Challenges and Therapeutic Opportunities of Autophagy in Cancer Therapy
Abstract
Simple Summary
Abstract
1. Introduction
2. Cellular and Molecular Aspects of Autophagy
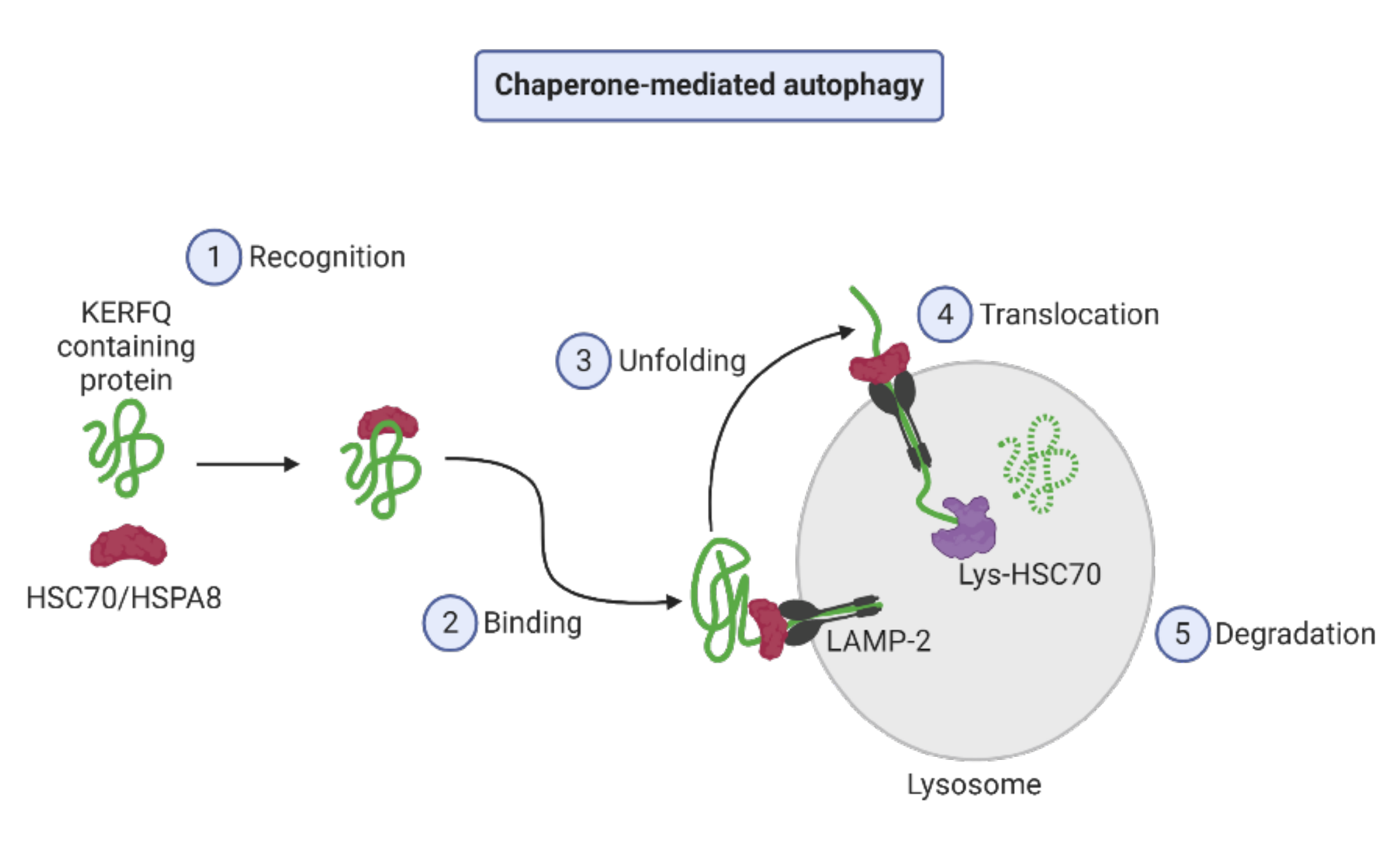
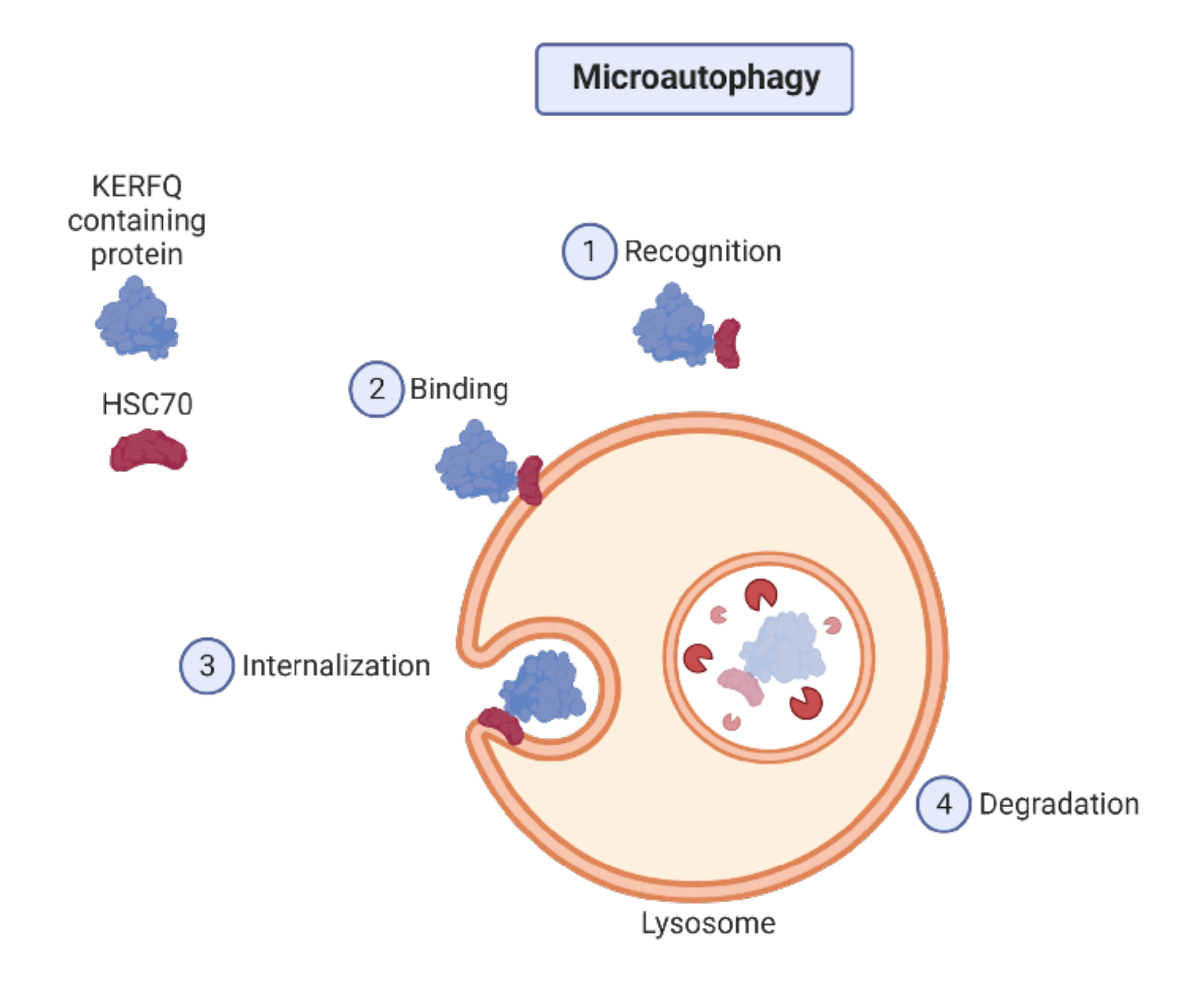
2.1. Autophagy Types
2.2. Autophagy Regulation
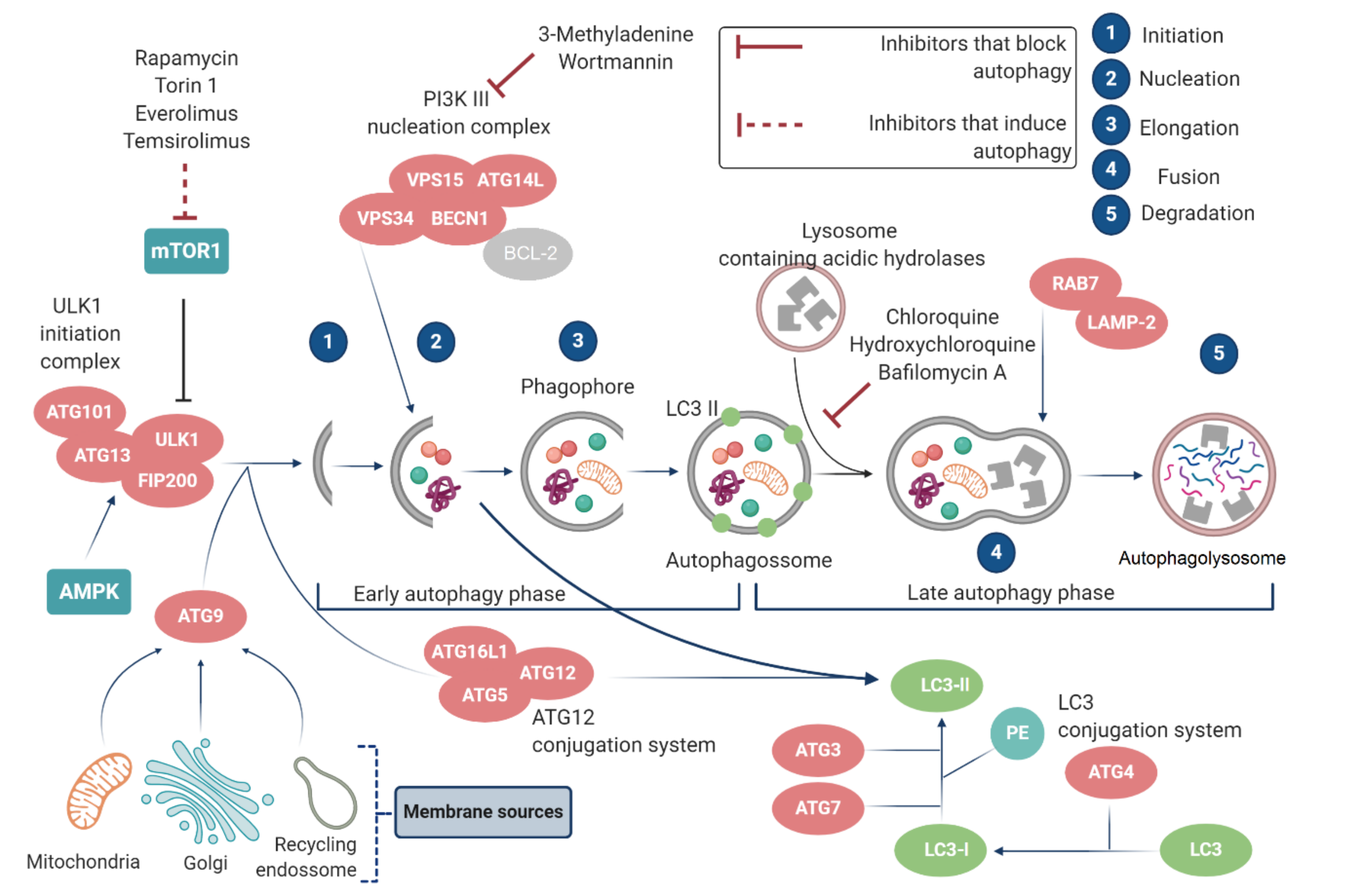
2.2.1. mTOR and ATGs
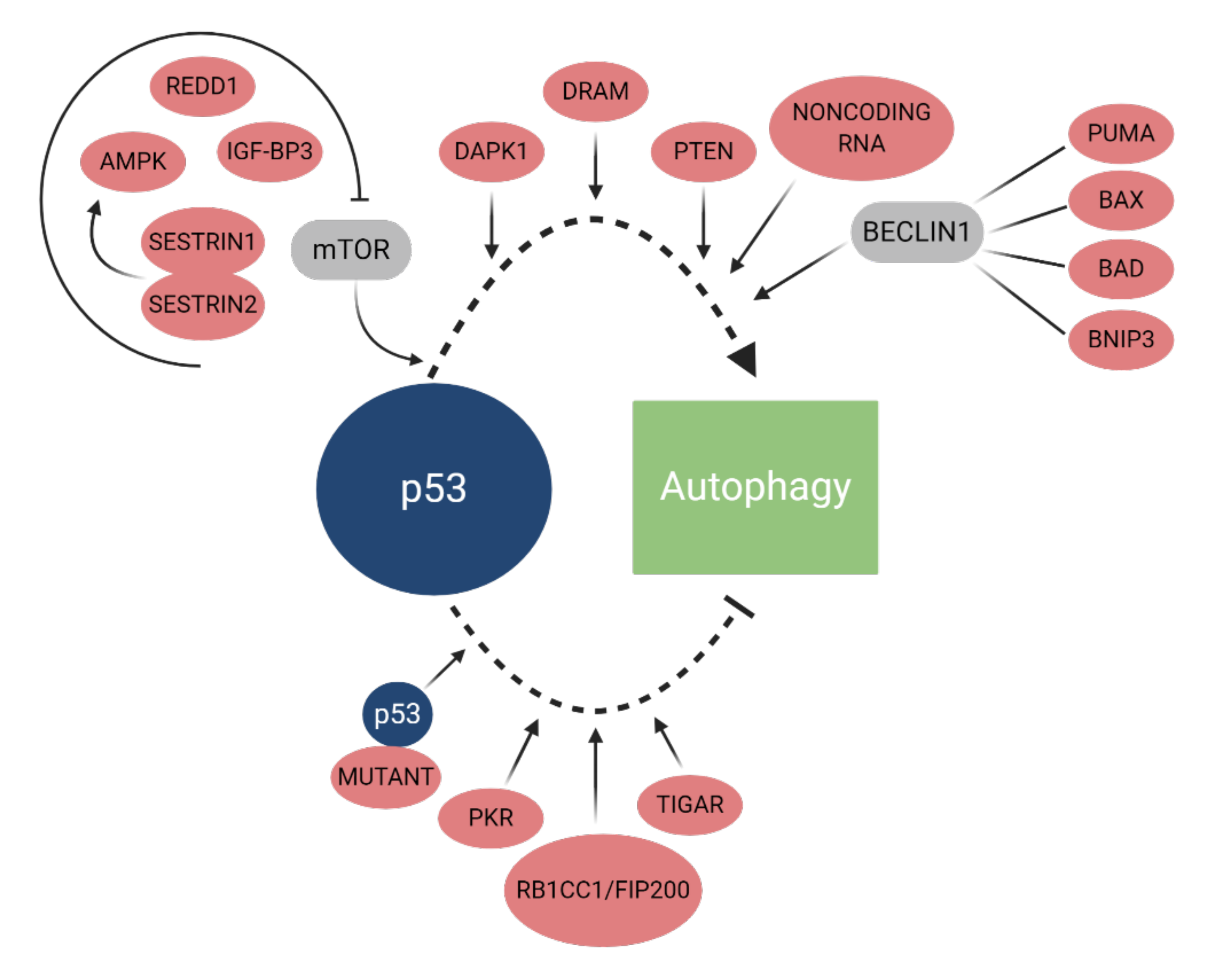
2.2.2. p53
2.2.3. Non-Coding RNAs
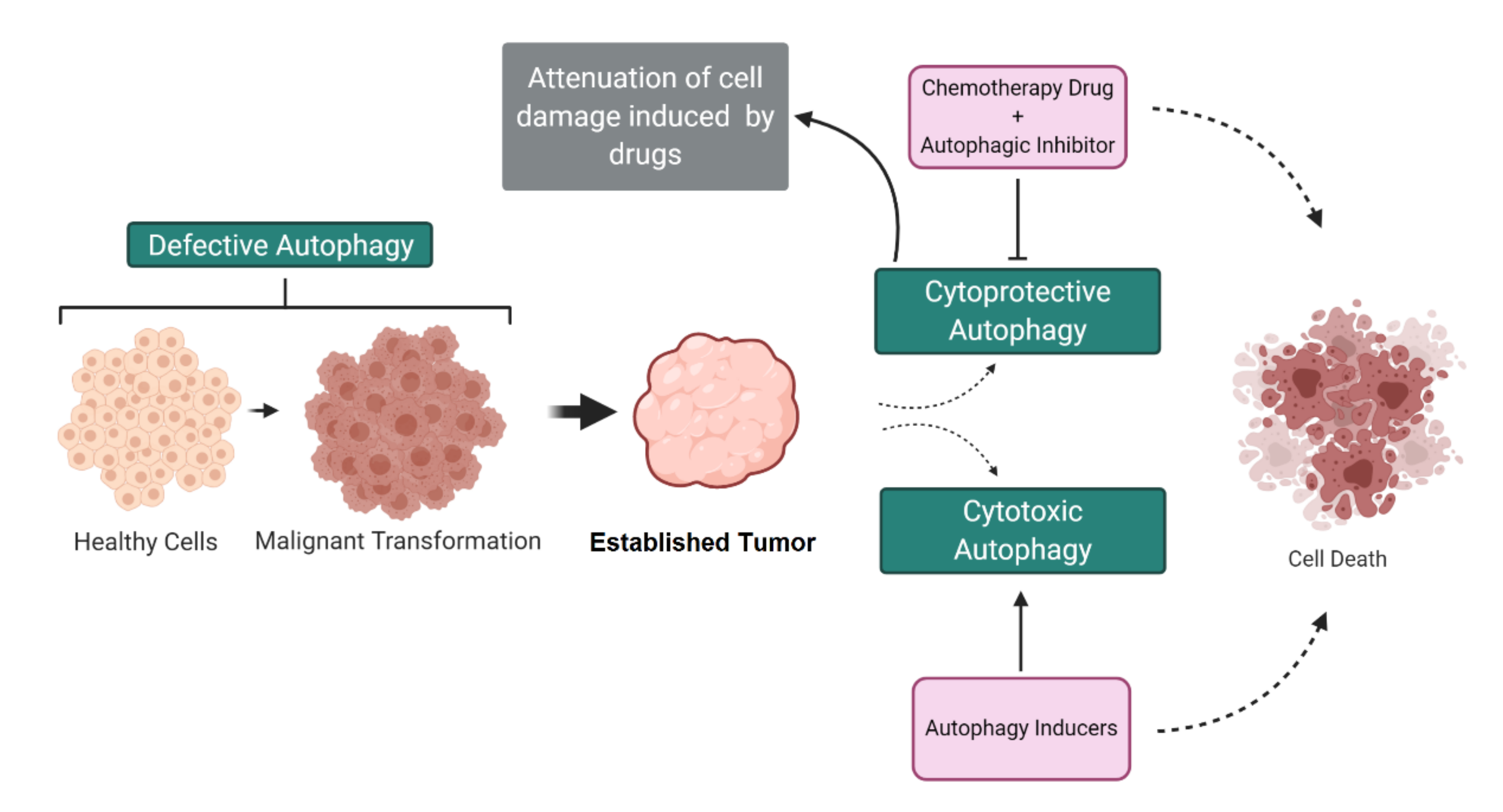
3. Dual Role of Autophagy in Cancer
4. Autophagy-Mediated Resistance in Cancer Cells
4.1. Cisplatin and Paclitaxel
4.2. Tyrosine Kinase Inhibitors
4.3. Trametinib
4.4. Gemcitabine
4.5. 5-Fluorouracil
4.6. Cytarabine
4.7. Tamoxifen
4.8. Temsirolimus and Everolimus
5. Autophagy-Mediated Cell Death in Cancer Cells
5.1. Sorafenib
5.2. Itraconazole
5.3. Clozapine
5.4. SU11274
5.5. Temozolomide and Arsenic Trioxide
5.6. Natural Products
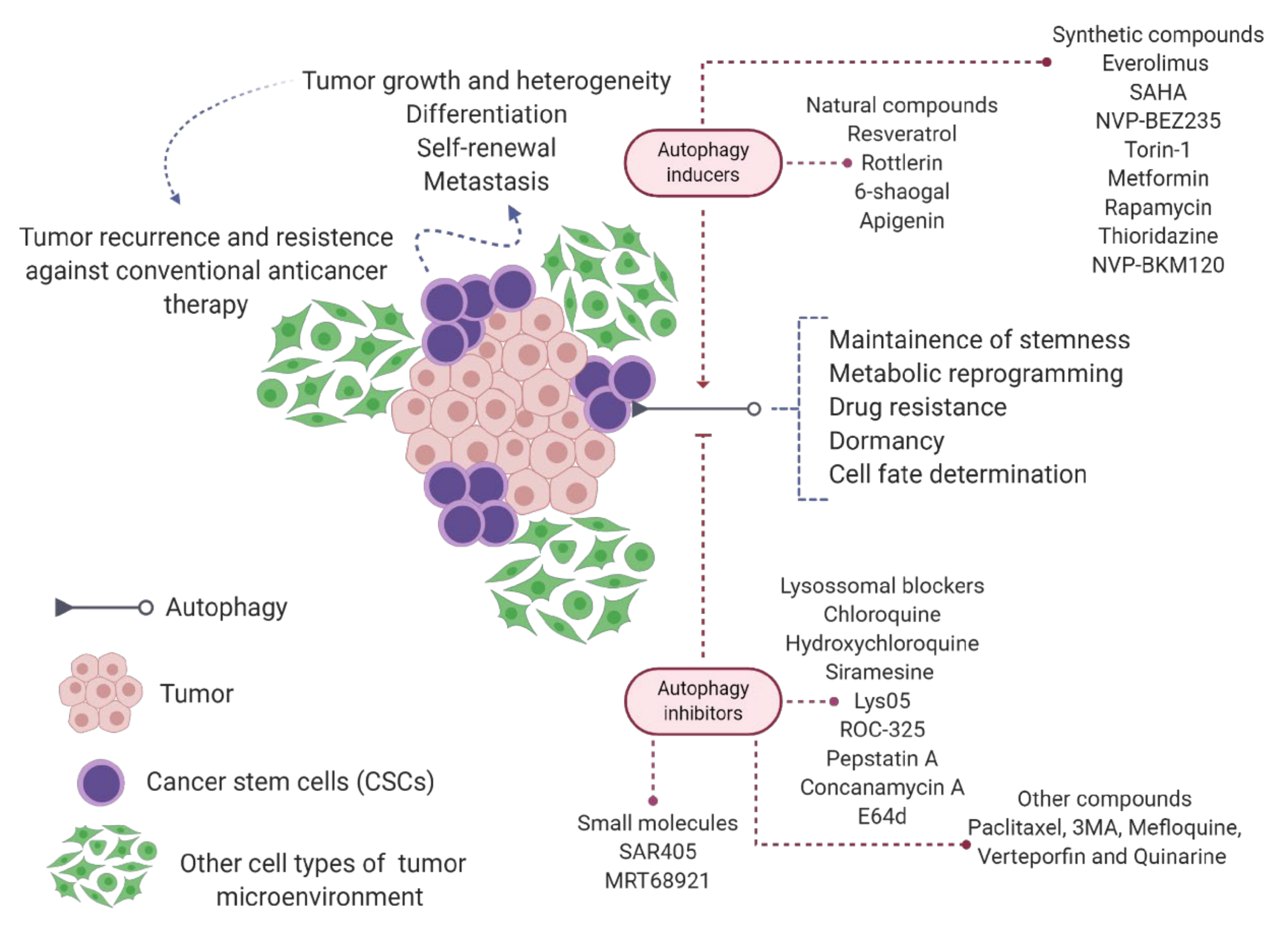
6. Autophagy in Cancer Stem Cell Biology
6.1. Glioblastoma
6.2. Liver and Pancreatic Cancers
6.3. Breast Cancer
6.4. Gastric, Lung, and Colon Cancers
6.5. Acute Myeloid Leukemia
7. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
References
- Levine, B.; Kroemer, G. Autophagy in the pathogenesis of disease. Cell 2008, 132, 27–42. [Google Scholar] [CrossRef]
- Galluzzi, L.; Vitale, I.; Abrams, J.M.; Alnemri, E.S.; Baehrecke, E.H.; Blagosklonny, M.V.; Dawson, T.M.; Dawson, V.L.; El-Deiry, W.S.; Fulda, S.; et al. Molecular definitions of cell death subroutines: Recommendations of the Nomenclature Committee on Cell Death 2012. Cell Death Differ. 2012, 19, 107–120. [Google Scholar] [CrossRef]
- Levine, B.; Kroemer, G. Biological Functions of Autophagy Genes: A Disease Perspective. Cell 2019, 176, 11–42. [Google Scholar] [CrossRef]
- Singh, S.S.; Vats, S.; Chia, A.Y.; Tan, T.Z.; Deng, S.; Ong, M.S.; Arfuso, F.; Yap, C.T.; Goh, B.C.; Sethi, G.; et al. Dual role of autophagy in hallmarks of cancer. Oncogene 2018, 37, 1142–1158. [Google Scholar] [CrossRef] [PubMed]
- Holohan, C.; Van Schaeybroeck, S.; Longley, D.B.; Johnston, P.G. Cancer drug resistance: An evolving paradigm. Nat. Rev. Cancer 2013, 13, 714–726. [Google Scholar] [CrossRef] [PubMed]
- Cufí, S.; Vazquez-Martin, A.; Oliveras-Ferraros, C.; Martin-Castillo, B.; Vellon, L.; Menendez, J.A. Autophagy positively regulates the CD44(+) CD24(-/low) breast cancer stem-like phenotype. Cell Cycle 2011, 10, 3871–3885. [Google Scholar] [CrossRef] [PubMed]
- Burada, F.; Nicoli, E.R.; Ciurea, M.E.; Uscatu, D.C.; Ioana, M.; Gheonea, D.I. Autophagy in colorectal cancer: An important switch from physiology to pathology. World J. Gastroint. Oncol. 2015, 7, 271–284. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, S.; Kanaseki, T.; Mizushima, N.; Mizuta, T.; Arakawa-Kobayashi, S.; Thompson, C.B.; Tsujimoto, Y. Role of Bcl-2 family proteins in a non-apoptotic programmed cell death dependent on autophagy genes. Nat. Cell Biol. 2004, 6, 1221–1228. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Chang, H.; Peng, X.; Bai, Q.; Yi, L.; Zhou, Y.; Zhu, J.; Mi, M. Resveratrol inhibits breast cancer stem-like cells and induces autophagy via suppressing Wnt/β-catenin signaling pathway. PLoS ONE 2014, 9, e102535. [Google Scholar] [CrossRef]
- Shimizu, S. Autophagic Cell Death and Cancer Chemotherapeutics. In Innovative Medicine: Basic Research and Development; Nakao, K., Minato, N., Uemoto, S., Eds.; Springer: Tokyo, Japan, 2015; pp. 219–226. [Google Scholar] [CrossRef]
- Liu, Y.; Levine, B. Autosis and autophagic cell death: The dark side of autophagy. Cell Death Differ. 2015, 22, 367–376. [Google Scholar] [CrossRef]
- Boya, P.; Reggiori, F.; Codogno, P. Emerging regulation and functions of autophagy. Nat. Cell Biol. 2013, 15, 713–720. [Google Scholar] [CrossRef] [PubMed]
- Russell, R.C.; Yuan, H.X.; Guan, K.L. Autophagy regulation by nutrient signaling. Cell Res. 2014, 24, 42–57. [Google Scholar] [CrossRef] [PubMed]
- Ueno, T.; Komatsu, M. Autophagy in the liver, functions in health and disease. Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 170–184. [Google Scholar] [CrossRef] [PubMed]
- Deter, R.L.; De Duve, C. Influence of glucagon, an inducer of cellular autophagy, on some physical properties of rat liver lysosomes. J. Cell Biol. 1967, 33, 437–449. [Google Scholar] [CrossRef] [PubMed]
- Rocchi, A.; He, C. Emerging roles of autophagy in metabolism and metabolic disorders. Front. Biol. 2015, 10, 154–164. [Google Scholar] [CrossRef] [PubMed]
- Chinchwadkar, S.; Padmanabhan, S.; Mishra, P.; Singh, S.; Suresh, S.N.; Vats, S.; Barve, G.; Ammanathan, V.; Manjithaya, R. Multifaceted Housekeeping Functions of Autophagy. J. Indian Inst. Sci. 2017, 97, 79–94. [Google Scholar] [CrossRef]
- Fujikake, N.; Shin, M.; Shimizu, S. Association Between Autophagy and Neurodegenerative Diseases. Front. Neurosci. 2018, 12, 255. [Google Scholar] [CrossRef]
- Matsuzawa-Ishimoto, Y.; Hwang, S.; Cadwell, K. Autophagy and Inflammation. Annu. Rev. Immunol. 2018, 36, 73–101. [Google Scholar] [CrossRef]
- Gatica, D.; Chiong, M.; Lavandero, S.; Klionsky, D.J. Molecular Mechanisms of Autophagy in the Cardiovascular System. Circ. Res. 2015, 116, 456–467. [Google Scholar] [CrossRef]
- Tan, P.; Ye, Y.; Mao, J.; He, L. Autophagy and Immune-Related Diseases. Adv. Exp. Med. Biol. 2019, 1209, 167–179. [Google Scholar] [CrossRef]
- Yun, C.W.; Lee, S.H. The Roles of Autophagy in Cancer. Int. J. Mol. Sci. 2018, 19, 3466. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.H.; Kleeman, L.K.; Jiang, H.H.; Gordon, G.; Goldman, J.E.; Berry, G.; Herman, B.; Levine, B. Protection against fatal Sindbis virus encephalitis by beclin, a novel Bcl-2-interacting protein. J. Virol. 1998, 72, 8586–8596. [Google Scholar] [CrossRef] [PubMed]
- Aita, V.M.; Liang, X.H.; Murty, V.V.; Pincus, D.L.; Yu, W.; Cayanis, E.; Kalachikov, S.; Gilliam, T.C.; Levine, B. Cloning and genomic organization of beclin 1, a candidate tumor suppressor gene on chromosome 17q21. Genomics 1999, 59, 59–65. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.H.; Jackson, S.; Seaman, M.; Brown, K.; Kempkes, B.; Hibshoosh, H.; Levine, B. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature 1999, 402, 672–676. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.H.; Yu, J.; Brown, K.; Levine, B. Beclin 1 contains a leucine-rich nuclear export signal that is required for its autophagy and tumor suppressor function. Cancer Res. 2001, 61, 3443–3449. [Google Scholar]
- Marino, G.; Salvador-Montoliu, N.; Fueyo, A.; Knecht, E.; Mizushima, N.; Lopez-Otin, C. Tissue-specific autophagy alterations and increased tumorigenesis in mice deficient in Atg4C/autophagin-3. J. Biol. Chem. 2007, 282, 18573–18583. [Google Scholar] [CrossRef]
- Hara, T.; Nakamura, K.; Matsui, M.; Yamamoto, A.; Nakahara, Y.; Suzuki-Migishima, R.; Yokoyama, M.; Mishima, K.; Saito, I.; Okano, H.; et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature 2006, 441, 885–889. [Google Scholar] [CrossRef]
- Nakai, A.; Yamaguchi, O.; Takeda, T.; Higuchi, Y.; Hikoso, S.; Taniike, M.; Omiya, S.; Mizote, I.; Matsumura, Y.; Asahi, M.; et al. The role of autophagy in cardiomyocytes in the basal state and in response to hemodynamic stress. Nat. Med. 2007, 13, 619–624. [Google Scholar] [CrossRef]
- Qu, X.; Yu, J.; Bhagat, G.; Furuya, N.; Hibshoosh, H.; Troxel, A.; Rosen, J.; Eskelinen, E.L.; Mizushima, N.; Ohsumi, Y.; et al. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J. Clin. Invest. 2003, 112, 1809–1820. [Google Scholar] [CrossRef]
- Yue, Z.; Jin, S.; Yang, C.; Levine, A.J.; Heintz, N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc. Natl. Acad. Sci. USA 2003, 100, 15077–15082. [Google Scholar] [CrossRef]
- Zhu, H.; Tannous, P.; Johnstone, J.L.; Kong, Y.; Shelton, J.M.; Richardson, J.A.; Le, V.; Levine, B.; Rothermel, B.A.; Hill, J.A. Cardiac autophagy is a maladaptive response to hemodynamic stress. J. Clin. Investig. 2007, 117, 1782–1793. [Google Scholar] [CrossRef] [PubMed]
- Matsui, Y.; Takagi, H.; Qu, X.; Abdellatif, M.; Sakoda, H.; Asano, T.; Levine, B.; Sadoshima, J. Distinct roles of autophagy in the heart during ischemia and reperfusion. Circ. Res. 2007, 100, 914–922. [Google Scholar] [CrossRef] [PubMed]
- Komatsu, M.; Waguri, S.; Ueno, T.; Iwata, J.; Murata, S.; Tanida, I.; Ezaki, J.; Mizushima, N.; Ohsumi, Y.; Uchiyama, Y.; et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J. Cell Biol. 2005, 169, 425–434. [Google Scholar] [CrossRef] [PubMed]
- Komatsu, M.; Waguri, S.; Chiba, T.; Murata, S.; Iwata, J.; Tanida, I.; Ueno, T.; Koike, M.; Uchiyama, Y.; Kominami, E.; et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature 2006, 441, 880–884. [Google Scholar] [CrossRef]
- Iwata, J.; Ezaki, J.; Komatsu, M.; Yokota, S.; Ueno, T.; Tanida, I.; Chiba, T.; Tanaka, K.; Kominami, E. Excess peroxisomes are degraded by autophagic machinery in mammals. J. Biol. Chem. 2006, 281, 4035–4041. [Google Scholar] [CrossRef]
- Khandia, R.; Dadar, M.; Munjal, A.; Dhama, K.; Karthik, K.; Tiwari, R.; Yatoo, M.I.; Iqbal, H.; Singh, K.P.; Joshi, S.K.; et al. A Comprehensive Review of Autophagy and Its Various Roles in Infectious, Non-Infectious, and Lifestyle Diseases: Current Knowledge and Prospects for Disease Prevention, Novel Drug Design, and Therapy. Cells 2019, 8, 674. [Google Scholar] [CrossRef]
- Yin, Z.; Pascual, C.; Klionsky, D.J. Autophagy: Machinery and regulation. Microb. Cell 2016, 3, 588–596. [Google Scholar] [CrossRef]
- Fang, Y.; Tan, J.; Zhang, Q. Signaling pathways and mechanisms of hypoxia-induced autophagy in the animal cells. Cell Biol. Int. 2015, 39, 891–898. [Google Scholar] [CrossRef]
- Lee, W.S.; Sung, M.S.; Lee, E.G.; Yoo, H.G.; Cheon, Y.H.; Chae, H.J.; Yoo, W.H. A pathogenic role for ER stress-induced autophagy and ER chaperone GRP78/BiP in T lymphocyte systemic lupus erythematosus. J. Leukoc. Biol. 2015, 97, 425–433. [Google Scholar] [CrossRef]
- Soria, L.R.; Brunetti-Pierri, N. Ammonia and autophagy: An emerging relationship with implications for disorders with hyperammonemia. J. Inh. Met. Dis. 2019, 42, 1097–1104. [Google Scholar] [CrossRef]
- Lin, S.Y.; Li, T.Y.; Liu, Q.; Zhang, C.; Li, X.; Chen, Y.; Zhang, S.M.; Lian, G.; Liu, Q.; Ruan, K.; et al. GSK3-TIP60-ULK1 Signaling Pathway Links Growth Factor Deprivation to Autophagy. Science 2012, 336, 477–481. [Google Scholar] [CrossRef] [PubMed]
- Gao, Q. Oxidative Stress and Autophagy. In Autophagy: Biology and Diseases; Qin, Z.H., Ed.; Springer: Singapore, 2019. [Google Scholar] [CrossRef]
- Paik, S.; Kim, J.K.; Chung, C.; Jo, E. Autophagy: A new strategy for host-directed therapy of tuberculosis. Virulence 2019, 10, 448–459. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, H. Regulation of Autophagy by mTOR Signaling Pathway. Adv. Exp. Med. Biol. 2019, 1206, 67–83. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Klionsky, D.J. Mammalian autophagy: Core molecular machinery and signaling regulation. Curr. Opin. Cell Biol. 2010, 22, 124–131. [Google Scholar] [CrossRef] [PubMed]
- Mizushima, N.; Yoshimori, T.; Ohsumi, Y. The role of Atg proteins in autophagosome formation. Annu. Rev. Cell Dev. Biol. 2011, 27, 107–132. [Google Scholar] [CrossRef]
- Munz, C. The macroautophagy machinery in endo- and exocytosis. J. Mol. Biol. 2017, 429, 473–485. [Google Scholar] [CrossRef]
- Huber, L.A.; Teis, D. Lysosomal signaling in control of degradation pathways. Curr. Opin. Cell Biol. 2016, 39, 8–14. [Google Scholar] [CrossRef]
- Yang, Q.; Wang, R.; Zhu, L. Chaperone-Mediated Autophagy. Adv. Exp. Med. Biol. 2019, 1206, 435–452. [Google Scholar] [CrossRef]
- Hjerpe, R.; Bett, J.S.; Keuss, M.J.; Solovyova, A.; McWilliams, T.G.; Johnson, C.; Sahu, I.; Varghese, J.; Wood, N.; Wightman, M.; et al. UBQLN2 Mediates Autophagy-Independent Protein Aggregate Clearance by the Proteasome. Cell 2016, 166, 935–949. [Google Scholar] [CrossRef]
- Alfaro, I.E.; Albornoz, A.; Molina, A.; Moreno, J.; Cordero, K.; Criollo, A.; Budini, M. Chaperone Mediated Autophagy in the Crosstalk of Neurodegenerative Diseases and Metabolic Disorders. Front. Endocrinol. 2019, 9, 778. [Google Scholar] [CrossRef]
- Kaushik, S.; Cuervo, A.M. The coming of age of chaperone-mediated autophagy. Nat. Rev. 2018, 19, 365–381. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Hochstrasser, M. Microautophagy regulates proteasome homeostasis. Cur. Genet. 2020, 66, 683–687. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Li, J.; Bao, J. Microautophagy: Lesser-known self-eating. Cell. Mol. Life Sci. 2012, 69, 1125–1136. [Google Scholar] [CrossRef] [PubMed]
- Parzych, K.R.; Klionsky, D.J. An overview of autophagy: Morphology, mechanism, and regulation. Antioxid. Redox Signal 2014, 20, 460–473. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Chen, Y.; Tooze, S.A. Autophagy pathway: Cellular and molecular mechanisms. Autophagy 2018, 14, 207–215. [Google Scholar] [CrossRef] [PubMed]
- Foot, N.; Henshall, T.; Kumar, S. Ubiquitination and the Regulation of Membrane Proteins. Physiol. Rev. 2017, 97, 253–281. [Google Scholar] [CrossRef]
- Allen, E.A.; Baehrecke, E.H. Autophagy in animal development. Cell Death Differ. 2020, 27, 903–918. [Google Scholar] [CrossRef]
- Zaffagnini, G.; Martens, S. Mechanisms of Selective Autophagy. J. Mol. Biol. 2016, 428, 1714–1724. [Google Scholar] [CrossRef]
- Sharma, V.; Verma, S.; Seranova, E.; Sarkar, S.; Kumar, D. Selective autophagy and xenophagy in infection and disease. Front. Cell Dev. Biol. 2018, 6, 147. [Google Scholar] [CrossRef]
- Bo Otto, F.; Thumm, M. Nucleophagy-Implications for Microautophagy and Health. Int. J. Mol. Sci. 2020, 21, 4506. [Google Scholar] [CrossRef]
- Kissová, I.; Salin, B.; Schaeffer, J.; Bhatia, S.; Manon, S.; Camougrand, N. Selective and non-selective autophagic degradation of mitochondria in yeast. Autophagy 2007, 3, 329–336. [Google Scholar] [CrossRef] [PubMed]
- Tong, J.; Yan, X.; Yu, L. The late stage of autophagy: Cellular events and molecular regulation. Protein Cell 2010, 1, 907–915. [Google Scholar] [CrossRef] [PubMed]
- Mercer, T.J.; Gubas, A.; Tooze, S.A. A molecular perspective of mammalian autophagosome biogenesis. J. Biol. Chem. 2018, 293, 5386–5395. [Google Scholar] [CrossRef] [PubMed]
- Galluzzi, L.; Vitale, I.; Aaronson, S. Molecular mechanisms of cell death: Recommendations of the Nomenclature Committee on Cell Death 2018. Cell Death Differ. 2018, 25, 486–541. [Google Scholar] [CrossRef]
- Ohsumi, Y. Historical landmarks of autophagy research. Cell Res. 2014, 24, 9–23. [Google Scholar] [CrossRef]
- Luo, Y.; Jiang, C.; Yu, L.; Yang, A. Chemical Biology of Autophagy-Related Proteins with Posttranslational Modifications: From Chemical Synthesis to Biological Applications. Front. Chem. 2020, 8, 233. [Google Scholar] [CrossRef]
- Paquette, M.; El-Houjeiri, L.; Pause, A. mTOR Pathways in Cancer and Autophagy. Cancers 2018, 10, 18. [Google Scholar] [CrossRef]
- Nazio, F.; Strappazzon, F.; Antonioli, M.; Bielli, P.; Cianfanelli, V.; Bordi, M.; Gretzmeier, C.; Dengjel, J.; Piacentini, M.; Fimia, G.M.; et al. Mtor inhibits autophagy by controlling ULK1 ubiquitylation, self-association and function through AMBRA1 and TRAF6. Nat. Cell Biol. 2013, 15, 406–416. [Google Scholar] [CrossRef]
- Kim, J.; Kundu, M.; Viollet, B.; Guan, K.L. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat. Cell Biol. 2011, 13, 132–141. [Google Scholar] [CrossRef]
- Lin, M.G.; Hurley, J.H. Structure and function of the ULK1 complex in autophagy. Curr. Opin. Cell Biol. 2016, 39, 61–68. [Google Scholar] [CrossRef]
- Mizushima, N. The role of the Atg1/ULK1 complex in autophagy regulation. Curr. Opin. Cell Biol. 2010, 22, 132–139. [Google Scholar] [CrossRef] [PubMed]
- Burman, C.; Ktistakis, N.T. Regulation of autophagy by phosphatidylinositol 3-phosphate. FEBS Lett. 2010, 584, 1302–1312. [Google Scholar] [CrossRef] [PubMed]
- Cebollero, E.; van der Vaart, A.; Zhao, M.; Rieter, E.; Klionsky, D.J.; Helms, J.B.; Reggiori, F. Phosphatidylinositol-3-phosphate clearance plays a key role in autophagosome completion. Curr. Biol. 2012, 22, 1545–1553. [Google Scholar] [CrossRef] [PubMed]
- Arakawa, S.; Honda, S.; Yamaguchi, H.; Shimizu, S. Molecular mechanisms and physiological roles of Atg5/Atg7-independent alternative autophagy. Proc. Jpn. Acad. Ser. B 2017, 93, 378–385. [Google Scholar] [CrossRef]
- Mizushima, N.; Komatsu, M. Autophagy: Renovation of Cells and Tissues. Cell 2011, 147, 728. [Google Scholar] [CrossRef]
- Glick, D.; Barth, S.; Macleod, K.F. Autophagy: Cellular and molecular mechanisms. J. Pathol. 2010, 221, 3–12. [Google Scholar] [CrossRef]
- Runwal, G.; Stamatakou, E.; Siddiqi, F.H.; Puri, C.; Zhu, Y.; Rubinsztein, D.C. LC3-positive structures are prominent in autophagy-deficient cells. Sci. Rep. 2019, 9, 10147. [Google Scholar] [CrossRef]
- Harada, K.; Kotani, T.; Kirisako, H.; Sakoh-Nakatogawa, M.; Oikawa, Y.; Kimura, Y.; Hirano, H.; Yamamoto, H.; Ohsumi, Y.; Nakatogawa, H. Two distinct mechanisms target the autophagy-related E3 complex to the pre-autophagosomal structure. eLife 2019, 8, e43088. [Google Scholar] [CrossRef]
- Kaur, J.; Debnath, J. Autophagy at the crossroads of catabolism and anabolism. Nat. Rev. 2015, 16, 461–472. [Google Scholar] [CrossRef]
- Bader, C.A.; Shandala, T.; Ng, Y.S.; Johnson, I.R.; Brooks, D.A. Atg9 is required for intraluminal vesicles in amphisomes and autolysosomes. Biol. Open 2015, 4, 1345–1355. [Google Scholar] [CrossRef]
- Kaiser, S.E.; Mao, K.; Taherbhoy, A.M.; Yu, S.; Olszewski, J.L.; Duda, D.M.; Kurinov, I.; Deng, A.; Fenn, T.D.; Klionsky, D.J.; et al. Noncanonical E2 recruitment by the autophagy E1 revealed by Atg7-Atg3 and Atg7-Atg10 structures. Nat. Struct. Mol. Biol. 2012, 19, 1242–1249. [Google Scholar] [CrossRef] [PubMed]
- Bento, C.F.; Renna, M.; Ghislat, G.; Puri, C.; Ashkenazi, A.; Vicinanza, M.; Menzies, F.M.; Rubinsztein, D.C. Mammalian Autophagy: How Does It Work? Annu. Rev. Biochem. 2016, 85, 685–713. [Google Scholar] [CrossRef] [PubMed]
- Frudd, K.; Burgoyne, T.; Burgoyne, J.R. Oxidation of Atg3 and Atg7 mediates inhibition of autophagy. Nat. Communic. 2018, 9, 95. [Google Scholar] [CrossRef] [PubMed]
- Stolz, A.; Ernst, A.; Dikic, I. Cargo recognition and trafficking in selective autophagy. Nat. Cell Biol. 2014, 16, 495–501. [Google Scholar] [CrossRef]
- Liu, W.J.; Ye, L.; Huang, W.F.; Guo, L.J.; Xu, Z.G.; Wu, H.L.; Yang, C.; Liu, H.F. p62 links the autophagy pathway and the ubiqutin-proteasome system upon ubiquitinated protein degradation. Cell. Mol. Biol. Lett. 2016, 21, 29. [Google Scholar] [CrossRef]
- Noda, N.N.; Inagaki, F. Mechanisms of autophagy. Annu. Rev. Biophys. 2015, 44, 101–122. [Google Scholar] [CrossRef]
- Itakura, E.; Kishi-Itakura, C.; Mizushima, N. The hairpin-type tail-anchored SNARE syntaxin 17 targets to autophagosomes for fusion with endosomes/lysosomes. Cell 2012, 151, 1256–1269. [Google Scholar] [CrossRef]
- Diao, J.; Liu, R.; Rong, Y.; Zhao, M.; Zhang, J.; Lai, Y.; Zhou, Q.; Wilz, L.M.; Li, J.; Vivona, S.; et al. ATG14 promotes membrane tethering and fusion of autophagosomes to endolysosomes. Nature 2015, 520, 563–566. [Google Scholar] [CrossRef]
- Hyttinen, J.M.; Niittykoski, M.; Salminen, A.; Kaarniranta, K. Maturation of autophagosomes and endosomes: A key role for Rab7. Biochim. Biophys. Acta 2013, 1833, 503–510. [Google Scholar] [CrossRef]
- Jiang, P.; Nishimura, T.; Sakamaki, Y.; Itakura, E.; Hatta, T.; Natsume, T.; Mizushima, N. The HOPS complex mediates autophagosome-lysosome fusion through interaction with syntaxin 17. Mol. Biol. Cell 2014, 25, 1327–1337. [Google Scholar] [CrossRef]
- Chun, Y.; Kim, J. Autophagy: An Essential Degradation Program for Cellular Homeostasis and Life. Cells 2018, 7, 278. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.Y.; Luo, Y.; Zhu, Y.M.; Liu, Z.H.; Kent, T.A.; Rong, J.G.; Li, W.; Qiao, S.G.; Li, M.; Ni, Y.; et al. Inhibition of autophagy blocks cathepsins-tBid-mitochondrial apoptotic signaling pathway via stabilization of lysosomal membrane in ischemic astrocytes. Cell Death Dis. 2017, 8, e2618. [Google Scholar] [CrossRef] [PubMed]
- Yoshii, S.R.; Mizushima, N. Monitoring and Measuring Autophagy. Int. J. Mol. Sci. 2017, 18, 1865. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.J.; Chen, S.; Huang, K.X.; Le, W.D. Why should autophagic flux be assessed? Acta Pharm. Sin. 2013, 34, 595–599. [Google Scholar] [CrossRef] [PubMed]
- Bresciani, A.; Spiezia, M.C.; Boggio, R.; Cariulo, C.; Nordheim, A.; Altobelli, R.; Kuhlbrodt, K.; Dominguez, C.; Munoz-Sanjuan, I.; Wityak, J.; et al. Quantifying autophagy using novel LC3B and p62 TR-FRET assays. PLoS ONE 2018, 13, e0194423. [Google Scholar] [CrossRef]
- Zhan, L.; Chen, S.; Li, K.; Liang, D.; Zhu, X.; Liu, L.; Lu, Z.; Sun, W.; Xu, E. Autophagosome maturation mediated by Rab7 contributes to neuroprotection of hypoxic preconditioning against global cerebral ischemia in rats. Cell Death Dis. 2017, 8, e2949. [Google Scholar] [CrossRef]
- Tasdemir, E.; Maiuri, M.C.; Galluzzi, L.; Vitale, I.; Djavaheri-Mergny, M.; D’Amelio, M.; Criollo, A.; Morselli, E.; Zhu, C.; Harper, F.; et al. Regulation of autophagy by cytoplasmic p53. Nat. Cell Biol. 2008, 10, 676–687. [Google Scholar] [CrossRef]
- Broz, D.K.; Mello, S.S.; Bieging, K.T.; Jiang, D.; Dusek, R.L.; Brady, C.A.; Sidow, A.; Attardi, L.D. Global genomic profiling reveals an extensive p53-regulated autophagy program contributing to key p53 responses. Genes Dev. 2013, 27, 1016–1031. [Google Scholar] [CrossRef]
- Mrakovcic, M.; Fröhlich, L.F. P53-mediated molecular control of autophagy in tumor cells. Biomolecules 2018, 8, 14. [Google Scholar] [CrossRef]
- Kruse, J.P.; Gu, W. Modes of p53 regulation. Cell 2009, 137, 609–622. [Google Scholar] [CrossRef]
- Deisenroth, C.; Franklin, D.A.; Zhang, Y. The Evolution of the Ribosomal Protein–MDM2–p53 Pathway. Cold Spring Harb. Perspect. Med. 2016, 6, a026138. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.; Chen, S.; Thorne, R.F.; Wu, M. TP53, TP53 Target Genes (DRAM, TIGAR), and Autophagy. Adv. Exp. Med. Biol. 2019, 1206, 127–149. [Google Scholar] [CrossRef] [PubMed]
- Vadysirisack, D.D.; Baenke, F.; Ory, B.; Lei, K.; Ellisen, L.W. Feedback Control of p53 Translation by REDD1 and mTORC1 Limits the p53-Dependent DNA Damage Response. Mol. Cell Biol. 2011, 31, 4356–4365. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Wang, N.; Liu, P.; Xie, X. AMPK and Cancer. AMP-Act. Protein Kinase 2016, 203–226. [Google Scholar] [CrossRef]
- Lee, J.H.; Budanov, A.V.; Park, E.J.; Birse, R.; Kim, T.E.; Guy, A.; Ocorr, K.; Ellisman, M.H.; Bodmer, R.; Bier, E. Sestrin as a feedback inhibitor of TOR that prevents age-related pathologies. Science 2010, 327, 1223–1228. [Google Scholar] [CrossRef]
- Crighton, D.; Wilkinson, S.; Ryan, K.M. DRAM links autophagy to p53 and programmed cell death. Autophagy 2007, 3, 72–74. [Google Scholar] [CrossRef]
- Singh, P.; Ravanan, P.; Talwar, T. Death Associated Protein Kinase 1 (DAPK1): A Regulator of Apoptosis and Autophagy. Front. Mol. Neurosci. 2016, 9, 46. [Google Scholar] [CrossRef]
- Decuypere, J.P.; Parys, J.B.; Bultynck, G. Regulation of the Autophagic Bcl-2/Beclin 1 Interaction. Cells 2012, 1, 284–312. [Google Scholar] [CrossRef]
- Menon, M.B.; Dhamija, S. Beclin 1 Phosphorylation-at the Center of Autophagy Regulation. Front. Cell Dev. Biol. 2018, 6, 137. [Google Scholar] [CrossRef]
- Erlich, S.; Mizrachy, L.; Segev, O.; Lindenboim, L.; Zmira, O.; Adi-Harel, S.; Hirsch, J.A.; Stein, R.; Pinkas-Kramarski, R. Differential interactions between Beclin 1 and Bcl-2 family members. Autophagy 2014, 3, 561–568. [Google Scholar] [CrossRef]
- Sohn, E.J.; Park, H.T. Natural agents mediated autophagic signal networks in cancer. Cancer Cell Int. 2017, 17, 110. [Google Scholar] [CrossRef] [PubMed]
- Kang, R.; Zeh, H.J.; Lotze, M.T.; Tang, D. The Beclin 1 network regulates autophagy and apoptosis. Cell Death Differ. 2011, 18, 571–580. [Google Scholar] [CrossRef] [PubMed]
- Yoon, C.H.; Lee, E.S.; Lim, D.S.; Bae, Y.S. PKR, a p53 target gene, plays a crucial role in the tumor-suppressor function of p53. Proc. Natl. Acad. Sci. USA 2009, 106, 7852–7857. [Google Scholar] [CrossRef] [PubMed]
- Morselli, E.; Shen, S.; Ruckenstuhl, C.; Bauer, M.A.; Mariño, G.; Galluzzi, L.; Criollo, A.; Michaud, M.; Maiuri, M.C.; Chano, T.; et al. p53 Inhibits Autophagy by Interacting with the Human Ortholog of Yeast Atg17, RB1CC1/FIP200. Cell Cycle 2011, 10, 2763–2769. [Google Scholar] [CrossRef] [PubMed]
- Bensaad, K.; Tsuruta, A.; Selak, M.A.; Nieves Calvo Vidal, M.; Katsunori, N.; Bartrons, R.; Gottlieb, E.; Vousen, K.H. TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell 2006, 126, 107–120. [Google Scholar] [CrossRef]
- Xie, J.M.; Li, B.; Yu, H.P.; Gao, Q.G.; Li, W.; Wu, H.R.; Qin, Z.H. TIGAR Has a Dual Role in Cancer Cell Survival Through Regulating Apoptosis and Autophagy. Cancer Res. 2014, 15, 5127–5138. [Google Scholar] [CrossRef]
- Bensaad, K.; Cheung, E.C.; Vousden, K.H. Modulation of intracellular ROS levels by TIGAR controls autophagy. EMBO J. 2009, 28, 3015–3026. [Google Scholar] [CrossRef]
- Zhao, Y.; Wang, Z.; Zhang, W.; Zhang, L. Non-coding RNAs regulate autophagy process via influencing the expression of associated protein. Prog. Bioph. Mol. Biol. 2020, 151, 32–39. [Google Scholar] [CrossRef]
- Fan, Y.X.; Dai, Y.Z.; Wang, X.L.; Ren, Y.Q.; Han, J.J.; Zhang, H. MiR-18a upregulation enhances autophagy in triple negative cancer cells via inhibiting mTOR signaling pathway. Eur. Rev. Med. Pharmacol. Sci. 2016, 20, 2194–2200. [Google Scholar]
- Yu, G.; Jia, Z.; Dou, Z. miR-24-3p regulates bladder cancer cell proliferation, migration, invasion and autophagy by targeting DEDD. Oncol. Rep. 2017, 37, 1123–1131. [Google Scholar] [CrossRef]
- Seca, H.; Lima, R.T.; Lopes-Rodrigues, V.; Guimaraes, J.E.; Almeida, G.M.; Vasconcelos, M.H. Targeting miR-21 induces autophagy and chemosensitivity of leukemia cells. Curr. Drug Targets 2013, 14, 1135–1143. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Guo, Q.; Zhao, Y.; Chen, J.; Wang, S.; Hu, J.; Sun, Y. BRAF-activated long non-coding RNA contributes to cell proliferation and activates autophagy in papillary thyroid carcinoma. Oncol. Lett. 2014, 8, 1947–1952. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Wu, D.; Sang, X.B.; Wang, L.L.; Zong, Z.H.; Sun, K.X.; Liu, B.L.; Zhao, Y. The lncRNA HULC functions as an oncogene by targeting ATG7 and ITGB1 in epithelial ovarian carcinoma. Cell Death Dis. 2017, 8, e3118. [Google Scholar] [CrossRef] [PubMed]
- Park, S.M.; Ou, J.; Chamberlain, L.; Simone, T.M.; Yang, H.; Virbasius, C.M.; Green, M.R. U2AF35(S34F) Promotes Transformation by Directing Aberrant ATG7 Pre-mRNA 3′ End Formation. Mol. Cell. 2016, 62, 479–490. [Google Scholar] [CrossRef] [PubMed]
- Dikic, I.; Elazar, Z. Mechanism and medical implications of mammalian autophagy. Nat. Rev. Mol. Cell Biol. 2018, 19, 349–364. [Google Scholar] [CrossRef]
- Kisen, G.O.; Tessitore, L.; Costelli, P.; Gordon, P.B.; Schwarze, P.E.; Baccino, F.M.; Seglen, P.O. Reduced autophagic activity in primary rat hepatocellular carcinoma and ascites hepatoma cells. Carcinogenesis 1993, 14, 2501–2505. [Google Scholar] [CrossRef] [PubMed]
- Takamura, A.; Komatsu, M.; Hara, T.; Sakamoto, A.; Kishi, C.; Waguri, S.; Eishi, Y.; Hino, O.; Tanaka, K.; Mizushima, N. Autophagy-deficient mice develop multiple liver tumors. Genes Dev. 2011, 25, 795–800. [Google Scholar] [CrossRef]
- Takahashi, Y.; Coppola, D.; Matsushita, N.; Cualing, H.D.; Sun, M.; Sato, Y.; Liang, C.; Jung, J.U.; Cheng, J.Q.; Mulé, J.J.; et al. Bif-1 interacts with Beclin 1 through UVRAG and regulates autophagy and tumorigenesis. Nat. Cell Biol. 2007, 9, 1142–1151. [Google Scholar] [CrossRef]
- Arico, S.; Petiot, A.; Bauvy, C.; Dubbelhuis, P.F.; Meijer, A.J.; Codogno, P.; Ogier-Denis, E. The tumor suppressor PTEN positively regulates macroautophagy by inhibiting the phosphatidylinositol 3-kinase/protein kinase B pathway. J. Biol. Chem. 2001, 276, 35243–35246. [Google Scholar] [CrossRef]
- Pattingre, S.; Tassa, A.; Qu, X.; Garuti, R.; Liang, X.H.; Mizushima, N.; Packer, M.; Schneider, M.D.; Levine, B. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell 2005, 122, 927–939. [Google Scholar] [CrossRef]
- Ma, L.; Chen, Z.; Erdjument-Bromage, H.; Tempst, P.; Pandolfi, P.P. Phosphorylation and functional inactivation of TSC2 by Erk implications for tuberous sclerosis and cancer pathogenesis. Cell 2005, 121, 179–193. [Google Scholar] [CrossRef] [PubMed]
- Kung, C.P.; Budina, A.; Balaburski, G.; Bergenstock, M.K.; Murphy, M. Autophagy in tumor suppression and cancer therapy. Crit. Rev. Eukaryot. Gene Expr. 2011, 21, 71–100. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.T.; Liu, H.; Mao, M.J.; Tan, Y.; Mo, X.Q.; Meng, X.J.; Cao, M.T.; Zhong, C.Y.; Liu, Y.; Shan, H.; et al. Crosstalk between autophagy and epithelial-mesenchymal transition and its application in cancer therapy. Mol. Cancer 2019, 18, 101. [Google Scholar] [CrossRef] [PubMed]
- Singla, M.; Bhattacharyya, S. Autophagy as a potential therapeutic target during epithelial to mesenchymal transition in renal cell carcinoma: An in vitro study. Biomed. Pharmacother. 2017, 94, 332–340. [Google Scholar] [CrossRef] [PubMed]
- Catalano, M.; D’Alessandro, G.; Lepore, F.; Corazzari, M.; Caldarola, S.; Valacca, C.; Faienza, F.; Esposito, V.; Limatola, C.; Cecconi, F.; et al. Autophagy induction impairs migration and invasion by reversing EMT in glioblastoma cells. Mol. Oncol. 2015, 9, 1612–1625. [Google Scholar] [CrossRef] [PubMed]
- Panda, P.K.; Mukhopadhyay, S.; Das, D.N.; Sinha, N.; Naik, P.P.; Bhutia, S.K. Mechanism of autophagic regulation in carcinogenesis and cancer therapeutics. Semin. Cell Dev. Biol. 2015, 39, 43–55. [Google Scholar] [CrossRef]
- Tang, J.C.; Feng, Y.L.; Liang, X.; Cai, X.J. Autophagy in 5-Fluorouracil Therapy in Gastrointestinal Cancer: Trends and Challenges. Chin. Med. J. 2016, 129, 456–463. [Google Scholar] [CrossRef]
- Galluzzi, L.; Pietrocola, F.; Bravo-San Pedro, J.M.; Amaravadi, R.K.; Baehrecke, E.H.; Cecconi, F.; Codogno, P.; Debnath, J.; Gewirtz, D.A.; Karantza, V.; et al. Autophagy in malignant transformation and cancer progression. EMBO J. 2015, 34, 856–880. [Google Scholar] [CrossRef]
- Kinsey, C.G.; Camolotto, S.A.; Boespflug, A.M.; Guillen, K.P.; Foth, M.; Truong, A.; Schuman, S.S.; Shea, J.E.; Seipp, M.T.; Yap, J.T.; et al. Protective autophagy elicited by RAF→ MEK→ ERKinhibition suggests a treatment strategy for RAS-driven cancers. Nat. Med. 2019, 25, 620–627. [Google Scholar] [CrossRef]
- Bryant, K.L.; Stalnecker, C.A.; Zeitouni, D.; Klomp, J.E.; Peng, S.; Tikunov, A.P.; Gunda, V.; Pierobon, M.; Waters, A.M.; George, S.D.; et al. Combination of ERK and autophagy inhibition as a treatment approach for pancreatic cancer. Nat. Med. 2019, 25, 628–640. [Google Scholar] [CrossRef]
- Guo, J.Y.; Chen, H.Y.; Mathew, R.; Fan, J.; Strohecker, A.M.; Karsli-Uzunbas, G.; Kamphorst, J.J.; Chen, G.; Lemons, J.M.; Karantza, V.; et al. Activated Ras requires autophagy to maintain oxidative metabolism and tumorigenesis. Genes Dev. 2011, 25, 460–470. [Google Scholar] [CrossRef]
- Yang, S.; Wang, X.; Contino, G.; Liesa, M.; Sahin, E.; Ying, H.; Bause, A.; Li, Y.; Stommel, J.M.; Dell’antonio, G.; et al. Pancreatic cancers require autophagy for tumor growth. Genes Dev. 2011, 25, 717–729. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Lai, Z.L.; Chen, H.F.; Zhang, M.; Wang, A.; Jia, T.; Sun, W.Q.; Zhu, X.M.; Chen, X.F.; Zhao, Z.; et al. Curcumin synergizes with 5-fluorouracil by impairing AMPK/ULK1-dependent autophagy, AKT activity and enhancing apoptosis in colon cancer cells with tumor growth inhibition in xenograft mice. J. Exp. Clin. Can. Res. 2017, 36, 190. [Google Scholar] [CrossRef] [PubMed]
- Shuhua, W.; Chenbo, S.; Yangyang, L.; Xiangqian, G.; Shuang, H.; Tangyue, L.; Dong, T. Autophagy-related genes Raptor, Rictor, and Beclin1 expression and relationship with multidrug resistance in colorectal carcinoma. Human Pathol. 2015, 46, 1752–1759. [Google Scholar] [CrossRef] [PubMed]
- Grimaldi, A.; Santini, D.; Zappavigna, S.; Lombardi, A.; Misso, G.; Boccellino, M.; Desiderio, V.; Vitiello, P.P.; Di Lorenzo, G.; Zoccoli, A.; et al. Antagonistic effects of chloroquine on autophagy occurrence potentiate the anticancer effects of everolimus on renal cancer cells. Can. Biol. Ther. 2015, 16, 567–579. [Google Scholar] [CrossRef] [PubMed]
- Fukuda, T.; Oda, K.; Wada-Hiraike, O.; Sone, K.; Inaba, K.; Ikeda, Y.; Miyasaka, A.; Kashiyama, T.; Tanikawa, M.; Arimoto, T.; et al. The anti-malarial chloroquine suppresses proliferation and overcomes cisplatin resistance of endometrial cancer cells via autophagy inhibition. Gynecol. Oncol. 2015, 137, 538–545. [Google Scholar] [CrossRef] [PubMed]
- Koehler, B.C.; Jassowicz, A.; Scherr, A.L.; Lorenz, S.; Radhakrishnan, P.; Kautz, N.; Elssner, C.; Weiss, J.; Jaeger, D.; Schneider, M.; et al. Pan-Bcl-2 inhibitor Obatoclax is a potent late stage autophagy inhibitor in colorectal cancer cells independent of canonical autophagy signaling. BMC Cancer 2015, 15, 919. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Wu, W.K.; Gu, C.; Zhong, D.; Zhao, X.; Kong, Y.; Lin, Q.; Chan, M.T.; Zhou, Z.; Liu, S. Obatoclax impairs lysosomal function to block autophagy in cisplatin-sensitive and -resistant esophageal cancer cells. Oncotarget 2016, 7, 14693–14707. [Google Scholar] [CrossRef]
- Monneret, C. Platinum anticancer drugs. From serendipity to rational design. Ann. Pharmac. Franc. 2011, 69, 286–295. [Google Scholar] [CrossRef]
- Wang, J.; Wu, G.S. Role of autophagy in cisplatin resistance in ovarian cancer cells. J. Biol. Chem. 2014, 289, 17163–17173. [Google Scholar] [CrossRef]
- Zhang, S.F.; Wang, X.Y.; Fu, Z.Q.; Peng, Q.H.; Zhang, J.Y.; Ye, F.; Fu, Y.F.; Zhou, C.Y.; Lu, W.G.; Cheng, X.D.; et al. TXNDC17 promotes paclitaxel resistance via inducing autophagy in ovarian cancer. Autophagy 2015, 11, 225–238. [Google Scholar] [CrossRef] [PubMed]
- Jiménez-Guerrero, R.; Gasca, J.; Flores, M.L.; Pérez-Valderrama, B.; Tejera-Parrado, C.; Medina, R.; Tortolero, M.; Romero, F.; Japón, M.A.; Sáez, C. Obatoclax and Paclitaxel Synergistically Induce Apoptosis and Overcome Paclitaxel Resistance in Urothelial Cancer Cells. Cancers 2018, 10, 490. [Google Scholar] [CrossRef] [PubMed]
- Gorzalczany, Y.; Gilad, Y.; Amihai, D.; Hammel, I.; Sagi-Eisenberg, R.; Merimsky, O. Combining an EGFR directed tyrosine kinase inhibitor with autophagy-inducing drugs: A beneficial strategy to combat non-small cell lung cancer. Cancer Lett. 2011, 310, 207–215. [Google Scholar] [CrossRef] [PubMed]
- Fung, C.; Chen, X.; Grandis, J.R.; Duvvuri, U. EGFR tyrosine kinase inhibition induces autophagy in cancer cells. Cancer Biol. Ther. 2012, 13, 1417–1424. [Google Scholar] [CrossRef] [PubMed]
- Han, W.; Pan, H.; Chen, Y.; Sun, J.; Wang, Y.; Li, J.; Ge, W.; Feng, L.; Lin, X.; Wang, X.; et al. EGFR tyrosine kinase inhibitors activate autophagy as a cytoprotective response in human lung cancer cells. PLoS ONE 2011, 6, e18691. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Zou, Z.; Becker, N.; Anderson, M.; Sumpter, R.; Xiao, G.; Kinch, L.; Koduru, P.; Christudass, C.S.; Veltri, R.W.; et al. EGFR-mediated Beclin 1 phosphorylation in autophagy suppression, tumor progression, and tumor chemoresistance. Cell 2013, 154, 1269–1284. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.V.; Bell, D.W.; Settleman, J.; Haber, D.A. Epidermal growth factor receptor mutations in lung cancer. Nat. Rev. Cancer. 2007, 7, 169–181. [Google Scholar] [CrossRef] [PubMed]
- Lynch, T.J.; Bell, D.W.; Sordella, R.; Gurubhagavatula, S.; Okimoto, R.A.; Brannigan, B.W.; Harris, P.L.; Haserlat, S.M.; Supko, J.G.; Haluska, F.G.; et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N. Engl. J. Med. 2004, 350, 2129–2139. [Google Scholar] [CrossRef]
- Qi, M.; Tian, Y.; Li, W.; Li, D.; Zhao, T.; Yang, Y.; Li, Q.; Chen, S.; Yang, Y.; Zhang, Z.; et al. ERK inhibition represses gefitinib resistance in non-small cell lung cancer cells. Oncotarget 2018, 9, 12020–12034. [Google Scholar] [CrossRef]
- Zhu, Y.; He, W.; Gao, X.; Li, B.; Mei, C.; Xu, R.; Chen, H. Resveratrol overcomes gefitinib resistance by increasing the intracellular gefitinib concentration and triggering apoptosis, autophagy and senescence in PC9/G NSCLC cells. Sci. Rep. 2015, 5, 17730. [Google Scholar] [CrossRef]
- Tang, Q.; Li, G.; Wei, X.; Zhang, J.; Chiu, J.F.; Hasenmayer, D.; Zhang, D.; Zhang, H. Resveratrol-induced apoptosis is enhanced by inhibition of autophagy in esophageal squamous cell carcinoma. Cancer Lett. 2013, 336, 325–337. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zhu, X.; Qiao, H.; Ye, M.; Lai, X.; Yu, S.; Ding, L.; Wen, A.; Zhang, J. Protective autophagy promotes the resistance of HER2-positive breast cancer cells to lapatinib. Tumour Biol. 2016, 7, 2321–2331. [Google Scholar] [CrossRef] [PubMed]
- Jiao, Q.; Bi, L.; Ren, Y.; Song, S.; Wang, Q.; Wang, Y.S. Advances in studies of tyrosine kinase inhibitors and their acquired resistance. Mol. Cancer 2018, 17, 36. [Google Scholar] [CrossRef] [PubMed]
- Dent, P.; Booth, L.; Poklepovic, A.; Martinez, J.; Hoff, D.V.; Hancock, J.F. Neratinib degrades MST4 via autophagy that reduces membrane stiffness and is essential for the inactivation of PI3K, ERK1/2, and YAP/TAZ signaling. J. Cell Physiol. 2020, 235, 7889–7899. [Google Scholar] [CrossRef]
- Shimizu, S.; Takehara, T.; Hikita, H.; Kodama, T.; Tsunematsu, H.; Miyagi, T.; Hosui, A.; Ishida, H.; Tatsumi, T.; Kanto, T.; et al. Inhibition of autophagy potentiates the antitumor effect of the multikinase inhibitor sorafenib in hepatocellular carcinoma. Int. J. Cancer 2012, 13, 548–557. [Google Scholar] [CrossRef]
- Zhai, B.; Hu, F.; Jiang, X.; Xu, J.; Zhao, D.; Liu, B.; Pan, S.; Dong, X.; Tan, G.; Wei, Z.; et al. Inhibition of Akt reverses the acquired resistance to sorafenib by switching protective autophagy to autophagic cell death in hepatocellular carcinoma. Mol. Can. Therap. 2014, 13, 1589–1598. [Google Scholar] [CrossRef]
- Sun, T.; Liu, H.; Ming, L. Multiple Roles of Autophagy in the Sorafenib Resistance of Hepatocellular Carcinoma. Cell Physiol. Biochem. 2017, 44, 716–727. [Google Scholar] [CrossRef]
- Liu, R.; Li, J.; Zhang, T.; Zou, L.; Chen, Y.; Wang, K.; Lei, Y.; Yuan, K.; Li, Y.; Liu, R.; et al. Gankyrin drives metabolic reprogramming to promote tumorigenesis, metastasis and drug resistance through activating β-catenin/c-Myc signaling in human hepatocellular carcinoma. Cancer Lett. 2019, 443, 34–46. [Google Scholar] [CrossRef]
- Luo, T.; Fu, J.; Xu, A.; Su, B.; Ren, Y.; Li, N.; Zhu, J.; Zhao, X.; Dai, R.; Cao, J.; et al. PSMD10/gankyrin induces autophagy to promote tumor progression through cytoplasmic interaction with ATG7 and nuclear transactivation of ATG7 expression. Autophagy 2016, 12, 1355–1371. [Google Scholar] [CrossRef]
- Hashimoto, D.; Bläuer, M.; Hirota, M.; Ikonen, N.H.; Sand, J.; Laukkarinen, J. Autophagy is needed for the growth of pancreatic adenocarcinoma and has a cytoprotective effect against anticancer drugs. Eur. J. Can. 2014, 50, 1382–1390. [Google Scholar] [CrossRef]
- Boone, B.A.; Bahary, N.; Zureikat, A.H.; Moser, A.J.; Normolle, D.P.; Wu, W.C.; Singhi, A.D.; Bao, P.; Bartlett, D.L.; Liotta, L.A.; et al. Safety and Biologic Response of Pre-operative Autophagy Inhibition in Combination with Gemcitabine in Patients with Pancreatic Adenocarcinoma. Annu. Surg. Oncol. 2015, 22, 4402–4410. [Google Scholar] [CrossRef] [PubMed]
- Karasic, T.B.; O’Hara, M.H.; Loaiza-Bonilla, A.; Reiss, K.A.; Teitelbaum, U.R.; Borazanci, E.; De Jesus-Acosta, A.; Redlinger, C.; Burrell, J.A.; Laheru, D.A.; et al. Effect of Gemcitabine and nab-Paclitaxel With or Without Hydroxychloroquine on Patients With Advanced Pancreatic Cancer: A Phase 2 Randomized Clinical Trial. JAMA Oncol. 2019, 5, 993–998. [Google Scholar] [CrossRef] [PubMed]
- Windon, A.L.; Loaiza-Bonilla, A.; Jensen, C.E.; Randall, M.; Morrissette, J.; Shroff, S.G. A KRAS wild type mutational status confers a survival advantage in pancreatic ductal adenocarcinoma. J. Gastroint. Oncol. 2018, 9, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.M.; Shao, L.J.; Jiang, Z.F.; Liu, R.Y. Gemcitabine-Induced Autophagy Protects Human Lung Cancer Cells from Apoptotic Death. Lung 2016, 194, 959–966. [Google Scholar] [CrossRef]
- Chen, M.; He, M.; Song, Y.; Chen, L.; Xiao, P.; Wan, X.; Dai, F.; Shen, P. The cytoprotective role of gemcitabine-induced autophagy associated with apoptosis inhibition in triple-negative MDA-MB-231 breast cancer cells. Int. J. Mol. Med. 2014, 34, 276–282. [Google Scholar] [CrossRef]
- Vogel, R.D.; Hofheinz, R.D.; Kubicka, S.; Arnold, D. Treatment decisions in metastatic colorectal cancer - Beyond first and second line combination therapies. Cancer Treat. Rev. 2017, 59, 54–60. [Google Scholar] [CrossRef]
- Zhao, G.; Han, X.; Zheng, S.; Li, Z.; Sha, Y.; Ni, J.; Sun, Z.; Qiao, S.; Song, Z. Curcumin induces autophagy, inhibits proliferation and invasion by downregulating AKT/mTOR signaling pathway in human melanoma cells. Oncol. Rep. 2016, 35, 1065–1074. [Google Scholar] [CrossRef]
- Li, J.; Hou, N.; Faried, A.; Tsutsumi, S.; Takeuchi, T.; Kuwano, H. Inhibition of autophagy by 3-MA enhances the effect of 5-FU-induced apoptosis in colon cancer cells. Ann. Surg. Oncol. 2009, 16, 761–771. [Google Scholar] [CrossRef]
- Lamba, J.K. Genetic factors influencing cytarabine therapy. Pharmacogen. 2009, 10, 1657–1674. [Google Scholar] [CrossRef]
- Burger, J.A.; Kipps, T.J. CXCR4: A key receptor in the crosstalk between tumor cells and their microenvironment. Blood 2006, 107, 1761–1767. [Google Scholar] [CrossRef]
- Du, W.; Lu, C.; Zhu, X.; Hu, D.; Chen, X.; Li, J.; Liu, W.; Zhu, J.; He, Y.; Yao, J. Prognostic significance of CXCR4 expression in acute myeloid leukemia. Cancer Med. 2019, 8, 6595–6603. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Mei, S.; Meng, W.; Xue, S.; Jiang, L.; Yang, Y.; Hui, L.; Chen, Y.; Guan, M.X. CXCR4-mediated signaling regulates autophagy and influences acute myeloid leukemia cell survival and drug resistance. Cancer Lett. 2018, 425, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Bosnjak, M.; Ristic, B.; Arsikin, K.; Mircic, A.; Suzin-Zivkovic, V.; Perovic, V.; Bogdanovic, A.; Paunovic, V.; Markovic, I.; Bumbasirevic, V.; et al. Inhibition of mTOR-dependent autophagy sensitizes leukemic cells to cytarabine-induced apoptotic death. PLoS ONE 2014, 9, e94374. [Google Scholar] [CrossRef] [PubMed]
- Sumitomo, Y.; Koya, J.; Nakazaki, K.; Kataoka, K.; Tsuruta-Kishino, T.; Morita, K.; Sato, T.; Kurokawa, M. Cytoprotective autophagy maintains leukemia-initiating cells in murine myeloid leukemia. Blood 2016, 128, 1614–1624. [Google Scholar] [CrossRef]
- Santos, C.P.; Pereira, G.J.; Barbosa, C.M.; Jurkiewicz, A.; Smaili, S.S.; Bincoletto, C. Comparative study of autophagy inhibition by 3MA and CQ on Cytarabine induced death of leukaemia cells. J. Can. Res. Clin. Oncol. 2014, 140, 909–920. [Google Scholar] [CrossRef]
- Sen, N.; Gui, B.; Kumar, R. Role of MTA1 in cancer progression and metastasis. Cancer Metastasis Rev. 2014, 33, 879–889. [Google Scholar] [CrossRef]
- Lee, M.H.; Koh, D.; Na, H.; Ka, N.L.; Kim, S.; Kim, H.J.; Hong, S.; Shin, Y.K.; Seong, J.K.; Lee, M.O. MTA1 is a novel regulator of autophagy that induces tamoxifen resistance in breast cancer cells. Autophagy 2018, 14, 812–824. [Google Scholar] [CrossRef]
- Qadir, M.A.; Kwok, B.; Dragowska, W.H.; To, K.H.; Le, D.; Bally, M.B.; Gorski, S.M. Macroautophagy inhibition sensitizes tamoxifen-resistant breast cancer cells and enhances mitochondrial depolarization. Breast Cancer Res. Treat. 2008, 112, 389–403. [Google Scholar] [CrossRef]
- Cook, K.L.; Shajahan, A.N.; Clarke, R. Autophagy and endocrine resistance in breast cancer. Exp. Rev. Anticancer Ther. 2011, 11, 1283–1294. [Google Scholar] [CrossRef]
- Leignadier, J.; Dalenc, F.; Poirot, M.; Silvente-Poirot, S. Improving the efficacy of hormone therapy in breast cancer: The role of cholesterol metabolism in SERM-mediated autophagy, cell differentiation and death. Biochem. Pharmacol. 2017, 144, 18–28. [Google Scholar] [CrossRef]
- Das, C.K.; Parekh, A.; Parida, P.K.; Bhutia, S.K.; Mandal, M. Lactate dehydrogenase A regulates autophagy and tamoxifen resistance in breast cancer. Biochim. Biophys. Acta 2019, 1866, 1004–1018. [Google Scholar] [CrossRef] [PubMed]
- Mishra, D.; Banerjee, D. Lactate Dehydrogenases as Metabolic Links between Tumor and Stroma in the Tumor Microenvironment. Cancers 2019, 11, 750. [Google Scholar] [CrossRef] [PubMed]
- Le, A.; Cooper, C.R.; Gouw, A.M.; Dinavahi, R.; Maitra, A.; Deck, L.M.; Royer, R.E.; Vander Jagt, D.L.; Semenza, G.L.; Dang, C.V. Inhibition of lactate dehydrogenase A induces oxidative stress and inhibits tumor progression. Proc. Natl. Acad. Sci. USA 2010, 107, 2037–2042. [Google Scholar] [CrossRef] [PubMed]
- Bray, K.; Mathew, R.; Lau, A.; Kamphorst, J.J.; Fan, J.; Chen, J.; Chen, H.Y.; Ghavami, A.; Stein, M.; DiPaola, R.S.; et al. Autophagy suppresses RIP kinase-dependent necrosis enabling survival to mTOR inhibition. PLoS ONE 2012, 7, e41831. [Google Scholar] [CrossRef] [PubMed]
- Rangwala, R.; Chang, Y.C.; Hu, J.; Algazy, K.M.; Evans, T.L.; Fecher, L.A.; Schuchter, L.M.; Torigian, D.A.; Panosian, J.T.; Troxel, A.B.; et al. Combined MTOR and autophagy inhibition: Phase I trial of hydroxychloroquine and temsirolimus in patients with advanced solid tumors and melanoma. Autophagy 2014, 10, 1391–1402. [Google Scholar] [CrossRef]
- Motzer, R.J.; Escudier, B.; Oudard, S.; Hutson, T.E.; Porta, C.; Bracarda, S.; Grünwald, V.; Thompson, J.A.; Figlin, R.A.; Hollaender, N.; et al. RECORD-1 Study Group. Efficacy of everolimus in advanced renal cell carcinoma: A double-blind, randomised, placebo-controlled phase III trial. Lancet 2008, 372, 449–456. [Google Scholar] [CrossRef]
- Haas, N.B.; Appleman, L.J.; Stein, M.; Redlinger, M.; Wilks, M.; Xu, X.; Onorati, A.; Kalavacharla, A.; Kim, T.; Zhen, C.J.; et al. Autophagy Inhibition to Augment mTOR Inhibition: A Phase I/II Trial of Everolimus and Hydroxychloroquine in Patients with Previously Treated Renal Cell Carcinoma. Clin. Cancer Res. 2019, 25, 2080–2087. [Google Scholar] [CrossRef]
- Shen, H.M.; Codogno, P. Autophagic cell death: Loch Ness monster or endangered species? Autophagy 2011, 7, 457–465. [Google Scholar] [CrossRef]
- Galluzzi, L.; Baehrecke, E.H.; Ballabio, A.; Boya, P.; Bravo-San Pedro, J.M.; Cecconi, F.; Choi, A.M.; Chu, C.T.; Codogno, P.; Colombo, M.I.; et al. Molecular definitions of autophagy and related processes. EMBO J. 2017, 36, 1811–1836. [Google Scholar] [CrossRef]
- Klionsky, D.J.; Abdelmohsen, K.; Abe, A.; Abedin, M.J.; Abeliovich, H.; Acevedo Arozena, A. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy 2016, 12, 1–222. [Google Scholar] [CrossRef]
- Serrano-Oviedo, L.; Ortega-Muelas, M.; García-Cano, J.; Valero, M.L.; Cimas, F.J.; Pascual-Serra, R.; Fernandez-Aroca, D.M.; Roche, O.; Ruiz-Hidalgo, M.J.; Belandia, B.; et al. Autophagic cell death associated to Sorafenib in renal cell carcinoma is mediated through Akt inhibition in an ERK1/2 independent fashion. PLoS ONE 2018, 13, e0200878. [Google Scholar] [CrossRef] [PubMed]
- Xue, H.; Li, J.; Xie, H.; Wang, Y. Review of Drug Repositioning Approaches and Resources. Int. J. Biol. Sci. 2018, 14, 1232–1244. [Google Scholar] [CrossRef] [PubMed]
- Petroni, G.; Bagni, G.; Iorio, J.; Duranti, C.; Lottini, T.; Stefanini, M.; Kragol, G.; Becchetti, A.; Arcangeli, A. Clarithromycin inhibits autophagy in colorectal cancer by regulating the hERG1 potassium channel interaction with PI3K. Cell Death Dis. 2020, 11, 161. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.F.; Tsai, K.L.; Hsu, C.J.; Tsai, W.L.; Cheng, J.S.; Chang, H.W.; Shiau, C.W.; Goan, Y.G.; Tseng, H.H.; Wu, C.H.; et al. Drug Repurposing Screening Identifies Tioconazole as an ATG4 Inhibitor that Suppresses Autophagy and Sensitizes Cancer Cells to Chemotherapy. Theranostics 2018, 8, 830–845. [Google Scholar] [CrossRef]
- Wei, X.; Liu, W.; Wang, J.Q.; Tang, Z. “Hedgehog pathway”: A potential target of itraconazole in the treatment of cancer. J. Cancer Res. Clin. Oncol. 2020, 146, 297–304. [Google Scholar] [CrossRef]
- Wang, X.; Wei, S.; Zhao, Y.; Shi, C.; Liu, P.; Zhang, C.; Lei, Y.; Zhang, B.; Bai, B.; Huang, Y.; et al. Anti-proliferation of breast cancer cells with itraconazole: Hedgehog pathway inhibition induces apoptosis and autophagic cell death. Cancer Lett. 2017, 385, 128–136. [Google Scholar] [CrossRef]
- Antonarakis, E.S.; Heath, E.I.; Smith, D.C.; Rathkopf, D.; Blackford, A.L.; Danila, D.C.; King, S.; Frost, A.; Ajiboye, A.S.; Zhao, M.; et al. Repurposing itraconazole as a treatment for advanced prostate cancer: A noncomparative randomized phase II trial in men with metastatic castration-resistant prostate cancer. Oncologist 2013, 18, 163–173. [Google Scholar] [CrossRef]
- Yin, Y.C.; Lin, C.C.; Chen, T.T.; Chen, J.Y.; Tsai, H.J.; Wang, C.Y.; Chen, S.Y. Clozapine induces autophagic cell death in non-small cell lung cancer cells. Cell. Physiol. Biochem. 2015, 35, 945–956. [Google Scholar] [CrossRef]
- Liu, R.; Li, J.; Zhang, T.; Zou, L.; Chen, Y.; Wang, K.; Lei, Y.; Yuan, K.; Li, Y.; Lan, J.; et al. Itraconazole suppresses the growth of glioblastoma through induction of autophagy: Involvement of abnormal cholesterol trafficking. Autophagy 2014, 10, 1241–1255. [Google Scholar] [CrossRef]
- Kim, S.H.; Park, S.; Yu, H.S.; Ko, K.H.; Park, H.G.; Kim, Y.S. The antipsychotic agent clozapine induces autophagy via the AMPK-ULK1-Beclin1 signaling pathway in the rat frontal cortex. Prog. Neuro-Psychopharm. Biolog. Psych. 2018, 81, 96–104. [Google Scholar] [CrossRef]
- Chou, F.H.; Tsai, K.Y.; Su, C.Y.; Lee, C.C. The incidence and relative risk factors for developing cancer among patients with schizophrenia: A nine-year follow-up study. Schizophr. Res. 2011, 129, 97–103. [Google Scholar] [CrossRef] [PubMed]
- Ziaaddini, H.; Kheradmand, A.; Vahabi, M. Prevalence of cigarette smoking in schizophrenic patients compared to other hospital admitted psychiatric patients. Addict. Health 2009, 1, 38–42. [Google Scholar] [PubMed]
- Liu, Y.; Yang, Y.; Ye, Y.C.; Shi, Q.F.; Chai, K.; Tashiro, S.; Onodera, S.; Ikejima, T. Activation of ERK-p53 and ERK-mediated phosphorylation of Bcl-2 are involved in autophagic cell death induced by the c-Met inhibitor SU11274 in human lung cancer A549 cells. J. Pharmacol. Sci. 2012, 118, 423–432. [Google Scholar] [CrossRef] [PubMed]
- Puchalski, R.B.; Shah, N.; Miller, J.; Dalley, R.; Nomura, S.R.; Yoon, J.G.; Smith, K.A.; Lankerovich, M.; Bertagnolli, D.; Bickley, K.; et al. An anatomic transcriptional atlas of human glioblastoma. Science 2018, 360, 660–663. [Google Scholar] [CrossRef]
- Johnson, D.R.; Chang, S.M. Recent medical management of glioblastoma. Adv. Exp. Med. Biol. 2012, 746, 26–40. [Google Scholar] [CrossRef]
- Lefranc, F.; Kiss, R. Autophagy, the Trojan horse to combat glioblastomas. Neurosurg. Focus 2006, 20, E7. [Google Scholar] [CrossRef]
- Li, C.; Liu, Y.; Liu, H.; Zhang, W.; Shen, C.; Cho, K.; Chen, X.; Peng, F.; Bi, Y.; Hou, X.; et al. Impact of autophagy inhibition at different stages on cytotoxic effect of autophagy inducer in glioblastoma cells. Cell. Physiol. Biochem. 2015, 35, 1303–1316. [Google Scholar] [CrossRef]
- Kanzawa, T.; Kondo, Y.; Ito, H.; Kondo, S.; Germano, I. Induction of autophagic cell death in malignant glioma cells by arsenic trioxide. Cancer Res. 2003, 63, 2103–2108. [Google Scholar]
- Goussetis, D.J.; Altman, J.K.; Glaser, H.; McNeer, J.L.; Tallman, M.S.; Platanias, L.C. Autophagy is a critical mechanism for the induction of the antileukemic effects of arsenic trioxide. J. Biol. Chem. 2010, 285, 29989–29997. [Google Scholar] [CrossRef]
- Rosenfeld, M.R.; Ye, X.; Supko, J.G.; Desideri, S.; Grossman, S.A.; Brem, S.; Mikkelson, T.; Wang, D.; Chang, Y.C.; Hu, J.; et al. A phase I/II trial of hydroxychloroquine in conjunction with radiation therapy and concurrent and adjuvant temozolomide in patients with newly diagnosed glioblastoma multiforme. Autophagy 2014, 10, 1359–1368. [Google Scholar] [CrossRef]
- Kanzawa, T.; Germano, I.M.; Komata, T.; Ito, H.; Kondo, Y.; Kondo, S. Role of autophagy in temozolomide-induced cytotoxicity for malignant glioma cells. Cell Death Differ. 2004, 11, 448–457. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Luo, W.; Zeng, C.; Zhang, Y.; Wang, L.; Yao, W.; Nie, C. PP2A mediates apoptosis or autophagic cell death in multiple myeloma cell lines. Oncotarget 2017, 8, 80770–80789. [Google Scholar] [CrossRef] [PubMed]
- Nie, C.; Luo, Y.; Zhao, X.; Luo, N.; Tong, A.; Liu, X.; Yuan, Z.; Wang, C.; Wei, Y. Caspase-9 mediates Puma activation in UCN-01-induced apoptosis. Cell Death Dis. 2014, 5, e1495. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Yang, L.J.; Chen, Y.; He, J.; Yi, S.; Wen, L.; Zhao, J.; Zhang, B.P.; Cui, G.H. Betulinic acid inhibits autophagic flux and induces apoptosis in human multiple myeloma cells in vitro. Acta Pharmacol. Sin. 2012, 33, 1542–1548. [Google Scholar] [CrossRef] [PubMed]
- Salazar, M.; Carracedo, A.; Salanueva, I.J.; Hernández-Tiedra, S.; Lorente, M.; Egia, A.; Vázquez, P.; Blázquez, C.; Torres, S.; García, S.; et al. Cannabinoid action induces autophagy-mediated cell death through stimulation of ER stress in human glioma cells. J. Clin. Investig. 2009, 119, 1359–1372. [Google Scholar] [CrossRef]
- Armstrong, J.L.; Hill, D.S.; McKee, C.S.; Hernandez-Tiedra, S.; Lorente, M.; Lopez-Valero, I.; Eleni Anagnostou, M.; Babatunde, F.; Corazzari, M.; Redfern, C.; et al. Exploiting cannabinoid-induced cytotoxic autophagy to drive melanoma cell death. J. Invest. Dermatol. 2015, 135, 1629–1637. [Google Scholar] [CrossRef]
- Jeong, S.; Kim, B.G.; Kim, D.Y.; Kim, B.R.; Kim, J.L.; Park, S.H.; Na, Y.J.; Jo, M.J.; Yun, H.K.; Jeong, Y.A.; et al. Cannabidiol Overcomes Oxaliplatin Resistance by Enhancing NOS3- and SOD2-Induced Autophagy in Human Colorectal Cancer Cells. Cancers 2019, 11, 781. [Google Scholar] [CrossRef]
- Mohammadinejad, R.; Ahmadi, Z.; Tavakol, S.; Ashrafizadeh, M. Berberine as a potential autophagy modulator. J. Cell. Physiol. 2019, 234, 14914–14926. [Google Scholar] [CrossRef]
- Jin, Y.; Khadka, D.B.; Cho, W.J. Pharmacological effects of berberine and its derivatives: A patent update. Exp. Opin. Therap. Patents 2016, 26, 229–243. [Google Scholar] [CrossRef]
- Feng, X.; Sureda, A.; Jafari, S.; Memariani, Z.; Tewari, D.; Annunziata, G.; Barrea, L.; Hassan, S.; Šmejkal, K.; Malaník, M.; et al. Berberine in Cardiovascular and Metabolic Diseases: From Mechanisms to Therapeutics. Theranostics 2019, 9, 1923–1951. [Google Scholar] [CrossRef]
- La, X.; Zhang, L.; Li, Z.; Yang, P.; Wang, Y. Berberine-induced autophagic cell death by elevating GRP78 levels in cancer cells. Oncotarget 2017, 8, 20909–20924. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Wang, Y.; Newton, I.P.; Zhang, L.; Ji, P.; Li, Z. GRP78 is implicated in the modulation of tumor aerobic glycolysis by promoting autophagic degradation of IKKβ. Cell. Signal. 2015, 27, 1237–1245. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Zhang, C.; Bao, J.; Jia, X.; Liang, Y.; Wang, X.; Chen, M.; Su, H.; Li, P.; Wan, J.B.; et al. Synergistic chemopreventive effects of curcumin and berberine on human breast cancer cells through induction of apoptosis and autophagic cell death. Sci. Rep. 2016, 6, 26064. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Qi, Q.; Feng, Z.; Zhang, X.; Huang, B.; Chen, A.; Prestegarden, L.; Li, X.; Wang, J. Berberine induces autophagy in glioblastoma by targeting the AMPK/mTOR/ULK1-pathway. Oncotarget 2016, 7, 66944–66958. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.; Huang, H.P.; Wang, Y.; Jin, J.; Long, W.G.; Chen, K.; Zhao, X.H.; Chen, C.G.; Li, J. Curcumin overcome primary gefitinib resistance in non-small-cell lung cancer cells through inducing autophagy-related cell death. J. Exp. Clin. Cancer Res. 2019, 38, 254. [Google Scholar] [CrossRef]
- Li, J.; Hou, N.; Faried, A.; Tsutsumi, S.; Takeuchi, T.; Kuwano, H.; Li, W.; Zhou, Y.; Yang, J.; Li, H.; et al. Curcumin induces apoptotic cell death and protective autophagy in human gastric cancer cells. Oncol. Rep. 2017, 37, 3459–3466. [Google Scholar] [CrossRef]
- Sun, J.; Feng, Y.; Wang, Y.; Ji, Q.; Cai, G.; Shi, L.; Wang, Y.; Huang, Y.; Zhang, J.; Li, Q. α-hederin induces autophagic cell death in colorectal cancer cells through reactive oxygen species dependent AMPK/mTOR signaling pathway activation. Int. J. Oncol. 2019, 54, 1601–1612. [Google Scholar] [CrossRef]
- Law, B.; Michelangeli, F.; Qu, Y.Q.; Xu, S.W.; Han, Y.; Mok, S.; Dias, I.; Javed, M.U.; Chan, W.K.; Xue, W.W.; et al. Neferine induces autophagy-dependent cell death in apoptosis-resistant cancers via ryanodine receptor and Ca2+-dependent mechanism. Sci. Rep. 2019, 9, 20034. [Google Scholar] [CrossRef]
- Lou, J.S.; Bi, W.C.; Chan, G.; Jin, Y.; Wong, C.W.; Zhou, Z.Y.; Wang, H.Y.; Yao, P.; Dong, T.; Tsim, K. Ginkgetin induces autophagic cell death through p62/SQSTM1-mediated autolysosome formation and redox setting in non-small cell lung cancer. Oncotarget 2017, 8, 93131–93148. [Google Scholar] [CrossRef]
- Bezerra, D.P.; Militão, G.C.; de Castro, F.O.; Pessoa, C.; de Moraes, M.O.; Silveira, E.R.; Lima, M.A.; Elmiro, F.J.; Costa-Lotufo, L.V. Piplartine induces inhibition of leukemia cell proliferation triggering both apoptosis and necrosis pathways. Toxicol. Vitro. 2007, 21, 1–8. [Google Scholar] [CrossRef]
- Bezerra, D.P.; Pessoa, C.; de Moraes, M.O.; Saker-Neto, N.; Silveira, E.R.; Costa-Lotufo, L.V. Overview of the therapeutic potential of piplartine (piperlongumine). Eur. J. Pharmac. Sci. 2013, 48, 453–463. [Google Scholar] [CrossRef] [PubMed]
- Adams, D.J.; Dai, M.; Pellegrino, G.; Wagner, B.K.; Stern, A.M.; Shamji, A.F.; Schreiber, S.L. Synthesis, cellular evaluation, and mechanism of action of piperlongumine analogs. Proc. Natl. Acad. Sci. USA 2012, 109, 15115–15120. [Google Scholar] [CrossRef] [PubMed]
- Costa, C.O.D.; Araujo Neto, J.H.; Baliza, I.; Dias, R.B.; Valverde, L.F.; Vidal, M.; Sales, C.; Rocha, C.; Moreira, D.; Soares, M.; et al. Novel piplartine-containing ruthenium complexes: Synthesis, cell growth inhibition, apoptosis induction and ROS production on HCT116 cells. Oncotarget 2017, 8, 104367–104392. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, M.S.; Barbosa, M.; de Souza, T.B.; Moreira, D.; Martins, F.T.; Villarreal, W.; Machado, R.P.; Doriguetto, A.C.; Soares, M.; Bezerra, D.P. A novel platinum complex containing a piplartine derivative exhibits enhanced cytotoxicity, causes oxidative stress and triggers apoptotic cell death by ERK/p38 pathway in human acute promyelocytic leukemia HL-60 cells. Redox Biol. 2019, 20, 182–194. [Google Scholar] [CrossRef]
- Baliza, I.; Silva, S.; Santos, L.S.; Neto, J.; Dias, R.B.; Sales, C.; Rocha, C.; Soares, M.; Batista, A.A.; Bezerra, D.P. Ruthenium Complexes with Piplartine Cause Apoptosis Through MAPK Signaling by a p53-Dependent Pathway in Human Colon Carcinoma Cells and Inhibit Tumor Development in a Xenograft Model. Front. Oncol. 2019, 9, 582. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, J.W.; Xiao, X.; Shan, Y.; Xue, B.; Jiang, G.; He, Q.; Chen, J.; Xu, H.G.; Zhao, R.X.; et al. Piperlongumine induces autophagy by targeting p38 signaling. Cell Death Dis. 2013, 4, e824. [Google Scholar] [CrossRef]
- Wang, F.; Mao, Y.; You, Q.; Hua, D.; Cai, D. Piperlongumine induces apoptosis and autophagy in human lung cancer cells through inhibition of PI3K/Akt/mTOR pathway. Int. J. Immunopathol. Pharmacol. 2015, 28, 362–373. [Google Scholar] [CrossRef]
- Makhov, P.; Golovine, K.; Teper, E.; Kutikov, A.; Mehrazin, R.; Corcoran, A.; Tulin, A.; Uzzo, R.G.; Kolenko, V.M. Piperlongumine promotes autophagy via inhibition of Akt/mTOR signalling and mediates cancer cell death. Br. J. Cancer 2014, 110, 899–907. [Google Scholar] [CrossRef]
- Berger, A.; Venturelli, S.; Kallnischkies, M.; Böcker, A.; Busch, C.; Weiland, T.; Noor, S.; Leischner, C.; Weiss, T.S.; Lauer, U.M.; et al. Kaempferol, a new nutrition-derived pan-inhibitor of human histone deacetylases. J. Nutri. Biochem. 2013, 24, 977–985. [Google Scholar] [CrossRef]
- Kim, T.W.; Lee, S.Y.; Kim, M.; Cheon, C.; Ko, S.G. Kaempferol induces autophagic cell death via IRE1-JNK-CHOP pathway and inhibition of G9a in gastric cancer cells. Cell Death Dis. 2018, 9, 875. [Google Scholar] [CrossRef]
- Han, B.; Yu, Y.Q.; Yang, Q.L.; Shen, C.Y.; Wang, X.J. Kaempferol induces autophagic cell death of hepatocellular carcinoma cells via activating AMPK signaling. Oncotarget 2017, 8, 86227–86239. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Pan, W.; Zhu, M.; Zhang, M.; Hao, X.; Liang, G.; Feng, Y. Fangchinoline induces autophagic cell death via p53/sestrin2/AMPK signalling in human hepatocellular carcinoma cells. Br. J. Pharmacol. 2011, 164, 731–742. [Google Scholar] [CrossRef] [PubMed]
- Lei, Y.; Zhang, D.; Yu, J.; Dong, H.; Zhang, J.; Yang, S. Targeting autophagy in cancer stem cells as an anticancer therapy. Cancer Lett. 2017, 393, 33–39. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Shi, P.; Zhao, G.; Xu, J.; Peng, W.; Zhang, J.; Zhang, G.; Wang, X.; Dong, Z.; Chen, F.; et al. Targeting cancer stem cell pathways for cancer therapy. Sig. Transduct. Target Ther. 2020, 5, 8. [Google Scholar] [CrossRef]
- Mandhair, H.K.; Arambasic, M.; Novak, U.; Radpour, R. Molecular modulation of autophagy: New venture to target resistant cancer stem cells. World J. Stem Cells 2020, 12, 303–322. [Google Scholar] [CrossRef]
- Rahman, M.A.; Saha, S.K.; Rahman, M.S.; Uddin, M.J.; Uddin, M.S.; Pang, M.G.; Rhim, H.; Cho, S.G. Molecular Insights into Therapeutic Potential of Autophagy Modulation by Natural Products for Cancer Stem Cells. Front. Cell Dev. Biol. 2020, 8, 283. [Google Scholar] [CrossRef]
- Ramos, E.K.; Hoffmann, A.D.; Gerson, S.L.; Liu, H. New Opportunities and Challenges to Defeat Cancer Stem Cells. Trends Cancer 2017, 3, 780–796. [Google Scholar] [CrossRef]
- Shibue, T.; Weinberg, R.A. EMT, CSCs, and drug resistance: The mechanistic link and clinical implications. Nat. Rev. 2017, 14, 611–629. [Google Scholar] [CrossRef]
- Smith, A.G.; Macleod, K.F. Autophagy, cancer stem cells and drug resistance. J. Pathol. 2019, 247, 708–718. [Google Scholar] [CrossRef]
- Pérez-Hernández, M.; Arias, A.; Martínez-García, D.; Pérez-Tomás, R.; Quesada, R.; Soto-Cerrato, V. Targeting Autophagy for Cancer Treatment and Tumor Chemosensitization. Cancers 2019, 11, 1599. [Google Scholar] [CrossRef]
- Han, Y.; Fan, S.; Qin, T.; Yang, J.; Sun, Y.; Lu, Y.; Mao, J.; Li, L. Role of autophagy in breast cancer and breast cancer stem cells (Review). Int. J. Oncol. 2018, 52, 1057–1070. [Google Scholar] [CrossRef] [PubMed]
- Nazio, F.; Bordi, M.; Cianfanelli, V.; Locatelli, F.; Cecconi, F. Autophagy and cancer stem cells: Molecular mechanisms and therapeutic applications. Cell Death Differ. 2019, 26, 690–702. [Google Scholar] [CrossRef] [PubMed]
- Kondapuram, S.K.; Sarvagalla, S.; Coumar, M.S. Targeting autophagy with small molecules for cancer therapy. J. Cancer Metastasis Treat. 2019, 5, 32. [Google Scholar] [CrossRef]
- Jensen, S.S.; Petterson, S.A.; Halle, B.; Aaberg-Jessen, C.; Kristensen, B.W. Effects of the lysosomal destabilizing drug siramesine on glioblastoma in vitro and in vivo. BMC Cancer 2017, 17, 178. [Google Scholar] [CrossRef]
- Buccarelli, M.; Marconi, M.; Pacioni, S.; De Pascalis, I.; D’Alessandris, Q.G.; Martini, M.; Ascione, B.; Malorni, W.; Larocca, L.M.; Pallini, R.; et al. Inhibition of autophagy increases susceptibility of glioblastoma stem cells to temozolomide by igniting ferroptosis. Cell Death Dis. 2018, 9, 841. [Google Scholar] [CrossRef]
- Hegazy, A.M.; Yamada, D.; Kobayashi, M.; Kohno, S.; Ueno, M.; Ali, M.A.; Ohta, K.; Tadokoro, Y.; Ino, Y.; Todo, T.; et al. Therapeutic Strategy for Targeting Aggressive Malignant Gliomas by Disrupting Their Energy Balance. J. Biolog. Chem. 2016, 291, 21496–21509. [Google Scholar] [CrossRef]
- Chiao, M.T.; Cheng, W.Y.; Yang, Y.C.; Shen, C.C.; Ko, J.L. Suberoylanilide hydroxamic acid (SAHA) causes tumor growth slowdown and triggers autophagy in glioblastoma stem cells. Autophagy 2013, 9, 1509–1526. [Google Scholar] [CrossRef]
- Wang, W.J.; Long, L.M.; Yang, N.; Zhang, Q.Q.; Ji, W.J.; Zhao, J.H.; Qin, Z.H.; Wang, Z.; Chen, G.; Liang, Z.Q. NVP-BEZ235, a novel dual PI3K/mTOR inhibitor, enhances the radiosensitivity of human glioma stem cells in vitro. Acta Pharmacol. Sin. 2013, 34, 681–690. [Google Scholar] [CrossRef]
- Cheng, H.W.; Liang, Y.H.; Kuo, Y.L.; Chuu, C.P.; Lin, C.Y.; Lee, M.H.; Wu, A.T.; Yeh, C.T.; Chen, E.I.; Whang-Peng, J.; et al. Identification of thioridazine, an antipsychotic drug, as an antiglioblastoma and anticancer stem cell agent using public gene expression data. Cell Death Dis. 2015, 6, e1753. [Google Scholar] [CrossRef]
- Chu, C.W.; Ko, H.J.; Chou, C.H.; Cheng, T.S.; Cheng, H.W.; Liang, Y.H.; Lai, Y.L.; Lin, C.Y.; Wang, C.; Loh, J.K.; et al. Thioridazine Enhances P62-Mediated Autophagy and Apoptosis Through Wnt/β-Catenin Signaling Pathway in Glioma Cells. Int. J. Mol. Sci. 2019, 20, 473. [Google Scholar] [CrossRef]
- Lin, C.M.; Chen, H.H.; Lin, C.A.; Wu, H.C.; Sheu, J.J.; Chen, H.J. Apigenin-induced lysosomal degradation of β-catenin in Wnt/β-catenin signaling. Sci. Rep. 2017, 7, 372. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.J.; Zhang, S.S.; Guo, X.L.; Sun, K.; Han, Z.P.; Li, R.; Zhao, Q.D.; Deng, W.J.; Xie, X.Q.; Zhang, J.W.; et al. Autophagy contributes to the survival of CD133+ liver cancer stem cells in the hypoxic and nutrient-deprived tumor microenvironment. Cancer Lett. 2013, 339, 70–81. [Google Scholar] [CrossRef] [PubMed]
- Balic, A.; Sørensen, M.D.; Trabulo, S.M.; Sainz, B., Jr.; Cioffi, M.; Vieira, C.R.; Miranda-Lorenzo, I.; Hidalgo, M.; Kleeff, J.; Erkan, M.; et al. Chloroquine targets pancreatic cancer stem cells via inhibition of CXCR4 and hedgehog signaling. Mol. Cancer Therap. 2014, 13, 1758–1771. [Google Scholar] [CrossRef] [PubMed]
- Ray, A.; Vasudevan, S.; Sengupta, S. 6-Shogaol Inhibits Breast Cancer Cells and Stem Cell-Like Spheroids by Modulation of Notch Signaling Pathway and Induction of Autophagic Cell Death. PLoS ONE 2015, 10, e0137614. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.H.; Lee, C.Y.; Lu, C.C.; Tsai, F.J.; Hsu, Y.M.; Tsao, J.W.; Juan, Y.N.; Chiu, H.Y.; Yang, J.S.; Wang, C.C. Resveratrol-induced autophagy and apoptosis in cisplatin-resistant human oral cancer CAR cells: A key role of AMPK and Akt/mTOR signaling. Int. J. Oncol. 2017, 50, 873–882. [Google Scholar] [CrossRef]
- García-Zepeda, S.P.; García-Villa, E.; Díaz-Chávez, J.; Hernández-Pando, R.; Gariglio, P. Resveratrol induces cell death in cervical cancer cells through apoptosis and autophagy. Eur. J. Cancer Prev. 2013, 22, 577–584. [Google Scholar] [CrossRef]
- Selvaraj, S.; Sun, Y.; Sukumaran, P.; Singh, B.B. Resveratrol activates autophagic cell death in prostate cancer cells via downregulation of STIM1 and the mTOR pathway. Mol. Carcinog. 2016, 55, 818–831. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, S.; Liu, Y.; Liu, J.; Ma, Y.; Zhu, Y.; Zhang, J. Effects of the combination of RAD001 and docetaxel on breast cancer stem cells. Eur. J. Cancer. 2012, 48, 1581–1592. [Google Scholar] [CrossRef]
- Yu, F.; Zhao, J.; Hu, Y.; Zhou, Y.; Guo, R.; Bai, J.; Zhang, S.; Zhang, H.; Zhang, J. The combination of NVP-BKM120 with trastuzumab or RAD001 synergistically inhibits the growth of breast cancer stem cells in vivo. Oncol. Rep. 2016, 36, 356–364. [Google Scholar] [CrossRef]
- Liang, D.H.; Choi, D.S.; Ensor, J.E.; Kaipparettu, B.A.; Bass, B.L.; Chang, J.C. The autophagy inhibitor chloroquine targets cancer stem cells in triple negative breast cancer by inducing mitochondrial damage and impairing DNA break repair. Cancer Lett. 2016, 376, 249–258. [Google Scholar] [CrossRef]
- Li, L.Q.; Pan, D.; Zhang, S.W.; Y-Xie, D.; Zheng, X.L.; Chen, H. Autophagy regulates chemoresistance of gastric cancer stem cells via the Notch signaling pathway. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 3402–3407. [Google Scholar] [CrossRef] [PubMed]
- Hao, C.; Liu, G.; Tian, G. Autophagy inhibition of cancer stem cells promotes the efficacy of cisplatin against non-small cell lung carcinoma. Therap. Adv. Resp. Dis. 2019, 13, 1753466619866097. [Google Scholar] [CrossRef] [PubMed]
- Francipane, M.G.; Lagasse, E. Selective targeting of human colon cancer stem-like cells by the mTOR inhibitor Torin-1. Oncotarget 2013, 4, 1948–1962. [Google Scholar] [CrossRef] [PubMed]
- Silvente-Poirot, S.; Segala, G.; Poirot, M.C.; Poirot, M. Ligand-dependent transcriptional induction of lethal autophagy: A new perspective for cancer treatment. Autophagy 2018, 14, 555–557. [Google Scholar] [CrossRef]
- Segala, G.; David, M.; de Medina, P.; Poirot, M.C.; Serhan, N.; Vergez, F.; Mougel, A.; Saland, E.; Carayon, K.; Leignadier, J.; et al. Dendrogenin A drives LXR to trigger lethal autophagy in cancers. Nat. Commun. 2017, 8, 1903. [Google Scholar] [CrossRef]
- Folkerts, H.; Hilgendorf, S.; Wierenga, A.; Jaques, J.; Mulder, A.B.; Coffer, P.J.; Schuringa, J.J.; Vellenga, E. Inhibition of autophagy as a treatment strategy for p53 wild-type acute myeloid leukemia. Cell Death Dis. 2017, 8, e2927. [Google Scholar] [CrossRef]
- Lee, S.W.; Kim, H.K.; Lee, N.H.; Yi, H.Y.; Kim, H.S.; Hong, S.H.; Hong, Y.K.; Joe, Y.A. The synergistic effect of combination temozolomide and chloroquine treatment is dependent on autophagy formation and p53 status in glioma cells. Cancer Lett. 2015, 360, 195–204. [Google Scholar] [CrossRef]
- Baquero, P.; Dawson, A.; Mukhopadhyay, A.; Kuntz, E.M.; Mitchell, R.; Olivares, O.; Ianniciello, A.; Scott, M.T.; Dunn, K.; Nicastri, M.C.; et al. Targeting quiescent leukemic stem cells using second generation autophagy inhibitors. Leukemia 2019, 33, 981–994. [Google Scholar] [CrossRef]
- Ondrej, M.; Cechakova, L.; Fabrik, I.; Klimentova, J.; Tichy, A. Lys05—A Promising Autophagy Inhibitor in the Radiosensitization Battle: Phosphoproteomic Perspective. Cancer Genom. Proteom. 2020, 17, 369–382. [Google Scholar] [CrossRef]
- Takeda, M.; Koseki, J.; Takahashi, H.; Miyoshi, N.; Nishida, N.; Nishimura, J.; Hata, T.; Matsuda, C.; Mizushima, T.; Yamamoto, H.; et al. Disruption of Endolysosomal RAB5/7 Efficiently Eliminates Colorectal Cancer Stem Cells. Cancer Res. 2019, 79, 1426–1437. [Google Scholar] [CrossRef]
- An, H.K.; Kim, K.S.; Lee, J.W.; Park, M.H.; Moon, H.I.; Park, S.J.; Baik, J.S.; Kim, C.H.; Lee, Y.C. Mimulone-induced autophagy through p53-mediated AMPK/mTOR pathway increases caspase-mediated apoptotic cell death in A549 human lung cancer cells. PLoS ONE 2014, 9, e114607. [Google Scholar] [CrossRef] [PubMed]
- Tan, Q.; Joshua, A.M.; Saggar, J.K.; Yu, M.; Wang, M.; Kanga, N.; Zhang, J.Y.; Chen, X.; Wouters, B.G.; Tannock, I.F. Effect of pantoprazole to enhance activity of docetaxel against human tumour xenografts by inhibiting autophagy. Br. J. Cancer. 2015, 112, 832–840. [Google Scholar] [CrossRef] [PubMed]
- Kumar, D.; Shankar, S.; Srivastava, R.K. Rottlerin induces autophagy and apoptosis in prostate cancer stem cells via PI3K/Akt/mTOR signaling pathway. Cancer Lett. 2014, 343, 179–189. [Google Scholar] [CrossRef] [PubMed]
| Drugs | Action | Number of Clinical Trial * |
|---|---|---|
| α-Hederin | Induces autophagy-mediated cell death in colorectal cancer cells [239] | - |
| 6-Shogaol | Induces autophagy-mediated cell death in breast cancer cells [276] | - |
| Apigenin | Induces autophagy-mediated cell death in embryonic cancer cells [273] | 1 |
| Arsenic trioxide | Induces autophagy-mediated cell death in glioblastoma cells [220] | 125 |
| Berberine | Induces autophagy-mediated cell death in colon and liver cancer cells [233] | 8 |
| Induces autophagy-mediated cell death in breast cancer cells [235] | ||
| Betulinic acid | Induces autophagy-mediated cell death in myeloma cells [224] | 1 |
| Cannabinoid | Induces autophagy-mediated cell death in glioma [227] | 22 |
| Induces autophagy-mediated cell death in melanoma cells [228] | ||
| Induces autophagy-mediated cell death in colorectal cancer cells [229] | ||
| Chloroquine | Inhibits cytoprotective autophagy in liver cancer cells [274] | 23 |
| Inhibits cytoprotective autophagy in breast cancer cells [282] | ||
| Inhibits cytoprotective autophagy in gastric cancer cells [283] | ||
| Inhibits cytoprotective autophagy in glioma [289] | ||
| Clarithromycin | Inhibits cytoprotective autophagy in colorectal cancer cells [205] | 60 |
| Clozapine | Induces autophagy-mediated cell death in lung cancer cells [210] | 2 |
| Curcumin | Inhibits cytoprotective autophagy in colon cancer cells [145] | 68 |
| Inhibits cytoprotective autophagy in melanoma cells [179] | ||
| Dendrogenin A | Induces autophagy-mediated cell death in acute myeloid leukemia and melanoma [287] | 2 |
| Everolimus | Induces autophagy-mediated cell death in breast cancer cells [280] | 604 |
| Fangchinoline | Induces autophagy-mediated cell death in liver cancer cells [254] | - |
| Ginkgetin | Induces autophagy-mediated cell death in lung cancer cells [241] | - |
| Obatoclax (GX15-070) | Inhibits cytoprotective autophagy in colorectal cancer cells [149] | 19 |
| Inhibits cytoprotective autophagy in esophageal bladder cancer [150] | ||
| Hydroxychloroquine | Inhibits cytoprotective autophagy in acute myeloid leukemia [288] | 81 |
| Itraconazole | Induces autophagy-mediated cell death in breast cancer cells [208] | 56 |
| Kaempferol | Induces autophagy-mediated cell death in gastric cancer cells [252] | - |
| Induces autophagy-mediated cell death in liver cancer cells [253] | ||
| Lys05 | Inhibits cytoprotective autophagy in chronic myeloid leukemia [290] | - |
| Inhibits cytoprotective autophagy in lung cancer cells [291] | ||
| Mefloquine | Inhibits cytoprotective autophagy in colorectal cancer cells [292] | 1 |
| Mimulone | Induces autophagy-mediated cell death in lung cancer cells [293] | - |
| Neferin | Induces autophagy-mediated cell death in cervical cancer cells [240] | - |
| Nigericin | Induces autophagy-mediated cell death in glioblastoma cells [268] | - |
| Dactolisib (NVP-BEZ235) | Induces autophagy-mediated cell death in glioma [270] | 22 |
| Buparlisib (NVP-BKM120) | Induces autophagy-mediated cell death in breast cancer cells [281] | 86 |
| Pantoprazole | Inhibits cytoprotective autophagy in breast cancer cells, vulvar epidermoid carcinoma and prostate cancer cells [294] | 16 |
| Piplartine/piperlongumine | Induces autophagy-mediated cell death in osteosarcoma cells [248] | - |
| Quinacrine | Inhibits cytoprotective autophagy in glioblastoma cells [267] | 4 |
| Resveratrol | Induces autophagy-mediated cell death in breast cancer cells [9] | 16 |
| Induces autophagy-mediated cell death in oral cancer cells [273] | ||
| Rottlerin | Induces autophagy-mediated cell death in prostate cancer cells [295] | - |
| Siramesine | Inhibits cytoprotective autophagy in glioblastoma cells [266] | - |
| Sorafenib | Induces autophagy-mediated cell death in renal carcinoma cells [203] | 836 |
| SU11274 | Induces autophagy-mediated cell death in lung cancer cells [215] | - |
| Suberoylanilide hydroxamic acid (Vorinostat/SAHA) | Induces autophagy-mediated cell death in glioblastoma cells [269] | 256 |
| Temozolomide | Induces autophagy-mediated cell death in glioblastoma cells [219] | 953 |
| Induces cytoprotective autophagy in glioblastoma cells [223] | ||
| Thioridazine | Induces autophagy-mediated cell death in glioblastoma cells [272] | 1 |
| Tioconazole | Inhibits cytoprotective autophagy in glioblastoma, colorectal and breast cancer cells [206] | - |
| Torin-1 | Induces autophagy-mediated cell death in colorectal cancer cells [285] | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Silva, V.R.; Neves, S.P.; Santos, L.d.S.; Dias, R.B.; Bezerra, D.P. Challenges and Therapeutic Opportunities of Autophagy in Cancer Therapy. Cancers 2020, 12, 3461. https://doi.org/10.3390/cancers12113461
Silva VR, Neves SP, Santos LdS, Dias RB, Bezerra DP. Challenges and Therapeutic Opportunities of Autophagy in Cancer Therapy. Cancers. 2020; 12(11):3461. https://doi.org/10.3390/cancers12113461
Chicago/Turabian StyleSilva, Valdenizia R., Sara P. Neves, Luciano de S. Santos, Rosane B. Dias, and Daniel P. Bezerra. 2020. "Challenges and Therapeutic Opportunities of Autophagy in Cancer Therapy" Cancers 12, no. 11: 3461. https://doi.org/10.3390/cancers12113461
APA StyleSilva, V. R., Neves, S. P., Santos, L. d. S., Dias, R. B., & Bezerra, D. P. (2020). Challenges and Therapeutic Opportunities of Autophagy in Cancer Therapy. Cancers, 12(11), 3461. https://doi.org/10.3390/cancers12113461






