DNAJA3/Tid1 Is Required for Mitochondrial DNA Maintenance and Regulates Migration and Invasion of Human Gastric Cancer Cells
Abstract
Simple Summary
Abstract
1. Introduction
2. Results
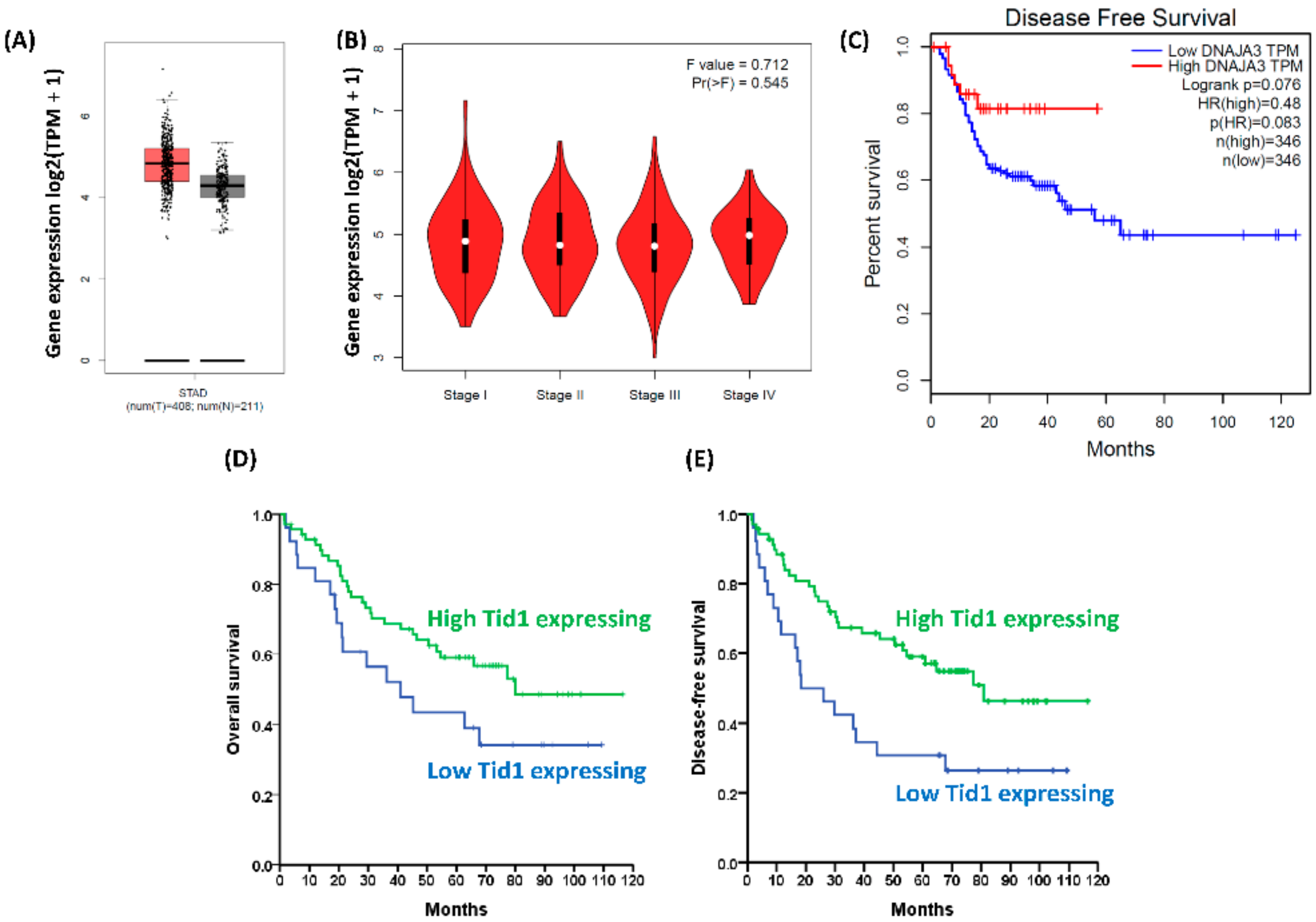
2.1. Low Tid1 Protein-Expressing Gastric Cancer Patients Have Poorer Prognosis than High Tid1-Expressing Patients
2.2. Tid1 Does Not Affect Cell Proliferation, Colony Formation, Tumor Sphere Formation, or Chemotherapy Resistance of Human Gastric Cancer Cells
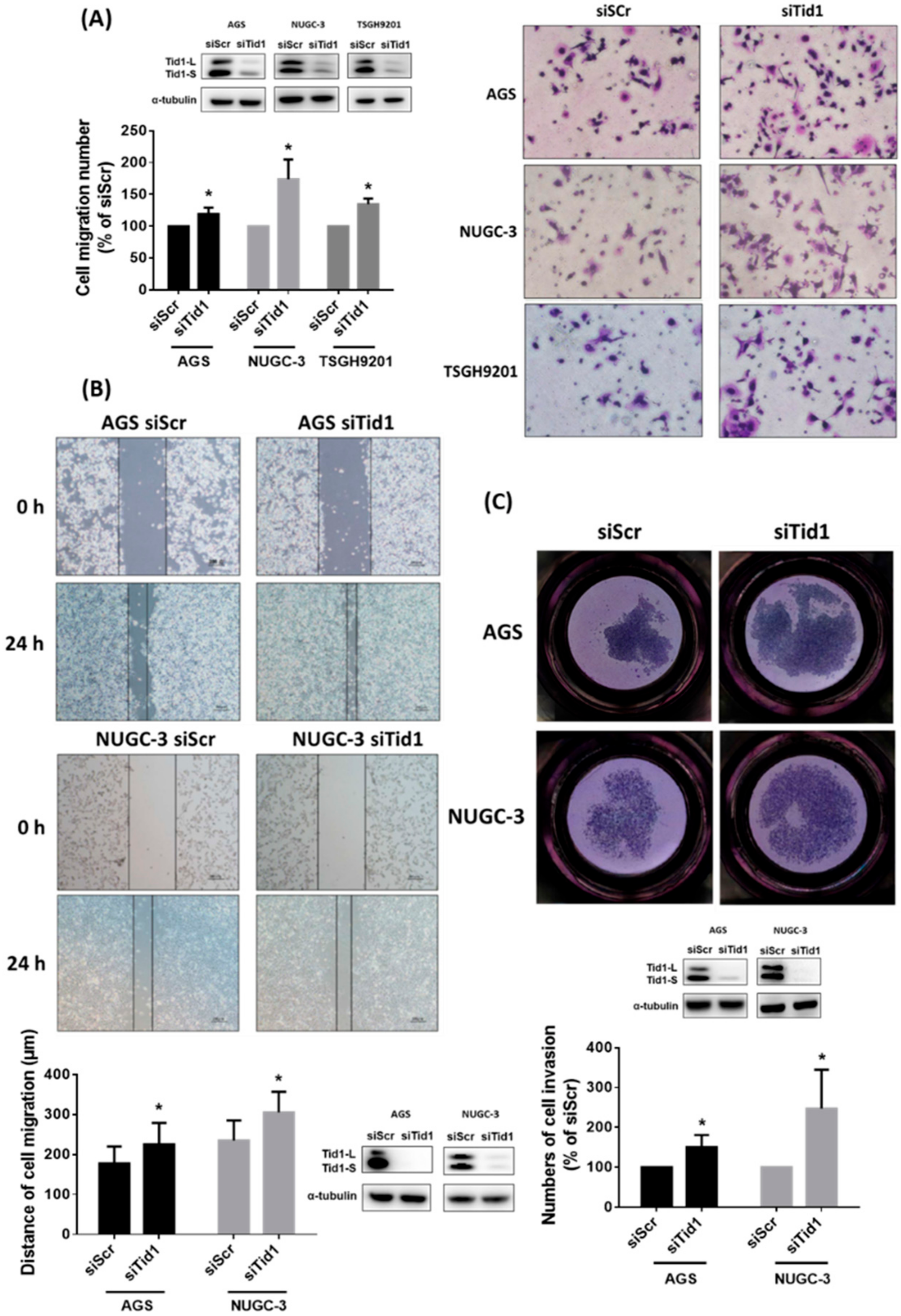
2.3. Tid1 Knockdown Increases Cell Migration and Invasion of Gastric Cancer Cells
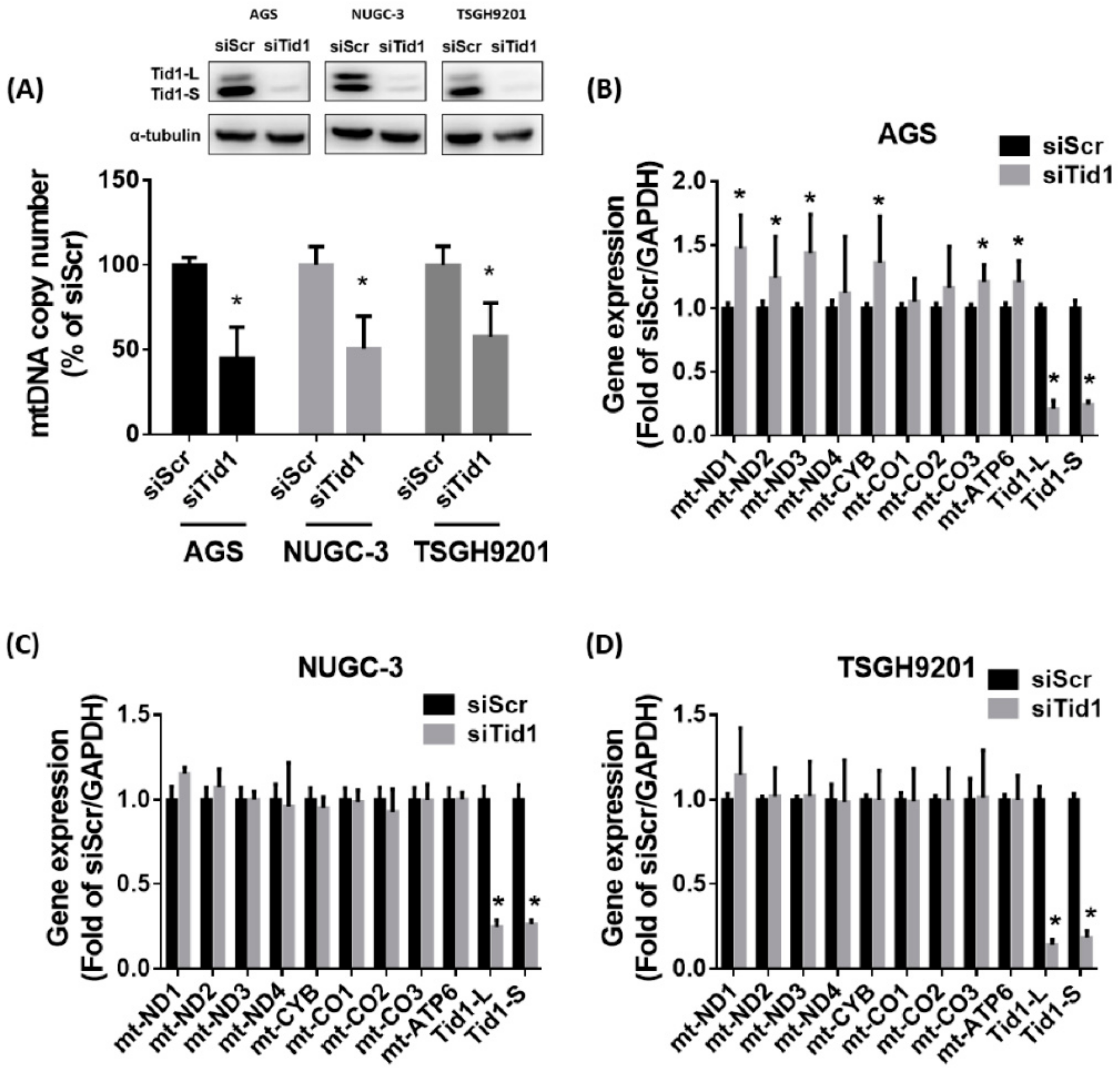
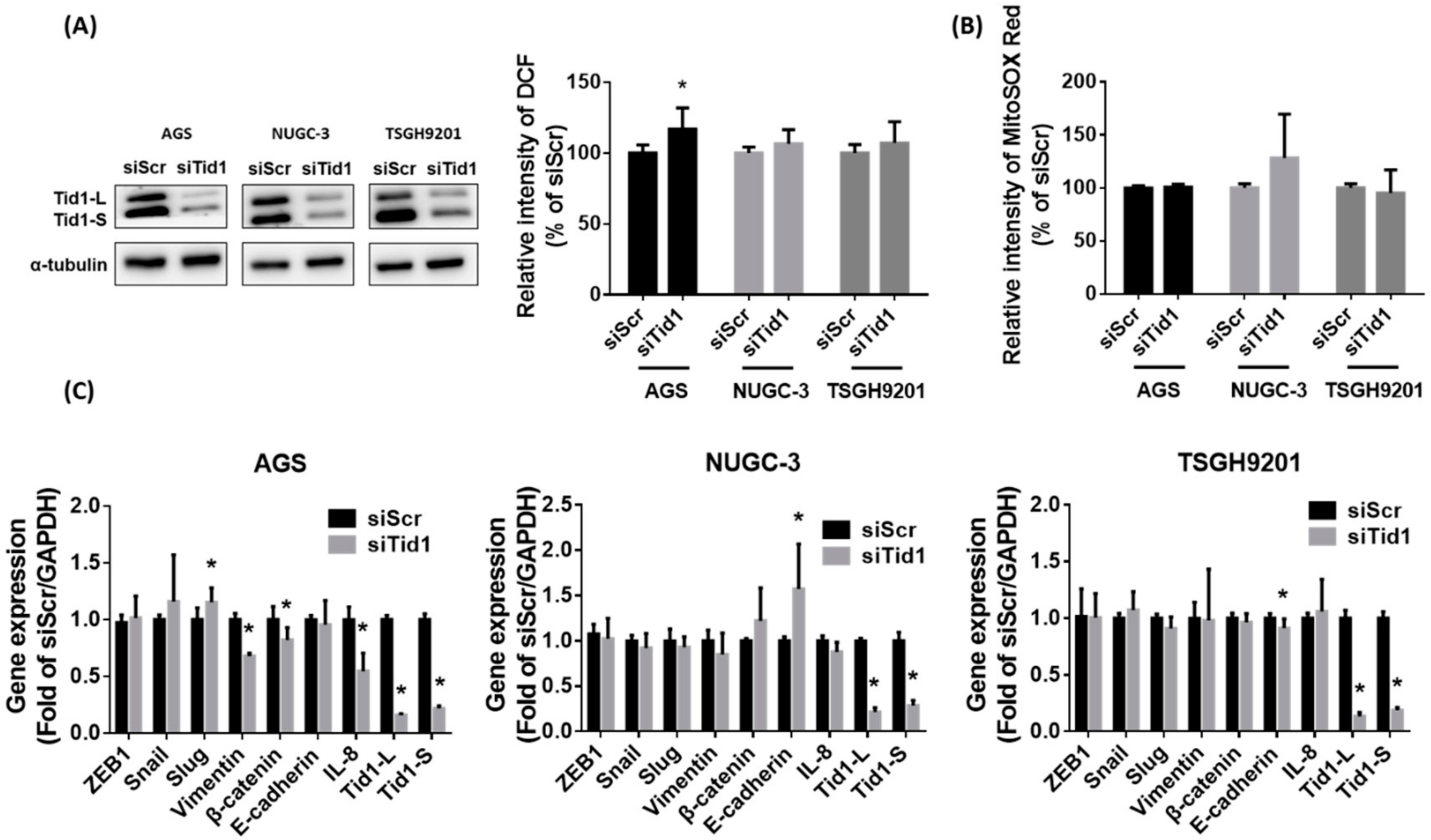
2.4. Tid1 Is Required for mtDNA Maintenance, but Tid1 Knockdown Might Not Consistently Interfere with Mitochondrial Gene Expression, Content, and Respiratory Function in Gastric Cancer Cells
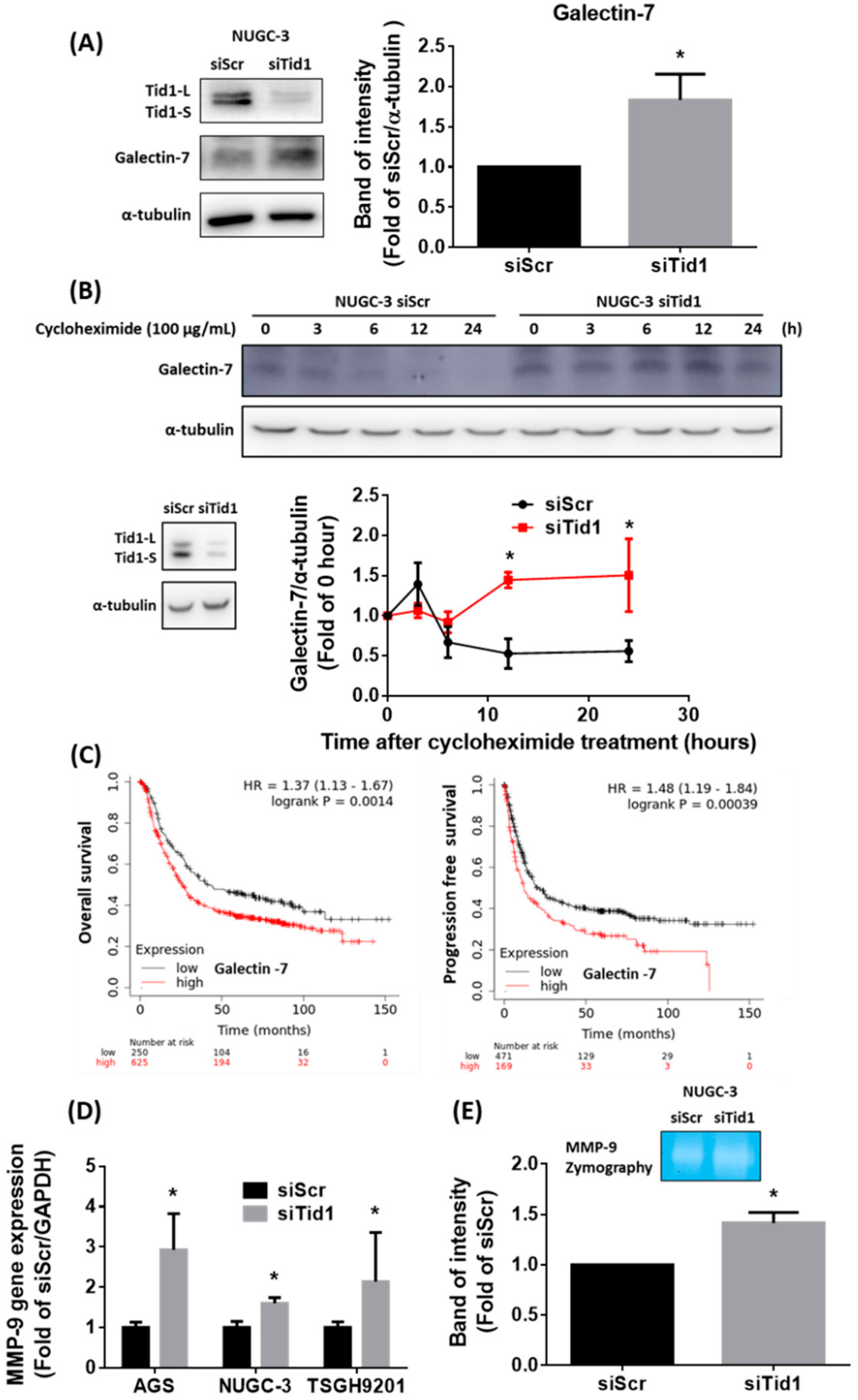
2.5. Galectin-7-Matrix Metallopeptidase 9 (MMP-9) Axis Is Associated with the Tid1 Knockdown-Increased Cell Migration and Invasion of Gastric Cancer Cells
3. Discussion
4. Materials and Methods
4.1. Clinical Gene Expression and Survival Analyses
4.2. Patient Sample Collection and Follow-Up
4.3. Immunohistochemical (IHC) Staining Procedures and Interpretations
4.4. Cell Culture
4.5. siRNA Transfection
4.6. Western Blot Analyses
4.7. Sulforhodamine B (SRB) Cell Proliferation Assay and Chemotherapy Sensitivity Assay
4.8. Soft Agar Colony Formation Assay
4.9. Tumor Sphere Formation Assay
4.10. Wound Healing Migration Assay
4.11. Transwell Migration Assay
4.12. Transwell Invasion Assay
4.13. mtDNA Copy Number Assay
4.14. RNA Extraction and Quantitative Real-Time PCR
4.15. Analysis of Mitochondrial Mass and Mitochondrial Membrane Potential
4.16. Mitochondrial Oxygen Consumption (OCR) Assay
4.17. Intracellular and Mitochondrial ROS Analyses
4.18. Gelatin Zymography
4.19. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| ATP | Adenosine triphosphate |
| ROS | reactive oxygen species |
| OXPHOS | oxidative phosphorylation |
| mtDNA | mitochondrial DNA |
| Tid1 | Tumorous imaginal disc 1 |
| DNAJA3 | DnaJ homolog subfamily A member 3 |
| Hsp | heat shock protein |
| AJCC | American Joint Committee on Cancer |
| DFS | disease free survival |
| OS | overall survival |
| SRB | sulforhodamine B |
| OCR | oxygen consumption rate |
| MMP-9 | matrix metallopeptidase 9 |
| EMT | epithelial to mesenchymal transition |
| IL-8 | interleukin-8 |
| HER2 | human epidermal growth factor receptor 2 |
| STAD | stomach adenocarcinoma |
| CT | computed tomography |
| TNM | tumor-node-metastasis |
| IHC | immunohistochemical |
| PVDF | polyvinylidene difluoride membranes |
| NAO | nonyl acridine orange |
| DCFH-dA | dichlorodihydro-fluorescein diacetate |
| EGFR | epidermal growth factor receptor |
References
- Zhu, A.L.; Sonnenberg, A. Is gastric cancer again rising? J. Clin. Gastroenterol. 2012, 46, 804–806. [Google Scholar] [CrossRef] [PubMed]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2020. CA Cancer J. Clin. 2020, 70, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Ko, K.P.; Shin, A.; Cho, S.; Park, S.K.; Yoo, K.Y. Environmental contributions to gastrointestinal and liver cancer in the Asia-Pacific region. J. Gastroenterol. Hepatol. 2018, 33, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Wagner, A.D.; Grothe, W.; Haerting, J.; Kleber, G.; Grothey, A.; Fleig, W.E. Chemotherapy in advanced gastric cancer: A systematic review and meta-analysis based on aggregate data. J. Clin. Oncol. 2006, 24, 2903–2909. [Google Scholar] [CrossRef]
- Smyth, E.C.; Nilsson, M.; Grabsch, H.I.; van Grieken, N.C.; Lordick, F. Gastric cancer. Lancet 2020, 396, 635–648. [Google Scholar] [CrossRef]
- Digklia, A.; Wagner, A.D. Advanced gastric cancer: Current treatment landscape and future perspectives. World J. Gastroenterol. 2016, 22, 2403–2414. [Google Scholar] [CrossRef]
- Tan, Z. Recent advances in the surgical treatment of advanced gastric cancer: A review. Med Sci Monit. 2019, 25, 3537–3541. [Google Scholar] [CrossRef]
- Leiting, J.L.; Grotz, T.E. Advancements and challenges in treating advanced gastric cancer in the West. World J. Gastrointe. Oncol. 2019, 11, 652–664. [Google Scholar] [CrossRef]
- Youle, R.J. Mitochondria-Striking a balance between host and endosymbiont. Science 2019, 365, eaaw9855. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Warburg, O. On respiratory impairment in cancer cells. Science 1956, 124, 269–270. [Google Scholar] [PubMed]
- Wang, S.F.; Chen, S.; Tseng, L.M.; Lee, H.C. Role of the mitochondrial stress response in human cancer progression. Exp. Biol. Med. 2020, 245, 861–878. [Google Scholar] [CrossRef] [PubMed]
- Taanman, J.W. The mitochondrial genome: Structure, transcription, translation and replication. Biochimica Biophysica Acta Bioenerg. 1999, 1410, 103–123. [Google Scholar] [CrossRef]
- Wu, C.W.; Yin, P.H.; Hung, W.Y.; Li, A.F.; Li, S.H.; Chi, C.W.; Wei, Y.H.; Lee, H.C. Mitochondrial DNA mutations and mitochondrial DNA depletion in gastric cancer. Genes Chromosomes Cancer 2005, 44, 19–28. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.C.; Wei, Y.H. Mitochondrial DNA instability and metabolic shift in human cancers. Int. J. Mol. Sci. 2009, 10, 674–701. [Google Scholar] [CrossRef]
- Lee, H.C.; Yin, P.H.; Lin, J.C.; Wu, C.C.; Chen, C.Y.; Wu, C.W.; Chi, C.W.; Tam, T.N.; Wei, Y.H. Mitochondrial genome instability and mtDNA depletion in human cancers. Ann. N. Y. Acad. Sci. 2005, 1042, 109–122. [Google Scholar] [CrossRef]
- Lee, H.C.; Huang, K.H.; Yeh, T.S.; Chi, C.W. Somatic alterations in mitochondrial DNA and mitochondrial dysfunction in gastric cancer progression. World J. Gastroenterol. 2014, 20, 3950–3959. [Google Scholar] [CrossRef]
- Liu, T.; Daniels, C.K.; Cao, S. Comprehensive review on the HSC70 functions, interactions with related molecules and involvement in clinical diseases and therapeutic potential. Pharmacol. Ther. 2012, 136, 354–374. [Google Scholar] [CrossRef]
- Syken, J.; De-Medina, T.; Munger, K. TID1, a human homolog of the Drosophila tumor suppressor I(2)tid, encodes two mitochondrial modulators of apoptosis with opposing functions. Proc. Natl. Acad. Sci. USA 1999, 96, 8499–8504. [Google Scholar] [CrossRef]
- Lu, B.; Garrido, N.; Spelbrink, J.N.; Suzuki, C.K. Tid1 isoforms are mitochondrial DnaJ-like chaperones with unique carboxyl termini that determine cytosolic fate. J. Biol. Chem. 2006, 281, 13150–13158. [Google Scholar] [CrossRef] [PubMed]
- Ng, A.C.; Baird, S.D.; Screaton, R.A. Essential role of TID1 in maintaining mitochondrial membrane potential homogeneity and mitochondrial DNA integrity. Mol. Cell. Biol. 2014, 34, 1427–1437. [Google Scholar] [CrossRef] [PubMed]
- Lo, J.F.; Hayashi, M.; Woo-Kim, S.; Tian, B.; Huang, J.F.; Fearns, C.; Takayama, S.; Zapata, J.M.; Yang, Y.; Lee, J.D.; et al. Tid1, a cochaperone of the heat shock 70 protein and the mammalian counterpart of the Drosophila tumor suppressor l(2)tid, is critical for early embryonic development and cell survival. Mol. Cell. Biol. 2004, 24, 2226–2236. [Google Scholar] [CrossRef] [PubMed]
- Cheng, L.H.; Hung, K.F.; Lee, T.C.; Huang, C.Y.; Chiu, W.T.; Lo, J.F.; Huang, T.F. Mitochondrial co-chaperone protein Tid1 is required for energy homeostasis during skeletal myogenesis. Stem Cell Res. Ther. 2016, 7, 185. [Google Scholar] [CrossRef]
- Elwi, A.N.; Lee, B.; Meijndert, H.C.; Braun, J.E.; Kim, S.W. Mitochondrial chaperone DnaJA3 induces Drp1-dependent mitochondrial fragmentation. Int. J. Biochem. Cell Biol. 2012, 44, 1366–1376. [Google Scholar] [CrossRef]
- Trinh, D.L.; Elwi, A.N.; Kim, S.W. Direct interaction between p53 and Tid1 proteins affects p53 mitochondrial localization and apoptosis. Oncotarget 2010, 1, 396–404. [Google Scholar] [CrossRef]
- Ahn, B.Y.; Trinh, D.L.; Zajchowski, L.D.; Lee, B.; Elwi, A.N.; Kim, S.W. Tid1 is a new regulator of p53 mitochondrial translocation and apoptosis in cancer. Oncogene 2010, 29, 1155–1166. [Google Scholar] [CrossRef]
- Choi, J.H.; Choi, D.K.; Sohn, K.C.; Kwak, S.S.; Suk, J.; Lim, J.S.; Shin, I.; Kim, S.W.; Lee, J.H.; Joe, C.O.; et al. Absence of a human DnaJ protein hTid-1S correlates with aberrant actin cytoskeleton organization in lesional psoriatic skin. J. Biol. Chem. 2012, 287, 25954–25963. [Google Scholar] [CrossRef]
- Tarunina, M.; Alger, L.; Chu, G.; Munger, K.; Gudkov, A.; Jat, P.S. Functional genetic screen for genes involved in senescence: Role of Tid1, a homologue of the Drosophila tumor suppressor l(2)tid, in senescence and cell survival. Mol. Cell Biol. 2004, 24, 10792–10801. [Google Scholar] [CrossRef]
- Chen, C.Y.; Chiou, S.H.; Huang, C.Y.; Jan, C.I.; Lin, S.C.; Hu, W.Y.; Chou, S.H.; Liu, C.J.; Lo, J.F. Tid1 functions as a tumour suppressor in head and neck squamous cell carcinoma. J. Pathol. 2009, 219, 347–355. [Google Scholar] [CrossRef]
- Jan, C.I.; Yu, C.C.; Hung, M.C.; Harn, H.J.; Nieh, S.; Lee, H.S.; Lou, M.A.; Wu, Y.C.; Chen, C.Y.; Huang, C.Y.; et al. Tid1, CHIP and ErbB2 interactions and their prognostic implications for breast cancer patients. J. Pathol. 2011, 225, 424–437. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.Y.; Jan, C.I.; Lo, J.F.; Yang, S.C.; Chang, Y.L.; Pan, S.H.; Wang, W.L.; Hong, T.M.; Yang, P.C. Tid1-L inhibits EGFR signaling in lung adenocarcinoma by enhancing EGFR Ubiquitinylation and degradation. Cancer Res. 2013, 73, 4009–4019. [Google Scholar] [CrossRef] [PubMed]
- Kurzik-Dumke, U.; Horner, M.; Czaja, J.; Nicotra, M.R.; Simiantonaki, N.; Koslowski, M.; Natali, P.G. Progression of colorectal cancers correlates with overexpression and loss of polarization of expression of the htid-1 tumor suppressor. Int. J. Mol. Med. 2008, 21, 19–31. [Google Scholar] [CrossRef]
- Wang, T.H.; Lin, Y.H.; Yang, S.C.; Chang, P.C.; Wang, T.C.; Chen, C.Y. Tid1-S regulates the mitochondrial localization of EGFR in non-small cell lung carcinoma. Oncogenesis 2017, 6, e361. [Google Scholar] [CrossRef]
- American Joint Committee on Cancer. AJCC Cancer Staging Manual, 8th ed.; Springer: New York, NY, USA, 2017. [Google Scholar]
- Edwards, K.M.; Munger, K. Depletion of physiological levels of the human TID1 protein renders cancer cell lines resistant to apoptosis mediated by multiple exogenous stimuli. Oncogene 2004, 23, 8419–8431. [Google Scholar] [CrossRef] [PubMed]
- Hung, W.Y.; Huang, K.H.; Wu, C.W.; Chi, C.W.; Kao, H.L.; Li, A.F.; Yin, P.H.; Lee, H.C. Mitochondrial dysfunction promotes cell migration via reactive oxygen species-enhanced beta5-integrin expression in human gastric cancer SC-M1 cells. Biochim. Biophys. Acta Gen. Subj. 2012, 1820, 1102–1110. [Google Scholar] [CrossRef]
- Hung, W.Y.; Wu, C.W.; Yin, P.H.; Chang, C.J.; Li, A.F.; Chi, C.W.; Wei, Y.H.; Lee, H.C. Somatic mutations in mitochondrial genome and their potential roles in the progression of human gastric cancer. Biochim. Biophys. Acta Gen. Subj 2010, 1800, 264–270. [Google Scholar] [CrossRef]
- Wang, S.F.; Chen, M.S.; Chou, Y.C.; Ueng, Y.F.; Yin, P.H.; Yeh, T.S.; Lee, H.C. Mitochondrial dysfunction enhances cisplatin resistance in human gastric cancer cells via the ROS-activated GCN2-eIF2alpha-ATF4-xCT pathway. Oncotarget 2016, 7, 74132–74151. [Google Scholar] [CrossRef]
- Senturk, S.; Yao, Z.; Camiolo, M.; Stiles, B.; Rathod, T.; Walsh, A.M.; Nemajerova, A.; Lazzara, M.J.; Altorki, N.K.; Krainer, A.; et al. p53Psi is a transcriptionally inactive p53 isoform able to reprogram cells toward a metastatic-like state. Proc. Natl. Acad. Sci. USA 2014, 111, E3287–E3296. [Google Scholar] [CrossRef]
- Kim, S.W.; Hayashi, M.; Lo, J.F.; Fearns, C.; Xiang, R.; Lazennec, G.; Yang, Y.; Lee, J.D. Tid1 negatively regulates the migratory potential of cancer cells by inhibiting the production of interleukin-8. Cancer Res. 2005, 65, 8784–8791. [Google Scholar] [CrossRef]
- Wu, Y.; Zhou, B.P. New insights of epithelial-mesenchymal transition in cancer metastasis. Acta Biochim. Biophys. Sinica 2008, 40, 643–650. [Google Scholar] [CrossRef] [PubMed]
- Green, A.R.; Green, V.L.; White, M.C.; Speirs, V. Expression of cytokine messenger RNA in normal and neoplastic human breast tissue: Identification of interleukin-8 as a potential regulatory factor in breast tumours. Int. J. Cancer 1997, 72, 937–941. [Google Scholar] [CrossRef]
- De Larco, J.E.; Wuertz, B.R.; Rosner, K.A.; Erickson, S.A.; Gamache, D.E.; Manivel, J.C.; Furcht, L.T. A potential role for interleukin-8 in the metastatic phenotype of breast carcinoma cells. Am. J. Pathol. 2001, 158, 639–646. [Google Scholar] [CrossRef]
- Bendre, M.S.; Gaddy-Kurten, D.; Mon-Foote, T.; Akel, N.S.; Skinner, R.A.; Nicholas, R.W.; Suva, L.J. Expression of interleukin 8 and not parathyroid hormone-related protein by human breast cancer cells correlates with bone metastasis in vivo. Cancer Res. 2002, 62, 5571–5579. [Google Scholar]
- Chen, Y.S.; Chang, C.W.; Tsay, Y.G.; Huang, L.Y.; Wu, Y.C.; Cheng, L.H.; Yang, C.C.; Wu, C.H.; Teo, W.H.; Hung, K.F.; et al. HSP40 co-chaperone protein Tid1 suppresses metastasis of head and neck cancer by inhibiting Galectin-7-TCF3-MMP9 axis signaling. Theranostics 2018, 8, 3841–3855. [Google Scholar] [CrossRef]
- Szasz, A.M.; Lanczky, A.; Nagy, A.; Forster, S.; Hark, K.; Green, J.E.; Boussioutas, A.; Busuttil, R.; Szabo, A.; Gyorffy, B.; et al. Cross-validation of survival associated biomarkers in gastric cancer using transcriptomic data of 1065 patients. Oncotarget 2016, 7, 49322–49333. [Google Scholar] [CrossRef]
- Silver, P.A.; Way, J.C. Eukaryotic DnaJ homologs and the specificity of Hsp70 activity. Cell 1993, 74, 5–6. [Google Scholar] [CrossRef]
- Georgopoulos, C.; Welch, W.J. Role of the major heat shock proteins as molecular chaperones. Annu. Rev. Cell Biol. 1993, 9, 601–634. [Google Scholar] [CrossRef]
- Cyr, D.M.; Langer, T.; Douglas, M.G. DnaJ-like proteins: Molecular chaperones and specific regulators of Hsp70. Trends Biochem. Sci. 1994, 19, 176–181. [Google Scholar] [CrossRef]
- Pfanner, N.; Craig, E.A.; Meijer, M. The protein import machinery of the mitochondrial inner membrane. Trends Biochem. Sci. 1994, 19, 368–372. [Google Scholar] [CrossRef]
- Stuart, R.A.; Cyr, D.M.; Craig, E.A.; Neupert, W. Mitochondrial molecular chaperones: Their role in protein translocation. Trends Biochem. Sci. 1994, 19, 87–92. [Google Scholar] [CrossRef]
- Bae, M.K.; Jeong, J.W.; Kim, S.H.; Kim, S.Y.; Kang, H.J.; Kim, D.M.; Bae, S.K.; Yun, I.; Trentin, G.A.; Rozakis-Adcock, M.; et al. Tid-1 interacts with the von Hippel-Lindau protein and modulates angiogenesis by destabilization of HIF-1alpha. Cancer Res. 2005, 65, 2520–2525. [Google Scholar] [CrossRef] [PubMed]
- Cody, C.C.; Higuchi, M. The awakening of an advanced malignant cancer: An insult to the mitochondrial genome. Biochim. Biophys. Acta Gen. Subj. 2012, 1820, 652–662. [Google Scholar]
- Li, X.; Zhong, Y.; Lu, J.; Axcrona, K.; Eide, L.; Syljuåsen, R.G.; Peng, Q.; Wang, J.; Zhang, H.; Goscinski, M.A.; et al. MtDNA depleted PC3 cells exhibit Warburg effect and cancer stem cell features. Oncotarget 2016, 7, 40297. [Google Scholar] [CrossRef]
- Naito, A.; Cook, C.C.; Mizumachi, T.; Wang, M.; Xie, C.H.; Evans, T.T.; Kelly, T.; Higuchi, M. Progressive tumor features accompany epithelial-mesenchymal transition induced in mitochondrial DNA-depleted cells. Cancer Sci. 2008, 99, 1584–1588. [Google Scholar] [CrossRef]
- Singh, K.K.; Ayyasamy, V.; Owens, K.M.; Koul, M.S.; Vujcic, M. Mutations in mitochondrial DNA polymerase-gamma promote breast tumorigenesis. J. Hum. Genet. 2009, 54, 516–524. [Google Scholar] [CrossRef]
- Castro, J.P.; Wardelmann, K.; Grune, T.; Kleinridders, A. Mitochondrial chaperones in the brain: Safeguarding brain health and metabolism? Front. Endocrinol. 2018, 9, 196. [Google Scholar] [CrossRef]
- Lee, B.; Ahn, Y.; Kang, S.M.; Park, Y.; Jeon, Y.J.; Rho, J.M.; Kim, S.W. Stoichiometric expression of mtHsp40 and mtHsp70 modulates mitochondrial morphology and cristae structure via Opa1L cleavage. Mol. Biol. Cell 2015, 26, 2156–2167. [Google Scholar] [CrossRef]
- Hayashi, M.; Imanaka-Yoshida, K.; Yoshida, T.; Wood, M.; Fearns, C.; Tatake, R.J.; Lee, J.D. A crucial role of mitochondrial Hsp40 in preventing dilated cardiomyopathy. Nat. Med. 2006, 12, 128–132. [Google Scholar] [CrossRef]
- Kim, S.J.; Kwon, M.C.; Ryu, M.J.; Chung, H.K.; Tadi, S.; Kim, Y.K.; Kim, J.M.; Lee, S.H.; Park, J.H.; Kweon, G.R.; et al. CRIF1 is essential for the synthesis and insertion of oxidative phosphorylation polypeptides in the mammalian mitochondrial membrane. Cell Metab. 2012, 16, 274–283. [Google Scholar] [CrossRef]
- Isomoto, H.; Oka, M.; Yano, Y.; Kanazawa, Y.; Soda, H.; Terada, R.; Yasutake, T.; Nakayama, T.; Shikuwa, S.; Takeshima, F.; et al. Expression of heat shock protein (Hsp) 70 and Hsp 40 in gastric cancer. Cancer Lett. 2003, 198, 219–228. [Google Scholar] [CrossRef]
- Castle, P.E.; Ashfaq, R.; Ansari, F.; Muller, C.Y. Immunohistochemical evaluation of heat shock proteins in normal and preinvasive lesions of the cervix. Cancer Lett. 2005, 229, 245–252. [Google Scholar] [CrossRef] [PubMed]
- Oka, M.; Sato, S.; Soda, H.; Fukuda, M.; Kawabata, S.; Nakatomi, K.; Shiozawa, K.; Nakamura, Y.; Ohtsuka, K.; Kohno, S.; et al. Autoantibody to heat shock protein Hsp40 in sera of lung cancer patients. Jpn. J. Cancer Res. 2001, 92, 316–320. [Google Scholar] [CrossRef] [PubMed]
- Kanazawa, Y.; Isomoto, H.; Oka, M.; Yano, Y.; Soda, H.; Shikuwa, S.; Takeshima, F.; Omagari, K.; Mizuta, Y.; Murase, K.; et al. Expression of heat shock protein (Hsp) 70 and Hsp 40 in colorectal cancer. Med. Oncol. 2003, 20, 157–164. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.W.; Chao, T.H.; Xiang, R.; Lo, J.F.; Campbell, M.J.; Fearns, C.; Lee, J.D. Tid1, the human homologue of a Drosophila tumor suppressor, reduces the malignant activity of ErbB-2 in carcinoma cells. Cancer Res. 2004, 64, 7732–7739. [Google Scholar] [CrossRef] [PubMed]
- Copeland, E.; Balgobin, S.; Lee, C.M.; Rozakis-Adcock, M. hTID-1 defines a novel regulator of c-Met Receptor signaling in renal cell carcinomas. Oncogene 2011, 30, 2252–2263. [Google Scholar] [CrossRef]
- Dhennin-Duthille, I.; Nyga, R.; Yahiaoui, S.; Gouilleux-Gruart, V.; Regnier, A.; Lassoued, K.; Gouilleux, F. The tumor suppressor hTid1 inhibits STAT5b activity via functional interaction. J. Biol. Chem. 2011, 286, 5034–5042. [Google Scholar] [CrossRef]
- Qian, J.; Perchiniak, E.M.; Sun, K.; Groden, J. The mitochondrial protein hTID-1 partners with the caspase-cleaved adenomatous polyposis cell tumor suppressor to facilitate apoptosis. Gastroenterology 2010, 138, 1418–1428. [Google Scholar] [CrossRef]
- Advedissian, T.; Deshayes, F.; Viguier, M. Galectin-7 in epithelial homeostasis and carcinomas. Int. J. Mol. Sci. 2017, 18, 2760. [Google Scholar] [CrossRef]
- Liu, F.T.; Rabinovich, G.A. Galectins as modulators of tumour progression. Nat. Rev. Cancer 2005, 5, 29–41. [Google Scholar] [CrossRef]
- Vladoiu, M.C.; Labrie, M.; St-Pierre, Y. Intracellular galectins in cancer cells: Potential new targets for therapy (Review). Int. J. Oncol. 2014, 44, 1001–1014. [Google Scholar] [CrossRef] [PubMed]
- Balan, V.; Nangia-Makker, P.; Raz, A. Galectins as cancer biomarkers. Cancers 2010, 2, 592–610. [Google Scholar] [CrossRef] [PubMed]
- Kaur, M.; Kaur, T.; Kamboj, S.S.; Singh, J. Roles of galectin-7 in cancer. Asian Pac. J. Cancer Prev. 2016, 17, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.J.; Hwang, J.A.; Ro, J.Y.; Lee, Y.S.; Chun, K.H. Galectin-7 is epigenetically-regulated tumor suppressor in gastric cancer. Oncotarget 2013, 4, 1461–1471. [Google Scholar] [CrossRef]
- St-Pierre, Y.; Campion, C.G.; Grosset, A.A. A distinctive role for galectin-7 in cancer? Front. Biosci. 2012, 17, 438–450. [Google Scholar] [CrossRef]
- Sengupta, N.; MacDonald, T.T. The role of matrix metalloproteinases in stromal/epithelial interactions in the gut. Physiology 2007, 22, 401–409. [Google Scholar] [CrossRef]
- Al-Batran, S.E.; Pauligk, C.; Wirtz, R.; Werner, D.; Steinmetz, K.; Homann, N.; Schmalenberg, H.; Hofheinz, R.D.; Hartmann, J.T.; Atmaca, A.; et al. The validation of matrix metalloproteinase-9 mRNA gene expression as a predictor of outcome in patients with metastatic gastric cancer. Ann. Oncol. 2012, 23, 1699–1705. [Google Scholar] [CrossRef]
- Yang, H.; Wang, B.; Yan, J.; Wang, T.; Zhou, X.N.; Wen, H.Y.; Zhu, X.M. Toll-like receptor 2 promotes invasion by SGC-7901 human gastric carcinoma cells and is associated with gastric carcinoma metastasis. Ann. Clin. Lab Sci. 2014, 44, 158–166. [Google Scholar]
- Shan, Y.Q.; Ying, R.C.; Zhou, C.H.; Zhu, A.K.; Ye, J.; Zhu, W.; Ju, T.F.; Jin, H.C. MMP-9 is increased in the pathogenesis of gastric cancer by the mediation of HER2. Cancer Gene Ther. 2015, 22, 101–107. [Google Scholar] [CrossRef]
- Tang, Z.; Li, C.; Kang, B.; Gao, G.; Li, C.; Zhang, Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017, 45, W98–W102. [Google Scholar] [CrossRef]
- Huang, K.H.; Hsu, C.C.; Fang, W.L.; Chi, C.W.; Sung, M.T.; Kao, H.L.; Li, A.F.; Yin, P.H.; Yang, M.H.; Lee, H.C.; et al. SIRT3 expression as a biomarker for better prognosis in gastric cancer. World J. Surg. 2014, 38, 910–917. [Google Scholar] [CrossRef] [PubMed]
- Ming, S.C. Gastric carcinoma. A pathobiological classification. Cancer 1977, 39, 2475–2485. [Google Scholar] [CrossRef]


| Characteristic | Tid1 Low Expression (0,1) (n = 30) | Tid1 High Expression (2,3) (n = 70) | p |
|---|---|---|---|
| Age (mean ± SD) | 67.80 ± 12.0 | 67.49 ± 12.6 | 0.908 |
| Gender | |||
| Male | 22 | 57 | |
| Female | 8 | 13 | 0.362 |
| Tumor size | |||
| <4 cm | 5 | 19 | |
| 4–8 cm | 17 | 38 | |
| >8 cm | 8 | 13 | 0.441 |
| Tumor location | |||
| Upper stomach | 5 | 15 | |
| Middle stomach | 15 | 25 | |
| Lower stomach | 10 | 27 | |
| Whole stomach | 0 | 3 | 0.548 |
| Cell grade | |||
| Poor differentiated | 18 | 35 | |
| Moderate differentiated | 11 | 34 | |
| Well differentiated | 1 | 1 | 0.381 |
| Gross appearance | |||
| Superficial type | 1 | 0 | |
| Borrmann type I & II | 7 | 25 | |
| Borrmann type III & IV | 22 | 45 | 0.177 |
| Stromal reaction type | |||
| Medullary type | 2 | 4 | |
| Intermediate type | 15 | 51 | |
| Scirrhous type | 13 | 15 | 0.058 |
| Lauren’s histology | |||
| Intestinal type | 8 | 27 | |
| Diffuse type | 14 | 19 | |
| Mixed type | 8 | 24 | 0.161 |
| Ming’s histology | |||
| Expanding | 2 | 7 | |
| Infiltrating | 28 | 63 | 0.720 |
| Lymphovascular invasion | |||
| No | 8 | 19 | |
| Yes | 22 | 51 | 0.133 |
| TNM pathological T category | |||
| T1 | 10 | 35 | |
| T2 | 7 | 15 | |
| T3 | 11 | 14 | |
| T4 | 2 | 6 | 0.289 |
| TNM pathological N category | |||
| N0 | 4 | 19 | |
| N1 | 5 | 20 | |
| N2 | 7 | 17 | |
| N3 | 14 | 14 | 0.041 * |
| TNM Stage | |||
| I | 1 | 9 | |
| II | 6 | 18 | |
| III | 23 | 43 | 0.274 |
| Overall survival rate (5 years) | 43.3% | 58.5% | 0.082 |
| Disease-free survival rate (5 years) | 36.7% | 57.1% | 0.008 * |
| Gene | Forward Primer Sequence | Reverse Primer Sequence |
|---|---|---|
| GAPDH | 5′-CCGTCTAGAAAAACCTGCC-3′ | 5′-GCCAAATTCGTTGTCATACC-3′ |
| Tid1-L | 5′-GTTCCAAAGAGGCTAACGAG-3′ | 5′-TTGCTTCCTGCGGAGCTATC-3 |
| Tid1-S | 5′-GTTCCAAAGAGGCTAACGAG-3′ | 5′-GGCCTAGTTTCCAGTGGATC-3′ |
| mt-ND1 | 5′-CCCTAAAACCCGCCACATCT-3′ | 5′-GGCTAGAATAAATAGGAGGCCTAGGT-3′ |
| mt-ND2 | 5′-AACCCGTCATCTACTCTACCATCT-3′ | 5′-GCTTCTGTGGAACGAGGGTTTATTT-3′ |
| mt-ND3 | 5′-CCACAACTCAACGGCTACATAGAAA-3′ | 5′-GGGTAAAAGGAGGGCAATTTCTAGA-3′ |
| mt-ND4 | 5′-TCACAACACCCTAGGCTCACTAA-3′ | 5′-GGGAGTCATAAGTGGAGTCCGT-3′ |
| mt-CYB | 5′-TCACCTCCCATTCCGATAAAATCAC-3′ | 5′-GGGTTGGCTAGGGTATAATTGTCTG-3′ |
| mt-CO1 | 5′-CAGCAGTCCTACTTCTCCTATCTCT-3′ | 5′-GGGTCGAAGAAGGTGGTGTT-3′ |
| mt-CO2 | 5′-GGGTCGAAGAAGGTGGTGTT-3 | 5′-GTAAAGGATGCGTAGGGATGGG-3′ |
| mt-CO3 | 5′-TCACCTGAGCTCACCATAGTCTAAT-3′ | 5′-GCCGTCGGAAATGGTGAAG-3′ |
| mt-ATP | 5′-CGTACGCCTAACCGCTAACATT-3′ | 5 -GCGACAGCGATTTCTAGGATAGT-3′ |
| Snail | 5′-ACATCCGAAGCCACACGCTGC-3′ | 5′-CGCAGGTTGGAGCGGTCAGC-3′ |
| Slu | 5′-TGTGTGGACTACCGCTGC-3′ | 5′-TCCGGAAAGAGGAGAGAGG-3′ |
| Vimentin | 5′-CCTTGAACGCAAAGTGGAATC-3′ | 5′-GACATGCTGTTCCTGAATCTGAG-3′ |
| β-catenin | 5′-CCAGCCGACACCAAGAAG-3′ | 5′-CGAATCAATCCAACAGTAGCC-3′ |
| E-cadherin | 5′-CCCACCACGTACAAGGGTC-3′ | 5′-CTGGGGTATTGGGGGCATC-3′ |
| ZEB1 | 5’- TTCAAACCCATAGTGGTTGCT-3’ | 5’- TGGGAGATACCAAACCAACTG-3’ |
| IL-8 | 5′-AATTGAGGCCAAGGGCCAAGAG-3′ | 5′-AGGACTTGTGGATCCTGGCTAG-3′ |
| MMP-9 | 5′-TTGACAGCGACAAGAAGTGG-3′ | 5′-CCCTCAGTGAAGCGGTACAT-3′ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, S.-F.; Huang, K.-H.; Tseng, W.-C.; Lo, J.-F.; Li, A.F.-Y.; Fang, W.-L.; Chen, C.-F.; Yeh, T.-S.; Chang, Y.-L.; Chou, Y.-C.; et al. DNAJA3/Tid1 Is Required for Mitochondrial DNA Maintenance and Regulates Migration and Invasion of Human Gastric Cancer Cells. Cancers 2020, 12, 3463. https://doi.org/10.3390/cancers12113463
Wang S-F, Huang K-H, Tseng W-C, Lo J-F, Li AF-Y, Fang W-L, Chen C-F, Yeh T-S, Chang Y-L, Chou Y-C, et al. DNAJA3/Tid1 Is Required for Mitochondrial DNA Maintenance and Regulates Migration and Invasion of Human Gastric Cancer Cells. Cancers. 2020; 12(11):3463. https://doi.org/10.3390/cancers12113463
Chicago/Turabian StyleWang, Sheng-Fan, Kuo-Hung Huang, Wei-Chuan Tseng, Jeng-Fan Lo, Anna Fen-Yau Li, Wen-Liang Fang, Chian-Feng Chen, Tien-Shun Yeh, Yuh-Lih Chang, Yueh-Ching Chou, and et al. 2020. "DNAJA3/Tid1 Is Required for Mitochondrial DNA Maintenance and Regulates Migration and Invasion of Human Gastric Cancer Cells" Cancers 12, no. 11: 3463. https://doi.org/10.3390/cancers12113463
APA StyleWang, S.-F., Huang, K.-H., Tseng, W.-C., Lo, J.-F., Li, A. F.-Y., Fang, W.-L., Chen, C.-F., Yeh, T.-S., Chang, Y.-L., Chou, Y.-C., Hung, H.-H., & Lee, H.-C. (2020). DNAJA3/Tid1 Is Required for Mitochondrial DNA Maintenance and Regulates Migration and Invasion of Human Gastric Cancer Cells. Cancers, 12(11), 3463. https://doi.org/10.3390/cancers12113463






