Comprehensive Analysis of Tumour Sub-Volumes for Radiomic Risk Modelling in Locally Advanced HNSCC
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Characteristics of Patient Cohorts
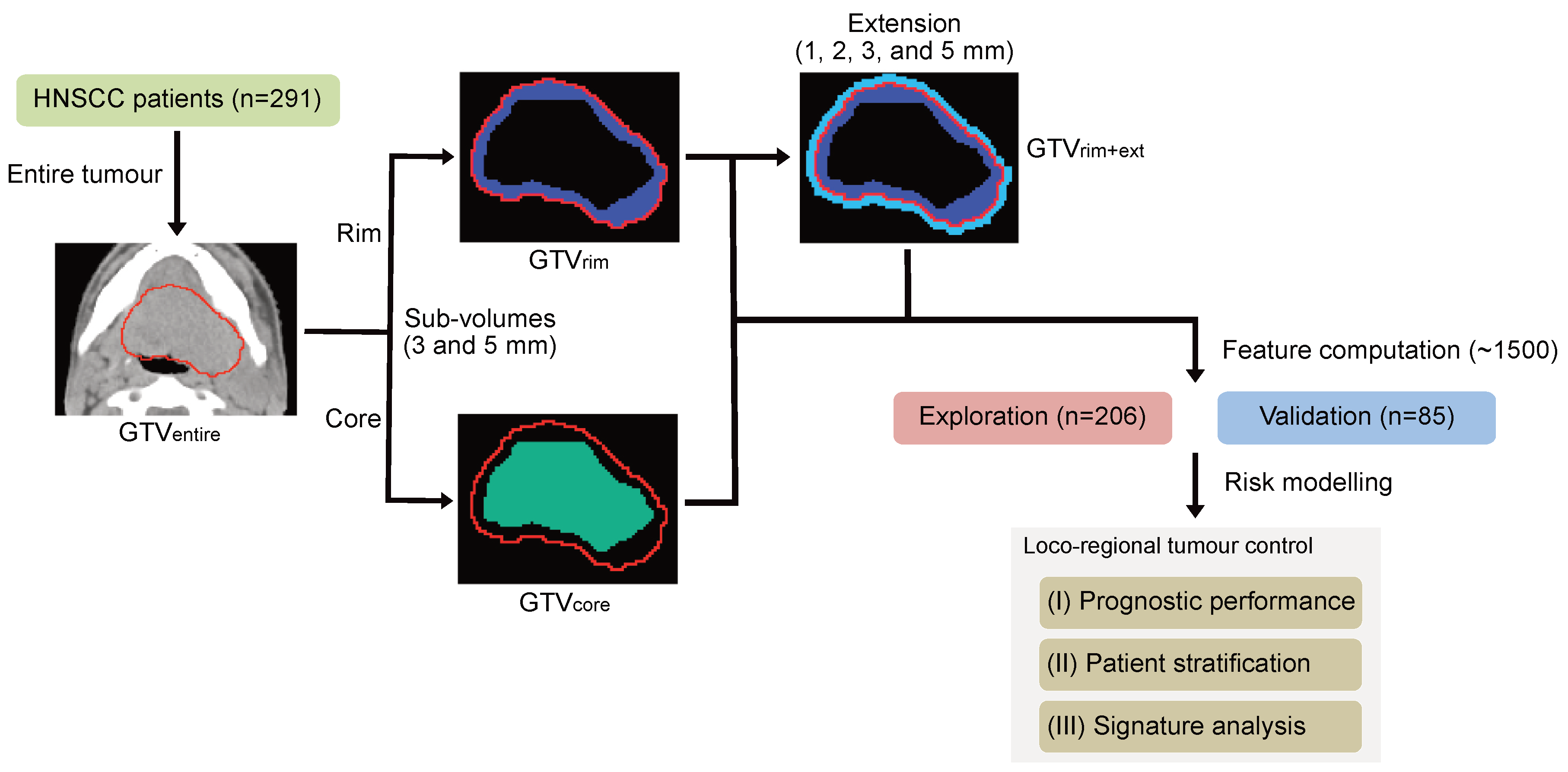
2.2. Tumour Sub-Volume Definition and Feature Computation
2.3. Radiomic Risk Modelling
2.4. Performance Assessments
3. Results
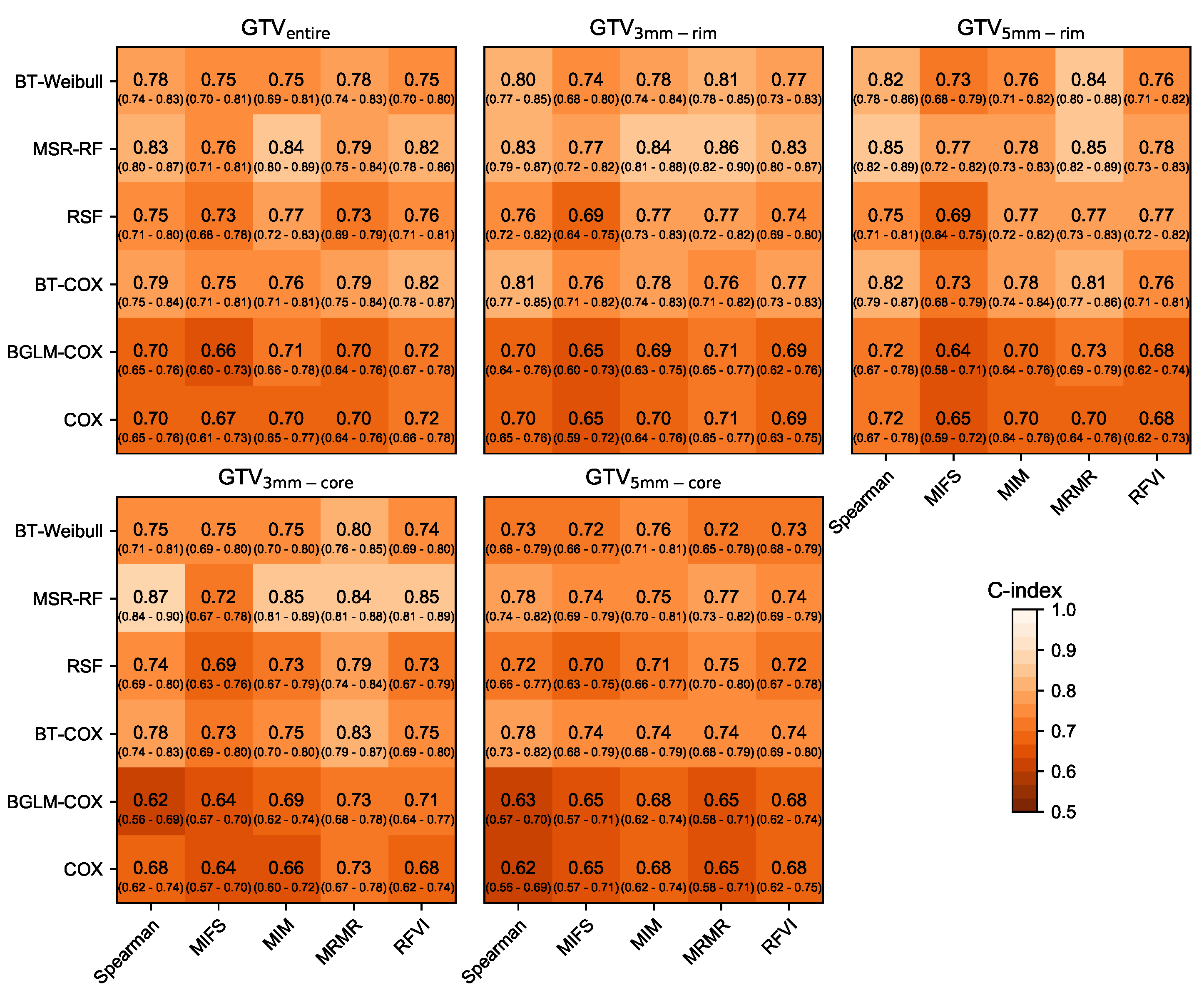
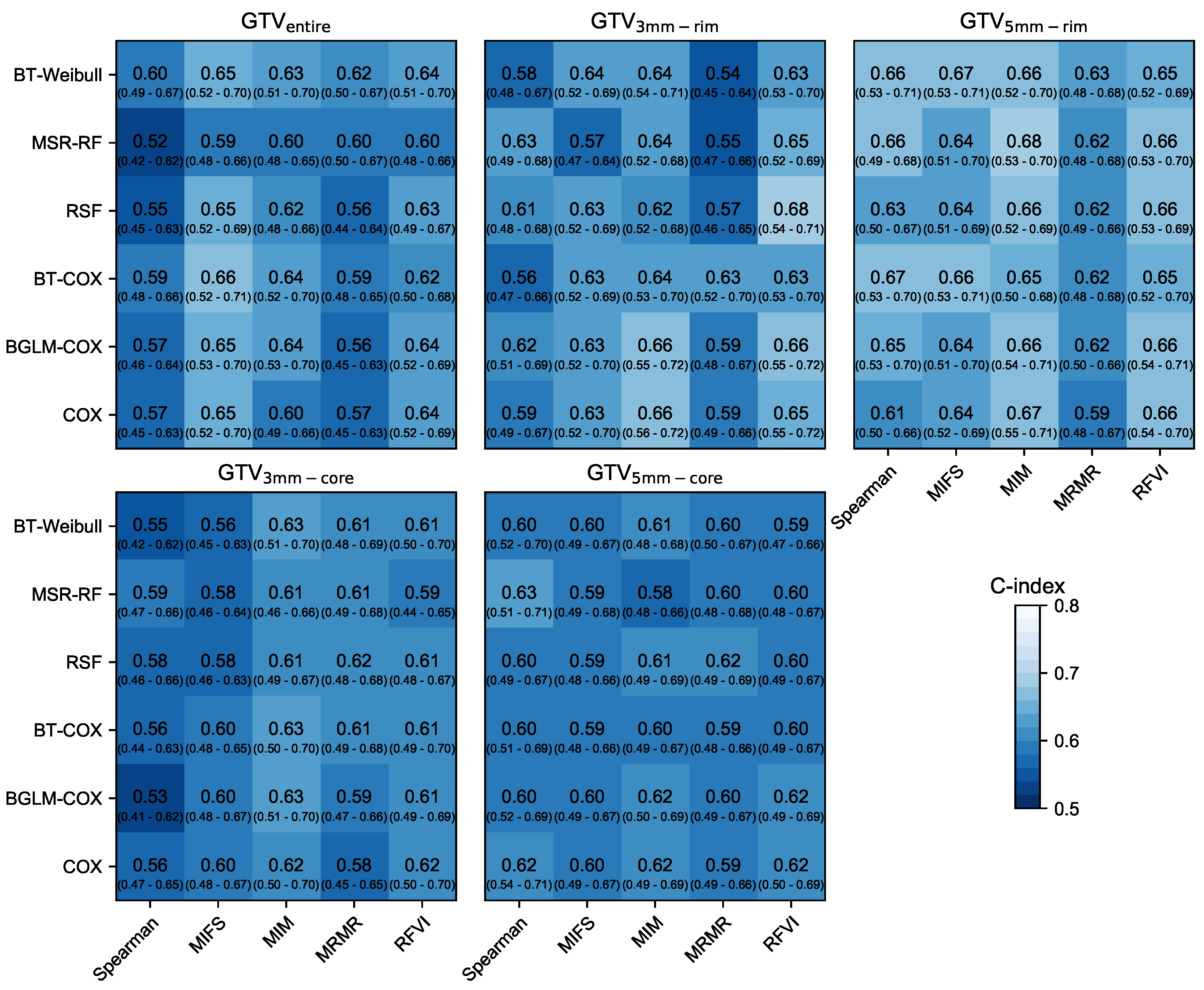
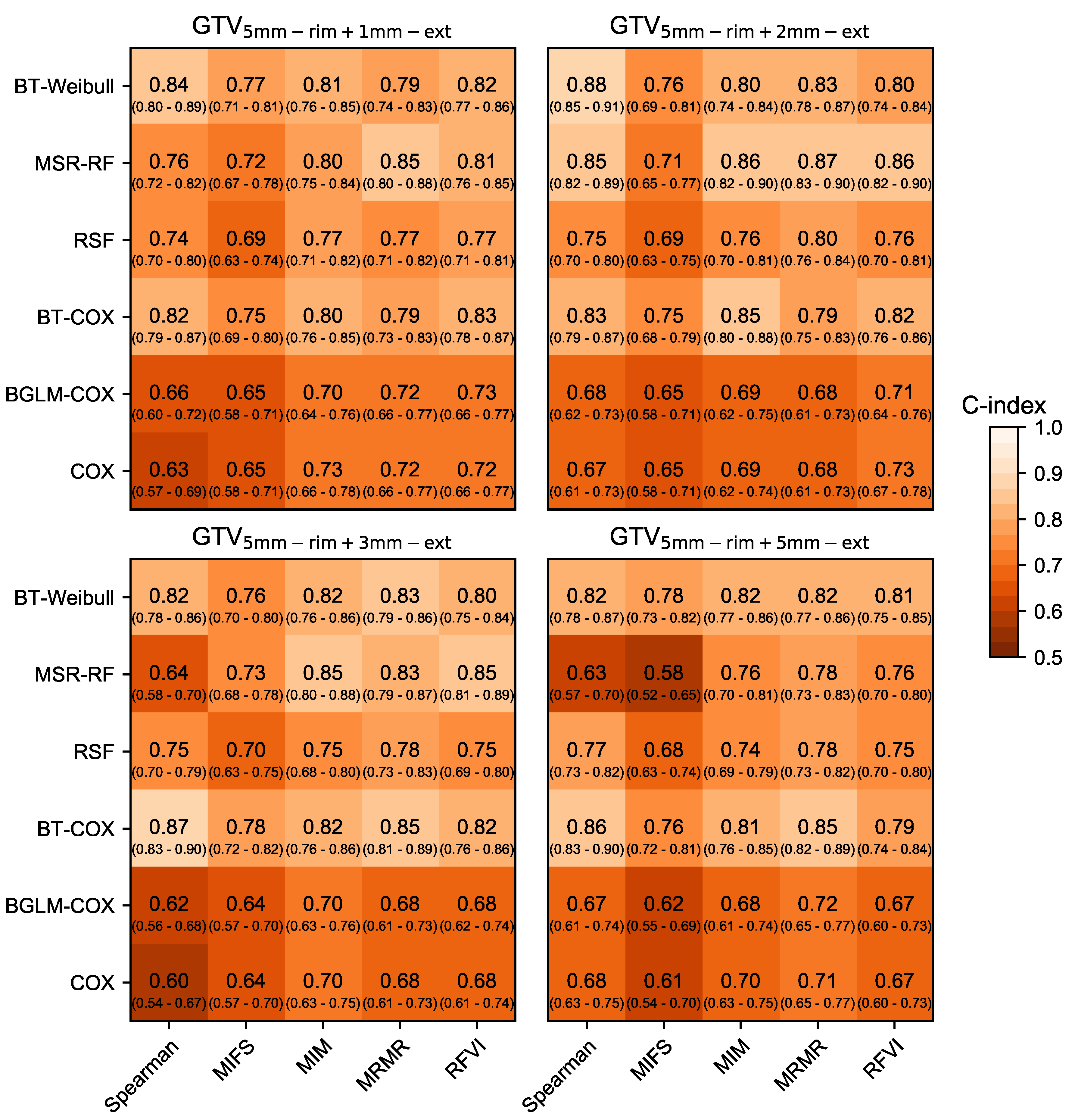
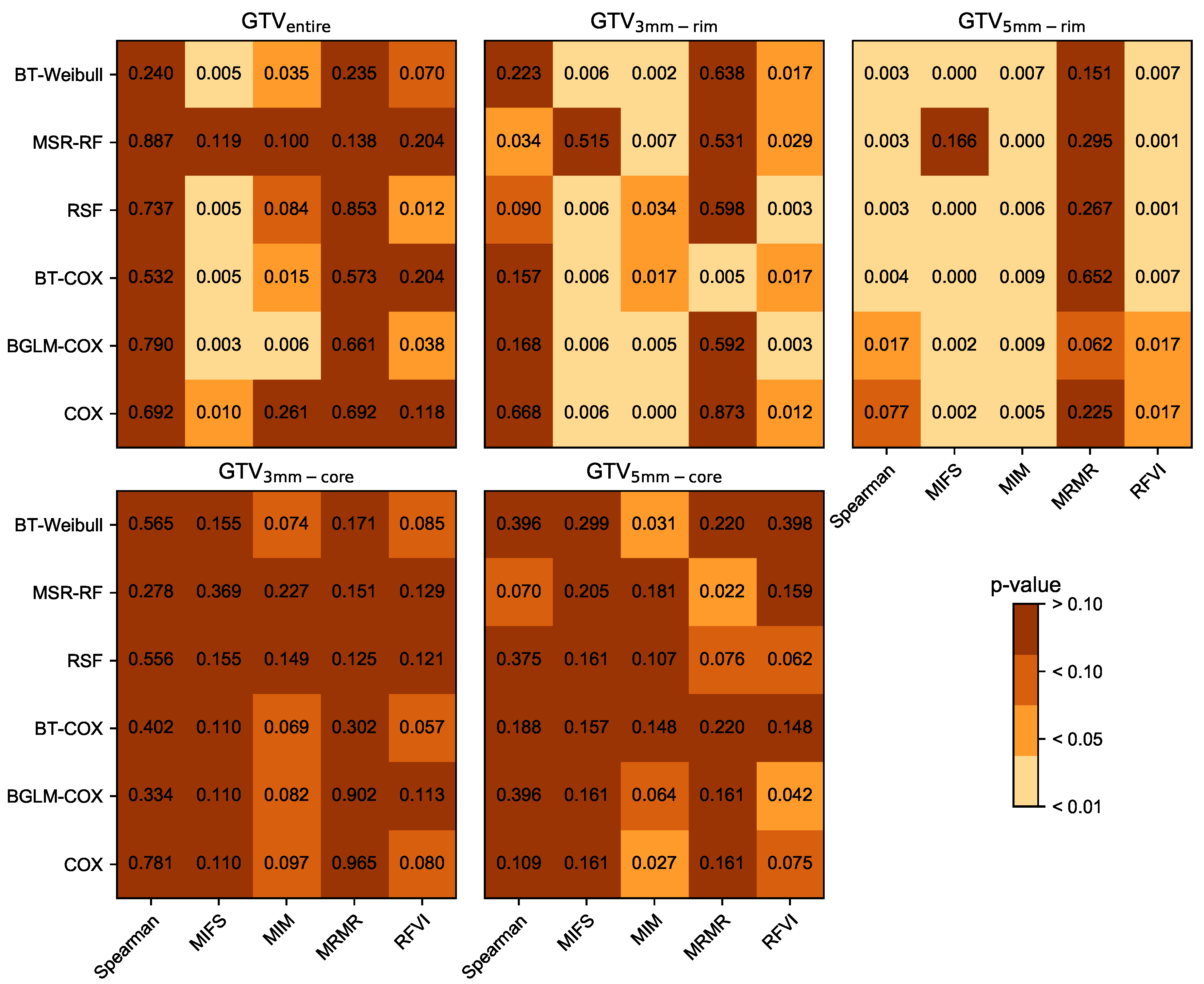
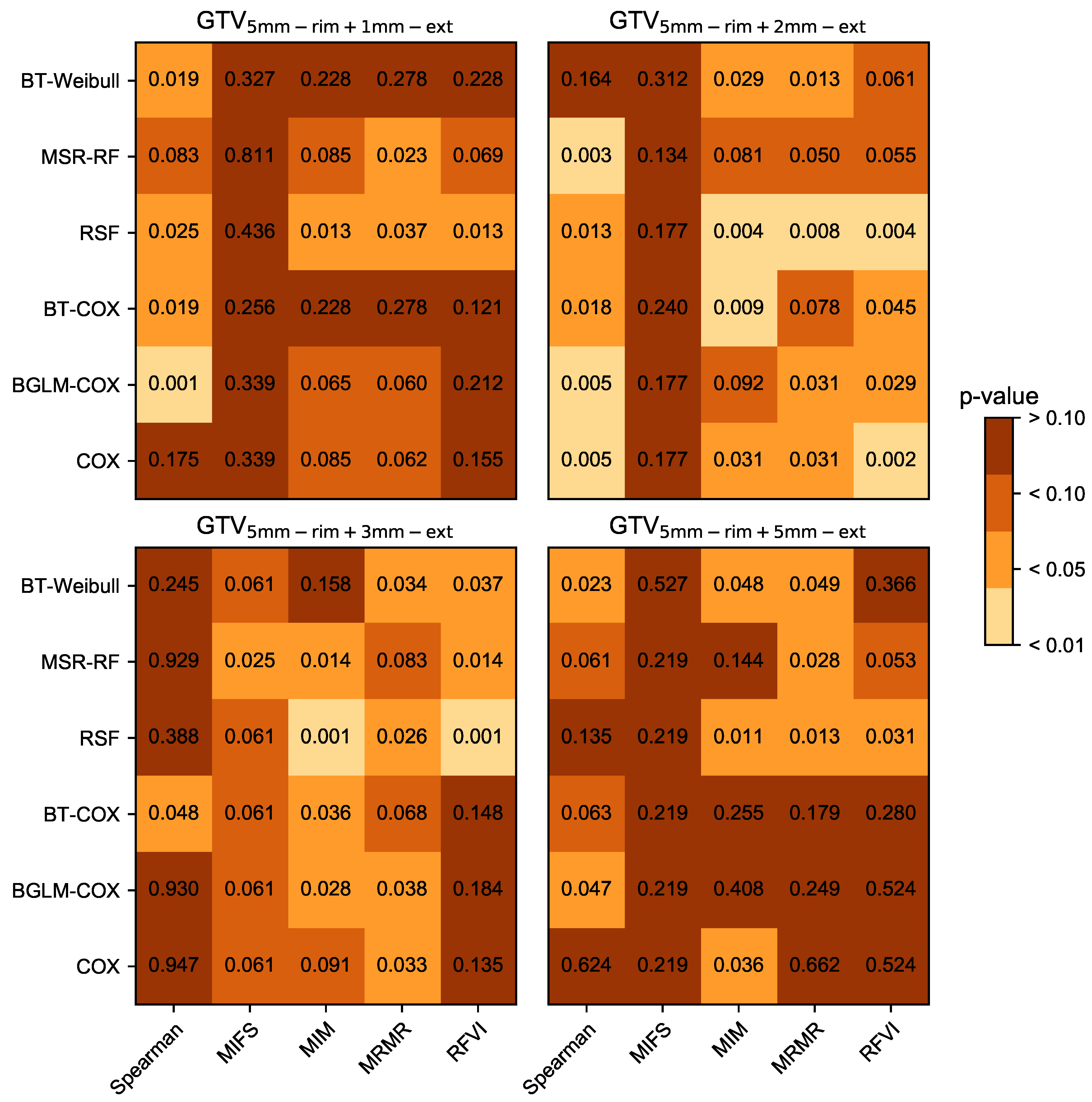
3.1. Prognostic Performance
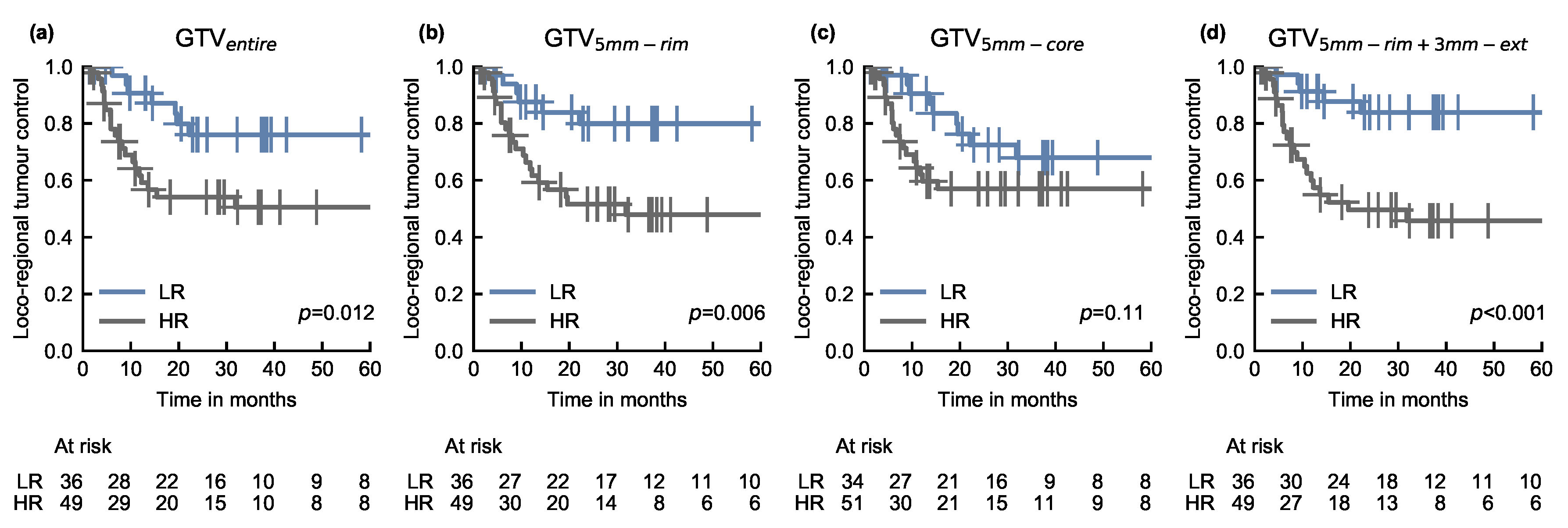
3.2. Risk-Based Patient Stratification
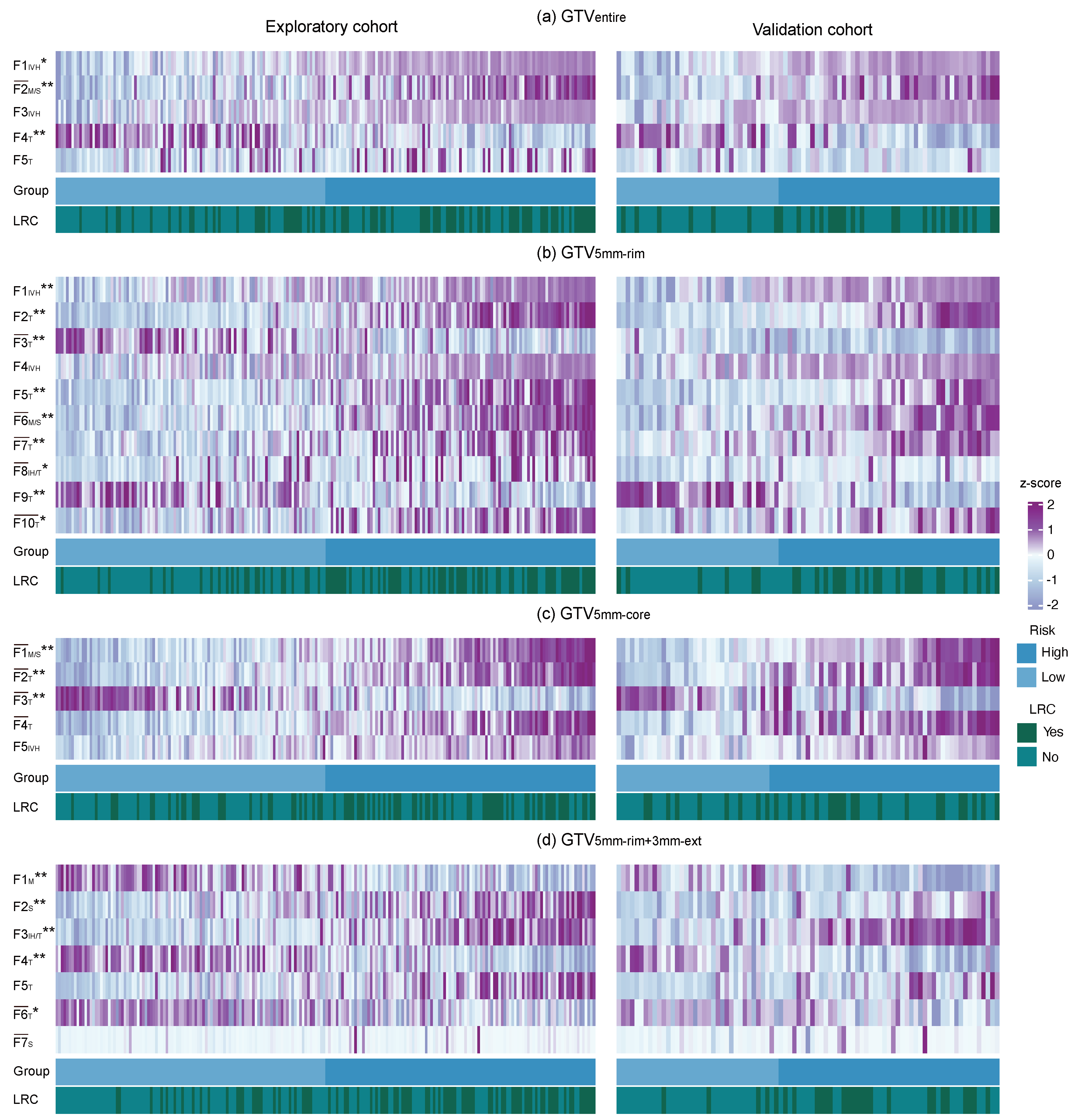
3.3. Radiomic Signature Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| BGLM-Cox | boosted gradient linear model-Cox |
| BT-Cox | boosted tree-Cox |
| C-index | concordance index |
| Cox | Cox proportional hazard model |
| CT | computed tomography |
| DKTK-ROG | German Cancer Consortium Radiation Oncology Group |
| FDG | F-fluorodeoxyglucose |
| FMISO | F-fluoromisonidazole |
| GTV | gross tumour volume |
| HNSCC | head and neck squamous cell carcinoma |
| MIFS | mutual information feature selection |
| MIM | mutual information maximisation |
| MLA | multi-level model approach |
| MRI | magnetic resonance imaging |
| MRMR | minimum redundancy maximum relevance |
| MSR-RF | maximally selected rank statistics random forest |
| OS | overall survival |
| PET | positron emission tomography |
| RCT | radio-chemotherapy |
| RFVI | random forest variable importance |
| RSF | random survival forest |
| UKD | University Hospital Dresden |
Appendix A
Appendix A.1. Multi-Level Model
| Image Acquisition Parameters | Exploratory Cohort (n = 206) | Validation Cohort (n = 85) | |
|---|---|---|---|
| Voxel spacing in-plane (mm) | |||
| 0.85/0.87/0.88/0.90 | 1/2/1/1 | 0/0/0/0 | |
| 0.92/0.93/0.94/0.96 | 1/1/3/2 | 0/0/0/0 | |
| 0.97/0.98/1.17/1.27/1.36 | 3/113/21/26/29 | 0/13/0/14/58 | |
| Spacing in z-direction (mm) | |||
| 2.0/2.5/3.0/3.75/5.0 | 36/22/74/1/63 | 0/0/27/0/58 | |
| Reconstruction kernel | |||
| B10s/B20f/s/B30f/s/B31f/s | 20/3/1/2/29/19/16 | 1/51/1/0/0/12/0 | |
| B40f/s/B50s/Missing | 1/1/9/12/93 | 0/0/0/0/20 | |
| Mean exposure (mA) | 181.3 (Missing: 59) | 76.8 (Missing: 14) | |
| Mean exposure time (ms) | 733.8 (Missing: 59) | 508.8 (Missing: 14) | |
| Tube voltage (kV) | |||
| 120/130/140/Missing | 86/9/16/95 | 71/0/0/14 |

| Configuration Settings | Configuration Value(s) |
|---|---|
| Image interpolation | |
| Interpolation method | Cubic spline |
| Voxel dimensions | 1.0 × 1.0 × 1.0 mm |
| Anti-aliasing smoothing parameter | 0.98 |
| RoI interpolation | |
| Interpolation method | Cubic spline |
| Inclusion threshold | 0.5 |
| Discretisation | |
| Discretisation method | Fixed Bin Number (FBN) of 32 bins |
| Intensity Volume Histogram discretisation method | Fixed Bin Number (FBN) of 1000 bins |
| Image transformation | |
| Wavelet | coiflet-1 |
| Mean-Intensity Laplacian of Gaussian | kernel widths: 1.0, 2.0, 3.0, 5.0, 6.0 mm |
| Texture matrices | |
| Grey-level Run Length Matrix (GLRLM) | Calculation method: 3D |
| Merge method: volume merge | |
| Grey-level Size Zone Matrix (GLSZM) | Calculation method: 3D |
| Neighbourhood Grey Tone Difference Matrix (NGTDM) | Calculation method: 3D |
| Neighbourhood Grey Level Dependence Matrix (NGLDM) | Distance for neighborhood: 1.8 voxels |
| Difference level: 0.0 | |
| Calculation method: 3D | |
| Grey Level Co-occurrence Matrix (GLCM) | Distance for neighborhood: 1.0 voxels |
| Calculation method: 3D | |
| Merge method: volume merge | |
| Grey Level Distance Zone Matrix (GLDZM) | Calculation method: 3D |
| Algorithm | Hyper-Parameter Name | Hyper-Parameter Value(s) |
|---|---|---|
| Cox proportional | ||
| hazard model | Signature size | 2, 3, 4, 5, 7, 10 |
| Boosted tree and | ||
| boosted gradient models | Signature size: | 2, 3, 4, 5, 7, 10 |
| : | 0.001, 0.01, 0.05 | |
| : | lambda.min , lambda.1se | |
| mStop: | 200 | |
| Random survival forest | Signature size: | 2, 3, 4, 5, 7, 10 |
| ntree: | 2000, 5000 | |
| mtry: | 100 | |
| node-Size: | 25–50, step size 1 | |
| maxDepth: | 10, 15, 20, 25, 40 | |
| nSplit: | 1, 2, 100 | |
| splitRule: | logrank, logrankscore | |
| Maximally selected rank | ||
| statistics random forest | Signature size: | 2, 3, 4, 5, 7, 10 |
| ntree: | 2000, 5000 | |
| mtry: | 100 | |
| node-Size: | 25–50, step size 1 | |
| minprop: | 0.1 | |
| : | 0.1, 0.5 | |
| splitRule: | C, maxstat | |
| Minimum redundancy maximum relevance | topFeatures | 100 |
| RelativeImportanceThreshold | 0 | |
| Mutual information feature selection | topFeatures | 100 |
| RelativeImportanceThreshold | 0.05 | |
| Random forest variable importance | topFeatures | 20 |
| nTree | 1000 | |
| K | 2 | |
| nRepetition | 50 | |
| nSteps | 2 | |
| nSplits | 1 | |
| splitrule | logrank | |
| nodeSize | 45 | |
| mTry | 500 | |
| nVariables | 10 |
| Sub-Volume | Hyper-Parameter Value(s) |
|---|---|
| GTV | |
| RSF-RFVI | Signature size: 5 |
| ntree: 5000 | |
| mtry: 100 | |
| node-Size: 10 | |
| maxDepth: 25 | |
| nSplit: 1 | |
| splitRule: logrankscore | |
| GTV | |
| RSF-MIM | Signature size: 10 |
| ntree: 5000 | |
| mtry: 100 | |
| node-Size: 5 | |
| maxDepth: 40 | |
| nSplit: 1 | |
| splitRule: logrank | |
| GTV | |
| RSF-MIM | Signature size: 5 |
| ntree: 5000 | |
| mtry: 100 | |
| nodeSize: 7 | |
| maxDepth: 40 | |
| nSplit: 1 | |
| splitRule: logrank | |
| GTV | |
| RSF-RFVI | Signature size: 7 |
| ntree: 5000 | |
| mtry: 100 | |
| node-Size: 5 | |
| maxDepth: 15 | |
| nSplit: 1 | |
| splitRule: logrankscore |
| Tumour Sub-Volume | Feature Name | Synonym | Image | Cluster |
|---|---|---|---|---|
| GTV | ivh_diff_v10_v90 | wav_coif1_llh | no | |
| morph_integ_int, stat_energy | base, wav_coif1_lll | yes | ||
| ivh_v10 | wav_coif1_llh | no | ||
| rlm_gl_var_3d_avg | wav_coif1_lhl | no | ||
| rlm_srhge_3d_avg | base | no | ||
| GTV | ivh_diff_v10_v90 | wav_coif1_llh | no | |
| dzm_ldhge_3d | base | no | ||
| dzm_lgze_3d, dzm_sdlge_3d, | base | yes | ||
| szm_lgze_3d, szm_szlge_3d | ||||
| ivh_v10 | wav_coif1_llh | no | ||
| dzm_ldhge_3d | wav_coif1_lll | no | ||
| morph_integ_int, stat_energy | base, wav_coif1_lll | yes | ||
| dzm_hgze_3d, szm_hgze_3d, szm_szhge_3d | base | yes | ||
| cm_auto_corr_d1_3d, cm_joint_avg_d1_3d, | ||||
| cm_sum_avg_d1_3d, ih_mean, | base | yes | ||
| ngl_hgce_d1_a0.0_3d, rlm_hgre_3d_avg | ||||
| cm_info_corr2_d1_3d_avg | base | no | ||
| ngt_strength_3d, rlm_rlnu_3d_avg | wav_coif1_llh | yes | ||
| GTV | morph_integ_int, stat_energy | base, wav_coif1_lll | yes | |
| ngt_strength_3d, rlm_rlnu_3d_avg | wav_coif1_llh | yes | ||
| dzm_gl_var_3d, szm_gl_var_3d | wav_coif1_hhl | yes | ||
| dzm_glnu_3d, szm_glnu_3d | wav_coif1_llh | yes | ||
| ivh_i50 | wav_coif1_lhl | no | ||
| GTV | morph_moran_i | base | no | |
| stat_energy | wav_coif1_hhl | no | ||
| ih_kurt,rlm_glnu_norm_3d_avg, stat_kurt | wav_coif1_lhl | yes | ||
| rlm_gl_var_3d_avg | wav_coif1_lhh | no | ||
| stat_max, stat_min, stat_range | wav_coif1_hhl | yes | ||
| ngl_dc_var_d1_a0.0_3d | wav_coif1_lhh | no | ||
| stat_cov | base, wav_coif1_lll | yes |
References
- Baumann, M.; Krause, M.; Overgaard, J.; Debus, J.; Bentzen, S.M.; Daartz, J.; Richter, C.; Zips, D.; Bortfeld, T. Radiation oncology in the era of precision medicine. Nat. Rev. Cancer 2016, 16, 234. [Google Scholar] [CrossRef] [PubMed]
- Aerts, H.; Velazquez, E.; Leijenaar, R.; Parmar, C.; Grossmann, P.; Carvalho, S.; Bussink, J.; Monshouwer, R.; Haibe-Kains, B.; Rietveld, D.; et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat. Commun. 2014, 5, 4006. [Google Scholar] [CrossRef]
- Vallières, M.; Kay-Rivest, E.; Perrin, L.; Liem, X.; Furstoss, C.; Aerts, H.; Khaouam, N.; Nguyen-Tan, P.; Wang, C.; Sultanem, K.; et al. Radiomics strategies for risk assessment of tumour failure in head-and-neck cancer. Sci. Rep. 2017, 7, 10117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leger, S.; Zwanenburg, A.; Pilz, K.; Lohaus, F.; Linge, A.; Zöphel, K.; Kotzerke, J.; Schreiber, A.; Tinhofer, I.; Budach, C.; et al. A comparative study of machine learning methods for time-to-event survival data for radiomics risk modelling. Sci. Rep. 2017, 7, 13206. [Google Scholar] [CrossRef] [PubMed]
- Leger, S.; Zwanenburg, A.; Pilz, K.; Zschaeck, S.; Zöphel, K.; Kotzerke, J.; Schreiber, A.; Zips, D.; Krause, M.; Baumann, M.; et al. CT imaging during treatment improves radiomic models for patients with locally advanced head and neck cancer. Radiother. Oncol. 2019, 130, 10–17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, J.; Gong, G.; Cui, Y.; Li, R. Intratumor partitioning and texture analysis of dynamic contrast-enhanced (DCE)-MRI identifies relevant tumor subregions to predict pathological response of breast cancer to neoadjuvant chemotherapy. J. Magn. Reson. Imaging 2016, 44, 1107–1115. [Google Scholar] [CrossRef]
- Serganova, I.; Doubrovin, M.; Vider, J.; Ponomarev, V.; Soghomonyan, S.; Beresten, T.; Ageyeva, L.; Serganov, A.; Cai, S.; Balatoni, J.; et al. Molecular imaging of temporal dynamics and spatial heterogeneity of hypoxia-inducible factor-1 signal transduction activity in tumors in living mice. Cancer Res. 2004, 64, 6101–6108. [Google Scholar] [CrossRef] [Green Version]
- Troost, E.G.; Laverman, P.; Philippens, M.E.; Lok, J.; van der Kogel, A.J.; Oyen, W.J.; Boerman, O.C.; Kaanders, J.H.; Bussink, J. Correlation of [18 F] FMISO autoradiography and pimonodazole immunohistochemistry in human head and neck carcinoma xenografts. Eur. J. Nucl. Med. Mol. Imaging 2008, 35, 1803–1811. [Google Scholar] [CrossRef]
- Zips, D.; Zöphel, K.; Abolmaali, N.; Perrin, R.; Abramyuk, A.; Haase, R.; Appold, S.; Steinbach, J.; Kotzerke, J.; Baumann, M. Exploratory prospective trial of hypoxia-specific PET imaging during radiochemotherapy in patients with locally advanced head-and-neck cancer. Radiother. Oncol. 2012, 105, 21–28. [Google Scholar] [CrossRef] [Green Version]
- Gatenby, R.; Grove, O.; Gillies, R. Quantitative imaging in cancer evolution and ecology. Radiology 2013, 269, 8–15. [Google Scholar] [CrossRef] [Green Version]
- O’Connor, J.; Rose, C.; Waterton, J.; Carano, R.; Parker, G.; Jackson, A. Imaging intratumor heterogeneity: Role in therapy response, resistance, and clinical outcome. Clin. Cancer Res. 2015, 21, 249–257. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Löck, S.; Perrin, R.; Seidlitz, A.; Bandurska-Luque, A.; Zschaeck, S.; Zöphel, K.; Krause, M.; Steinbach, J.; Kotzerke, J.; Zips, D.; et al. Residual tumour hypoxia in head-and-neck cancer patients undergoing primary radiochemotherapy, final results of a prospective trial on repeat FMISO-PET imaging. Radiother. Oncol. 2017, 124, 533–540. [Google Scholar] [CrossRef]
- Schütze, C.; Bergmann, R.; Yaromina, A.; Hessel, F.; Kotzerke, J.; Steinbach, J.; Baumann, M.; Beuthien-Baumann, B. Effect of increase of radiation dose on local control relates to pre-treatment FDG uptake in FaDu tumours in nude mice. Radiother. Oncol. 2007, 83, 311–315. [Google Scholar] [CrossRef]
- Schütze, C.; Bergmann, R.; Brüchner, K.; Mosch, B.; Yaromina, A.; Zips, D.; Hessel, F.; Krause, M.; Thames, H.; Kotzerke, J.; et al. Effect of [18F] FMISO stratified dose-escalation on local control in FaDu hSCC in nude mice. Radiother. Oncol. 2014, 111, 81–87. [Google Scholar] [CrossRef]
- Peltanova, B.; Raudenska, M.; Masarik, M. Effect of tumor microenvironment on pathogenesis of the head and neck squamous cell carcinoma: A systematic review. Mol. Cancer 2019, 18, 63. [Google Scholar] [CrossRef] [PubMed]
- Alsahafi, E.; Begg, K.; Amelio, I.; Raulf, N.; Lucarelli, P.; Sauter, T.; Tavassoli, M. Clinical update on head and neck cancer: Molecular biology and ongoing challenges. Cell Death Dis. 2019, 10, 1–17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Algohary, A.; Shiradkar, R.; Pahwa, S.; Purysko, A.; Verma, S.; Moses, D.; Shnier, R.; Haynes, A.M.; Delprado, W.; Thompson, J.; et al. Combination of Peri-Tumoral and Intra-Tumoral Radiomic Features on Bi-Parametric MRI Accurately Stratifies Prostate Cancer Risk: A Multi-Site Study. Cancers 2020, 12, 2200. [Google Scholar] [CrossRef] [PubMed]
- Grove, O.; Berglund, A.; Schabath, M.; Aerts, H.; Dekker, A.; Wang, H.; Rios Velazquez, E.; Lambin, P.; Gu, Y.; Balagurunathan, Y.; et al. Quantitative computed tomographic descriptors associate tumor shape complexity and intratumor heterogeneity with prognosis in lung adenocarcinoma. PLoS ONE 2015, 10, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, J.; Gensheimer, M.; Dong, X.; Rubin, D.; Napel, S.; Diehn, M.; Loo, B.; Li, R. Robust Intratumor Partitioning to Identify High-Risk Subregions in Lung Cancer: A Pilot Study. Int. J. Radiat. Oncol. Biol. Phys. 2016, 95, 1504–1512. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leger, S. Radiomics Risk Modelling Using Machine Learning Algorithms for Personalised Radiation Oncology. Ph.D. Thesis, Faculty of Medicine and University Hospital Carl Gustav Carus, Technische Universität Dresden, 2018. [Google Scholar]
- Leger, S.; Zwanenburg, A.; Pilz, K.; Lohaus, F.; Linge, A.; Zöphel, K.; Kotzerke, J.; Schreiber, A.; Tinhofer, I.; Budach, C.; et al. Identification of tumour sub-volumes for improved radiomic risk modelling in locally advanced HNSCC. Radiother. Oncol. 2018, 127, 263–264. [Google Scholar] [CrossRef]
- Apolle, R.; Rehm, M.; Bortfeld, T.; Baumann, M.; Troost, E.G. The clinical target volume in lung, head-and-neck, and esophageal cancer: Lessons from pathological measurement and recurrence analysis. Clin. Transl. Radiat. Oncol. 2017, 3, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Linge, A.; Lohaus, F.; Löck, S.; Nowak, A.; Gudziol, V.; Valentini, C.; von Neubeck, C.; Jütz, M.; Tinhofer, I.; Budach, V.; et al. HPV status, cancer stem cell marker expression, hypoxia gene signatures and tumour volume identify good prognosis subgroups in patients with HNSCC after primary radiochemotherapy: A multicentre retrospective study of the German Cancer Consortium Radiation Oncology Group (DKTK-ROG). Radiother. Oncol. 2016, 121, 364–373. [Google Scholar] [PubMed]
- Shafiq-UI-Hassan, G.G.; Latifi, K.; Ullah, G.; Hunt, D.; Balagurunathan, Y.; Abdalah, M.; Matthew, B.; Goldgof, D.; Mackin, D.; Court, L.; et al. Intrinsic dependencies of CT radiomic features on voxel size and number of gray levels. Med. Phys. 2017, 44, 1050–1062. [Google Scholar] [CrossRef] [PubMed]
- Zwanenburg, A.; Vallières, M.; Abdalah, M.A.; Aerts, H.J.; Andrearczyk, V.; Apte, A.; Ashrafinia, S.; Bakas, S.; Beukinga, R.J.; Boellaard, R.; et al. The image biomarker standardization initiative: Standardized quantitative radiomics for high-throughput image-based phenotyping. Radiology 2020, 295, 328–338. [Google Scholar] [CrossRef] [Green Version]
- Harrell, F.E., Jr.; Lee, K.L.; Mark, D.B. Multivariable prognostic models: Issues in developing models, evaluating assumptions and adequacy, and measuring and reducing errors. Stat. Med. 1996, 15, 361–387. [Google Scholar] [CrossRef]
- Pencina, M.J.; D’Agostino, R.B. Overall C as a measure of discrimination in survival analysis: Model specific population value and confidence interval estimation. Stat. Med. 2004, 23, 2109–2123. [Google Scholar] [CrossRef]
- El Naqa, I.; Grigsby, P.; Apte, A.; Kidd, E.; Donnelly, E.; Khullar, D.; Chaudhari, S.; Yang, D.; Schmitt, M.; Laforest, R.; et al. Exploring feature-based approaches in PET images for predicting cancer treatment outcomes. Pattern Recognit. 2009, 42, 1162–1171. [Google Scholar] [CrossRef] [Green Version]
- Dou, T.; Aerts, H.; Coroller, T.; Mak, R. Radiomic-Based Phenotyping of Tumor Core and Rim to Predict Survival in Nonsmall Cell Lung Cancer. Int. J. Radiat. Oncol. Biol. Phys. 2017, 99, S84. [Google Scholar] [CrossRef] [Green Version]
- Dou, T.H.; Coroller, T.P.; van Griethuysen, J.J.; Mak, R.H.; Aerts, H.J. Peritumoral radiomics features predict distant metastasis in locally advanced NSCLC. PloS ONE 2018, 13, e0206108. [Google Scholar] [CrossRef] [Green Version]
- Hosny, A.; Parmar, C.; Coroller, T.P.; Grossmann, P.; Zeleznik, R.; Kumar, A.; Bussink, J.; Gillies, R.J.; Mak, R.H.; Aerts, H.J. Deep learning for lung cancer prognostication: A retrospective multi-cohort radiomics study. PLoS Med. 2018, 15, e1002711. [Google Scholar] [CrossRef] [Green Version]
- Keek, S.; Sanduleanu, S.; Wesseling, F.; de Roest, R.; van den Brekel, M.; van der Heijden, M.; Vens, C.; Giuseppina, C.; Licitra, L.; Scheckenbach, K.; et al. Computed tomography-derived radiomic signature of head and neck squamous cell carcinoma (peri) tumoral tissue for the prediction of locoregional recurrence and distant metastasis after concurrent chemo-radiotherapy. PLoS ONE 2020, 15, e0232639. [Google Scholar]
- Campbell, S.; Poon, I.; Markel, D.; Vena, D.; Higgins, K.; Enepekides, D.; Rapheal, S.; Wong, J.; Allo, G.; Morgen, E.; et al. Evaluation of microscopic disease in oral tongue cancer using whole-mount histopathologic techniques: Implications for the management of head-and-neck cancers. Int. J. Radiat. Oncol. Biol. Phys. 2012, 82, 574–581. [Google Scholar] [CrossRef] [PubMed]
- Apolle, R.; Bijl, H.P.; Blanchard, P.; Laprie, A.; Madani, I.; Ruffier, A.; Van Elmpt, W.; Troost, E.G. Target volume delineation for adaptive treatment in HNSCC is highly variable among experts. Radiother. Oncol. 2019, 133, S655–S656. [Google Scholar] [CrossRef]
- Zwanenburg, A.; Leger, S.; Agolli, L.; Pilz, K.; Troost, E.G.; Richter, C.; Löck, S. Assessing robustness of radiomic features by image perturbation. Sci. Rep. 2019, 9, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Haarburger, C.; Müller-Franzes, G.; Weninger, L.; Kuhl, C.; Truhn, D.; Merhof, D. Radiomics feature reproducibility under inter-rater variability in segmentations of CT images. Sci. Rep. 2020, 10, 1–10. [Google Scholar] [CrossRef]
- Pavic, M.; Bogowicz, M.; Würms, X.; Glatz, S.; Finazzi, T.; Riesterer, O.; Roesch, J.; Rudofsky, L.; Friess, M.; Veit-Haibach, P.; et al. Influence of inter-observer delineation variability on radiomics stability in different tumor sites. Acta Oncol. 2018, 57, 1070–1074. [Google Scholar] [CrossRef] [Green Version]
- Vaupel, P.; Kallinowski, F.; Okunieff, P. Blood flow, oxygen and nutrient supply, and metabolic microenvironment of human tumors: A review. Cancer Res. 1989, 49, 6449–6465. [Google Scholar]
- Ljungkvist, A.S.; Bussink, J.; Rijken, P.F.; Raleigh, J.A.; Denekamp, J.; Van Der Kogel, A.J. Changes in tumor hypoxia measured with a double hypoxic marker technique. Int. J. Radiat. Oncol. Biol. Phys. 2000, 48, 1529–1538. [Google Scholar] [CrossRef]
- Kanungo, T.; Mount, D.; Netanyahu, N.; Piatko, C.; Silverman, R.; Wu, A. An efficient k-means clustering algorithm: Analysis and implementation. IEEE Trans. Pattern Anal. Mach. Intell. 2002, 24, 881–892. [Google Scholar] [CrossRef]
- Euser, A.M.; Zoccali, C.; Jager, K.J.; Dekker, F.W. Cohort studies: Prospective versus retrospective. Nephron Clin. Pract. 2009, 113, c214–c217. [Google Scholar] [CrossRef] [Green Version]
- Clarke, L.P.; Nordstrom, R.J.; Zhang, H.; Tandon, P.; Zhang, Y.; Redmond, G.; Farahani, K.; Kelloff, G.; Henderson, L.; Shankar, L.; et al. The quantitative imaging network: NCI’s historical perspective and planned goals. Transl. Oncol. 2014, 7, 1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buckler, A.J.; Bresolin, L.; Dunnick, N.R.; Sullivan, D.C.; Group. A collaborative enterprise for multi-stakeholder participation in the advancement of quantitative imaging. Radiology 2011, 258, 906–914. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buckler, A.J.; Bresolin, L.; Dunnick, N.R.; Sullivan, D.C.; Group. Quantitative imaging test approval and biomarker qualification: Interrelated but distinct activities. Radiology 2011, 259, 875–884. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| Clinical Variable | Exploratory Cohort | Validation Cohort | p-Value |
|---|---|---|---|
| Number of patients | 206 | 85 | - |
| Gender | |||
| male | 174 | 74 | 0.70 |
| female | 32 | 11 | |
| Age in years | |||
| median | 59.0 | 55.0 | 0.023 |
| range | 39.2–84.5 | 37.0–76.0 | - |
| cTN staging | |||
| T stage 1/2/3/4 | 2/23/51/130 | 2/9/30/44 | 0.21 |
| N stage 0/1/2/3/missing | 30/7/154/15/0 | 9/8/64/3/1 | 0.097 |
| UICC stage 2010 | |||
| I/II/III/IV | 0/0/15/191 | 1/2/9/73 | 0.039 |
| GTV (cm) | |||
| median | 29.1 | 40.6 | 0.067 |
| range | 4.5–321.7 | 2.7–239.1 | - |
| Tumour site | |||
| oropharynx/oral cavity/ | |||
| hypopharynx/larynx | 93/51/62/0 | 29/23/28/5 | 0.003 |
| p16 status | |||
| negative/positive/missing | 148/28/30 | 52/5/28 | 0.26 |
| Loco-regional tumour recurrence | 84 (41%) | 28 (33%) | 0.26 |
| Follow up time (months) | |||
| median | 21.2 | 24.3 | - |
| range | 1.2–131.9 | 1.3–107.2 | 0.64 |
| Tumour Sub-Volume | Exploratory Cohort | Validation Cohort | ||||
|---|---|---|---|---|---|---|
| All | All | 20 cm | 20 cm | |||
| (n = 206) | (n = 85) | (n = 20) | (n = 65) | |||
| GTV | 0.75 ± 0.05 | 0.61 ± 0.04 | 0.61 ± 0.07 | 0.59 ± 0.02 | ||
| GTV | 0.76 ± 0.06 | 0.63 ± 0.03 | 0.62 ± 1.00 | 0.61 ± 0.02 | ||
| GTV | 0.74 ± 0.06 | 0.60 ± 0.02 | 0.63 ± 0.05 | 0.57 ± 0.02 | ||
| GTV | 0.76 ± 0.06 | 0.65 ± 0.02 | 0.67 ± 0.07 | 0.61 ± 0.01 | ||
| GTV | 0.72 ± 0.04 | 0.59 ± 0.01 | 0.69 ± 0.07 | 0.57 ± 0.04 | ||
| GTV | 0.76 ± 0.06 | 0.62 ± 0.03 | 0.58 ± 0.07 | 0.65 ± 0.03 | ||
| GTV | 0.76 ± 0.07 | 0.63 ± 0.03 | 0.67 ± 0.04 | 0.66 ± 0.04 | ||
| GTV | 0.75 ± 0.08 | 0.64 ± 0.05 | 0.61 ± 0.05 | 0.65 ± 0.05 | ||
| GTV | 0.75 ± 0.07 | 0.63 ± 0.04 | 0.65 ± 0.06 | 0.62 ± 0.05 | ||
| Representative Model | Exploratory Cohort | Validation Cohort | ||||
|---|---|---|---|---|---|---|
| (n = 206) | (n = 85) | |||||
| C-Index | 95% CI | p-Value | C-Index | 95% CI | p-Value | |
| GTV | ||||||
| RSF-RFVI | 0.75 | [0.71–0.81] | <0.001 | 0.63 | [0.49–0.67] | 0.012 |
| GTV | ||||||
| BT-Cox-MRMR | 0.76 | [0.71–0.82] | <0.001 | 0.63 | [0.52–0.70] | 0.005 |
| GTV | ||||||
| BT-Cox-MIM | 0.75 | [0.70–0.80] | <0.001 | 0.63 | [0.50–0.70] | 0.069 |
| GTV | ||||||
| RSF-MIM | 0.77 | [0.72–0.82] | <0.001 | 0.66 | [0.52–0.69] | 0.006 |
| GTV | ||||||
| RSF-MIM | 0.71 | [0.66–0.77] | <0.001 | 0.61 | [0.49–0.69] | 0.11 |
| GTV | ||||||
| RSF-RFVI | 0.77 | [0.69–0.80] | <0.001 | 0.67 | [0.60–0.77] | <0.001 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Leger, S.; Zwanenburg, A.; Leger, K.; Lohaus, F.; Linge, A.; Schreiber, A.; Kalinauskaite, G.; Tinhofer, I.; Guberina, N.; Guberina, M.; et al. Comprehensive Analysis of Tumour Sub-Volumes for Radiomic Risk Modelling in Locally Advanced HNSCC. Cancers 2020, 12, 3047. https://doi.org/10.3390/cancers12103047
Leger S, Zwanenburg A, Leger K, Lohaus F, Linge A, Schreiber A, Kalinauskaite G, Tinhofer I, Guberina N, Guberina M, et al. Comprehensive Analysis of Tumour Sub-Volumes for Radiomic Risk Modelling in Locally Advanced HNSCC. Cancers. 2020; 12(10):3047. https://doi.org/10.3390/cancers12103047
Chicago/Turabian StyleLeger, Stefan, Alex Zwanenburg, Karoline Leger, Fabian Lohaus, Annett Linge, Andreas Schreiber, Goda Kalinauskaite, Inge Tinhofer, Nika Guberina, Maja Guberina, and et al. 2020. "Comprehensive Analysis of Tumour Sub-Volumes for Radiomic Risk Modelling in Locally Advanced HNSCC" Cancers 12, no. 10: 3047. https://doi.org/10.3390/cancers12103047
APA StyleLeger, S., Zwanenburg, A., Leger, K., Lohaus, F., Linge, A., Schreiber, A., Kalinauskaite, G., Tinhofer, I., Guberina, N., Guberina, M., Balermpas, P., von der Grün, J., Ganswindt, U., Belka, C., Peeken, J. C., Combs, S. E., Boeke, S., Zips, D., Richter, C., ... Löck, S. (2020). Comprehensive Analysis of Tumour Sub-Volumes for Radiomic Risk Modelling in Locally Advanced HNSCC. Cancers, 12(10), 3047. https://doi.org/10.3390/cancers12103047





