Comparison of Mutated KRAS and Methylated HOXA9 Tumor-Specific DNA in Advanced Lung Adenocarcinoma
Abstract
:Simple Summary
Abstract
1. Introduction
2. Results
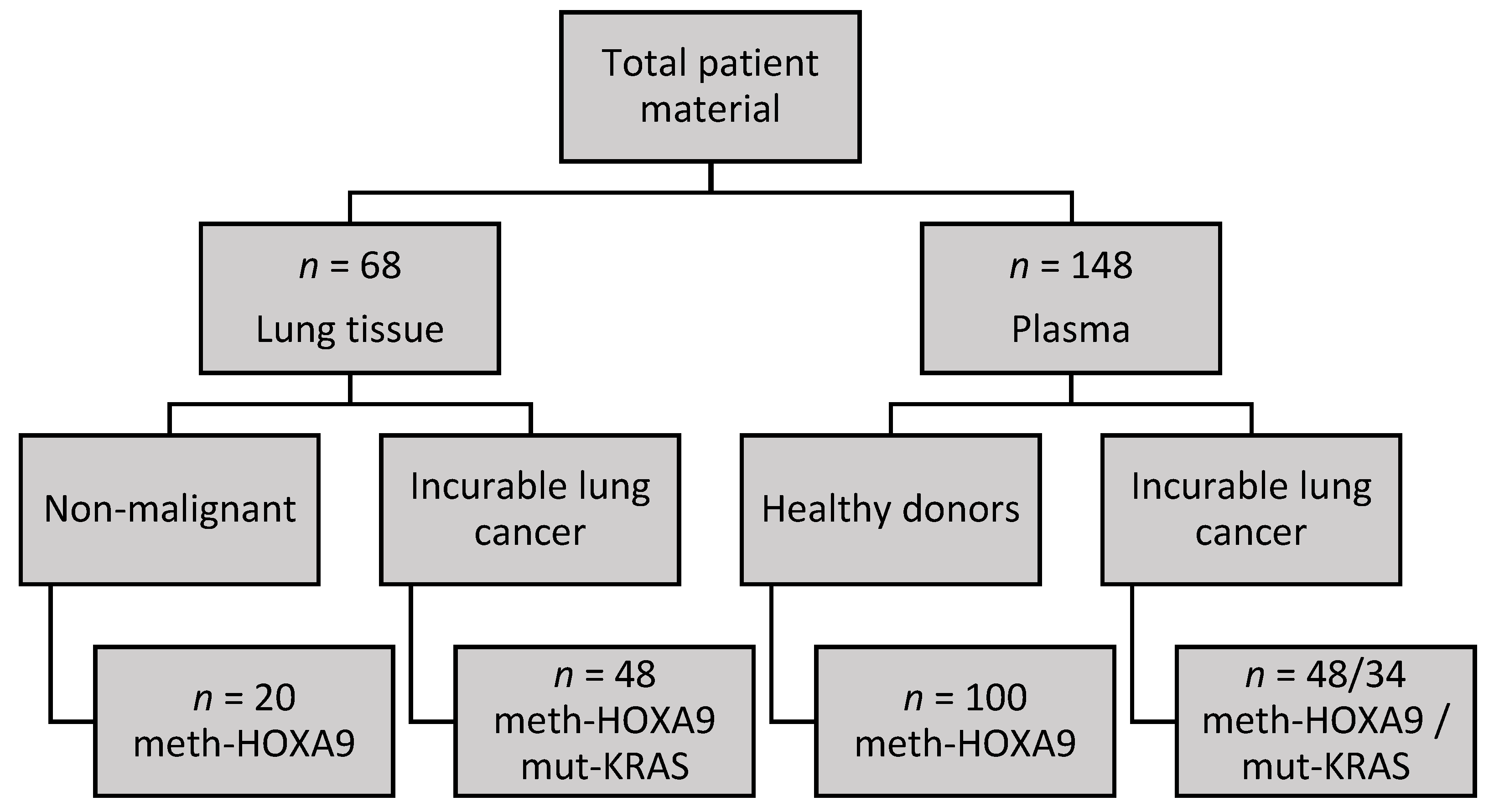
2.1. Cohort Characteristics
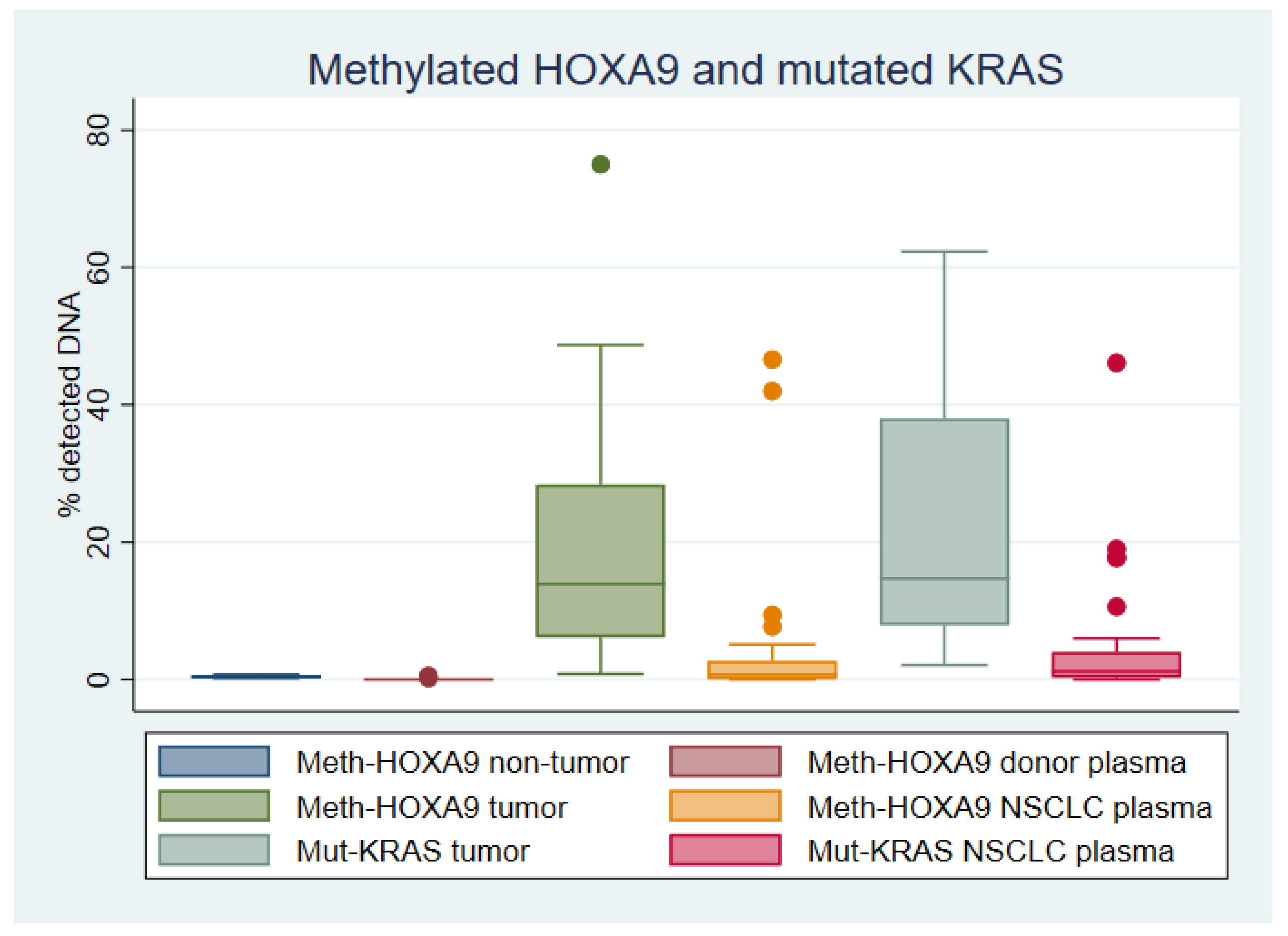
2.2. HOXA9 Methylation in Lung Tissue
2.3. HOXA9 Methylation in Plasma
2.4. KRAS Mutations in Lung Adenocarcinoma Tissue
2.5. KRAS Mutations in Plasma
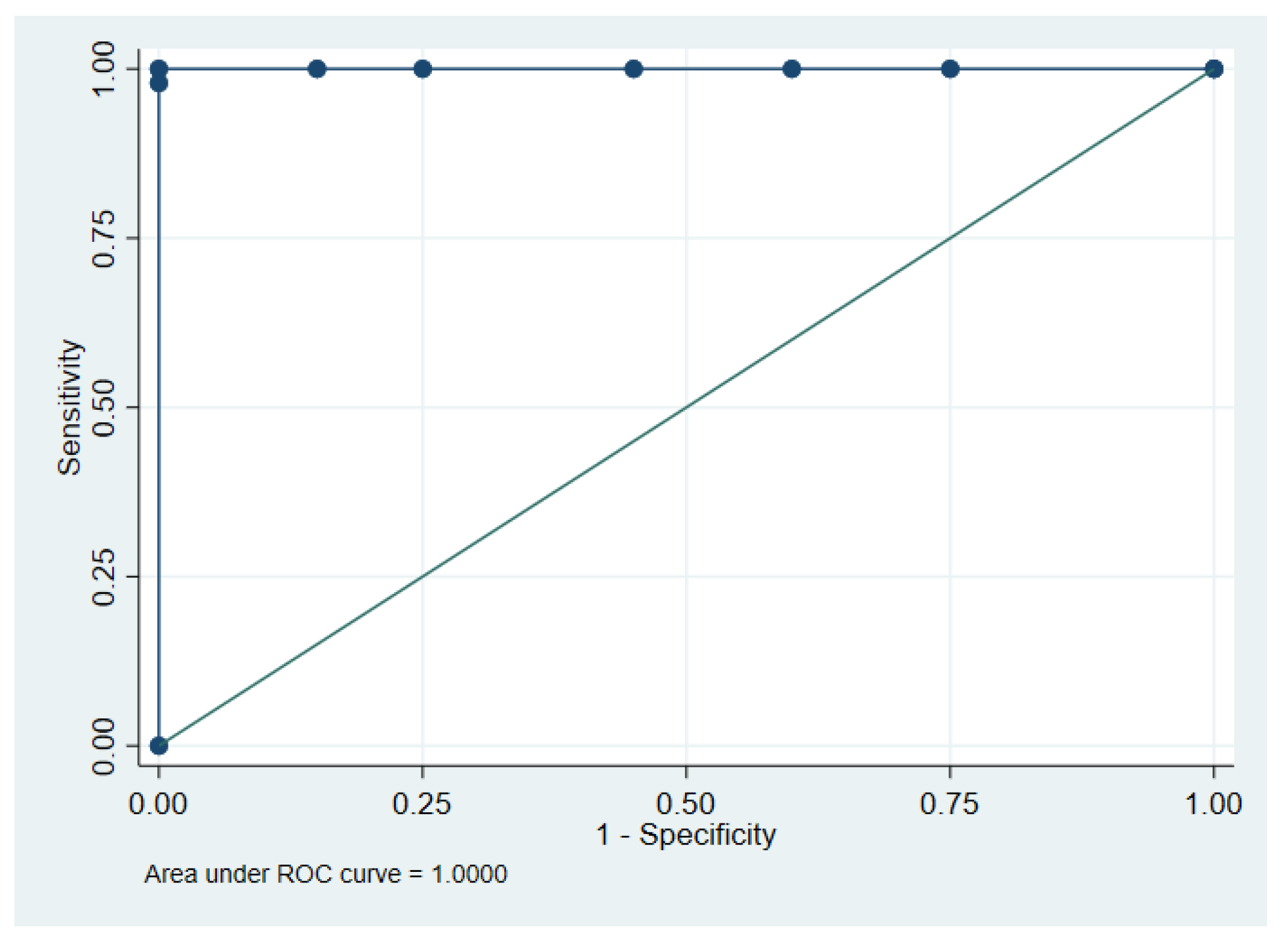
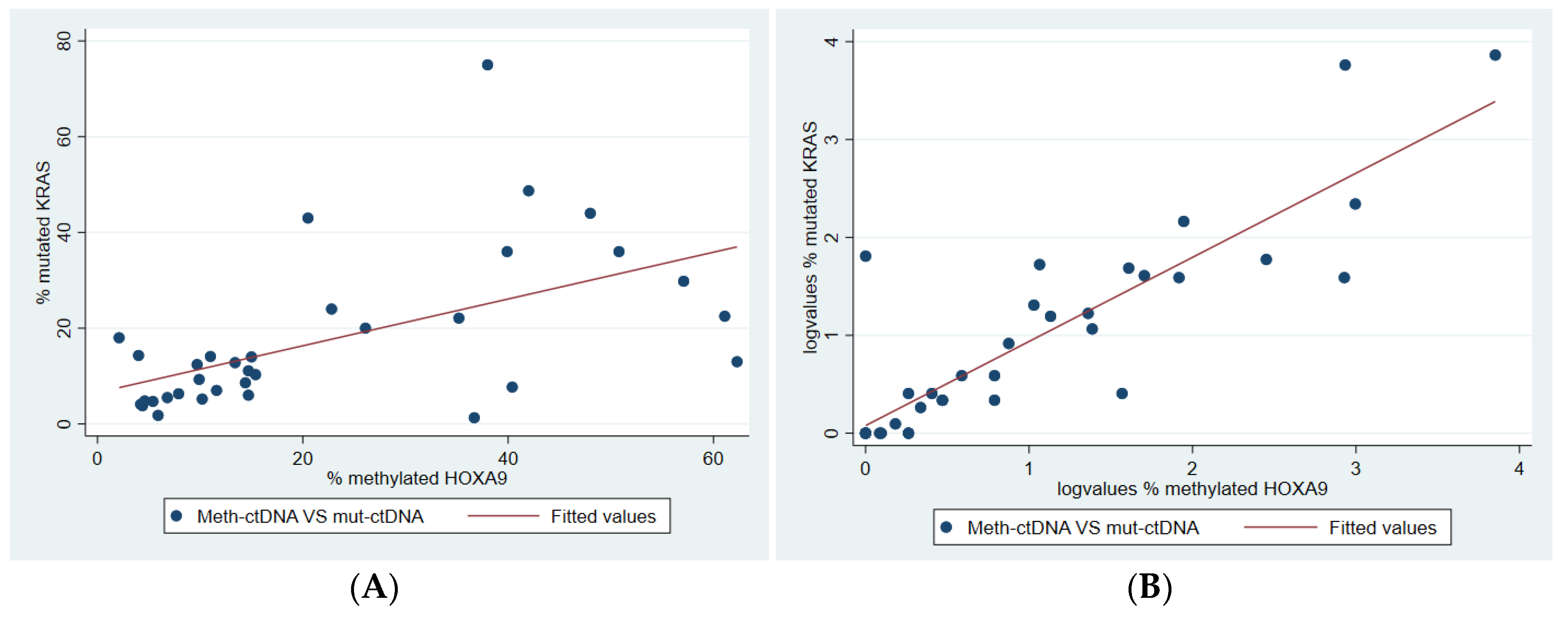
2.6. Correlation Between Methylated and Mutated ctDNA
3. Discussion
4. Materials and Methods
4.1. Participants
4.2. Samples
4.3. DNA Extraction
4.4. HOXA9 Methylation Analysis
4.5. Methylated HOXA9 Limit of Blank and Limit of Detection
4.6. KRAS Mutations Analyzed by NGS
4.7. KRAS Mutations Analyzed by ddPCR
4.8. KRAS Mutation Limit of Blank and Limit of Detection
4.9. Statistical Analyses
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [Green Version]
- Jakobsen, E.; Rasmussen, T.R.; Green, A. Mortality and survival of lung cancer in Denmark: Results from the Danish Lung Cancer Group 2000–2012. Acta Oncol. 2016, 55, 2–9. [Google Scholar] [CrossRef] [Green Version]
- Howlader, N.; Noone, A.M.; Krapcho, M.; Miller, D.; Bishop, K.; Kosary, C.L.; Yu, M.; Ruhl, J.; Tatalovich, Z.; Mariotto, A.; et al. SEER Cancer Statistics Review, 1975–2014, National Cancer Institute; Bethesda: Montgomery, MD, USA, 2017; based on November 2016 SEER data submission, posted to the SEER web site, April 2017. Available online: https://seer.cancer.gov/csr/1975_2014/ (accessed on 1 July 2020).
- Hulbert, A.; Jusue-Torres, I.; Stark, A.; Chen, C.; Rodgers, K.; Lee, B.; Griffin, C.; Yang, A.; Huang, P.; Wrangle, J.; et al. Early Detection of Lung Cancer Using DNA Promoter Hypermethylation in Plasma and Sputum. Clin. Cancer Res. 2017, 23, 1998–2005. [Google Scholar] [CrossRef] [Green Version]
- Roncarati, R.; Lupini, L.; Miotto, E.; Saccenti, E.; Mascetti, S.; Morandi, L.; Bassi, C.; Rasio, D.; Callegari, E.; Conti, V.; et al. Molecular testing on bronchial washings for the diagnosis and predictive assessment of lung cancer. Mol. Oncol. 2020, 14, 2163–2175. [Google Scholar] [CrossRef]
- Liu, B.; Filho, J.R.; Mallisetty, A.; Villani, C.; Kottorou, A.E.; Rodgers, K.P.; Chen, C.; Ito, T.; Holmes, K.; Gastala, N.; et al. Detection of Promoter DNA Methylation in Urine and Plasma Aids the Detection of Non–Small Cell Lung Cancer. Clin. Cancer Res. 2020, 26, 4339–4348. [Google Scholar] [CrossRef]
- Merker, J.D.; Oxnard, G.R.; Compton, C.; Diehn, M.; Hurley, P.; Lazar, A.J.; Lindeman, N.; Lockwood, C.M.; Rai, A.J.; Schilsky, R.L.; et al. Circulating Tumor DNA Analysis in Patients With Cancer: American Society of Clinical Oncology and College of American Pathologists Joint Review. J. Clin. Oncol. 2018, 36, 1631–1641. [Google Scholar] [CrossRef]
- Siravegna, G.; Mussolin, B.; Venesio, T.; Marsoni, S.; Seoane, J.; Dive, C.; Papadopoulos, N.; Kopetz, S.; Corcoran, R.; Siu, L.; et al. How liquid biopsies can change clinical practice in oncology. Ann. Oncol. 2019, 30, 1580–1590. [Google Scholar] [CrossRef] [Green Version]
- McGranahan, N.; Favero, F.; De Bruin, E.C.; Birkbak, N.J.; Szallasi, Z.I.; Swanton, C. Clonal status of actionable driver events and the timing of mutational processes in cancer evolution. Sci. Transl. Med. 2015, 7, 283ra54. [Google Scholar] [CrossRef] [Green Version]
- De Bruin, E.C.; McGranahan, N.; Mitter, R.; Salm, M.; Wedge, D.C.; Yates, L.; Jamal-Hanjani, M.; Shafi, S.; Murugaesu, N.; Rowan, A.J.; et al. Spatial and temporal diversity in genomic instability processes defines lung cancer evolution. Science 2014, 346, 251–263. [Google Scholar] [CrossRef] [Green Version]
- Nelson, M.A.; Wymer, J.; Clements, N. Detection of K-ras gene mutations in non-neoplastic lung tissue and lung cancers. Cancer Lett. 1996, 103, 115–121. [Google Scholar] [CrossRef]
- Suzuki, Y.; Orita, M.; Shiraishi, M.; Hayashi, K.; Sekiya, T. Detection of ras gene mutations in human lung cancers by single-strand conformation polymorphism analysis of polymerase chain reaction products. Oncogene 1990, 5, 1037–1043. [Google Scholar] [PubMed]
- Westcott, P.M.K.; To, M.D. The genetics and biology of KRAS in lung cancer. Chin. J. Cancer 2013, 32, 63–70. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bouras, E.; Karakioulaki, M.; Bougioukas, K.I.; Aivaliotis, M.; Tzimagiorgis, G.; Chourdakis, M. Gene promoter methylation and cancer: An umbrella review. Gene 2019, 710, 333–340. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Kelly, T.K.; Jones, P.A. Epigenetics in cancer. Carcinogenesis 2010, 31, 27–36. [Google Scholar] [CrossRef] [PubMed]
- Baylin, S.B. DNA methylation and gene silencing in cancer. Nat. Clin. Pract. Oncol. 2005, 2 (Suppl. 1), S4–S11. [Google Scholar] [CrossRef] [PubMed]
- Garrigou, S.; Perkins, G.; Garlan, F.; Normand, C.; Didelot, A.; Le Corre, D.; Peyvandi, S.; Mulot, C.; Niarra, R.; Aucouturier, P.; et al. A Study of Hypermethylated Circulating Tumor DNA as a Universal Colorectal Cancer Biomarker. Clin. Chem. 2016, 62, 1129–1139. [Google Scholar] [CrossRef] [Green Version]
- Fackler, M.J.; Bujanda, Z.L.; Umbricht, C.; Teo, W.W.; Cho, S.; Zhang, Z.; Visvanathan, K.; Jeter, S.; Argani, P.; Wang, C.; et al. Novel Methylated Biomarkers and a Robust Assay to Detect Circulating Tumor DNA in Metastatic Breast Cancer. Cancer Res. 2014, 74, 2160–2170. [Google Scholar] [CrossRef] [Green Version]
- Bhatlekar, S.; Fields, J.Z.; Boman, B.M. HOX genes and their role in the development of human cancers. J. Mol. Med. 2014, 92, 811–823. [Google Scholar] [CrossRef]
- Makiyama, K.; Hamada, J.-I.; Takada, M.; Murakawa, K.; Takahashi, Y.; Tada, M.; Tamoto, E.; Shindo, G.; Matsunaga, A.; Teramoto, K.-I.; et al. Aberrant expression of HOX genes in human invasive breast carcinoma. Oncol. Rep. 2005, 13, 673–679. [Google Scholar] [CrossRef] [Green Version]
- Singh, A.; Gupta, S.; Badarukhiya, J.A.; Sachan, M. Detection of aberrant methylation of HOXA9 and HIC1 through multiplex MethyLight assay in serum DNA for the early detection of epithelial ovarian cancer. Int. J. Cancer 2020, 147, 1740–1752. [Google Scholar] [CrossRef]
- Hwang, J.-A.; Bin Lee, B.; Kim, Y.; Hong, S.-H.; Kim, Y.-H.; Han, J.; Shim, Y.M.; Yoon, C.-Y.; Lee, Y.-S.; Kim, D.-H. HOXA9 inhibits migration of lung cancer cells and its hypermethylation is associated with recurrence in non-small cell lung cancer. Mol. Carcinog. 2014, 54, E72–E80. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.-L.; Lee, D.C.; Sohn, H.A.; Lee, S.-Y.; Jeon, H.S.; Lee, J.H.; Park, C.G.; Lee, H.Y.; Yeom, Y.I.; Son, J.W.; et al. Homeobox A9 directly targeted by miR-196b regulates aggressiveness through nuclear Factor-kappa B activity in non-small cell lung cancer cells. Mol. Carcinog. 2015, 55, 1915–1926. [Google Scholar] [CrossRef] [PubMed]
- Nawaz, I.; Qiu, X.; Wu, H.; Li, Y.; Fan, Y.; Hu, L.-F.; Zhou, Q.; Ernberg, I. Development of a multiplex methylation specific PCR suitable for (early) detection of non-small cell lung cancer. Epigenetics 2014, 9, 1138–1148. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wrangle, J.; Machida, E.O.; Danilova, L.; Hulbert, A.; Franco, N.; Zhang, W.; Glöckner, S.C.; Tessema, M.; Van Neste, L.; Easwaran, H.P.; et al. Functional identification of cancer-specific methylation of CDO1, HOXA9, and TAC1 for the diagnosis of lung cancer. Clin. Cancer Res. 2014, 20, 1856–1864. [Google Scholar] [CrossRef] [Green Version]
- Ooki, A.; Maleki, Z.; Tsay, J.-C.J.; Goparaju, C.; Brait, M.; Turaga, N.; Nam, H.-S.; Rom, W.N.; Pass, H.I.; Sidransky, D.; et al. A Panel of Novel Detection and Prognostic Methylated DNA Markers in Primary Non–Small Cell Lung Cancer and Serum DNA. Clin. Cancer Res. 2017, 23, 7141–7152. [Google Scholar] [CrossRef] [Green Version]
- Ricciuti, B.; Leonardi, G.C.; Metro, G.; Grignani, F.; Paglialunga, L.; Bellezza, G.; Baglivo, S.; Mencaroni, C.; Baldi, A.; Zicari, D.; et al. Targeting the KRAS variant for treatment of non-small cell lung cancer: Potential therapeutic applications. Expert Rev. Respir. Med. 2015, 10, 53–68. [Google Scholar] [CrossRef]
- Lampignano, R.; Neumann, M.H.D.; Weber, S.; Kloten, V.; Herdean, A.; Voss, T.; Groelz, D.; Babayan, A.; Tibbesma, M.; Schlumpberger, M.; et al. Multicenter Evaluation of Circulating Cell-Free DNA Extraction and Downstream Analyses for the Development of Standardized (Pre)analytical Work Flows. Clin. Chem. 2019, 66, 149–160. [Google Scholar] [CrossRef]
- Milbury, C.A.; Zhong, Q.; Lin, J.; Williams, M.; Olson, J.; Link, D.R.; Hutchison, B.J. Determining lower limits of detection of digital PCR assays for cancer-related gene mutations. Biomol. Detect. Quantif. 2014, 1, 8–22. [Google Scholar] [CrossRef] [Green Version]
- Cho, M.-S.; Park, C.H.; Lee, S.; Park, H.S. Clinicopathological parameters for circulating tumor DNA shedding in surgically resected non-small cell lung cancer with EGFR or KRAS mutation. PLoS ONE 2020, 15, e0230622. [Google Scholar] [CrossRef] [Green Version]
- Thomsen, C.B.; Andersen, R.F.; Lindebjerg, J.; Hansen, T.F.; Jensen, L.H.; Jakobsen, A. Correlation Between Tumor-Specific Mutated and Methylated DNA in Colorectal Cancer. JCO Precis. Oncol. 2019, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Thomsen, C.B.; Andersen, R.F.; Steffensen, K.D.; Adimi, P.; Jakobsen, A. Delta tocotrienol in recurrent ovarian cancer. A phase II trial. Pharmacol. Res. 2019, 141, 392–396. [Google Scholar] [CrossRef]
- Rusan, M.; Andersen, R.F.; Jakobsen, A.; Steffensen, K.D. Circulating HOXA9-methylated tumour DNA: A novel biomarker of response to poly (ADP-ribose) polymerase inhibition in BRCA-mutated epithelial ovarian cancer. Eur. J. Cancer 2020, 125, 121–129. [Google Scholar] [CrossRef]
- Pallisgaard, N.; Spindler, K.-L.G.; Andersen, R.F.; Brandslund, I.; Jakobsen, A. Controls to validate plasma samples for cell free DNA quantification. Clin. Chim. Acta 2015, 446, 141–146. [Google Scholar] [CrossRef]
- Bio-Rad. Droplet Digital PCR Applications Guide. 2019. Available online: http://www.bio-rad.com/webroot/web/pdf/lsr/literature/Bulletin_6407.pdf (accessed on 27 November 2019).
- Illumina. TruSight Tumor 15 reference guide, Document # 1000000001245 v07. 2020. Available online: https://emea.support.illumina.com/content/dam/illumina-support/documents/documentation/chemistry_documentation/trusight/trusight-tumor-15-reference-guide-1000000001245-07.pdf (accessed on 10 December 2020).
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wen, S.W.C.; Andersen, R.F.; Petersen, L.M.S.; Hager, H.; Hilberg, O.; Jakobsen, A.; Hansen, T.F. Comparison of Mutated KRAS and Methylated HOXA9 Tumor-Specific DNA in Advanced Lung Adenocarcinoma. Cancers 2020, 12, 3728. https://doi.org/10.3390/cancers12123728
Wen SWC, Andersen RF, Petersen LMS, Hager H, Hilberg O, Jakobsen A, Hansen TF. Comparison of Mutated KRAS and Methylated HOXA9 Tumor-Specific DNA in Advanced Lung Adenocarcinoma. Cancers. 2020; 12(12):3728. https://doi.org/10.3390/cancers12123728
Chicago/Turabian StyleWen, Sara W. C., Rikke F. Andersen, Lena Marie S. Petersen, Henrik Hager, Ole Hilberg, Anders Jakobsen, and Torben F. Hansen. 2020. "Comparison of Mutated KRAS and Methylated HOXA9 Tumor-Specific DNA in Advanced Lung Adenocarcinoma" Cancers 12, no. 12: 3728. https://doi.org/10.3390/cancers12123728
APA StyleWen, S. W. C., Andersen, R. F., Petersen, L. M. S., Hager, H., Hilberg, O., Jakobsen, A., & Hansen, T. F. (2020). Comparison of Mutated KRAS and Methylated HOXA9 Tumor-Specific DNA in Advanced Lung Adenocarcinoma. Cancers, 12(12), 3728. https://doi.org/10.3390/cancers12123728







