1. Introduction
There is a need for new prognostic options and treatment principles for nasopharyngeal cancer (NPC), a malignancy frequently associated with Epstein–Barr virus (EBV), and measures targeting the immune system may offer such possibilities. However, this requires detailed knowledge about the cancer and its local microenvironment, particularly on the presence and distribution of intralesional immune cells and their targets and functions. In a recent study by Wang et al., the importance of the immune status of NPC was underscored: focusing on immune checkpoints PD-L1 and B7-H4 on tumor cells and PD-L1, B7-H3, B7-H4, IDO-1, VISTA, ICOS, and OX40 on intralesional immune cells, specific signatures were demonstrated to predict survival [
1]. Interestingly, the association was particularly strong for patients with high levels of EBV DNA in plasma, suggesting the importance of EBV-antigen presence to the cancer and immune system interaction and indicating a need to monitor EBV status or even antigen levels in studies focusing on aspects of the immune system in NPC. Furthermore, in an even more recent study, Chen et al. demonstrated that specific gene signatures of macrophages, plasmacytoid dendritic cells (DCs), CLEC9A+ DCs, natural killer cells, and plasma cells were associated with improved progression-free survival [
2].
With regard to interactions between the immune system and NPC, tissue-infiltrating lymphocytes (TILs) are of importance. The density/distribution of TILs has been investigated and identified as an independent positive prognostic factor [
3]. Arguably, this lymphocyte population includes T-cells that, if appropriately instructed by DCs (e.g., via adjuvant targets such as C-lectin receptors), can facilitate antigen cross-presentation and can produce cytotoxic T-cell (CTL) responses [
4,
5]. Furthermore, the presence and distribution of lymphocytes, notably T-cells, as demonstrated for other cancers and indicated to predict, e.g., response to immunotherapy [
6], may allow for classification of NPC into specific immune phenotypes. In addition to T-cell observations, following morphological findings suggesting a presence of DCs in NPC lesions [
7,
8,
9,
10,
11,
12], we recently reported on intralesional DC subsets and particularly highlighted CD1c+ myeloid cells expressing the C-lectin receptor CD207 [
13]. However, taken together, data on the presence of T-cells, in particular CD8+ T-cells, and CD207+ DCs in NPC are scarce. Furthermore, it is unknown whether immune phenotypes exist in NPC and, if so, whether they influence survival.
In this study, biopsy material from patients with NPC, of which we have previously reported data on intralesional levels of EBV DNA and survival [
14], were revisited. In order to assess aspects of tumor heterogeneity, overall histology was examined focusing on cancer cells, cytokeratin, and Epstein–Barr virus-encoded small RNAs (EBERs). Based on the presence and distribution of lymphocytes (notably CD8+ cells), the material was classified into immune phenotypes. In addition, using a digital image technique, CD8+ and CD207+ cells were quantitatively assessed. The impact on survival of immune phenotypes as well as of CD8+ T-cells and CD207+ DCs was determined. The data were also analyzed in relation to intralesional EBV DNA. Finally, data on gene expression in the context of NPC and healthy control tissue were retrieved from a public database [
15,
16] and analyzed in silico focusing on CD8+ low and CD8+ high T-cell subgroups of NPC.
3. Discussion
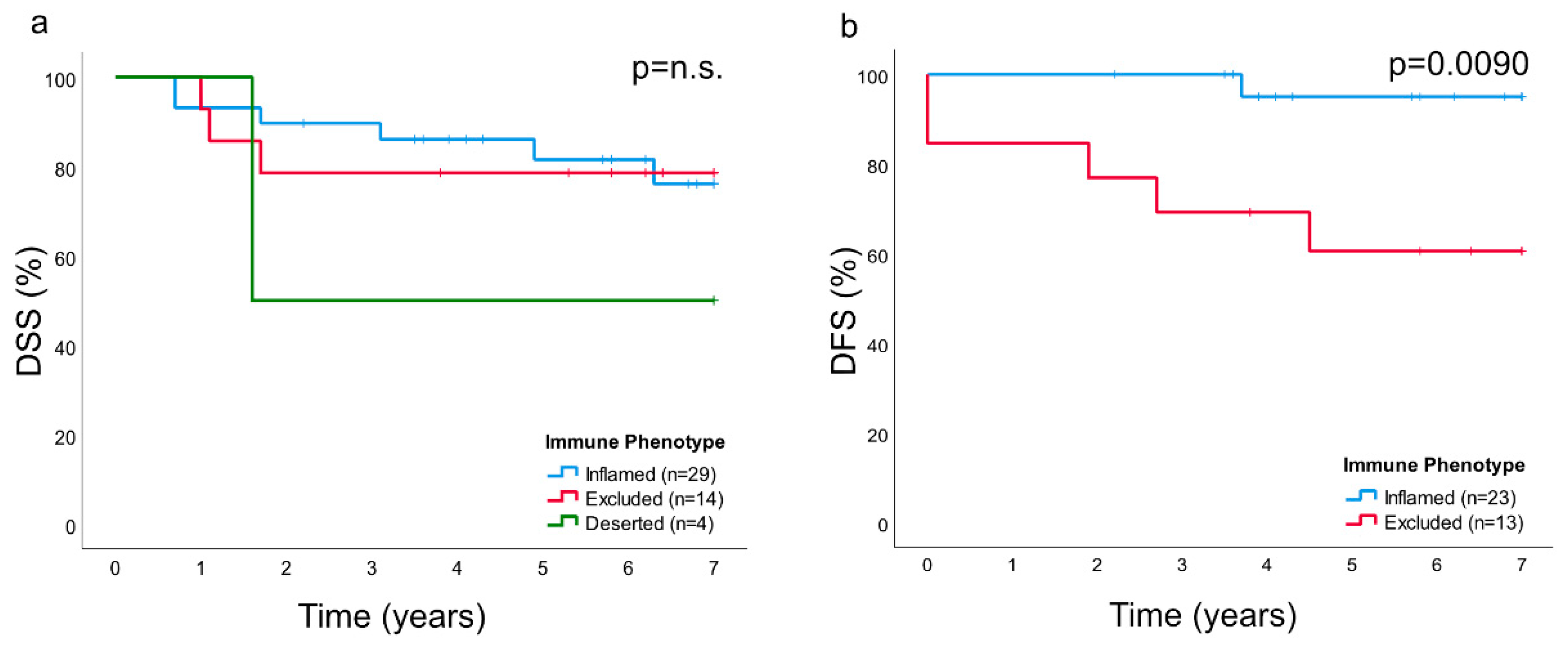
In this study, primary biopsies from patients with NPC were analyzed. A marked inter- and intraindividual variation was observed with regard to growth pattern of cancer cells as well as to presentation of cytokeratin, EBERs, and immune cells. Based on presence and distribution of lymphocytes as established for other cancers [
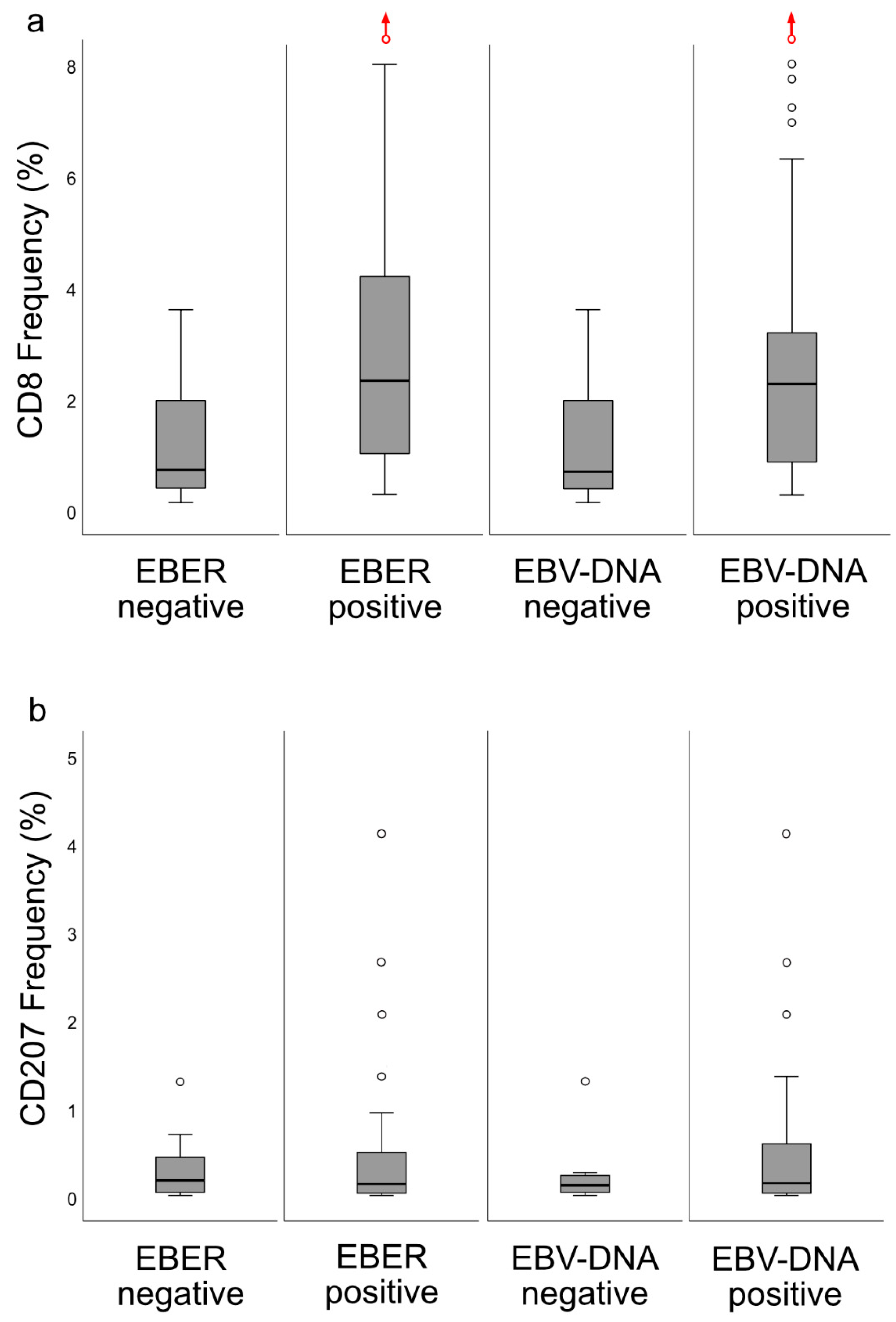
6], the lesions were classified into specific immune subsets, which carried prognostic information: DFS was better for the “inflamed” than for the “excluded” phenotype, while the “deserted” phenotype, arguably with the worst prognosis, was not eligible for analysis due to a majority of cases presenting with spread disease. CD8+ cells were present in areas of cancer cells and in the surrounding stroma, whereas CD207+ cells were observed largely in areas of cancer cells. The ratio of CD8+ cells were higher for EBV-positive
cf. EBV-negative NPC. In contrast, no such differences were observed for CD207. Gene expression analysis revealed differences between NPC and control tissue (e.g., with regard to interferon activity) as well as between subgroups of NPC based on CD8 expression (high vs. low). Taken together, the observations may be of relevance to prognostication of NPC as well as for explorations into the field of immunotherapy.
The notion that head and neck cancer lesions, including NPC, are heterogeneous in nature (e.g., Wang et al. [
3]), was confirmed by this study. With regard to patterns of cancer cell growth, this was evident for inter- as well as intraindividual comparisons. Inferentially, a similar heterogeneity was observed for cytokeratin (which in this context may be viewed as a cancer cell marker), which is typically expressed by NPC cells [
24]. These observations, in combination with occasional findings of diffuse EBER patterns, indirectly suggest the possibility that intralesional cancer-associated antigen-levels, as previously suggested through analysis of EBV-DNA [
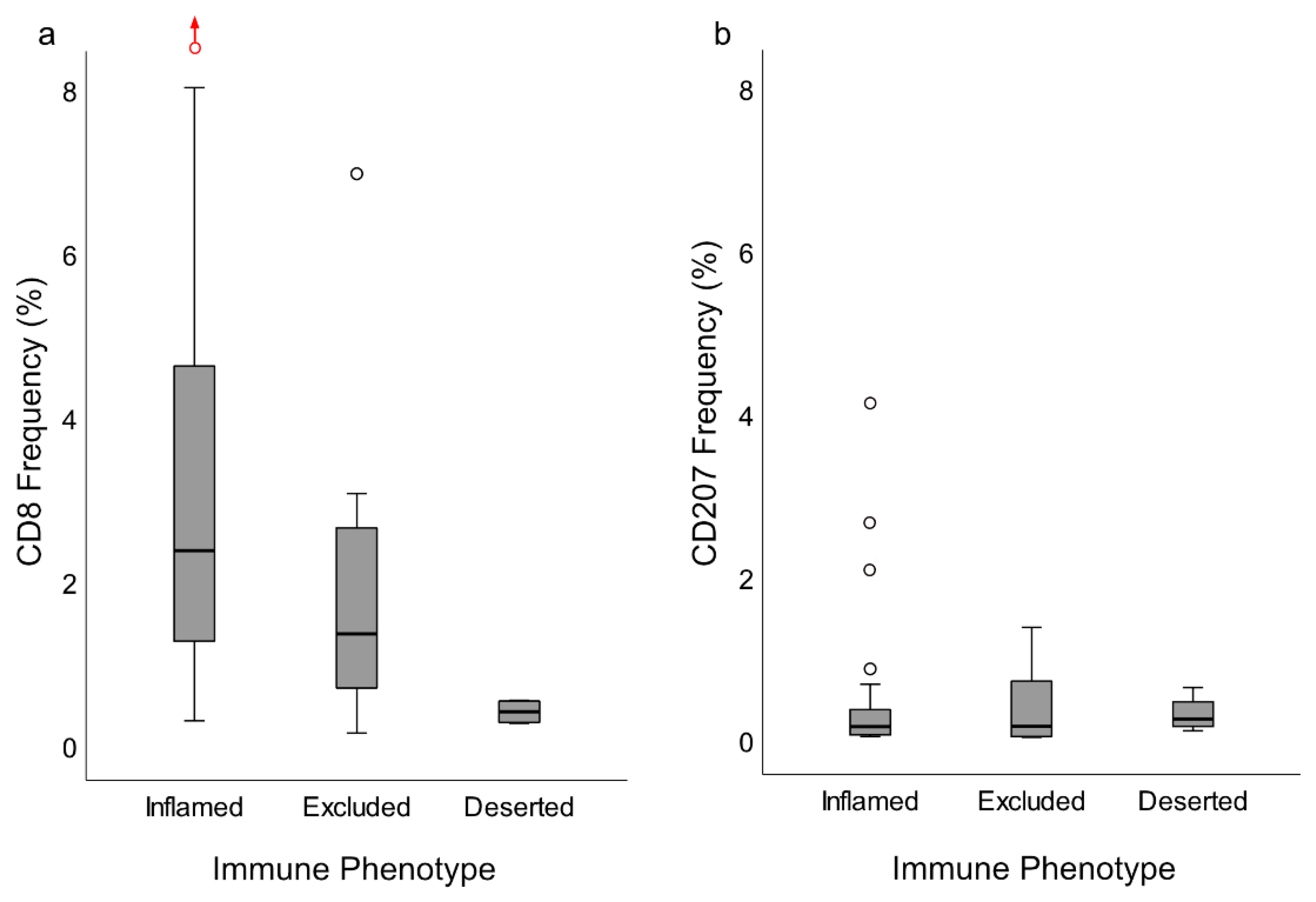
14], and immune responses may also vary considerably. In this study, such a marked variability was observed for the presence and distribution of CD8+ T-cells and CD207+ DCs. Taken together, our observations suggest that NPC heterogeneity, including immune cell aspects, must be taken into account when, e.g., prognostic information is explored and candidate treatment targets are selected for this condition. Arguably, even individual information may be of importance to future immunological treatment possibilities. The use of digital image “microdissection” and quantitative digital analysis in this study represents efforts in that direction. However, a limitation of the technique is that it does not allow for quantitation of CD8+ cells in cancer nodules due to difficulties in discriminating between areas of cancer cells infiltrated by lymphocytes and the lymphocyte-rich surrounding stroma.
In this study, as previously suggested for other cancers [
6,
25], the presence and distribution of lymphocytes (notably CD8+ T-cells) was utilized to classify NPC into specific immune phenotypes. The majority of NPC lesions were of an immune “inflamed” nature (61.7%), with lymphocytes infiltrating areas of cancer cells, while the minority was either immune “excluded” (29.8%) or “deserted” (8.5%), a classification that was verified by quantitative analysis of CD8+ T-cells. Of the immune phenotypes, the “deserted” subset was associated with the poorest prognosis. A high frequency of spread disease at diagnosis excluded this subset from an analysis of DFS. However, a clear difference in DFS was observed between the “inflamed” and the “excluded” phenotypes (in favor of the former). DSS showed a similar pattern, but the differences failed to reach statistical significance. We suggest that immune phenotype information should be evaluated further as a prognostic marker for NPC and in the context of treatment selection, for example, whether to combine first-line treatment (i.e., radiotherapy) with chemotherapy for the “deserted phenotype” even for low-stage disease or to offer checkpoint inhibitor immunotherapy to patients with NPC manifesting the “inflamed” phenotype, since observations in other types of cancers suggest that this phenotype is associated with a positive treatment response to such interventions [
26,
27]. Our observations extend previous work in the field: for example, the impact of TILs in NPC and their potential as prognostic markers [
3]. However, a limitation of our study is its sample size, which does not allow for a comprehensive analysis of survival.
The DC is specialized in antigen presentation and for a successful adaptive immune response to occur; either by the immune system through its own capacity or facilitated by “vaccinations”, this cell type needs to be present and active. Through this study, as CD207 is considered a selective DC marker [
28,
29], our previous observation that NPC lesions feature CD207+ DCs was confirmed (likely reflecting CD1c+ myeloid cells) [
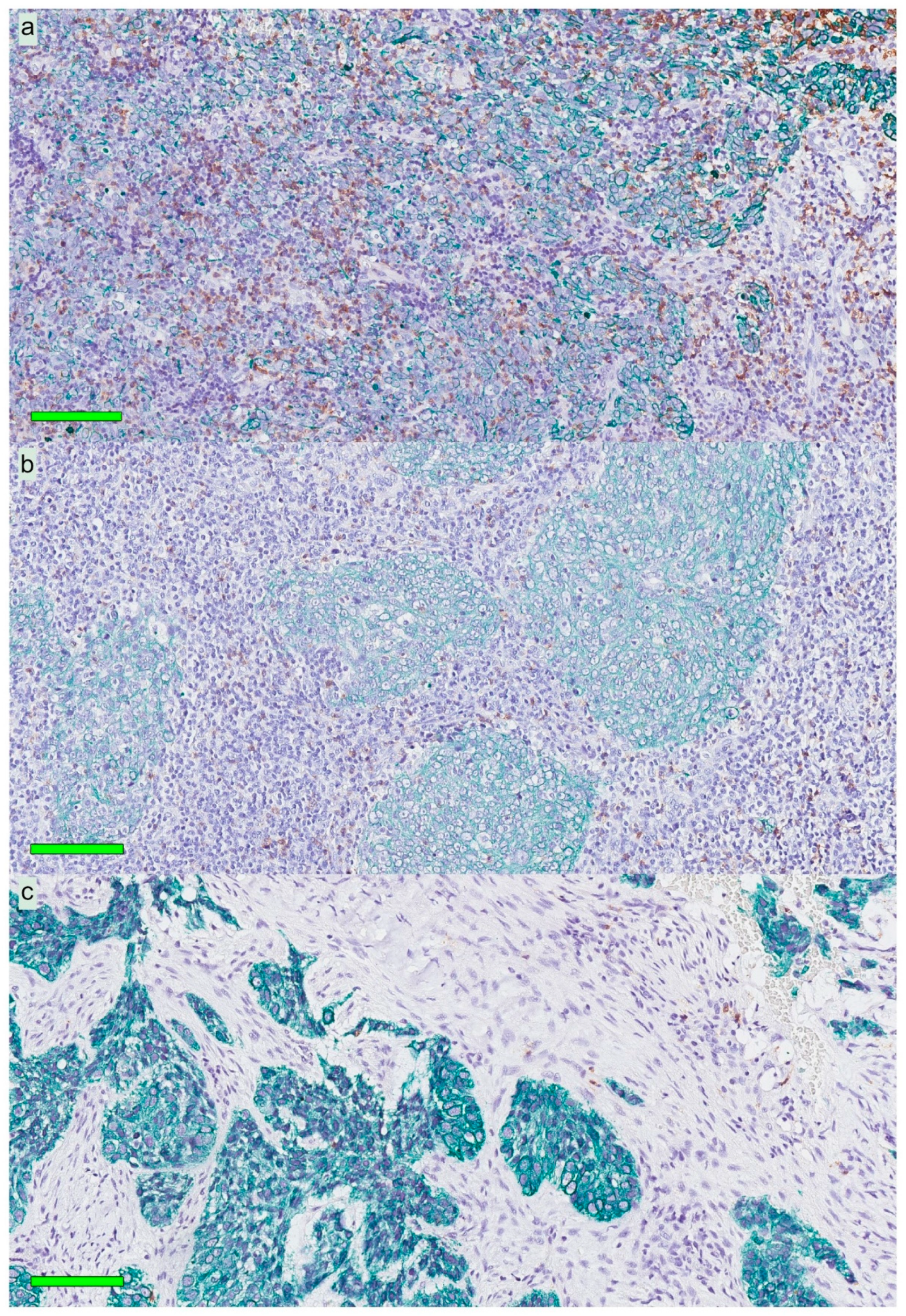
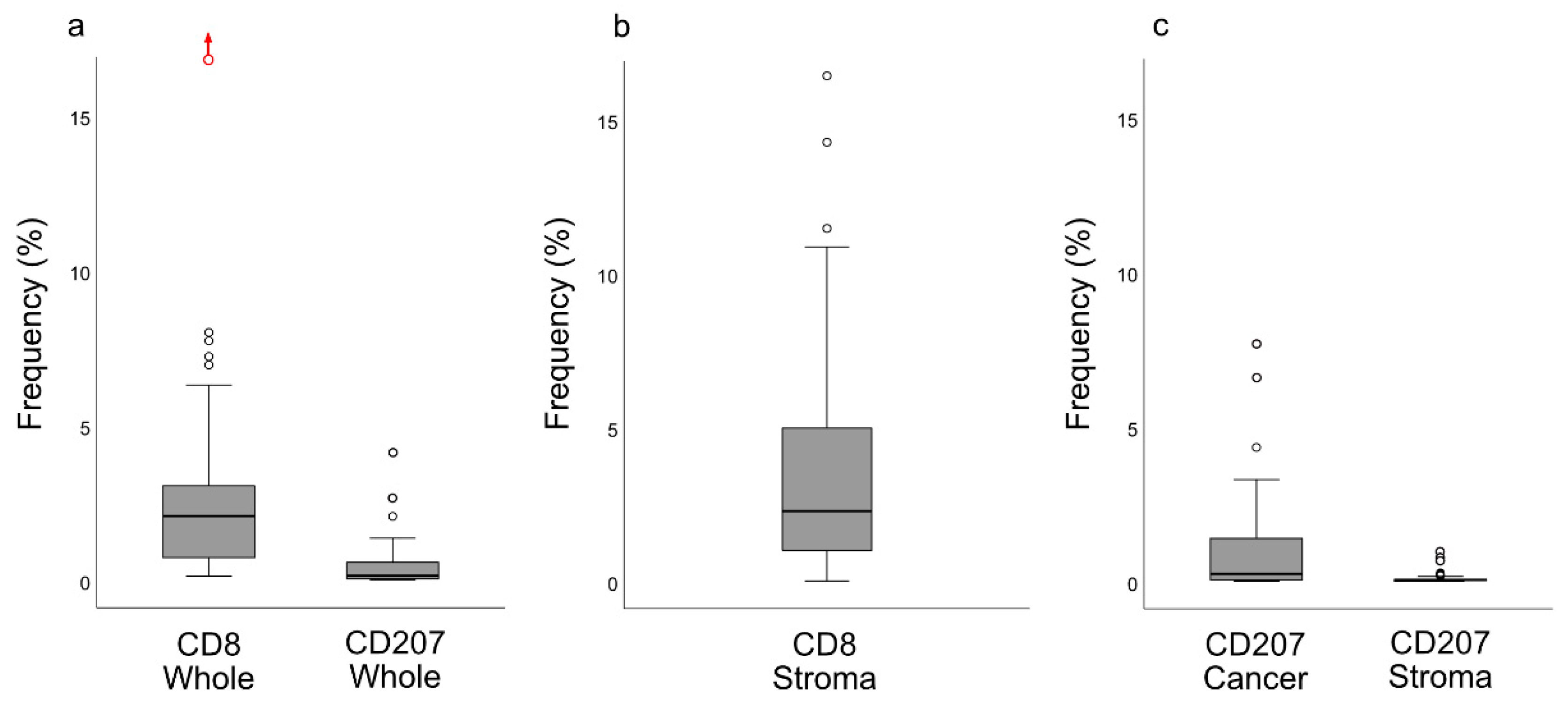
13]. Importantly, our present observations added the information that these cells were largely distributed within the areas of cancer cells, i.e., the frequency of CD207+ cells was 8-fold greater in this compartment
cf. the surrounding stroma. However, while CD207+ DCs were present in close relation to cancer cells, this was apparently not enough to induce a meaningful immunological response targeting cancer antigens, which would prevent NPC from occurring or to be killed off. We suggest that the DC presence in NPC represents treatment possibilities and that the C-lectin receptor CD207 and potentially other pattern recognition receptors known to facilitate cross-presentation of antigen [
4,
5] may be adjuvant targets. However, additional DC subsets must also be considered, and any target and its function must be considered in relation to the overall milieu of the NPC lesion, including aspects that exert immunosuppressive actions that may prevent antigen presentation. Furthermore, whether CD207 has a role in EBV-specific T-cell responses remains to be shown.
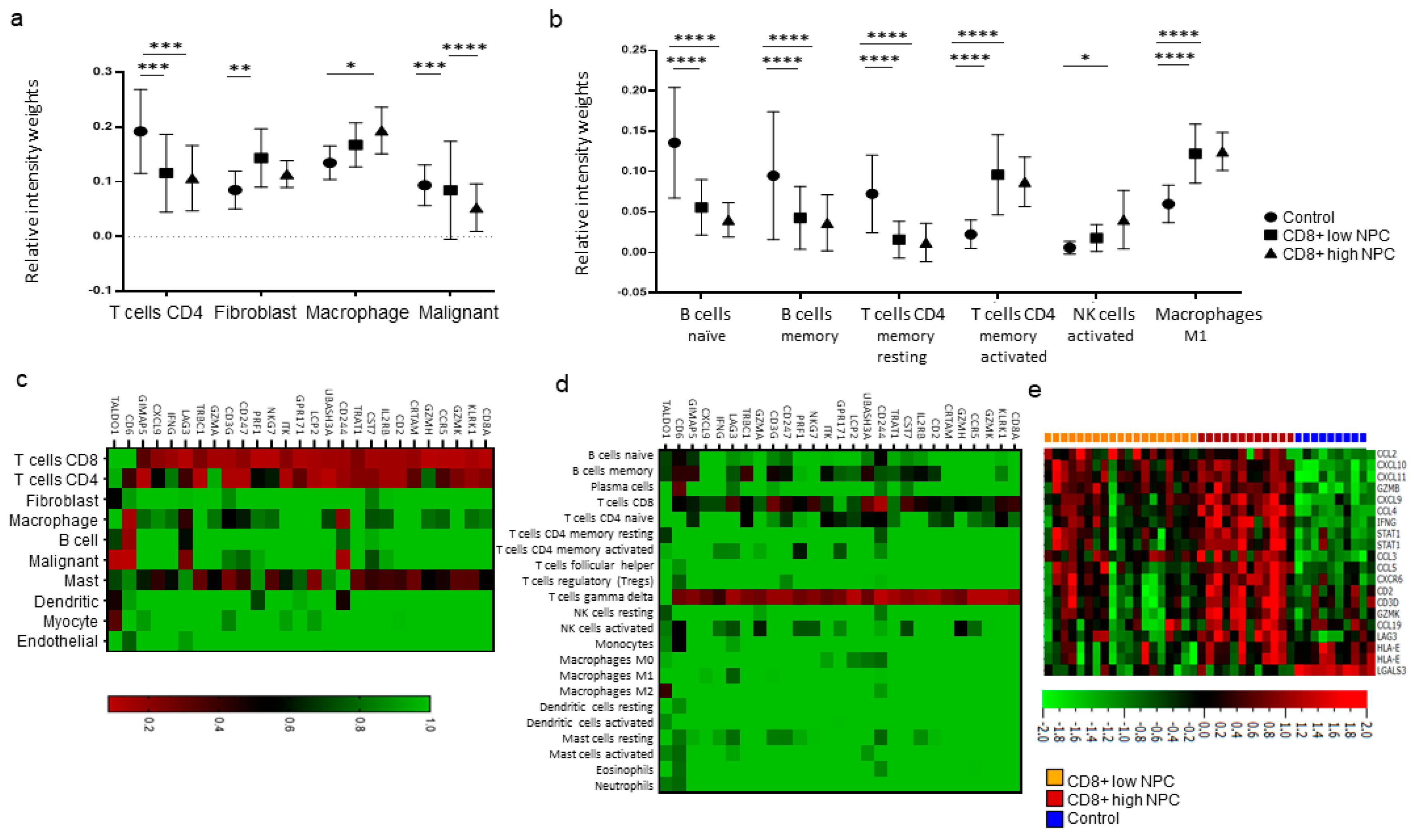
The interplay between the immune system and cancer cells in NPC is not sufficient to eradicate the disease despite the fact that antigens are present and that key immune cells such as DCs and T-cells are available. More information is needed, and the present gene expression analysis, performed on available transcriptional data from EBV-positive NPC and control tissue [
17,
18], highlighted some immune features associated with NPC. High expressions of signatures related to interferon activity, M1 macrophages, and CD4+ memory activated T-cells were observed for NPC (
cf. controls), albeit with marked heterogeneity between NPC samples. When comparing high vs. low CD8 vs. controls, high fibroblasts scores for CD8 low NPC were revealed. Furthermore, a high activated Natural Killer (NK) cell profile for CD8 high NPC was observed. (DCs could not be determined for NPC
cf. control tissue.) Taken together, our observations suggest that there are immunological subsets of NPC and distinctions between NPC rich in CD8 (likely representing an “inflamed” phenotype) and low in CD8 (“excluded” and particularly “deserted” phenotypes). However, the gene expression data are difficult to put into a greater context given the lack of synchronous morphological assessment and clinical data, reflecting a general problem with gene expression data extracted from public databases.
In a previous study, we explored the association between intralesional EBV DNA and survival [
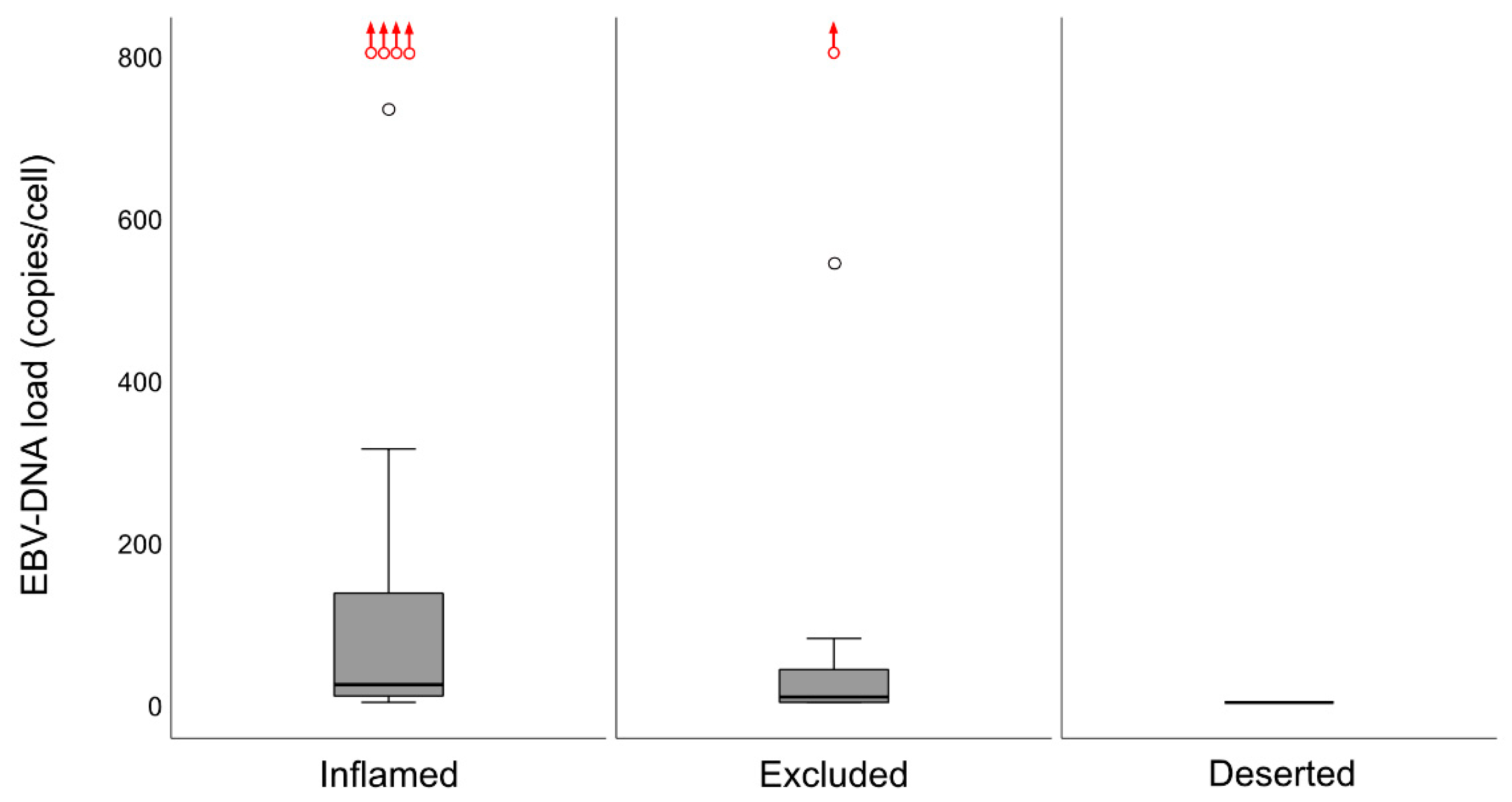
14]. When the material was split at a level of 70 copies of EBV DNA per cell, higher levels predicted a greater DFS. In agreement, in this study, when intralesional EBV DNA load was assessed in relation to immune phenotypes, high loads were associated with the “inflamed” phenotype and low loads with the “deserted” phenotype. For reasons yet to be defined, the associations to immune phenotypes appeared stronger when focusing on EBER than EBV DNA. Similarly, the association between CD8 ratios and “EBV status” was stronger for EBER than EBV DNA. Taken together, our observations confirm that overall immune features of NPC, notably the presence of CD8+ T-cells, depend on the presence of EBV. We suggest that analyses of EBV should be included whenever immune features of NPC are examined, and that intralesional EBV DNA should be complemented by analysis of EBER.
4. Materials and Methods
4.1. Study Design and Patients
The study was of a retrospective design and involved an analysis of formalin-fixated paraffin-embedded (FFPE) primary tumor tissue from a well-defined population of 48 patients with NPC diagnosed between 2001 and 2015. Data from this material have been reported previously focusing on intralesional EBV DNA and survival [
14]. Through immunohistochemistry and based on lymphocyte presence and distribution, the tumors were classified into immune phenotypes. Furthermore, CD8+ and CD207+ cells were assessed using a quantitative digital image technique. The data were analyzed in relation to EBER, intralesional EBV DNA, clinical stage, and survival. Approval was granted by the Ethics Committee at Lund University (ref. no. 2014/117). In accordance with the approval, informed consent was not required, but the study was advertised in printed media with a possibility to opt out. Separately, gene expression data in NPC and control tissue were retrieved from a public database and analyzed in silico.
4.2. Clinical Characteristics
Patient characteristics have been previously reported [
14]. Briefly, out of the 48 available NPC patients, one was lost to follow-up, and the median follow-up of the remaining 47 patients was 6.4 years; 31% of the patients were diagnosed with T1 lesions, 85% were N-positive, and 19% featured distant disease at the time of diagnosis (UICC’s TNM classification system, 7th version). The 5- and 7-year overall survival (OS) was 75% and 65%, respectively. At histopathological examination, 75% of the tumors were EBER-positive non-keratinizing cancers. Eighty-three percent of the patients presented EBV DNA-positive lesions. Clinical data on the material retrieved for gene expression analysis in silico was restricted to diagnosis, EBER status (all were EBER positive), and stage (UICC’s TNM classification system, 6th version). In the latter material, comprising data on 31 NPC patients, 21 featured stage I and II disease, 10 stage III disease, and none stage IV disease.
4.3. Immunohistochemistry
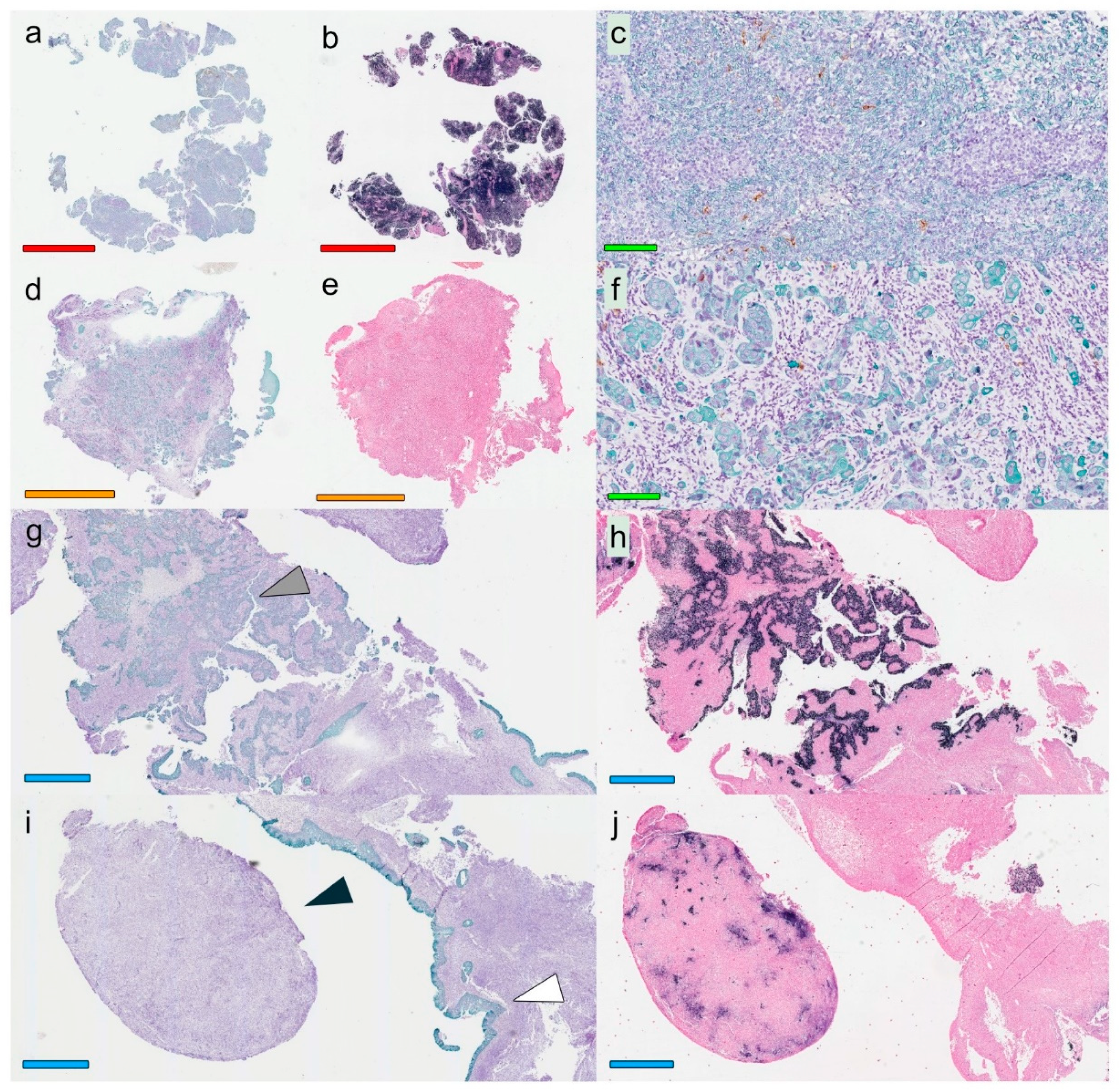
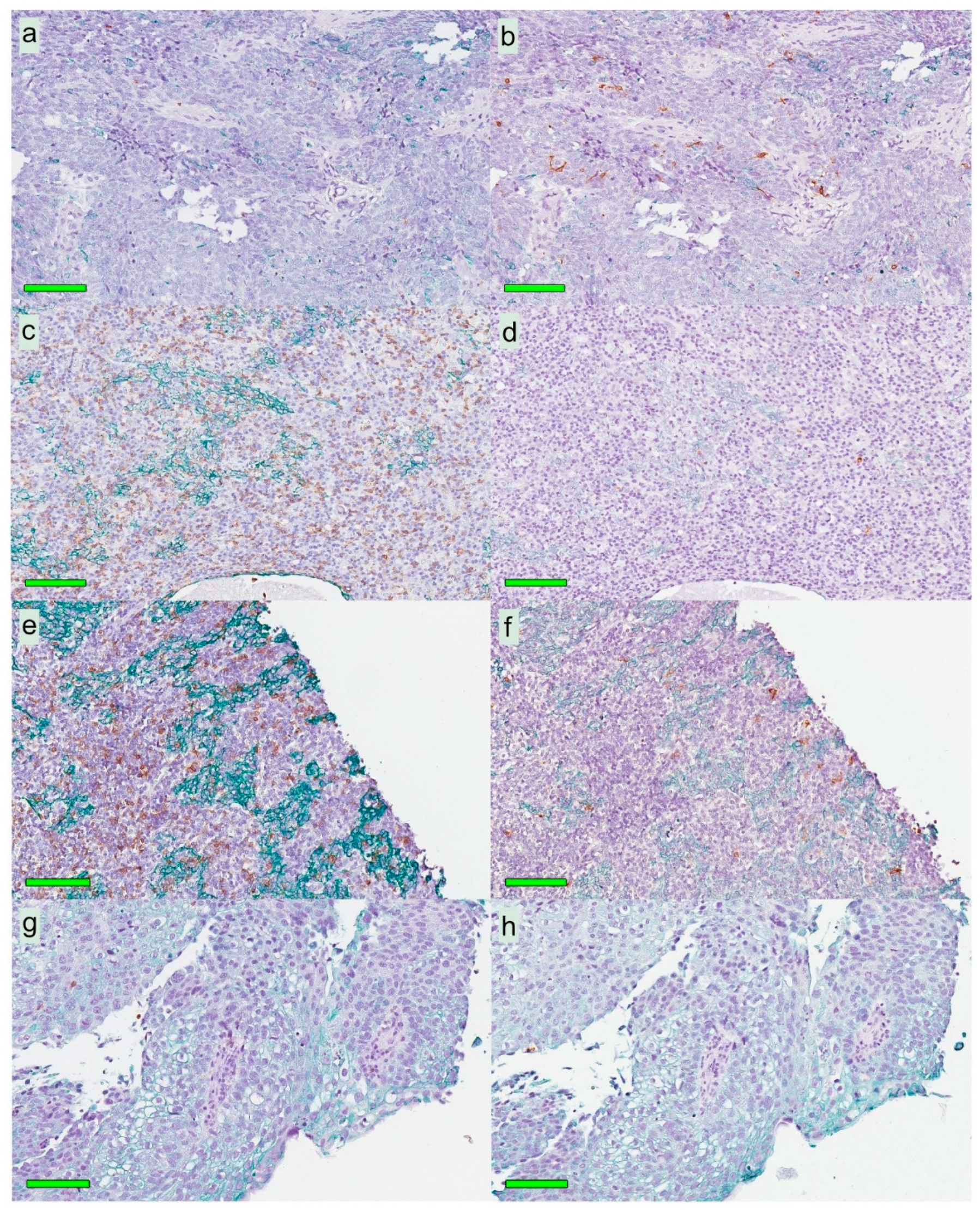
A modified double-staining immunohistochemistry protocol (EnVision Doublestain, Dako/Agilent, Glostrup, Denmark) was applied in order to simultaneously identify cytokeratin-positive cancer cells together with CD207+ DCs or CD8+ T cells. Briefly, 4-µm rehydrated sections were subjected to double-staining immunohistochemistry in the automated Autostainer IHC-robot (Dako/Agilent) using the EnVision Doublestain System kit K5361 (Dako/Agilent). Prior to immunohistochemistry, an antigen retrieval procedure was performed in a PT-link machine for heat-induced epitope retrieval (HIER) using low pH (pH 6) retrieval buffer. Endogenous peroxidase activity was blocked with H
2O
2. Sections were then incubated with an anti-CD207 (clone 12D6, Novocastra/Leica, Newcastle, UK; dilution 1:300) or anti-CD8 (clone C8/144B, Dako/Agilent, Glostrup, Denmark; dilution 1:400) primary antibody for 1 h at room temperature. After subsequent incubation with polymer/HRP-linked secondary antibodies for 30 min, the immunoreactivity was visualized using diaminobenzidine 3.3 (DAB) HRP chromogen (resulting in a brown opaque staining). Next, sections were incubated with Dako Double Stain Blocking Reagent (Dako/Agilent) to prevent additional binding of secondary antibodies to the first primary antibody. Sections were then incubated with an anti-cytokeratin primary antibody (clone CK AE1/AE3, Novocastra/Leica, dilution 1:300). This second-round immunoreactivity was also visualized with a polymer/HRP-linked secondary antibody (Dako/Agilent, dilution 1:300), but the visualization was performed with Vina Green HRP chromogen (Biocare Medical, cat BRR807A, Pacheco, CA, USA). Finally, sections were counterstained with Mayer’s hematoxylin, air-dried, and mounted with Pertex (Histolab Products, Gothenburg, Sweden). In addition to the protocols above, previously stained slides for EBER evaluation (EBER-ISH for EBER1 and EBER2) were retrieved [
14]. All slides were digitalized using the automated Scanscope XT digital slide scanner (Aperio Technologies, Vista, CA, USA). Evaluation and quantification were performed using the Aperio Imagescope Software (Aperio Technologies).
4.4. Immunoprofiling
Based on overall lymphocyte presence, and with special regards to CD8+ T-cell distribution (i.e., the assessment was performed on hematoxylin-slides stained for CD8), the lesions were classified according to immune phenotypes defined as “inflamed” (lymphocytes infiltrating cancer cell areas), “excluded” (lymphocytes in the surrounding stroma with no, or exceedingly few, infiltrating lymphocytes within cancer cell areas), and “deserted” (no or exceedingly few lymphocytes both in cancer cell areas and the surrounding stroma). In cases of highly heterogeneous lesions, the dominating pattern was chosen for the classification. This assessment was performed temporarily blinded to clinical data (J.S.N) and verified totally blinded by a second observer (S.S.): EBER slides were not used as support in the analysis.
4.5. Quantification of CD8 and CD207 Immunoreactivity
Slides stained for CD8 and CD207 were reviewed in a standardized manner utilizing cytokeratin (all slides were stained for cytokeratin) with EBER slides as support. Through digital image dissection, nonrelevant regions were excluded. These comprised physical artefacts (e.g., occasionally folded sections), normal epithelium, germinal centers, and gland structures. The dissections were performed on consecutive slides for CD8 and CD207, enabling a consequent and representative exclusion strategy. The Positive Pixel 9-algorithm (Aperio Technologies) was used to automatically segment out tissue background area as well as positive staining (i.e., brown DAB chromogen) by color threshold values, and then the ratio of the total analyzed tissue area with staining-positive pixels was calculated. Accordingly, this ratio represented the quantities of CD8 and CD207 in the target area. Thresholds to define staining (chromogen) positivity were the same for CD8 and CD207.
In addition, areas (minimum 0.1 mm2 after exclusion of nonrelevant regions as defined above) were selected to represent areas of cancer cells and areas of surrounding stroma (in direct proximity to cancer cells). CD207 positivity was quantitated for both locations. However, CD8 positivity was quantitated for the surrounding stroma only. The latter was due to a very high lymphocyte presence, which in combination with a variable degree of lymphocyte infiltration, rendered it impossible to quantitate CD8 within cancer cell areas in an objective and robust manner. When the presence of the marker (CD8 and DC207) was different between various areas of the tumor, the area with more pronounced presence was chosen. The selected areas were assessed for each marker following the same digital image quantification process as described above. All quantification procedures were performed blinded to the clinical data.
4.6. Gene Expression Data Analysis
Normalized mRNA data, generated with the Affymetrix Human Genome U133 Plus 2.0 Array, were obtained from the GEO database (
https://www.ncbi.nlm.nih.gov/geo/: accession number GSE12452) [
15,
16]. The analyses were performed using Qlucore Omics Explorer 3.6 (Qlucore, Lund, Sweden). Data were divided into two subsets: control tissue (
n = 10), comprising 4 cases of nasopharyngeal tissue adjacent to but separated from NPC lesions and 6 cases of nasopharyngeal tissue from non-NPC subjects, and NPC tissue (
n = 31, all EBER-positive).
Utilizing CIBERSORTX [
30], the gene expression data was immunoprofiled using signatures identified in two independent studies: one including the overall distribution of 10 cell populations using data from single-cell analysis of head and neck cancers [
18] and the other defining 22 general tumor-associated immune cell populations [
17]. The scoring from both studies for CD8+ T-cell signatures were used to divide the NPC samples into CD8+ high and low. Also, the material was analyzed for CD8A-correlated genes and expression levels based on an interferon signature (39 genes). Heat map clustering was performed to understand the relationship between CD8A-correlated genes and individual cell populations [
17,
18].
4.7. Statistics
For the principal data set subjected to digital imaging, statistical analyses were performed using SPSS version 25 (IBM, Armonk, NY). Data were presented as medians with interquartile ranges (IQR). For comparisons of CD8 and CD207 ratios, respectively, between immune phenotypes of NPC and between tumor stages, an analysis of variance (the Kruskal–Wallis test) was followed by the Mann Whitney U-test. For comparisons of EBV DNA load between immune phenotypes, the Kruskal–Wallis test was followed by the Mann Whitney U-test. χ2-tests were performed to explore associations between CD8 and CD207 (grouped by median values) and EBER (present or not), EBV DNA (present or not), immune phenotypes, and stage. Correlations between CD8 and CD207 ratios and between these ratios and EBV-DNA were explored using the Spearman test. Comparisons of ratios for CD207 between areas of cancer cells and surrounding stroma was explored using the Wilcoxon Signed Ranks test. Survival, i.e., DSS and DFS in relation to CD8 and CD207, respectively, as well as to immune phenotypes, was described using Kaplan–Meyer curves, and significance levels were determined by the log-rank test. A Cox regression analysis was not performed due to the restricted sample size. The explicit p-value was provided when the p-value was <0.05 but >0.0001, whereas lower values were provided as <0.0001.
CD8A-correlated gene markers and the interferon signatures were examined using the limma multigroup analysis (edgeR) [
31]. A Pearson correlation of r > 0.7 and adjusted
p-values less than 0.01 were considered statistically significant. For gene expression data analyzed in silico, statistical analyses were performed using GraphPad version 8.4.3 (GraphPad Software, La Jolla, CA, USA). The intensity fractions obtained were analyzed for differences in the immune cell populations using the two-way ANOVA, Tukey method.