Cancer Alters the Metabolic Fingerprint of Extracellular Vesicles
Simple Summary
Abstract
1. Introduction
2. Results
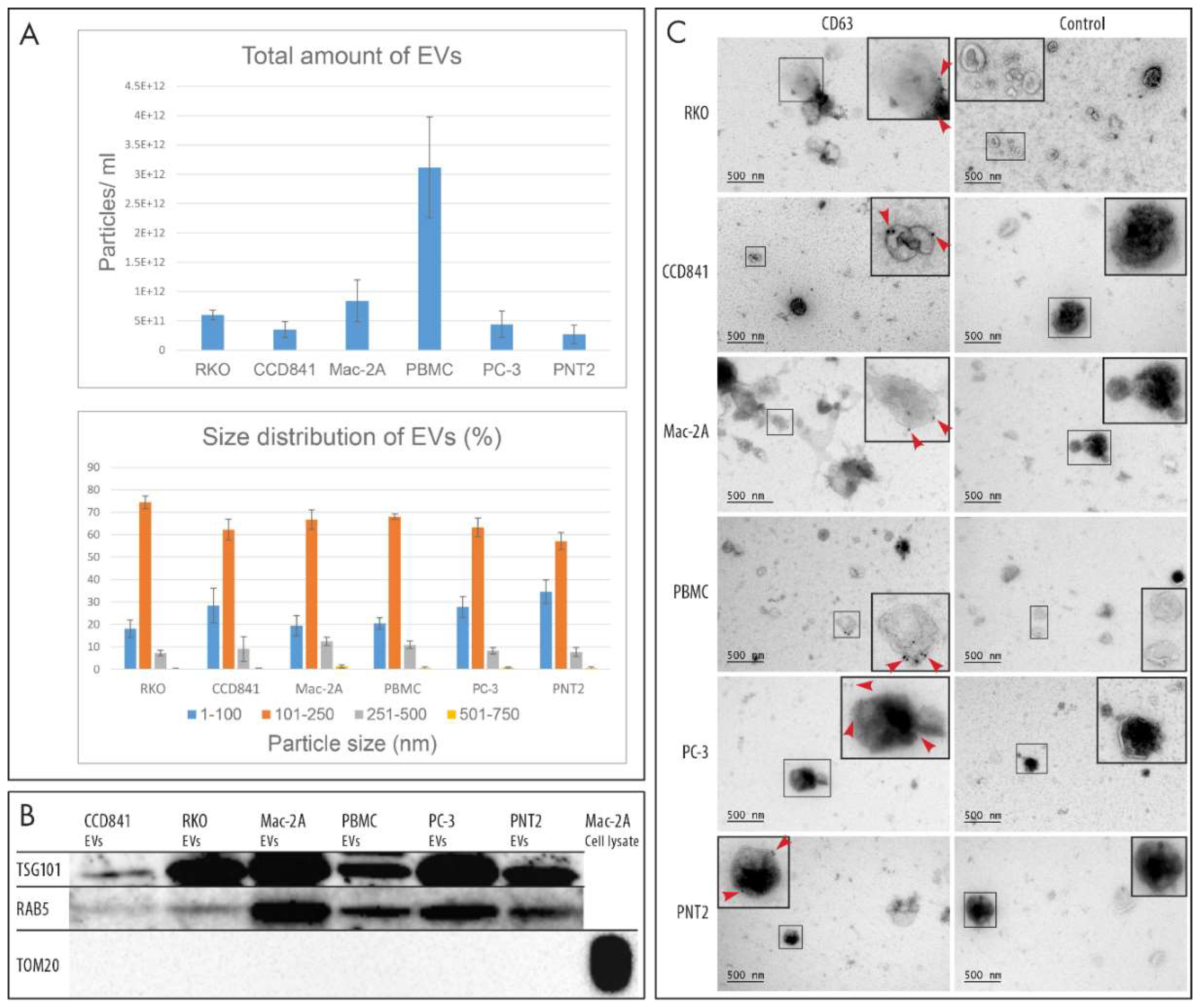
2.1. Characterization of EVs
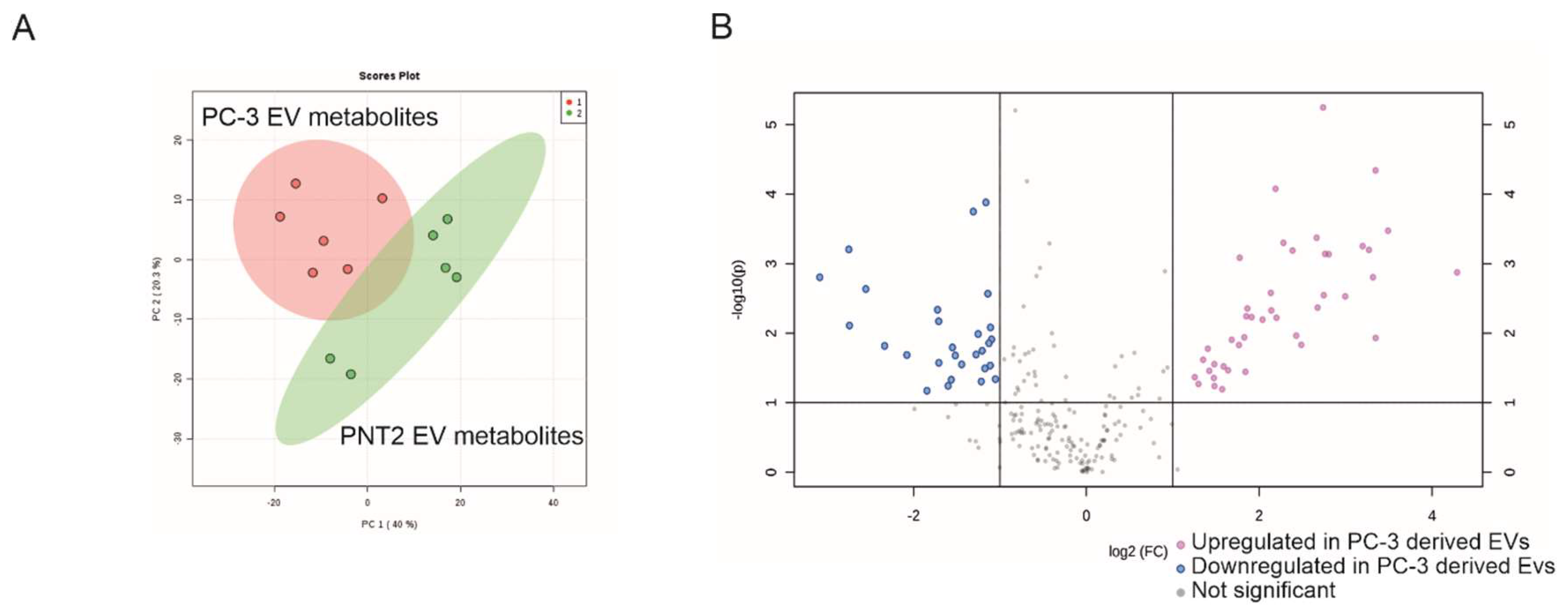
2.2. Non-Targeted Metabolite Profile Analysis of EVs from Prostate Cancer PC-3 and Normal Prostate Epithelial PNT2 Cell Lines
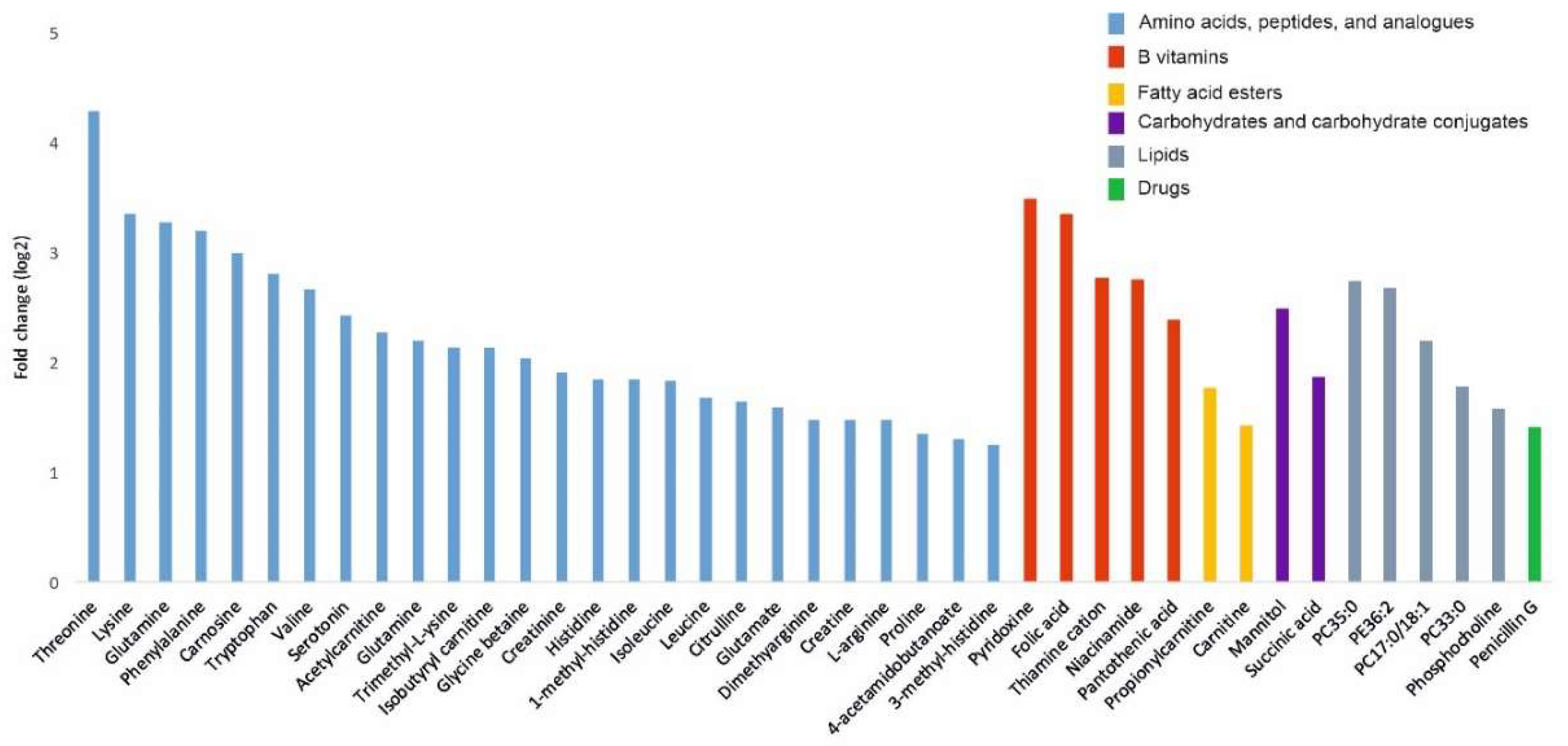
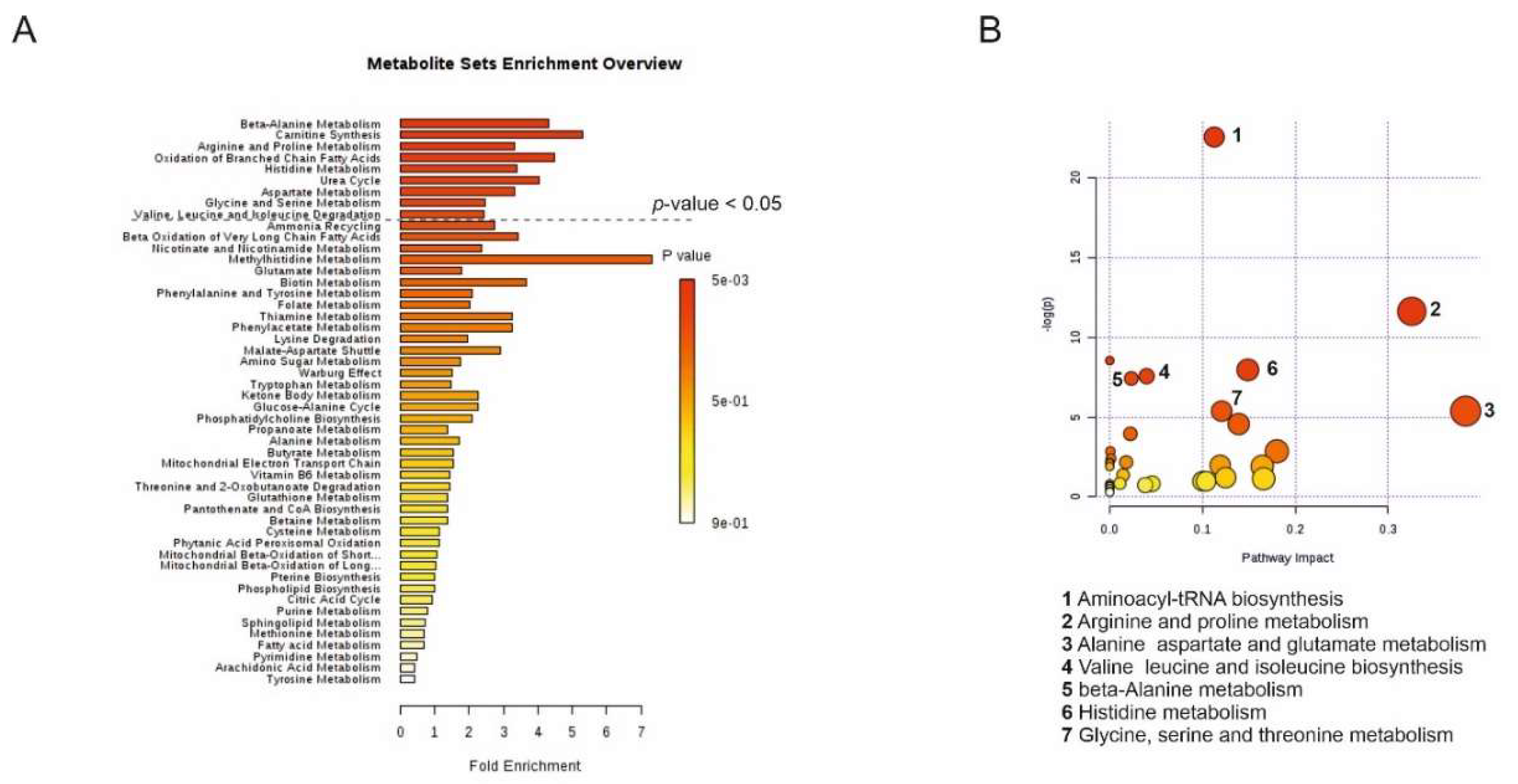
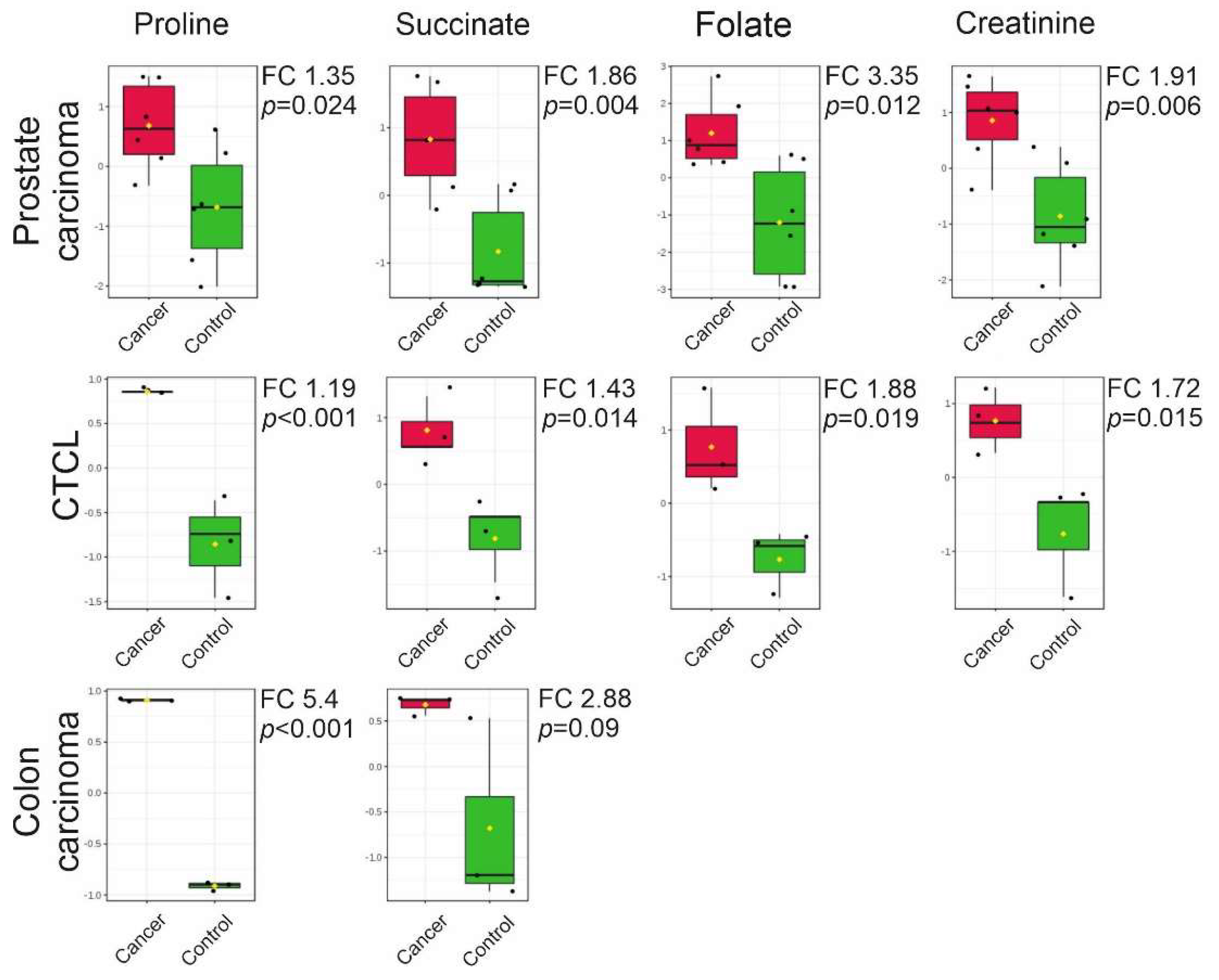
2.3. Targeted LC-MS Metabolite Profile Analysis of Cancer EVs
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. EV Isolation Using Differential Ultracentrifugation
4.3. Nanoparticle Tracking Analysis
4.4. Western Blotting
4.5. Immunoelectron Microscopy
4.6. Non-Targeted Liquid Chromatography-Mass Spectrometry Metabolite Analysis
4.7. Targeted Liquid Chromatography-Mass Spectrometry Metabolite Analysis
4.8. Statistical Analysis
4.9. Metabolic Pathway Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Cairns, R.A.; Harris, I.S.; Mak, T.W. Regulation of cancer cell metabolism. Nat. Rev. Cancer 2011, 11, 85–95. [Google Scholar] [CrossRef]
- DeBerardinis, R.J.; Chandel, N.S. Fundamentals of cancer metabolism. Sci. Adv. 2016, 2, e1600200. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Vander Heiden, M.G.; Cantley, L.C.; Thompson, C.B. Understanding the Warburg effect: The metabolic requirements of cell proliferation. Science 2009, 324, 1029–1033. [Google Scholar] [CrossRef] [PubMed]
- Yong, C.; Stewart, G.; Frezza, C. Oncometabolites in renal cancer: Warburg’s hypothesis re-examined. Nat. Rev. Nephrol. 2020, 16, 156–172. [Google Scholar] [CrossRef] [PubMed]
- Sciacovelli, M.; Frezza, C. Oncometabolites: Unconventional triggers of oncogenic signalling cascades. Free Radic. Biol. Med. 2016, 100, 175–181. [Google Scholar] [CrossRef] [PubMed]
- Yáñez-Mó, M.; Siljander, P.R.-M.; Andreu, Z.; Bedina Zavec, A.; Borràs, F.E.; Buzas, E.I.; Buzas, K.; Casal, E.; Cappello, F.; Carvalho, J. Biological properties of extracellular vesicles and their physiological functions. J. Extracell. Vesicles 2015, 4, 27066. [Google Scholar] [CrossRef] [PubMed]
- Théry, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef] [PubMed]
- Palviainen, M.; Saraswat, M.; Varga, Z.; Kitka, D.; Neuvonen, M.; Puhka, M.; Joenväärä, S.; Renkonen, R.; Nieuwland, R.; Takatalo, M. Extracellular vesicles from human plasma and serum are carriers of extravesicular cargo—Implications for biomarker discovery. PLoS ONE 2020, 15, e0236439. [Google Scholar] [CrossRef]
- Maia, J.; Caja, S.; Strano Moraes, M.C.; Couto, N.; Costa-Silva, B. Exosome-based cell-cell communication in the tumor microenvironment. Front. Cell Dev. Biol. 2018, 6, 18. [Google Scholar] [CrossRef]
- Webber, J.; Yeung, V.; Clayton, A. Extracellular vesicles as modulators of the cancer microenvironment. In Seminars in Cell & Developmental Biology; Academic Press: Cambridge, MA, USA, 2015; pp. 27–34. [Google Scholar]
- Becker, A.; Thakur, B.K.; Weiss, J.M.; Kim, H.S.; Peinado, H.; Lyden, D. Extracellular vesicles in cancer: Cell-to-cell mediators of metastasis. Cancer Cell 2016, 30, 836–848. [Google Scholar] [CrossRef] [PubMed]
- Hoshino, A.; Kim, H.S.; Bojmar, L.; Gyan, K.E.; Cioffi, M.; Hernandez, J.; Zambirinis, C.P.; Rodrigues, G.; Molina, H.; Heissel, S. Extracellular vesicle and particle biomarkers define multiple human cancers. Cell 2020, 182, 1044–1061. [Google Scholar] [CrossRef]
- Williams, C.; Palviainen, M.; Reichardt, N.-C.; Siljander, P.R.-M.; Falcón-Pérez, J.M. Metabolomics applied to the study of extracellular vesicles. Metabolites 2019, 9, 276. [Google Scholar] [CrossRef]
- Altadill, T.; Campoy, I.; Lanau, L.; Gill, K.; Rigau, M.; Gil-Moreno, A.; Reventos, J.; Byers, S.; Colas, E.; Cheema, A.K. Enabling metabolomics based biomarker discovery studies using molecular phenotyping of exosome-like vesicles. PLoS ONE 2016, 11, e0151339. [Google Scholar] [CrossRef] [PubMed]
- Puhka, M.; Takatalo, M.; Nordberg, M.-E.; Valkonen, S.; Nandania, J.; Aatonen, M.; Yliperttula, M.; Laitinen, S.; Velagapudi, V.; Mirtti, T. Metabolomic profiling of extracellular vesicles and alternative normalization methods reveal enriched metabolites and strategies to study prostate cancer-related changes. Theranostics 2017, 7, 3824. [Google Scholar] [CrossRef]
- Clos-Garcia, M.; Loizaga-Iriarte, A.; Zuñiga-Garcia, P.; Sánchez-Mosquera, P.; Rosa Cortazar, A.; González, E.; Torrano, V.; Alonso, C.; Pérez-Cormenzana, M.; Ugalde-Olano, A. Metabolic alterations in urine extracellular vesicles are associated to prostate cancer pathogenesis and progression. J. Extracell. Vesicles 2018, 7, 1470442. [Google Scholar] [CrossRef]
- Zhang, J.; Lu, S.; Zhou, Y.; Meng, K.; Chen, Z.; Cui, Y.; Shi, Y.; Wang, T.; He, Q.Y. Motile hepatocellular carcinoma cells preferentially secret sugar metabolism regulatory proteins via exosomes. Proteomics 2017, 17, 1700103. [Google Scholar] [CrossRef]
- Wang, X.I.; Kruithof-de Julio, M.; Economides, K.D.; Walker, D.; Yu, H.; Halili, M.V.; Hu, Y.P.; Price, S.M.; Abate-Shen, C.; Shen, M.M. A luminal epithelial stem cell that is a cell of origin for prostate cancer. Nature 2009, 461, 495–500. [Google Scholar] [CrossRef] [PubMed]
- Peterson, E.; Weed, J.; Sicco, K.L.; Latkowski, J.-A. Cutaneous T Cell Lymphoma: A Difficult Diagnosis Demystified. Dermatol. Clin. 2019, 37, 455–469. [Google Scholar] [CrossRef]
- Fleming, M.; Ravula, S.; Tatishchev, S.F.; Wang, H.L. Colorectal carcinoma: Pathologic aspects. J. Gastrointest. Oncol. 2012, 3, 153. [Google Scholar]
- Tretter, L.; Patocs, A.; Chinopoulos, C. Succinate, an intermediate in metabolism, signal transduction, ROS, hypoxia, and tumorigenesis. Biochim. Biophys. Acta (BBA)-Bioenerg. 2016, 1857, 1086–1101. [Google Scholar] [CrossRef]
- Tanner, J.J.; Fendt, S.-M.; Becker, D.F. The proline cycle as a potential cancer therapy target. Biochemistry 2018, 57, 3433–3444. [Google Scholar] [CrossRef]
- Krupenko, S.A.; Krupenko, N.I. Loss of ALDH1L1 folate enzyme confers a selective metabolic advantage for tumor progression. Chemico-Biol. Interact. 2019, 302, 149–155. [Google Scholar] [CrossRef]
- Kazak, L.; Cohen, P. Creatine metabolism: Energy homeostasis, immunity and cancer biology. Nat. Rev. Endocrinol. 2020, 16, 421–436. [Google Scholar] [CrossRef]
- Lázaro-Ibáñez, E.; Sanz-Garcia, A.; Visakorpi, T.; Escobedo-Lucea, C.; Siljander, P.; Ayuso-Sacido, Á.; Yliperttula, M. Different gDNA content in the subpopulations of prostate cancer extracellular vesicles: Apoptotic bodies, microvesicles, and exosomes. Prostate 2014, 74, 1379–1390. [Google Scholar] [CrossRef]
- Lázaro-Ibáñez, E.; Lunavat, T.R.; Jang, S.C.; Escobedo-Lucea, C.; Oliver-De La Cruz, J.; Siljander, P.; Lötvall, J.; Yliperttula, M. Distinct prostate cancer-related mRNA cargo in extracellular vesicle subsets from prostate cell lines. BMC Cancer 2017, 17, 1–11. [Google Scholar] [CrossRef]
- Palviainen, M.; Saari, H.; Kärkkäinen, O.; Pekkinen, J.; Auriola, S.; Yliperttula, M.; Puhka, M.; Hanhineva, K.; Siljander, P.R.-M. Metabolic signature of extracellular vesicles depends on the cell culture conditions. J. Extracell. Vesicles 2019, 8, 1596669. [Google Scholar] [CrossRef]
- Lane, R.E.; Korbie, D.; Trau, M.; Hill, M.M. Purification protocols for extracellular vesicles. In Extracellular Vesicles; Springer: Berlin/Heidelberg, Germany, 2017; pp. 111–130. [Google Scholar]
- Cvjetkovic, A.; Lötvall, J.; Lässer, C. The influence of rotor type and centrifugation time on the yield and purity of extracellular vesicles. J. Extracell. Vesicles 2014, 3, 23111. [Google Scholar] [CrossRef]
- Fouad, Y.A.; Aanei, C. Revisiting the hallmarks of cancer. Am. J. Cancer Res. 2017, 7, 1016. [Google Scholar]
- Frezza, C. Metabolism and Cancer: The Future is Now. Br. J. Cancer 2020, 122, 133–135. [Google Scholar] [CrossRef] [PubMed]
- Dias, A.S.; Almeida, C.R.; Helguero, L.A.; Duarte, I.F. Metabolic crosstalk in the breast cancer microenvironment. Eur. J. Cancer 2019, 121, 154–171. [Google Scholar] [CrossRef]
- Lima, A.R.; Pinto, J.; Barros-Silva, D.; Jerónimo, C.; Henrique, R.; Bastos, M.; Carvalho, M. New findings on urinary prostate cancer metabolome through combined GC–MS and 1H NMR analytical platforms. Metabolomics 2020, 16, 70. [Google Scholar] [CrossRef]
- Li, H.; Feng, Z.; He, M.-L. Lipid metabolism alteration contributes to and maintains the properties of cancer stem cells. Theranostics 2020, 10, 7053. [Google Scholar] [CrossRef] [PubMed]
- Jasbi, P.; Wang, D.; Cheng, S.L.; Fei, Q.; Cui, J.Y.; Liu, L.; Wei, Y.; Raftery, D.; Gu, H. Breast cancer detection using targeted plasma metabolomics. J. Chromatogr. B 2019, 1105, 26–37. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.-Y.; Huang, T.-W.; Hsieh, Y.-T.; Wang, Y.-F.; Yen, C.-C.; Lee, G.-L.; Yeh, C.-C.; Peng, Y.-J.; Kuo, Y.-Y.; Wen, H.-T. Cancer-derived succinate promotes macrophage polarization and cancer metastasis via succinate receptor. Mol. Cell 2020, 77, 213–227.e5. [Google Scholar] [CrossRef]
- Asante, I.; Chui, D.; Pei, H.; Zhou, E.; De Giovanni, C.; Conti, D.; Louie, S. Alterations in folate-dependent one-carbon metabolism as colon cell transition from normal to cancerous. J. Nutr. Biochem. 2019, 69, 1–9. [Google Scholar] [CrossRef]
- Park, J.; Shin, Y.; Kim, T.H.; Kim, D.-H.; Lee, A. Urinary Metabolites as Biomarkers for Diagnosis of Breast Cancer: A Preliminary Study. J. Breast Dis. 2019, 7, 44–51. [Google Scholar] [CrossRef]
- Kumar, N.; Shahjaman, M.; Mollah, M.N.H.; Islam, S.S.; Hoque, M.A. Serum and plasma metabolomic biomarkers for lung cancer. Bioinformation 2017, 13, 202. [Google Scholar] [CrossRef]
- Kim, D.J.; Cho, E.J.; Yu, K.-S.; Jang, I.-J.; Yoon, J.-H.; Park, T.; Cho, J.-Y. Comprehensive metabolomic search for biomarkers to differentiate early stage hepatocellular carcinoma from cirrhosis. Cancers 2019, 11, 1497. [Google Scholar] [CrossRef]
- Vsiansky, V.; Svobodova, M.; Gumulec, J.; Cernei, N.; Sterbova, D.; Zitka, O.; Kostrica, R.; Smilek, P.; Plzak, J.; Betka, J. Prognostic significance of serum free amino acids in head and neck cancers. Cells 2019, 8, 428. [Google Scholar] [CrossRef]
- Plewa, S.; Horała, A.; Dereziński, P.; Nowak-Markwitz, E.; Matysiak, J.; Kokot, Z.J. Wide spectrum targeted metabolomics identifies potential ovarian cancer biomarkers. Life Sci. 2019, 222, 235–244. [Google Scholar] [CrossRef] [PubMed]
- Phang, J.M. Proline metabolism in cell regulation and cancer biology: Recent advances and hypotheses. Antioxid. Redox Signal. 2019, 30, 635–649. [Google Scholar] [CrossRef]
- Phang, J.M.; Liu, W.; Zabirnyk, O. Proline metabolism and microenvironmental stress. Annu. Rev. Nutr. 2010, 30, 441–463. [Google Scholar] [CrossRef]
- D’aniello, C.; Patriarca, E.J.; Phang, J.M.; Minchiotti, G. Proline Metabolism in Tumor Growth and Metastatic Progression. Front. Oncol. 2020, 10, 776. [Google Scholar] [CrossRef]
- Huynh, T.Y.L.; Zareba, I.; Baszanowska, W.; Lewoniewska, S.; Palka, J. Understanding the role of key amino acids in regulation of proline dehydrogenase/proline oxidase (prodh/pox)-dependent apoptosis/autophagy as an approach to targeted cancer therapy. Mol. Cell. Biochem. 2020, 466, 35–44. [Google Scholar] [CrossRef] [PubMed]
- Elia, I.; Broekaert, D.; Christen, S.; Boon, R.; Radaelli, E.; Orth, M.F.; Verfaillie, C.; Grünewald, T.G.; Fendt, S.-M. Proline metabolism supports metastasis formation and could be inhibited to selectively target metastasizing cancer cells. Nat. Commun. 2017, 8, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Ding, J.; Kuo, M.-L.; Su, L.; Xue, L.; Luh, F.; Zhang, H.; Wang, J.; Lin, T.G.; Zhang, K.; Chu, P. Human mitochondrial pyrroline-5-carboxylate reductase 1 promotes invasiveness and impacts survival in breast cancers. Carcinogenesis 2017, 38, 519–531. [Google Scholar] [CrossRef]
- Fan, Y.; Zhou, X.; Xia, T.-S.; Chen, Z.; Li, J.; Liu, Q.; Alolga, R.N.; Chen, Y.; Lai, M.-D.; Li, P. Human plasma metabolomics for identifying differential metabolites and predicting molecular subtypes of breast cancer. Oncotarget 2016, 7, 9925. [Google Scholar] [CrossRef]
- Khan, I.; Nam, M.; Kwon, M.; Seo, S.-S.; Jung, S.; Han, J.S.; Hwang, G.-S.; Kim, M.K. LC/MS-based polar metabolite profiling identified unique biomarker signatures for cervical cancer and cervical intraepithelial neoplasia using global and targeted metabolomics. Cancers 2019, 11, 511. [Google Scholar] [CrossRef]
- Dando, I.; Pozza, E.D.; Ambrosini, G.; Torrens-Mas, M.; Butera, G.; Mullappilly, N.; Pacchiana, R.; Palmieri, M.; Donadelli, M. Oncometabolites in cancer aggressiveness and tumour repopulation. Biol. Rev. 2019, 94, 1530–1546. [Google Scholar] [CrossRef]
- Moosavi, B.; Zhu, X.-L.; Yang, W.-C.; Yang, G.-F. Molecular pathogenesis of tumorigenesis caused by succinate dehydrogenase defect. Eur. J. Cell Biol. 2020, 99, 151057. [Google Scholar] [CrossRef]
- Mu, X.; Zhao, T.; Xu, C.; Shi, W.; Geng, B.; Shen, J.; Zhang, C.; Pan, J.; Yang, J.; Hu, S. Oncometabolite succinate promotes angiogenesis by upregulating VEGF expression through GPR91-mediated STAT3 and ERK activation. Oncotarget 2017, 8, 13174. [Google Scholar] [CrossRef]
- Dalla Pozza, E.; Dando, I.; Pacchiana, R.; Liboi, E.; Scupoli, M.T.; Donadelli, M.; Palmieri, M. Regulation of succinate dehydrogenase and role of succinate in cancer. Semin. Cell Dev. Biol. 2020, 98, 4–14. [Google Scholar] [CrossRef]
- Gong, Z.-G.; Zhao, W.; Zhang, J.; Wu, X.; Hu, J.; Yin, G.-C.; Xu, Y.-J. Metabolomics and eicosanoid analysis identified serum biomarkers for distinguishing hepatocellular carcinoma from hepatitis B virus-related cirrhosis. Oncotarget 2017, 8, 63890. [Google Scholar] [CrossRef] [PubMed]
- Newman, A.C.; Maddocks, O.D. One-carbon metabolism in cancer. Br. J. Cancer 2017, 116, 1499–1504. [Google Scholar] [CrossRef] [PubMed]
- Brasky, T.M.; White, E.; Chen, C.-L. Long-term, supplemental, one-carbon metabolism–related vitamin B use in relation to lung cancer risk in the Vitamins and Lifestyle (VITAL) Cohort. J. Clin. Oncol. 2017, 35, 3440. [Google Scholar] [CrossRef] [PubMed]
- Houghton, S.C.; Eliassen, A.H.; Zhang, S.M.; Selhub, J.; Rosner, B.A.; Willett, W.C.; Hankinson, S.E. Plasma B-vitamins and one-carbon metabolites and the risk of breast cancer in younger women. Breast Cancer Res. Treat. 2019, 176, 191–203. [Google Scholar] [CrossRef]
- Myte, R.; Gylling, B.; Häggström, J.; Schneede, J.; Ueland, P.M.; Hallmans, G.; Johansson, I.; Palmqvist, R.; Van Guelpen, B. Untangling the role of one-carbon metabolism in colorectal cancer risk: A comprehensive Bayesian network analysis. Sci. Rep. 2017, 7, 43434. [Google Scholar] [CrossRef]
- Farran, B.; Pavitra, E.; Kasa, P.; Peela, S.; Raju, G.S.R.; Nagaraju, G.P. Folate-targeted immunotherapies: Passive and active strategies for cancer. Cytokine Growth Factor Rev. 2019, 45, 45–52. [Google Scholar] [CrossRef]
- Scaranti, M.; Cojocaru, E.; Banerjee, S.; Banerji, U. Exploiting the folate receptor α in oncology. Nat. Rev. Clin. Oncol. 2020, 17, 349–359. [Google Scholar] [CrossRef]
- Wyss, M.; Kaddurah-Daouk, R. Creatine and creatinine metabolism. Physiol. Rev. 2000, 80, 1107–1213. [Google Scholar] [CrossRef] [PubMed]
- Weinstein, S.J.; Mackrain, K.; Stolzenberg-Solomon, R.Z.; Selhub, J.; Virtamo, J.; Albanes, D. Serum creatinine and prostate cancer risk in a prospective study. Cancer Epidemiol. Prev. Biomark. 2009, 18, 2643–2649. [Google Scholar] [CrossRef]
- Chiong, E.; Wong, A.F.W.; Chan, Y.H.; Chin, C.M. Review of clinical manifestations of biochemically-advanced prostate cancer cases. Asian J. Surg. 2005, 28, 202–206. [Google Scholar] [CrossRef]
- Schwameis, R.; Postl, M.; Bekos, C.; Hefler, L.; Reinthaller, A.; Seebacher, V.; Grimm, C.; Polterauer, S.; Helmy-Bader, S. Prognostic value of serum creatine level in patients with vulvar cancer. Sci. Rep. 2019, 9, 1–7. [Google Scholar] [CrossRef]
- Panotopoulos, J.; Posch, F.; Funovics, P.T.; Willegger, M.; Scharrer, A.; Lamm, W.; Brodowicz, T.; Windhager, R.; Ay, C. Elevated serum creatinine and low albumin are associated with poor outcomes in patients with liposarcoma. J. Orthop. Res. 2016, 34, 533–538. [Google Scholar] [CrossRef]
- Lafleur, J.; Hefler-Frischmuth, K.; Grimm, C.; Schwameis, R.; Gensthaler, L.; Reiser, E.; Hefler, L.A. Prognostic Value of Serum Creatinine Levels in Patients with Epithelial Ovarian Cancer. Anticancer Res. 2018, 38, 5127–5130. [Google Scholar] [CrossRef]
- Cussenot, O.; Berthon, P.; Berger, R.; Mowszowicz, I.; Faille, A.; Hojman, F.; Teillac, P.; Le Duc, A.; Calvo, F. Immortalization of human adult normal prostatic epithelial cells by liposomes containing large T-SV40 gene. J. Urol. 1991, 146, 881–886. [Google Scholar] [CrossRef]
- Berthon, P.; Cussenot, O.; Hopwood, L.; Leduc, A.; Maitland, N.J. Functional expression of sv40 in normal human prostatic epithelial and fibroblastic cells-differentiation pattern of nontumorigenic cell-lines. Int. J. Oncol. 1995, 6, 333–343. [Google Scholar] [CrossRef]
- Davis, T.H.; Morton, C.C.; Miller-Cassman, R.; Balk, S.P.; Kadin, M.E. Hodgkin’s disease, lymphomatoid papulosis, and cutaneous T-cell lymphoma derived from a common T-cell clone. N. Engl. J. Med. 1992, 326, 1115–1122. [Google Scholar] [CrossRef]
- Wishart, D.S.; Feunang, Y.D.; Marcu, A.; Guo, A.C.; Liang, K.; Vázquez-Fresno, R.; Sajed, T.; Johnson, D.; Li, C.; Karu, N. HMDB 4.0: The human metabolome database for 2018. Nucleic Acids Res. 2018, 46, D608–D617. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Chong, J.; Wishart, D.S.; Xia, J. Using MetaboAnalyst 4.0 for comprehensive and integrative metabolomics data analysis. Curr. Protoc. Bioinform. 2019, 68, e86. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Palviainen, M.; Laukkanen, K.; Tavukcuoglu, Z.; Velagapudi, V.; Kärkkäinen, O.; Hanhineva, K.; Auriola, S.; Ranki, A.; Siljander, P. Cancer Alters the Metabolic Fingerprint of Extracellular Vesicles. Cancers 2020, 12, 3292. https://doi.org/10.3390/cancers12113292
Palviainen M, Laukkanen K, Tavukcuoglu Z, Velagapudi V, Kärkkäinen O, Hanhineva K, Auriola S, Ranki A, Siljander P. Cancer Alters the Metabolic Fingerprint of Extracellular Vesicles. Cancers. 2020; 12(11):3292. https://doi.org/10.3390/cancers12113292
Chicago/Turabian StylePalviainen, Mari, Kirsi Laukkanen, Zeynep Tavukcuoglu, Vidya Velagapudi, Olli Kärkkäinen, Kati Hanhineva, Seppo Auriola, Annamari Ranki, and Pia Siljander. 2020. "Cancer Alters the Metabolic Fingerprint of Extracellular Vesicles" Cancers 12, no. 11: 3292. https://doi.org/10.3390/cancers12113292
APA StylePalviainen, M., Laukkanen, K., Tavukcuoglu, Z., Velagapudi, V., Kärkkäinen, O., Hanhineva, K., Auriola, S., Ranki, A., & Siljander, P. (2020). Cancer Alters the Metabolic Fingerprint of Extracellular Vesicles. Cancers, 12(11), 3292. https://doi.org/10.3390/cancers12113292






