Biological Functions of the ING Proteins
Abstract
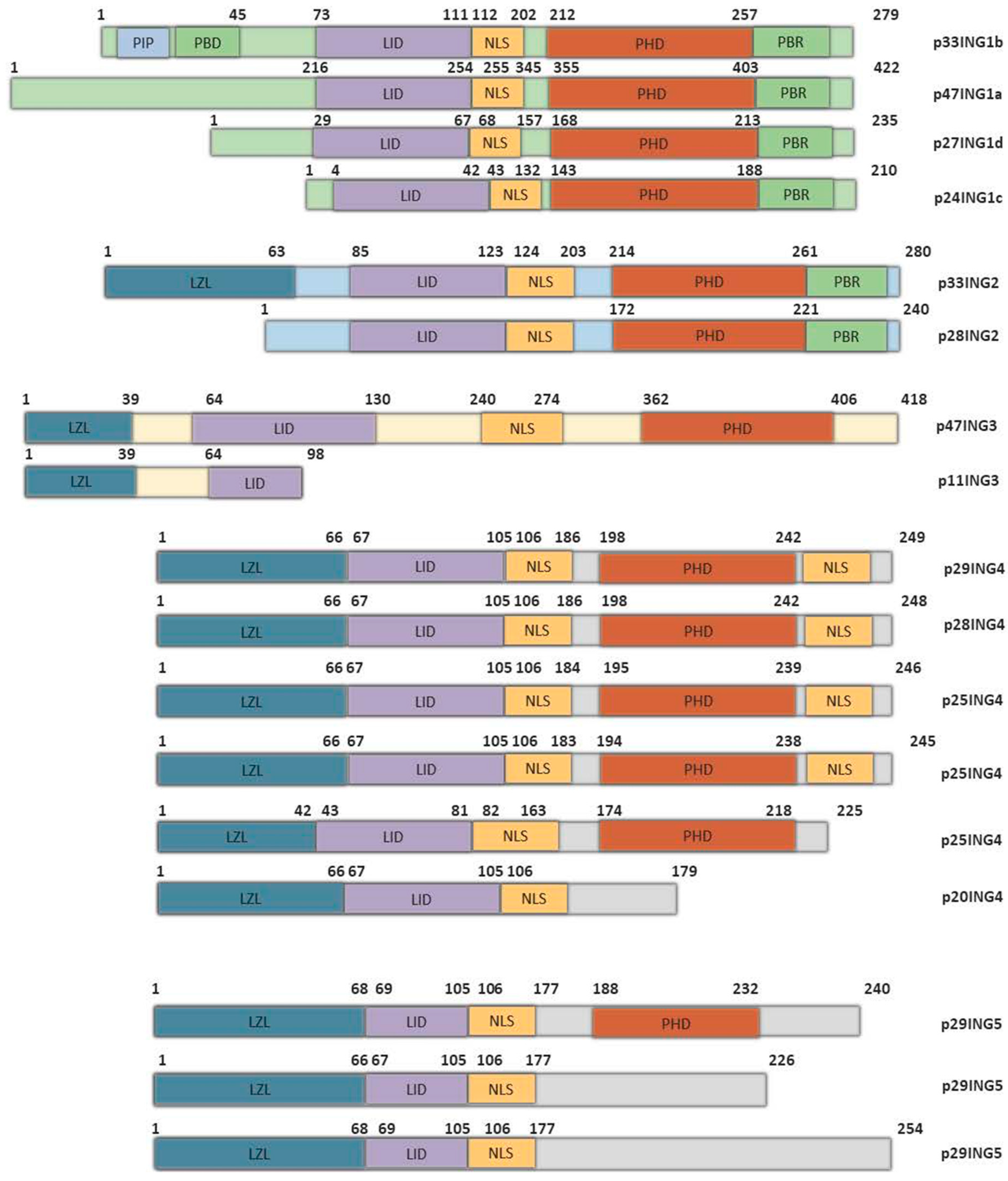
:1. Introduction
2. A PHD for RegulatING Histone Acetylation
3. ING1: PreventING Abnormal Growth
4. ING2: ControlING Spermatogenesis and Tumor Growth
5. ING3: A distINGuished Member of the ING Family
6. ING4: SupressING NF-κB
7. ING5: DifferentiatING Stem Cells
8. Discussion and Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Toh, T.B.; Lim, J.J.; Chow, E.K.H. Epigenetics in cancer stem cells. Mol. Cancer 2017, 16, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Qureshi, I.A.; Mehler, M.F. Advances in epigenetics and epigenomics for neurodegenerative diseases. Curr. Neurol. Neurosci. Rep. 2011, 11, 464–473. [Google Scholar] [CrossRef] [PubMed]
- Rosenquist, R.; Esteller, M.; Plass, C. Introduction: Epigenetics in cancer. Semin. Cancer Biol. 2018, 51, iv–v. [Google Scholar] [CrossRef] [PubMed]
- Nicoglou, A.; Merlin, F. Epigenetics: A way to bridge the gap between biological fields. Stud. Hist. Philos. Biol. Biomed. Sci. 2017, 66, 73–82. [Google Scholar] [CrossRef] [PubMed]
- Dawson, M.A.; Kouzarides, T. Review Cancer Epigenetics: From Mechanism to Therapy. Cell 2012, 150, 12–27. [Google Scholar] [CrossRef]
- Jones, P.A.; Baylin, S.B. The fundamental role of epigenetic events in cancer. Nat. Rev. Genet. 2002, 3, 415–428. [Google Scholar] [CrossRef]
- Tallen, G.; Riabowol, K. Keep-ING balance: Tumor suppression by epigenetic regulation. FEBS Lett. 2014, 588, 2728–2742. [Google Scholar] [CrossRef]
- Turner, B.M. Defining an epigenetic code. Nat. Cell Biol. 2007, 9, 2–6. [Google Scholar] [CrossRef]
- Perrera, V.; Martello, G. How Does Reprogramming to Pluripotency Affect Genomic Imprinting? Front. Cell Dev. Biol. 2019, 7, 1–16. [Google Scholar] [CrossRef]
- Bostick, M.; Kin, J.K.; Esteve, P.-O.; Clark, A.; Pradhan, S.; Jacobsen, S. UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science 2007, 317, 1760–1765. [Google Scholar] [CrossRef]
- Kaneda, M.; Okano, M.; Hata, K.; Sado, T.; Tsujimoto, H.; Li, E.; Sasaki, H. Essential role for de novo DNA methyltransferase Dnmt3a in paternal and maternal imprinting. Nature 2004, 429, 900–903. [Google Scholar] [CrossRef] [PubMed]
- Di Lorenzo, A.; Bedford, M.T. Histone arginine methylation. FEBS Lett. 2011, 585, 2024–2031. [Google Scholar] [CrossRef] [PubMed]
- Qin, J.; Wen, B.; Liang, Y.; Yu, W.; Li, H. Histone Modifications and their Role in Colorectal Cancer (Review). Pathol. Oncol. Res. 2019. [Google Scholar] [CrossRef] [PubMed]
- Berger, S.L. The complex language of chromatin regulation during transcription. Nature 2007, 447, 407–412. [Google Scholar] [CrossRef] [PubMed]
- Trisciuoglio, D.; Di Martile, M.; Del Bufalo, D. Emerging Role of Histone Acetyltransferase in Stem Cells and Cancer. Stem Cells Int. 2018, 2018, 1–11. [Google Scholar] [CrossRef]
- Guérillon, C.; Larrieu, D.; Pedeux, R. ING1 and ING2: Multifaceted tumor suppressor genes. Cell. Mol. Life Sci. 2013, 70, 3753–3772. [Google Scholar] [CrossRef]
- Feng, X.; Bonni, S.; Riabowol, K. HSP70 Induction by ING Proteins Sensitizes Cells to Tumor Necrosis Factor Alpha Receptor-Mediated Apoptosis. Mol. Cell. Biol. 2006, 26, 9244–9255. [Google Scholar] [CrossRef]
- Mulder, K.W.; Wang, X.; Escriu, C.; Ito, Y.; Schwarz, R.F.; Gillis, J.; Sirokmány, G.; Donati, G.; Uribe-lewis, S.; Pavlidis, P.; et al. Diverse epigenetic strategies interact to control epidermal differentiation. Nat. Cell Biol. 2012, 14, 753–763. [Google Scholar] [CrossRef]
- Bertschmann, J.; Thalappilly, S.; Riabowol, K. The ING1a model of rapid cell senescence. Mech. Ageing Dev. 2019, 177, 109–117. [Google Scholar] [CrossRef]
- Chen, J.; Tran, U.M.; Rajarajacholan, U.; Thalappilly, S.; Riabowol, K. ING1b-inducible microRNA203 inhibits cell proliferation. Br. J. Cancer 2013, 108, 1143–1148. [Google Scholar] [CrossRef]
- Gournay, M.; Paineau, M.; Archambeau, J.; Pedeux, R. Regulat-INGs in tumors and diseases: Focus on ncRNAs. Cancer Lett. 2019, 447, 66–74. [Google Scholar] [CrossRef] [PubMed]
- Doyon, Y.; Cayrou, C.; Ullah, M.; Landry, A.-J.; Côté, V.; Selleck, W.; Lane, W.S.; Tan, S.; Yang, X.-J.; Côté, J. ING tumor suppressor proteins are critical regulators of chromatin acetylation required for genome expression and perpetuation. Mol. Cell 2006, 21, 51–64. [Google Scholar] [CrossRef] [PubMed]
- Bose, P.; Thakur, S.; Thalappilly, S.; Ahn, B.Y.; Satpathy, S.; Feng, X.; Suzuki, K.; Kim, S.W.; Riabowol, K. ING1 induces apoptosis through direct effects at the mitochondria. Cell Death Dis. 2013, 4, e788. [Google Scholar] [CrossRef] [PubMed]
- Taverna, S.D.; Ilin, S.; Rogers, R.S.; Tanny, J.C.; Lavender, H.; Li, H.; Baker, L.; Boyle, J.; Blair, L.P.; Chait, B.T.T.; et al. Yng1 PHD Finger Binding to H3 Trimethylated at K4 Promotes NuA3 HAT Activity at K14 of H3 and Transcription at a Subset of Targeted ORFs. Mol. Cell 2006, 24, 785–796. [Google Scholar] [CrossRef]
- Peña, P.V.; Davrazou, F.; Shi, X.; Walter, K.L.; Verkhusha, V.V.; Gozani, O.; Zhao, R.; Kutateladze, T.G. Molecular mechanism of histone H3K4me3 recognition by plant homeodomain of ING2. Nature 2006, 442, 100–103. [Google Scholar] [CrossRef]
- Kuzmichev, A.; Zhang, Y.; Erdjument-Bromage, H.; Tempst, P.; Reinberg, D. Role of the Sin3-histone deacetylase complex in growth regulation by the candidate tumor suppressor p33(ING1). Mol. Cell. Biol. 2002, 22, 835–848. [Google Scholar] [CrossRef]
- Li, H.; Ilin, S.; Wang, W.; Duncan, E.M.; Wysocka, J.; Allis, C.D.; Patel, D.J. Molecular basis for site-specific read-out of histone H3K4me3 by the BPTF PHD finger of NURF. Nature 2006, 442, 91–95. [Google Scholar] [CrossRef]
- Mishima, Y.; Miyagi, S.; Saraya, A.; Negishi, M.; Endoh, M.; Endo, T.A.; Toyoda, T.; Shinga, J.; Katsumoto, T.; Chiba, T.; et al. The Hbo1-Brd1/Brpf2 complex is responsible for global acetylation of H3K14 and required for fetal liver erythropoiesis. Blood 2011, 118, 2443–2453. [Google Scholar] [CrossRef]
- Doyon, Y.; Selleck, W.; Lane, W.S.; Tan, S.; Côté, J.; Co, J. Structural and Functional Conservation of the NuA4 Histone Acetyltransferase Complex from Yeast to Humans. Mol. Cell. Biol. 2004, 24, 1884–1896. [Google Scholar] [CrossRef]
- He, G.H.Y.; Helbing, C.C.; Wagner, M.J.; Sensen, C.W.; Riabowol, K. Phylogenetic analysis of the ING family of PHD finger proteins. Mol. Biol. Evol. 2005, 22, 104–116. [Google Scholar] [CrossRef]
- Han, X.; Feng, X.; Rattner, J.B.; Smith, H.; Bose, P.; Suzuki, K.; Soliman, M.A.; Scott, M.S.; Burke, B.E.; Riabowol, K. Tethering by lamin A stabilizes and targets the ING1 tumour suppressor. Nat. Cell Biol. 2008, 10, 1333–1340. [Google Scholar] [CrossRef] [PubMed]
- Eapen, S.A.; Netherton, S.J.; Sarker, K.P.; Deng, L.; Chan, A.; Riabowol, K.; Bonni, S. Identification of a novel function for the chromatin remodeling protein ING2 in muscle differentiation. PLoS ONE 2012, 7, e40684. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaadige, M.R.; Ayer, D.E. The Polybasic Region That Follows the Plant Homeodomain Zinc Finger 1 of Pf1 Is Necessary and Sufficient for Specific Phosphoinositide Binding. J. Biol. Chem. 2006, 281, 28831–28836. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Satpathy, S.; Nabbi, A.; Riabowol, K. RegulatING chromatin regulators: Post-translational modification of the ING family of epigenetic regulators. Biochem. J. 2013, 450, 433–442. [Google Scholar] [CrossRef] [Green Version]
- Garkavtsev, I.; Kazarov, A.; Gudkov, A.; Riabowol, K. Suppression of the novel growth inhibitor p33ING1 promotes neoplastic transformation. Nat. Genet. 1996, 14, 415–420. [Google Scholar] [CrossRef]
- Soliman, M.A.; Berardi, P.; Pastyryeva, S.; Bonnefin, P.; Feng, X.; Colina, A.; Young, D.; Riabowol, K. ING1a expression increases during replicative senescence and induces a senescent phenotype. Aging Cell 2008, 7, 783–794. [Google Scholar] [CrossRef]
- Soliman, M.A.; Riabowol, K. After a decade of study-ING, a PHD for a versatile family of proteins. Trends Biochem. Sci. 2007, 32, 509–519. [Google Scholar] [CrossRef]
- Scott, M.; Bonnefin, P.; Vieyra, D.; Boisvert, F.M.; Young, D.; Bazett-Jones, D.P.; Riabowol, K. UV-induced binding of ING1 to PCNA regulates the induction of apoptosis. J. Cell Sci. 2001, 114, 3455–3462. [Google Scholar]
- Russell, M.; Berardi, P.; Gong, W.; Riabowol, K. Grow-ING, Age-ING and Die-ING: ING proteins link cancer, senescence and apoptosis. Exp. Cell Res. 2006, 312, 951–961. [Google Scholar] [CrossRef]
- Gozani, O.; Karuman, P.; Jones, D.R.; Ivanov, D.; Cha, J.; Lugovskoy, A.A.; Baird, C.L.; Zhu, H.; Field, S.J.; Lessnick, S.L.; et al. The PHD Finger of the Chromatin-Associated Protein ING2 Functions as a Nuclear Phosphoinositide Receptor. Cell 2003, 114, 99–111. [Google Scholar] [CrossRef]
- Thakur, S.; Feng, X.; Qiao Shi, Z.; Ganapathy, A.; Kumar Mishra, M.; Atadja, P.; Morris, D.; Riabowol, K. ING1 and 5-azacytidine act synergistically to block breast cancer cell growth. PLoS ONE 2012, 7, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goeman, F.; Thormeyer, D.; Abad, M.; Serrano, M.; Schmidt, O.; Palmero, I.; Baniahmad, A. Growth Inhibition by the Tumor Suppressor p33ING1 in Immortalized and Primary Cells: Involvement of Two Silencing Domains and Effect of Ras. Mol. Cell. Biol. 2005, 25, 422–431. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kichina, J.V.; Zeremski, M.; Aris, L.; Gurova, K.V.; Walker, E.; Franks, R.; Nikitin, A.Y.; Kiyokawa, H.; Gudkov, A.V. Targeted disruption of the mouse ing1 locus results in reduced body size, hypersensitivity to radiation and elevated incidence of lymphomas. Oncogene 2006, 25, 857–866. [Google Scholar] [CrossRef] [PubMed]
- Coles, A.H.; Liang, H.; Zhu, Z.; Marfella, C.G.A.; Kang, J.; Imbalzano, A.N.; Jones, S.N. Deletion of p37Ing1 in mice reveals a p53-independent role for Ing1 in the suppression of cell proliferation, apoptosis, and tumorigenesis. Cancer Res. 2007, 67, 2054–2061. [Google Scholar] [CrossRef] [Green Version]
- Coles, A.H.; Marfella, C.G.A.; Imbalzano, A.N.; Steinman, H.A.; Garlick, D.S.; Gerstein, R.M.; Jones, S.N. p37Ing1b regulates B-cell proliferation and cooperates with p53 to suppress diffuse large B-cell lymphomagenesis. Cancer Res. 2008, 68, 8705–8714. [Google Scholar] [CrossRef] [Green Version]
- Shah, S.; Smith, H.; Feng, X.; Rancourt, D.E.; Riabowol, K. ING function in apoptosis in diverse model systemsThis paper is one of a selection of papers published in this Special Issue, entitled CSBMCB’s 51st Annual Meeting–Epigenetics and Chromatin Dynamics, and has undergone the Journal’s usual peer review pro. Biochem. Cell Biol. 2009, 87, 117–125. [Google Scholar] [CrossRef]
- Nagashima, M.; Shiseki, M.; Miura, K.; Hagiwara, K.; Linke, S.P.; Pedeux, R.; Wang, X.W.; Yokota, J.; Riabowol, K.; Harris, C.C. DNA damage-inducible gene p33ING2 negatively regulates cell proliferation through acetylation of p53. Proc. Natl. Acad. Sci. USA 2001, 98, 9671–9676. [Google Scholar] [CrossRef] [Green Version]
- Pedeux, R.; Sengupta, S.; Shen, J.C.; Demidov, O.N.; Saito, S.; Onogi, H.; Kumamoto, K.; Wincovitch, S.; Garfield, S.H.; McMenamin, M.; et al. ING2 Regulates the Onset of Replicative Senescence by Induction of p300-Dependent p53 Acetylation. Mol. Cell. Biol. 2005, 25, 6639–6648. [Google Scholar] [CrossRef] [Green Version]
- Kumamoto, K.; Spillare, E.A.; Fujita, K.; Horikawa, I.; Yamashita, T.; Appella, E.; Nagashima, M.; Takenoshita, S.; Yokota, J.; Harris, C.C. Nutlin-3a activates p53 to both down-regulate inhibitor of growth 2 and up-regulate mir-34a, mir-34b, and mir-34c expression, and induce senescence. Cancer Res. 2008, 68, 3193–3203. [Google Scholar] [CrossRef] [Green Version]
- Kumamoto, K.; Fujita, K.; Kurotani, R.; Saito, M.; Unoki, M.; Hagiwara, N.; Shiga, H.; Bowman, E.D.; Yanaihara, N.; Okamura, S.; et al. ING2 is upregulated in colon cancer and increases invasion by enhanced MMP13 expression. Int. J. Cancer 2009, 125, 1306–1315. [Google Scholar] [CrossRef] [Green Version]
- Smith, K.T.; Martin-Brown, S.A.; Florens, L.; Washburn, M.P.; Workman, J.L. Deacetylase Inhibitors Dissociate the Histone-Targeting ING2 Subunit from the Sin3 Complex. Chem. Biol. 2010, 17, 65–74. [Google Scholar] [CrossRef] [Green Version]
- Zhong, J.; Yang, L.; Liu, N.; Zheng, J.; Lin, C.Y. Knockdown of inhibitor of growth protein 2 inhibits cell invasion and enhances chemosensitivity to 5-fu in human gastric cancer cells. Dig. Dis. Sci. 2013, 58, 3189–3197. [Google Scholar] [CrossRef] [PubMed]
- Saito, M.; Kumamoto, K.; Robles, A.I.; Horikawa, I.; Furusato, B.; Okamura, S.; Goto, A.; Yamashita, T.; Nagashima, M.; Lee, T.L.; et al. Targeted disruption of ing2 results in defective spermatogenesis and development of soft-tissue sarcomas. PLoS ONE 2010, 5, e15541. [Google Scholar] [CrossRef] [PubMed]
- Binda, O.; Nassif, C.; Branton, P.E. SIRT1 negatively regulates HDAC1-dependent transcriptional repression by the RBP1 family of proteins. Oncogene 2008, 27, 3384–3392. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Squatrito, M.; Gorrini, C.; Amati, B. Tip60 in DNA damage response and growth control: Many tricks in one HAT. Trends Cell Biol. 2006, 16, 433–442. [Google Scholar] [CrossRef] [PubMed]
- Mouche, A.; Archambeau, J.; Ricordel, C.; Chaillot, L.; Bigot, N.; Guillaudeux, T.; Grenon, M.; Pedeux, R. ING3 is required for ATM signaling and DNA repair in response to DNA double strand breaks. Cell Death Differ. 2019, 26, 2344–2357. [Google Scholar] [CrossRef]
- Zhao, S.; Wang, L.; Zhang, C.; Deng, Y.; Zhao, B.; Ren, Y.; Fu, Y.; Meng, X. Inhibitor of growth 3 induces cell death by regulating cell proliferation, apoptosis and cell cycle arrest by blocking the PI3K/AKT pathway. Cancer Gene Ther. 2018, 25, 240–247. [Google Scholar] [CrossRef]
- McClurg, U.L.; Nabbi, A.; Ricordel, C.; Korolchuk, S.; McCracken, S.; Heer, R.; Wilson, L.; Butler, L.M.; Irving-Hooper, B.K.; Pedeux, R.; et al. Human ex vivo prostate tissue model system identifies ING3 as an oncoprotein. Br. J. Cancer 2018, 118, 713–726. [Google Scholar] [CrossRef] [Green Version]
- Nabbi, A.; McClurg, U.L.; Thalappilly, S.; Almami, A.; Mobahat, M.; Bismar, T.A.; Binda, O.; Riabowol, K.T. ING3 promotes prostate cancer growth by activating the androgen receptor. BMC Med. 2017, 15, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Cekaite, L.; Clancy, T.; Sioud, M. Increased miR-21 expression during human monocyte differentiation into DCs. Front. Biosci. 2010, E2, 818–828. [Google Scholar] [CrossRef] [Green Version]
- Fink, D.; Yau, T.Y.; Nabbi, A.; Wagner, B.; Wagner, C.; Misa Hu, S.T.; Lang, V.; Handschuh, S.; Riabowol, K.; Rülicke, T. Loss of Ing3 expression results in growth retardation and embryonic death. In Proceedings of the ECAR Meeting, VetMeduni, Vienna, Austria, 4–6 July 2019. [Google Scholar]
- Feng, X.; Hara, Y.; Riabowol, K. Different HATS of the ING1 gene family. Trends Cell Biol. 2002, 12, 532–538. [Google Scholar] [CrossRef]
- Palacios, A.; Moreno, A.; Oliveira, B.L.; Rivera, T.; Prieto, J.; García, P.; Fernández-Fernández, M.R.; Bernadó, P.; Palmero, I.; Blanco, F.J. The dimeric structure and the bivalent recognition of H3K4me3 by the tumor suppressor ING4 suggests a mechanism for enhanced targeting of the HBO1 complex to chromatin. J. Mol. Biol. 2010, 396, 1117–1127. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.; Cheng, Y.; Su, G. The essential role of tumor suppressor gene ING4 in various human cancers and non-neoplastic disorders. Biosci. Rep. 2019, 39, BSR20180773. [Google Scholar] [CrossRef] [PubMed]
- Saksouk, N.; Avvakumov, N.; Champagne, K.S.; Hung, T.; Doyon, Y.; Cayrou, C.; Paquet, E.; Ullah, M.; Landry, A.J.; Côté, V.; et al. HBO1 HAT Complexes Target Chromatin throughout Gene Coding Regions via Multiple PHD Finger Interactions with Histone H3 Tail. Mol. Cell 2009, 33, 257–265. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wright, D.G.; Marchal, C.; Hoang, K.; Ankney, J.A.; Nguyen, S.T.; Rushing, A.W.; Polakowski, N.; Miotto, B.; Lemasson, I. Human T-cell leukemia virus type-1-encoded protein HBZ represses p53 function by inhibiting the acetyltransferase activity of p300/CBP and HBO1. Oncotarget 2016, 7, 1687–1706. [Google Scholar] [CrossRef] [Green Version]
- Iizuka, M.; Sarmento, O.F.; Sekiya, T.; Scrable, H.; Allis, C.D.; Smith, M.M. Hbo1 Links p53-Dependent Stress Signaling to DNA Replication Licensing. Mol. Cell. Biol. 2008, 28, 140–153. [Google Scholar] [CrossRef] [Green Version]
- Contzler, R.; Regamey, A.; Favre, B.; Roger, T.; Hohl, D.; Huber, M. Histone acetyltransferase HBO1 inhibits NF-κB activity by coactivator sequestration. Biochem. Biophys. Res. Commun. 2006, 350, 208–213. [Google Scholar] [CrossRef]
- Lu, L.; Li, J.; Le, Y.; Jiang, H. Inhibitor of growth 4 (ING4) inhibits hypoxia-induced EMT by decreasing HIF-1α and snail in HK2 cells. Acta Histochem. 2019, 121, 695–703. [Google Scholar] [CrossRef]
- Kuphal, S.; Bosserhoff, A. Reduced expression and novel splice variants of ING4 in human gastric adenocarcinoma. J. Pathol. 2009, 219, 400–409. [Google Scholar] [CrossRef]
- Li, J.; Li, G. Cell cycle regulator ING4 is a suppressor of melanoma angiogenesis that is regulated by the metastasis suppressor BRMS1. Cancer Res. 2010, 70, 10445–10453. [Google Scholar] [CrossRef] [Green Version]
- Berger, P.L.; Winn, M.E.; Miranti, C.K. Miz1, a Novel Target of ING4, Can Drive Prostate Luminal Epithelial Cell Differentiation. Prostate 2017, 59, 49–59. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Zhang, H.; Sheng, W.; Xiang, J. Adenovirus-mediated ING4 expression suppresses lung carcinoma cell growth via induction of cell cycle alteration and apoptosis and inhibition of tumor invasion and angiogenesis. Cancer Lett. 2008, 271, 105–116. [Google Scholar] [CrossRef] [PubMed]
- Lou, C.; Jiang, S.; Guo, X. ING4 is negatively correlated with microvessel density in colon cancer. Tumor Biol. 2012, 33, 2357–2364. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Yu, L.; Wang, Y. Expression of tumor suppressor gene ING4 in ovarian carcinoma is correlated with microvessel density. J. Cancer Res. Clin. Oncol. 2012, 138, 647–655. [Google Scholar] [CrossRef]
- Ren, Y.; Zhao, S.; Chen, H.; Fu, Y.M.; Zhao, B. Association between the expression of inhibitor of growth family member 4 and the progression of clear cell renal carcinoma. Oncol. Lett. 2017, 14, 2453–2457. [Google Scholar] [CrossRef] [Green Version]
- Berger, P.L.; Frank, S.B.; Schulz, V.V.; Eric, A.N.; Edick, M.J.; Holly, B.; Chang, T.-T.A.; Hostetter, G.; Kim, S.; Miranti, C.K. Transient Induction of ING4 by MYC Drives Prostate Epithelial Cell Differentiation and its Disruption Drives Prostate Tumorigenesis. Cancer Res. 2014, 74, 3357–3368. [Google Scholar] [CrossRef] [Green Version]
- Garkavtsev, I.; Kozin, S.V.; Chernova, O.; Xu, L. The candidate tumour suppressor protein ING4 regulates brain tumour growth and angiogenesis. Nature 2004, 428, 328–332. [Google Scholar] [CrossRef]
- Nozell, S.; Laver, T.; Moseley, D.; Nowoslawski, L.; Devos, M.; Atkinson, G.P.; Harrison, K.; Nabors, L.B.; Benveniste, E.N. The ING4 Tumor Suppressor Attenuates NF- KB Activity at the Promoters of Target Genes. Mol. Cell. Biol. 2008, 28, 6632–6645. [Google Scholar] [CrossRef] [Green Version]
- Coles, A.H.; Gannon, H.; Cerny, A.; Kurt-Jones, E.; Jones, S.N. Inhibitor of growth-4 promotes IkappaB promoter activation to suppress NF-kappaB signaling and innate immunity. Proc. Natl. Acad. Sci. USA 2010, 107, 11423–11428. [Google Scholar] [CrossRef] [Green Version]
- Nashun, B.; Hill, P.W.; Hajkova, P. Reprogramming of cell fate: Epigenetic memory and the erasure of memories past. Embo J. 2015, 34, 1296–1308. [Google Scholar] [CrossRef]
- Awe, J.P.; Byrne, J.A. Identifying candidate oocyte reprogramming factors using cross-species global transcriptional analysis. Cell. Reprogram. 2013, 15, 126–133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Huang, W.; Wu, Y.; Hou, J.; Nie, Y.; Gu, H.; Li, J.; Hu, S.; Zhang, H. MicroRNA-193 Pro-Proliferation Effects for Bone Mesenchymal Stem Cells After Low-Level Laser Irradiation Treatment Through Inhibitor of Growth Family, Member 5. Stem Cells Dev. 2012, 21, 2508–2519. [Google Scholar] [CrossRef] [PubMed]
- Tanis, S.E.J.; Jansen, P.W.T.C.; Zhou, H.; van Heeringen, S.J.; Vermeulen, M.; Kretz, M.; Mulder, K.W. Splicing and Chromatin Factors Jointly Regulate Epidermal Differentiation. Cell Rep. 2018, 25, 1292–1303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, F.; Wang, A.Y.; Chesnelong, C.; Yang, Y.; Nabbi, A.; Thalappilly, S.; Alekseev, V.; Riabowol, K. ING5 activity in self-renewal of glioblastoma stem cells via calcium and follicle stimulating hormone pathways. Oncogene 2017, 37, 286–301. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ullah, M.; Pelletier, N.; Xiao, L.; Zhao, S.P.; Wang, K.; Degerny, C.; Tahmasebi, S.; Cayrou, C.; Doyon, Y.; Goh, S.; et al. Molecular Architecture of Quartet MOZ/MORF Histone Acetyltransferase Complexes. Mol. Cell. Biol. 2008, 28, 6828–6843. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Katsumoto, T.; Aikawa, Y.; Iwama, A.; Ueda, S.; Ichikawa, H.; Ochiya, T.; Kitabayashi, I. MOZ is essential for maintenance of hematopoietic stem cells. Genes Dev. 2006, 20, 1321–1330. [Google Scholar] [CrossRef] [Green Version]
- Thomas, T.; Corcoran, L.M.; Gugasyan, R.; Dixon, M.P.; Brodnicki, T.; Nutt, S.L.; Metcalf, D.; Voss, A.K. Monocytic leukemia zinc finger protein is essential for the development of long-term reconstituting hematopoietic stem cells. Genes Dev. 2006, 20, 1175–1186. [Google Scholar] [CrossRef] [Green Version]
- Perez-Campo, F.M.; Costa, G.; Lie-a-Ling, M.; Stifani, S.; Kouskoff, V.; Lacaud, G. MOZ-mediated repression of p16(INK) is critical for the self-renewal of neural and hematopoietic stem cells. Stem Cells 2014, 32, 1591–1601. [Google Scholar] [CrossRef] [Green Version]
- Blanchard, K.L.; Liang, J.; Prouty, L.; Williams, B.J.; Dayton, M.A. Acute mixed lineage leukemia with an inv(8)(p11q13) resulting in fusion of the genes for MOZ and TIF2. Blood 1998, 92, 2118–2122. [Google Scholar]
- Cross, N.C.P.; Carapeti, M.; Aguiar, R.C.T.; Goldman, J.M. A novel fusion between MOZ and the nuclear receptor coactivator TIF2 in acute myeloid leukemia. Blood 1998, 91, 3127–3133. [Google Scholar]
- Huntly, B.J.P.; Deguchi, K.; Lee, B.H.; Duclos, N.; Rowan, R.; Amaral, S.; Curley, D.; Gilliland, D.G.; Shigematsu, H.; Mizuno, S.; et al. MOZ-TIF2, but not BCR-ABL, confers properties of leukemic stem cells to committed murine hematopoietic progenitors. Cancer Cell 2004, 6, 587–596. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deguchi, K.; Ayton, P.M.; Carapeti, M.; Kutok, J.L.; Snyder, C.S.; Williams, I.R.; Cross, N.C.P.; Glass, C.K.; Cleary, M.L.; Gilliland, D.G. MOZ-TIF2-induced acute myeloid leukemia requires the MOZ nucleosome binding motif and TIF2-mediated recruitment of CBP. Cancer Cell 2003, 3, 259–271. [Google Scholar] [CrossRef] [Green Version]
- Thomas, T.; Voss, A.K.; Chowdhury, K.; Gruss, P. Querkopf, a MYST family histone acetyltransferase, is required for normal cerebral cortex development. Development 2000, 127, 2537–2548. [Google Scholar] [PubMed]
- Sheikh, B.N.; Dixon, M.P.; Thomas, T.; Voss, A.K. Querkopf is a key marker of self-renewal and multipotency of adult neural stem cells. J. Cell Sci. 2012, 125, 295–309. [Google Scholar] [CrossRef] [Green Version]
- Ormaza, G.; Rodríguez, J.A.; Ibáñez de Opakua, A.; Merino, N.; Villate, M.; Gorroño, I.; Rábano, M.; Palmero, I.; Vilaseca, M.; Kypta, R.; et al. The Tumor Suppressor ING5 Is a Dimeric, Bivalent Recognition Molecule of the Histone H3K4me3 Mark. J. Mol. Biol. 2019, 431, 2298–2319. [Google Scholar] [CrossRef]
- Kueh, A.J.; Dixon, M.P.; Voss, A.K.; Thomas, T. HBO1 Is Required for H3K14 Acetylation and Normal Transcriptional Activity during Embryonic Development. Mol. Cell. Biol. 2011, 31, 845–860. [Google Scholar] [CrossRef] [Green Version]
- Rajarajacholan, U.K.; Thalappilly, S.; Riabowol, K. ING1 regulates rRNA levels by altering nucleolar chromatin structure and mTOR localization. Nucleic Acids Res. 2017, 45, 1776–1792. [Google Scholar] [CrossRef] [Green Version]
- Li, N.; Li, Q.; Cao, X.; Zhao, G.; Xue, L.; Tong, T. The tumor suppressor p33 ING1b upregulates p16 INK4a expression and induces cellular senescence. FEBS Lett. 2011, 585, 3106–3112. [Google Scholar] [CrossRef] [Green Version]
- Gómez-Cabello, D.; Callejas, S.; Benguría, A.; Moreno, A.; Alonso, J.; Palmero, I. Regulation of the MicroRNA Processor DGCR8 by the Tumor Suppressor ING1. Mol. Cell. Pathobiol. Regul. 2010, 70, 1866–1875. [Google Scholar] [CrossRef] [Green Version]
- Thalappilly, S.; Feng, X.; Pastyryeva, S.; Suzuki, K.; Muruve, D.; Larocque, D.; Richard, S.; Truss, M.; von Deimling, A.; Riabowol, K.; et al. The p53 tumor suppressor is stabilized by inhibitor of growth 1 (ING1) by blocking polyubiquitination. PLoS ONE 2011, 6, e21065. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Chin, M.Y.; Li, G. The novel tumor suppressor p33ING2 enhances nucleotide excision repair via inducement of histone H4 acetylation and chromatin relaxation. Cancer Res. 2006, 66, 1906–1911. [Google Scholar] [CrossRef] [PubMed]
- Bua, D.J.; Martin, G.M.; Binda, O.; Gozani, O. Nuclear phosphatidylinositol-5-phosphate regulates ING2 stability at discrete chromatin targets in response to DNA damage. Sci. Rep. 2013, 3, 2137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, M.; Chen, F.; Wang, Q.; Wang, K.; Pan, Q.; Zhang, X. Downregulation of inhibitor of growth 3 is correlated with tumorigenesis and progression of hepatocellular carcinoma. Oncol. Lett. 2012, 4, 47–52. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, H.Y.; Liu, H.L.; Tian, L.T.; Song, R.P.; Song, X.; Yin, D.L.; Liang, Y.J.; Qu, L.D.; Jiang, H.C.; Liu, J.R.; et al. Expression and prognostic value of ING3 in human primary hepatocellular carcinoma. Exp. Biol. Med. 2012, 237, 352–361. [Google Scholar] [CrossRef] [PubMed]
- Raho, G.; Miranda, C.; Tamborini, E.; Pierotti, M.A.; Greco, A. Detection of novel mRNA splice variants of human ING4 tumor suppressor gene. Oncogene 2007, 26, 5247–5257. [Google Scholar] [CrossRef] [Green Version]
- Shiseki, M.; Nagashima, M.; Pedeux, R.M.; Kitahama-Shiseki, M.; Miura, K.; Okamura, S.; Onogi, H.; Higashimoto, Y.; Appella, E.; Yokota, J.; et al. p29ING4 and p28ING5 bind to p53 and p300, and enhance p53 activity. Cancer Res. 2003, 63, 2373–2378. [Google Scholar]
- Li, X.; Kikuchi, K.; Takano, Y. ING Genes Work as Tumor Suppressor Genes in the Carcinogenesis of Head and Neck Squamous Cell Carcinoma. J. Oncol. 2011. [Google Scholar] [CrossRef] [Green Version]
- Xing, Y.N.; Yang, X.; Xu, X.Y.; Zheng, Y.; Xu, H.M.; Takano, Y.; Zheng, H.C. The altered expression of ING5 protein is involved in gastric carcinogenesis and subsequent progression. Hum. Pathol. 2011, 42, 25–35. [Google Scholar] [CrossRef]
- Zhang, F.; Bäumer, N.; Rode, M.; Ji, P.; Zhang, T.; Berdel, W.E.; Müller-Tidow, C. The inhibitor of growth protein 5 (ING5) depends on INCA1 as a co-factor for its antiproliferative effects. PLoS ONE 2011, 6, e21505. [Google Scholar] [CrossRef] [Green Version]
| ING Protein | Cellular Functions | Knockout Phenotype |
|---|---|---|
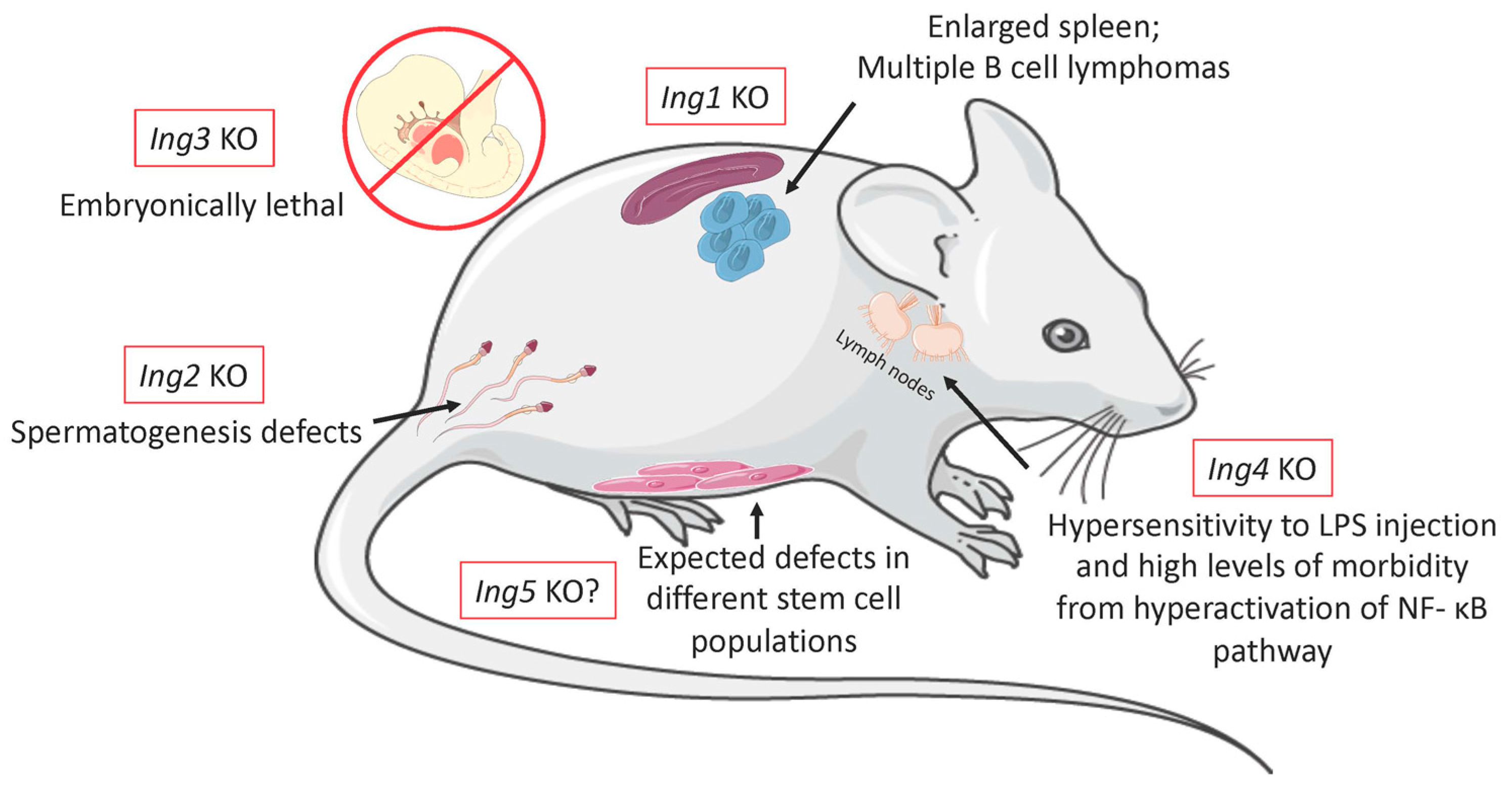
| ING1 | ING1b: Activation of apoptosis via interaction with p53 or via intrinsic apoptosis pathway 90 Downregulated in several cancers (leukemia, ovarian, gastric, colorectal, etc.) [7] Interaction with lamin-A to maintain nuclear morphology [98] Overexpression of human ING1 causes cell-cycle arrest in the G1 and S-phase [35,99] Regulates miRNA expression signatures [98,100] Acts at early stages of the DNA damage response activating a variety of repair mechanisms [38,101] Recruitment of the HDAC1 complex to the H3K4me3 mark [26] ING1a: Expression induced by stress-response, leads to senescence induction via Rb tumor suppressor pathway [36] | Reduction in body weight and size Reduction in progeny that did not follow the mendelian ratios, indicating possible role in development [43] Problems in DNA damage response Enlarged spleen with multiple B-cell lymphomas [45] Deletion of p53 along with ING1 KO greatly accelerated tumor formation and reduced lifespan [44] |
| ING2 | Downregulated in several cancers (hepatocellular carcinoma, lung cancer, and head and neck squamous cell carcinoma) [7] Upregulated in colon cancer and Burkitt lymphoma [7,50] Downregulation is caused by p53 binding to its promoter [47] Recruitment of the HDAC1 complex to the H3K4me3 mark [22] Important to muscle differentiation 24 Required for the initial DNA damage sensing and chromatin regulation in the nucleotide excision repair process [102,103] | Adult mice had no visible abnormalities Slight deviation from the mendelian ratio Spermatogenesis defects Activation of p53-dependent and independent mechanisms of apoptosis in the testes Males showed higher incidences of soft tissue sarcomas [53] |
| ING3 | Member of the NuA4-Tip60 HAT Complex [29] Silenced in some cancers like ovarian and head and neck squamous cell carcinoma [7,104,105] Upregulated in prostate cancer, important for the signaling of the androgen receptor pathway [59] Required for ATM signaling in double strand breaks response [56] | KO is embryonically lethal at day 10.5, most likely due to abnormal brain development [61] Severe growth retardation and are half the size of heterozygous and wild type embryos Ectopic expression was able to rescue WT normal phenotype [61] |
| ING4 | Downregulated in several tumor (breast, gastrointestinal, lung, etc.) [64,106] Member of the HB01-JADE-hEAF6 complex [63] Induces p53 mediated apoptosis [66] Required for cells to progress through the S phase [107] Disruption of ING4 caused prostate epithelial differentiation and oncogenesis [72,77] | Increased tumor vascularization in transplanted SCID mice No visible phenotype, and the Mendelian ratio was observed Do not form spontaneous tumors upon aging Hypersensitivity to LPS injection exhibiting high levels of morbidity through hyperactivation of the NF-κB pathway [80] |
| ING5 | Downregulated in several tumors (HNSCC, acute myeloid leukemia) [7,108] Upregulated in gastric and colon cancers [108,109] Component of both the HBO1-MYST and the MOZ/MORF [22,107] Induces cell cycle arrest and apoptosis when overexpressed in some cancer cells [110] Essential for epithelial stem cells differentiation [18,84] Regulates brain-tumor-initiating cell differentiation [85] | There are no knockout animals for ING5 yet We speculate that a KO animal for ING5 would present defects on different stem cell populations Due to the known activities of ING5 on epithelial stem cells and its associated complexes on different stem cell types |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dantas, A.; Al Shueili, B.; Yang, Y.; Nabbi, A.; Fink, D.; Riabowol, K. Biological Functions of the ING Proteins. Cancers 2019, 11, 1817. https://doi.org/10.3390/cancers11111817
Dantas A, Al Shueili B, Yang Y, Nabbi A, Fink D, Riabowol K. Biological Functions of the ING Proteins. Cancers. 2019; 11(11):1817. https://doi.org/10.3390/cancers11111817
Chicago/Turabian StyleDantas, Arthur, Buthaina Al Shueili, Yang Yang, Arash Nabbi, Dieter Fink, and Karl Riabowol. 2019. "Biological Functions of the ING Proteins" Cancers 11, no. 11: 1817. https://doi.org/10.3390/cancers11111817
APA StyleDantas, A., Al Shueili, B., Yang, Y., Nabbi, A., Fink, D., & Riabowol, K. (2019). Biological Functions of the ING Proteins. Cancers, 11(11), 1817. https://doi.org/10.3390/cancers11111817





