Focus-ING on DNA Integrity: Implication of ING Proteins in Cell Cycle Regulation and DNA Repair Modulation
Abstract
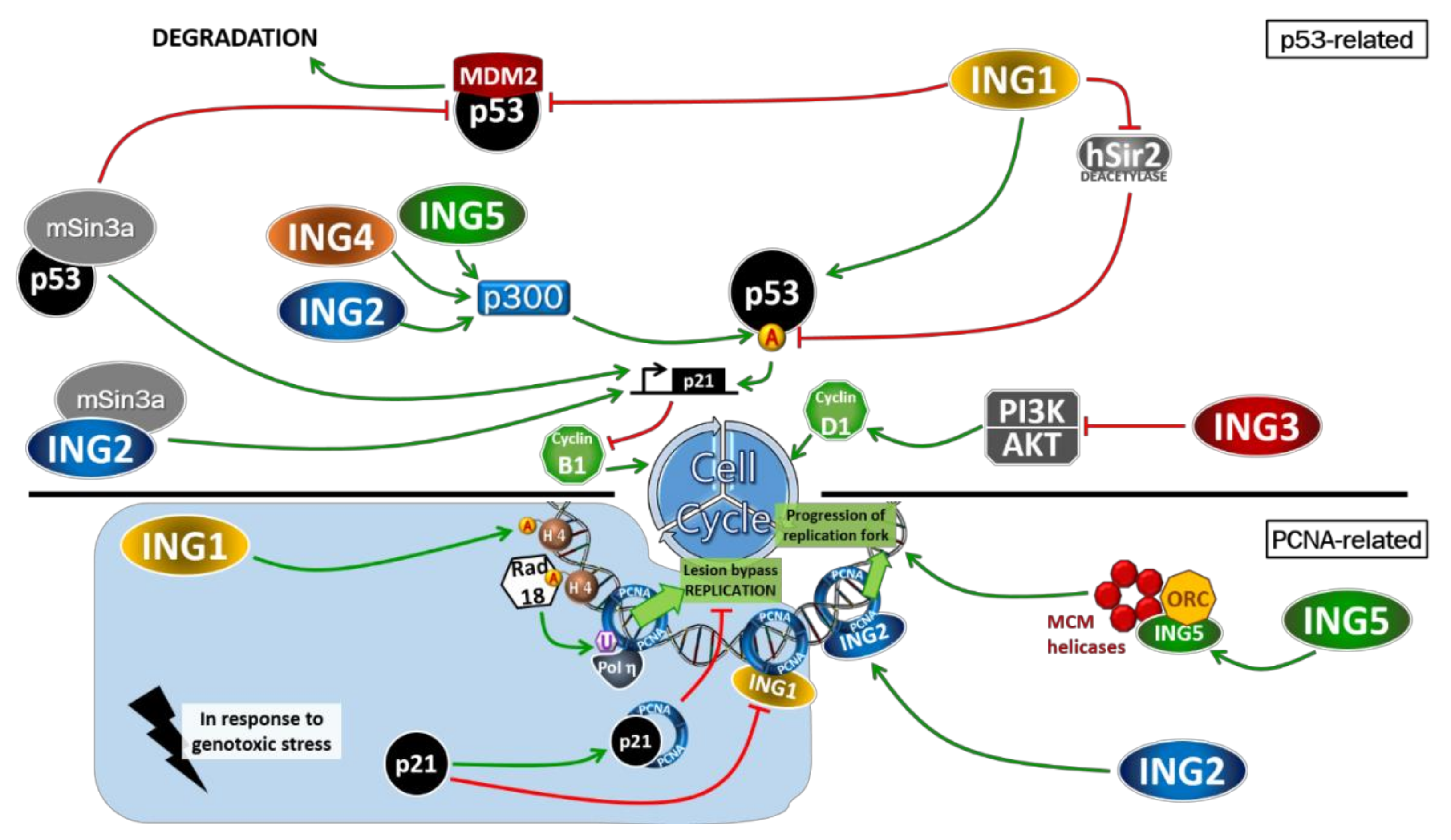
1. Introduction
2. Chromatin Remodeling
2.1. INGs Belong to Histone-Modifying Complexes
2.2. ING1 Participates in H3 and H4 Acetylation
2.3. The Dual Role of ING2 in Chromatin Remodeling
2.4. ING3 in Complex with hNuA4 Promotes Histones Acetylation
2.5. ING4 and ING5 Enhance Histones Acetylation
3. Cell Cycle Regulation
3.1. ING1 in Cell Cycle Regulation
3.2. ING2 in Cell Cycle Regulation
3.3. ING3 in Cell Cycle Regulation
3.4. ING4 and ING5 in Cell Cycle Regulation
4. DNA Repair Regulation
4.1. ING1 and ING2 Participate in UV-Induced Repair
4.2. DSB Repair (DSBR) Is Modulated by ING Proteins
4.3. ING2 Can Regulate DSBR
4.4. ING3 Modulates DSBR
4.5. ING4 and ING5 Involvement in DDR
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Garkavtsev, I.; Kazarov, A.; Gudkov, A.; Riabowol, K. Suppression of the novel growth inhibitor p33ING1 promotes neoplastic transformation. Nat. Genet. 1996, 14, 415–420. [Google Scholar] [CrossRef] [PubMed]
- Dantas, A.; Al Shueili, B.; Yang, Y.; Nabbi, A.; Fink, D.; Riabowol, K. Biological Functions of the ING Proteins. Cancers 2019, 11, 1817. [Google Scholar] [CrossRef] [PubMed]
- He, G.H.Y.; Helbing, C.C.; Wagner, M.J.; Sensen, C.W.; Riabowol, K. Phylogenetic Analysis of the ING Family of PHD Finger Proteins. Mol. Biol. Evol. 2005, 22, 104–116. [Google Scholar] [CrossRef] [PubMed]
- Mjelle, R.; Hegre, S.A.; Aas, P.A.; Slupphaug, G.; Drabløs, F.; Sætrom, P.; Krokan, H.E. Cell cycle regulation of human DNA repair and chromatin remodeling genes. DNA Repair 2015, 30, 53–67. [Google Scholar] [CrossRef]
- Aasland, R.; Gibson, T.J.; Stewart, A.F. The PHD finger: Implications for chromatin-mediated transcriptional regulation. Trends Biochem. Sci. 1995, 20, 56–59. [Google Scholar] [CrossRef]
- Wysocka, J.; Swigut, T.; Xiao, H.; Milne, T.A.; Kwon, S.Y.; Landry, J.; Kauer, M.; Tackett, A.J.; Chait, B.T.; Badenhorst, P.; et al. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature 2006, 442, 86–90. [Google Scholar] [CrossRef]
- Shi, X.; Hong, T.; Walter, K.L.; Ewalt, M.; Michishita, E.; Hung, T.; Carney, D.; Peña, P.; Lan, F.; Kaadige, M.R.; et al. ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature 2006, 442, 96–99. [Google Scholar] [CrossRef]
- Ythier, D.; Larrieu, D.; Brambilla, C.; Brambilla, E.; Pedeux, R. The new tumor suppressor genes ING: Genomic structure and status in cancer. Int. J. Cancer 2008, 123, 1483–1490. [Google Scholar] [CrossRef]
- Guérillon, C.; Larrieu, D.; Pedeux, R. ING1 and ING2: Multifaceted tumor suppressor genes. Cell. Mol. Life Sci. 2013, 70, 3753–3772. [Google Scholar] [CrossRef]
- Culurgioni, S.; Muñoz, I.G.; Moreno, A.; Palacios, A.; Villate, M.; Palmero, I.; Montoya, G.; Blanco, F.J. Crystal structure of inhibitor of growth 4 (ING4) dimerization domain reveals functional organization of ING family of chromatin-binding proteins. J. Biol. Chem. 2012, 287, 10876–10884. [Google Scholar] [CrossRef]
- Ormaza, G.; Rodríguez, J.A.; Ibáñez de Opakua, A.; Merino, N.; Villate, M.; Gorroño, I.; Rábano, M.; Palmero, I.; Vilaseca, M.; Kypta, R.; et al. The Tumor Suppressor ING5 Is a Dimeric, Bivalent Recognition Molecule of the Histone H3K4me3 Mark. J. Mol. Biol. 2019, 431, 2298–2319. [Google Scholar] [CrossRef] [PubMed]
- Guérillon, C.; Bigot, N.; Pedeux, R. The ING tumor suppressor genes: Status in human tumors. Cancer Lett. 2014, 345, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Liu, Y.; Zhao, Y.; Tian, F.; Wang, G. miR-153-3p Suppresses Inhibitor of Growth Protein 2 Expression to Function as Tumor Suppressor in Acute Lymphoblastic Leukemia. Technol. Cancer Res. Treat. 2019, 18, 1533033819852990. [Google Scholar] [CrossRef] [PubMed]
- Gournay, M.; Paineau, M.; Archambeau, J.; Pedeux, R. Regulat-INGs in tumors and diseases: Focus on ncRNAs. Cancer Lett. 2019, 447, 66–74. [Google Scholar] [CrossRef]
- Epstein, R.J. A periodic table for cancer. Future Oncol. 2015, 11, 785–800. [Google Scholar] [CrossRef]
- Negrini, S.; Gorgoulis, V.G.; Halazonetis, T.D. Genomic instability an evolving hallmark of cancer. Nat. Rev. Mol. Cell Biol. 2010, 11, 220–228. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of Cancer: The Next Generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Fraga, M.F.; Esteller, M. Towards the human cancer epigenome: A first draft of histone modifications. Cell Cycle 2005, 4, 1377–1381. [Google Scholar] [CrossRef]
- Doyon, Y.; Cayrou, C.; Ullah, M.; Landry, A.J.; Côté, V.; Selleck, W.; Lane, W.S.; Tan, S.; Yang, X.J.; Côté, J. ING tumor suppressor proteins are critical regulators of chromatin acetylation required for genome expression and perpetuation. Mol. Cell 2006, 21, 51–64. [Google Scholar] [CrossRef]
- Ogiwara, H.; Ui, A.; Otsuka, A.; Satoh, H.; Yokomi, I.; Nakajima, S.; Yasui, A.; Yokota, J.; Kohno, T. Histone acetylation by CBP and p300 at double-strand break sites facilitates SWI/SNF chromatin remodeling and the recruitment of non-homologous end joining factors. Oncogene 2011, 30, 2135–2146. [Google Scholar] [CrossRef]
- Nair, N.; Shoaib, M.; Sørensen, C.S. Chromatin dynamics in genome stability: Roles in suppressing endogenous DNA damage and facilitating DNA repair. Int. J. Mol. Sci. 2017, 18, 1486. [Google Scholar] [CrossRef] [PubMed]
- Takata, H.; Hanafusa, T.; Mori, T.; Shimura, M.; Iida, Y.; Ishikawa, K.; Yoshikawa, K.; Yoshikawa, Y.; Maeshima, K. Chromatin Compaction Protects Genomic DNA from Radiation Damage. PLoS ONE 2013, 8, e75622. [Google Scholar] [CrossRef] [PubMed]
- Groselj, B.; Sharma, N.L.; Hamdy, F.C.; Kerr, M.; Kiltie, A.E. Histone deacetylase inhibitors as radiosensitisers: Effects on DNA damage signalling and repair. Br. J. Cancer 2013, 108, 748–754. [Google Scholar] [CrossRef] [PubMed]
- Vieyra, D.; Loewith, R.; Scott, M.; Bonnefin, P.; Boisvert, F.M.; Cheema, P.; Pastyryeva, S.; Meijer, M.; Johnston, R.N.; Bazett-Jones, D.P.; et al. Human ING1 proteins differentially regulate histone acetylation. J. Biol. Chem. 2002, 277, 29832–29839. [Google Scholar] [CrossRef] [PubMed]
- Soliman, M.A.; Berardi, P.; Pastyryeva, S.; Bonnefin, P.; Feng, X.; Colina, A.; Young, D.; Riabowol, K. ING1a expression increases during replicative senescence and induces a senescent phenotype. Aging Cell 2008, 7, 783–794. [Google Scholar] [CrossRef] [PubMed]
- Rajarajacholan, U.K.; Thalappilly, S.; Riabowol, K. The ING1a Tumor Suppressor Regulates Endocytosis to Induce Cellular Senescence Via the Rb-E2F Pathway. PLoS Biol. 2013, 11, e1001502. [Google Scholar] [CrossRef]
- Kuo, W.H.W.; Wang, Y.; Wong, R.P.C.; Campos, E.I.; Li, G. The ING1b tumor suppressor facilitates nucleotide excision repair by promoting chromatin accessibility to XPA. Exp. Cell Res. 2007, 313, 1628–1638. [Google Scholar] [CrossRef]
- Sardiu, M.E.; Smith, K.T.; Groppe, B.D.; Gilmore, J.M.; Saraf, A.; Egidy, R.; Peak, A.; Seidel, C.W.; Florens, L.; Workman, J.L.; et al. Suberoylanilide hydroxamic acid (SAHA)-Induced dynamics of a human histone deacetylase protein interaction network. Mol. Cell. Proteom. 2014, 13, 3114–3125. [Google Scholar] [CrossRef]
- Pedeux, R.; Sengupta, S.; Shen, J.C.; Demidov, O.N.; Saito, S.; Onogi, H.; Kumamoto, K.; Wincovitch, S.; Garfield, S.H.; McMenamin, M.; et al. ING2 Regulates the Onset of Replicative Senescence by Induction of p300-Dependent p53 Acetylation. Mol. Cell. Biol. 2005, 25, 6639–6648. [Google Scholar] [CrossRef]
- Goeman, F.; Otto, K.; Kyrylenko, S.; Schmidt, O.; Baniahmad, A. ING2 recruits histone methyltransferase activity with methylation site specificity distinct from histone H3 lysines 4 and 9. Biochim. Biophys. Acta Mol. Cell Res. 2008, 1783, 1673–1680. [Google Scholar] [CrossRef][Green Version]
- Smith, K.T.; Martin-Brown, S.A.; Florens, L.; Washburn, M.P.; Workman, J.L. Deacetylase Inhibitors Dissociate the Histone-Targeting ING2 Subunit from the Sin3 Complex. Chem. Biol. 2010, 17, 65–74. [Google Scholar] [CrossRef] [PubMed]
- Noguchi, C.; Singh, T.; Ziegler, M.A.; Peake, J.D.; Khair, L.; Aza, A.; Nakamura, T.M.; Noguchi, E. The NuA4 acetyltransferase and histone H4 acetylation promote replication recovery after topoisomerase I-poisoning. Epigenet. Chromatin 2019, 12, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Ayrapetov, M.K.; Xu, C.; Gursoy-Yuzugullu, O.; Hu, Y.; Price, B.D. Histone H2A.Z Controls a Critical Chromatin Remodeling Step Required for DNA Double-Strand Break Repair. Mol. Cell 2012, 48, 723–733. [Google Scholar] [CrossRef] [PubMed]
- Murr, R.; Loizou, J.I.; Yang, Y.G.; Cuenin, C.; Li, H.; Wang, Z.Q.; Herceg, Z. Histone acetylation by Trrap–Tip60 modulates loading of repair proteins and repair of DNA double-strand breaks. Nat. Cell Biol. 2006, 8, 91–99. [Google Scholar] [CrossRef]
- Choy, J.S.; Tobe, B.T.D.; Huh, J.H.; Kron, S.J. Yng2p-dependent NuA4 Histone H4 Acetylation Activity Is Required for Mitotic and Meiotic Progression. J. Biol. Chem. 2001, 276, 43653–43662. [Google Scholar] [CrossRef]
- Doyon, Y.; Selleck, W.; Lane, W.S.; Tan, S.; Cote, J. Structural and Functional Conservation of the NuA4 Histone Acetyltransferase Complex from Yeast to Humans. Mol. Cell. Biol. 2004, 24, 1884–1896. [Google Scholar] [CrossRef]
- Downs, J.A.; Allard, S.; Jobin-Robitaille, O.; Javaheri, A.; Auger, A.; Bouchard, N.; Kron, S.J.; Jackson, S.P.; Côté, J. Binding of chromatin-modifying activities to phosphorylated histone H2A at DNA damage sites. Mol. Cell 2004, 16, 979–990. [Google Scholar] [CrossRef]
- Bennett, G.; Peterson, C.L. SWI/SNF recruitment to a DNA double-strand break by the NuA4 and Gcn5 histone acetyltransferases. DNA Repair 2015, 30, 38–45. [Google Scholar] [CrossRef]
- Lee, H.S.; Park, J.H.; Kim, S.J.; Kwon, S.J.; Kwon, J. A cooperative activation loop among SWI/SNF, γ-H2AX and H3 acetylation for DNA double-strand break repair. EMBO J. 2010, 29, 1434–1445. [Google Scholar] [CrossRef]
- Zhang, T.; Meng, J.; Liu, X.; Zhang, X.; Peng, X.; Cheng, Z. ING5 differentially regulates protein lysine acetylation and promotes p300 autoacetylation. Oncotarget 2018, 9, 1617–1629. [Google Scholar] [CrossRef]
- Zhang, R.; Erler, J.; Langowski, J. Histone Acetylation Regulates Chromatin Accessibility: Role of H4K16 in Inter-nucleosome Interaction. Biophys. J. 2017, 112, 450–459. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Gaya, V.; Casaní-Galdón, S.; Ugidos, M.; Kuang, Z.; Mellor, J.; Conesa, A.; Tarazona, S. Elucidating the Role of Chromatin State and Transcription Factors on the Regulation of the Yeast Metabolic Cycle: A Multi-Omic Integrative Approach. Front. Genet. 2018, 9, 578. [Google Scholar] [CrossRef] [PubMed]
- Ohzeki, J.; Shono, N.; Otake, K.; Martins, N.M.C.; Kugou, K.; Kimura, H.; Nagase, T.; Larionov, V.; Earnshaw, W.C.; Masumoto, H. KAT7/HBO1/MYST2 Regulates CENP-A Chromatin Assembly by Antagonizing Suv39h1-Mediated Centromere Inactivation. Dev. Cell 2016, 37, 413–427. [Google Scholar] [CrossRef] [PubMed]
- Rivera, C.; Gurard-Levin, Z.A.; Almouzni, G.; Loyola, A. Histone lysine methylation and chromatin replication. Biochim. Biophys. Acta Gene Regul. Mech. 2014, 1839, 1433–1439. [Google Scholar] [CrossRef]
- Champagne, K.S.; Saksouk, N.; Peña, P.V.; Johnson, K.; Ullah, M.; Yang, X.J.; Côté, J.; Kutateladze, T.G. The crystal structure of the ING5 PHD finger in complex with an H3K4me3 histone peptide. Proteins 2008, 72, 1371–1376. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. The hallmarks of cancer. Cell 2000, 100, 57–70. [Google Scholar] [CrossRef]
- Helbing, C.C.; Veillette, C.; Riabowol, K.; Johnston, R.N.; Garkavtsev, I. A novel candidate tumor suppressor, ING1, is involved in the regulation of apoptosis. Cancer Res. 1997, 57, 1255–1258. [Google Scholar]
- Saito, M.; Kumamoto, K.; Robles, A.I.; Horikawa, I.; Furusato, B.; Okamura, S.; Goto, A.; Yamashita, T.; Nagashima, M.; Lee, T.L.; et al. Targeted Disruption of Ing2 Results in Defective Spermatogenesis and Development of Soft-Tissue Sarcomas. PLoS ONE 2010, 5, e15541. [Google Scholar] [CrossRef]
- Garkavtsev, I.; Riabowol, K. Extension of the replicative life span of human diploid fibroblasts by inhibition of the p33ING1 candidate tumor suppressor. Mol. Cell. Biol. 1997, 17, 2014–2019. [Google Scholar] [CrossRef]
- Nagashima, M.; Shiseki, M.; Miura, K.; Hagiwara, K.; Linke, S.P.; Pedeux, R.; Wang, X.W.; Yokota, J.; Riabowol, K.; Harris, C.C. DNA damage-inducible gene p33ING2 negatively regulates cell proliferation through acetylation of p53. Proc. Natl. Acad. Sci. USA 2001, 98, 9671–9676. [Google Scholar] [CrossRef]
- Wong, R.P.C.; Lin, H.; Khosravi, S.; Piche, B.; Jafarnejad, S.M.; Chen, D.W.C.; Li, G. Tumour suppressor ING1b maintains genomic stability upon replication stress. Nucl. Acids Res. 2011, 39, 3632–3642. [Google Scholar] [CrossRef] [PubMed]
- Leung, K.M.; Po, L.S.; Tsang, F.C.; Siu, W.Y.; Lau, A.; Ho, H.T.B.; Poon, R.Y.C. The candidate tumor suppressor ING1b can stabilize p53 by disrupting the regulation of p53 by MDM2. Cancer Res. 2002, 62, 4890–4893. [Google Scholar] [PubMed]
- Kataoka, H.; Bonnefin, P.; Vieyra, D.; Feng, X.; Hara, Y.; Miura, Y.; Joh, T.; Nakabayashi, H.; Vaziri, H.; Harris, C.C.; et al. ING1 represses transcription by direct DNA binding and through effects on p53. Cancer Res. 2003, 63, 5785–5792. [Google Scholar] [PubMed]
- Tsang, F.C.; Po, L.S.; Leung, K.M.; Lau, A.; Siu, W.Y.; Poon, R.Y.C. ING1b decreases cell proliferation through p53-dependent and -independent mechanisms. FEBS Lett. 2003, 553, 277–285. [Google Scholar] [CrossRef]
- Thalappilly, S.; Feng, X.; Pastyryeva, S.; Suzuki, K.; Muruve, D.; Larocque, D.; Richard, S.; Truss, M.; von Deimling, A.; Riabowol, K.; et al. The p53 tumor suppressor is stabilized by inhibitor of growth 1 (ING1) by blocking polyubiquitination. PLoS ONE 2011, 6, e21065. [Google Scholar] [CrossRef][Green Version]
- González, L.; Freije, J.M.P.; Cal, S.; López-Otín, C.; Serrano, M.; Palmero, I. A functional link between the tumour suppressors ARF and p33ING1. Oncogene 2006, 25, 5173–5179. [Google Scholar] [CrossRef][Green Version]
- Zilfou, J.T.; Hoffman, W.H.; Sank, M.; George, D.L.; Murphy, M. The Corepressor mSin3a Interacts with the Proline-Rich Domain of p53 and Protects p53 from Proteasome-Mediated Degradation. Mol. Cell. Biol. 2001, 21, 3974–3985. [Google Scholar] [CrossRef]
- Coles, A.H.; Liang, H.; Zhu, Z.; Marfella, C.G.A.; Kang, J.; Imbalzano, A.N.; Jones, S.N. Deletion of p37Ing1 in mice reveals a p53-independent role for Ing1 in the suppression of cell proliferation, apoptosis, and tumorigenesis. Cancer Res. 2007, 67, 2054–2061. [Google Scholar] [CrossRef]
- Larrieu, D.; Ythier, D.; Brambilla, C.; Pedeux, R. ING2 controls the G 1 to S-phase transition by regulating p21 expression. Cell Cycle 2010, 9, 3984–3990. [Google Scholar] [CrossRef]
- Campos, E.I.; Chin, M.Y.; Kuo, W.H.; Li, G. Biological functions of the ING family tumor suppressors. Cell. Mol. Life Sci. 2004, 61, 2597–2613. [Google Scholar] [CrossRef]
- Liu, L.; Scolnick, D.M.; Trievel, R.C.; Zhang, H.B.; Marmorstein, R.; Halazonetis, T.D.; Berger, S.L. p53 sites acetylated in vitro by PCAF and p300 are acetylated in vivo in response to DNA damage. Mol. Cell. Biol. 1999, 19, 1202–1209. [Google Scholar] [CrossRef] [PubMed]
- Ohkouchi, C.; Kumamoto, K.; Saito, M.; Ishigame, T.; Suzuki, S.I.; Takenoshita, S.; Harris, C.C. ING2, a tumor associated gene, enhances PAI-1 and HSPA1A expression with HDAC1 and mSin3A through the PHD domain and C-terminal. Mol. Med. Rep. 2017, 16, 7367–7374. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Gozani, O.; Karuman, P.; Jones, D.R.; Ivanov, D.; Cha, J.; Lugovskoy, A.A.; Baird, C.L.; Zhu, H.; Field, S.J.; Lessnick, S.L.; et al. The PHD finger of the chromatin-associated protein ING2 functions as a nuclear phosphoinositide receptor. Cell 2003, 114, 99–111. [Google Scholar] [CrossRef]
- Kaadige, M.R.; Ayer, D.E. The polybasic region that follows the plant homeodomain zinc finger 1 of Pf1 is necessary and sufficient for specific phosphoinositide binding. J. Biol. Chem. 2006, 281, 28831–28836. [Google Scholar] [CrossRef] [PubMed]
- Larrieu, D.; Ythier, D.; Binet, R.; Brambilla, C.; Brambilla, E.; Sengupta, S.; Pedeux, R. ING2 controls the progression of DNA replication forks to maintain genome stability. EMBO Rep. 2009, 10, 1168–1174. [Google Scholar] [CrossRef] [PubMed]
- Sarker, K.P.; Kataoka, H.; Chan, A.; Netherton, S.J.; Pot, I.; Huynh, M.A.; Feng, X.; Bonni, A.; Riabowol, K.; Bonni, S. ING2 as a Novel Mediator of Transforming Growth Factor-β-dependent Responses in Epithelial Cells. J. Biol. Chem. 2008, 283, 13269–13279. [Google Scholar] [CrossRef]
- Nagashima, M.; Shiseki, M.; Pedeux, R.M.; Okamura, S.; Kitahama-Shiseki, M.; Miura, K.; Yokota, J.; Harris, C.C. A novel PHD-finger motif protein, p47ING3, modulates p53-mediated transcription, cell cycle control, and apoptosis. Oncogene 2003, 22, 343–350. [Google Scholar] [CrossRef]
- Chen, G.; Wang, Y.; Garate, M.; Zhou, J.; Li, G. The tumor suppressor ING3 is degraded by SCFSkp2-mediated ubiquitin–proteasome system. Oncogene 2010, 29, 1498–1508. [Google Scholar] [CrossRef]
- Suzuki, S.; Nozawa, Y.; Tsukamoto, S.; Kaneko, T.; Imai, H.; Minami, N. ING3 Is Essential for Asymmetric Cell Division during Mouse Oocyte Maturation. PLoS ONE 2013, 8, e74749. [Google Scholar] [CrossRef]
- McClurg, U.L.; Nabbi, A.; Ricordel, C.; Korolchuk, S.; McCracken, S.; Heer, R.; Wilson, L.; Butler, L.M.; Irving-Hooper, B.K.; Pedeux, R.; et al. Human ex vivo prostate tissue model system identifies ING3 as an oncoprotein. Br. J. Cancer 2018, 118, 713–726. [Google Scholar] [CrossRef]
- Lu, M.; Chen, F.; Wang, Q.; Wang, K.; Pan, Q.; Zhang, X. Downregulation of inhibitor of growth 3 is correlated with tumorigenesis and progression of hepatocellular carcinoma. Oncol. Lett. 2012, 4, 47–52. [Google Scholar] [CrossRef] [PubMed]
- Gunduz, M.; Beder, L.B.; Gunduz, E.; Nagatsuka, H.; Fukushima, K.; Pehlivan, D.; Cetin, E.; Yamanaka, N.; Nishizaki, K.; Shimizu, K.; et al. Downregulation of ING3 mRNA expression predicts poor prognosis in head and neck cancer. Cancer Sci. 2008, 29, 531–538. [Google Scholar] [CrossRef] [PubMed]
- Nourani, A.; Doyon, Y.; Utley, R.T.; Allard, S.; Lane, W.S.; Cote, J. Role of an ING1 Growth Regulator in Transcriptional Activation and Targeted Histone Acetylation by the NuA4 Complex. Mol. Cell. Biol. 2001, 21, 7629–7640. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Wang, L.; Zhang, C.; Deng, Y.; Zhao, B.; Ren, Y.; Fu, Y.; Meng, X. Inhibitor of growth 3 induces cell death by regulating cell proliferation, apoptosis and cell cycle arrest by blocking the PI3K/AKT pathway. Cancer Gene Ther. 2018, 25, 240–247. [Google Scholar] [CrossRef]
- Kawabe, T.; Suganuma, M.; Ando, T.; Kimura, M.; Hori, H.; Okamoto, T. Cdc25C interacts with PCNA at G2/M transition. Oncogene 2002, 21, 1717–1726. [Google Scholar] [CrossRef]
- Ando, T.; Kawabe, T.; Ohara, H.; Ducommun, B.; Itoh, M.; Okamoto, T. Involvement of the Interaction between p21 and Proliferating Cell Nuclear Antigen for the Maintenance of G2/M Arrest after DNA Damage. J. Biol. Chem. 2001, 276, 42971–42977. [Google Scholar] [CrossRef]
- Dai, W.; Dong, P.; Liu, J.; Gao, Y.; Hu, Y.; Lin, H.; Song, Y.; Mei, Q. Euscaphic acid inhibits proliferation and promotes apoptosis of nasopharyngeal carcinoma cells by silencing the PI3K/AKT/mTOR signaling pathway. Am. J. Transl. Res. 2019, 11, 2090–2098. [Google Scholar]
- Rassidakis, G.Z.; Feretzaki, M.; Atwell, C.; Grammatikakis, I.; Lin, Q.; Lai, R.; Claret, F.X.; Medeiros, L.J.; Amin, H.M. Inhibition of Akt increases p27Kip1 levels and induces cell cycle arrest in anaplastic large cell lymphoma. Blood 2005, 105, 827–829. [Google Scholar] [CrossRef]
- Oh, S.Y.; Park, K.S.; Kim, J.A.; Choi, K.Y. Differential modulation of zinc-stimulated p21(Cip/WAF1) and cyclin D1 induction by inhibition of PI3 kinase in HT-29 colorectal cancer cells. Exp. Mol. Med. 2002, 34, 27–31. [Google Scholar] [CrossRef]
- Klein, E.A.; Assoian, R.K. Transcriptional regulation of the cyclin D1 gene at a glance. J. Cell Sci. 2008, 121, 3853–3857. [Google Scholar] [CrossRef]
- Nabbi, A.; Almami, A.; Thakur, S.; Suzuki, K.; Boland, D.; Bismar, T.A.; Riabowol, K. ING3 protein expression profiling in normal human tissues suggest its role in cellular growth and self-renewal. Eur. J. Cell Biol. 2015, 94, 214–222. [Google Scholar] [CrossRef] [PubMed]
- Mouche, A.; Archambeau, J.; Ricordel, C.; Chaillot, L.; Bigot, N.; Guillaudeux, T.; Grenon, M.; Pedeux, R. ING3 is required for ATM signaling and DNA repair in response to DNA double strand breaks. Cell Death Differ. 2019. [Google Scholar] [CrossRef] [PubMed]
- Nabbi, A.; McClurg, U.L.; Thalappilly, S.; Almami, A.; Mobahat, M.; Bismar, T.A.; Binda, O.; Riabowol, K.T. ING3 promotes prostate cancer growth by activating the androgen receptor. BMC Med. 2017, 15, 103. [Google Scholar] [CrossRef] [PubMed]
- Shiseki, M.; Nagashima, M.; Pedeux, R.M.; Kitahama-Shiseki, M.; Miura, K.; Okamura, S.; Onogi, H.; Higashimoto, Y.; Appella, E.; Yokota, J.; et al. p29ING4 and p28ING5 bind to p53 and p300, and enhance p53 activity. Cancer Res. 2003, 63, 2373–2378. [Google Scholar]
- Li, X.; Cai, L.; Liang, M.; Wang, Y.; Yang, J.; Zhao, Y. Ing4 induces Cell Growth Inhibition in Human Lung Adenocarcinoma A549 Cells by Means of Wnt-1/β-Catenin Signaling Pathway. Anat. Rec. 2008, 291, 593–600. [Google Scholar] [CrossRef]
- Liu, N.; Wang, J.; Wang, J.; Wang, R.; Liu, Z.; Yu, Y.; Lu, H. ING5 is a Tip60 cofactor that acetylates p53 in response to DNA damage. Cancer Res. 2013, 73, 3749–3760. [Google Scholar] [CrossRef]
- Guo, Q.; Fast, W. Citrullination of Inhibitor of Growth 4 (ING4) by Peptidylarginine Deminase 4 (PAD4) Disrupts the Interaction between ING4 and p53. J. Biol. Chem. 2011, 286, 17069–17078. [Google Scholar] [CrossRef]
- Cuthbert, G.L.; Daujat, S.; Snowden, A.W.; Erdjument-Bromage, H.; Hagiwara, T.; Yamada, M.; Schneider, R.; Gregory, P.D.; Tempst, P.; Bannister, A.J.; et al. Histone Deimination Antagonizes Arginine Methylation. Cell 2004, 118, 545–553. [Google Scholar] [CrossRef]
- Cazzalini, O.; Sommatis, S.; Tillhon, M.; Dutto, I.; Bachi, A.; Rapp, A.; Nardo, T.; Scovassi, A.I.; Necchi, D.; Cardoso, M.C.; et al. CBP and p300 acetylate PCNA to link its degradation with nucleotide excision repair synthesis. Nucl. Acids Res. 2014, 42, 8433–8448. [Google Scholar] [CrossRef]
- Goldstein, M.; Kastan, M.B. The DNA Damage Response: Implications for Tumor Responses to Radiation and Chemotherapy. Annu. Rev. Med. 2015, 66, 129–143. [Google Scholar] [CrossRef]
- Scott, M.; Bonnefin, P.; Vieyra, D.; Boisvert, F.M.; Young, D.; Bazett-Jones, D.P.; Riabowol, K. UV-induced binding of ING1 to PCNA regulates the induction of apoptosis. J. Cell Sci. 2001, 114, 3455–3462. [Google Scholar] [PubMed]
- Scott, M.; Boisvert, F.M.; Vieyra, D.; Johnston, R.N.; Bazett-Jones, D.P.; Riabowol, K. UV induces nucleolar translocation of ING1 through two distinct nucleolar targeting sequences. Nucl. Acids Res. 2001, 29, 2052–2058. [Google Scholar] [CrossRef] [PubMed]
- Sugitani, N.; Sivley, R.M.; Perry, K.E.; Capra, J.A.; Chazin, W.J. XPA: A key scaffold for human nucleotide excision repair. DNA Repair 2016, 44, 123–135. [Google Scholar] [CrossRef] [PubMed]
- Krasikova, Y.S.; Rechkunova, N.I.; Maltseva, E.A.; Lavrik, O.I. RPA and XPA interaction with DNA structures mimicking intermediates of the late stages in nucleotide excision repair. PLoS ONE 2018, 13, e0190782. [Google Scholar] [CrossRef]
- Scharer, O.D. Nucleotide Excision Repair in Eukaryotes. Cold Spring Harb. Perspect. Biol. 2013, 5, a012609. [Google Scholar] [CrossRef]
- Kichina, J.V.; Zeremski, M.; Aris, L.; Gurova, K.V.; Walker, E.; Franks, R.; Nikitin, A.Y.; Kiyokawa, H.; Gudkov, A.V. Targeted disruption of the mouse ing1 locus results in reduced body size, hypersensitivity to radiation and elevated incidence of lymphomas. Oncogene 2006, 25, 857–866. [Google Scholar] [CrossRef]
- Ceruti, J.M.; Ogara, M.F.; Menéndez, C.; Palmero, I.; Cánepa, E.T. Inhibitor of growth 1 (ING1) acts at early steps of multiple DNA repair pathways. Mol. Cell. Biochem. 2013, 378, 117–126. [Google Scholar] [CrossRef]
- Gong, W.; Russell, M.; Suzuki, K.; Riabowol, K. Subcellular Targeting of p33ING1b by Phosphorylation-Dependent 14-3-3 Binding Regulates p21WAF1 Expression. Mol. Cell. Biol. 2006, 26, 2947–2954. [Google Scholar] [CrossRef][Green Version]
- Garate, M.; Campos, E.I.; Bush, J.A.; Xiao, H.; Li, G. Phosphorylation of the tumor suppressor p33 ING1b at Ser-126 influences its protein stability and proliferation of melanoma cells. FASEB J. 2007, 21, 3705–3716. [Google Scholar] [CrossRef]
- Laherty, C.D.; Billin, A.N.; Lavinsky, R.M.; Yochum, G.S.; Bush, A.C.; Sun, J.M.; Mullen, T.M.; Davie, J.R.; Rose, D.W.; Glass, C.K.; et al. SAP30, a component of the mSin3 corepressor complex involved in N-CoR-mediated repression by specific transcription factors. Mol. Cell 1998, 2, 33–42. [Google Scholar] [CrossRef]
- Wang, J.; Chin, M.Y.; Li, G. The Novel Tumor Suppressor p33ING2 Enhances Nucleotide Excision Repair via Inducement of Histone H4 Acetylation and Chromatin Relaxation. Cancer Res. 2006, 66, 1906–1911. [Google Scholar] [CrossRef] [PubMed]
- Adamson, B.; Smogorzewska, A.; Sigoillot, F.D.; King, R.W.; Elledge, S.J. A genome-wide homologous recombination screen identifies the RNA-binding protein RBMX as a component of the DNA-damage response. Nat. Cell Biol. 2012, 14, 318–328. [Google Scholar] [CrossRef] [PubMed]
- Bua, D.J.; Martin, G.M.; Binda, O.; Gozani, O. Nuclear phosphatidylinositol-5-phosphate regulates ING2 stability at discrete chromatin targets in response to DNA damage. Sci. Rep. 2013, 3, 2137. [Google Scholar] [CrossRef] [PubMed]
- Oh, J.; Symington, L.S. Role of the Mre11 complex in preserving genome integrity. Genes 2018, 9, 589. [Google Scholar] [CrossRef]
- Van der Linden, E.; Sanchez, H.; Kinoshita, E.; Kanaar, R.; Wyman, C. RAD50 and NBS1 form a stable complex functional in DNA binding and tethering. Nucl. Acids Res. 2009, 37, 1580–1588. [Google Scholar] [CrossRef]
- Williams, G.J.; Lees-Miller, S.P.; Tainer, J.A. Mre11–Rad50–Nbs1 conformations and the control of sensing, signaling, and effector responses at DNA double-strand breaks. DNA Repair 2010, 9, 1299–1306. [Google Scholar] [CrossRef]
- Deshpande, R.A.; Williams, G.J.; Limbo, O.; Williams, R.S.; Kuhnlein, J.; Lee, J.H.; Classen, S.; Guenther, G.; Russell, P.; Tainer, J.A.; et al. ATP-driven Rad50 conformations regulate DNA tethering, end resection, and ATM checkpoint signaling. EMBO J. 2014, 33, 482–500. [Google Scholar] [CrossRef]
- You, Z.; Chahwan, C.; Bailis, J.; Hunter, T.; Russell, P. ATM Activation and Its Recruitment to Damaged DNA Require Binding to the C Terminus of Nbs1. Mol. Cell. Biol. 2005, 25, 5363–5379. [Google Scholar] [CrossRef]
- Bakkenist, C.J.; Kastan, M.B. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature 2003, 421, 499–506. [Google Scholar] [CrossRef]
- Limbo, O.; Yamada, Y.; Russell, P. Mre11-Rad50-dependent activity of ATM/Tel1 at DNA breaks and telomeres in the absence of Nbs1. Mol. Biol. Cell 2018, 29, 1389–1399. [Google Scholar] [CrossRef]
- Sun, Y.; Jiang, X.; Xu, Y.; Ayrapetov, M.K.; Moreau, L.A.; Whetstine, J.R.; Price, B.D. Histone H3 methylation links DNA damage detection to activation of the tumour suppressor Tip60. Nat. Cell Biol. 2009, 11, 1376–1382. [Google Scholar] [CrossRef] [PubMed]
- Rogakou, E.P.; Pilch, D.R.; Orr, A.H.; Ivanova, V.S.; Bonner, W.M. DNA Double-stranded Breaks Induce Histone H2AX Phosphorylation on Serine 139. J. Biol. Chem. 1998, 273, 5858–5868. [Google Scholar] [CrossRef] [PubMed]
- Bonner, W.M.; Redon, C.E.; Dickey, J.S.; Nakamura, A.J.; Sedelnikova, O.A.; Solier, S.; Pommier, Y. GammaH2AX and cancer. Nat. Rev. Cancer 2008, 8, 957–967. [Google Scholar] [CrossRef] [PubMed]
- Lou, Z.; Minter-Dykhouse, K.; Franco, S.; Gostissa, M.; Rivera, M.A.; Celeste, A.; Manis, J.P.; van Deursen, J.; Nussenzweig, A.; Paull, T.T.; et al. MDC1 Maintains Genomic Stability by Participating in the Amplification of ATM-Dependent DNA Damage Signals. Mol. Cell 2006, 21, 187–200. [Google Scholar] [CrossRef] [PubMed]
- Huen, M.S.Y.; Grant, R.; Manke, I.; Minn, K.; Yu, X.; Yaffe, M.B.; Chen, J. RNF8 Transduces the DNA-Damage Signal via Histone Ubiquitylation and Checkpoint Protein Assembly. Cell 2007, 131, 901–914. [Google Scholar] [CrossRef] [PubMed]
- Doil, C.; Mailand, N.; Bekker-Jensen, S.; Menard, P.; Larsen, D.H.; Pepperkok, R.; Ellenberg, J.; Panier, S.; Durocher, D.; Bartek, J.; et al. RNF168 Binds and Amplifies Ubiquitin Conjugates on Damaged Chromosomes to Allow Accumulation of Repair Proteins. Cell 2009, 136, 435–446. [Google Scholar] [CrossRef] [PubMed]
- Stewart, G.S.; Panier, S.; Townsend, K.; Al-Hakim, A.K.; Kolas, N.K.; Miller, E.S.; Nakada, S.; Ylanko, J.; Olivarius, S.; Mendez, M.; et al. The RIDDLE Syndrome Protein Mediates a Ubiquitin-Dependent Signaling Cascade at Sites of DNA Damage. Cell 2009, 136, 420–434. [Google Scholar] [CrossRef]
- Larrieu, D. Identification de Nouvelles Fonctions Suppressives de Tumeurs Pour ING2: Implication Dans la Réplication et la Réponse Aux Dommages à l’ADN. 2010. Available online: http://www.theses.fr (accessed on 18 September 2019).
- Guerillon, C.; Larrieu, D.; Mourcin, F.; Brambilla, C.; Sengupta, S.; Pedeux, R. 532 The Tumor Suppressive Protein ING2 is Required for DNA Damage Response Proteins Recruitment and Promotes NHEJ. Eur. J. Cancer 2012, 48, S126. [Google Scholar] [CrossRef]
- Mattiroli, F.; Vissers, J.H.A.; van Dijk, W.J.; Ikpa, P.; Citterio, E.; Vermeulen, W.; Marteijn, J.A.; Sixma, T.K. RNF168 Ubiquitinates K13-15 on H2A/H2AX to Drive DNA Damage Signaling. Cell 2012, 150, 1182–1195. [Google Scholar] [CrossRef]
- Bohgaki, M.; Bohgaki, T.; El Ghamrasni, S.; Srikumar, T.; Maire, G.; Panier, S.; Fradet-Turcotte, A.; Stewart, G.S.; Raught, B.; Hakem, A.; et al. RNF168 ubiquitylates 53BP1 and controls its response to DNA double-strand breaks. Proc. Natl. Acad. Sci. USA 2013, 110, 20982–20987. [Google Scholar] [CrossRef]
- Fradet-Turcotte, A.; Canny, M.D.; Escribano-Díaz, C.; Orthwein, A.; Leung, C.C.Y.; Huang, H.; Landry, M.C.; Kitevski-Leblanc, J.; Noordermeer, S.M.; Sicheri, F.; et al. 53BP1 is a reader of the DNA-damage-induced H2A Lys 15 ubiquitin mark. Nature 2013, 499, 50–54. [Google Scholar] [CrossRef] [PubMed]
- Jacquet, K.; Fradet-Turcotte, A.; Avvakumov, N.; Lambert, J.P.; Roques, C.; Pandita, R.K.; Paquet, E.; Herst, P.; Gingras, A.C.; Pandita, T.K.; et al. The TIP60 Complex Regulates Bivalent Chromatin Recognition by 53BP1 through Direct H4K20me Binding and H2AK15 Acetylation. Mol. Cell 2016, 62, 409–421. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Lee, J.H.; Jiang, W.; Crowe, J.L.; Zha, S.; Paull, T.T. Regulation of the DNA Damage Response by DNA-PKcs Inhibitory Phosphorylation of ATM. Mol. Cell 2017, 65, 91–104. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Gong, Y.; Peng, B.; Shi, R.; Fan, D.; Zhao, H.; Zhu, M.; Zhang, H.; Lou, Z.; Zhou, J.; et al. MRE11 UFMylation promotes ATM activation. Nucl. Acids Res. 2019, 47, 4124–4135. [Google Scholar] [CrossRef] [PubMed]
- Qin, B.; Yu, J.; Nowsheen, S.; Wang, M.; Tu, X.; Liu, T.; Li, H.; Wang, L.; Lou, Z. UFL1 promotes histone H4 ufmylation and ATM activation. Nat. Commun. 2019, 10, 1242. [Google Scholar] [CrossRef]
- Cazzalini, O.; Perucca, P.; Savio, M.; Necchi, D.; Bianchi, L.; Stivala, L.A.; Ducommun, B.; Scovassi, A.I.; Prosperi, E. Interaction of p21 CDKN1A with PCNA regulates the histone acetyltransferase activity of p300 in nucleotide excision repair. Nucl. Acids Res. 2008, 36, 1713–1722. [Google Scholar] [CrossRef]



© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Archambeau, J.; Blondel, A.; Pedeux, R. Focus-ING on DNA Integrity: Implication of ING Proteins in Cell Cycle Regulation and DNA Repair Modulation. Cancers 2020, 12, 58. https://doi.org/10.3390/cancers12010058
Archambeau J, Blondel A, Pedeux R. Focus-ING on DNA Integrity: Implication of ING Proteins in Cell Cycle Regulation and DNA Repair Modulation. Cancers. 2020; 12(1):58. https://doi.org/10.3390/cancers12010058
Chicago/Turabian StyleArchambeau, Jérôme, Alice Blondel, and Rémy Pedeux. 2020. "Focus-ING on DNA Integrity: Implication of ING Proteins in Cell Cycle Regulation and DNA Repair Modulation" Cancers 12, no. 1: 58. https://doi.org/10.3390/cancers12010058
APA StyleArchambeau, J., Blondel, A., & Pedeux, R. (2020). Focus-ING on DNA Integrity: Implication of ING Proteins in Cell Cycle Regulation and DNA Repair Modulation. Cancers, 12(1), 58. https://doi.org/10.3390/cancers12010058




