TCA Cycle Rewiring as Emerging Metabolic Signature of Hepatocellular Carcinoma
Abstract
1. Introduction
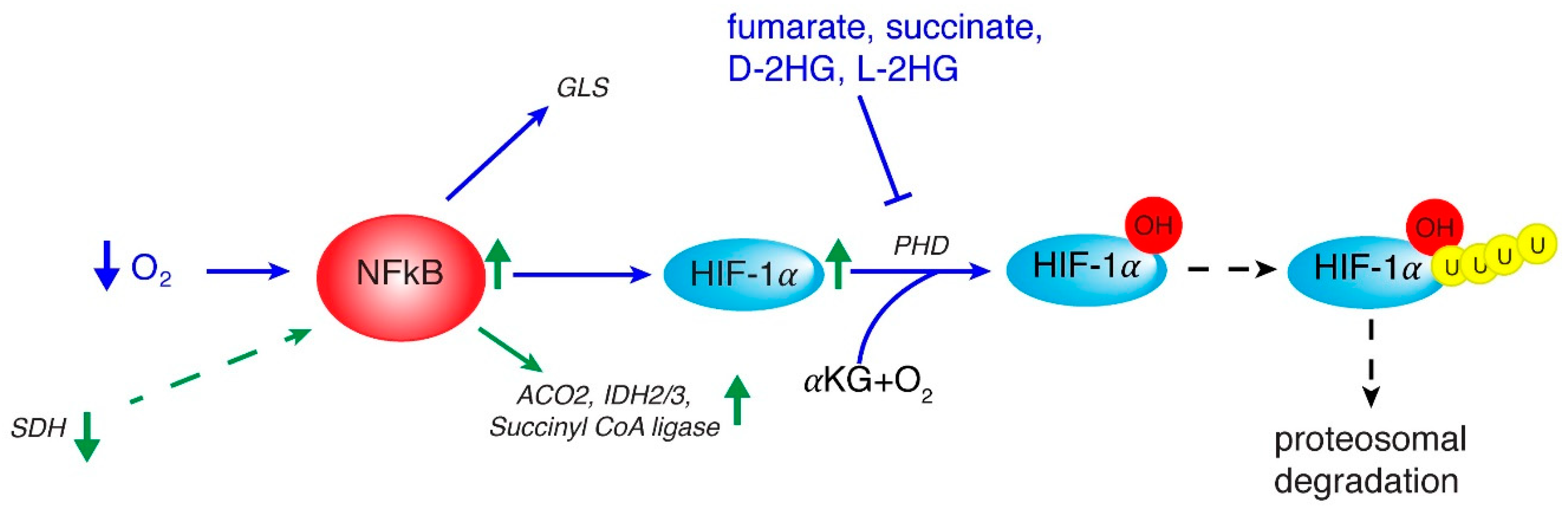
2. Inflammation and Role of NF-κB
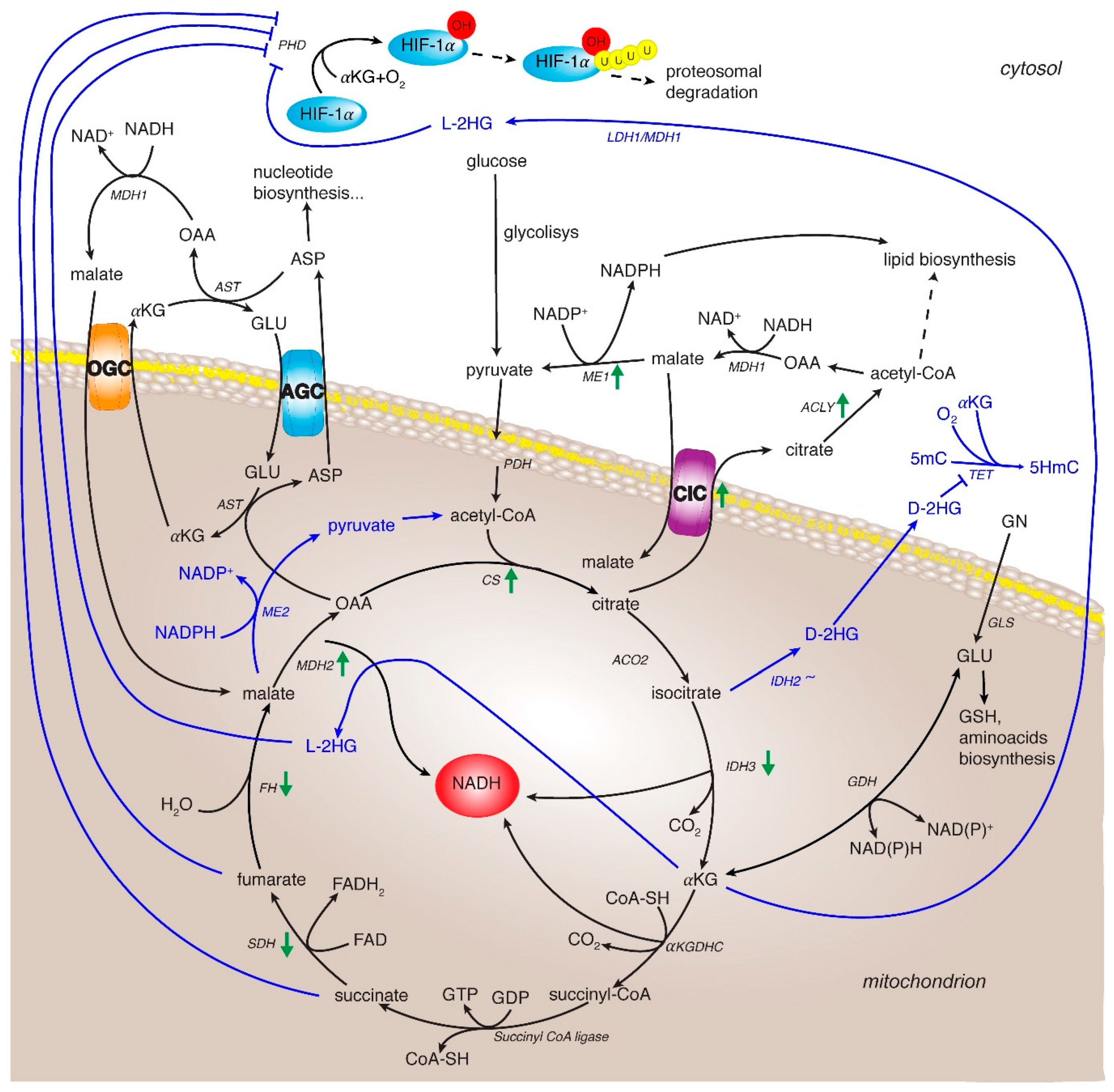
3. HIF in Regulating Metabolic Rewiring
4. TCA Cycle and Related Enzymes in HCC
4.1. Citrate Synthase
4.2. Isocitrate Dehydrogenase
4.3. α-Ketoglutarate Dehydrogenase Complex
4.4. Succinate Dehydrogenase
4.5. Fumarate Hydratase
4.6. Malate Dehydrogenase
4.7. Citrate/Pyruvate Shuttle
4.8. The Malate/Aspartate Shuttle (MAS)
4.9. Glutamine Metabolism
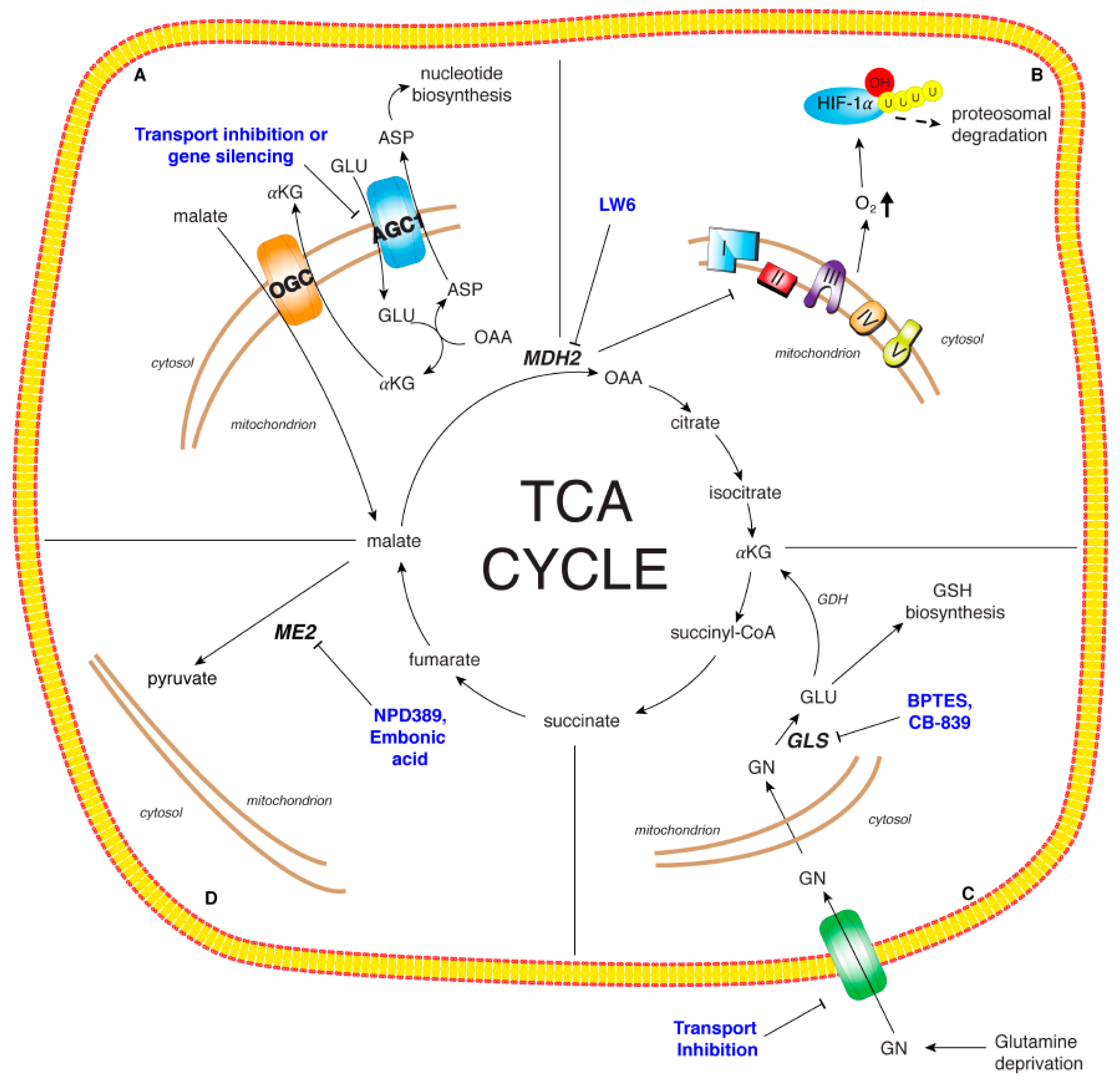
5. TCA Cycle and Related Enzymes as New Potential Therapeutic Targets
5.1. Targeting MDH2 and Malic Enzymes
5.2. Targeting MAS
5.3. Targeting Glutamine
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Mazzanti, R.; Gramantieri, L.; Bolondi, L. Hepatocellular carcinoma: Epidemiology and clinical aspects. Mol. Asp. Med. 2008, 29, 130–143. [Google Scholar] [CrossRef] [PubMed]
- Cornella, H.; Alsinet, C.; Villanueva, A. Molecular pathogenesis of hepatocellular carcinoma. Alcohol. Clin. Exp. Res. 2011, 35, 821–825. [Google Scholar] [CrossRef] [PubMed]
- Llovet, J.M.; Di Bisceglie, A.M.; Bruix, J.; Kramer, B.S.; Lencioni, R.; Zhu, A.X.; Sherman, M.; Schwartz, M.; Lotze, M.; Talwalkar, J.; et al. Design and endpoints of clinical trials in hepatocellular carcinoma. J. Natl. Cancer Inst. 2008, 100, 698–711. [Google Scholar] [CrossRef] [PubMed]
- Bruix, J.; Sherman, M. Management of hepatocellular carcinoma. Hepatology 2005, 42, 1208–1236. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Warburg, O.; Wind, F.; Negelein, E. The metabolism of tumors in the body. J. Gen. Physiol. 1927, 8, 519–530. [Google Scholar] [CrossRef]
- Almuhaideb, A.; Papathanasiou, N.; Bomanji, J. 18f-fdg pet/ct imaging in oncology. Ann. Saudi Med. 2011, 31, 3–13. [Google Scholar] [CrossRef]
- Pavlova, N.N.; Thompson, C.B. The emerging hallmarks of cancer metabolism. Cell Metab. 2016, 23, 27–47. [Google Scholar] [CrossRef]
- Warburg, O. On the origin of cancer cells. Science 1956, 123, 309–314. [Google Scholar] [CrossRef]
- Tan, A.S.; Baty, J.W.; Dong, L.F.; Bezawork-Geleta, A.; Endaya, B.; Goodwin, J.; Bajzikova, M.; Kovarova, J.; Peterka, M.; Yan, B.; et al. Mitochondrial genome acquisition restores respiratory function and tumorigenic potential of cancer cells without mitochondrial DNA. Cell Metab. 2015, 21, 81–94. [Google Scholar] [CrossRef]
- Eagle, H. The minimum vitamin requirements of the l and hela cells in tissue culture, the production of specific vitamin deficiencies, and their cure. J. Exp. Med. 1955, 102, 595–600. [Google Scholar] [CrossRef] [PubMed]
- Rivera, S.; Azcon-Bieto, J.; Lopez-Soriano, F.J.; Miralpeix, M.; Argiles, J.M. Amino acid metabolism in tumour-bearing mice. Biochem. J. 1988, 249, 443–449. [Google Scholar] [CrossRef] [PubMed]
- Yuneva, M.O.; Fan, T.W.; Allen, T.D.; Higashi, R.M.; Ferraris, D.V.; Tsukamoto, T.; Mates, J.M.; Alonso, F.J.; Wang, C.; Seo, Y.; et al. The metabolic profile of tumors depends on both the responsible genetic lesion and tissue type. Cell Metab. 2012, 15, 157–170. [Google Scholar] [CrossRef] [PubMed]
- Venneti, S.; Dunphy, M.P.; Zhang, H.; Pitter, K.L.; Zanzonico, P.; Campos, C.; Carlin, S.D.; La Rocca, G.; Lyashchenko, S.; Ploessl, K.; et al. Glutamine-based pet imaging facilitates enhanced metabolic evaluation of gliomas in vivo. Sci. Transl. Med. 2015, 7, 274ra17. [Google Scholar] [CrossRef]
- DeBerardinis, R.J.; Mancuso, A.; Daikhin, E.; Nissim, I.; Yudkoff, M.; Wehrli, S.; Thompson, C.B. Beyond aerobic glycolysis: Transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc. Natl. Acad. Sci. USA 2007, 104, 19345–19350. [Google Scholar] [CrossRef]
- Metallo, C.M.; Gameiro, P.A.; Bell, E.L.; Mattaini, K.R.; Yang, J.; Hiller, K.; Jewell, C.M.; Johnson, Z.R.; Irvine, D.J.; Guarente, L.; et al. Reductive glutamine metabolism by idh1 mediates lipogenesis under hypoxia. Nature 2011, 481, 380–384. [Google Scholar] [CrossRef]
- Menendez, J.A.; Lupu, R. Fatty acid synthase and the lipogenic phenotype in cancer pathogenesis. Nat. Rev. Cancer 2007, 7, 763–777. [Google Scholar] [CrossRef]
- Rysman, E.; Brusselmans, K.; Scheys, K.; Timmermans, L.; Derua, R.; Munck, S.; Van Veldhoven, P.P.; Waltregny, D.; Daniels, V.W.; Machiels, J.; et al. De novo lipogenesis protects cancer cells from free radicals and chemotherapeutics by promoting membrane lipid saturation. Cancer Res. 2010, 70, 8117–8126. [Google Scholar] [CrossRef]
- Kaplan, R.S.; Morris, H.P.; Coleman, P.S. Kinetic characteristics of citrate influx and efflux with mitochondria from morris hepatomas 3924a and 16. Cancer Res. 1982, 42, 4399–4407. [Google Scholar]
- Catalina-Rodriguez, O.; Kolukula, V.K.; Tomita, Y.; Preet, A.; Palmieri, F.; Wellstein, A.; Byers, S.; Giaccia, A.J.; Glasgow, E.; Albanese, C.; et al. The mitochondrial citrate transporter, cic, is essential for mitochondrial homeostasis. Oncotarget 2012, 3, 1220–1235. [Google Scholar] [CrossRef]
- Pope, E.D., 3rd; Kimbrough, E.O.; Vemireddy, L.P.; Surapaneni, P.K.; Copland, J.A., 3rd; Mody, K. Aberrant lipid metabolism as a therapeutic target in liver cancer. Expert Opin. Ther. Targets 2019, 23, 473–483. [Google Scholar] [CrossRef] [PubMed]
- Palmieri, F. The mitochondrial transporter family (slc25): Physiological and pathological implications. Pflug. Arch. Eur. J. Physiol. 2004, 447, 689–709. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, L.B.; Luengo, A.; Danai, L.V.; Bush, L.N.; Diehl, F.F.; Hosios, A.M.; Lau, A.N.; Elmiligy, S.; Malstrom, S.; Lewis, C.A.; et al. Aspartate is an endogenous metabolic limitation for tumour growth. Nat. Cell Biol. 2018, 20, 782–788. [Google Scholar] [CrossRef] [PubMed]
- Tannahill, G.M.; Curtis, A.M.; Adamik, J.; Palsson-McDermott, E.M.; McGettrick, A.F.; Goel, G.; Frezza, C.; Bernard, N.J.; Kelly, B.; Foley, N.H.; et al. Succinate is an inflammatory signal that induces il-1beta through HIF-1alpha. Nature 2013, 496, 238–242. [Google Scholar] [CrossRef]
- Myllyla, R.; Tuderman, L.; Kivirikko, K.I. Mechanism of the prolyl hydroxylase reaction. 2. Kinetic analysis of the reaction sequence. Eur. J. Biochem. 1977, 80, 349–357. [Google Scholar] [CrossRef]
- Selak, M.A.; Armour, S.M.; MacKenzie, E.D.; Boulahbel, H.; Watson, D.G.; Mansfield, K.D.; Pan, Y.; Simon, M.C.; Thompson, C.B.; Gottlieb, E. Succinate links tca cycle dysfunction to oncogenesis by inhibiting HIF-α prolyl hydroxylase. Cancer Cell 2005, 7, 77–85. [Google Scholar] [CrossRef]
- Karin, M. NF-κB as a critical link between inflammation and cancer. Cold Spring Harb. Perspect. Biol. 2009, 1, a000141. [Google Scholar] [CrossRef]
- Merle, P.; Trepo, C. Molecular mechanisms underlying hepatocellular carcinoma. Viruses 2009, 1, 852–872. [Google Scholar] [CrossRef]
- Qian, B.Z.; Pollard, J.W. Macrophage diversity enhances tumor progression and metastasis. Cell 2010, 141, 39–51. [Google Scholar] [CrossRef]
- Grivennikov, S.I.; Greten, F.R.; Karin, M. Immunity, inflammation, and cancer. Cell 2010, 140, 883–899. [Google Scholar] [CrossRef]
- Pollard, J.W. Tumour-educated macrophages promote tumour progression and metastasis. Nat. Rev. Cancer 2004, 4, 71–78. [Google Scholar] [CrossRef] [PubMed]
- Dolcet, X.; Llobet, D.; Pallares, J.; Matias-Guiu, X. NF-κB in development and progression of human cancer. Virchows Arch. Int. J. Pathol. 2005, 446, 475–482. [Google Scholar] [CrossRef] [PubMed]
- Gilmore, T.D. Multiple mutations contribute to the oncogenicity of the retroviral oncoprotein v-rel. Oncogene 1999, 18, 6925–6937. [Google Scholar] [CrossRef] [PubMed]
- Cabannes, E.; Khan, G.; Aillet, F.; Jarrett, R.F.; Hay, R.T. Mutations in the ikba gene in hodgkin’s disease suggest a tumour suppressor role for IκBα. Oncogene 1999, 18, 3063–3070. [Google Scholar] [CrossRef]
- Migliazza, A.; Lombardi, L.; Rocchi, M.; Trecca, D.; Chang, C.C.; Antonacci, R.; Fracchiolla, N.S.; Ciana, P.; Maiolo, A.T.; Neri, A. Heterogeneous chromosomal aberrations generate 3’ truncations of the nfkb2/lyt-10 gene in lymphoid malignancies. Blood 1994, 84, 3850–3860. [Google Scholar] [CrossRef]
- Houldsworth, J.; Mathew, S.; Rao, P.H.; Dyomina, K.; Louie, D.C.; Parsa, N.; Offit, K.; Chaganti, R.S. Rel proto-oncogene is frequently amplified in extranodal diffuse large cell lymphoma. Blood 1996, 87, 25–29. [Google Scholar] [CrossRef]
- Mukhopadhyay, T.; Roth, J.A.; Maxwell, S.A. Altered expression of the p50 subunit of the NF-κB transcription factor complex in non-small cell lung carcinoma. Oncogene 1995, 11, 999–1003. [Google Scholar]
- Romieu-Mourez, R.; Kim, D.W.; Shin, S.M.; Demicco, E.G.; Landesman-Bollag, E.; Seldin, D.C.; Cardiff, R.D.; Sonenshein, G.E. Mouse mammary tumor virus c-rel transgenic mice develop mammary tumors. Mol. Cell. Biol. 2003, 23, 5738–5754. [Google Scholar] [CrossRef]
- Nair, A.; Venkatraman, M.; Maliekal, T.T.; Nair, B.; Karunagaran, D. NF-κB is constitutively activated in high-grade squamous intraepithelial lesions and squamous cell carcinomas of the human uterine cervix. Oncogene 2003, 22, 50–58. [Google Scholar] [CrossRef]
- Sasaki, N.; Morisaki, T.; Hashizume, K.; Yao, T.; Tsuneyoshi, M.; Noshiro, H.; Nakamura, K.; Yamanaka, T.; Uchiyama, A.; Tanaka, M.; et al. Nuclear factor-kappab p65 (rela) transcription factor is constitutively activated in human gastric carcinoma tissue. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2001, 7, 4136–4142. [Google Scholar]
- Elsharkawy, A.M.; Mann, D.A. Nuclear factor-kappab and the hepatic inflammation-fibrosis-cancer axis. Hepatology 2007, 46, 590–597. [Google Scholar] [CrossRef] [PubMed]
- Mollica, M.P.; Lionetti, L.; Putti, R.; Cavaliere, G.; Gaita, M.; Barletta, A. From chronic overfeeding to hepatic injury: Role of endoplasmic reticulum stress and inflammation. Nutr. Metab. Cardiovasc. Dis. Nmcd 2011, 21, 222–230. [Google Scholar] [CrossRef] [PubMed]
- Forbes, S.J.; Parola, M. Liver fibrogenic cells. Best Pract. Res. Clin. Gastroenterol. 2011, 25, 207–217. [Google Scholar] [CrossRef] [PubMed]
- Racanelli, V.; Rehermann, B. The liver as an immunological organ. Hepatology 2006, 43, S54–S62. [Google Scholar] [CrossRef] [PubMed]
- Thirunavukkarasu, C.; Watkins, S.C.; Gandhi, C.R. Mechanisms of endotoxin-induced NO, IL-6, and TNF-α production in activated rat hepatic stellate cells: Role of p38 mapk. Hepatology 2006, 44, 389–398. [Google Scholar] [CrossRef] [PubMed]
- Cani, P.D.; Bibiloni, R.; Knauf, C.; Waget, A.; Neyrinck, A.M.; Delzenne, N.M.; Burcelin, R. Changes in gut microbiota control metabolic endotoxemia-induced inflammation in high-fat diet-induced obesity and diabetes in mice. Diabetes 2008, 57, 1470–1481. [Google Scholar] [CrossRef] [PubMed]
- Cai, D.; Yuan, M.; Frantz, D.F.; Melendez, P.A.; Hansen, L.; Lee, J.; Shoelson, S.E. Local and systemic insulin resistance resulting from hepatic activation of ikk-beta and NF-κB. Nat. Med. 2005, 11, 183–190. [Google Scholar] [CrossRef]
- Tan, C.E.; Wang, K.W.; Chew, L.S.; Rajasoorya, C.; Lim, H.S. Manifestations and outcome of treatment of thyrotoxicosis: A clinical study. Ann. Acad. Med. Singap. 1990, 19, 798–801. [Google Scholar]
- Johnson, R.F.; Perkins, N.D. Nuclear factor-kappab, p53, and mitochondria: Regulation of cellular metabolism and the warburg effect. Trends Biochem. Sci. 2012, 37, 317–324. [Google Scholar] [CrossRef]
- Koong, A.C.; Chen, E.Y.; Giaccia, A.J. Hypoxia causes the activation of nuclear factor kappa b through the phosphorylation of IκBα on tyrosine residues. Cancer Res. 1994, 54, 1425–1430. [Google Scholar]
- Van Uden, P.; Kenneth, N.S.; Rocha, S. Regulation of hypoxia-inducible factor-1α by NF-κB. Biochem J. 2008, 412, 477–484. [Google Scholar] [CrossRef] [PubMed]
- Kawauchi, K.; Araki, K.; Tobiume, K.; Tanaka, N. P53 regulates glucose metabolism through an IKK-NF-κB pathway and inhibits cell transformation. Nat. Cell Biol. 2008, 10, 611–618. [Google Scholar] [CrossRef] [PubMed]
- Zhou, F.; Xu, X.; Wu, J.; Wang, D.; Wang, J. NF-κB controls four genes encoding core enzymes of tricarboxylic acid cycle. Gene 2017, 621, 12–20. [Google Scholar] [CrossRef] [PubMed]
- Fitzpatrick, S.F. Immunometabolism and sepsis: A role for hif? Front. Mol. Biosci. 2019, 6, 85. [Google Scholar] [CrossRef]
- Lisy, K.; Peet, D.J. Turn me on: Regulating hif transcriptional activity. Cell Death Differ. 2008, 15, 642–649. [Google Scholar] [CrossRef] [PubMed]
- Gordan, J.D.; Simon, M.C. Hypoxia-inducible factors: Central regulators of the tumor phenotype. Curr. Opin. Genet. Dev. 2007, 17, 71–77. [Google Scholar] [CrossRef]
- McKeown, S.R. Defining normoxia, physoxia and hypoxia in tumours-implications for treatment response. Br. J. Radiol. 2014, 87, 20130676. [Google Scholar] [CrossRef]
- Semenza, G.L. Hypoxia-inducible factors in physiology and medicine. Cell 2012, 148, 399–408. [Google Scholar] [CrossRef]
- Kim, J.W.; Tchernyshyov, I.; Semenza, G.L.; Dang, C.V. Hif-1-mediated expression of pyruvate dehydrogenase kinase: A metabolic switch required for cellular adaptation to hypoxia. Cell Metab. 2006, 3, 177–185. [Google Scholar] [CrossRef]
- McFate, T.; Mohyeldin, A.; Lu, H.; Thakar, J.; Henriques, J.; Halim, N.D.; Wu, H.; Schell, M.J.; Tsang, T.M.; Teahan, O.; et al. Pyruvate dehydrogenase complex activity controls metabolic and malignant phenotype in cancer cells. J. Biol. Chem. 2008, 283, 22700–22708. [Google Scholar] [CrossRef]
- Zhong, H.; Chiles, K.; Feldser, D.; Laughner, E.; Hanrahan, C.; Georgescu, M.M.; Simons, J.W.; Semenza, G.L. Modulation of hypoxia-inducible factor 1alpha expression by the epidermal growth factor/phosphatidylinositol 3-kinase/pten/akt/frap pathway in human prostate cancer cells: Implications for tumor angiogenesis and therapeutics. Cancer Res. 2000, 60, 1541–1545. [Google Scholar] [PubMed]
- Dai, C.X.; Gao, Q.; Qiu, S.J.; Ju, M.J.; Cai, M.Y.; Xu, Y.F.; Zhou, J.; Zhang, B.H.; Fan, J. Hypoxia-inducible factor-1 alpha, in association with inflammation, angiogenesis and myc, is a critical prognostic factor in patients with hcc after surgery. BMC Cancer 2009, 9, 418. [Google Scholar] [CrossRef] [PubMed]
- Xiang, Z.L.; Zeng, Z.C.; Fan, J.; Tang, Z.Y.; He, J.; Zeng, H.Y.; Chang, J.Y. The expression of hif-1alpha in primary hepatocellular carcinoma and its correlation with radiotherapy response and clinical outcome. Mol. Biol. Rep. 2012, 39, 2021–2029. [Google Scholar] [CrossRef] [PubMed]
- Guaragnella, N.; Giannattasio, S.; Moro, L. Mitochondrial dysfunction in cancer chemoresistance. Biochem. Pharmacol. 2014, 92, 62–72. [Google Scholar] [CrossRef]
- Hamaguchi, T.; Iizuka, N.; Tsunedomi, R.; Hamamoto, Y.; Miyamoto, T.; Iida, M.; Tokuhisa, Y.; Sakamoto, K.; Takashima, M.; Tamesa, T.; et al. Glycolysis module activated by hypoxia-inducible factor 1alpha is related to the aggressive phenotype of hepatocellular carcinoma. Int. J. Oncol. 2008, 33, 725–731. [Google Scholar]
- Wiegand, G.; Remington, S.J. Citrate synthase: Structure, control, and mechanism. Annu. Rev. Biophys. Biophys. Chem. 1986, 15, 97–117. [Google Scholar] [CrossRef]
- Chen, L.; Liu, T.; Zhou, J.; Wang, Y.; Wang, X.; Di, W.; Zhang, S. Citrate synthase expression affects tumor phenotype and drug resistance in human ovarian carcinoma. PLoS ONE 2014, 9, e115708. [Google Scholar] [CrossRef]
- Schlichtholz, B.; Turyn, J.; Goyke, E.; Biernacki, M.; Jaskiewicz, K.; Sledzinski, Z.; Swierczynski, J. Enhanced citrate synthase activity in human pancreatic cancer. Pancreas 2005, 30, 99–104. [Google Scholar] [CrossRef]
- El-Ashmawy, N.E.; El-Bahrawy, H.A.; Shamloula, M.M.; El-Feky, O.A. Biochemical/metabolic changes associated with hepatocellular carcinoma development in mice. Tumour Biol. J. Int. Soc. Oncodev. Biol. Med. 2014, 35, 5459–5466. [Google Scholar] [CrossRef]
- Zhang, J.; Baddoo, M.; Han, C.; Strong, M.J.; Cvitanovic, J.; Moroz, K.; Dash, S.; Flemington, E.K.; Wu, T. Gene network analysis reveals a novel 22-gene signature of carbon metabolism in hepatocellular carcinoma. Oncotarget 2016, 7, 49232–49245. [Google Scholar] [CrossRef]
- Gao, R.; Cheng, J.; Fan, C.; Shi, X.; Cao, Y.; Sun, B.; Ding, H.; Hu, C.; Dong, F.; Yan, X. Serum metabolomics to identify the liver disease-specific biomarkers for the progression of hepatitis to hepatocellular carcinoma. Sci. Rep. 2015, 5, 18175. [Google Scholar] [CrossRef] [PubMed]
- Stoddard, B.L.; Dean, A.; Koshland, D.E. Structure of isocitrate dehydrogenase with isocitrate, nicotinamide adenine dinucleotide phosphate, and calcium at 2.5-a resolution: A pseudo-michaelis ternary complex. Biochemistry 1993, 32, 9310–9316. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Zhao, J.; Xu, Z.; Peng, B.; Huang, Q.; Arnold, E.; Ding, J. Structures of human cytosolic nadp-dependent isocitrate dehydrogenase reveal a novel self-regulatory mechanism of activity. J. Biol. Chem. 2004, 279, 33946–33957. [Google Scholar] [CrossRef] [PubMed]
- Ramachandran, N.; Colman, R.F. Chemical characterization of distinct subunits of pig heart dpn-specific isocitrate dehydrogenase. J. Biol. Chem. 1980, 255, 8859–8864. [Google Scholar] [PubMed]
- Anderson, N.M.; Mucka, P.; Kern, J.G.; Feng, H. The emerging role and targetability of the tca cycle in cancer metabolism. Protein Cell 2018, 9, 216–237. [Google Scholar] [CrossRef]
- Yen, K.E.; Bittinger, M.A.; Su, S.M.; Fantin, V.R. Cancer-associated idh mutations: Biomarker and therapeutic opportunities. Oncogene 2010, 29, 6409–6417. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhang, L.; Zhang, Y.; Meng, B.; Ying, W.; Qian, X. Identification of hedgehog signaling as a potential oncogenic driver in an aggressive subclass of human hepatocellular carcinoma: A reanalysis of the tcga cohort. Sci. China Life Sci. 2019, 62, 1481–1491. [Google Scholar] [CrossRef]
- Xu, W.; Yang, H.; Liu, Y.; Yang, Y.; Wang, P.; Kim, S.H.; Ito, S.; Yang, C.; Wang, P.; Xiao, M.T.; et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of alpha-ketoglutarate-dependent dioxygenases. Cancer Cell 2011, 19, 17–30. [Google Scholar] [CrossRef]
- Cervello, M.; Bachvarov, D.; Lampiasi, N.; Cusimano, A.; Azzolina, A.; McCubrey, J.A.; Montalto, G. Novel combination of sorafenib and celecoxib provides synergistic anti-proliferative and pro-apoptotic effects in human liver cancer cells. PLoS ONE 2013, 8, e65569. [Google Scholar] [CrossRef]
- Laurenti, G.; Tennant, D.A. Isocitrate dehydrogenase (idh), succinate dehydrogenase (sdh), fumarate hydratase (fh): Three players for one phenotype in cancer? Biochem. Soc. Trans. 2016, 44, 1111–1116. [Google Scholar] [CrossRef]
- Lee, S.M.; Koh, H.J.; Park, D.C.; Song, B.J.; Huh, T.L.; Park, J.W. Cytosolic nadp(+)-dependent isocitrate dehydrogenase status modulates oxidative damage to cells. Free Radic. Biol. Med. 2002, 32, 1185–1196. [Google Scholar] [CrossRef]
- Wagenknecht, T.; Francis, N.; DeRosier, D.J. Alpha-ketoglutarate dehydrogenase complex may be heterogeneous in quaternary structure. J. Mol. Biol. 1983, 165, 523–539. [Google Scholar] [CrossRef]
- Sheu, K.F.; Blass, J.P. The alpha-ketoglutarate dehydrogenase complex. Ann. N. Y. Acad. Sci. 1999, 893, 61–78. [Google Scholar] [CrossRef] [PubMed]
- Lawlis, V.B.; Roche, T.E. Regulation of bovine kidney alpha-ketoglutarate dehydrogenase complex by calcium ion and adenine nucleotides. Effects on s0.5 for alpha-ketoglutarate. Biochemistry 1981, 20, 2512–2518. [Google Scholar] [CrossRef] [PubMed]
- McCormack, J.G.; Denton, R.M. The effects of calcium ions and adenine nucleotides on the activity of pig heart 2-oxoglutarate dehydrogenase complex. Biochem J. 1979, 180, 533–544. [Google Scholar] [CrossRef] [PubMed]
- Hamada, M.; Koike, K.; Nakaula, Y.; Hiraoka, T.; Koike, M. A kinetic study of the alpha-keto acid dehydrogenase complexes from pig heart mitochondria. J. Biochem. 1975, 77, 1047–1056. [Google Scholar] [CrossRef] [PubMed]
- Kiselevsky, Y.V.; Ostrovtsova, S.A.; Strumilo, S.A. Kinetic characterization of the pyruvate and oxoglutarate dehydrogenase complexes from human heart. Acta Biochim. Pol. 1990, 37, 135–139. [Google Scholar]
- Schoolwerth, A.C.; LaNoue, K.F. Control of ammoniagenesis by alpha-ketoglutarate in rat kidney mitochondria. Am. J. Physiol. 1983, 244, F399–F408. [Google Scholar] [CrossRef]
- Burr, S.P.; Costa, A.S.; Grice, G.L.; Timms, R.T.; Lobb, I.T.; Freisinger, P.; Dodd, R.B.; Dougan, G.; Lehner, P.J.; Frezza, C.; et al. Mitochondrial protein lipoylation and the 2-oxoglutarate dehydrogenase complex controls hif1alpha stability in aerobic conditions. Cell Metab 2016, 24, 740–752. [Google Scholar] [CrossRef]
- Vatrinet, R.; Leone, G.; De Luise, M.; Girolimetti, G.; Vidone, M.; Gasparre, G.; Porcelli, A.M. The alpha-ketoglutarate dehydrogenase complex in cancer metabolic plasticity. Cancer Metab. 2017, 5, 3. [Google Scholar] [CrossRef]
- Oldham, W.M.; Clish, C.B.; Yang, Y.; Loscalzo, J. Hypoxia-mediated increases in l-2-hydroxyglutarate coordinate the metabolic response to reductive stress. Cell Metab. 2015, 22, 291–303. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Guo, Y.R.; Liu, K.; Yin, Z.; Liu, R.; Xia, Y.; Tan, L.; Yang, P.; Lee, J.H.; Li, X.J.; et al. Kat2a coupled with the alpha-kgdh complex acts as a histone h3 succinyltransferase. Nature 2017, 552, 273–277. [Google Scholar] [CrossRef] [PubMed]
- Rasheed, M.; Tarjan, G. Succinate dehydrogenase complex: An updated review. Arch. Pathol. Lab. Med. 2018, 142, 1564–1570. [Google Scholar] [CrossRef] [PubMed]
- Baysal, B.E.; Ferrell, R.E.; Willett-Brozick, J.E.; Lawrence, E.C.; Myssiorek, D.; Bosch, A.; van der Mey, A.; Taschner, P.E.; Rubinstein, W.S.; Myers, E.N.; et al. Mutations in sdhd, a mitochondrial complex ii gene, in hereditary paraganglioma. Science 2000, 287, 848–851. [Google Scholar] [CrossRef]
- Gottlieb, E.; Tomlinson, I.P. Mitochondrial tumour suppressors: A genetic and biochemical update. Nat. Rev. Cancer 2005, 5, 857–866. [Google Scholar] [CrossRef]
- Bardella, C.; Pollard, P.J.; Tomlinson, I. Sdh mutations in cancer. Biochim. Biophys. Acta 2011, 1807, 1432–1443. [Google Scholar] [CrossRef]
- Shimizu, T.; Inoue, K.; Hachiya, H.; Shibuya, N.; Shimoda, M.; Kubota, K. Frequent alteration of the protein synthesis of enzymes for glucose metabolism in hepatocellular carcinomas. J. Gastroenterol. 2014, 49, 1324–1332. [Google Scholar] [CrossRef]
- Tseng, P.L.; Wu, W.H.; Hu, T.H.; Chen, C.W.; Cheng, H.C.; Li, C.F.; Tsai, W.H.; Tsai, H.J.; Hsieh, M.C.; Chuang, J.H.; et al. Decreased succinate dehydrogenase b in human hepatocellular carcinoma accelerates tumor malignancy by inducing the warburg effect. Sci. Rep. 2018, 8, 3081. [Google Scholar] [CrossRef]
- Li, J.; Liang, N.; Long, X.; Zhao, J.; Yang, J.; Du, X.; Yang, T.; Yuan, P.; Huang, X.; Zhang, J.; et al. Sdhc-related deficiency of sdh complex activity promotes growth and metastasis of hepatocellular carcinoma via ros/nfkappab signaling. Cancer Lett. 2019, 461, 44–55. [Google Scholar] [CrossRef]
- Cho, E.H. Succinate as a regulator of hepatic stellate cells in liver fibrosis. Front. Endocrinol. 2018, 9, 455. [Google Scholar] [CrossRef]
- Mann, P.J.; Woolf, B. The action of salts on fumarase. I. Biochem J. 1930, 24, 427–434. [Google Scholar] [CrossRef] [PubMed]
- Stepinski, J.; Bizon, D.; Piec, G.; Angielski, S. The purine nucleotide cycle activity in renal cortex and medulla. Am. J. Kidney Dis. Off. J. Natl. Kidney Found. 1989, 14, 307–309. [Google Scholar] [CrossRef]
- Isaacs, J.S.; Jung, Y.J.; Mole, D.R.; Lee, S.; Torres-Cabala, C.; Chung, Y.L.; Merino, M.; Trepel, J.; Zbar, B.; Toro, J.; et al. Hif overexpression correlates with biallelic loss of fumarate hydratase in renal cancer: Novel role of fumarate in regulation of hif stability. Cancer Cell 2005, 8, 143–153. [Google Scholar] [CrossRef] [PubMed]
- Pollard, P.J.; Briere, J.J.; Alam, N.A.; Barwell, J.; Barclay, E.; Wortham, N.C.; Hunt, T.; Mitchell, M.; Olpin, S.; Moat, S.J.; et al. Accumulation of krebs cycle intermediates and over-expression of hif1alpha in tumours which result from germline fh and sdh mutations. Hum. Mol. Genet. 2005, 14, 2231–2239. [Google Scholar] [CrossRef] [PubMed]
- Jones, P.A.; Baylin, S.B. The epigenomics of cancer. Cell 2007, 128, 683–692. [Google Scholar] [CrossRef] [PubMed]
- Wong, C.C.; Qian, Y.; Yu, J. Interplay between epigenetics and metabolism in oncogenesis: Mechanisms and therapeutic approaches. Oncogene 2017, 36, 3359–3374. [Google Scholar] [CrossRef]
- Letouze, E.; Martinelli, C.; Loriot, C.; Burnichon, N.; Abermil, N.; Ottolenghi, C.; Janin, M.; Menara, M.; Nguyen, A.T.; Benit, P.; et al. Sdh mutations establish a hypermethylator phenotype in paraganglioma. Cancer Cell 2013, 23, 739–752. [Google Scholar] [CrossRef]
- Leshets, M.; Silas, Y.B.H.; Lehming, N.; Pines, O. Fumarase: From the tca cycle to DNA damage response and tumor suppression. Front. Mol. Biosci. 2018, 5, 68. [Google Scholar] [CrossRef]
- Yogev, O.; Yogev, O.; Singer, E.; Shaulian, E.; Goldberg, M.; Fox, T.D.; Pines, O. Fumarase: A mitochondrial metabolic enzyme and a cytosolic/nuclear component of the DNA damage response. PLoS Biol. 2010, 8, e1000328. [Google Scholar] [CrossRef]
- Lee, W.C.; Chou, H.S.; Wu, T.J.; Lee, C.F.; Hsu, P.Y.; Hsu, H.Y.; Wu, T.H.; Chan, K.M. Down-regulation of metabolic proteins in hepatocellular carcinoma with portal vein thrombosis. Clin. Proteom. 2017, 14, 29. [Google Scholar] [CrossRef]
- Mullinax, T.R.; Mock, J.N.; McEvily, A.J.; Harrison, J.H. Regulation of mitochondrial malate dehydrogenase. Evidence for an allosteric citrate-binding site. J. Biol. Chem. 1982, 257, 13233–13239. [Google Scholar] [PubMed]
- Fahien, L.A.; Kmiotek, E.H.; MacDonald, M.J.; Fibich, B.; Mandic, M. Regulation of malate dehydrogenase activity by glutamate, citrate, alpha-ketoglutarate, and multienzyme interaction. J. Biol. Chem. 1988, 263, 10687–10697. [Google Scholar] [PubMed]
- Gelpi, J.L.; Dordal, A.; Montserrat, J.; Mazo, A.; Cortes, A. Kinetic studies of the regulation of mitochondrial malate dehydrogenase by citrate. Biochem. J. 1992, 283, 289–297. [Google Scholar] [CrossRef] [PubMed]
- Morgunov, I.; Srere, P.A. Interaction between citrate synthase and malate dehydrogenase. Substrate channeling of oxaloacetate. J. Biol. Chem. 1998, 273, 29540–29544. [Google Scholar] [CrossRef] [PubMed]
- Mansouri, S.; Shahriari, A.; Kalantar, H.; Moini Zanjani, T.; Haghi Karamallah, M. Role of malate dehydrogenase in facilitating lactate dehydrogenase to support the glycolysis pathway in tumors. Biomed. Rep. 2017, 6, 463–467. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Harvey, C.T.; Geng, H.; Xue, C.; Chen, V.; Beer, T.M.; Qian, D.Z. Malate dehydrogenase 2 confers docetaxel resistance via regulations of jnk signaling and oxidative metabolism. Prostate 2013, 73, 1028–1037. [Google Scholar] [CrossRef]
- Lee, K.; Ban, H.S.; Naik, R.; Hong, Y.S.; Son, S.; Kim, B.K.; Xia, Y.; Song, K.B.; Lee, H.S.; Won, M. Identification of malate dehydrogenase 2 as a target protein of the hif-1 inhibitor lw6 using chemical probes. Angew. Chem. 2013, 52, 10286–10289. [Google Scholar] [CrossRef]
- Iacobazzi, V.; Infantino, V. Citrate—new functions for an old metabolite. Biol. Chem. 2014, 395, 387–399. [Google Scholar] [CrossRef]
- Iacobazzi, V.; Infantino, V.; Palmieri, F. Epigenetic mechanisms and sp1 regulate mitochondrial citrate carrier gene expression. Biochem. Biophys. Res. Commun. 2008, 376, 15–20. [Google Scholar] [CrossRef]
- Infantino, V.; Pierri, C.L.; Iacobazzi, V. Metabolic routes in inflammation: The citrate pathway and its potential as therapeutic target. In Current Medicinal Chemistry; BenthamScience: Sharjah, UAE, 2018. [Google Scholar]
- Iacobazzi, V.; Infantino, V.; Bisaccia, F.; Castegna, A.; Palmieri, F. Role of foxa in mitochondrial citrate carrier gene expression and insulin secretion. Biochem. Biophys. Res. Commun. 2009, 385, 220–224. [Google Scholar] [CrossRef]
- Infantino, V.; Convertini, P.; Cucci, L.; Panaro, M.A.; Di Noia, M.A.; Calvello, R.; Palmieri, F.; Iacobazzi, V. The mitochondrial citrate carrier: A new player in inflammation. Biochem. J. 2011, 438, 433–436. [Google Scholar] [CrossRef] [PubMed]
- Infantino, V.; Iacobazzi, V.; Menga, A.; Avantaggiati, M.L.; Palmieri, F. A key role of the mitochondrial citrate carrier (slc25a1) in tnfalpha- and ifngamma-triggered inflammation. Biochim. Biophys. Acta 2014, 1839, 1217–1225. [Google Scholar] [CrossRef] [PubMed]
- Infantino, V.; Iacobazzi, V.; Palmieri, F.; Menga, A. Atp-citrate lyase is essential for macrophage inflammatory response. Biochem. Biophys. Res. Commun. 2013, 440, 105–111. [Google Scholar] [CrossRef] [PubMed]
- Lewis, C.A.; Parker, S.J.; Fiske, B.P.; McCloskey, D.; Gui, D.Y.; Green, C.R.; Vokes, N.I.; Feist, A.M.; Vander Heiden, M.G.; Metallo, C.M. Tracing compartmentalized nadph metabolism in the cytosol and mitochondria of mammalian cells. Mol. Cell 2014, 55, 253–263. [Google Scholar] [CrossRef] [PubMed]
- Fan, J.; Ye, J.; Kamphorst, J.J.; Shlomi, T.; Thompson, C.B.; Rabinowitz, J.D. Quantitative flux analysis reveals folate-dependent nadph production. Nature 2014, 510, 298–302. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Sun, S.; Neufeld, C.I.; Perez-Ramirez, B.; Xu, Q. Reactive oxygen species-responsive protein modification and its intracellular delivery for targeted cancer therapy. Angew. Chem. 2014, 53, 13444–13448. [Google Scholar] [CrossRef]
- Wen, D.; Liu, D.; Tang, J.; Dong, L.; Liu, Y.; Tao, Z.; Wan, J.; Gao, D.; Wang, L.; Sun, H.; et al. Malic enzyme 1 induces epithelial-mesenchymal transition and indicates poor prognosis in hepatocellular carcinoma. Tumour Biol. J. Int. Soc. Oncodev. Biol. Med. 2015, 36, 6211–6221. [Google Scholar] [CrossRef]
- Lu, Y.X.; Ju, H.Q.; Liu, Z.X.; Chen, D.L.; Wang, Y.; Zhao, Q.; Wu, Q.N.; Zeng, Z.L.; Qiu, H.B.; Hu, P.S.; et al. Me1 regulates nadph homeostasis to promote gastric cancer growth and metastasis. Cancer Res. 2018, 78, 1972–1985. [Google Scholar] [CrossRef]
- Zheng, F.J.; Ye, H.B.; Wu, M.S.; Lian, Y.F.; Qian, C.N.; Zeng, Y.X. Repressing malic enzyme 1 redirects glucose metabolism, unbalances the redox state, and attenuates migratory and invasive abilities in nasopharyngeal carcinoma cell lines. Chin. J. Cancer 2012, 31, 519–531. [Google Scholar] [CrossRef]
- Chakrabarti, G. Mutant kras associated malic enzyme 1 expression is a predictive marker for radiation therapy response in non-small cell lung cancer. Radiat. Oncol. 2015, 10, 145. [Google Scholar] [CrossRef]
- Vander Heiden, M.G.; DeBerardinis, R.J. Understanding the intersections between metabolism and cancer biology. Cell 2017, 168, 657–669. [Google Scholar] [CrossRef] [PubMed]
- Palmieri, F. The mitochondrial transporter family slc25: Identification, properties and physiopathology. Mol. Asp. Med. 2013, 34, 465–484. [Google Scholar] [CrossRef] [PubMed]
- Del Arco, A.; Morcillo, J.; Martinez-Morales, J.R.; Galian, C.; Martos, V.; Bovolenta, P.; Satrustegui, J. Expression of the aspartate/glutamate mitochondrial carriers aralar1 and citrin during development and in adult rat tissues. Eur. J. Biochem. 2002, 269, 3313–3320. [Google Scholar] [CrossRef] [PubMed]
- Greenhouse, W.V.; Lehninger, A.L. Occurrence of the malate-aspartate shuttle in various tumor types. Cancer Res. 1976, 36, 1392–1396. [Google Scholar] [PubMed]
- Convertini, P.; Todisco, S.; De Santis, F.; Pappalardo, I.; Iacobazzi, D.; Castiglione Morelli, M.A.; Fondufe-Mittendorf, Y.N.; Martelli, G.; Palmieri, F.; Infantino, V. Transcriptional regulation factors of the human mitochondrial aspartate/glutamate carrier gene, isoform 2 (slc25a13): Usf1 as basal factor and foxa2 as activator in liver cells. Int. J. Mol. Sci. 2019, 20, 1888. [Google Scholar] [CrossRef] [PubMed]
- Infantino, V.; Dituri, F.; Convertini, P.; Santarsiero, A.; Palmieri, F.; Todisco, S.; Mancarella, S.; Giannelli, G.; Iacobazzi, V. Epigenetic upregulation and functional role of the mitochondrial aspartate/glutamate carrier isoform 1 in hepatocellular carcinoma. Biochim. Biophys. Acta Mol. Basis Dis. 2019, 1865, 38–47. [Google Scholar] [CrossRef]
- Fernandez-Alvarez, A.; Llorente-Izquierdo, C.; Mayoral, R.; Agra, N.; Bosca, L.; Casado, M.; Martin-Sanz, P. Evaluation of epigenetic modulation of cyclooxygenase-2 as a prognostic marker for hepatocellular carcinoma. Oncogenesis 2012, 1, e23. [Google Scholar] [CrossRef]
- Tian, Y.; Wong, V.W.; Chan, H.L.; Cheng, A.S. Epigenetic regulation of hepatocellular carcinoma in non-alcoholic fatty liver disease. Semin. Cancer Biol. 2013, 23, 471–482. [Google Scholar] [CrossRef]
- Patel, D.; Menon, D.; Bernfeld, E.; Mroz, V.; Kalan, S.; Loayza, D.; Foster, D.A. Aspartate rescues s-phase arrest caused by suppression of glutamine utilization in kras-driven cancer cells. J. Biol. Chem. 2016, 291, 9322–9329. [Google Scholar] [CrossRef]
- Schoors, S.; Bruning, U.; Missiaen, R.; Queiroz, K.C.; Borgers, G.; Elia, I.; Zecchin, A.; Cantelmo, A.R.; Christen, S.; Goveia, J.; et al. Fatty acid carbon is essential for dntp synthesis in endothelial cells. Nature 2015, 520, 192–197. [Google Scholar] [CrossRef]
- Lopez-Rios, F.; Sanchez-Arago, M.; Garcia-Garcia, E.; Ortega, A.D.; Berrendero, J.R.; Pozo-Rodriguez, F.; Lopez-Encuentra, A.; Ballestin, C.; Cuezva, J.M. Loss of the mitochondrial bioenergetic capacity underlies the glucose avidity of carcinomas. Cancer Res. 2007, 67, 9013–9017. [Google Scholar] [CrossRef] [PubMed]
- Weinberg, S.E.; Chandel, N.S. Targeting mitochondria metabolism for cancer therapy. Nat. Chem. Biol. 2015, 11, 9–15. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, L.B.; Gui, D.Y.; Hosios, A.M.; Bush, L.N.; Freinkman, E.; Vander Heiden, M.G. Supporting aspartate biosynthesis is an essential function of respiration in proliferating cells. Cell 2015, 162, 552–563. [Google Scholar] [CrossRef] [PubMed]
- Amoedo, N.D.; Punzi, G.; Obre, E.; Lacombe, D.; De Grassi, A.; Pierri, C.L.; Rossignol, R. Agc1/2, the mitochondrial aspartate-glutamate carriers. Biochim. Biophys. Acta 2016, 1863, 2394–2412. [Google Scholar] [CrossRef]
- Palmieri, L.; Pardo, B.; Lasorsa, F.M.; del Arco, A.; Kobayashi, K.; Iijima, M.; Runswick, M.J.; Walker, J.E.; Saheki, T.; Satrustegui, J.; et al. Citrin and aralar1 are Ca2+-stimulated aspartate/glutamate transporters in mitochondria. EMBO J. 2001, 20, 5060–5069. [Google Scholar] [CrossRef]
- Rabinovich, S.; Adler, L.; Yizhak, K.; Sarver, A.; Silberman, A.; Agron, S.; Stettner, N.; Sun, Q.; Brandis, A.; Helbling, D.; et al. Diversion of aspartate in ass1-deficient tumours fosters de novo pyrimidine synthesis. Nature 2015, 527, 379–383. [Google Scholar] [CrossRef]
- Alkan, H.F.; Walter, K.E.; Luengo, A.; Madreiter-Sokolowski, C.T.; Stryeck, S.; Lau, A.N.; Al-Zoughbi, W.; Lewis, C.A.; Thomas, C.J.; Hoefler, G.; et al. Cytosolic aspartate availability determines cell survival when glutamine is limiting. Cell Metab. 2018, 28, 706–720. [Google Scholar] [CrossRef]
- Fuchs, B.C.; Bode, B.P. Stressing out over survival: Glutamine as an apoptotic modulator. J. Surg. Res. 2006, 131, 26–40. [Google Scholar] [CrossRef]
- Fuchs, B.C.; Bode, B.P. Amino acid transporters asct2 and lat1 in cancer: Partners in crime? Semin. Cancer Biol. 2005, 15, 254–266. [Google Scholar] [CrossRef]
- Mullen, A.R.; Wheaton, W.W.; Jin, E.S.; Chen, P.H.; Sullivan, L.B.; Cheng, T.; Yang, Y.; Linehan, W.M.; Chandel, N.S.; DeBerardinis, R.J. Reductive carboxylation supports growth in tumour cells with defective mitochondria. Nature 2011, 481, 385–388. [Google Scholar] [CrossRef]
- Chen, J.Q.; Russo, J. Dysregulation of glucose transport, glycolysis, tca cycle and glutaminolysis by oncogenes and tumor suppressors in cancer cells. Biochim. Biophys. Acta 2012, 1826, 370–384. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Cui, H. Targeting glutamine induces apoptosis: A cancer therapy approach. Int. J. Mol. Sci. 2015, 16, 22830–22855. [Google Scholar] [CrossRef] [PubMed]
- Xiang, Y.; Stine, Z.E.; Xia, J.; Lu, Y.; O’Connor, R.S.; Altman, B.J.; Hsieh, A.L.; Gouw, A.M.; Thomas, A.G.; Gao, P.; et al. Targeted inhibition of tumor-specific glutaminase diminishes cell-autonomous tumorigenesis. J. Clin. Investig. 2015, 125, 2293–2306. [Google Scholar] [CrossRef] [PubMed]
- DeWaal, D.; Nogueira, V.; Terry, A.R.; Patra, K.C.; Jeon, S.M.; Guzman, G.; Au, J.; Long, C.P.; Antoniewicz, M.R.; Hay, N. Hexokinase-2 depletion inhibits glycolysis and induces oxidative phosphorylation in hepatocellular carcinoma and sensitizes to metformin. Nat. Commun. 2018, 9, 446. [Google Scholar] [CrossRef] [PubMed]
- Bjornson, E.; Mukhopadhyay, B.; Asplund, A.; Pristovsek, N.; Cinar, R.; Romeo, S.; Uhlen, M.; Kunos, G.; Nielsen, J.; Mardinoglu, A. Stratification of hepatocellular carcinoma patients based on acetate utilization. Cell Rep. 2015, 13, 2014–2026. [Google Scholar] [CrossRef] [PubMed]
- Baggetto, L.G. Deviant energetic metabolism of glycolytic cancer cells. Biochimie 1992, 74, 959–974. [Google Scholar] [CrossRef]
- Guay, C.; Madiraju, S.R.; Aumais, A.; Joly, E.; Prentki, M. A role for atp-citrate lyase, malic enzyme, and pyruvate/citrate cycling in glucose-induced insulin secretion. J. Biol. Chem. 2007, 282, 35657–35665. [Google Scholar] [CrossRef]
- Sauer, L.A.; Dauchy, R.T.; Nagel, W.O.; Morris, H.P. Mitochondrial malic enzymes. Mitochondrial nad(p)+-dependent malic enzyme activity and malate-dependent pyruvate formation are progression-linked in morris hepatomas. J. Biol. Chem. 1980, 255, 3844–3848. [Google Scholar]
- Wen, Y.; Xu, L.; Chen, F.L.; Gao, J.; Li, J.Y.; Hu, L.H.; Li, J. Discovery of a novel inhibitor of nad(p)(+)-dependent malic enzyme (me2) by high-throughput screening. Acta Pharmacol. Sin. 2014, 35, 674–684. [Google Scholar] [CrossRef]
- Gao, X.; Sheng, Y.; Yang, J.; Wang, C.; Zhang, R.; Zhu, Y.; Zhang, Z.; Zhang, K.; Yan, S.; Sun, H.; et al. Osteopontin alters DNA methylation through up-regulating DNMT1 and sensitizes CD133+/CD44+ cancer stem cells to 5 azacytidine in hepatocellular carcinoma. J. Exp. Clin. Cancer Res. 2018, 37, 179. [Google Scholar] [CrossRef]
- Xie, C.R.; Li, Z.; Sun, H.G.; Wang, F.Q.; Sun, Y.; Zhao, W.X.; Zhang, S.; Zhao, W.X.; Wang, X.M.; Yin, Z.Y. Mutual regulation between chd5 and ezh2 in hepatocellular carcinoma. Oncotarget 2015, 6, 40940–40952. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Lv, L.Z.; Cai, Q.C.; Jiang, Y. Potential roles of ezh2, bmi-1 and mir-203 in cell proliferation and invasion in hepatocellular carcinoma cell line hep3b. World J. Gastroenterol. 2015, 21, 13268–13276. [Google Scholar] [CrossRef] [PubMed]
- Bricambert, J.; Miranda, J.; Benhamed, F.; Girard, J.; Postic, C.; Dentin, R. Salt-inducible kinase 2 links transcriptional coactivator p300 phosphorylation to the prevention of chrebp-dependent hepatic steatosis in mice. J. Clin. Investig. 2010, 120, 4316–4331. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Luo, R.Z.; Chen, J.W.; Cao, Y.; Lu, J.B.; He, J.H.; Wu, Q.L.; Cai, M.Y. High expression of transcriptional coactivator p300 correlates with aggressive features and poor prognosis of hepatocellular carcinoma. J. Transl. Med. 2011, 9, 5. [Google Scholar] [CrossRef]
- Dierks, T.; Stappen, R.; Salentin, A.; Kramer, R. Probing the active site of the reconstituted aspartate/glutamate carrier from bovine heart mitochondria: Carbodiimide-catalyzed acylation of a functional lysine residue. Biochim. Biophys. Acta 1992, 1103, 13–24. [Google Scholar] [CrossRef]
- Fuchs, B.C.; Perez, J.C.; Suetterlin, J.E.; Chaudhry, S.B.; Bode, B.P. Inducible antisense rna targeting amino acid transporter atb0/asct2 elicits apoptosis in human hepatoma cells. Am. J. Physiol. Gastrointest. Liver Physiol. 2004, 286, G467–G478. [Google Scholar] [CrossRef]
- Cardaci, S.; Rizza, S.; Filomeni, G.; Bernardini, R.; Bertocchi, F.; Mattei, M.; Paci, M.; Rotilio, G.; Ciriolo, M.R. Glutamine deprivation enhances antitumor activity of 3-bromopyruvate through the stabilization of monocarboxylate transporter-1. Cancer Res. 2012, 72, 4526–4536. [Google Scholar] [CrossRef]
- Wang, Q.; Hardie, R.A.; Hoy, A.J.; van Geldermalsen, M.; Gao, D.; Fazli, L.; Sadowski, M.C.; Balaban, S.; Schreuder, M.; Nagarajah, R.; et al. Targeting asct2-mediated glutamine uptake blocks prostate cancer growth and tumour development. J. Pathol. 2015, 236, 278–289. [Google Scholar] [CrossRef]
- Imai, H.; Kaira, K.; Oriuchi, N.; Shimizu, K.; Tominaga, H.; Yanagitani, N.; Sunaga, N.; Ishizuka, T.; Nagamori, S.; Promchan, K.; et al. Inhibition of l-type amino acid transporter 1 has antitumor activity in non-small cell lung cancer. Anticancer Res. 2010, 30, 4819–4828. [Google Scholar]
- Xu, P.; Oosterveer, M.H.; Stein, S.; Demagny, H.; Ryu, D.; Moullan, N.; Wang, X.; Can, E.; Zamboni, N.; Comment, A.; et al. Lrh-1-dependent programming of mitochondrial glutamine processing drives liver cancer. Genes Dev. 2016, 30, 1255–1260. [Google Scholar] [CrossRef]
- Gross, M.I.; Demo, S.D.; Dennison, J.B.; Chen, L.; Chernov-Rogan, T.; Goyal, B.; Janes, J.R.; Laidig, G.J.; Lewis, E.R.; Li, J.; et al. Antitumor activity of the glutaminase inhibitor cb-839 in triple-negative breast cancer. Mol. Cancer Ther. 2014, 13, 890–901. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Todisco, S.; Convertini, P.; Iacobazzi, V.; Infantino, V. TCA Cycle Rewiring as Emerging Metabolic Signature of Hepatocellular Carcinoma. Cancers 2020, 12, 68. https://doi.org/10.3390/cancers12010068
Todisco S, Convertini P, Iacobazzi V, Infantino V. TCA Cycle Rewiring as Emerging Metabolic Signature of Hepatocellular Carcinoma. Cancers. 2020; 12(1):68. https://doi.org/10.3390/cancers12010068
Chicago/Turabian StyleTodisco, Simona, Paolo Convertini, Vito Iacobazzi, and Vittoria Infantino. 2020. "TCA Cycle Rewiring as Emerging Metabolic Signature of Hepatocellular Carcinoma" Cancers 12, no. 1: 68. https://doi.org/10.3390/cancers12010068
APA StyleTodisco, S., Convertini, P., Iacobazzi, V., & Infantino, V. (2020). TCA Cycle Rewiring as Emerging Metabolic Signature of Hepatocellular Carcinoma. Cancers, 12(1), 68. https://doi.org/10.3390/cancers12010068




