Circulating Cell-Free Tumour DNA for Early Detection of Pancreatic Cancer
Abstract
Simple Summary
Abstract
1. Introduction
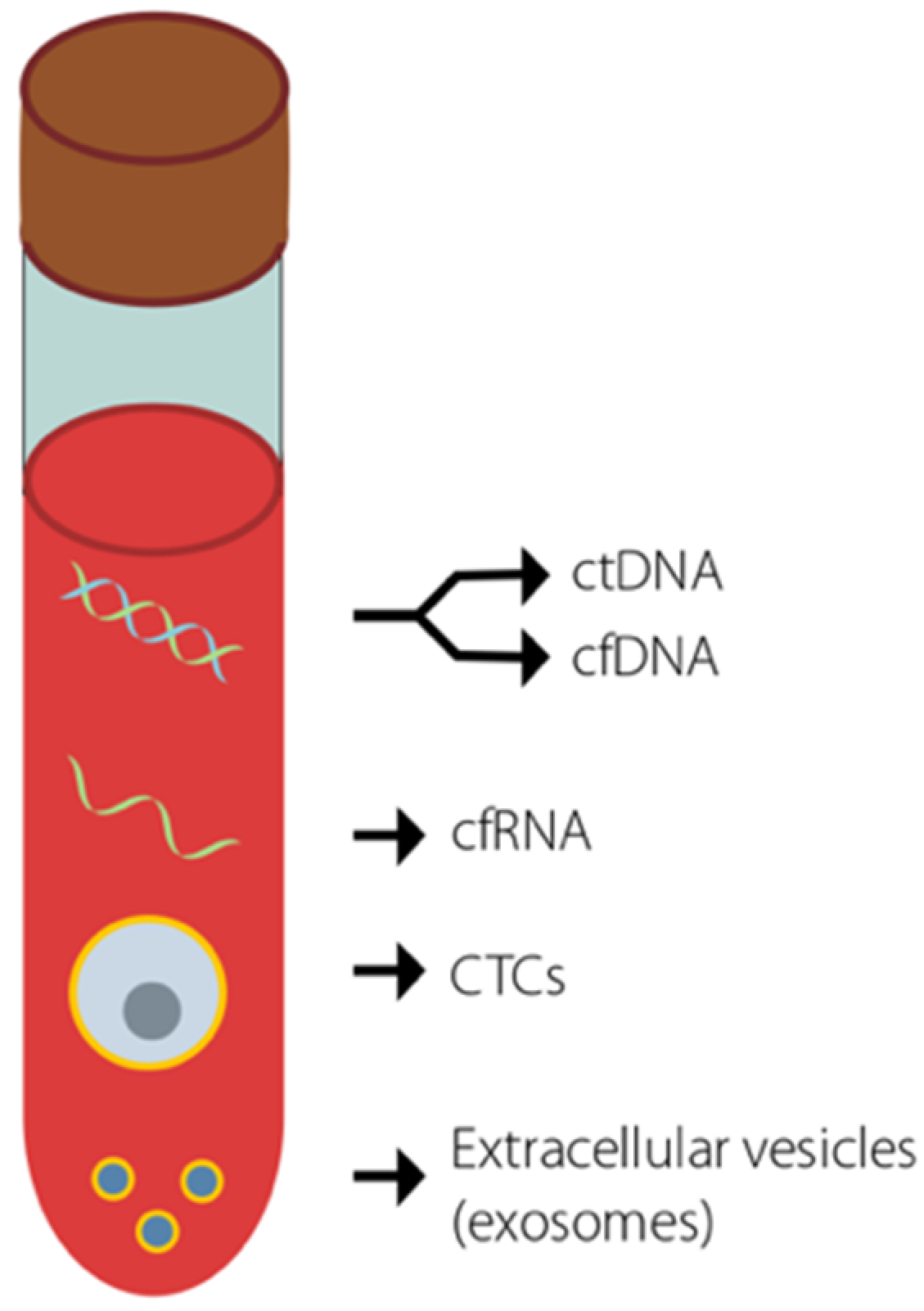
2. Liquid Biopsies for ctDNA
3. Detection of ctDNA
4. ctDNA in Pancreatic Cancer
5. The Use of ctDNA to Detect Early Stage Pancreatic Cancer
6. Methylation Analysis in Cancer Diagnosis Based on Liquid Biopsy
7. Technical Advances Facilitate the Detection and Analysis of ctDNA and Also Unravel Its Properties
8. Future Directions
9. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Rahib, L.; Smith, B.D.; Aizenberg, R.; Rosenzweig, A.B.; Fleshman, J.M.; Matrisian, L.M. Projecting cancer incidence and deaths to 2030: The unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 2014, 74, 2913–2921. [Google Scholar] [CrossRef] [PubMed]
- Font-Burgada, J.; Sun, B.; Karin, M. Obesity and Cancer: The Oil that Feeds the Flame. Cell Metab. 2016, 23, 48–62. [Google Scholar] [CrossRef] [PubMed]
- Cancer Statistics. Available online: https://seer.cancer.gov/statistics/ (accessed on 19 November 2020).
- Cancer Research UK. Available online: https://www.cancerresearchuk.org/ (accessed on 19 November 2020).
- Werner, J.; Combs, S.E.; Springfeld, C.; Hartwig, W.; Hackert, T.; Büchler, M.W. Advanced-stage pancreatic cancer: Therapy options. Nat. Rev. Clin. Oncol. 2013, 10, 323–333. [Google Scholar] [CrossRef]
- Conroy, T.; Hammel, P.; Hebbar, M.; Ben Abdelghani, M.; Wei, A.C.; Raoul, J.-L.; Choné, L.; Francois, E.; Artru, P.; Biagi, J.J.; et al. FOLFIRINOX or Gemcitabine as Adjuvant Therapy for Pancreatic Cancer. N. Engl. J. Med. 2018, 379, 2395–2406. [Google Scholar] [CrossRef]
- Ghaneh, P.; Palmer, D.H.; Cicconi, S.; Halloran, C.; Psarelli, E.E.; Rawcliffe, C.L.; Sripadam, R.; Mukherjee, S.; Wadsley, J.; Al-Mukhtar, A.; et al. ESPAC-5F: Four-arm, prospective, multicenter, international randomized phase II trial of immediate surgery compared with neoadjuvant gemcitabine plus capecitabine (GEMCAP) or FOLFIRINOX or chemoradiotherapy (CRT) in patients with borderline resectable pancreatic cancer. J. Clin. Oncol. 2020, 38, 4505. [Google Scholar] [CrossRef]
- Sohal, D.; Lew, D.L.; Ahmad, S.A.; Gandhi, N.; Beg, M.S.; Wang-Gillam, A.; Wade, J.L.; Guthrie, K.A.; Lowy, A.M.; Philip, P.A.; et al. SWOG S1505: Initial findings on eligibility and neoadjuvant chemotherapy experience with mFOLFIRINOX versus gemcitabine/nab-paclitaxel for resectable pancreatic adenocarcinoma. J. Clin. Oncol. 2019, 37, 4137. [Google Scholar] [CrossRef]
- Unno, M.; Motoi, F.; Matsuyama, Y.; Satoi, S.; Matsumoto, I.; Aosasa, S.; Shirakawa, H.; Wada, K.; Fujii, T.; Yoshitomi, H.; et al. Randomized phase II/III trial of neoadjuvant chemotherapy with gemcitabine and S-1 versus upfront surgery for resectable pancreatic cancer (Prep-02/JSAP-05). J. Clin. Oncol. 2019, 37, 189. [Google Scholar] [CrossRef]
- O’Reilly, E.M.; Perelshteyn, A.; Jarnagin, W.R.; Schattner, M.; Gerdes, H.; Capanu, M.; Tang, L.H.; LaValle, J.; Winston, C.; DeMatteo, R.P.; et al. A Single-Arm, Nonrandomized Phase II Trial of Neoadjuvant Gemcitabine and Oxaliplatin in Patients with Resectable Pancreas Adenocarcinoma. Ann. Surg. 2014, 260, 142–148. [Google Scholar] [CrossRef]
- Tajima, H.; Ohta, T.; Kitagawa, H.; Okamoto, K.; Sakai, S.; Makino, I.; Kinoshita, J.; Furukawa, H.; Nakamura, K.; Hayashi, H.; et al. Pilot study of neoadjuvant chemotherapy with gemcitabine and oral S-1 for resectable pancreatic cancer. Exp. Ther. Med. 2012, 3, 787–792. [Google Scholar] [CrossRef]
- Heinrich, S.; Pestalozzi, B.C.; Schäfer, M.; Weber, A.; Bauerfeind, P.; Knuth, A.; Clavien, P.-A. Prospective Phase II Trial of Neoadjuvant Chemotherapy with Gemcitabine and Cisplatin for Resectable Adenocarcinoma of the Pancreatic Head. J. Clin. Oncol. 2008, 26, 2526–2531. [Google Scholar] [CrossRef] [PubMed]
- Oba, A.; Ho, F.; Bao, Q.R.; Al-Musawi, M.H.; Schulick, R.D.; del Chiaro, M. Neoadjuvant Treatment in Pancreatic Cancer. Front. Oncol. 2020, 10, 245. [Google Scholar] [CrossRef] [PubMed]
- Corcoran, R.B.; Chabner, B.A. Application of Cell-free DNA Analysis to Cancer Treatment. N. Engl. J. Med. 2018, 379, 1754–1765. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.-E.; Lee, K.T.; Lee, J.K.; Paik, S.W.; Rhee, J.C.; Choi, K.W. Clinical usefulness of carbohydrate antigen 19-9 as a screening test for pancreatic cancer in an asymptomatic population. J. Gastroenterol. Hepatol. 2004, 19, 182–186. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Liu, F. Diagnostic value of serum carbohydrate antigen 19-9 in pancreatic cancer: A meta-analysis. Tumor Biol. 2014, 35, 7459–7465. [Google Scholar] [CrossRef] [PubMed]
- Goonetilleke, K.; Siriwardena, A.K. Systematic review of carbohydrate antigen (CA 19-9) as a biochemical marker in the diagnosis of pancreatic cancer. Eur. J. Surg. Oncol. (EJSO) 2007, 33, 266–270. [Google Scholar] [CrossRef] [PubMed]
- Frohlich, J.J. Serum Tumor Markers in Breast Cancer: Are They of Clinical Value? Clin. Chem. 2006, 52, 345–351. [Google Scholar] [CrossRef]
- Singh, N.; Gupta, S.; Pandey, R.M.; Chauhan, S.S.; Saraya, A. High Levels of Cell-Free Circulating Nucleic Acids in Pancreatic Cancer are Associated With Vascular Encasement, Metastasis and Poor Survival. Cancer Investig. 2015, 33, 78–85. [Google Scholar] [CrossRef]
- Fazel, R.; Krumholz, H.M.; Wang, Y.; Ross, J.S.; Chen, J.; Ting, H.H.; Shah, N.D.; Nasir, K.; Einstein, A.J.; Nallamothu, B.K. Exposure to Low-Dose Ionizing Radiation from Medical Imaging Procedures. N. Engl. J. Med. 2009, 361, 849–857. [Google Scholar] [CrossRef]
- Martini, V.; Timme, S.; Fichtner-Feigl, S.; Hoeppner, J.; Kulemann, B. Circulating Tumor Cells in Pancreatic Cancer: Current Perspectives. Cancers 2019, 11, 1659. [Google Scholar] [CrossRef]
- Dawson, S.-J.; Tsui, D.W.; Murtaza, M.; Biggs, H.; Rueda, O.M.; Chin, S.-F.; Dunning, M.J.; Gale, D.; Forshew, T.; Mahler-Araujo, B.; et al. Analysis of Circulating Tumor DNA to Monitor Metastatic Breast Cancer. N. Engl. J. Med. 2013, 368, 1199–1209. [Google Scholar] [CrossRef] [PubMed]
- Bettegowda, C.; Sausen, M.; Leary, R.J.; Kinde, I.; Wang, Y.; Agrawal, N.; Bartlett, B.R.; Wang, H.; Luber, B.; Alani, R.M.; et al. Detection of Circulating Tumor DNA in Early- and Late-Stage Human Malignancies. Sci. Transl. Med. 2014, 6, 224ra24. [Google Scholar] [CrossRef] [PubMed]
- Sausen, M.; Phallen, J.; Adleff, V.; Jones, S.; Leary, R.J.; Barrett, M.T.; Anagnostou, V.; Parpart-Li, S.; Murphy, D.; Li, Q.K.; et al. Clinical implications of genomic alterations in the tumour and circulation of pancreatic cancer patients. Nat. Commun. 2015, 6, 7686. [Google Scholar] [CrossRef]
- Pietrasz, D.; Pécuchet, N.; Garlan, F.; Didelot, A.; Dubreuil, O.; Doat, S.; Imbert-Bismut, F.; Karoui, M.; Vaillant, J.-C.; Taly, V.; et al. Plasma Circulating Tumor DNA in Pancreatic Cancer Patients Is a Prognostic Marker. Clin. Cancer Res. 2017, 23, 116–123. [Google Scholar] [CrossRef] [PubMed]
- Yi, X.; Ma, J.; Guan, Y.; Chen, R.; Yang, L.; Xia, X. The feasibility of using mutation detection in ctDNA to assess tumor dynamics. Int. J. Cancer 2017, 140, 2642–2647. [Google Scholar] [CrossRef]
- Abbosh, C.; The TRACERx Consortium; Birkbak, N.J.; Wilson, G.A.; Jamal-Hanjani, M.; Constantin, T.; Salari, R.; Le Quesne, J.; Moore, D.A.; Veeriah, S.; et al. Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution. Nat. Cell Biol. 2017, 545, 446–451. [Google Scholar] [CrossRef]
- Warton, K.; Mahon, K.L.; Samimi, G. Methylated circulating tumor DNA in blood: Power in cancer prognosis and response. Endocr. Relat. Cancer 2016, 23, R157–R171. [Google Scholar] [CrossRef]
- Mandel, P.; Metais, P. Les acides nucléiques du plasma sanguin chez l’homme. Compt. Ren. Sean. Soc. Biol. 1948, 142, 241–243. [Google Scholar]
- Leon, S.A.; Shapiro, B.; Sklaroff, D.M.; Yaros, M.J. Free DNA in the serum of cancer patients and the effect of therapy. Cancer Res. 1977, 37, 646–650. [Google Scholar]
- Filho, E.M.R.; Simon, D.; Ikuta, N.; Klovan, C.; Dannebrock, F.A.; De Oliveira, C.O.; Regner, A. Elevated Cell-Free Plasma DNA Level as an Independent Predictor of Mortality in Patients with Severe Traumatic Brain Injury. J. Neurotrauma 2014, 31, 1639–1646. [Google Scholar] [CrossRef]
- Tsai, N.-W.; Lin, T.-K.; Chen, S.-D.; Chang, W.-N.; Wang, H.-C.; Yang, T.-M.; Lin, Y.-J.; Jan, C.-R.; Huang, C.-R.; Liou, C.-W.; et al. The value of serial plasma nuclear and mitochondrial DNA levels in patients with acute ischemic stroke. Clin. Chim. Acta 2011, 412, 476–479. [Google Scholar] [CrossRef] [PubMed]
- Breitbach, S.; Sterzing, B.; Magallanes, C.; Tug, S.; Simon, P. Direct measurement of cell-free DNA from serially collected capillary plasma during incremental exercise. J. Appl. Physiol. 2014, 117, 119–130. [Google Scholar] [CrossRef] [PubMed]
- De Vlaminck, I.; Valantine, H.A.; Snyder, T.M.; Strehl, C.; Cohen, G.; Luikart, H.; Neff, N.F.; Okamoto, J.; Bernstein, D.; Weisshaar, D.; et al. Circulating Cell-Free DNA Enables Noninvasive Diagnosis of Heart Transplant Rejection. Sci. Transl. Med. 2014, 6, 241ra77. [Google Scholar] [CrossRef] [PubMed]
- Lo, Y.M.D.; Corbetta, N.; Chamberlain, P.F.; Rai, V.; Sargent, I.L.; Redman, C.W.; Wainscoat, J.S. Presence of fetal DNA in maternal plasma and serum. Lancet 1997, 350, 485–487. [Google Scholar] [CrossRef]
- Lo, Y.M.D.; Lun, F.M.F.; Chan, K.C.A.; Tsui, N.B.Y.; Chong, K.C.; Lau, T.K.; Leung, T.Y.; Zee, B.C.Y.; Cantor, C.R.; Chiu, R.W.K. Digital PCR for the molecular detection of fetal chromosomal aneuploidy. Proc. Natl. Acad. Sci. USA 2007, 104, 13116–13121. [Google Scholar] [CrossRef]
- Allyse, M.; Minear, M.A.; Rote, M.; Hung, A.; Chandrasekharan, S.; Berson, E.; Sridhar, S. Non-invasive prenatal testing: A review of international implementation and challenges. Int. J. Women’s Heal. 2015, 7, 113–126. [Google Scholar] [CrossRef]
- Hill, M.; Wright, D.; Daley, R.; Lewis, C.; McKay, F.; Mason, S.; Lench, N.J.; Howarth, A.; Boustred, C.; Lo, K.; et al. Evaluation of non-invasive prenatal testing (NIPT) for aneuploidy in an NHS setting: A reliable accurate prenatal non-invasive diagnosis (RAPID) protocol. BMC Pregnancy Childbirth 2014, 14, 229. [Google Scholar] [CrossRef]
- Hyett, J.A.; Gardener, G.; Stojilkovic-Mikic, T.; Finning, K.M.; Martin, P.G.; Rodeck, C.H.; Chitty, L.S. Reduction in diagnostic and therapeutic interventions by non-invasive determination of fetal sex in early pregnancy. Prenat. Diagn. 2005, 25, 1111–1116. [Google Scholar] [CrossRef]
- Stroun, M.; Anker, P.; Maurice, P.; Lyautey, J.; Lederrey, C.; Beljanski, M. Neoplastic Characteristics of the DNA Found in the Plasma of Cancer Patients. Oncology 1989, 46, 318–322. [Google Scholar] [CrossRef]
- Fan, H.C.; Blumenfeld, Y.J.; Chitkara, U.; Hudgins, L.; Quake, S.R. Analysis of the Size Distributions of Fetal and Maternal Cell-Free DNA by Paired-End Sequencing. Clin. Chem. 2010, 56, 1279–1286. [Google Scholar] [CrossRef]
- Snyder, M.W.; Kircher, M.; Hill, A.J.; Daza, R.M.; Shendure, J. Cell-free DNA Comprises an In Vivo Nucleosome Footprint that Informs Its Tissues-Of-Origin. Cell 2016, 164, 57–68. [Google Scholar] [CrossRef] [PubMed]
- Sun, K.; Jiang, P.; Chan, K.C.A.; Wong, J.; Cheng, Y.K.Y.; Liang, R.H.S.; Chan, W.-K.; Ma, E.S.K.; Chan, S.L.; Cheng, S.H.; et al. Plasma DNA tissue mapping by genome-wide methylation sequencing for noninvasive prenatal, cancer, and transplantation assessments. Proc. Natl. Acad. Sci. USA 2015, 112, E5503–E5512. [Google Scholar] [CrossRef] [PubMed]
- Tie, J.; Kinde, I.; Wang, Y.; Wong, H.L.; Roebert, J.; Christie, M.; Tacey, M.; Wong, R.; Singh, M.; Karapetis, C.S.; et al. Circulating tumor DNA as an early marker of therapeutic response in patients with metastatic colorectal cancer. Ann. Oncol. 2015, 26, 1715–1722. [Google Scholar] [CrossRef] [PubMed]
- Xi, L.; Pham, T.H.-T.; Payabyab, E.C.; Sherry, R.M.; Rosenberg, S.A.; Raffeld, M. Circulating Tumor DNA as an Early Indicator of Response to T-cell Transfer Immunotherapy in Metastatic Melanoma. Clin. Cancer Res. 2016, 22, 5480–5486. [Google Scholar] [CrossRef]
- Riediger, A.L.; Dietz, S.; Schirmer, U.; Meister, M.; Heinzmann-Groth, I.; Schneider, M.; Muley, T.; Thomas, M.; Sültmann, H. Mutation analysis of circulating plasma DNA to determine response to EGFR tyrosine kinase inhibitor therapy of lung adenocarcinoma patients. Sci. Rep. 2016, 6, 33505. [Google Scholar] [CrossRef]
- Diehl, F.; Schmidt, K.; Choti, M.A.; Romans, K.; Goodman, S.; Li, M.; Thornton, K.; Agrawal, N.; Sokoll, L.; Szabo, S.A.; et al. Circulating mutant DNA to assess tumor dynamics. Nat. Med. 2008, 14, 985–990. [Google Scholar] [CrossRef]
- Tug, S.; Helmig, S.; Deichmann, E.R.; Schmeier-Jürchott, A.; Wagner, E.; Zimmermann, T.; Radsak, M.; Giacca, M.; Simon, P. Exercise-induced increases in cell free DNA in human plasma originate predominantly from cells of the haematopoietic lineage. Exerc. Immunol. Rev. 2015, 21, 164–173. [Google Scholar]
- Lo, Y.M.D.; Zhang, J.; Leung, T.N.; Lau, T.K.; Chang, A.M.; Hjelm, N.M. Rapid Clearance of Fetal DNA from Maternal Plasma. Am. J. Hum. Genet. 1999, 64, 218–224. [Google Scholar] [CrossRef]
- El Messaoudi, S.; Rolet, F.; Mouliere, F.; Thierry, A.R. Circulating cell free DNA: Preanalytical considerations. Clin. Chim. Acta 2013, 424, 222–230. [Google Scholar] [CrossRef]
- Swinkels, D.W.; Wiegerinck, E.; Steegers, E.A.; De Kok, J.B. Effects of Blood-Processing Protocols on Cell-free DNA Quantification in Plasma. Clin. Chem. 2003, 49, 525–526. [Google Scholar] [CrossRef]
- Alidousty, C.; Brandes, D.; Heydt, C.; Wagener, S.; Wittersheim, M.; Schäfer, S.C.; Holz, B.; Merkelbach-Bruse, S.; Büttner, R.; Fassunke, J.; et al. Comparison of Blood Collection Tubes from Three Different Manufacturers for the Collection of Cell-Free DNA for Liquid Biopsy Mutation Testing. J. Mol. Diagn. 2017, 19, 801–804. [Google Scholar] [CrossRef] [PubMed]
- Risberg, B.; Tsui, D.W.Y.; Biggs, H.; De Almagro, A.R.-V.M.; Dawson, S.-J.; Hodgkin, C.; Jones, L.; Parkinson, C.; Piskorz, A.; Marass, F.; et al. Effects of Collection and Processing Procedures on Plasma Circulating Cell-Free DNA from Cancer Patients. J. Mol. Diagn. 2018, 20, 883–892. [Google Scholar] [CrossRef] [PubMed]
- Warton, K.; Yuwono, N.; Cowley, M.J.; McCabe, M.J.; So, A.; Ford, C.E. Evaluation of Streck BCT and PAXgene Stabilised Blood Collection Tubes for Cell-Free Circulating DNA Studies in Plasma. Mol. Diagn. Ther. 2017, 21, 563–570. [Google Scholar] [CrossRef] [PubMed]
- Diaz, I.M.; Nocon, A.; Mehnert, D.H.; Fredebohm, J.; Diehl, F.; Holtrup, F. Performance of Streck cfDNA Blood Collection Tubes for Liquid Biopsy Testing. PLoS ONE 2016, 11, e0166354. [Google Scholar] [CrossRef]
- Razavi, P.; Li, B.T.; Brown, D.N.; Jung, B.; Hubbell, E.; Shen, R.; Abida, W.; Juluru, K.; De Bruijn, I.; Hou, C.; et al. High-intensity sequencing reveals the sources of plasma circulating cell-free DNA variants. Nat. Med. 2019, 25, 1928–1937. [Google Scholar] [CrossRef]
- Liu, M.; Oxnard, G.; Klein, E.; Swanton, C.; Seiden, M.; Cummings, S.R.; Absalan, F.; Alexander, G.; Allen, B.; Amini, H.; et al. Sensitive and specific multi-cancer detection and localization using methylation signatures in cell-free DNA. Ann. Oncol. 2020, 31, 745–759. [Google Scholar] [CrossRef]
- Diehl, F.; Li, M.; He, Y.; Kinzler, K.W.; Vogelstein, B.; Dressman, D. BEAMing: Single-molecule PCR on microparticles in water-in-oil emulsions. Nat. Methods 2006, 3, 551–559. [Google Scholar] [CrossRef]
- Forshew, T.; Murtaza, M.; Parkinson, C.; Gale, D.; Tsui, D.W.Y.; Kaper, F.; Dawson, S.-J.; Piskorz, A.M.; Jimenez-Linan, M.; Bentley, D.; et al. Noninvasive Identification and Monitoring of Cancer Mutations by Targeted Deep Sequencing of Plasma DNA. Sci. Transl. Med. 2012, 4, 136ra68. [Google Scholar] [CrossRef]
- Taly, V.; Pekin, D.; Benhaim, L.; Kotsopoulos, S.K.; Le Corre, D.; Li, X.; Atochin, I.; Link, D.R.; Griffiths, A.D.; Pallier, K.; et al. Multiplex Picodroplet Digital PCR to Detect KRAS Mutations in Circulating DNA from the Plasma of Colorectal Cancer Patients. Clin. Chem. 2013, 59, 1722–1731. [Google Scholar] [CrossRef]
- Hindson, B.J.; Ness, K.D.; Masquelier, D.A.; Belgrader, P.; Heredia, N.J.; Makarewicz, A.J.; Bright, I.J.; Lucero, M.Y.; Hiddessen, A.L.; Legler, T.C.; et al. High-Throughput Droplet Digital PCR System for Absolute Quantitation of DNA Copy Number. Anal. Chem. 2011, 83, 8604–8610. [Google Scholar] [CrossRef]
- Adalsteinsson, V.A.; Ha, G.; Freeman, S.S.; Choudhury, A.D.; Stover, D.; Parsons, H.A.; Gydush, G.; Reed, S.C.; Rotem, D.; Rhoades, J.; et al. Scalable whole-exome sequencing of cell-free DNA reveals high concordance with metastatic tumors. Nat. Commun. 2017, 8, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Murtaza, M.; Dawson, S.-J.; Tsui, D.W.Y.; Gale, D.; Forshew, T.; Piskorz, A.M.; Parkinson, C.; Chin, S.-F.; Kingsbury, Z.; Wong, A.S.C.; et al. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nat. Cell Biol. 2013, 497, 108–112. [Google Scholar] [CrossRef] [PubMed]
- Kinde, I.; Wu, J.; Papadopoulos, N.; Kinzler, K.W.; Vogelstein, B. Detection and quantification of rare mutations with massively parallel sequencing. Proc. Natl. Acad. Sci. USA 2011, 108, 9530–9535. [Google Scholar] [CrossRef] [PubMed]
- Newman, A.M.; Lovejoy, A.F.; Klass, D.M.; Kurtz, D.M.; Chabon, J.J.; Scherer, F.; Stehr, H.; Liu, C.L.; Bratman, S.V.; Say, C.; et al. Integrated digital error suppression for noninvasive detection of circulating tumor DNA in NSCLC. J. Clin. Oncol. 2016, 34, e20500. [Google Scholar] [CrossRef]
- Shapiro, B.; Chakrabarty, M.; Cohn, E.M.; Leon, S.A. Determination of circulating DNA levels in patients with benign or malignant gastrointestinal disease. Cancer 1983, 51, 2116–2120. [Google Scholar] [CrossRef]
- Sorenson, G.D.; Pribish, D.M.; Valone, F.H.; Memoli, V.A.; Bzik, D.J.; Yao, S.L. Soluble normal and mutated DNA sequences from single-copy genes in human blood. Cancer Epidemiology Biomarkers Prev. 1994, 3, 67–71. [Google Scholar]
- Kim, J.; Reber, H.A.; Dry, S.M.; Elashoff, D.; Chen, S.L.; Umetani, N.; Kitago, M.; Hines, O.J.; Kazanjian, K.K.; Hiramatsu, S.; et al. Unfavourable prognosis associated with K-ras gene mutation in pancreatic cancer surgical margins. Gut 2006, 55, 1598–1605. [Google Scholar] [CrossRef]
- Talar-Wojnarowska, R.; Gasiorowska, A.; Smolarz, B.; Romanowicz-Makowska, H.; Strzelczyk, J.; Janiak, A.; Kulig, A.; Malecka-Panas, E. Usefulness of p16 and K-ras mutation in pancreatic adenocarcinoma and chronic pancreatitis differential diagnosis. J. Physiol. Pharmacol. Off. J. Pol. Physiol. Soc. 2004, 55, 129–138. [Google Scholar]
- Maire, F.; Micard, S.; Hammel, P.; Voitot, H.; Levy, P.E.; Cugnenc, P.-H.; Ruszniewski, P.; Puig, P.L. Differential diagnosis between chronic pancreatitis and pancreatic cancer: Value of the detection of KRAS2 mutations in circulating DNA. Br. J. Cancer 2002, 87, 551–554. [Google Scholar] [CrossRef]
- Feng, D.-X.; Shengdao, Z.; Tianquan, H.; Yu, J.; Ruoqing, L.; Zurong, Y.; Xuezhi, W. A Prospective Study of Detection of Pancreatic Carcinoma by Combined Plasma K -ras Mutations and Serum CA19-9 Analysis. Pancreas 2002, 25, 336–341. [Google Scholar] [CrossRef]
- Tjensvoll, K.; Lapin, M.; Buhl, T.; Oltedal, S.; Berry, K.S.-O.; Gilje, B.; Søreide, J.A.; Javle, M.; Nordgård, O.; Smaaland, R. Clinical relevance of circulating KRAS mutated DNA in plasma from patients with advanced pancreatic cancer. Mol. Oncol. 2015, 10, 635–643. [Google Scholar] [CrossRef] [PubMed]
- Berger, A.W.; Schwerdel, D.; Costa, I.G.; Hackert, T.; Strobel, O.; Lam, S.; Barth, T.F.; Schröppel, B.; Meining, A.; Büchler, M.W.; et al. Detection of Hot-Spot Mutations in Circulating Cell-Free DNA From Patients With Intraductal Papillary Mucinous Neoplasms of the Pancreas. Gastroenterology 2016, 151, 267–270. [Google Scholar] [CrossRef] [PubMed]
- Brychta, N.; Krahn, T.; Von Ahsen, O. Detection of KRAS Mutations in Circulating Tumor DNA by Digital PCR in Early Stages of Pancreatic Cancer. Clin. Chem. 2016, 62, 1482–1491. [Google Scholar] [CrossRef] [PubMed]
- Calvez-Kelm, F.; Foll, M.; Wozniak, M.B.; Delhomme, T.M.; Durand, G.; Chopard, P.; Pertesi, M.; Fabianova, E.; Adamcakova, Z.; Holcatova, I.; et al. KRAS mutations in blood circulating cell-free DNA: A pancreatic cancer case-control. Oncotarget 2016, 7, 78827–78840. [Google Scholar] [CrossRef] [PubMed]
- Takai, E.; Totoki, Y.; Nakamura, H.; Kato, M.; Shibata, T.; Yachida, S. Clinical Utility of Circulating Tumor DNA for Molecular Assessment and Precision Medicine in Pancreatic Cancer. Adv. Exp. Med. Biol. 2016, 924, 13–17. [Google Scholar] [CrossRef] [PubMed]
- Cohen, J.D.; Javed, A.A.; Thoburn, C.; Wong, F.; Tie, J.; Gibbs, P.; Schmidt, C.M.; Yip-Schneider, M.T.; Allen, P.J.; Schattner, M.; et al. Combined circulating tumor DNA and protein biomarker-based liquid biopsy for the earlier detection of pancreatic cancers. Proc. Natl. Acad. Sci. USA 2017, 114, 10202–10207. [Google Scholar] [CrossRef] [PubMed]
- Cheng, H.; Liu, C.; Jiang, J.; Luo, G.; Lu, Y.; Jin, K.; Guo, M.; Zhang, Z.; Xu, J.; Liu, L.; et al. Analysis of ctDNA to predict prognosis and monitor treatment responses in metastatic pancreatic cancer patients. Int. J. Cancer 2017, 140, 2344–2350. [Google Scholar] [CrossRef]
- Cohen, J.D.; Li, L.; Wang, Y.; Thoburn, C.; Afsari, B.; Danilova, L.; Douville, C.; Javed, A.A.; Wong, F.; Mattox, A.; et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science 2018, 359, 926–930. [Google Scholar] [CrossRef]
- Chen, L.; Zhang, Y.; Cheng, Y.; Zhang, D.; Zhu, S.; Ma, X. Prognostic value of circulating cell-free DNA in patients with pancreatic cancer: A systemic review and meta-analysis. Gene 2018, 679, 328–334. [Google Scholar] [CrossRef]
- Bernard, V.; Kim, D.U.; Lucas, F.A.S.; Castillo, J.; Allenson, K.; Mulu, F.C.; Stephens, B.M.; Huang, J.; Semaan, A.; Guerrero, P.A.; et al. Circulating Nucleic Acids Are Associated With Outcomes of Patients With Pancreatic Cancer. Gastroenterology 2019, 156, 108–118.e4. [Google Scholar] [CrossRef]
- Berger, A.W.; Schwerdel, D.; Reinacher-Schick, A.; Uhl, W.; Algül, H.; Friess, H.; Janssen, K.-P.; König, A.; Ghadimi, M.; Gallmeier, E.; et al. A Blood-Based Multi Marker Assay Supports the Differential Diagnosis of Early-Stage Pancreatic Cancer. Theranostics 2019, 9, 1280–1287. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Liu, L.; Ji, Y.; Li, C.; Wei, T.; Yang, X.; Zhang, Y.; Cai, X.; Gao, Y.; Xu, W.; et al. Enrichment of short mutant cell-free DNA fragments enhanced detection of pancreatic cancer. EBioMedicine 2019, 41, 345–356. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.C.; Maddala, T.; Aravanis, A.; Hubbell, E.; Beausang, J.F.; Filippova, D.; Gross, S.; Jamshidi, A.; Kurtzman, K.; Shen, L.; et al. Breast cancer cell-free DNA (cfDNA) profiles reflect underlying tumor biology: The Circulating Cell-Free Genome Atlas (CCGA) study. J. Clin. Oncol. 2018, 36, 536. [Google Scholar] [CrossRef]
- Fernandez-Cuesta, L.; Perdomo, S.; Avogbe, P.H.; Leblay, N.; Delhomme, T.M.; Gaborieau, V.; Abedi-Ardekani, B.; Chanudet, E.; Olivier, M.; Zaridze, D.; et al. Identification of Circulating Tumor DNA for the Early Detection of Small-cell Lung Cancer. EBioMedicine 2016, 10, 117–123. [Google Scholar] [CrossRef]
- Fiala, C.; Diamandis, E. Utility of circulating tumor DNA in cancer diagnostics with emphasis on early detection. BMC Med. 2018, 16, 1–10. [Google Scholar] [CrossRef]
- Zhou, W.; Sokoll, L.J.; Bruzek, D.J.; Zhang, L.; Velculescu, V.E.; Goldin, S.B.; Hruban, R.H.; Kern, S.E.; Hamilton, S.R.; Chan, D.W.; et al. Identifying markers for pancreatic cancer by gene expression analysis. Cancer Epidemiology Biomarkers Prev. 1998, 7, 109–112. [Google Scholar]
- Capello, M.; Bantis, L.E.; Scelo, G.; Zhao, Y.; Li, P.; Dhillon, D.S.; Patel, N.J.; Kundnani, D.L.; Wang, H.; Abbruzzese, J.L.; et al. Sequential Validation of Blood-Based Protein Biomarker Candidates for Early-Stage Pancreatic Cancer. J. Natl. Cancer Inst. 2017, 109. [Google Scholar] [CrossRef]
- Young, R.P.; Christmas, T.; Hopkins, R.J. Multi-analyte assays and early detection of common cancers. J. Thorac. Dis. 2018, 10, S2165–S2167. [Google Scholar] [CrossRef]
- Pannala, R.; Leirness, J.B.; Bamlet, W.R.; Basu, A.; Petersen, G.M.; Chari, S.T. Prevalence and Clinical Profile of Pancreatic Cancer–Associated Diabetes Mellitus. Gastroenterology 2008, 134, 981–987. [Google Scholar] [CrossRef]
- Kirkegård, J.; Mortensen, F.V.; Cronin-Fenton, D. Chronic Pancreatitis and Pancreatic Cancer Risk: A Systematic Review and Meta-analysis. Am. J. Gastroenterol. 2017, 112, 1366–1372. [Google Scholar] [CrossRef]
- Bamlet, W.R.; Chaffee, K.G.; Olswold, C.; De Andrade, M.; Petersen, G.M. Risk of malignancy in first-degree relatives of patients with pancreatic carcinoma. Cancer 2005, 104, 388–394. [Google Scholar] [CrossRef]
- Ben, Q.; Xu, M.; Ning, X.; Liu, J.; Hong, S.; Huang, W.; Zhang, H.; Li, Z. Diabetes mellitus and risk of pancreatic cancer: A meta-analysis of cohort studies. Eur. J. Cancer 2011, 47, 1928–1937. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.; Hart, S.N.; Polley, E.C.; Gnanaolivu, R.; Shimelis, H.; Lee, K.Y.; Lilyquist, J.; Na, J.; Moore, R.M.; Antwi, S.O.; et al. Association Between Inherited Germline Mutations in Cancer Predisposition Genes and Risk of Pancreatic Cancer. JAMA 2018, 319, 2401–2409. [Google Scholar] [CrossRef] [PubMed]
- Kanwal, R.; Gupta, S. Epigenetic modifications in cancer. Clin. Genet. 2012, 81, 303–311. [Google Scholar] [CrossRef]
- Grønbæk, K.; Hother, C.; Jones, P.A. Epigenetic changes in cancer. APMIS 2007, 115, 1039–1059. [Google Scholar] [CrossRef]
- Sharma, S.; Kelly, T.K.; Jones, P.A. Epigenetics in cancer. Carcinogenesis 2009, 31, 27–36. [Google Scholar] [CrossRef]
- Baylin, S.B.; Jones, P.A. A decade of exploring the cancer epigenome—Biological and translational implications. Nat. Rev. Cancer 2011, 11, 726–734. [Google Scholar] [CrossRef]
- Montavon, C.; Gloss, B.S.; Warton, K.; Barton, C.A.; Statham, A.L.; Scurry, J.P.; Tabor, B.; Nguyen, T.V.; Qu, W.; Samimi, G.; et al. Prognostic and diagnostic significance of DNA methylation patterns in high grade serous ovarian cancer. Gynecol. Oncol. 2012, 124, 582–588. [Google Scholar] [CrossRef]
- Henriksen, S.D.; Madsen, P.H.; Larsen, A.C.; Johansen, M.B.; Drewes, A.M.; Pedersen, I.S.; Krarup, H.B.; Thorlacius-Ussing, O. Cell-free DNA promoter hypermethylation in plasma as a diagnostic marker for pancreatic adenocarcinoma. Clin. Epigenetics 2016, 8, 1–12. [Google Scholar] [CrossRef]
- Liggett, T.; Melnikov, A.; Yi, Q.-L.; Replogle, C.; Brand, R.; Kaul, K.; Talamonti, M.; Abrams, R.A.; Levenson, V. Differential methylation of cell-free circulating DNA among patients with pancreatic cancer versus chronic pancreatitis. Cancer 2010, 116, 1674–1680. [Google Scholar] [CrossRef]
- Eissa, M.A.L.; Lerner, L.; Abdelfatah, E.; Shankar, N.; Canner, J.K.; Hasan, N.M.; Yaghoobi, V.; Huang, B.; Kerner, Z.; Takaesu, F.; et al. Promoter methylation of ADAMTS1 and BNC1 as potential biomarkers for early detection of pancreatic cancer in blood. Clin. Epigenetics 2019, 11, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Chan, K.C.A.; Jiang, P.; Chan, C.W.M.; Sun, K.; Wong, J.; Hui, E.P.; Chan, S.L.; Chan, W.C.; Hui, D.S.C.; Ng, S.S.M.; et al. Noninvasive detection of cancer-associated genome-wide hypomethylation and copy number aberrations by plasma DNA bisulfite sequencing. Proc. Natl. Acad. Sci. USA 2013, 110, 18761–18768. [Google Scholar] [CrossRef] [PubMed]
- Grunau, C. Bisulfite genomic sequencing: Systematic investigation of critical experimental parameters. Nucleic Acids Res. 2001, 29, e65. [Google Scholar] [CrossRef] [PubMed]
- Shen, S.Y.; Singhania, R.; Fehringer, G.; Chakravarthy, A.; Roehrl, M.H.A.; Chadwick, D.; Zuzarte, P.C.; Borgida, A.; Wang, T.T.; Li, T.; et al. Sensitive tumour detection and classification using plasma cell-free DNA methylomes. Nat. Cell Biol. 2018, 563, 579–583. [Google Scholar] [CrossRef]
- Suzuki, M.M.; Bird, A. DNA methylation landscapes: Provocative insights from epigenomics. Nat. Rev. Genet. 2008, 9, 465–476. [Google Scholar] [CrossRef]
- Ibn Sina, A.A.; Carrascosa, L.G.; Liang, Z.; Grewal, Y.S.; Wardiana, A.; Shiddiky, M.J.A.; Gardiner, R.A.; Samaratunga, H.; Gandhi, M.K.; Scott, R.J.; et al. Epigenetically reprogrammed methylation landscape drives the DNA self-assembly and serves as a universal cancer biomarker. Nat. Commun. 2018, 9, 1–13. [Google Scholar] [CrossRef]
- Cho, H.; Mariotto, A.B.; Schwartz, L.M.; Luo, J.; Woloshin, S. When Do Changes in Cancer Survival Mean Progress? The Insight from Population Incidence and Mortality. J. Natl. Cancer Inst. Monogr. 2014, 2014, 187–197. [Google Scholar] [CrossRef]
| Author | Cancer Type | Detection Method | Interpretation | Number of Patients (n) | Sensitivity | Specificity | Diagnostic Targets |
|---|---|---|---|---|---|---|---|
| Sorenson et al., 1994 [68] | Different stages of PDAC | PCR | KRAS mutation found in blood of patients with PC, the mutations were identical with those in tumour biopsy | N/A | N/A | N/A | N/A |
| Maire et al., 2002 [71] | Late stages of PC | PCR | KRAS2 mutation detected in 47% patients with PC, and in 13% of chronic pancreatitis; analysis of KRAS2 + CA19.9 increased sensitivity to 98% and specificity to 77% | 47 PC, 31 ctrl | 47%/98% with CA19–9 | 13% in ctrl (specificity of 77% with CA19–9) | KRAS2 |
| Dianxu et al., 2002 [72] | Early stages of PC | PCR-RFLP | KRAS mutation found in 70.7% of patients with PC but not in 21 controls. Combined with CA19–9, proportion increased to 90.2% | 58 + 21 ctrl | 70.7%/90.2% with CA19–9 | N/A | KRAS/KRAS + CA19–9 |
| Singh et al., 2015 [20] | All stages of PC | PCR-RFLP | ctDNA exceeding 62 ng/mL linked to lower OS and metastasis | N/A | N/A | N/A | N/A |
| Sausen et al., 2015 [25] | Stage II of PC | NGS, ddPCR | ctDNA detected in 43% of patients, ctDNA linked to an adverse prognosis and predicted relapse 6,5 months before CT detection | whole-exome analyses of 24 tumours, targeted genomic analyses of 77 | N/A | N/A | N/A |
| Tjensvoll et al., 2016 [73] | All stages of PC | PNA-clamp PCR | KRAS mutation detected in 71% patients, ctDNA corresponded to CT results and CA19–9 levels | 14 patients with several blood samples | 71% | N/A | KRAS mutations |
| Berger et al., 2016 [74] | Metastatic PC | ddPCR | KRAS mutation in 41.7% of patients, 0% in control population | 21 IPMN patients; 38 controls; 24 metastatic PDAC, 26 resected SCA; 16 borderline IPMN | 41.7% | 84.2% | KRASG12.D and KRASG12.V |
| Brychta et al., 2016 * [75] | Early stage pancreatic cancer (mostly stage I & II) | ChIP-based digital PCR | Detection rates varied between 0% and 50% for specific mutations. KRAS mutation not detected in healthy patients | 50 (82% of stage I & II) | 72% (based on both liquid and standard biopsy) | N/A | KRASG12.D, KRASG12.V, and KRASG12.C mutations in blood and tumour samples |
| Le Calvez-Kelm et al., 2016 * [76] | All stages of PC | NGS | KRAS cfDNA mutations detected in 21.1% of cancers. No improvement over CA19–9 | 437 PC cases, 141 chronic pancreatitis subjects, 394 healthy controls | 21.1% | 96.3% | KRAS, CA 19–9 |
| Takai et al., 2016 * [77] | All stages of PC | ddPCR and NGS | ddPCR detected KRAS mutation in 58.9% of non-resectable PC | 259 | 58.8% in inoperable tumours | N/A | KRAS mutations |
| Cohen et al., 2017 * [78] | Resectable PDAC | PCR-based test and protein biomarkers | KRAS mutation detected in 30% of PC patients (66/221), in 66% when combined with protein biomarkers | 221 with resectable PC, 182 controls | 30% (only KRAS), 66%: KRAS + four protein biomarkers | N/A | KRAS mutations and protein biomarkers |
| Cheng et al., 2017 [79] | Metastatic PC | ddPCR, NGS | 72.3% of PC patients presented with ctDNA-detected KRAS mutation | 10: exome seq, 188 ddPCR, | 76.9% | N/A | 60 genes screened |
| Pietrasz et al., 2017 [26] | All stages of PC | NGS, ddPCR | ctDNA found in 48% of patients with PC, the presence of ctDNA was a predictor of an adverse prognosis | 135, 31 resectable | 48% | N/A | N/A |
| Cohen et al., 2018 * [80] | I–III stages of PC | NGS | Highly efficient multi-analyte test used | 1005 patients of 8 cancer types, stage I–III | 76% (any stage) | 99% | CancerSEEK tested for 8 cancer types; 8 protein biomarkers and mutations in 1933 distinct genomic positions |
| Chen et al., 2018 [81] | All stages of PC | Meta-analysis of literature; significant in predicting OS and PFS | The presence of ctDNA or elevated cfDNA linked to poor prognosis | 1243 from 18 articles | N/A | N/A | N/A |
| Bernard et al., 2019 [82] | Localised or metastatic PC | ddPCR from ctDNA and exosomes | ctDNA showed no correlation with outcomes, as opposed to exosome levels. However, detection of ctDNA post-resection correlated with lower PFS and OS | 194 (overall receiving treatment, also with metastasis), 34 with resectable PC | N/A | N/A | N/A |
| Berger et al., 2019 * [83] | Resectable PC | Fluorimetry (HS Assay for cfDNA quantification) | CA19–9, THBS2 and cfDNA levels in combination were a better PC biomarker (c-statistics 0.90) than any of those separately | 52 | 90% | N/A | thrombospondin-2 (THBS2), CA19–9 |
| Liu et al., 2019 * [84] | Mostly stage I and II | hybrid-capture-based cfDNA sequencing (SLHC-seq) | ctDNA fragmentation pattern may affect the detection of early PC; cancer-specific mutations found in 88% patients, KRAS hotspots in 70% | 112 | 88% | 8 mutations detected in 28 heathy controls | 791 cancer-specific mutations |
| Author | Cancer Type | Detection Method | Interpretation | Number of Patients (n) | Sensitivity | Specificity | Diagnostic Targets |
|---|---|---|---|---|---|---|---|
| Liggett et al., 2010 [102] | All stages of PC, chronic pancreatitis | Methylation-specific PCR | 17 gene promoters were indicated as informative to differentiate between chronic pancreatitis and PC | 30 overall | 91.2% | 90.8% | 56 fragments in each sample (MethDet56) |
| Henriksen et al., 2016 [101] | All stages of PC | Methylation-specific PCR | The number of tested methylated genes was significantly (p < 0.001) higher in cancer patients than in control | 97 PDAC | 76% | 83% | 28 gene panel |
| Eissa et al., 2019 [103] | All stages of PC | Methylation on beads | Methylation of ADAMTS1 and BNC1 is a reliable marker for early detection of PC | 39 | 97.3% | 91.6% | ADAMTS1 and BNC1 |
| Liu et al., 2020 [58] | All stages, data shown here for stage I | Bisulfite sequencing | ctDNA is a good diagnosis method for early stage PDAC and for identification of cancer origin | 20 | 63% | 99% | panel of > 100,000 methylation regions |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jaworski, J.J.; Morgan, R.D.; Sivakumar, S. Circulating Cell-Free Tumour DNA for Early Detection of Pancreatic Cancer. Cancers 2020, 12, 3704. https://doi.org/10.3390/cancers12123704
Jaworski JJ, Morgan RD, Sivakumar S. Circulating Cell-Free Tumour DNA for Early Detection of Pancreatic Cancer. Cancers. 2020; 12(12):3704. https://doi.org/10.3390/cancers12123704
Chicago/Turabian StyleJaworski, Jedrzej J., Robert D. Morgan, and Shivan Sivakumar. 2020. "Circulating Cell-Free Tumour DNA for Early Detection of Pancreatic Cancer" Cancers 12, no. 12: 3704. https://doi.org/10.3390/cancers12123704
APA StyleJaworski, J. J., Morgan, R. D., & Sivakumar, S. (2020). Circulating Cell-Free Tumour DNA for Early Detection of Pancreatic Cancer. Cancers, 12(12), 3704. https://doi.org/10.3390/cancers12123704





