo, R.; Moriconi, C.; Pagliuca, A.; Mottolese, M.; Federici, G.; Di Benedetto, A.; Todaro, M.; Stassi, G.; Sperati, F.; et al. TAZ is required for metastatic activity and chemoresistance of breast cancer stem cells. Oncogene 2015, 34, 681–690. [Google Scholar] [CrossRef]
- Li, W.; Ma, H.; Zhang, J.; Zhu, L.; Wang, C.; Yang, Y. Unraveling the roles of CD44/CD24 and ALDH1 as cancer stem cell markers in tumorigenesis and metastasis. Sci. Rep. 2017, 7, 13856. [Google Scholar] [CrossRef]
- Yin, H.; Glass, J. The phenotypic radiation resistance of CD44+/CD24− or low breast cancer cells is mediated through the enhanced activation of ATM signaling. PLoS ONE 2011, 6, e24080. [Google Scholar] [CrossRef]
- Palomeras, S.; Ruiz-Martínez, S.; Puig, T. Targeting breast cancer stem cells to overcome treatment resistance. Molecules 2018, 23, 2193. [Google Scholar] [CrossRef]
- Lawson, D.A.; Bhakta, N.R.; Kessenbrock, K.; Prummel, K.D.; Yu, Y.; Takai, K. Single-cell analysis reveals a stem-cell program in human metastatic breast cancer cells. Nature 2015, 526, 131–135. [Google Scholar] [CrossRef]
- Van Pham, P.; Phan, N.L.-C.; Nguyen, N.T.; Truong, N.H.; Duong, T.T.; Le, D.V.; Truong, K.D.; Phan, N.K. Differentiation of breast cancer stem cells by knockdown of CD44: Promising differentiation therapy. J. Transl. Med. 2011, 9, 209. [Google Scholar] [CrossRef]
- Crabtree, J.S.; Miele, L. Breast cancer stem cells. Biomedicines 2018, 6, 77. [Google Scholar] [CrossRef]
- Bensimon, J.; Altmeyer-Morel, S.; Benjelloun, H.; Chevillard, S.; Lebeau, J. CD24−/low stem-like breast cancer marker defines the radiation-resistant cells involved in memorization and transmission of radiation-induced genomic instability. Oncogene 2012, 32, 251–258. [Google Scholar] [CrossRef]
- Muzio, G.; Maggiora, M.; Paiuzzi, E.; Oraldi, M.; Canuto, R. Aldehyde dehydrogenases and cell proliferation. Free Radic. Biol. Med. 2012, 52, 735–746. [Google Scholar] [CrossRef]
- Lu, H.; Clauser, K.R.; Tam, W.L.; Fröse, J.; Ye, X.; Eaton, E.N.; Reinhardt, F.; Donnenberg, V.S.; Bhargava, R.; Carr, S.A.; et al. A breast cancer stem cell niche supported by juxtacrine signalling from monocytes and macrophages. Nat. Cell Biol. 2014, 16, 1105–1117. [Google Scholar] [CrossRef]
- Liu, T.J.; Sun, B.C.; Zhao, X.L.; Zhao, X.M.; Sun, T.; Gu, Q.; Yao, Z.; Dong, X.Y.; Zhao, N.; Liu, N. CD133+ cells with cancer stem cell characteristics associates with vasculogenic mimicry in triple-negative breast cancer. Oncogene 2013, 32, 544–553. [Google Scholar] [CrossRef]
- Joseph, C.; Arshad, M.; Kurozomi, S.; Althobiti, M.; Miligy, I.; Al-Izzi, S.; Toss, M.S.; Goh, F.Q.; Johnston, S.J.; Martin, S.G.; et al. Overexpression of the cancer stem cell marker CD133 confers a poor prognosis in invasive breast cancer. Breast Cancer Res. Treat. 2019, 174, 387–399. [Google Scholar] [CrossRef]
- Desgrosellier, J.S.; Lesperance, J.; Seguin, L.; Gozo, M.; Kato, S.; Franovic, A. Integrin alphavbeta3 drives slug activation and stemness in the pregnant and neoplastic mammary gland. Dev. Cell 2014, 30, 295–308. [Google Scholar] [CrossRef]
- Lo, P.K.; Kanojia, D.; Liu, X.; Singh, U.P.; Berger, F.G.; Wang, Q. CD49f and CD61 identify Her2/neu-induced mammary tumor-initiating cells that are potentially derived from luminal progenitors and maintained by the integrin-TGFbeta signaling. Oncogene 2012, 31, 2614–2626. [Google Scholar] [CrossRef]
- Zhou, J.; Chen, Q.; Zou, Y.; Chen, H.; Qi, L.; Chen, Y. Stem cells and cellular origins of breast cancer: Updates in the rationale, controversies, and therapeutic implications. Front. Oncol. 2019, 9, 820. [Google Scholar] [CrossRef]
- Liu, S.; Cong, Y.; Wang, D.; Sun, Y.; Deng, L.; Liu, Y.; Martin-Trevino, R.; Shang, L.; McDermott, S.P.; Landis, M.D.; et al. Breast cancer stem cells transition between epithelial and mesenchymal states reflective of their normal counterparts. Stem Cell Rep. 2014, 2, 78–91. [Google Scholar] [CrossRef]
- Brabletz, T. EMT and MET in metastasis: Where are the cancer stem cells? Cancer Cell 2012, 22, 699–701. [Google Scholar] [CrossRef]
- Croker, A.K.; Goodale, D.; Chu, J.; Postenka, C.; Hedley, B.D.; Hess, D.A.; Allan, A.L. High aldehyde dehydrogenase and expression of cancer stem cell markers selects for breast cancer cells with enhanced malignant and metastatic ability. J. Cell. Mol. Med. 2008, 13, 2236–2252. [Google Scholar] [CrossRef]
- Idowu, M.O.; Kmieciak, M.; Dumur, C.; Burton, R.S.; Grimes, M.M.; Powers, C.N. CD44(+)/CD24(−/low) cancer stem/progenitor cells are more abundant in triple-negative invasive breast carcinoma phenotype and are associated with poor outcome. Hum. Pathol. 2012, 43, 364–373. [Google Scholar] [CrossRef]
- Yang, F.; Cao, L.; Sun, Z.; Jin, J.; Fang, H.; Zhang, W.; Guan, X. Evaluation of breast cancer stem cells and intratumor stemness heterogeneity in triple-negative breast cancer as prognostic factors. Int. J. Biol. Sci. 2016, 12, 1568–1577. [Google Scholar] [CrossRef]
- Elbaiomy, M.A.; Akl, T.; Atwan, N.; Elsayed, A.A.; Elzaafarany, M.; Shamaa, S. Clinical impact of breast cancer stem cells in metastatic breast cancer patients. J. Oncol. 2020, 2020, 1–8. [Google Scholar] [CrossRef]
- Camerlingo, R.; Ferraro, G.A.; De Francesco, F.; Romano, M.; Nicoletti, G.; Di Bonito, M. The role of CD44+/CD24-/low biomarker for screening, diagnosis and monitoring of breast cancer. Oncol. Rep. 2014, 31, 1127–1132. [Google Scholar] [CrossRef]
- Kim, H.J.; Kim, M.-J.; Lee, J.W.; Son, B.H.; Kim, S.-B.; Ahn, J.H.; Noh, W.C.; Gong, G. Different prognostic significance of CD24 and CD44 expression in breast cancer according to hormone receptor status. Breast 2011, 20, 78–85. [Google Scholar] [CrossRef]
- Charafe-Jauffret, E.; Ginestier, C.; Tarpin, C.; Diebel, M.; Esterni, B.; Houvenaeghel, G.; Extra, J.-M.; Bertucci, F.; Jacquemier, J.; Iovino, F.; et al. Aldehyde Dehydrogenase 1-positive cancer stem cells mediate metastasis and poor clinical outcome in inflammatory breast cancer. Clin. Cancer Res. 2010, 16, 45–55. [Google Scholar] [CrossRef]
- Li, H.; Ma, F.; Wang, H.; Lin, C.; Fan, Y.; Zhang, X.; Qian, H.; Xu, B. Stem cell marker Aldehyde Dehydrogenase 1 (ALDH1)-expressing cells are enriched in triple-negative breast cancer. Int. J. Biol. Markers 2013, 28, 357–364. [Google Scholar] [CrossRef]
- Ma, F.; Li, H.; Wang, H.; Shi, X.; Fan, Y.; Ding, X.; Lin, C.; Zhan, Q.; Qian, H.; Xu, B. Enriched CD44+/CD24− population drives the aggressive phenotypes presented in triple-negative breast cancer (TNBC). Cancer Lett. 2014, 353, 153–159. [Google Scholar] [CrossRef]
- Park, S.-Y.; Choi, J.-H.; Nam, J.-S. Targeting cancer stem cells in triple-negative breast cancer. Cancers 2019, 11, 965. [Google Scholar] [CrossRef]
- Yehiely, F.; Moyano, J.V.; Evans, J.R.; Nielsen, T.O.; Cryns, V.L. Deconstructing the molecular portrait of basal-like breast cancer. Trends Mol. Med. 2006, 12, 537–544. [Google Scholar] [CrossRef]
- Yu, K.-D.; Zhu, R.; Zhan, M.; Rodriguez, A.A.; Yang, W.; Wong, S.; Makris, A.; Lehmann, B.D.; Chen, X.; Mayer, I.; et al. Identification of prognosis-relevant subgroups in patients with chemoresistant triple-negative breast cancer. Clin. Cancer Res. 2013, 19, 2723–2733. [Google Scholar] [CrossRef]
- Brooks, M.D.; Burness, M.L.; Wicha, M.S. Therapeutic implications of cellular heterogeneity and plasticity in breast cancer. Cell Stem Cell 2015, 17, 260–271. [Google Scholar] [CrossRef]
- Jia, Z. Expression of HER2 in breast cancer promotes a massive reorganization of gene activity and suggests a role for epigenetic regulation. J. Data Min. Genom. Proteom. 2012, 3. [Google Scholar] [CrossRef] [PubMed]
- Ithimakin, S.; Day, K.C.; Malik, F.; Zen, Q.; Dawsey, S.J.; Bersano-Begey, T.F.; Quraishi, A.A.; Ignatoski, K.W.; Daignault, S.; Davis, A.; et al. HER2 drives luminal breast cancer stem cells in the absence of HER2 amplification: Implications for efficacy of adjuvant trastuzumab. Cancer Res. 2013, 73, 1635–1646. [Google Scholar] [CrossRef] [PubMed]
- Duru, N.; Fan, M.; Candas, D.; Menaa, C.; Liu, H.-C.; Nantajit, D.; Wen, Y.; Xiao, K.; Eldridge, A.; Chromy, B.A.; et al. HER2-associated radioresistance of breast cancer stem cells isolated from HER2-negative breast cancer cells. Clin. Cancer Res. 2012, 18, 6634–6647. [Google Scholar] [CrossRef]
- Oak, P.S.; Kopp, F.; Thakur, C.; Ellwart, J.W.; Rapp, U.R.; Ullrich, A.; Wagner, E.; Knyazev, P.G.; Roidl, A. Combinatorial treatment of mammospheres with trastuzumab and salinomycin efficiently targets HER2-positive cancer cells and cancer stem cells. Int. J. Cancer 2012, 131, 2808–2819. [Google Scholar] [CrossRef] [PubMed]
- Duan, J.-J.; Qiu, W.; Xu, S.-L.; Wang, B.; Ye, X.-Z.; Ping, Y.-F.; Zhang, X.; Bian, X.-W.; Yu, S.-C. Strategies for isolating and enriching cancer stem cells: Well begun is half done. Stem Cells Dev. 2013, 22, 2221–2239. [Google Scholar] [CrossRef] [PubMed]
- Jiao, X.R.A.; Cristofanilli, M.; Miftakhova, R.; Prestell, R. Breast cancer stem cell isolation. Breast cancer: Methods and protocols. Methods Mol. Biol. 2016, 1406, 15. [Google Scholar]
- Cammareri, P.; Lombardo, Y.; Francipane, M.G.; Bonventre, S.; Todaro, M.; Stassi, G.; Bonventre, S. Isolation and culture of colon cancer stem cells. Methods Cell Biol. 2008, 86, 311–324. [Google Scholar] [CrossRef] [PubMed]
- Jeon, Y.-K.; Kim, S.-H.; Choi, S.-H.; Kim, K.-H.; Yoo, B.-C.; Ku, J.-L.; Park, J.-G. Promoter hypermethylation and loss ofCD133gene expression in colorectal cancers. World J. Gastroenterol. 2010, 16, 3153–3160. [Google Scholar] [CrossRef]
- Ding, X.-W.; Wu, J.-H.; Jiang, C.-P. ABCG2: A potential marker of stem cells and novel target in stem cell and cancer therapy. Life Sci. 2010, 86, 631–637. [Google Scholar] [CrossRef]
- Keysar, S.B.; Jimeno, A. More than markers: Biological significance of cancer stem cell-defining molecules. Mol. Cancer Ther. 2010, 9, 2450–2457. [Google Scholar] [CrossRef]
- Zhou, S.; Schuetz, J.D.; Bunting, K.D.; Colapietro, A.M.; Sampath, J.; Morris, J.J. The ABC transporter Bcrp1/ABCG2 is expressed in a wide variety of stem cells and is a molecular determinant of the side-population phenotype. Nat. Med. 2001, 7, 1028–1034. [Google Scholar] [CrossRef]
- Christgen, M.; Ballmaier, M.; Bruchhardt, H.; Von Wasielewski, R.; Kreipe, H.; Lehmann, U. Identification of a distinct side population of cancer cells in the Cal-51 human breast carcinoma cell line. Mol. Cell. Biochem. 2007, 306, 201–212. [Google Scholar] [CrossRef]
- Greve, B.; Kelsch, R.; Spaniol, K.; Eich, H.T.; Götte, M. Flow cytometry in cancer stem cell analysis and separation. Cytom. Part A 2012, 81, 284–293. [Google Scholar] [CrossRef] [PubMed]
- Hiraga, T.; Ito, S.; Nakamura, H. Side population in MDA-MB-231 human breast cancer cells exhibits cancer stem cell-like properties without higher bone-metastatic potential. Oncol. Rep. 2011, 25, 289–296. [Google Scholar] [CrossRef] [PubMed]
- Haraguchi, N.; Utsunomiya, T.; Inoue, H.; Tanaka, F.; Mimori, K.; Barnard, G.F.; Mori, M. Characterization of a side population of cancer cells from human gastrointestinal system. Stem Cells 2006, 24, 506–513. [Google Scholar] [CrossRef]
- Ho, M.M.; Ng, A.V.; Lam, S.; Hung, J.Y. Side population in human lung cancer cell lines and tumors is enriched with stem-like cancer cells. Cancer Res. 2007, 67, 4827–4833. [Google Scholar] [CrossRef] [PubMed]
- Patrawala, L.; Calhoun, T.; Schneider-Broussard, R.; Zhou, J.; Claypool, K.; Tang, D.G. Side population is enriched in tumorigenic, stem-like cancer cells, whereas ABCG2+and ABCG2−cancer cells are similarly tumorigenic. Cancer Res. 2005, 65, 6207–6219. [Google Scholar] [CrossRef]
- Wu, C.; Alman, B.A. Side population cells in human cancers. Cancer Lett. 2008, 268, 1–9. [Google Scholar] [CrossRef]
- Wu, C.; Wei, Q.; Utomo, V.; Nadesan, P.; Whetstone, H.; Kandel, R.; Wunder, J.S.; Alman, B.A. Side population cells isolated from mesenchymal neoplasms have tumor initiating potential. Cancer Res. 2007, 67, 8216–8222. [Google Scholar] [CrossRef]
- Goodell, M.A.; Brose, K.; Paradis, G.; Conner, A.S.; Mulligan, R.C. Isolation and functional properties of murine hematopoietic stem cells that are replicating in vivo. J. Exp. Med. 1996, 183, 1797–1806. [Google Scholar] [CrossRef]
- Wolf, N.S.; Koné, A.; Priestley, G.V.; Bartelmez, S.H. In vivo and in vitro characterization of long-term repopulating primitive hematopoietic cells isolated by sequential Hoechst 33342-rhodamine 123 FACS selection. Exp. Hematol. 1993, 21, 614–622. [Google Scholar]
- Britton, K.M.; Kirby, J.A.; Lennard, T.W.J.; Meeson, A. Cancer stem cells and side population cells in breast cancer and metastasis. Cancers 2011, 3, 2106–2130. [Google Scholar] [CrossRef]
- Alvi, A.J.; Clayton, H.; Joshi, C.; Enver, T.; Ashworth, A.; Vivanco, M.; Dale, T.C.; Smalley, M.J. Functional and molecular characterisation of mammary side population cells. Breast Cancer Res. 2002, 5, R1–R8. [Google Scholar] [CrossRef] [PubMed]
- Clayton, H.; Titley, I.; Vivanco, M. Growth and differentiation of progenitor/stem cells derived from the human mammary gland. Exp. Cell Res. 2004, 297, 444–460. [Google Scholar] [CrossRef] [PubMed]
- Nakanishi, T.; Chumsri, S.; Khakpour, N.; Brodie, A.H.; Leylandjones, B.; Hamburger, A.W.; Ross, D.D.; Burger, A.M. Side-population cells in luminal-type breast cancer have tumour-initiating cell properties, and are regulated by HER2 expression and signalling. Br. J. Cancer 2010, 102, 815–826. [Google Scholar] [CrossRef]
- Zhou, J.; Zhang, H.; Gu, P.; Margolick, J.B.; Yin, D.; Zhang, Y. Cancer stem/progenitor cell active compound 8-quinolinol in combination with paclitaxel achieves an improved cure of breast cancer in the mouse model. Breast Cancer Res. Treat. 2009, 115, 269–277. [Google Scholar] [CrossRef] [PubMed]
- Woodward, W.A.; Chen, M.S.; Behbod, F.; Alfaro, M.P.; Buchholz, T.A.; Rosen, J.M. WNT/beta-catenin mediates radiation resistance of mouse mammary progenitor cells. Proc. Natl. Acad. Sci. USA 2007, 104, 618–623. [Google Scholar] [CrossRef] [PubMed]
- Steiniger, S.C.; Coppinger, J.A.; Kruger, J.A.; Yates, J.; Janda, K.D. Quantitative mass spectrometry identifies drug targets in cancer stem cell-containing side population. Stem Cells 2008, 26, 3037–3046. [Google Scholar] [CrossRef]
- Yin, L.; Castagnino, P.; Assoian, R.K. ABCG2 expression and side population abundance regulated by a transforming growth factor beta-directed epithelial-mesenchymal transition. Cancer Res. 2008, 68, 800–807. [Google Scholar] [CrossRef]
- Zhou, J.; Wulfkuhle, J.; Zhang, H.; Gu, P.; Yang, Y.; Deng, J. Activation of the PTEN/mTOR/STAT3 pathway in breast cancer stem-like cells is required for viability and maintenance. Proc. Natl. Acad. Sci. USA 2007, 104, 16158–16163. [Google Scholar] [CrossRef]
- Brown, H.K.; Tellez-Gabriel, M.; Heymann, D. Cancer stem cells in osteosarcoma. Cancer Lett. 2017, 386, 189–195. [Google Scholar] [CrossRef]
- Diestra, J.E.; Scheffer, G.L.; Català, I.; Maliepaard, M.; Schellens, J.H.M.; Scheper, R.J.; Germà-Lluch, J.R.; Izquierdo, M.A. Frequent expression of the multi-drug resistance-associated protein BCRP/MXR/ABCP/ABCG2 in human tumours detected by the BXP-21 monoclonal antibody in paraffin-embedded material. J. Pathol. 2002, 198, 213–219. [Google Scholar] [CrossRef]
- Jonker, J.W.; Freeman, J.; Bolscher, E.; Musters, S.; Alvi, A.J.; Titley, I.; Schinkel, A.H.; Dale, T.C. Contribution of the ABC transporters Bcrp1 and Mdr1a/1b to the side population phenotype in mammary gland and bone marrow of mice. Stem Cells 2005, 23, 1059–1065. [Google Scholar] [CrossRef]
- Kim, M.; Turnquist, H.; Jackson, J.; Sgagias, M.; Yan, Y.; Gong, M.; Dean, M.; Sharp, J.G.; Cowan, K. The multidrug resistance transporter ABCG2 (breast cancer resistance protein 1) effluxes Hoechst 33342 and is overexpressed in hematopoietic stem cells. Clin. Cancer Res. 2002, 8, 22–28. [Google Scholar] [PubMed]
- Martin, C.M.; Meeson, A.P.; Robertson, S.M.; Hawke, T.J.; Richardson, J.A.; Bates, S.; Goetsch, S.C.; Gallardo, T.D.; Garry, D.J. Persistent expression of the ATP-binding cassette transporter, Abcg2, identifies cardiac SP cells in the developing and adult heart. Dev. Biol. 2004, 265, 262–275. [Google Scholar] [CrossRef] [PubMed]
- Scharenberg, C.W.; Harkey, M.A.; Torok-Storb, B. The ABCG2 transporter is an efficient Hoechst 33342 efflux pump and is preferentially expressed by immature human hematopoietic progenitors. Blood 2002, 99, 507–512. [Google Scholar] [CrossRef] [PubMed]
- Golebiewska, A.; Brons, N.H.; Bjerkvig, R.; Niclou, S.P. Critical appraisal of the side population assay in stem cell and cancer stem cell research. Cell Stem Cell 2011, 8, 136–147. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, C.A.; Ross, A.H. Cancer stem cells: Cell culture, markers, and targets for new therapies. J. Cell. Biochem. 2009, 108, 1031–1038. [Google Scholar] [CrossRef] [PubMed]
- Chute, J.P.; Muramoto, G.G.; Whitesides, J.; Colvin, M.; Safi, R.; Chao, N.J.; McDonnell, N.P. Inhibition of aldehyde dehydrogenase and retinoid signaling induces the expansion of human hematopoietic stem cells. Proc. Natl. Acad. Sci. USA 2006, 103, 11707–11712. [Google Scholar] [CrossRef]
- Khan, M.I.; Czarnecka, A.M.; Helbrecht, I.; Bartnik, E.; Lian, F.; Szczylik, C. Current approaches in identification and isolation of human renal cell carcinoma cancer stem cells. Stem Cell Res. Ther. 2015, 6, 1–11. [Google Scholar] [CrossRef]
- Marcato, P.; Dean, C.A.; Giacomantonio, C.A.; Lee, P.W. Aldehyde dehydrogenase: Its role as a cancer stem cell marker comes down to the specific isoform. Cell Cycle 2011, 10, 1378–1384. [Google Scholar] [CrossRef]
- Charafe-Jauffret, E.; Ginestier, C.; Bertucci, F.; Cabaud, O.; Wicinski, J.; Finetti, P.; Josselin, E.; Adélaïde, J.; Nguyen, T.-T.; Monville, F.; et al. ALDH1-positive cancer stem cells predict engraftment of primary breast tumors and are governed by a common stem cell program. Cancer Res. 2013, 73, 7290–7300. [Google Scholar] [CrossRef]
- Storms, R.W.; Trujillo, A.P.; Springer, J.B.; Shah, L.; Colvin, O.M.; Ludeman, S.M.; Smith, C. Isolation of primitive human hematopoietic progenitors on the basis of aldehyde dehydrogenase activity. Proc. Natl. Acad. Sci. USA 1999, 96, 9118–9123. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, B.A.; Weiss, S. Generation of neurons and astrocytes from isolated cells of the adult mammalian central nervous system. Science 1992, 255, 1707–1710. [Google Scholar] [CrossRef]
- Cariati, M.; Naderi, A.; Brown, J.P.; Smalley, M.J.; Pinder, S.E.; Caldas, C.; Purushotham, A.D. Alpha-6 integrin is necessary for the tumourigenicity of a stem cell-like subpopulation within the MCF7 breast cancer cell line. Int. J. Cancer 2008, 122, 298–304. [Google Scholar] [CrossRef] [PubMed]
- Eramo, A.; Lotti, F.; Sette, G.; Pilozzi, E.; Biffoni, M.; Di Virgilio, A.; Conticello, C.; Ruco, L.; Peschle, C.; De Maria, R. Identification and expansion of the tumorigenic lung cancer stem cell population. Cell Death Differ. 2008, 15, 504–514. [Google Scholar] [CrossRef] [PubMed]
- Lobo, N.A.; Shimono, Y.; Qian, D.; Clarke, M.F. The biology of cancer stem cells. Annu. Rev. Cell Dev. Biol. 2007, 23, 675–699. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.; Clarke, I.D.; Terasaki, M.; Bonn, E.V.; Hawkins, C.E.; Squire, J.; Dirks, P. Identification of a cancer stem cell in human brain tumors. Cancer Res. 2003, 63, 5821–5828. [Google Scholar]
- Pastrana, E.; Silva-Vargas, V.; Doetsch, F. Eyes wide open: A critical review of sphere-formation as an assay for stem cells. Cell Stem Cell 2011, 8, 486–498. [Google Scholar] [CrossRef]
- Abbaszadegan, M.R.; Bagheri, V.; Razavi, M.S.; Momtazi, A.A.; Sahebkar, A.; Gholamin, M. Isolation, identification, and characterization of cancer stem cells: A review. J. Cell. Physiol. 2017, 232, 2008–2018. [Google Scholar] [CrossRef]
- Suslov, O.N.; Kukekov, V.G.; Ignatova, T.N.; Steindler, D.A. Neural stem cell heterogeneity demonstrated by molecular phenotyping of clonal neurospheres. Proc. Natl. Acad. Sci. USA 2002, 99, 14506–14511. [Google Scholar] [CrossRef]
- Stingl, J. Detection and analysis of mammary gland stem cells. J. Pathol. 2009, 217, 229–241. [Google Scholar] [CrossRef]
- Bahmad, H.F.; Cheaito, K.; Chalhoub, R.M.; Hadadeh, O.; Monzer, A.; Ballout, F.; El-Hajj, A.; Mukherji, D.; Liu, Y.-N.; Daoud, G.; et al. Sphere-formation assay: Three-dimensional in vitro culturing of prostate cancer stem/progenitor sphere-forming cells. Front. Oncol. 2018, 8, 347. [Google Scholar] [CrossRef] [PubMed]
- Clevers, H. Modeling development and disease with organoids. Cell 2016, 165, 1586–1597. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.-L.; Fu, D.; Ma, Y.; Shen, X.-Z. The power and the promise of liver cancer stem cell markers. Stem Cells Dev. 2011, 20, 2023–2030. [Google Scholar] [CrossRef] [PubMed]
- Broadley, K.W.R.; Hunn, M.K.; Farrand, K.J.; Price, K.M.; Grasso, C.; Miller, R.J.; Hermans, I.F.; McConnell, M.J. Side population is not necessary or sufficient for a cancer stem cell phenotype in glioblastoma multiforme. Stem Cells 2011, 29, 452–461. [Google Scholar] [CrossRef] [PubMed]
- Pearce, D.J.; Bonnet, D. The combined use of Hoechst efflux ability and aldehyde dehydrogenase activity to identify murine and human hematopoietic stem cells. Exp. Hematol. 2007, 35, 1437–1446. [Google Scholar] [CrossRef] [PubMed]
- Pierre-Louis, O.; Clay, D.; De La Grange, P.B.; Blazsek, I.; Desterke, C.; Guerton, B.; Blondeau, C.; Malfuson, J.-V.; Prat, M.; Bennaceur-Griscelli, A.; et al. Dual SP/ALDH functionalities refine the human hematopoietic Lin−CD34+CD38−stem/progenitor cell compartment. Stem Cells 2009, 27, 2552–2562. [Google Scholar] [CrossRef]
- Liu, M.; Liu, Y.; Deng, L.; Wang, D.; He, X.; Zhou, L.; Wicha, M.S.; Bai, F.; Liu, S. Transcriptional profiles of different states of cancer stem cells in triple-negative breast cancer. Mol. Cancer 2018, 17, 1–6. [Google Scholar] [CrossRef]
- Pires, B.R.B.; De Amorim, Í.S.S.; Souza, E.; Rodrigues, J.A.; Mencalha, A. Targeting cellular signaling pathways in breast cancer stem cells and its implication for cancer treatment. Anticancer Res. 2016, 36, 5681–5692. [Google Scholar] [CrossRef]
- Yang, L.; Shi, P.; Zhao, G.; Xu, J.; Peng, W.; Zhang, J. Targeting cancer stem cell pathways for cancer therapy. Signal Transduct. Target. Ther. 2020, 5, 8. [Google Scholar] [CrossRef]
- Zhou, W.; Wang, G.; Guo, S. Regulation of angiogenesis via Notch signaling in breast cancer and cancer stem cells. Biochim. Biophys. Acta 2013, 1836, 304–320. [Google Scholar] [CrossRef]
- Butti, R.; Gunasekaran, V.P.; Kumar, T.V.; Banerjee, P.; Kundu, G.C. Breast cancer stem cells: Biology and therapeutic implications. Int. J. Biochem. Cell Biol. 2019, 107, 38–52. [Google Scholar] [CrossRef] [PubMed]
- Bouras, T.; Pal, B.; Vaillant, F.; Harburg, G.; Asselin-Labat, M.-L.; Oakes, S.R.; Lindeman, G.J.; Visvader, E.J. Notch signaling regulates mammary stem cell function and luminal cell-fate commitment. Cell Stem Cell 2008, 3, 429–441. [Google Scholar] [CrossRef] [PubMed]
- Farnie, G.; Clarke, R.B. Mammary stem cells and breast cancer—Role of Notch signalling. Stem Cell Rev. Rep. 2007, 3, 169–175. [Google Scholar] [CrossRef] [PubMed]
- D’Angelo, R.C.; Ouzounova, M.; Davis, A.; Choi, D.; Tchuenkam, S.M.; Kim, G.; Luther, T.; Quraishi, A.A.; Senbabaoglu, Y.; Conley, S.J.; et al. Notch reporter activity in breast cancer cell lines identifies a subset of cells with stem cell activity. Mol. Cancer Ther. 2015, 14, 779–787. [Google Scholar] [CrossRef] [PubMed]
- Mamaeva, V.; Niemi, R.; Beck, M.; Ozliseli, E.; Desai, D.; Landor, S. Inhibiting notch activity in breast cancer stem cells by glucose functionalized nanoparticles carrying gamma-secretase inhibitors. Mol. Ther. 2016, 24, 926–936. [Google Scholar] [CrossRef] [PubMed]
- Rizzo, P.; Miao, H.; D’Souza, G.; Osipo, C.; Yun, J.; Zhao, H.; Mascarenhas, J.; Wyatt, D.; Antico, G.; Hao, L.; et al. Cross-talk between notch and the estrogen receptor in breast cancer suggests novel therapeutic approaches. Cancer Res. 2008, 68, 5226–5235. [Google Scholar] [CrossRef]
- Schott, A.F.; Landis, M.D.; Dontu, G.; Griffith, K.A.; Layman, R.M.; Krop, I.; Paskett, L.A.; Wong, H.; Dobrolecki, L.E.; Lewis, M.T.; et al. Preclinical and clinical studies of gamma secretase inhibitors with docetaxel on human breast tumors. Clin. Cancer Res. 2013, 19, 1512–1524. [Google Scholar] [CrossRef]
- Kumar, S.; Srivastav, R.K.; Wilkes, D.W.; Ross, T.; Kim, S.; Kowalski, J.; Chatla, S.; Zhang, Q.; Nayak, A.; Guha, M.; et al. Estrogen-dependent DLL1-mediated Notch signaling promotes luminal breast cancer. Oncogene 2019, 38, 2092–2107. [Google Scholar] [CrossRef]
- Hossain, F.; Sorrentino, C.; Ucar, D.A.; Peng, Y.; Matossian, M.; Wyczechowska, D. Notch signaling regulates mitochondrial metabolism and NF-kappaB activity in triple-negative breast cancer cells via IKKalpha-dependent non-canonical pathways. Front Oncol. 2018, 8, 575. [Google Scholar] [CrossRef]
- Papadakos, K.S.; Bartoschek, M.; Cupello, C.R.; Gialeli, C.; Jin, S.-B.; Lendahl, U.; Pietras, K.; Blom, A.M. Cartilage Oligomeric Matrix Protein initiates cancer stem cells through activation of Jagged1-Notch3 signaling. Matrix Biol. 2019, 81, 107–121. [Google Scholar] [CrossRef]
- Li, Y.; Welm, B.; Podsypanina, K.; Huang, S.; Chamorro, M.; Zhang, X.; Rowlands, T.; Egeblad, M.; Cowin, P.; Werb, Z.; et al. Evidence that transgenes encoding components of the Wnt signaling pathway preferentially induce mammary cancers from progenitor cells. Proc. Natl. Acad. Sci. USA 2003, 100, 15853–15858. [Google Scholar] [CrossRef]
- Liu, B.Y.; McDermott, S.P.; Khwaja, S.S.; Alexander, C.M. The transforming activity of Wnt effectors correlates with their ability to induce the accumulation of mammary progenitor cells. Proc. Natl. Acad. Sci. USA 2004, 101, 4158–4163. [Google Scholar] [CrossRef] [PubMed]
- Korkaya, H.; Paulson, A.; Charafe-Jauffret, E.; Ginestier, C.; Brown, M.; Dutcher, J. Regulation of mammary stem/progenitor cells by PTEN/Akt/beta-catenin signaling. PLoS Biol. 2009, 7, e1000121. [Google Scholar] [CrossRef] [PubMed]
- Sulaiman, A.; McGarry, S.; Li, L.; Jia, D.; Ooi, S.; Addison, C.; Dimitroulakos, J.; Arnaout, A.; Nessim, C.; Yao, Z.; et al. Dual inhibition of Wnt and Yes-associated protein signaling retards the growth of triple-negative breast cancer in both mesenchymal and epithelial states. Mol. Oncol. 2018, 12, 423–440. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.M.; Wu, J.F.; Luo, Q.C.; Liu, Q.F.; Wu, Q.W.; Ye, G.D. Pygo2 activates MDR1 expression and mediates chemoresistance in breast cancer via the Wnt/beta-catenin pathway. Oncogene 2016, 35, 4787–4797. [Google Scholar] [CrossRef] [PubMed]
- Jang, G.B.; Kim, J.Y.; Cho, S.D.; Park, K.S.; Jung, J.Y.; Lee, H.Y. Blockade of Wnt/beta-catenin signaling suppresses breast cancer metastasis by inhibiting CSC-like phenotype. Sci. Rep. 2015, 5, 12465. [Google Scholar] [CrossRef] [PubMed]
- Castagnoli, L.; Cancila, V.; Cordoba-Romero, S.L.; Faraci, S.; Talarico, G.; Belmonte, B.; Iorio, M.V.; Milani, M.; Volpari, T.; Chiodoni, C.; et al. WNT signaling modulates PD-L1 expression in the stem cell compartment of triple-negative breast cancer. Oncogene 2019, 38, 4047–4060. [Google Scholar] [CrossRef]
- Pei, Y.Y.; Li, G.C.; Ran, J.; Wan, X.H.; Wei, F.X.; Wang, L. Kinesin family member 11 enhances the self-renewal ability of breast cancer cells by participating in the Wnt/beta-catenin pathway. J. Breast Cancer 2019, 22, 522–532. [Google Scholar] [CrossRef]
- Siddharth, S.; Goutam, K.; Das, S.; Nayak, A.; Nayak, D.; Sethy, C. Nectin-4 is a breast cancer stem cell marker that induces WNT/beta-catenin signaling via Pi3k/Akt axis. Int. J. Biochem. Cell Biol. 2017, 89, 85–94. [Google Scholar] [CrossRef]
- McGinn, O.; Ward, A.V.; Fettig, L.M.; Riley, D.; Ivie, J.; Paul, K.V. Cytokeratin 5 alters beta-catenin dynamics in breast cancer cells. Oncogene 2020, 39, 2478–2492. [Google Scholar] [CrossRef]
- Yan, Y.; Liu, F.; Han, L.; Zhao, L.; Chen, J.; Olopade, O.I.; He, M.; Wei, M. HIF-2α promotes conversion to a stem cell phenotype and induces chemoresistance in breast cancer cells by activating Wnt and Notch pathways. J. Exp. Clin. Cancer Res. 2018, 37, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Briscoe, J.; Thérond, P.P. The mechanisms of Hedgehog signalling and its roles in development and disease. Nat. Rev. Mol. Cell Biol. 2013, 14, 416–429. [Google Scholar] [CrossRef] [PubMed]
- Riaz, S.K.; Khan, J.S.; Shah, S.T.A.; Wang, F.; Ye, L.; Jiang, W.; Malik, M.F.A. Involvement of hedgehog pathway in early onset, aggressive molecular subtypes and metastatic potential of breast cancer. Cell Commun. Signal. 2018, 16, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Monkkonen, T.; Lewis, M.T. New paradigms for the Hedgehog signaling network in mammary gland development and breast cancer. Biochim. Biophys. Acta 2017, 1868, 315–332. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Dontu, G.; Mantle, I.D.; Patel, S.; Ahn, N.-S.; Jackson, K.W.; Suri, P.; Wicha, M.S. Hedgehog signaling and Bmi-1 regulate self-renewal of normal and malignant human mammary stem cells. Cancer Res. 2006, 66, 6063–6071. [Google Scholar] [CrossRef] [PubMed]
- Cochrane, C.R.; Szczepny, A.; Watkins, D.N.; Cain, J.E. Hedgehog signaling in the maintenance of cancer stem cells. Cancers 2015, 7, 1554–1585. [Google Scholar] [CrossRef]
- Piccolo, S.; Dupont, S.; Cordenonsi, M. The Biology of YAP/TAZ: Hippo signaling and beyond. Physiol. Rev. 2014, 94, 1287–1312. [Google Scholar] [CrossRef]
- Maugeri-Saccà, M.; De Maria, R. Hippo pathway and breast cancer stem cells. Crit. Rev. Oncol. 2016, 99, 115–122. [Google Scholar] [CrossRef]
- Frangou, C.; Li, Y.-W.; Shen, H.; Yang, N.; Wilson, K.E.; Blijlevens, M.; Guo, J.; Nowak, N.J.; Zhang, J. Molecular profiling and computational network analysis of TAZ-mediated mammary tumorigenesis identifies actionable therapeutic targets. Oncotarget 2014, 5, 12166–12176. [Google Scholar] [CrossRef]
- Chang, C.; Goel, H.L.; Gao, H.; Pursell, B.; Shultz, L.D.; Greiner, D.L. A laminin 511 matrix is regulated by TAZ and functions as the ligand for the alpha6Bbeta1 integrin to sustain breast cancer stem cells. Genes Dev. 2015, 29, 1–6. [Google Scholar] [CrossRef]
- Zhang, H.; Lang, T.-Y.; Zou, D.-L.; Zhou, L.; Lou, M.; Liu, J.-S.; Li, Y.-Z.; Ding, D.-Y.; Li, Y.-C.; Zhang, N.; et al. miR-520b promotes breast cancer stemness through Hippo/YAP signaling pathway. OncoTargets Ther. 2019, 12, 11691–11700. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Bin, X.; Zhang, X.-P. Effects of miRNAs on functions of breast cancer stem cells and treatment of breast cancer. OncoTargets Ther. 2018, 11, 4263–4270. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, R.-U.; Miyazaki, H.; Ochiya, T. The Roles of MicroRNAs in breast cancer. Cancers 2015, 7, 598–616. [Google Scholar] [CrossRef] [PubMed]
- Fazal, F.M.; Chang, H.Y. lncRNA structure: Message to the heart. Mol. Cell 2016, 64, 1–2. [Google Scholar] [CrossRef] [PubMed]
- McHugh, C.A.; Chen, C.K.; Chow, A.; Surka, C.F.; Tran, C.; McDonel, P. The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature 2015, 521, 232–236. [Google Scholar] [CrossRef] [PubMed]
- Alves, C.P.; Fonseca, A.S.; Muys, B.R.; Bueno, R.D.B.E.L.; Bürger, M.C.; De Souza, J.E.S.; Valente, V.; Zago, M.A.; Júnior, W.A.D.S. Brief report: The lincRNA hotair is required for epithelial-to-mesenchymal transition and stemness maintenance of cancer cell lines. Stem Cells 2013, 31, 2827–2832. [Google Scholar] [CrossRef] [PubMed]
- Hou, P.; Zhao, Y.; Li, Z.; Yao, R.; Ma, M.; Gao, Y. LincRNA-ROR induces epithelial-to-mesenchymal transition and contributes to breast cancer tumorigenesis and metastasis. Cell Death Dis. 2014, 5, e1287. [Google Scholar] [CrossRef]
- Li, H.; Zhu, L.; Xu, L.; Qin, K.; Liu, C.; Yu, Y.; Su, D.; Wu, K.; Sheng, Y. Long noncoding RNA linc00617 exhibits oncogenic activity in breast cancer. Mol. Carcinog. 2015, 56, 3–17. [Google Scholar] [CrossRef]
- Zheng, A.; Song, X.; Zhang, L.; Zhao, L.; Mao, X.; Wei, M. Long non-coding RNA LUCAT1/miR-5582-3p/TCF7L2 axis regulates breast cancer stemness via Wnt/beta-catenin pathway. J. Exp. Clin. Cancer Res. 2019, 38, 305. [Google Scholar] [CrossRef]
- Vidovic, D.; Huynh, T.T.; Konda, P.; Dean, C.; Cruickshank, B.M.; Sultan, M.; Coyle, K.M.; Gujar, S.; Marcato, P. ALDH1A3-regulated long non-coding RNA NRAD1 is a potential novel target for triple-negative breast tumors and cancer stem cells. Cell Death Differ. 2019, 27, 363–378. [Google Scholar] [CrossRef]
- Shin, V.Y.; Chen, J.; Cheuk, I.W.-Y.; Siu, M.-T.; Ho, C.-W.; Wang, X.; Jin, H.; Kwong, A. Long non-coding RNA NEAT1 confers oncogenic role in triple-negative breast cancer through modulating chemoresistance and cancer stemness. Cell Death Dis. 2019, 10, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Shima, H.; Kida, K.; Adachi, S.; Yamada, A.; Sugae, S.; Narui, K.; Miyagi, Y.; Nishi, M.; Ryo, A.; Murata, S.; et al. Lnc RNA H19 is associated with poor prognosis in breast cancer patients and promotes cancer stemness. Breast Cancer Res. Treat. 2018, 170, 507–516. [Google Scholar] [CrossRef] [PubMed]
- Ma, F.; Liu, X.; Zhou, S.; Li, W.; Liu, C.; Chadwick, M. Long non-coding RNA FGF13-AS1 inhibits glycolysis and stemness properties of breast cancer cells through FGF13-AS1/IGF2BPs/Myc feedback loop. Cancer Lett. 2019, 450, 63–75. [Google Scholar] [CrossRef] [PubMed]
- Eshahryari, A.; Jazi, M.E.; Samaei, N.E.; Mowla, S.J. Long non-coding RNA SOX2OT: Expression signature, splicing patterns, and emerging roles in pluripotency and tumorigenesis. Front. Genet. 2015, 6, 196. [Google Scholar] [CrossRef]
- Zeng, L.; Cen, Y.; Chen, J. Long non-coding RNA MALAT-1 contributes to maintenance of stem cell-like phenotypes in breast cancer cells. Oncol. Lett. 2018, 15, 2117–2122. [Google Scholar] [CrossRef]
- Keshavarz, M.; Asadi, M.H. Long non-coding RNA ES1 controls the proliferation of breast cancer cells by regulating the Oct4/Sox2/miR-302 axis. FEBS J. 2019, 286, 2611–2623. [Google Scholar] [CrossRef]
- Dittmer, J. Breast cancer stem cells: Features, key drivers and treatment options. Semin. Cancer Biol. 2018, 53, 59–74. [Google Scholar] [CrossRef]
- Tanei, T.; Choi, D.S.; Rodriguez, A.A.; Liang, D.H.; Dobrolecki, L.E.; Ghosh, M.; Landis, M.D.; Chang, J.C. Antitumor activity of Cetuximab in combination with Ixabepilone on triple negative breast cancer stem cells. Breast Cancer Res. 2016, 18, 1–9. [Google Scholar] [CrossRef]
- Liu, J.; Pan, S.; Hsieh, M.H.; Ng, N.; Sun, F.; Wang, T.; Kasibhatla, S.; Schuller, A.G.; Li, A.G.; Cheng, D.; et al. Targeting Wnt-driven cancer through the inhibition of Porcupine by LGK974. Proc. Natl. Acad. Sci. USA 2013, 110, 20224–20229. [Google Scholar] [CrossRef]
- Kester, L.; van Oudenaarden, A. Single-cell transcriptomics meets lineage tracing. Cell Stem Cell 2018, 23, 166–179. [Google Scholar] [CrossRef]
- Scheel, C.; Weinberg, R.A. Cancer stem cells and epithelial–mesenchymal transition: Concepts and molecular links. Semin. Cancer Biol. 2012, 22, 396–403. [Google Scholar] [CrossRef] [PubMed]
- Bao, L.; Cardiff, R.D.; Steinbach, P.; Messer, K.; Ellies, L.G. Multipotent luminal mammary cancer stem cells model tumor heterogeneity. Breast Cancer Res. 2015, 17, 137. [Google Scholar] [CrossRef] [PubMed]
- Lim, E.; Vaillant, F.; Wu, D.; Forrest, N.C.; Pal, B.; Hart, A.H. Aberrant luminal progenitors as the candidate target population for basal tumor development in BRCA1 mutation carriers. Nat. Med. 2009, 15, 907–913. [Google Scholar] [CrossRef] [PubMed]
- Koren, S.; Reavie, L.; Couto, J.P.; De Silva, D.; Stadler, M.B.; Roloff, T. PIK3CA(H1047R) induces multipotency and multi-lineage mammary tumours. Nature 2015, 525, 114–118. [Google Scholar] [CrossRef] [PubMed]
- Lagadec, C.; Vlashi, E.; Della Donna, L.; Dekmezian, C.; Pajonk, F. Radiation-induced reprogramming of breast cancer cells. Stem Cells 2012, 30, 833–844. [Google Scholar] [CrossRef]
- Van Keymeulen, A.; Lee, M.Y.; Ousset, M.; Brohée, S.; Rorive, S.; Giraddi, R.R.; Wuidart, A.; Bouvencourt, G.; Dubois, C.; Salmon, I.; et al. Reactivation of multipotency by oncogenic PIK3CA induces breast tumour heterogeneity. Nat. Cell Biol. 2015, 525, 119–123. [Google Scholar] [CrossRef]
- Meyer, D.S.; Brinkhaus, H.; Müller, U.; Müller, M.; Cardiff, R.D.; Bentires-Alj, M. Luminal expression of PIK3CA mutant H1047R in the mammary gland induces heterogeneous tumors. Cancer Res. 2011, 71, 4344–4351. [Google Scholar] [CrossRef]
- Gupta, P.B.; Fillmore, C.M.; Jiang, G.; Shapira, S.D.; Tao, K.; Kuperwasser, C. Stochastic state transitions give rise to phenotypic equilibrium in populations of cancer cells. Cell 2011, 146, 633–644. [Google Scholar] [CrossRef]
- Rycaj, K.; Tang, D.G. Cell-of-origin of cancer versus cancer stem cells: Assays and interpretations. Cancer Res. 2015, 75, 4003–4011. [Google Scholar] [CrossRef]
- Frank, E.; Sanes, J.R. Lineage of neurons and glia in chick dorsal root ganglia: Analysis in vivo with a recombinant retrovirus. Development 1991, 111, 895–908. [Google Scholar]
- Turner, D.L.; Cepko, C.L. A common progenitor for neurons and glia persists in rat retina late in development. Nat. Cell Biol. 1987, 328, 131–136. [Google Scholar] [CrossRef] [PubMed]
- Raj, B.; Wagner, D.E.; McKenna, A.; Pandey, S.; Klein, A.M.; Shendure, J. Simultaneous single-cell profiling of lineages and cell types in the vertebrate brain. Nat. Biotechnol. 2018, 36, 442–450. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, Q.H.; Pervolarakis, N.; Blake, K.; Ma, D.; Davis, R.T.; James, N.; Phung, A.T.; Willey, E.; Kumar, R.; Jabart, E.; et al. Profiling human breast epithelial cells using single cell RNA sequencing identifies cell diversity. Nat. Commun. 2018, 9, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Bach, K.; Pensa, S.; Grzelak, M.; Hadfield, J.; Adams, D.J.; Marioni, J.C.; Khaled, W. Differentiation dynamics of mammary epithelial cells revealed by single-cell RNA sequencing. Nat. Commun. 2017, 8, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Gulati, G.S.; Sikandar, S.S.; Wesche, D.J.; Manjunath, A.; Bharadwaj, A.; Berger, M.J. Single-cell transcriptional diversity is a hallmark of developmental potential. Science 2020, 367, 405–411. [Google Scholar] [CrossRef]
- Giraddi, R.R.; Chung, C.-Y.; Heinz, R.E.; Balcioglu, O.; Novotny, M.; Trejo, C.L.; Dravis, C.; Hagos, B.M.; Mehrabad, E.M.; Rodewald, L.W.; et al. Single-cell transcriptomes distinguish stem cell state changes and lineage specification programs in early mammary gland development. Cell Rep. 2018, 24, 1653–1666.e7. [Google Scholar] [CrossRef]
- Regan, J.L.; Sourisseau, T.; Soady, K.; Kendrick, H.; McCarthy, A.; Tang, C. Aurora A kinase regulates mammary epithelial cell fate by determining mitotic spindle orientation in a Notch-dependent manner. Cell Rep. 2013, 4, 110–123. [Google Scholar] [CrossRef]
- Kebschull, J.M.; Zador, A.M. Cellular barcoding: Lineage tracing, screening and beyond. Nat. Methods 2018, 15, 871–879. [Google Scholar] [CrossRef]
- Lu, R.; Neff, N.F.; Quake, S.R.; Weissman, I.L. Tracking single hematopoietic stem cells in vivo using high-throughput sequencing in conjunction with viral genetic barcoding. Nat. Biotechnol. 2011, 29, 928–933. [Google Scholar] [CrossRef]
- Weissman, T.A.; Pan, Y.A. Brainbow: New resources and emerging biological applications for multicolor genetic labeling and analysis. Genetics 2015, 199, 293–306. [Google Scholar] [CrossRef]
- Jao, L.-E.; Wente, S.R.; Chen, W. Efficient multiplex biallelic zebrafish genome editing using a CRISPR nuclease system. Proc. Natl. Acad. Sci. USA 2013, 110, 13904–13909. [Google Scholar] [CrossRef] [PubMed]
- Varshney, G.K.; Pei, W.; LaFave, M.C.; Idol, J.; Xu, L.; Gallardo, V.; Carrington, B.; Bishop, K.; Jones, M.; Li, M.; et al. High-throughput gene targeting and phenotyping in zebrafish using CRISPR/Cas9. Genome Res. 2015, 25, 1030–1042. [Google Scholar] [CrossRef] [PubMed]
- Baron, C.S.; Van Oudenaarden, A. Unravelling cellular relationships during development and regeneration using genetic lineage tracing. Nat. Rev. Mol. Cell Biol. 2019, 20, 753–765. [Google Scholar] [CrossRef] [PubMed]
- Alemany, A.; Florescu, M.; Baron, C.S.; Peterson-Maduro, J.; van Oudenaarden, A. Whole-organism clone tracing using single-cell sequencing. Nature 2018, 556, 108–112. [Google Scholar] [CrossRef] [PubMed]
- Frieda, K.L.; Linton, J.M.; Hormoz, S.; Choi, J.; Chow, K.K.; Singer, Z.S. Synthetic recording and in situ readout of lineage information in single cells. Nature 2017, 541, 107–111. [Google Scholar] [CrossRef] [PubMed]
- Kalhor, R.; Kalhor, K.; Mejia, L.; Leeper, K.; Graveline, A.; Mali, P.; Church, G.M. Developmental barcoding of whole mouse via homing CRISPR. Science 2018, 361, eaat9804. [Google Scholar] [CrossRef]
- Spanjaard, B.; Hu, B.; Mitic, N.; Olivares-Chauvet, P.; Janjuha, S.; Ninov, N. Simultaneous lineage tracing and cell-type identification using CRISPR-Cas9-induced genetic scars. Nat. Biotechnol. 2018, 36, 469–473. [Google Scholar] [CrossRef]
- Yao, Z.; Mich, J.K.; Ku, S.; Menon, V.; Krostag, A.-R.; Martinez, R.A.; Furchtgott, L.; Mulholland, H.; Bort, S.; Fuqua, M.A.; et al. A single-cell roadmap of lineage bifurcation in human ESC models of embryonic brain development. Cell Stem Cell 2017, 20, 120–134. [Google Scholar] [CrossRef]
- Jaitin, D.A.; Kenigsberg, E.; Keren-Shaul, H.; Elefant, N.; Paul, F.; Zaretsky, I.; Mildner, A.; Cohen, N.; Jung, S.; Tanay, A.; et al. Massively parallel single-cell RNA-seq for marker-free decomposition of tissues into cell types. Science 2014, 343, 776–779. [Google Scholar] [CrossRef]
- Rodriguez-Fraticelli, A.E.; Wolock, S.L.; Weinreb, C.S.; Panero, R.; Patel, S.H.; Jankovic, M. Clonal analysis of lineage fate in native haematopoiesis. Nature 2018, 553, 212–226. [Google Scholar] [CrossRef]
- Laurenti, E.; Gottgens, B. From haematopoietic stem cells to complex differentiation landscapes. Nature 2018, 553, 418–426. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, X.; Powell, K.; Li, L. Breast Cancer Stem Cells: Biomarkers, Identification and Isolation Methods, Regulating Mechanisms, Cellular Origin, and Beyond. Cancers 2020, 12, 3765. https://doi.org/10.3390/cancers12123765
Zhang X, Powell K, Li L. Breast Cancer Stem Cells: Biomarkers, Identification and Isolation Methods, Regulating Mechanisms, Cellular Origin, and Beyond. Cancers. 2020; 12(12):3765. https://doi.org/10.3390/cancers12123765
Chicago/Turabian StyleZhang, Xiaoli, Kimerly Powell, and Lang Li. 2020. "Breast Cancer Stem Cells: Biomarkers, Identification and Isolation Methods, Regulating Mechanisms, Cellular Origin, and Beyond" Cancers 12, no. 12: 3765. https://doi.org/10.3390/cancers12123765
APA StyleZhang, X., Powell, K., & Li, L. (2020). Breast Cancer Stem Cells: Biomarkers, Identification and Isolation Methods, Regulating Mechanisms, Cellular Origin, and Beyond. Cancers, 12(12), 3765. https://doi.org/10.3390/cancers12123765





