Pharmacokinetic Analysis of Dynamic Contrast-Enhanced Magnetic Resonance Imaging at 7T for Breast Cancer Diagnosis and Characterization
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients
2.2. MRI
2.3. Data Analysis
2.4. Histopathology
2.5. Statistical Methods
3. Results
3.1. Lesion Characteristics
3.2. Differentiation of Benign and Malignant Tumors Using PK Analysis
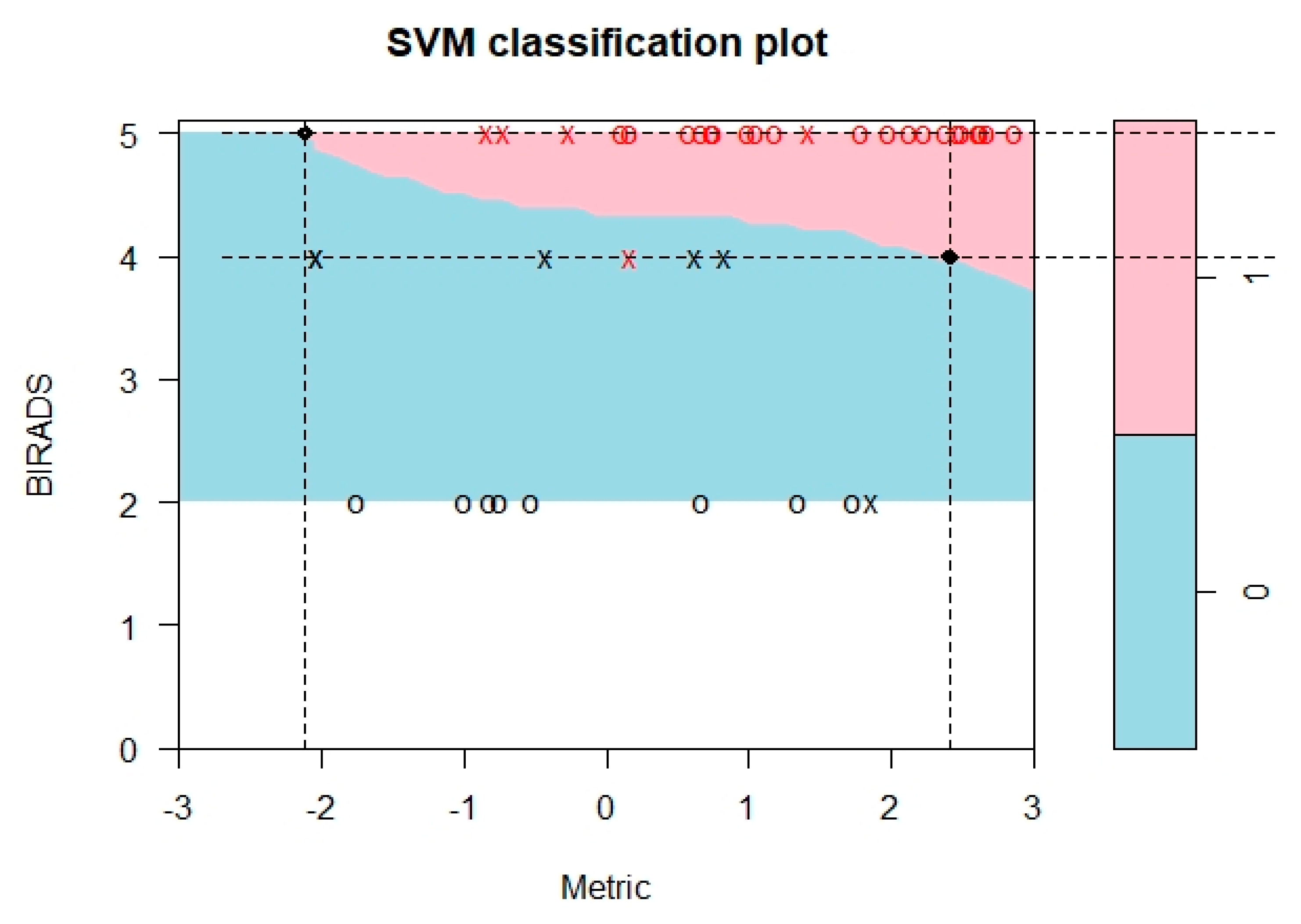
3.3. Differentiation of Benign and Malignant Tumors Using PK-Enhanced BI-RADS
3.4. Molecular Subtyping
3.5. Tumor Grading
3.6. Proliferation Rate
3.7. Inter-Reader Agreement
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Kuhl, C.; Weigel, S.; Schrading, S.; Arand, B.; Bieling, H.; Konig, R.; Tombach, B.; Leutner, C.; Rieber-Brambs, A.; Nordhoff, D.; et al. Prospective multicenter cohort study to refine management recommendations for women at elevated familial risk of breast cancer: The EVA trial. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2010, 28, 1450–1457. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marinovich, M.L.; Houssami, N.; Macaskill, P.; Sardanelli, F.; Irwig, L.; Mamounas, E.P.; von Minckwitz, G.; Brennan, M.E.; Ciatto, S. Meta-analysis of magnetic resonance imaging in detecting residual breast cancer after neoadjuvant therapy. J. Natl. Cancer Inst. 2013, 105, 321–333. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sardanelli, F.; Giuseppetti, G.M.; Panizza, P.; Bazzocchi, M.; Fausto, A.; Simonetti, G.; Lattanzio, V.; Del Maschio, A.; Italian Trial for Breast MR in Multifocal/Multicentric Cancer. Sensitivity of MRI versus mammography for detecting foci of multifocal, multicentric breast cancer in Fatty and dense breasts using the whole-breast pathologic examination as a gold standard. AJR Am. J. Roentgenol. 2004, 183, 1149–1157. [Google Scholar] [CrossRef] [PubMed]
- Grimm, L.J.; Johnson, K.S.; Marcom, P.K.; Baker, J.A.; Soo, M.S. Can breast cancer molecular subtype help to select patients for preoperative MR imaging? Radiology 2015, 274, 352–358. [Google Scholar] [CrossRef] [PubMed]
- Morrow, M.; Waters, J.; Morris, E. MRI for breast cancer screening, diagnosis, and treatment. Lancet 2011, 378, 1804–1811. [Google Scholar] [CrossRef]
- Pinker, K.; Helbich, T.H.; Morris, E.A. The potential of multiparametric MRI of the breast. Br. J. Radiol. 2017, 90, 20160715. [Google Scholar] [CrossRef] [Green Version]
- Gruber, S.; Pinker, K.; Zaric, O.; Minarikova, L.; Chmelik, M.; Baltzer, P.; Boubela, R.N.; Helbich, T.; Bogner, W.; Trattnig, S. Dynamic contrast-enhanced magnetic resonance imaging of breast tumors at 3 and 7 T: A comparison. Investig. Radiol. 2014, 49, 354–362. [Google Scholar] [CrossRef]
- Trattnig, S.; Bogner, W.; Gruber, S.; Szomolanyi, P.; Juras, V.; Robinson, S.; Zbyn, S.; Haneder, S. Clinical applications at ultrahigh field (7 T). Where does it make the difference? NMR Biomed. 2016, 29, 1316–1334. [Google Scholar] [CrossRef]
- Dietzel, M.; Baltzer, P.A.; Vag, T.; Groschel, T.; Gajda, M.; Camara, O.; Kaiser, W.A. Differential diagnosis of breast lesions 5 mm or less: Is there a role for magnetic resonance imaging? J. Comput. Assist. Tomogr. 2010, 34, 456–464. [Google Scholar] [CrossRef]
- Schlossbauer, T.; Leinsinger, G.; Wismuller, A.; Lange, O.; Scherr, M.; Meyer-Baese, A.; Reiser, M. Classification of small contrast enhancing breast lesions in dynamic magnetic resonance imaging using a combination of morphological criteria and dynamic analysis based on unsupervised vector-quantization. Investig. Radiol. 2008, 43, 56–64. [Google Scholar] [CrossRef]
- Onishi, N.; Kataoka, M.; Kanao, S.; Sagawa, H.; Iima, M.; Nickel, M.D.; Toi, M.; Togashi, K. Ultrafast dynamic contrast-enhanced mri of the breast using compressed sensing: Breast cancer diagnosis based on separate visualization of breast arteries and veins. J. Magn. Reson. Imaging 2017, 47, 97–104. [Google Scholar] [CrossRef] [PubMed]
- Korteweg, M.A.; Veldhuis, W.B.; Visser, F.; Luijten, P.R.; Mali, W.P.; van Diest, P.J.; van den Bosch, M.A.; Klomp, D.J. Feasibility of 7 Tesla breast magnetic resonance imaging determination of intrinsic sensitivity and high-resolution magnetic resonance imaging, diffusion-weighted imaging, and (1)H-magnetic resonance spectroscopy of breast cancer patients receiving neoadjuvant therapy. Investig. Radiol. 2011, 46, 370–376. [Google Scholar] [CrossRef]
- Kim, J.Y.; Kim, S.H.; Kim, Y.J.; Kang, B.J.; An, Y.Y.; Lee, A.W.; Song, B.J.; Park, Y.S.; Lee, H.B. Enhancement parameters on dynamic contrast enhanced breast MRI: Do they correlate with prognostic factors and subtypes of breast cancers? Magn. Reson. Imaging 2015, 33, 72–80. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Wang, K.; Sun, X.; Wang, K.; Sun, Y.; Zhang, G.; Shen, B. Parameters of dynamic contrast-enhanced MRI as imaging markers for angiogenesis and proliferation in human breast cancer. Med. Sci. Monit. 2015, 21, 376–382. [Google Scholar] [CrossRef] [Green Version]
- Tofts, P.S.K.; Allan, G. Measurement of the Blood-Brain Barrier Permeability and Leakage Space Using Dynamic MR Imaging. 1. Fundamental concepts. Magn. Reson. Med. 1989, 17, 357–367. [Google Scholar] [CrossRef]
- Tofts, P.S.; Berkowitz, B.; Schnall, M.D. Quantitative analysis of dynamic Gd-DTPA enhancement in breast tumors using a permeability model. Magn. Reson. Med. 1995, 33, 564–568. [Google Scholar] [CrossRef]
- Tofts, P.S.; Brix, G.; Buckley, D.L.; Evelhoch, J.L.; Henderson, E.; Knopp, M.V.; Larsson, H.B.; Lee, T.Y.; Mayr, N.A.; Parker, G.J.; et al. Estimating kinetic parameters from dynamic contrast-enhanced T(1)-weighted MRI of a diffusable tracer: Standardized quantities and symbols. J. Magn. Reson. Imaging 1999, 10, 223–232. [Google Scholar] [CrossRef]
- El Khouli, R.H.; Macura, K.J.; Kamel, I.R.; Jacobs, M.A.; Bluemke, D.A. 3-T dynamic contrast-enhanced MRI of the breast: Pharmacokinetic parameters versus conventional kinetic curve analysis. AJR Am. J. Roentgenol. 2011, 197, 1498–1505. [Google Scholar] [CrossRef] [Green Version]
- Jena, A.; Taneja, S.; Singh, A.; Negi, P.; Mehta, S.B.; Sarin, R. Role of pharmacokinetic parameters derived with high temporal resolution DCE MRI using simultaneous PET/MRI system in breast cancer: A feasibility study. Eur. J. Radiol. 2017, 86, 261–266. [Google Scholar] [CrossRef]
- Huang, W.; Tudorica, L.A.; Li, X.; Thakur, S.B.; Chen, Y.; Morris, E.A.; Tagge, I.J.; Korenblit, M.E.; Rooney, W.D.; Koutcher, J.A.; et al. Discrimination of benign and malignant breast lesions by using shutter-speed dynamic contrast-enhanced MR imaging. Radiology 2011, 261, 394–403. [Google Scholar] [CrossRef] [Green Version]
- Radjenovic, A.; Dall, B.J.; Ridgway, J.P.; Smith, M.A. Measurement of pharmacokinetic parameters in histologically graded invasive breast tumours using dynamic contrast-enhanced MRI. Br. J. Radiol. 2008, 81, 120–128. [Google Scholar] [CrossRef] [PubMed]
- Monti, S.; Aiello, M.; Incoronato, M.; Grimaldi, A.M.; Moscarino, M.; Mirabelli, P.; Ferbo, U.; Cavaliere, C.; Salvatore, M. DCE-MRI Pharmacokinetic-Based Phenotyping of Invasive Ductal Carcinoma: A Radiomic Study for Prediction of Histological Outcomes. Contrast Media Mol. Imaging 2018, 2018, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Zaric, O.; Pinker, K.; Zbyn, S.; Strasser, B.; Robinson, S.; Minarikova, L.; Gruber, S.; Farr, A.; Singer, C.; Helbich, T.H.; et al. Quantitative Sodium MR Imaging at 7 T: Initial Results and Comparison with Diffusion-weighted Imaging in Patients with Breast Tumors. Radiology 2016, 280, 39–48. [Google Scholar] [CrossRef] [PubMed]
- Pinker, K.; Baltzer, P.; Bogner, W.; Leithner, D.; Trattnig, S.; Zaric, O.; Dubsky, P.; Bago-Horvath, Z.; Rudas, M.; Gruber, S.; et al. Multiparametric MR Imaging with High-Resolution Dynamic Contrast-enhanced and Diffusion-weighted Imaging at 7 T Improves the Assessment of Breast Tumors: A Feasibility Study. Radiology 2015, 276, 360–370. [Google Scholar] [CrossRef] [PubMed]
- Bogner, W.; Pinker, K.; Zaric, O.; Baltzer, P.; Minarikova, L.; Porter, D.; Bago-Horvath, Z.; Dubsky, P.; Helbich, T.H.; Trattnig, S.; et al. Bilateral diffusion-weighted MR imaging of breast tumors with submillimeter resolution using readout-segmented echo-planar imaging at 7 T. Radiology 2015, 274, 74–84. [Google Scholar] [CrossRef] [PubMed]
- Pinker, K.; Bogner, W.; Baltzer, P.; Trattnig, S.; Gruber, S.; Abeyakoon, O.; Bernathova, M.; Zaric, O.; Dubsky, P.; Bago-Horvath, Z.; et al. Clinical application of bilateral high temporal and spatial resolution dynamic contrast-enhanced magnetic resonance imaging of the breast at 7 T. Eur. Radiol. 2014, 24, 913–920. [Google Scholar] [CrossRef]
- Kuhl, C.K.; Bieling, H.B.; Gieseke, J.; Kreft, B.P.; Sommer, T.; Lutterbey, G.; Schild, H.H. Healthy premenopausal breast parenchyma in dynamic contrast-enhanced MR imaging of the breast: Normal contrast medium enhancement and cyclical-phase dependency. Radiology 1997, 203, 137–144. [Google Scholar] [CrossRef]
- Amarosa, A.R.; McKellop, J.; Klautau Leite, A.P.; Moccaldi, M.; Clendenen, T.V.; Babb, J.S.; Zeleniuch-Jacquotte, A.; Moy, L.; Kim, S. Evaluation of the kinetic properties of background parenchymal enhancement throughout the phases of the menstrual cycle. Radiology 2013, 268, 356–365. [Google Scholar] [CrossRef] [Green Version]
- Berg, W.A.; Campassi, C.; Langenberg, P.; Sexton, M.J. ACR BI-RADS® Magnetic Resonance Imaging. In ACR BI-RADS® Atlas, Breast Imaging Reporting and Data System; American College of Radiology: Reston, VA, USA, 2013. [Google Scholar]
- Sung, K. DCE Tool Plugin. Version 2.2. Available online: http://kyungs.bol.ucla.edu/software/DCE_tool/DCE_tool.html (accessed on 19 August 2019).
- Rosset, A.; Spadola, L.; Ratib, O. OsiriX: An open-source software for navigating in multidimensional DICOM images. J. Digit. Imaging 2004, 17, 205–216. [Google Scholar] [CrossRef] [Green Version]
- Ma, Z.S.; Wang, D.W.; Sun, X.B.; Shi, H.; Pang, T.; Dong, G.Q.; Zhang, C.Q. Quantitative analysis of 3-Tesla magnetic resonance imaging in the differential diagnosis of breast lesions. Exp. Ther. Med. 2015, 9, 913–918. [Google Scholar] [CrossRef] [Green Version]
- Woolf, D.K.; Taylor, N.J.; Makris, A.; Tunariu, N.; Collins, D.J.; Li, S.P.; Ah-See, M.L.; Beresford, M.; Padhani, A.R. Arterial input functions in dynamic contrast-enhanced magnetic resonance imaging: Which model performs best when assessing breast cancer response? Br. J. Radiol. 2016, 89, 20150961. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bloom, H.J.; Richardson, W.W. Histological grading and prognosis in breast cancer; a study of 1409 cases of which 359 have been followed for 15 years. Br. J. Cancer 1957, 11, 359–377. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elston, C.W.; Ellis, I.O. Pathological prognostic factors in breast cancer. I. The value of histological grade in breast cancer: Experience from a large study with long-term follow-up. Histopathology 1991, 19, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Senn, H.J. St. Gallen consensus 2013: Optimizing and personalizing primary curative therapy of breast cancer worldwide. Breast Care 2013, 8, 101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goldhirsch, A.; Winer, E.P.; Coates, A.S.; Gelber, R.D.; Piccart-Gebhart, M.; Thurlimann, B.; Senn, H.J.; Panel, m. Personalizing the treatment of women with early breast cancer: Highlights of the St Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2013. Ann. Oncol. 2013, 24, 2206–2223. [Google Scholar] [CrossRef] [PubMed]
- Blows, F.M.; Driver, K.E.; Schmidt, M.K.; Broeks, A.; van Leeuwen, F.E.; Wesseling, J.; Cheang, M.C.; Gelmon, K.; Nielsen, T.O.; Blomqvist, C.; et al. Subtyping of breast cancer by immunohistochemistry to investigate a relationship between subtype and short and long term survival: A collaborative analysis of data for 10,159 cases from 12 studies. PLoS Med. 2010, 7, e1000279. [Google Scholar] [CrossRef]
- Dowsett, M.; Nielsen, T.O.; A’Hern, R.; Bartlett, J.; Coombes, R.C.; Cuzick, J.; Ellis, M.; Henry, N.L.; Hugh, J.C.; Lively, T.; et al. Assessment of Ki67 in breast cancer: Recommendations from the International Ki67 in Breast Cancer working group. J. Natl. Cancer Inst. 2011, 103, 1656–1664. [Google Scholar] [CrossRef] [Green Version]
- Kang, S.R.; Kim, H.W.; Kim, H.S. Evaluating the Relationship Between Dynamic Contrast-Enhanced MRI (DCE-MRI) Parameters and Pathological Characteristics in Breast Cancer. J. Magn. Reson. Imaging 2020, 52, 1360–1373. [Google Scholar] [CrossRef]
- Shin, J.K.; Kim, J.Y. Dynamic contrast-enhanced and diffusion-weighted MRI of estrogen receptor-positive invasive breast cancers: Associations between quantitative MR parameters and Ki-67 proliferation status. J. Magn. Reson. Imaging 2016, 45, 94–102. [Google Scholar] [CrossRef]
- Boser, B.E.; Guyon, I.M.; Vapnik, V.N. A training algorithm for optimal margin classifiers. In Proceedings of the Fifth Annual Workshop on Computational Learning Theory, Pittsburgh, PA, USA, 12 September 2020; pp. 144–152. [Google Scholar]
- R Documentation. Support Vector Machines. Available online: https://www.rdocumentation.org/packages/e1071/versions/1.7-3/topics/svm (accessed on 10 August 2020).
- Wu, C.; Pineda, F.; Hormuth, D.A., 2nd; Karczmar, G.S.; Yankeelov, T.E. Quantitative analysis of vascular properties derived from ultrafast DCE-MRI to discriminate malignant and benign breast tumors. Magn. Reson. Med. 2019, 81, 2147–2160. [Google Scholar] [CrossRef]
- Amarnath, J.; Sangeeta, T.; Mehta, S.B. Role of quantitative pharmacokinetic parameter (transfer constant: K(trans)) in the characterization of breast lesions on MRI. Indian J. Radiol. Imaging 2013, 23, 19–25. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Ai, T.; Hu, Y.; Yan, X.; Nickel, M.D.; Xu, X.; Xia, L. Application of whole-lesion histogram analysis of pharmacokinetic parameters in dynamic contrast-enhanced MRI of breast lesions with the CAIPIRINHA-Dixon-TWIST-VIBE technique. J. Magn. Reson. Imaging 2018, 47, 91–96. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Z.; Wu, Z.; Shi, G.; Yi, Z.; Xie, M.; Zeng, W.; Song, C.; Zheng, C.; Shen, J. Discrimination between benign and malignant breast lesions using volumetric quantitative dynamic contrast-enhanced MR imaging. Eur. Radiol. 2018, 28, 982–991. [Google Scholar] [CrossRef] [PubMed]
- Koo, H.R.; Cho, N.; Song, I.C.; Kim, H.; Chang, J.M.; Yi, A.; Yun, B.L.; Moon, W.K. Correlation of perfusion parameters on dynamic contrast-enhanced MRI with prognostic factors and subtypes of breast cancers. J. Magn. Reson. Imaging 2012, 36, 145–151. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.S.; Kim, S.H.; Kang, B.J.; Baek, J.E.; Song, B.J. Perfusion Parameters in Dynamic Contrast-enhanced MRI and Apparent Diffusion Coefficient Value in Diffusion-weighted MRI: Association with Prognostic Factors in Breast Cancer. Acad. Radiol. 2016, 23, 446–456. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Wang, M.; Li, H. Role of perfusion parameters on DCE-MRI and ADC values on DWMRI for invasive ductal carcinoma at 3.0 Tesla. World J. Surg. Oncol. 2018, 16, 239. [Google Scholar] [CrossRef] [PubMed]
- Morris, E.A.; Comstock, C.; Lee, C.; Lehman, C.D.; Ikeda, D.M.; Newstead, G.M.; Tozaki, M.; Hylton, N.; Hlbich, T.H.; Kuhl, C.; et al. ACR BI-RADS® Atlas, Breast Imaging Reporting and Data System, 5th ed.; American College of Radiology: Reston, VA, USA, 2013. [Google Scholar]
- Ioannidis, G.S.; Maris, T.G.; Nikiforaki, K.; Karantanas, A.; Marias, K. Investigating the Correlation of Ktrans With Semi-Quantitative MRI Parameters Towards More Robust and Reproducible Perfusion Imaging Biomarkers in Three Cancer Types. IEEE J. Biomed. Health Inform. 2018, 23, 1855–1862. [Google Scholar] [CrossRef] [PubMed]
- Brooks, G.P.; Johanson, G.A. Sample size considerations for multiple comparison procedures in ANOVA. J. Mod. Appl. Stat. Methods 2011, 10, 97–109. [Google Scholar] [CrossRef] [Green Version]


| Benign (n = 16) | ||||
| n | Mean size (mm) | Proliferation rate (n) | Molecular subtype (n) | |
| All benign lesions | 16 | 21.9 (8–45) | n/a | n/a |
| Histopathology | ||||
| Sclerosis adenosis | 3 | 20 (10–40) | n/a | n/a |
| Fibrosis | 1 | 13 | n/a | n/a |
| Fibroadenoma | 11 | 24.5 (10–45) | n/a | n/a |
| Papilloma | 1 | 8 mm | n/a | n/a |
| Malignant (n =27) | ||||
| n | Mean size (mm) | Proliferation rate (n) | Molecular subtype (n) | |
| All malignant lesions | 27 | 24.1 (6–95) | n/a | n/a |
| Histopathology | ||||
| Invasive ductal carcinoma | 23 | |||
| Grade 1 | 3 | 12.3 (11–15) | <15% (3), ≥ 15% (0) | Lum A (3) |
| Grade 2 | 10 | 23.7 (9–40) | <15% (2), ≥ 15% (8) | Lum A (2), Lum B (8) |
| Grade 3 | 10 | 32.7 (14–95) | <15% (0), ≥ 15% (10) | Lum B (6), HER2+ (2), TN (2) |
| Invasive lobular Carcinoma | 3 | |||
| Grade 1 | 0 | n/a | n/a | n/a |
| Grade 2 | 3 | 23 (6–30) | <15% (0), ≥ 15% (3) | Lum B (3) |
| Grade 3 | 0 | n/a | n/a | n/a |
| Carcinoma | 1 | |||
| Grade 1 | 0 | n/a | n/a | n/a |
| Grade 2 | 1 | 10 mm | <15% (0), ≥ 15% (1) | HER2+ (1) |
| Grade 3 | 0 | n/a | n/a | n/a |
| Benign vs. Malignant | Luminal A vs. Other Molecular Subtypes | Luminal A/B vs. Other Molecular Subtypes | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Benign (n = 16) 1 | Malignant (n = 27) 1 | p-Value 2 | Luminal A (n = 5) 1 | Others, (n = 22) 1 | p-Value 2 | Luminal A/B (n = 22) 1 | Others (n = 5) 1 | p-Value 2 | |
| Reader 1 | |||||||||
| KTrans-wtROI | 0.14 (0.06, 0.31) | 0.29 (0.21, 0.42) | 0.010 | 0.26 (0.18, 0.35) | 0.30 (0.22, 0.44) | 0.3 | 0.26 (0.20, 0.38) | 0.42 (0.35, 0.52) | 0.11 |
| KTrans-sROI | 0.21 (0.11, 0.37) | 0.38 (0.28, 0.51) | 0.005 | 0.36 (0.21, 0.39) | 0.39 (0.29, 0.52) | 0.2 | 0.35 (0.26, 0.40) | 0.52 (0.47, 0.52) | 0.086 |
| kep-wtROI | 0.19 (0.11, 0.45) | 0.43 (0.33, 0.63) | 0.005 | 0.41 (0.32, 0.43) | 0.48 (0.34, 0.69) | 0.4 | 0.40 (0.33, 0.56) | 0.52 (0.51, 0.70) | 0.3 |
| kep-sROI | 0.31 (0.13, 0.50) | 0.52 (0.43, 0.68) | 0.011 | 0.49 (0.44, 0.51) | 0.57 (0.44, 0.69) | 0.3 | 0.51 (0.43, 0.68) | 0.67 (0.61, 0.70) | 0.5 |
| Ve-wtROI | 0.90 (0.77, 1.00) | 0.77 (0.57, 0.86) | 0.015 | 0.81 (0.57, 0.82) | 0.76 (0.60, 0.86) | >0.9 | 0.78 (0.53, 0.86) | 0.75 (0.75, 0.83) | 0.7 |
| Ve-sROI | 0.84 (0.75, 0.95) | 0.75 (0.72, 0.81) | 0.095 | 0.79 (0.51, 0.79) | 0.75 (0.73, 0.81) | 0.8 | 0.75 (0.62, 0.81) | 0.77 (0.75, 0.80) | 0.3 |
| Reader 2 | |||||||||
| KTrans-wtROI | 0.12 (0.10, 0.35) | 0.28 (0.22, 0.41) | 0.032 | 0.24 (0.14, 0.32) | 0.28 (0.23, 0.43) | 0.2 | 0.25 (0.22, 0.35) | 0.42 (0.38, 0.52) | 0.033 |
| KTrans-sROI | 0.18 (0.10, 0.35) | 0.31 (0.24, 0.51) | 0.025 | 0.28 (0.14, 0.37) | 0.32 (0.26, 0.52) | 0.3 | 0.29 (0.20, 0.43) | 0.52 (0.34, 0.52) | 0.086 |
| kep-wtROI | 0.18 (0.12, 0.44) | 0.40 (0.29, 0.55) | 0.044 | 0.38 (0.28, 0.44) | 0.40 (0.30, 0.57) | 0.5 | 0.38 (0.27, 0.50) | 0.54 (0.48, 0.70) | 0.11 |
| kep-sROI | 0.24 (0.11, 0.50) | 0.50 (0.35, 0.68) | 0.032 | 0.46 (0.39, 0.50) | 0.53 (0.34, 0.69) | 0.4 | 0.48 (0.31, 0.63) | 0.68 (0.41, 0.70) | 0.3 |
| Ve-wtROI | 0.82 (0.74, 0.95) | 0.79 (0.75, 0.88) | 0.4 | 0.82 (0.60, 0.84) | 0.78 (0.75, 0.88) | 0.5 | 0.81 (0.72, 0.88) | 0.78 (0.75, 0.78) | 0.7 |
| Ve-sROI | 0.81 (0.75, 0.93) | 0.76 (0.68, 0.80) | 0.10 | 0.73 (0.61, 0.79) | 0.76 (0.72, 0.82) | 0.4 | 0.76 (0.62, 0.79) | 0.79 (0.75, 0.83) | 0.3 |
| Metric/Measure | Correlation | p-Value |
|---|---|---|
| KTrans-wtROI | 0.772476 | 3.20 × 10−13 |
| KTrans-sROI | 0.75388 | 1.10 × 10−12 |
| kep-wtROI | 0.702163 | 3.46 × 10−11 |
| kep-sROI | 0.606767 | 1.03 × 10−8 |
| Ve-wtROI | 0.527621 | 8.94 × 1−7 |
| Ve-sROI | 0.476563 | 7.70 × 10−6 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ochoa-Albiztegui, R.E.; Sevilimedu, V.; Horvat, J.V.; Thakur, S.B.; Helbich, T.H.; Trattnig, S.; Morris, E.A.; Reiner, J.S.; Pinker, K. Pharmacokinetic Analysis of Dynamic Contrast-Enhanced Magnetic Resonance Imaging at 7T for Breast Cancer Diagnosis and Characterization. Cancers 2020, 12, 3763. https://doi.org/10.3390/cancers12123763
Ochoa-Albiztegui RE, Sevilimedu V, Horvat JV, Thakur SB, Helbich TH, Trattnig S, Morris EA, Reiner JS, Pinker K. Pharmacokinetic Analysis of Dynamic Contrast-Enhanced Magnetic Resonance Imaging at 7T for Breast Cancer Diagnosis and Characterization. Cancers. 2020; 12(12):3763. https://doi.org/10.3390/cancers12123763
Chicago/Turabian StyleOchoa-Albiztegui, R. Elena, Varadan Sevilimedu, Joao V. Horvat, Sunitha B. Thakur, Thomas H. Helbich, Siegfried Trattnig, Elizabeth A. Morris, Jeffrey S. Reiner, and Katja Pinker. 2020. "Pharmacokinetic Analysis of Dynamic Contrast-Enhanced Magnetic Resonance Imaging at 7T for Breast Cancer Diagnosis and Characterization" Cancers 12, no. 12: 3763. https://doi.org/10.3390/cancers12123763
APA StyleOchoa-Albiztegui, R. E., Sevilimedu, V., Horvat, J. V., Thakur, S. B., Helbich, T. H., Trattnig, S., Morris, E. A., Reiner, J. S., & Pinker, K. (2020). Pharmacokinetic Analysis of Dynamic Contrast-Enhanced Magnetic Resonance Imaging at 7T for Breast Cancer Diagnosis and Characterization. Cancers, 12(12), 3763. https://doi.org/10.3390/cancers12123763








