Development of Sensitive Droplet Digital PCR Assays for Detecting Urinary TERT Promoter Mutations as Non-Invasive Biomarkers for Detection of Urothelial Cancer
Abstract
:Simple Summary
Abstract
1. Introduction
2. Results
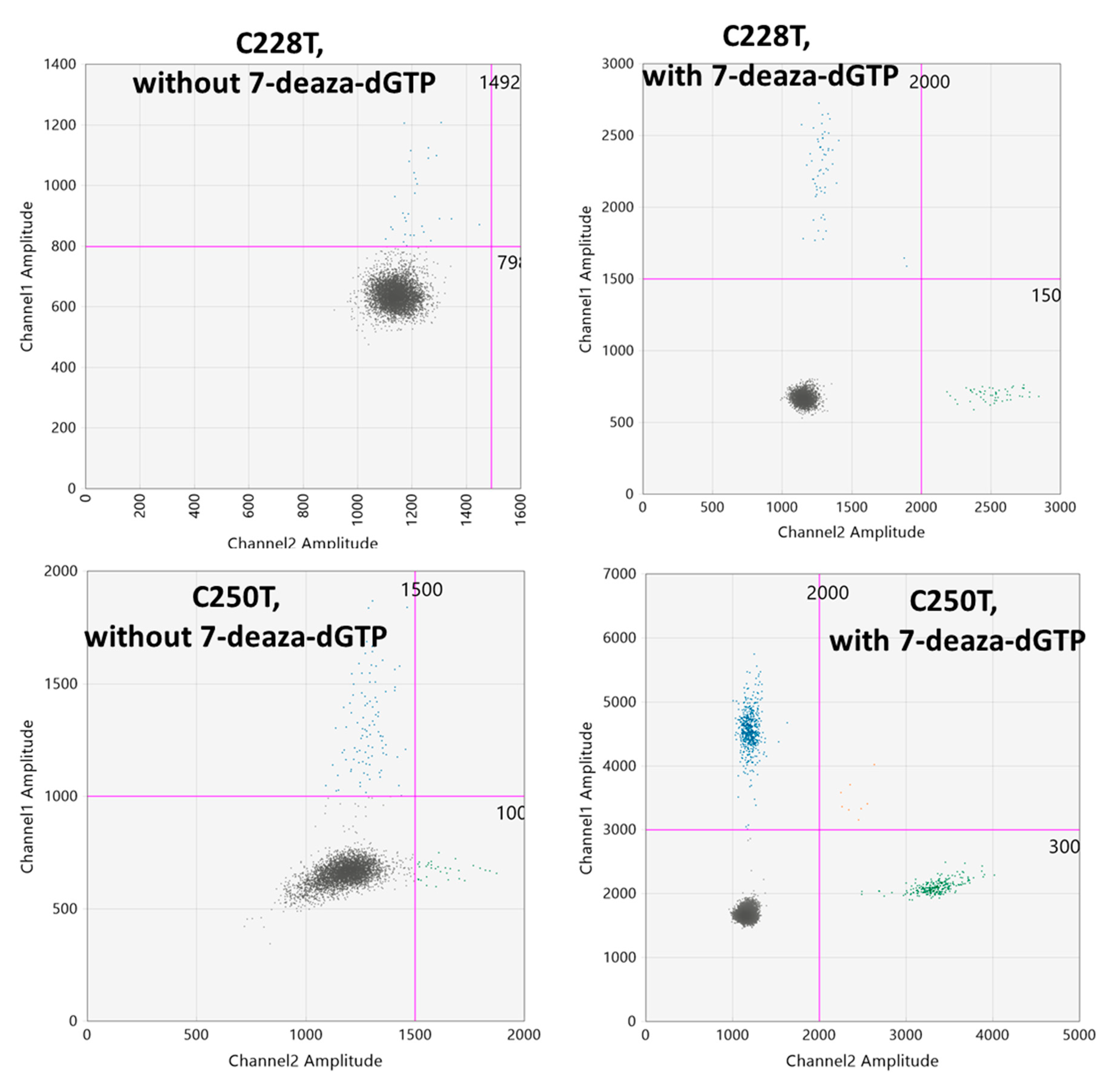
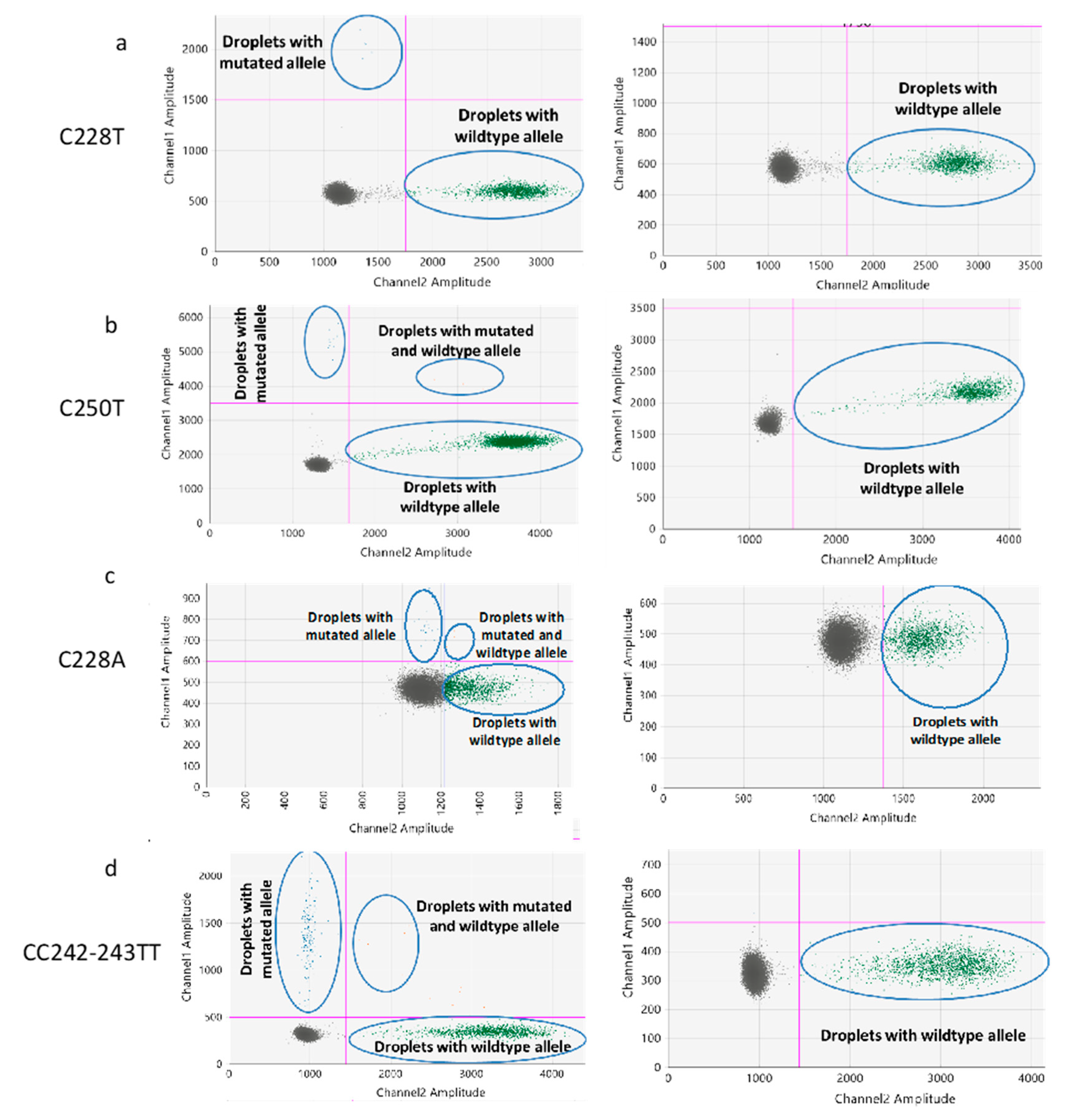
2.1. Optimization of the ddPCR Assays
2.2. Determination of the Threshold Number of Minimum Mutated Droplets to Call a Mutation
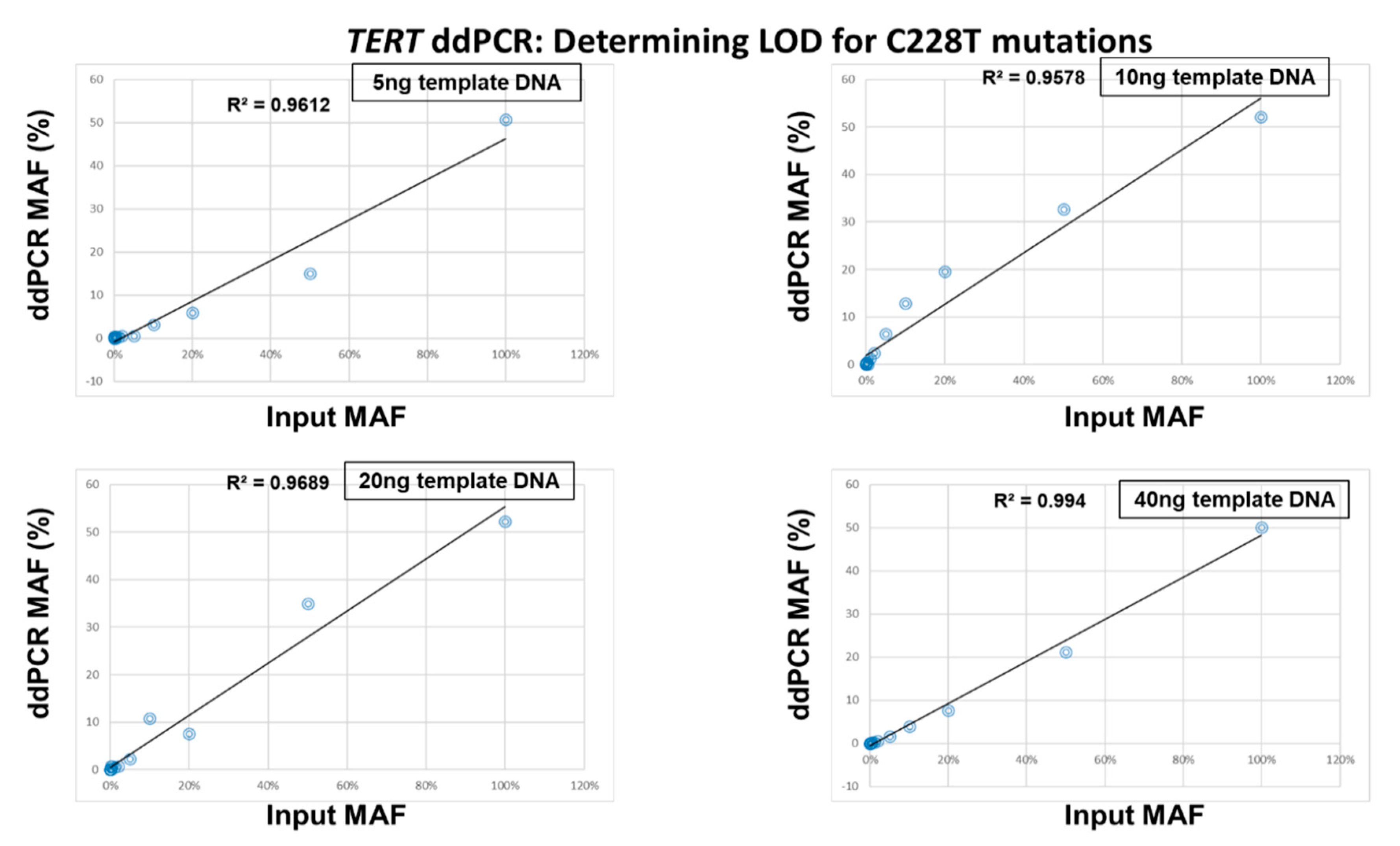
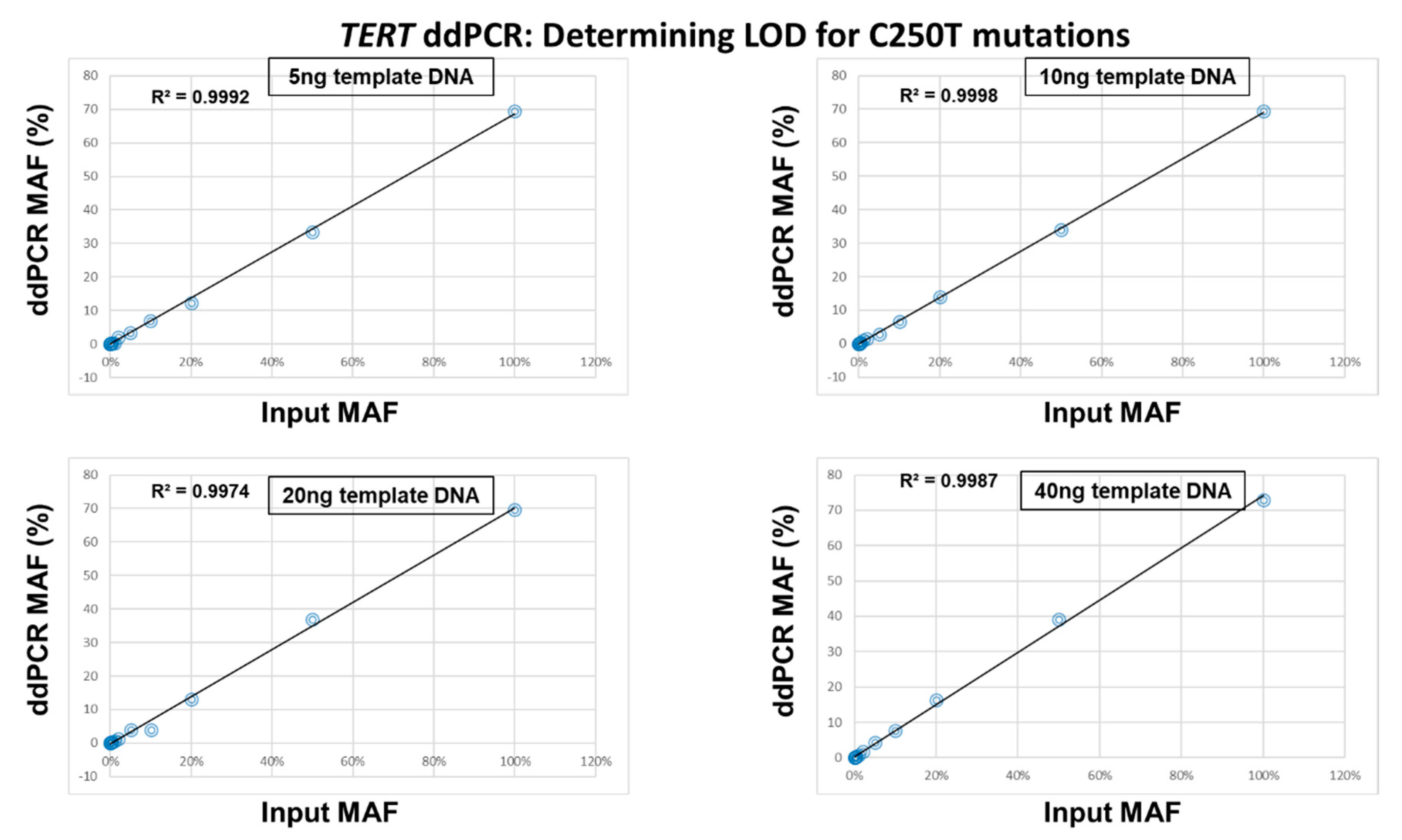
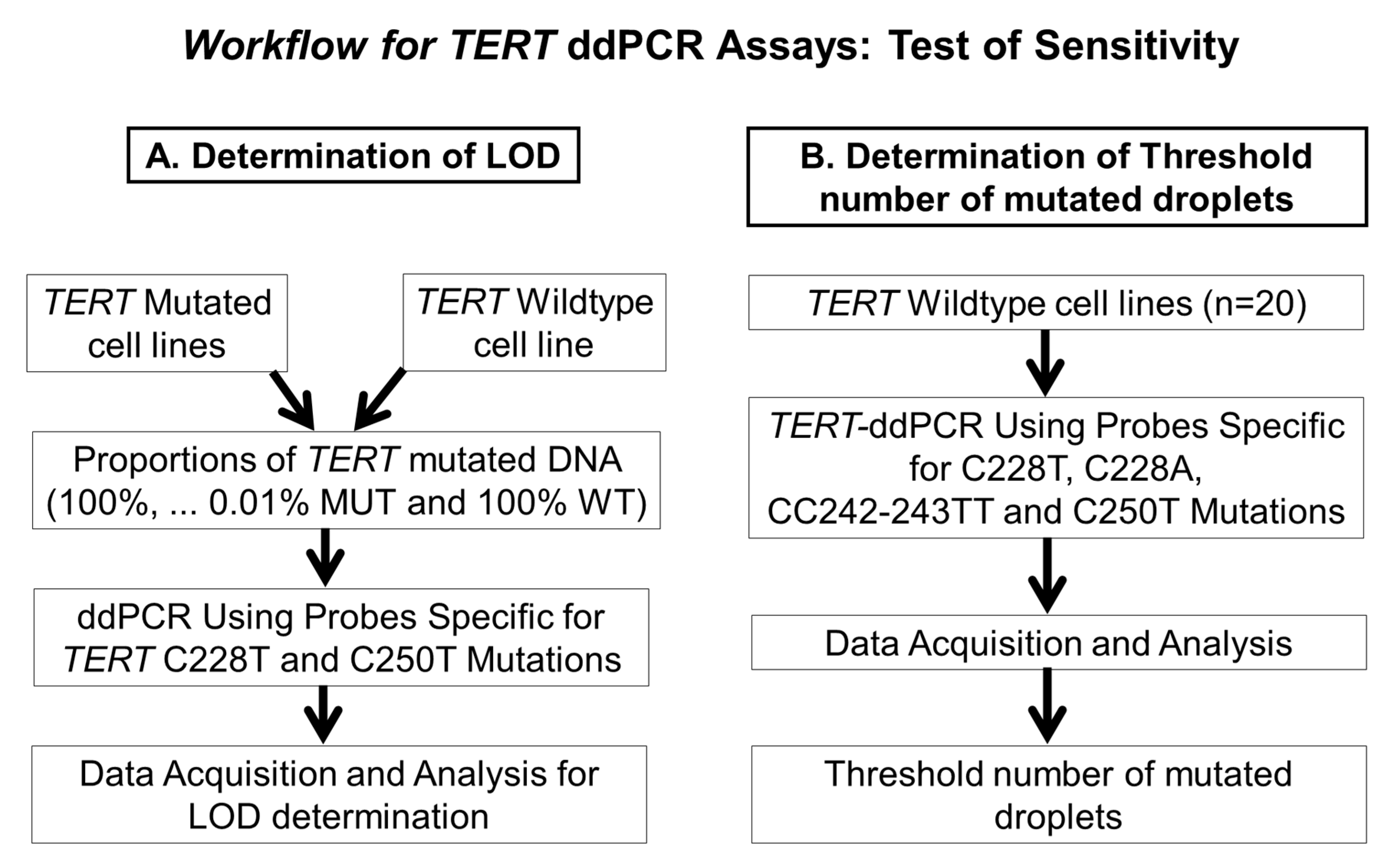
2.3. Determination of the Limit of Detection of the ddPCR Assays
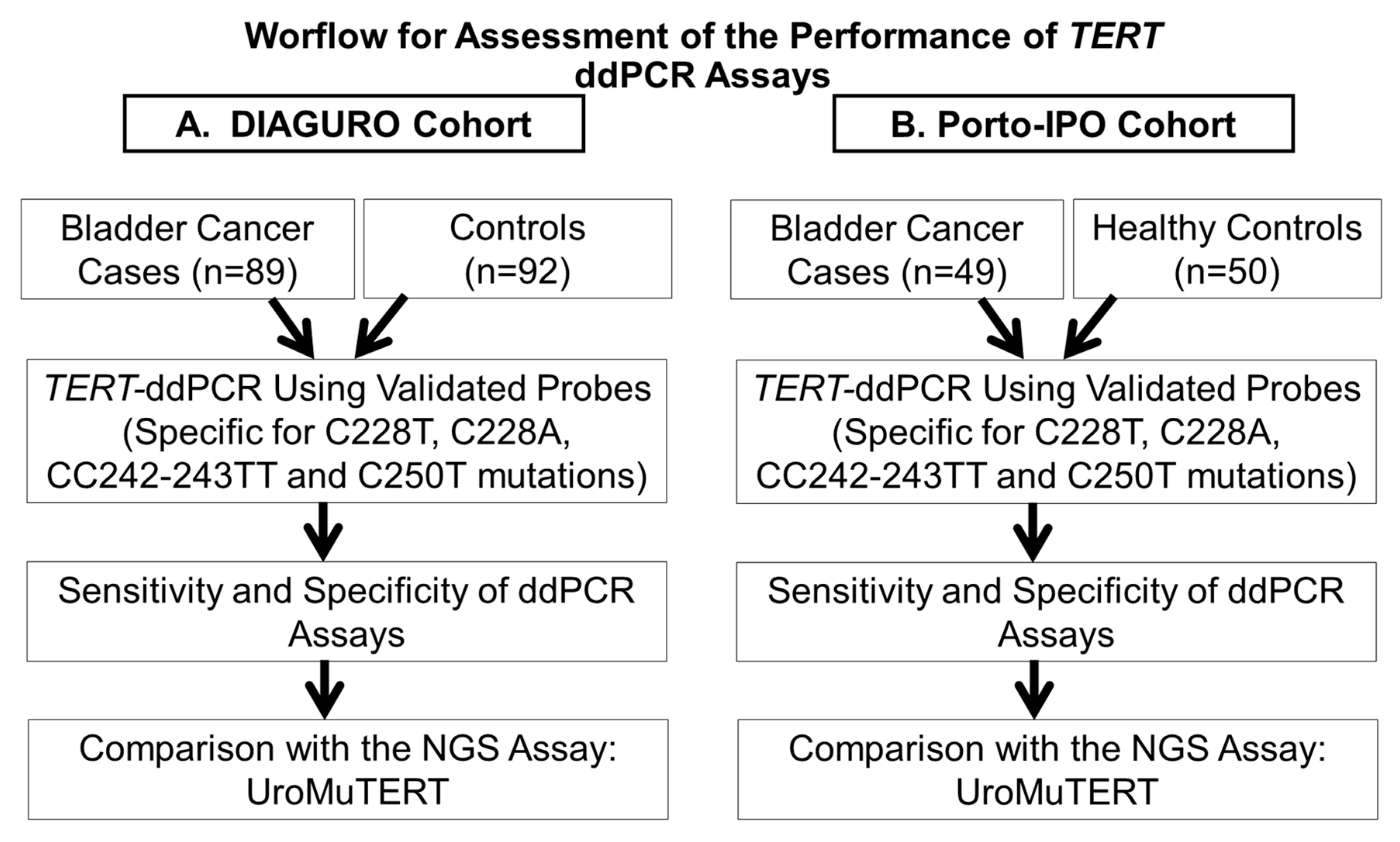
2.4. Comparison of the ddPCR and UroMuTERT Assays for the Detection of TERT Promoter Mutations in Urinary and Tumor DNA
2.5. Assessment of the Interrater Reliability of ddPCR and UroMuTERT Assays in Detecting Urinary TERT Promoter Mutations
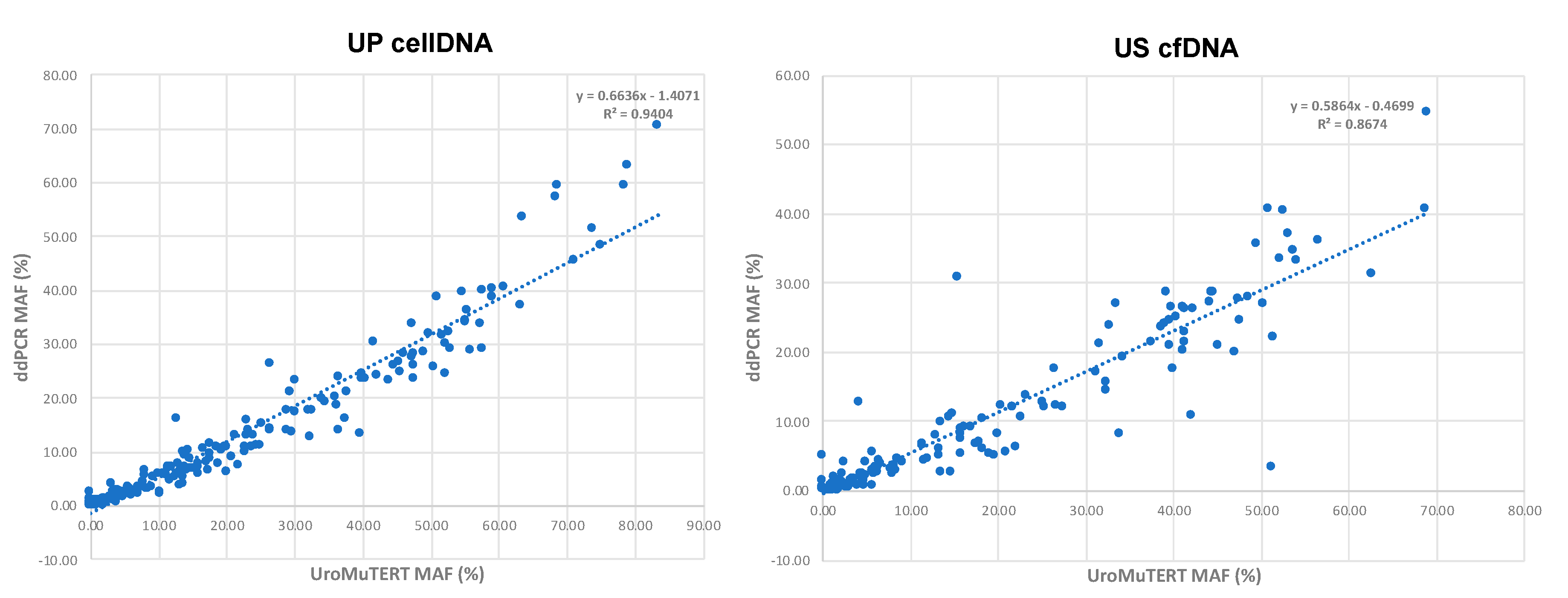
2.6. Correlation of MAFs between ddPCR and UroMuTERT Assays
3. Discussion
4. Materials and Methods
4.1. Study Population, Sample Collection and Processing
4.2. Detection of TERT Promoter Mutations Using ddPCR Assay
4.3. Determination of the False-Positive Measures and Threshold Number of Minimum Mutated Droplets to Call a Mutation
4.4. Determination of the Limit of Detection of the ddPCR Assays
4.5. Comparison of the ddPCR and UroMuTERT Assays for the Detection of TERT Promoter Mutations in the Urine Samples of UC Cases and Controls
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [Green Version]
- Knowles, M.A.; Hurst, C.D. Molecular biology of bladder cancer: New insights into pathogenesis and clinical diversity. Nat. Rev. Cancer 2015, 15, 25–41. [Google Scholar] [CrossRef]
- Yeung, C.; Dinh, T.; Lee, J. The health economics of bladder cancer: An updated review of the published literature. Pharmacoeconomics 2014, 32, 1093–1104. [Google Scholar] [CrossRef]
- Blick, C.G.; Nazir, S.A.; Mallett, S.; Turney, B.W.; Onwu, N.N.; Roberts, I.S.; Crew, J.P.; Cowan, N.C. Evaluation of diagnostic strategies for bladder cancer using computed tomography (CT) urography, flexible cystoscopy and voided urine cytology: Results for 778 patients from a hospital haematuria clinic. BJU Int. 2012, 110, 84–94. [Google Scholar] [CrossRef]
- Gonzalez, A.N.; Lipsky, M.J.; Li, G.; Rutman, M.P.; Cooper, K.L.; Weiner, D.M.; Badalato, G.; Decastro, G.J.; Wenske, S.; McKiernan, J.M.; et al. The Prevalence of Bladder Cancer During Cystoscopy for Asymptomatic Microscopic Hematuria. Urology 2019, 126, 34–38. [Google Scholar] [CrossRef]
- Kouba, E.; Lopez-Beltran, A.; Montironi, R.; Massari, F.; Huang, K.; Santoni, M.; Chovanec, M.; Cheng, M.; Scarpelli, M.; Zhang, J.; et al. Liquid biopsy in the clinical management of bladder cancer: Current status and future developments. Expert Rev. Mol. Diagn. 2020, 20, 255–264. [Google Scholar] [CrossRef]
- Christensen, E.; Birkenkamp-Demtröder, K.; Nordentoft, I.; Høyer, S.; van der Keur, K.; van Kessel, K.; Zwarthoff, E.; Agerbæk, M.; Ørntoft, T.F.; Jensen, J.B.; et al. Liquid Biopsy Analysis of FGFR3 and PIK3CA Hotspot Mutations for Disease Surveillance in Bladder Cancer. Eur. Urol. 2017, 71, 961–969. [Google Scholar] [CrossRef]
- Xylinas, E.; Kluth, L.A.; Rieken, M.; Karakiewicz, P.I.; Lotan, Y.; Shariat, S.F. Urine markers for detection and surveillance of bladder cancer. Urol. Oncol. 2014, 32, 222–229. [Google Scholar] [CrossRef]
- Hosen, I.; Rachakonda, P.S.; Heidenreich, B.; de Verdier, P.J.; Ryk, C.; Steineck, G.; Hemminki, K.; Kumar, R. Mutations in TERT promoter and FGFR3 and telomere length in bladder cancer. Int. J. Cancer 2015, 137, 1621–1629. [Google Scholar] [CrossRef]
- Kinde, I.; Munari, E.; Faraj, S.F.; Hruban, R.H.; Schoenberg, M.; Bivalacqua, T.; Allaf, M.; Springer, S.; Wang, Y.; Diaz, L.A., Jr.; et al. TERT promoter mutations occur early in urothelial neoplasia and are biomarkers of early disease and disease recurrence in urine. Cancer Res. 2013, 73, 7162–7167. [Google Scholar] [CrossRef] [Green Version]
- Avogbe, P.H.; Manel, A.; Vian, E.; Durand, G.; Forey, N.; Voegele, C.; Zvereva, M.; Hosen, M.I.; Meziani, S.; De Tilly, B.; et al. Urinary TERT promoter mutations as non-invasive biomarkers for the comprehensive detection of urothelial cancer. EBioMedicine 2019, 44, 431–438. [Google Scholar] [CrossRef] [Green Version]
- Hosen, M.I.; Sheikh, M.; Zvereva, M.; Scelo, G.; Forey, N.; Durand, G.; Voegele, C.; Poustchi, H.; Khoshnia, M.; Roshandel, G.; et al. Urinary TERT promoter mutations are detectable up to 10 years prior to clinical diagnosis of bladder cancer: Evidence from the Golestan Cohort Study. EBioMedicine 2020, 53, 102643. [Google Scholar] [CrossRef]
- Micheli, E.; Martufi, M.; Cacchione, S.; De Santis, P.; Savino, M. Self-organization of G-quadruplex structures in the hTERT core promoter stabilized by polyaminic side chain perylene derivatives. Biophys. Chem. 2010, 153, 43–53. [Google Scholar] [CrossRef]
- Monsen, R.C.; DeLeeuw, L.; Dean, W.L.; Gray, R.D.; Sabo, T.M.; Chakravarthy, S.; Chaires, J.B.; Trent, J.O. The hTERT core promoter forms three parallel G-quadruplexes. Nucleic Acids Res. 2020, 48, 5720–5734. [Google Scholar] [CrossRef]
- Colebatch, A.J.; Witkowski, T.; Waring, P.M.; McArthur, G.A.; Wong, S.Q.; Dobrovic, A. Optimizing Amplification of the GC-Rich TERT Promoter Region Using 7-Deaza-dGTP for Droplet Digital PCR Quantification of TERT Promoter Mutations. Clin. Chem. 2018, 64, 745–747. [Google Scholar] [CrossRef]
- Kouba, E.; Cheng, L. Clinical utility versus futility: A tipping point for liquid biopsies in bladder cancer. Future Oncol. 2019, 15, 3751–3753. [Google Scholar] [CrossRef] [Green Version]
- Russo, I.J.; Ju, Y.; Gordon, N.S.; Zeegers, M.P.; Cheng, K.K.; James, N.D.; Bryan, R.T.; Ward, D.G. Toward Personalised Liquid Biopsies for Urothelial Carcinoma: Characterisation of ddPCR and Urinary cfDNA for the Detection of the TERT 228G>A/T Mutation. Bladder Cancer 2018, 4, 41–48. [Google Scholar] [CrossRef] [Green Version]
- Satyal, U.; Srivastava, A.; Abbosh, P.H. Urine Biopsy-Liquid Gold for Molecular Detection and Surveillance of Bladder Cancer. Front. Oncol. 2019, 9, 1266. [Google Scholar] [CrossRef] [Green Version]
- Pritchard, J.J.G.; Hamilton, G.; Hurst, C.D.; Fraser, S.; Orange, C.; Knowles, M.A.; Jones, R.J.; Leung, H.Y.; Iwata, T. Monitoring of urothelial cancer disease status after treatment by digital droplet PCR liquid biopsy assays. Urol. Oncol. 2020, 38, 737.e1–737.e10. [Google Scholar] [CrossRef]
- Ge, J.; Liu, M.Y.; Li, L.; Deng, Q.; Liu, F.; Luo, Y.; Wang, L.; Yao, G.; Zhu, D.; Lu, H.; et al. Detection of IDH1 and TERT promoter mutations with droplet digital PCR in diffuse gliomas. Int. J. Clin. Exp. Pathol. 2020, 13, 230–238. [Google Scholar]
- McEvoy, A.C.; Calapre, L.; Pereira, M.R.; Giardina, T.; Robinson, C.; Khattak, M.A.; Meniawy, T.M.; Pritchard, A.L.; Hayward, N.K.; Amanuel, B.; et al. Sensitive droplet digital PCR method for detection of TERT promoter mutations in cell free DNA from patients with metastatic melanoma. Oncotarget 2017, 8, 78890–78900. [Google Scholar] [CrossRef] [Green Version]
- McEvoy, A.C.; Wood, B.A.; Ardakani, N.M.; Pereira, M.R.; Pearce, R.; Cowell, L.; Robinson, C.; Grieu-Iacopetta, F.; Spicer, A.J.; Amanuel, B.; et al. Droplet Digital PCR for Mutation Detection in Formalin-Fixed, Paraffin-Embedded Melanoma Tissues: A Comparison with Sanger Sequencing and Pyrosequencing. J. Mol. Diagn. 2018, 20, 240–252. [Google Scholar] [CrossRef] [Green Version]
- Hayashi, Y.; Fujita, K.; Matsuzaki, K.; Matsushita, M.; Kawamura, N.; Koh, Y.; Nakano, K.; Wang, C.; Ishizuya, Y.; Yamamoto, Y.; et al. Diagnostic potential of TERT promoter and FGFR3 mutations in urinary cell-free DNA in upper tract urothelial carcinoma. Cancer Sci. 2019, 110, 1771–1779. [Google Scholar] [CrossRef] [Green Version]
- Descotes, F.; Kara, N.; Decaussin-Petrucci, M.; Piaton, E.; Geiguer, F.; Rodriguez-Lafrasse, C.; Terrier, J.E.; Lopez, J.; Ruffion, A. Non-invasive prediction of recurrence in bladder cancer by detecting somatic TERT promoter mutations in urine. Br. J. Cancer 2017, 117, 583–587. [Google Scholar] [CrossRef] [PubMed]
- Akkari, Y.; Smith, T.; Westfall, J.; Lupo, S. Implementation of cancer next-generation sequencing testing in a community hospital. Cold Spring Harb. Mol. Case Stud. 2019, 5, a003707. [Google Scholar] [CrossRef] [Green Version]
- Olmedillas-López, S.; García-Arranz, M.; García-Olmo, D. Current and Emerging Applications of Droplet Digital PCR in Oncology. Mol. Diagn. Ther. 2017, 21, 493–510. [Google Scholar] [CrossRef]
- Perkins, G.; Lu, H.; Garlan, F.; Taly, V. Droplet-Based Digital PCR: Application in Cancer Research. Adv. Clin. Chem. 2017, 79, 43–91. [Google Scholar]
- Corless, B.C.; Chang, G.A.; Cooper, S.; Syeda, M.M.; Shao, Y.; Osman, I.; Karlin-Neumann, G.; Polsky, D. Development of Novel Mutation-Specific Droplet Digital PCR Assays Detecting TERT Promoter Mutations in Tumor and Plasma Samples. J. Mol. Diagn. 2019, 21, 274–285. [Google Scholar] [CrossRef] [Green Version]
- Sanmamed, M.F.; Fernández-Landázuri, S.; Rodríguez, C.; Zárate, R.; Lozano, M.D.; Zubiri, L.; Perez-Gracia, J.L.; Martín-Algarra, S.; González, A. Quantitative cell-free circulating BRAFV600E mutation analysis by use of droplet digital PCR in the follow-up of patients with melanoma being treated with BRAF inhibitors. Clin. Chem. 2015, 61, 297–304. [Google Scholar] [CrossRef] [Green Version]
- Zvereva, M.; Pisarev, E.; Hosen, I.; Kisil, O.; Matskeplishvili, S.; Kubareva, E.; Kamalov, D.; Tivtikyan, A.; Manel, A.; Vian, E.; et al. Activating Telomerase TERT Promoter Mutations and Their Application for the Detection of Bladder Cancer. Int. J. Mol. Sci. 2020, 21, 6034. [Google Scholar] [CrossRef]
- Burnham, P.; Dadhania, D.; Heyang, M.; Chen, F.; Westblade, L.F.; Suthanthiran, M.; Lee, J.R.; De Vlaminck, I. Urinary cell-free DNA is a versatile analyte for monitoring infections of the urinary tract. Nat. Commun. 2018, 9, 2412. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McHugh, M.L. Interrater reliability: The kappa statistic. Biochem. Med. 2012, 22, 276–282. [Google Scholar] [CrossRef]
- Meeks, J.J.; Al-Ahmadie, H.; Faltas, B.M.; Taylor, J.A.; Flaig, T.W.; DeGraff, D.J.; Christensen, E.; Woolbright, B.L.; McConkey, D.J.; Dyrskjøt, L. Genomic heterogeneity in bladder cancer: Challenges and possible solutions to improve outcomes. Nat. Rev. Urol. 2020, 17, 259–270. [Google Scholar] [CrossRef] [PubMed]
- Warrick, J.I.; Sjödahl, G.; Kaag, M.; Raman, J.D.; Merrill, S.; Shuman, L.; Chen, G.; Walter, V.; DeGraff, D.J. Intratumoral Heterogeneity of Bladder Cancer by Molecular Subtypes and Histologic Variants. Eur. Urol. 2019, 75, 18–22. [Google Scholar] [CrossRef]
- Pierson-Perry, J.F. EP17-A2: Evaluation of Detection Capability for Clinical Laboratory Measurement Procedures, 2nd ed.; Clinical and Laboratory Standards Institute (CLSI): Wayne, PA, USA, 2012; p. 80. [Google Scholar]
- Zieliński, W. The Shortest Clopper–Pearson Confidence Interval for Binomial Probability. Commun. Stat.-Simul. Comput. 2009, 39, 188–193. [Google Scholar] [CrossRef] [Green Version]
- Landis, J.R.; Koch, G.G. The Measurement of Observer Agreement for Categorical Data. Biometrics 1977, 33, 159–174. [Google Scholar] [CrossRef] [Green Version]

| C228T or C250T Mutations | ddPCR Assays | UroMuTERT Assay | ||||
|---|---|---|---|---|---|---|
| DIAGURO Cohort | PORTO Cohort | DIAGURO Cohort a | PORTO Cohort a | DIAGURO Cohort b | PORTO Cohort b | |
| US cfDNA or UP DNA | UP DNA | US cfDNA or UP DNA | UP DNA | US cfDNA or UP DNA | UP DNA | |
| Total Number analyzed | (n = 181) | (n = 99) | (n = 181) | (n = 99) | (n = 187) | (n = 100) |
| True Positive-no | 79 | 33 | 80 | 32 | 81 | 33 |
| True Negative-no | 85 | 50 | 88 | 50 | 89 | 50 |
| False Positive-no | 7 | 0 | 4 | 0 | 5 | 0 |
| False Negative-no | 10 | 16 | 9 | 17 | 12 | 17 |
| Sensitivity (95% CI)-% | 86.8 (80.3–94.5) | 67.4 (52.5–80.1) | 90.7 (83.1–95.7) | 65.3 (50.4–78.3) | 87.1 (78.6–93.2) | 66.0 (51.2–78.8) |
| Specificity (95% CI)-% | 92.4 (85.0–96.9) | 100.0 (92.9–100.0) | 95.6 (89.2–98.8) | 100.0 (92.9–100.0) | 94.7 (88.0–98.3) | 100.0 (92.9–100.0) |
| Accuracy (95% CI)-% | 91.3 (86.2–95.0) | 90.2 (82.6–95.3) | 93.1 (88.5–96.3) | 89.6 (81.8–94.8) | 92.4 (90.6–94.0) | 89.8 (87.8–91.6) |
| Sample Type | UroMuTERT-MUT | UroMuTERT-WT | Kappa Coefficient | ||
|---|---|---|---|---|---|
| ddPCR-MUT | ddPCR-WT | ddPCR-MUT | ddPCR-WT | ||
| n | n | n | n | ||
| All Follow-up cases (UP cellDNA and US cfDNA) | 173 | 10 | 15 | 196 | 0.87 |
| All US cfDNA Follow-up cases | 86 | 3 | 9 | 123 | 0.89 |
| All UP cellDNA Follow-up cases | 87 | 7 | 6 | 73 | 0.85 |
| Cases US/UP follow-up with MAF < 2% ddPCR | 64 | 10 | 14 | 73 | 0.78 |
| Cases US/UP follow-up with MAF > 2% ddPCR | 109 | 0 | 1 | 123 | 0.99 |
| Cases US/UP follow-up with MAF < 1% ddPCR | 18 | 7 | 4 | 73 | 0.70 |
| Cases US/UP follow-up with MAF > 1% ddPCR | 155 | 3 | 11 | 123 | 0.90 |
| Cases US/UP follow-up with MAF < 0.5% ddPCR | 3 | 7 | 3 | 73 | 0.32 |
| Cases US/UP follow-up with MAF > 0.5% ddPCR | 170 | 3 | 12 | 123 | 0.90 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. 2020 International Agency for Research on Cancer (IARC); Licensee MDPI, Basel, Switzerland. This is an open access article distributed under the terms of the Creative Commons Attribution IGO License (CC BY) (http://creativecommons.org/licenses/by/3.0/igo/legalcode), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. In any reproduction of this article there should not be any suggestion that IARC or this article endorse any specific organisation or products. The use of the IARC logo is not permitted. This notice should be preserved along with the article’s original URL.
Share and Cite
Hosen, M.I.; Forey, N.; Durand, G.; Voegele, C.; Bilici, S.; Avogbe, P.H.; Delhomme, T.M.; Foll, M.; Manel, A.; Vian, E.; et al. Development of Sensitive Droplet Digital PCR Assays for Detecting Urinary TERT Promoter Mutations as Non-Invasive Biomarkers for Detection of Urothelial Cancer. Cancers 2020, 12, 3541. https://doi.org/10.3390/cancers12123541
Hosen MI, Forey N, Durand G, Voegele C, Bilici S, Avogbe PH, Delhomme TM, Foll M, Manel A, Vian E, et al. Development of Sensitive Droplet Digital PCR Assays for Detecting Urinary TERT Promoter Mutations as Non-Invasive Biomarkers for Detection of Urothelial Cancer. Cancers. 2020; 12(12):3541. https://doi.org/10.3390/cancers12123541
Chicago/Turabian StyleHosen, Md Ismail, Nathalie Forey, Geoffroy Durand, Catherine Voegele, Selin Bilici, Patrice Hodonou Avogbe, Tiffany Myriam Delhomme, Matthieu Foll, Arnaud Manel, Emmanuel Vian, and et al. 2020. "Development of Sensitive Droplet Digital PCR Assays for Detecting Urinary TERT Promoter Mutations as Non-Invasive Biomarkers for Detection of Urothelial Cancer" Cancers 12, no. 12: 3541. https://doi.org/10.3390/cancers12123541
APA StyleHosen, M. I., Forey, N., Durand, G., Voegele, C., Bilici, S., Avogbe, P. H., Delhomme, T. M., Foll, M., Manel, A., Vian, E., Meziani, S., De Tilly, B., Polo, G., Lole, O., Francois, P., Boureille, A., Pisarev, E., Salas, A. R. O. S. E., Monteiro-Reis, S., ... Zvereva, M. (2020). Development of Sensitive Droplet Digital PCR Assays for Detecting Urinary TERT Promoter Mutations as Non-Invasive Biomarkers for Detection of Urothelial Cancer. Cancers, 12(12), 3541. https://doi.org/10.3390/cancers12123541








