Roles of TrkC Signaling in the Regulation of Tumorigenicity and Metastasis of Cancer
Abstract
1. Introduction
2. Incidence of TrkC Expression in Cancer Development
2.1. The Functional Role of Long Noncoding RNA and microRNA in TrkC Expression
2.2. Somatic Mutations of TrkC in Cancer
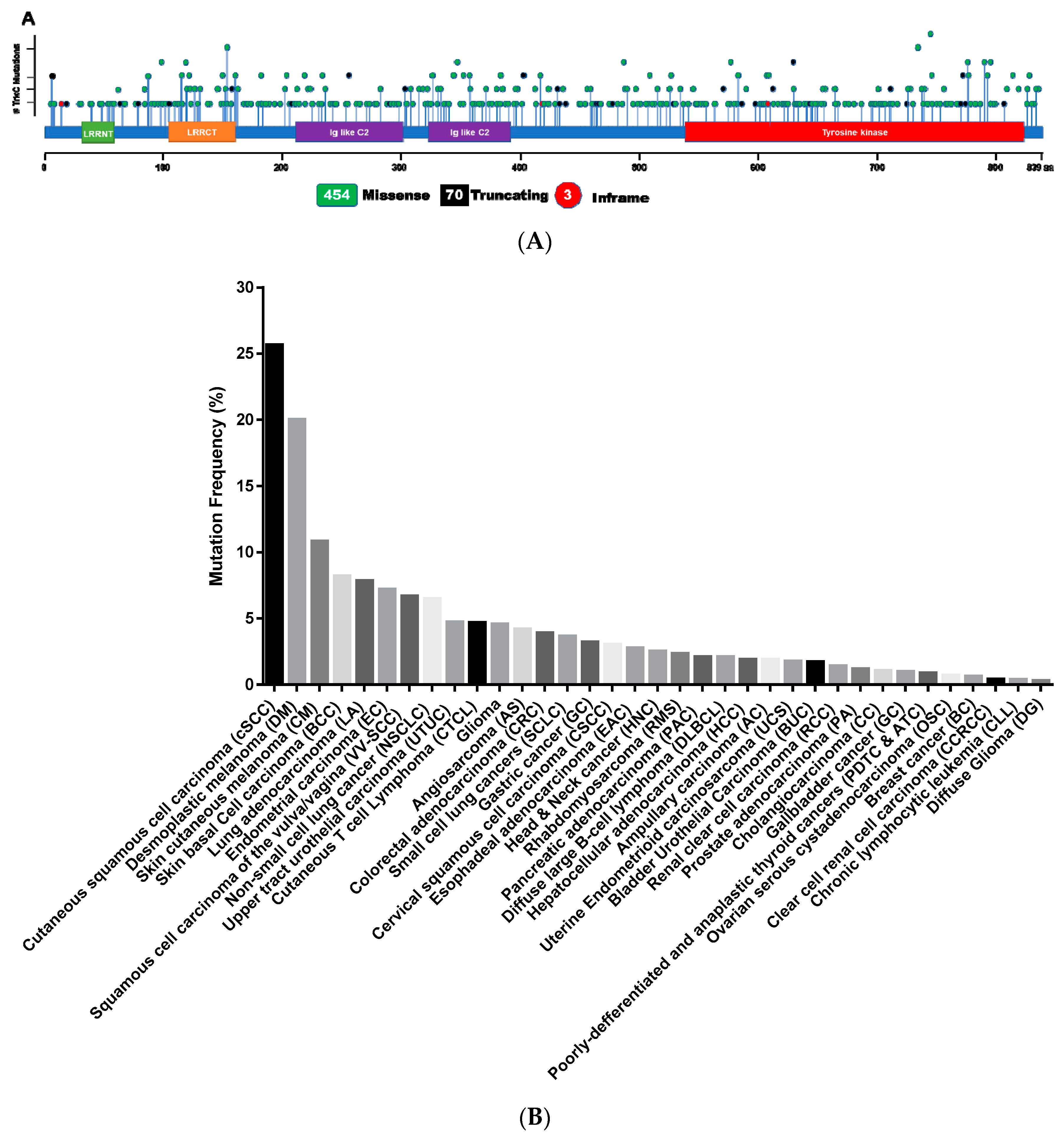
| Cancer Name | Frequency | Ref. |
|---|---|---|
| Neuroblastoma | 14/55 (25%) | [15] |
| Glioma | 215/234 (91.8%) | [17] |
| Medulloblastomas | 17/26 (85%) | [19] |
| Non-cerebellar primitive neuroectodermal (PNET) tumors | 21/31 (67.7%) | [19] |
| Breast cancer | 14/17 (82.4%) | [20] |
| Invasive ductal carcinoma | 118/236 (49.6%) | [71] |
| Hepatocellular carcinoma | 44/51 (86%) | [21] |
| Melanoma | 40/64 (62.5%) | [25] |
| Pancreatic cancer | 31/47 (66%) | [26,27] |
| Squamous cell carcinoma | 3.5/10 (35%) | [29] |
| Small cell lung cancers | 4.4/8 (55%) | [29] |
| Thyroid cancer | 21.8/25 (87%) | [32] |
| Cancer Name | Mutation (%) | Protein Change | Ref. |
|---|---|---|---|
| Ampullary Carcinoma (AC) | 1.88 | S136G, E211A, P796L | [62] |
| Angiosarcoma (AS) | 4.17 | P577L, G235E | TCGA |
| Oligodendroglioma (ODG) | 0.68 | Q808 * | [72] |
| Bladder Urothelial Carcinoma (BUG) | 2.94 | D609N, S477 *, L53F, E398K, G727 *, Q119E, E360K | TCGA |
| Breast Carcinoma (BC) | 0.75 | H349R, E810Q, G545A, L574V, E412K, N714K, P210T, A555V, K424T, G547R, V413M, Q177H, L108R, H254Y | [68] |
| Cervical Squamous Cell Carcinoma (CSCC) | 3.04 | A122T, F162S, L384M, D476N, L5V, R153Q, F162L | TCGA |
| Chronic Lymphocytic Leukemia (CLL) | 0.33 | A647T | [69] |
| Colorectal Adenocarcinoma (CRC) | 4.17 | F450L, L653I, S113A, R745Q, L115R, T777M, G608S, T149M, I759M, V217I, S117N, K181N, C782R, Q206H, T777M, T149M, K746R, K125N, R791Q, R745Q, Y456H, E86D, N191D, D624Y, Q159H, R201H, T730S, V97M, R745Q, G487D, R535M, R130H, T149M, A631D, D527G, Q586 *, V97M, R791W, G497R, Q145H, T332M, R130H, R791W, R814Q, A664T, K746T, R89H, Q119H, K346R | [58] |
| Cutaneous Melanoma (CM) | 20 | E762K, P509H, E211 *, H370N, P577S, L157F, E778K, D167N, R793 *, E318K, R542Q, E810K, H658Q, D697N, G235E, G356E, G633V, D584N, S359F, G356R, E590K, D576N, Y834N, R735C, G757R, R153L, G623E, R153L, W771 *, L299I, D242N, V217F, V37A, H423N, V726L, R116W, K768E, G178E, G727E, R735C, L152F, G339R, E819K, M667I, E86 *, Q773 *, W243L, Q255 *, P417L, D836H, T831I, P401S | TCGA |
| Cutaneous Squamous Cell Carcinoma (cSCC) | 25.64 | P577S, H264Y, D576N, Q773 *, E778K, G767E, A548V, D584N, L152F, D61G, G828E, K797E | [50] |
| Cutaneous T-Cell Lymphoma (CTCL) | 4.65 | P577S, S741N | [57] |
| Desmoplastic Melanoma (DM) | 20 | H128Y, P329L, E351K, S701F | [51] |
| Diffuse Glioma (DG) | 0.38 | I488T, L282R, I488V | [70] |
| Diffuse Large B-Cell Lymphoma (DLBCL) | 2.08 | D98N | TCGA |
| Endocervical Adenocarcinoma (EA) | 1.94 | E546K | TCGA |
| Esophageal Adenocarcinoma (EAC) | 3.97 | V687G, G487S, R46W, L115P, A826V | [59] |
| Gallbladder Cancer (GC) | 3.13 | I817M | [65] |
| Head and Neck Carcinoma (HNC) | 2.51 | H632Y, Q159K, S77 *, T253N, Q531R, A636V, H423N, Y705N, K367N, R326L, C362S, C231F, H729N, | TCGA |
| Hepatocellular Carcinoma (HCC) | 2.47 | G233S, Q145R, D527G, S701T, H622N, F395Y, E318D | TCGA |
| Cholangiocarcinoma (CC) | 2.78 | V451I | [64] |
| Lung Adenocarcinoma (LA) | 7.83 | L282M, P7R, H658N, H370N, L384M, N454S, S39R, Y188H, C320F, G545C, A380D, R121I, R814L, N718Y, P796S, G605L, G666C, G757V, N137K, P417H, R306H, M292I, T420S, H677Q, L639I, L827M, Y376C, P120H, S775 *, G487R, V97L, G67V, H84Y, G828V, K602N, P526Q, V779F, N382I, Y821F, K551N, N218H, E740K, R169L, P120H, T500S, S184R, D428H, S28Y, P330Q, T777K, K181N, A380D, F603I, R814L, E314D, S309I, P526Q, K397N, H394Q, K621N, G652V, G233V, I212T, P383A, R535M, N52K, F147L, V704F, G608C, V221L, W754C, E357D, Y604F, G463 *, E398Q, Y834F, R138L, Y821C, M202L, D240H, Q515H, K461R, V799L, Q773K, M464I, H729Y, E512K, P120H, V324A, Q172H, V273L, R343L, K346N, R121I, R459G, T506A, D495E, R343W, S741I, L364P, S4C, P509T, P612A, A435E, T230S, E314Q, H84N, G642 *, N338Y, T707K, D801N, V241A, G279A, G487S, F123L, S296R, L629F, G649C, A581P, Y744F, R343L, D635N, P738H, R735H, T563N | [53,55] |
| Pancreatic Adenocarcinoma (PA) | 1.83 | R153Q, V29M, R306H, K746T, E322K, Q643K, E223Q | [61] |
| Papillary Renal Cell Carcinoma (PRCC) | 1.41 | L270V, E179G, T490K, G104 * | TCGA |
| Plasma Cell Myeloma (PCM) | 0.98 | R745W, E351D | [73] |
| Prostate Adenocarcinoma (PA) | 1.13 | D609N, G497R, V640A, F747S, R793Q, P417L, T93M, P509S, T777M, T332M | [63] |
| Clear Cell Renal Cell Carcinoma (CCRCC) | 2.86 | D609N, R735H | TCGA |
| Rhabdomyosarcoma (RMS) | 2.3 | Y709F | [60] |
| Ovarian Serous Cancer (OSC) | 0.69 | P304L, L827F, D584E | TCGA |
| Skin Cancer, Non-Melanoma (Basal Cell Carcinoma; BCC) | 8.19 | S741N, S751R, G608I, E475K, R745W, Q673H, E590D, M99I, P467L, S117R, Q255 *, L760F, K381E, E154D, G696E, M202I, Q655R, R735C, K346R | [52] |
| Squamous Cell Carcinoma of the Vulva/Vagina (VV-SCC) | 6.67 | G437 * | [54] |
| Gastric cancer (GC) | 3.18 | H486N, H521N, K181R, T490M, L115P, R326H, R201H, D277G, W335R, S117T, R787H, R791W, L197F, A435E, L17 * | TCGA |
| Thyroid Cancer (TC) | 0.85 | R630W, N294T | [66,67] |
| Upper Tract Urothelial Carcinoma (UTUC) | 4.71 | D499N, D527Y, R153Q, R326H | [56] |
| Uterine Endometrioid Carcinosarcoma (UCS) | 7.18 | T261S, F617L, C523Y, T390I, M202I, K346N, R459W, P832S, A681T, D537Y, Y456H, R222 *, E598 *, R793Q, K111N, A96T, A580V, R222Q, E357D, H482Y, L187P, P55S, K125N, E556K, R47Q, S117N, A664T, G114R, G699D, I511T, S65N, K397N, I508T, E86D, N816S, E412K, V97M, G233D, G374D, Y352C, Q159H, A387T, E58K, D836A, V217A, D75N, M700I, R518C, E322K, D703N, S151N, F772L, V221I, L712P, E287D | TCGA |
| Esophageal Carcinoma (EC) | 2.43 | D277G, W335R, S117T, K181R, R791W, H486N, H521N, L197F, R787H, T490M | [59] |
2.3. TrkC Fusion in Cancer
2.4. The Biological Function of TrkC in Cancer
2.5. Targeted Therapies for Trk or Trk Fusion Protein
2.5.1. Larotrectinib

2.5.2. Entrectinib
2.5.3. Resistance to Larotrectinib and Entrectinib as Trk Inhibitor
2.6. Next-Generation of Trk Inhibitor
3. Conclusions
Funding
Conflicts of Interest
References
- Segal, R.A. Selectivity in neurotrophin signaling: Theme and variations. Annu. Rev. Neurosci. 2003, 26, 299–330. [Google Scholar] [CrossRef] [PubMed]
- Reichardt, L.F. Neurotrophin-regulated signalling pathways. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2006, 361, 1545–1564. [Google Scholar] [CrossRef] [PubMed]
- Urfer, R.; Tsoulfas, P.; O’Connell, L.; Hongo, J.A.; Zhao, W.; Presta, L.G. High resolution mapping of the binding site of TrkA for nerve growth factor and TrkC for neurotrophin-3 on the second immunoglobulin-like domain of the Trk receptors. J. Biol. Chem. 1998, 273, 5829–5840. [Google Scholar] [CrossRef] [PubMed]
- Lad, S.P.; Neet, K.E.; Mufson, E.J. Nerve growth factor: Structure, function and therapeutic implications for Alzheimer’s disease. Curr. Drug Targets CNS Neurol. Disord. 2003, 2, 315–334. [Google Scholar] [CrossRef]
- Wiesmann, C.; de Vos, A.M. Nerve growth factor: Structure and function. Cell. Mol. Life Sci. 2001, 58, 748–759. [Google Scholar] [CrossRef]
- Kaplan, D.R.; Miller, F.D. Neurotrophin signal transduction in the nervous system. Curr. Opin. Neurobiol. 2000, 10, 381–391. [Google Scholar] [CrossRef]
- Chao, M.V.; Bothwell, M. Neurotrophins: To cleave or not to cleave. Neuron 2002, 33, 9–12. [Google Scholar] [CrossRef]
- Salehi, A.; Verhaagen, J.; Dijkhuizen, P.A.; Swaab, D.F. Co-localization of high-affinity neurotrophin receptors in nucleus basalis of Meynert neurons and their differential reduction in Alzheimer’s disease. Neuroscience 1996, 75, 373–387. [Google Scholar] [CrossRef]
- Ginsberg, S.D.; Che, S.; Wuu, J.; Counts, S.E.; Mufson, E.J. Down regulation of trk but not p75NTR gene expression in single cholinergic basal forebrain neurons mark the progression of Alzheimer’s disease. J. Neurochem. 2006, 97, 475–487. [Google Scholar] [CrossRef]
- Savaskan, E.; Muller-Spahn, F.; Olivieri, G.; Bruttel, S.; Otten, U.; Rosenberg, C.; Hulette, C.; Hock, C. Alterations in trk A, trk B and trk C receptor immunoreactivities in parietal cortex and cerebellum in Alzheimer’s disease. Eur. Neurol. 2000, 44, 172–180. [Google Scholar] [CrossRef]
- Ginsberg, S.D.; Alldred, M.J.; Counts, S.E.; Cataldo, A.M.; Neve, R.L.; Jiang, Y.; Wuu, J.; Chao, M.V.; Mufson, E.J.; Nixon, R.A.; et al. Microarray analysis of hippocampal CA1 neurons implicates early endosomal dysfunction during Alzheimer’s disease progression. Biol. Psychiatry 2010, 68, 885–893. [Google Scholar] [CrossRef] [PubMed]
- Von Bohlen und Halbach, O.; Minichiello, L.; Unsicker, K. Haploinsufficiency for trkB and trkC receptors induces cell loss and accumulation of alpha-synuclein in the substantia Nigra. FASEB J. 2005, 19, 1740–1742. [Google Scholar] [CrossRef] [PubMed]
- Pacenta, H.L.; Macy, M.E. Entrectinib and other ALK/TRK inhibitors for the treatment of neuroblastoma. Drug Des. Dev. Ther. 2018, 12, 3549–3561. [Google Scholar] [CrossRef] [PubMed]
- Cheung, N.K.; Dyer, M.A. Neuroblastoma: Developmental biology, cancer genomics and immunotherapy. Nat. Rev. Cancer 2013, 13, 397–411. [Google Scholar] [CrossRef]
- Brodeur, G.M.; Nakagawara, A.; Yamashiro, D.J.; Ikegaki, N.; Liu, X.G.; Azar, C.G.; Lee, C.P.; Evans, A.E. Expression of TrkA, TrkB and TrkC in human neuroblastomas. J. Neuro Oncol. 1997, 31, 49–55. [Google Scholar] [CrossRef]
- Bouzas-Rodriguez, J.; Cabrera, J.R.; Delloye-Bourgeois, C.; Ichim, G.; Delcros, J.G.; Raquin, M.A.; Rousseau, R.; Combaret, V.; Benard, J.; Tauszig-Delamasure, S.; et al. Neurotrophin-3 production promotes human neuroblastoma cell survival by inhibiting TrkC-induced apoptosis. J. Clin. Investig. 2010, 120, 850–858. [Google Scholar] [CrossRef]
- Lawn, S.; Krishna, N.; Pisklakova, A.; Qu, X.; Fenstermacher, D.A.; Fournier, M.; Vrionis, F.D.; Tran, N.; Chan, J.A.; Kenchappa, R.S.; et al. Neurotrophin signaling via TrkB and TrkC receptors promotes the growth of brain tumor-initiating cells. J. Biol. Chem. 2015, 290, 3814–3824. [Google Scholar] [CrossRef]
- Calatozzolo, C.; Salmaggi, A.; Pollo, B.; Sciacca, F.L.; Lorenzetti, M.; Franzini, A.; Boiardi, A.; Broggi, G.; Marras, C. Expression of cannabinoid receptors and neurotrophins in human gliomas. Neurol. Sci. 2007, 28, 304–310. [Google Scholar] [CrossRef]
- Tajima, Y.; Molina, R.P., Jr.; Rorke, L.B.; Kaplan, D.R.; Radeke, M.; Feinstein, S.C.; Lee, V.M.; Trojanowski, J.Q. Neurotrophins and neuronal versus glial differentiation in medulloblastomas and other pediatric brain tumors. Acta Neuropathol. 1998, 95, 325–332. [Google Scholar] [CrossRef]
- Kim, M.S.; Jeong, J.; Seo, J.; Kim, H.S.; Kim, S.J.; Jin, W. Dysregulated JAK2 expression by TrkC promotes metastasis potential, and EMT program of metastatic breast cancer. Sci. Rep. 2016, 6, 33899. [Google Scholar] [CrossRef]
- Llovet, J.M.; Montal, R.; Sia, D.; Finn, R.S. Molecular therapies and precision medicine for hepatocellular carcinoma. Nat. Rev. Clin. Oncol. 2018, 15, 599–616. [Google Scholar] [CrossRef] [PubMed]
- Jin, W.; Lee, J.J.; Kim, M.S.; Son, B.H.; Cho, Y.K.; Kim, H.P. DNA methylation-dependent regulation of TrkA, TrkB, and TrkC genes in human hepatocellular carcinoma. Biochem. Biophys. Res. Commun. 2011, 406, 89–95. [Google Scholar] [CrossRef] [PubMed]
- Hisaoka, M.; Sheng, W.Q.; Tanaka, A.; Hashimoto, H. Gene expression of TrkC (NTRK3) in human soft tissue tumours. J. Pathol. 2002, 197, 661–667. [Google Scholar] [CrossRef] [PubMed]
- Truzzi, F.; Marconi, A.; Lotti, R.; Dallaglio, K.; French, L.E.; Hempstead, B.L.; Pincelli, C. Neurotrophins and their receptors stimulate melanoma cell proliferation and migration. J. Investig. Dermatol. 2008, 128, 2031–2040. [Google Scholar] [CrossRef]
- Xu, X.; Tahan, S.R.; Pasha, T.L.; Zhang, P.J. Expression of neurotrophin receptor Trk-C in nevi and melanomas. J. Cutan. Pathol. 2003, 30, 318–322. [Google Scholar] [CrossRef]
- Schneider, M.B.; Standop, J.; Ulrich, A.; Wittel, U.; Friess, H.; Andren-Sandberg, A.; Pour, P.M. Expression of nerve growth factors in pancreatic neural tissue and pancreatic cancer. J. Histochem. Cytochem. 2001, 49, 1205–1210. [Google Scholar] [CrossRef]
- Miknyoczki, S.J.; Lang, D.; Huang, L.; Klein-Szanto, A.J.; Dionne, C.A.; Ruggeri, B.A. Neurotrophins and Trk receptors in human pancreatic ductal adenocarcinoma: Expression patterns and effects on in vitro invasive behavior. Int. J. Cancer 1999, 81, 417–427. [Google Scholar] [CrossRef]
- Satoh, F.; Mimata, H.; Nomura, T.; Fujita, Y.; Shin, T.; Sakamoto, S.; Hamada, Y.; Nomura, Y. Autocrine expression of neurotrophins and their receptors in prostate cancer. Int. J. Urol. 2001, 8, S28–S34. [Google Scholar] [CrossRef]
- Ricci, A.; Greco, S.; Mariotta, S.; Felici, L.; Bronzetti, E.; Cavazzana, A.; Cardillo, G.; Amenta, F.; Bisetti, A.; Barbolini, G. Neurotrophins and neurotrophin receptors in human lung cancer. Am. J. Respir. Cell Mol. Biol. 2001, 25, 439–446. [Google Scholar] [CrossRef]
- Kim, M.S.; Kim, G.M.; Choi, Y.J.; Kim, H.J.; Kim, Y.J.; Jin, W. TrkC promotes survival and growth of leukemia cells through Akt-mTOR-dependent up-regulation of PLK-1 and Twist-1. Mol. Cells 2013, 36, 177–184. [Google Scholar] [CrossRef]
- Kamiya, A.; Inokuchi, M.; Otsuki, S.; Sugita, H.; Kato, K.; Uetake, H.; Sugihara, K.; Takagi, Y.; Kojima, K. Prognostic value of tropomyosin-related kinases A, B, and C in gastric cancer. Clin. Transl. Oncol. 2016, 18, 599–607. [Google Scholar] [CrossRef] [PubMed]
- McGregor, L.M.; McCune, B.K.; Graff, J.R.; McDowell, P.R.; Romans, K.E.; Yancopoulos, G.D.; Ball, D.W.; Baylin, S.B.; Nelkin, B.D. Roles of trk family neurotrophin receptors in medullary thyroid carcinoma development and progression. Proc. Natl. Acad. Sci. USA 1999, 96, 4540–4545. [Google Scholar] [CrossRef] [PubMed]
- Guidi, M.; Muinos-Gimeno, M.; Kagerbauer, B.; Marti, E.; Estivill, X.; Espinosa-Parrilla, Y. Overexpression of miR-128 specifically inhibits the truncated isoform of NTRK3 and upregulates BCL2 in SH-SY5Y neuroblastoma cells. BMC Mol. Biol. 2010, 11. [Google Scholar] [CrossRef] [PubMed]
- Laneve, P.; Di Marcotullio, L.; Gioia, U.; Fiori, M.E.; Ferretti, E.; Gulino, A.; Bozzoni, I.; Caffarelli, E. The interplay between microRNAs and the neurotrophin receptor tropomyosin-related kinase C controls proliferation of human neuroblastoma cells. Proc. Natl. Acad. Sci. USA 2007, 104, 7957–7962. [Google Scholar] [CrossRef] [PubMed]
- Ferretti, E.; De Smaele, E.; Po, A.; Di Marcotullio, L.; Tosi, E.; Espinola, M.S.B.; Di Rocco, C.; Riccardi, R.; Giangaspero, F.; Farcomeni, A.; et al. MicroRNA profiling in human medulloblastoma. Int. J. Cancer 2009, 124, 568–577. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Y.; Wu, X.C.; Li, Q.P.; Ge, X.X.; Wang, F.; Wu, P.Y.; Deng, X.T.; Miao, L. Long noncoding RNAs as potential biomarkers and therapeutic targets in gallbladder cancer: A systematic review and meta-analysis. Cancer Cell Int. 2019, 19. [Google Scholar] [CrossRef]
- Zhao, S.; Fan, N.F.; Chen, X.H.; Zhuo, C.H.; Xu, C.W.; Lin, R.B. Long noncoding RNA PVT1-214 enhances gastric cancer progression by upregulating TrkC expression in competitively sponging way. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 4173–4184. [Google Scholar] [CrossRef]
- Bu, J.Y.; Lv, W.Z.; Liao, Y.F.; Xiao, X.Y.; Lv, B.J. Long non-coding RNA LINC00978 promotes cell proliferation and tumorigenesis via regulating microRNA-497/NTRK3 axis in gastric cancer. Int. J. Biol. Macromol. 2019, 123, 1106–1114. [Google Scholar] [CrossRef]
- Xiong, D.M.; Sheng, Y.R.; Ding, S.J.; Chen, J.; Tan, X.X.; Zeng, T.; Qin, D.D.; Zhu, L.Y.; Huang, A.L.; Tang, H. LINC00052 regulates the expression of NTRK3 by miR-128 and miR-485-3p to strengthen HCC cells invasion and migration. Oncotarget 2016, 7, 47593–47608. [Google Scholar] [CrossRef]
- Martincorena, I.; Campbell, P.J. Somatic mutation in cancer and normal cells. Science 2015, 349, 1483–1489. [Google Scholar] [CrossRef]
- Stratton, M.R. Exploring the genomes of cancer cells: Progress and promise. Science 2011, 331, 1553–1558. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Bardelli, A.; Parsons, D.W.; Silliman, N.; Ptak, J.; Szabo, S.; Saha, S.; Markowitz, S.; Willson, J.K.; Parmigiani, G.; Kinzler, K.W.; et al. Mutational analysis of the tyrosine kinome in colorectal cancers. Science 2003, 300, 949. [Google Scholar] [CrossRef] [PubMed]
- Wood, L.D.; Calhoun, E.S.; Silliman, N.; Ptak, J.; Szabo, S.; Powell, S.M.; Riggins, G.J.; Wang, T.L.; Yan, H.; Gazdar, A.; et al. Somatic mutations of GUCY2F, EPHA3, and NTRK3 in human cancers. Hum. Mutat. 2006, 27, 1060–1061. [Google Scholar] [CrossRef] [PubMed]
- Jones, S.; Zhang, X.; Parsons, D.W.; Lin, J.C.; Leary, R.J.; Angenendt, P.; Mankoo, P.; Carter, H.; Kamiyama, H.; Jimeno, A.; et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science 2008, 321, 1801–1806. [Google Scholar] [CrossRef] [PubMed]
- Kubo, T.; Kuroda, Y.; Kokubu, A.; Hosoda, F.; Arai, Y.; Hiraoka, N.; Hirohashi, S.; Shibata, T. Resequencing analysis of the human tyrosine kinase gene family in pancreatic cancer. Pancreas 2009, 38, e200–e206. [Google Scholar] [CrossRef]
- Stephens, P.; Edkins, S.; Davies, H.; Greenman, C.; Cox, C.; Hunter, C.; Bignell, G.; Teague, J.; Smith, R.; Stevens, C.; et al. A screen of the complete protein kinase gene family identifies diverse patterns of somatic mutations in human breast cancer. Nat. Genet. 2005, 37, 590–592. [Google Scholar] [CrossRef]
- Davies, H.; Hunter, C.; Smith, R.; Stephens, P.; Greenman, C.; Bignell, G.; Teague, J.; Butler, A.; Edkins, S.; Stevens, C.; et al. Somatic mutations of the protein kinase gene family in human lung cancer. Cancer Res. 2005, 65, 7591–7595. [Google Scholar] [CrossRef]
- Kubo, T.; Kuroda, Y.; Shimizu, H.; Kokubu, A.; Okada, N.; Hosoda, F.; Arai, Y.; Nakamura, Y.; Taniguchi, H.; Yanagihara, K.; et al. Resequencing and copy number analysis of the human tyrosine kinase gene family in poorly differentiated gastric cancer. Carcinogenesis 2009, 30, 1857–1864. [Google Scholar] [CrossRef]
- Pickering, C.R.; Zhou, J.H.; Lee, J.J.; Drummond, J.A.; Peng, S.A.; Saade, R.E.; Tsai, K.Y.; Curry, J.L.; Tetzlaff, M.T.; Lai, S.Y.; et al. Mutational landscape of aggressive cutaneous squamous cell carcinoma. Clin. Cancer Res. 2014, 20, 6582–6592. [Google Scholar] [CrossRef]
- Shain, A.H.; Garrido, M.; Botton, T.; Talevich, E.; Yeh, I.; Sanborn, J.Z.; Chung, J.; Wang, N.J.; Kakavand, H.; Mann, G.J.; et al. Exome sequencing of desmoplastic melanoma identifies recurrent NFKBIE promoter mutations and diverse activating mutations in the MAPK pathway. Nat. Genet. 2015, 47, 1194–1199. [Google Scholar] [CrossRef] [PubMed]
- Bonilla, X.; Parmentier, L.; King, B.; Bezrukov, F.; Kaya, G.; Zoete, V.; Seplyarskiy, V.B.; Sharpe, H.J.; McKee, T.; Letourneau, A.; et al. Genomic analysis identifies new drivers and progression pathways in skin basal cell carcinoma. Nat. Genet. 2016, 48, 398–406. [Google Scholar] [CrossRef]
- Campbell, J.D.; Alexandrov, A.; Kim, J.; Wala, J.; Berger, A.H.; Pedamallu, C.S.; Shukla, S.A.; Guo, G.; Brooks, A.N.; Murray, B.A.; et al. Distinct patterns of somatic genome alterations in lung adenocarcinomas and squamous cell carcinomas. Nat. Genet. 2016, 48, 607–616. [Google Scholar] [CrossRef] [PubMed]
- Han, M.R.; Shin, S.; Park, H.C.; Kim, M.S.; Lee, S.H.; Jung, S.H.; Song, S.Y.; Lee, S.H.; Chung, Y.J. Mutational signatures and chromosome alteration profiles of squamous cell carcinomas of the vulva. Exp. Mol. Med. 2018, 50, e442. [Google Scholar] [CrossRef] [PubMed]
- Jordan, E.J.; Kim, H.R.; Arcila, M.E.; Barron, D.; Chakravarty, D.; Gao, J.; Chang, M.T.; Ni, A.; Kundra, R.; Jonsson, P.; et al. Prospective Comprehensive Molecular Characterization of Lung Adenocarcinomas for Efficient Patient Matching to Approved and Emerging Therapies. Cancer Discov. 2017, 7, 596–609. [Google Scholar] [CrossRef] [PubMed]
- Sfakianos, J.P.; Cha, E.K.; Iyer, G.; Scott, S.N.; Zabor, E.C.; Shah, R.H.; Ren, Q.; Bagrodia, A.; Kim, P.H.; Hakimi, A.A.; et al. Genomic Characterization of Upper Tract Urothelial Carcinoma. Eur. Urol. 2015, 68, 970–977. [Google Scholar] [CrossRef] [PubMed]
- Da Silva Almeida, A.C.; Abate, F.; Khiabanian, H.; Martinez-Escala, E.; Guitart, J.; Tensen, C.P.; Vermeer, M.H.; Rabadan, R.; Ferrando, A.; Palomero, T. The mutational landscape of cutaneous T cell lymphoma and Sezary syndrome. Nat. Genet. 2015, 47, 1465–1470. [Google Scholar] [CrossRef]
- Giannakis, M.; Mu, X.J.; Shukla, S.A.; Qian, Z.R.; Cohen, O.; Nishihara, R.; Bahl, S.; Cao, Y.; Amin-Mansour, A.; Yamauchi, M.; et al. Genomic Correlates of Immune-Cell Infiltrates in Colorectal Carcinoma. Cell Rep. 2016, 15, 857–865. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Research Network; Analysis Working Group: Asan University; BC Cancer Agency; Brigham and Women’s Hospital; Broad Institute; Brown University; Case Western Reserve University; Dana-Farber Cancer Institute; Duke University; Greater Poland Cancer Centre; et al. Integrated genomic characterization of oesophageal carcinoma. Nature 2017, 541, 169–175. [Google Scholar] [CrossRef]
- Shern, J.F.; Chen, L.; Chmielecki, J.; Wei, J.S.; Patidar, R.; Rosenberg, M.; Ambrogio, L.; Auclair, D.; Wang, J.; Song, Y.K.; et al. Comprehensive genomic analysis of rhabdomyosarcoma reveals a landscape of alterations affecting a common genetic axis in fusion-positive and fusion-negative tumors. Cancer Discov. 2014, 4, 216–231. [Google Scholar] [CrossRef]
- Bailey, P.; Chang, D.K.; Nones, K.; Johns, A.L.; Patch, A.M.; Gingras, M.C.; Miller, D.K.; Christ, A.N.; Bruxner, T.J.; Quinn, M.C.; et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature 2016, 531, 47–52. [Google Scholar] [CrossRef] [PubMed]
- Gingras, M.C.; Covington, K.R.; Chang, D.K.; Donehower, L.A.; Gill, A.J.; Ittmann, M.M.; Creighton, C.J.; Johns, A.L.; Shinbrot, E.; Dewal, N.; et al. Ampullary Cancers Harbor ELF3 Tumor Suppressor Gene Mutations and Exhibit Frequent WNT Dysregulation. Cell Rep. 2016, 14, 907–919. [Google Scholar] [CrossRef] [PubMed]
- Armenia, J.; Wankowicz, S.A.M.; Liu, D.; Gao, J.; Kundra, R.; Reznik, E.; Chatila, W.K.; Chakravarty, D.; Han, G.C.; Coleman, I.; et al. Publisher Correction: The long tail of oncogenic drivers in prostate cancer. Nat. Genet. 2019, 51, 1194. [Google Scholar] [CrossRef]
- Lowery, M.A.; Ptashkin, R.; Jordan, E.; Berger, M.F.; Zehir, A.; Capanu, M.; Kemeny, N.E.; O’Reilly, E.M.; El-Dika, I.; Jarnagin, W.R.; et al. Comprehensive Molecular Profiling of Intrahepatic and Extrahepatic Cholangiocarcinomas: Potential Targets for Intervention. Clin. Cancer Res. 2018, 24, 4154–4161. [Google Scholar] [CrossRef]
- Li, M.L.; Zhang, Z.; Li, X.G.; Ye, J.Y.; Wu, X.S.; Tan, Z.J.; Liu, C.; Shen, B.Y.; Wang, X.A.; Wu, W.G.; et al. Whole-exome and targeted gene sequencing of gallbladder carcinoma identifies recurrent mutations in the ErbB pathway. Nat. Genet. 2014, 46, 872–876. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Research Network. Integrated genomic characterization of papillary thyroid carcinoma. Cell 2014, 159, 676–690. [Google Scholar] [CrossRef]
- Landa, I.; Ibrahimpasic, T.; Boucai, L.; Sinha, R.; Knauf, J.A.; Shah, R.H.; Dogan, S.; Ricarte-Filho, J.C.; Krishnamoorthy, G.P.; Xu, B.; et al. Genomic and transcriptomic hallmarks of poorly differentiated and anaplastic thyroid cancers. J. Clin. Investig. 2016, 126, 1052–1066. [Google Scholar] [CrossRef] [PubMed]
- Razavi, P.; Chang, M.T.; Xu, G.; Bandlamudi, C.; Ross, D.S.; Vasan, N.; Cai, Y.; Bielski, C.M.; Donoghue, M.T.A.; Jonsson, P.; et al. The Genomic Landscape of Endocrine-Resistant Advanced Breast Cancers. Cancer Cell 2018, 34, 427–438. [Google Scholar] [CrossRef] [PubMed]
- Landau, D.A.; Tausch, E.; Taylor-Weiner, A.N.; Stewart, C.; Reiter, J.G.; Bahlo, J.; Kluth, S.; Bozic, I.; Lawrence, M.; Bottcher, S.; et al. Mutations driving CLL and their evolution in progression and relapse. Nature 2015, 526, 525–530. [Google Scholar] [CrossRef]
- Ceccarelli, M.; Barthel, F.P.; Malta, T.M.; Sabedot, T.S.; Salama, S.R.; Murray, B.A.; Morozova, O.; Newton, Y.; Radenbaugh, A.; Pagnotta, S.M.; et al. Molecular Profiling Reveals Biologically Discrete Subsets and Pathways of Progression in Diffuse Glioma. Cell 2016, 164, 550–563. [Google Scholar] [CrossRef]
- Zhang, W.; Lin, Z.C.; Zhang, T.X.; Liu, S.; Liu, X.; Liu, J.J.; Niu, Y. TrkC expression predicts favorable clinical outcome in invasive ductal carcinoma of breast independent of NT-3 expression. Am. J. Cancer Res. 2014, 4, 811–823. [Google Scholar] [CrossRef] [PubMed]
- Thomas, A.A.; Abrey, L.E.; Terziev, R.; Raizer, J.; Martinez, N.L.; Forsyth, P.; Paleologos, N.; Matasar, M.; Sauter, C.S.; Moskowitz, C.; et al. Multicenter phase II study of temozolomide and myeloablative chemotherapy with autologous stem cell transplant for newly diagnosed anaplastic oligodendroglioma. Neuro-Oncology 2017, 19, 1380–1390. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lohr, J.G.; Stojanov, P.; Carter, S.L.; Cruz-Gordillo, P.; Lawrence, M.S.; Auclair, D.; Sougnez, C.; Knoechel, B.; Gould, J.; Saksena, G.; et al. Widespread genetic heterogeneity in multiple myeloma: Implications for targeted therapy. Cancer Cell 2014, 25, 91–101. [Google Scholar] [CrossRef] [PubMed]
- Knezevich, S.R.; McFadden, D.E.; Tao, W.; Lim, J.F.; Sorensen, P.H. A novel ETV6-NTRK3 gene fusion in congenital fibrosarcoma. Nat. Genet. 1998, 18, 184–187. [Google Scholar] [CrossRef] [PubMed]
- Rubin, B.P.; Chen, C.J.; Morgan, T.W.; Xiao, S.; Grier, H.E.; Kozakewich, H.P.; Perez-Atayde, A.R.; Fletcher, J.A. Congenital mesoblastic nephroma t(12;15) is associated with ETV6-NTRK3 gene fusion: Cytogenetic and molecular relationship to congenital (infantile) fibrosarcoma. Am. J. Pathol. 1998, 153, 1451–1458. [Google Scholar] [CrossRef]
- Eguchi, M.; Eguchi-Ishimae, M.; Tojo, A.; Morishita, K.; Suzuki, K.; Sato, Y.; Kudoh, S.; Tanaka, K.; Setoyama, M.; Nagamura, F.; et al. Fusion of ETV6 to neurotrophin-3 receptor TRKC in acute myeloid leukemia with t(12;15)(p13;q25). Blood 1999, 93, 1355–1363. [Google Scholar] [CrossRef]
- Kralik, J.M.; Kranewitter, W.; Boesmueller, H.; Marschon, R.; Tschurtschenthaler, G.; Rumpold, H.; Wiesinger, K.; Erdel, M.; Petzer, A.L.; Webersinke, G. Characterization of a newly identified ETV6-NTRK3 fusion transcript in acute myeloid leukemia. Diagn. Pathol. 2011, 6, 19. [Google Scholar] [CrossRef]
- Knezevich, S.R.; Garnett, M.J.; Pysher, T.J.; Beckwith, J.B.; Grundy, P.E.; Sorensen, P.H. ETV6-NTRK3 gene fusions and trisomy 11 establish a histogenetic link between mesoblastic nephroma and congenital fibrosarcoma. Cancer Res. 1998, 58, 5046–5048. [Google Scholar]
- Tognon, C.; Garnett, M.; Kenward, E.; Kay, R.; Morrison, K.; Sorensen, P.H. The chimeric protein tyrosine kinase ETV6-NTRK3 requires both Ras-Erk1/2 and PI3-kinase-Akt signaling for fibroblast transformation. Cancer Res. 2001, 61, 8909–8916. [Google Scholar]
- Wai, D.H.; Knezevich, S.R.; Lucas, T.; Jansen, B.; Kay, R.J.; Sorensen, P.H. The ETV6-NTRK3 gene fusion encodes a chimeric protein tyrosine kinase that transforms NIH3T3 cells. Oncogene 2000, 19, 906–915. [Google Scholar] [CrossRef]
- Lae, M.; Freneaux, P.; Sastre-Garau, X.; Chouchane, O.; Sigal-Zafrani, B.; Vincent-Salomon, A. Secretory breast carcinomas with ETV6-NTRK3 fusion gene belong to the basal-like carcinoma spectrum. Mod. Pathol. 2009, 22, 291–298. [Google Scholar] [CrossRef] [PubMed]
- Tognon, C.; Knezevich, S.R.; Huntsman, D.; Roskelley, C.D.; Melnyk, N.; Mathers, J.A.; Becker, L.; Carneiro, F.; MacPherson, N.; Horsman, D.; et al. Expression of the ETV6-NTRK3 gene fusion as a primary event in human secretory breast carcinoma. Cancer Cell 2002, 2, 367–376. [Google Scholar] [CrossRef]
- Hechtman, J.F.; Zehir, A.; Yaeger, R.; Wang, L.; Middha, S.; Zheng, T.; Hyman, D.M.; Solit, D.; Arcila, M.E.; Borsu, L.; et al. Identification of Targetable Kinase Alterations in Patients with Colorectal Carcinoma That are Preferentially Associated with Wild-Type RAS/RAF. Mol. Cancer Res. 2016, 14, 296–301. [Google Scholar] [CrossRef] [PubMed]
- Hechtman, J.F.; Benayed, R.; Hyman, D.M.; Drilon, A.; Zehir, A.; Frosina, D.; Arcila, M.E.; Dogan, S.; Klimstra, D.S.; Ladanyi, M.; et al. Pan-Trk Immunohistochemistry Is an Efficient and Reliable Screen for the Detection of NTRK Fusions. Am. J. Surg. Pathol. 2017, 41, 1547–1551. [Google Scholar] [CrossRef]
- Wu, G.; Diaz, A.K.; Paugh, B.S.; Rankin, S.L.; Ju, B.; Li, Y.; Zhu, X.; Qu, C.; Chen, X.; Zhang, J.; et al. The genomic landscape of diffuse intrinsic pontine glioma and pediatric non-brainstem high-grade glioma. Nat. Genet. 2014, 46, 444–450. [Google Scholar] [CrossRef]
- Gatalica, Z.; Xiu, J.; Swensen, J.; Vranic, S. Molecular characterization of cancers with NTRK gene fusions. Mod. Pathol 2019, 32, 147–153. [Google Scholar] [CrossRef]
- Yeh, I.; Tee, M.K.; Botton, T.; Shain, A.H.; Sparatta, A.J.; Gagnon, A.; Vemula, S.S.; Garrido, M.C.; Nakamaru, K.; Isoyama, T.; et al. NTRK3 kinase fusions in Spitz tumours. J. Pathol. 2016, 240, 282–290. [Google Scholar] [CrossRef]
- Drilon, A.; Laetsch, T.W.; Kummar, S.; DuBois, S.G.; Lassen, U.N.; Demetri, G.D.; Nathenson, M.; Doebele, R.C.; Farago, A.F.; Pappo, A.S.; et al. Efficacy of Larotrectinib in TRK Fusion-Positive Cancers in Adults and Children. N. Engl. J. Med. 2018, 378, 731–739. [Google Scholar] [CrossRef]
- Nagasubramanian, R.; Wei, J.L.; Gordon, P.; Rastatter, J.C.; Cox, M.C.; Pappo, A. Infantile Fibrosarcoma With NTRK3-ETV6 Fusion Successfully Treated with the Tropomyosin-Related Kinase Inhibitor LOXO-101. Pediatr. Blood Cancer 2016, 63, 1468–1470. [Google Scholar] [CrossRef]
- Brenca, M.; Rossi, S.; Polano, M.; Gasparotto, D.; Zanatta, L.; Racanelli, D.; Valori, L.; Lamon, S.; Dei Tos, A.P.; Maestro, R. Transcriptome sequencing identifies ETV6-NTRK3 as a gene fusion involved in GIST. J. Pathol. 2016, 238, 543–549. [Google Scholar] [CrossRef]
- Leeman-Neill, R.J.; Kelly, L.M.; Liu, P.; Brenner, A.V.; Little, M.P.; Bogdanova, T.I.; Evdokimova, V.N.; Hatch, M.; Zurnadzy, L.Y.; Nikiforova, M.N.; et al. ETV6-NTRK3 is a common chromosomal rearrangement in radiation-associated thyroid cancer. Cancer 2014, 120, 799–807. [Google Scholar] [CrossRef] [PubMed]
- Van der Tuin, K.; Ventayol Garcia, M.; Corver, W.E.; Khalifa, M.N.; Ruano Neto, D.; Corssmit, E.P.M.; Hes, F.J.; Links, T.P.; Smit, J.W.A.; Plantinga, T.S.; et al. Targetable gene fusions identified in radioactive iodine refractory advanced thyroid carcinoma. Eur. J. Endocrinol. 2019, 180, 235–241. [Google Scholar] [CrossRef] [PubMed]
- Xu, B.; Aryeequaye, R.; Wang, L.; Katabi, N. Sinonasal Secretory Carcinoma of Salivary Gland with High Grade Transformation: A Case Report of this Under-Recognized Diagnostic Entity with Prognostic and Therapeutic Implications. Head Neck Pathol. 2018, 12, 274–278. [Google Scholar] [CrossRef] [PubMed]
- Stransky, N.; Cerami, E.; Schalm, S.; Kim, J.L.; Lengauer, C. The landscape of kinase fusions in cancer. Nat. Commun. 2014, 5, 4846. [Google Scholar] [CrossRef] [PubMed]
- Croce, S.; Hostein, I.; Longacre, T.A.; Mills, A.M.; Perot, G.; Devouassoux-Shisheboran, M.; Velasco, V.; Floquet, A.; Guyon, F.; Chakiba, C.; et al. Uterine and vaginal sarcomas resembling fibrosarcoma: A clinicopathological and molecular analysis of 13 cases showing common NTRK-rearrangements and the description of a COL1A1-PDGFB fusion novel to uterine neoplasms. Mod. Pathol. 2019, 32, 1008–1022. [Google Scholar] [CrossRef] [PubMed]
- Olson, N.; Rouhi, O.; Zhang, L.; Angeles, C.; Bridge, J.; Lopez-Terrada, D.; Royce, T.; Linos, K. A novel case of an aggressive superficial spindle cell sarcoma in an adult resembling fibrosarcomatous dermatofibrosarcoma protuberans and harboring an EML4-NTRK3 fusion. J. Cutan. Pathol. 2018, 45, 933–939. [Google Scholar] [CrossRef]
- Church, A.J.; Calicchio, M.L.; Nardi, V.; Skalova, A.; Pinto, A.; Dillon, D.A.; Gomez-Fernandez, C.R.; Manoj, N.; Haimes, J.D.; Stahl, J.A.; et al. Recurrent EML4-NTRK3 fusions in infantile fibrosarcoma and congenital mesoblastic nephroma suggest a revised testing strategy. Mod. Pathol. 2018, 31, 463–473. [Google Scholar] [CrossRef]
- Kao, Y.C.; Fletcher, C.D.M.; Alaggio, R.; Wexler, L.; Zhang, L.; Sung, Y.S.; Orhan, D.; Chang, W.C.; Swanson, D.; Dickson, B.C.; et al. Recurrent BRAF Gene Fusions in a Subset of Pediatric Spindle Cell Sarcomas: Expanding the Genetic Spectrum of Tumors with Overlapping Features with Infantile Fibrosarcoma. Am. J. Surg. Pathol. 2018, 42, 28–38. [Google Scholar] [CrossRef]
- Tannenbaum-Dvir, S.; Glade Bender, J.L.; Church, A.J.; Janeway, K.A.; Harris, M.H.; Mansukhani, M.M.; Nagy, P.L.; Andrews, S.J.; Murty, V.V.; Kadenhe-Chiweshe, A.; et al. Characterization of a novel fusion gene EML4-NTRK3 in a case of recurrent congenital fibrosarcoma. Mol. Case Stud. 2015, 1, a000471. [Google Scholar] [CrossRef]
- Kelly, L.M.; Barila, G.; Liu, P.Y.; Evdokimova, V.N.; Trivedi, S.; Panebianco, F.; Gandhi, M.; Carty, S.E.; Hodak, S.P.; Luo, J.H.; et al. Identification of the transforming STRN-ALK fusion as a potential therapeutic target in the aggressive forms of thyroid cancer. Proc. Natl. Acad. Sci. USA 2014, 111, 4233–4238. [Google Scholar] [CrossRef]
- Yang, Y.; Qin, S.K.; Zhu, J.; Wang, R.; Li, Y.M.; Xie, Z.Y.; Wu, Q. A Rare STRN-ALK Fusion in Lung Adenocarcinoma Identified Using Next-Generation Sequencing-Based Circulating Tumor DNA Profiling Exhibits Excellent Response to Crizotinib. Mayo Clin. Proc. Innov. Qual. Outcomes 2017, 1, 111–116. [Google Scholar] [CrossRef] [PubMed]
- Ling, Q.; Li, B.; Wu, X.; Wang, H.; Shen, Y.; Xiao, M.; Yang, Z.; Ma, R.; Chen, D.; Chen, H.; et al. 75P The landscape of NTRK fusions in Chinese patients with solid tumor. Ann. Oncol. 2018, 29 (Suppl. 8), mdy269.073. [Google Scholar] [CrossRef]
- Seethala, R.R.; Chiosea, S.I.; Liu, C.Z.; Nikiforova, M.; Nikiforov, Y.E. Clinical and Morphologic Features of ETV6-NTRK3 Translocated Papillary Thyroid Carcinoma in an Adult Population Without Radiation Exposure. Am. J. Surg. Pathol. 2017, 41, 446–457. [Google Scholar] [CrossRef] [PubMed]
- Skalova, A.; Vanecek, T.; Sima, R.; Laco, J.; Weinreb, I.; Perez-Ordonez, B.; Starek, I.; Geierova, M.; Simpson, R.H.; Passador-Santos, F.; et al. Mammary analogue secretory carcinoma of salivary glands, containing the ETV6-NTRK3 fusion gene: A hitherto undescribed salivary gland tumor entity. Am. J. Surg. Pathol. 2010, 34, 599–608. [Google Scholar] [CrossRef]
- Okamura, R.; Boichard, A.; Kato, S.; Sicklick, J.K.; Bazhenova, L.; Kurzrock, R. Analysis of NTRK Alterations in Pan-Cancer Adult and Pediatric Malignancies: Implications for NTRK-Targeted Therapeutics. JCO Precis. Oncol. 2018, 2018. [Google Scholar] [CrossRef]
- Taylor, J.; Pavlick, D.; Yoshimi, A.; Marcelus, C.; Chung, S.S.; Hechtman, J.F.; Benayed, R.; Cocco, E.; Durham, B.H.; Bitner, L.; et al. Oncogenic TRK fusions are amenable to inhibition in hematologic malignancies. J. Clin. Investig. 2018, 128, 3819–3825. [Google Scholar] [CrossRef]
- Tallegas, M.; Fraitag, S.; Binet, A.; Orbach, D.; Jourdain, A.; Reynaud, S.; Pierron, G.; Machet, M.C.; Maruani, A. Novel KHDRBS1-NTRK3 rearrangement in a congenital pediatric CD34-positive skin tumor: A case report. Virchows Arch. 2019, 474, 111–115. [Google Scholar] [CrossRef]
- Yoshihara, K.; Wang, Q.; Torres-Garcia, W.; Zheng, S.; Vegesna, R.; Kim, H.; Verhaak, R.G.W. The landscape and therapeutic relevance of cancer-associated transcript fusions. Oncogene 2015, 34, 4845–4854. [Google Scholar] [CrossRef]
- Wang, L.; Busam, K.J.; Benayed, R.; Cimera, R.; Wang, J.; Denley, R.; Rao, M.; Aryeequaye, R.; Mullaney, K.; Cao, L.; et al. Identification of NTRK3 Fusions in Childhood Melanocytic Neoplasms. J. Mol. Diagn. 2017, 19, 387–396. [Google Scholar] [CrossRef]
- Doebele, R.C.; Davis, L.E.; Vaishnavi, A.; Le, A.T.; Estrada-Bernal, A.; Keysar, S.; Jimeno, A.; Varella-Garcia, M.; Aisner, D.L.; Li, Y.; et al. An Oncogenic NTRK Fusion in a Patient with Soft-Tissue Sarcoma with Response to the Tropomyosin-Related Kinase Inhibitor LOXO-101. Cancer Discov. 2015, 5, 1049–1057. [Google Scholar] [CrossRef]
- Friedman, B.J.; Hernandez, S.; Fidai, C.; Jiang, A.; Shwayder, T.A.; Carskadon, S.; Andea, A.A.; Harms, P.W.; Chitale, D.; Palanisamy, N. A pediatric case of pigmented epithelioid melanocytoma with chromosomal copy number alterations in 15q and 17q and a novel NTRK3-SCAPER gene fusion. J. Cutan. Pathol. 2019. [Google Scholar] [CrossRef] [PubMed]
- Torre, M.; Jessop, N.; Hornick, J.L.; Alexandrescu, S. Expanding the spectrum of pediatric NTRK-rearranged fibroblastic tumors to the central nervous system: A case report with RBPMS-NTRK3 fusion. Neuropathology 2018, 38, 624–630. [Google Scholar] [CrossRef] [PubMed]
- Chiang, S.; Cotzia, P.; Hyman, D.M.; Drilon, A.; Tap, W.D.; Zhang, L.; Hechtman, J.F.; Frosina, D.; Jungbluth, A.A.; Murali, R.; et al. NTRK Fusions Define a Novel Uterine Sarcoma Subtype with Features of Fibrosarcoma. Am. J. Surg. Pathol. 2018, 42, 791–798. [Google Scholar] [CrossRef] [PubMed]
- Iyama, K.; Matsuse, M.; Mitsutake, N.; Rogounovitch, T.; Saenko, V.; Suzuki, K.; Ashizawa, M.; Ookouchi, C.; Suzuki, S.; Mizunuma, H.; et al. Identification of Three Novel Fusion Oncogenes, SQSTM1/NTRK3, AFAP1L2/RET, and PPFIBP2/RET, in Thyroid Cancers of Young Patients in Fukushima. Thyroid 2017, 27, 811–818. [Google Scholar] [CrossRef]
- Lu, Z.; Zhang, Y.; Feng, D.; Sheng, J.; Yang, W.; Liu, B. Targeted next generation sequencing identifies somatic mutations and gene fusions in papillary thyroid carcinoma. Oncotarget 2017, 8, 45784–45792. [Google Scholar] [CrossRef] [PubMed]
- Farago, A.F.; Taylor, M.S.; Doebele, R.C.; Zhu, V.W.; Kummar, S.; Spira, A.I.; Boyle, T.A.; Haura, E.B.; Arcila, M.E.; Benayed, R.; et al. Clinicopathologic Features of Non-Small-Cell Lung Cancer Harboring an NTRK Gene Fusion. JCO Precis. Oncol. 2018, 2018. [Google Scholar] [CrossRef] [PubMed]
- Yamazaki, F.; Nakatani, F.; Asano, N.; Wakai, S.; Sekimizu, M.; Mitani, S.; Kubo, T.; Kawai, A.; Ichikawa, H.; Yoshida, A. Novel NTRK3 Fusions in Fibrosarcomas of Adults. Am. J. Surg. Pathol. 2019, 43, 523–530. [Google Scholar] [CrossRef]
- Michal, M.; Hajkova, V.; Skalova, A.; Michal, M. STRN-NTRK3-rearranged Mesenchymal Tumor of the Uterus: Expanding the Morphologic Spectrum of Tumors with NTRK Fusions. Am. J. Surg. Pathol. 2019, 43, 1152–1154. [Google Scholar] [CrossRef]
- Wong, D.; Yip, S.; Sorensen, P.H. Methods for Identifying Patients with Tropomyosin Receptor Kinase (TRK) Fusion Cancer. Pathol. Oncol. Res. 2019. [Google Scholar] [CrossRef]
- Chmielecki, J.; Bailey, M.; He, J.; Elvin, J.; Vergilio, J.A.; Ramkissoon, S.; Suh, J.; Frampton, G.M.; Sun, J.X.; Morley, S.; et al. Genomic Profiling of a Large Set of Diverse Pediatric Cancers Identifies Known and Novel Mutations across Tumor Spectra. Cancer Res. 2017, 77, 509–519. [Google Scholar] [CrossRef]
- Joshi, S.K.; Davare, M.A.; Druker, B.J.; Tognon, C.E. Revisiting NTRKs as an emerging oncogene in hematological malignancies. Leukemia 2019, 33, 2563–2574. [Google Scholar] [CrossRef] [PubMed]
- Lemmon, M.A.; Schlessinger, J. Cell signaling by receptor tyrosine kinases. Cell 2010, 141, 1117–1134. [Google Scholar] [CrossRef] [PubMed]
- American Association for the Advancement of Science. Erratum for the Review “Somatic mutation in cancer and normal cells” by I. Martincorena and P. J. Campbell. Science 2016, 351. [Google Scholar] [CrossRef]
- Libermann, T.A.; Nusbaum, H.R.; Razon, N.; Kris, R.; Lax, I.; Soreq, H.; Whittle, N.; Waterfield, M.D.; Ullrich, A.; Schlessinger, J. Amplification, enhanced expression and possible rearrangement of EGF receptor gene in primary human brain tumours of glial origin. Nature 1985, 313, 144–147. [Google Scholar] [CrossRef]
- Sigismund, S.; Avanzato, D.; Lanzetti, L. Emerging functions of the EGFR in cancer. Mol. Oncol. 2018, 12, 3–20. [Google Scholar] [CrossRef]
- Du, Z.; Lovly, C.M. Mechanisms of receptor tyrosine kinase activation in cancer. Mol. Cancer 2018, 17, 58. [Google Scholar] [CrossRef]
- Carraway, K.L., 3rd; Sweeney, C. EGF receptor activation by heterologous mechanisms. Cancer Cell 2002, 1, 405–406. [Google Scholar] [CrossRef][Green Version]
- Kim, M.S.; Suh, K.W.; Hong, S.; Jin, W. TrkC promotes colorectal cancer growth and metastasis. Oncotarget 2017, 8, 41319–41333. [Google Scholar] [CrossRef]
- Pawson, T.; Gish, G.D.; Nash, P. SH2 domains, interaction modules and cellular wiring. Trends Cell Biol. 2001, 11, 504–511. [Google Scholar] [CrossRef]
- Jin, W.; Yun, C.; Hobbie, A.; Martin, M.J.; Sorensen, P.H.B.; Kim, S.J. Cellular transformation and activation of the phosphoinositide-3-kinase-Akt cascade by the ETV6-NTRK3 chimeric tyrosine kinase requires c-Src. Cancer Res. 2007, 67, 3192–3200. [Google Scholar] [CrossRef]
- Jin, W.; Yun, C.; Jeong, J.; Park, Y.; Lee, H.D.; Kim, S.J. c-Src is required for tropomyosin receptor kinase C (TrkC)-induced activation of the phosphatidylinositol 3-kinase (PI3K)-AKT pathway. J. Biol. Chem. 2008, 283, 1391–1400. [Google Scholar] [CrossRef] [PubMed]
- Lannon, C.L.; Martin, M.J.; Tognon, C.E.; Jin, W.; Kim, S.J.; Sorensen, P.H.B. A highly conserved NTRK3 C-terminal sequence in the ETV6-NTRK3 oncoprotein binds the phosphotyrosine binding domain of insulin receptor substrate-1. An essential interaction for transformation. J. Biol. Chem. 2004, 279, 15706. [Google Scholar] [CrossRef] [PubMed]
- Valastyan, S.; Weinberg, R.A. Tumor Metastasis: Molecular Insights and Evolving Paradigms. Cell 2011, 147, 275–292. [Google Scholar] [CrossRef] [PubMed]
- Polyak, K.; Weinberg, R.A. Transitions between epithelial and mesenchymal states: Acquisition of malignant and stem cell traits. Nat. Rev. Cancer 2009, 9, 265–273. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Kim, J.; Park, B.; Yang, K.M.; Sun, E.J.; Tognon, C.E.; Sorensen, P.H.; Kim, S.J. Novel identification of STAT1 as a crucial mediator of ETV6-NTRK3-induced tumorigenesis. Oncogene 2018, 37, 2270–2284. [Google Scholar] [CrossRef] [PubMed]
- Hix, L.M.; Karavitis, J.; Khan, M.W.; Shi, Y.H.; Khazaie, K.; Zhang, M. Tumor STAT1 transcription factor activity enhances breast tumor growth and immune suppression mediated by myeloid-derived suppressor cells. J. Biol. Chem. 2013, 288, 11676–11688. [Google Scholar] [CrossRef] [PubMed]
- Khodarev, N.N.; Minn, A.J.; Efimova, E.V.; Darga, T.E.; Labay, E.; Beckett, M.; Mauceri, H.J.; Roizman, B.; Weichselbaum, R.R. Signal transducer and activator of transcription 1 regulates both cytotoxic and prosurvival functions in tumor cells. Cancer Res. 2007, 67, 9214–9220. [Google Scholar] [CrossRef]
- Khodarev, N.N.; Beckett, M.; Labay, E.; Darga, T.; Roizman, B.; Weichselbaum, R.R. STAT1 is overexpressed in tumors selected for radioresistance and confers protection from radiation in transduced sensitive cells. Proc. Natl. Acad. Sci. USA 2004, 101, 1714–1719. [Google Scholar] [CrossRef]
- Weichselbaum, R.R.; Ishwaran, H.; Yoon, T.; Nuyten, D.S.A.; Baker, S.W.; Khodarev, N.; Su, A.W.; Shaikh, A.Y.; Roach, P.; Kreike, B.; et al. An interferon-related gene signature for DNA damage resistance is a predictive marker for chemotherapy and radiation for breast cancer. Proc. Natl. Acad. Sci. USA 2008, 105, 18490–18495. [Google Scholar] [CrossRef]
- Tripathi, V.; Sixt, K.M.; Gao, S.J.; Xu, X.; Huang, J.; Weigert, R.; Zhou, M.; Zhang, Y.E. Direct Regulation of Alternative Splicing by SMAD3 through PCBP1 Is Essential to the Tumor-Promoting Role of TGF-beta. Mol. Cell 2016, 64, 549–564. [Google Scholar] [CrossRef]
- Jin, W.; Yun, C.; Kwak, M.K.; Kim, T.A.; Kim, S.J. TrkC binds to the type II TGF-beta receptor to suppress TGF-beta signaling. Oncogene 2007, 26, 7684–7691. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Jin, W.; Kim, B.C.; Tognon, C.; Lee, H.J.; Patel, S.; Lannon, C.L.; Maris, J.M.; Triche, T.J.; Sorensen, P.H.B.; Kim, S.J. The ETV6-NTRK3 chimeric tyrosine kinase suppresses TGF-beta signaling by inactivating the TGF-beta type II receptor. Proc. Natl. Acad. Sci. USA 2005, 102, 16239–16244. [Google Scholar] [CrossRef] [PubMed]
- Jin, W.; Yun, C.; Kim, H.S.; Kim, S.J. TrkC binds to the bone morphogenetic protein type II receptor to suppress bone morphogenetic protein signaling. Cancer Res. 2007, 67, 9869–9877. [Google Scholar] [CrossRef] [PubMed]
- Epping, M.T.; Lunardi, A.; Nachmani, D.; Castillo-Martin, M.; Thin, T.H.; Cordon-Cardo, C.; Pandolfi, P.P. TSPYL2 is an essential component of the REST/NRSF transcriptional complex for TGF beta signaling activation. Cell Death Differ. 2015, 22, 1353–1362. [Google Scholar] [CrossRef] [PubMed]
- Wagoner, M.P.; Gunsalus, K.T.W.; Schoenike, B.; Richardson, A.L.; Friedl, A.; Roopra, A. The Transcription Factor REST Is Lost in Aggressive Breast Cancer. PLoS Genet. 2010, 6. [Google Scholar] [CrossRef] [PubMed]
- Scott, L.J. Larotrectinib: First Global Approval. Drugs 2019, 79, 201–206. [Google Scholar] [CrossRef]
- Cocco, E.; Scaltriti, M.; Drilon, A. NTRK fusion-positive cancers and TRK inhibitor therapy. Nat. Rev. Clin. Oncol. 2018, 15, 731–747. [Google Scholar] [CrossRef]
- Liu, Q.; Schwaller, J.; Kutok, J.; Cain, D.; Aster, J.C.; Williams, I.R.; Gilliland, D.G. Signal transduction and transforming properties of the TEL-TRKC fusions associated with t(12;15)(p13;q25) in congenital fibrosarcoma and acute myelogenous leukemia. EMBO J. 2000, 19, 1827–1838. [Google Scholar] [CrossRef]
- Khotskaya, Y.B.; Holla, V.R.; Farago, A.F.; Mills Shaw, K.R.; Meric-Bernstam, F.; Hong, D.S. Targeting TRK family proteins in cancer. Pharmacol. Ther. 2017, 173, 58–66. [Google Scholar] [CrossRef]
- Laetsch, T.W.; DuBois, S.G.; Mascarenhas, L. Larotrectinib for paediatric solid tumours harbouring NTRK gene fusions: Phase 1 results from a multicentre, open-label, phase 1/2 study. Lancet Oncol. 2018, 19, 705–714. [Google Scholar] [CrossRef]
- Geoerger, B.; van Tilburg, C.; DuBois, S.; Albert, C.; Federman, N.; Nagasubramanian, R.; Doz, F.; Orbach, D.; Bielack, S.; Shukla, N.; et al. Larotrectinib Efficacy and Safety in Pediatric Patients with TRK Fusion Cancer. Pediatr. Blood Cancer 2019, 66, S65–S66. [Google Scholar] [CrossRef]
- Shukla, N.; Roberts, S.S.; Baki, M.O.; Mushtaq, Q.; Goss, P.E.; Park, B.; Gundem, G.; Tian, K.; Geiger, H.; Redfield, K.; et al. Successful Targeted Therapy of Refractory Pediatric ETV6-NTRK3 Fusion-Positive Secretory Breast Carcinoma. JCO Precis. Oncol. 2017, 1. [Google Scholar] [CrossRef] [PubMed]
- Huang, E.J.; Reichardt, L.F. Trk receptors: Roles in neuronal signal transduction. Annu. Rev. Biochem. 2003, 72, 609–642. [Google Scholar] [CrossRef] [PubMed]
- Al-Salama, Z.T.; Keam, S.J. Entrectinib: First Global Approval. Drugs 2019, 79, 1477–1483. [Google Scholar] [CrossRef]
- De Braud, F.G.; Niger, M.; Damian, S.; Bardazza, B.; Martinetti, A.; Pelosi, G.; Marrapese, G.; Palmeri, L.; Cerea, G.; Valtorta, E.; et al. Alka-372-001: First-in-human, phase I study of entrectinib—An oral pan-trk, ROS1, and ALK inhibitor—In patients with advanced solid tumors with relevant molecular alterations. J. Clin. Oncol. 2015, 33. [Google Scholar] [CrossRef]
- Ricciuti, B.; Genova, C.; Crino, L.; Libra, M.; Leonardi, G.C. Antitumor activity of larotrectinib in tumors harboring NTRK gene fusions: A short review on the current evidence. OncoTargets Ther. 2019, 12, 3171–3179. [Google Scholar] [CrossRef]
- Drilon, A.; Li, G.; Dogan, S.; Gounder, M.; Shen, R.; Arcila, M.; Wang, L.; Hyman, D.M.; Hechtman, J.; Wei, G.; et al. What hides behind the MASC: Clinical response and acquired resistance to entrectinib after ETV6-NTRK3 identification in a mammary analogue secretory carcinoma (MASC). Ann. Oncol. 2016, 27, 920–926. [Google Scholar] [CrossRef]
- Russo, M.; Misale, S.; Wei, G.; Siravegna, G.; Crisafulli, G.; Lazzari, L.; Corti, G.; Rospo, G.; Novara, L.; Mussolin, B.; et al. Acquired Resistance to the TRK Inhibitor Entrectinib in Colorectal Cancer. Cancer Discov. 2016, 6, 36–44. [Google Scholar] [CrossRef]
- Drilon, A.; Ou, S.H.I.; Cho, B.C.; Kim, D.W.; Lees, J.; Lin, J.J.; Zhu, V.W.; Ahns, M.J.; Camidge, D.R.; Nguyen, J.; et al. Repotrectinib (TPX-0005) Is a Next-Generation ROS1/TRK/ALK Inhibitor That Potently Inhibit ROS1/TRK/ALK Solvent-Front Mutations. Cancer Discov. 2018, 8, 1227–1236. [Google Scholar] [CrossRef]
- Drilon, A.; Nagasubramanian, R.; Blake, J.F.; Ku, N.; Tuch, B.B.; Ebata, K.; Smith, S.; Lauriault, V.; Kolakowski, G.R.; Brandhuber, B.J.; et al. A Next-Generation TRK Kinase Inhibitor Overcomes Acquired Resistance to Prior TRK Kinase Inhibition in Patients with TRK Fusion-Positive Solid Tumors. Cancer Discov. 2017, 7, 963–972. [Google Scholar] [CrossRef]
- Hyman, D.; Kummar, S.; Farago, A.; Geoerger, B.; Mau-Sorensen, M.; Taylor, M.; Garralda, E.; Nagasubramanian, R.; Natheson, M.; Song, L.; et al. Abstract CT127: Phase I and expanded access experience of LOXO-195 (BAY 2731954), a selective next-generation TRK inhibitor (TRKi). Am. Assoc. Cancer Res. 2019. [Google Scholar] [CrossRef]

| Fusion Protein | Chromosomal Location | Recurrent Chromosomal Translocation | Tumor Type |
|---|---|---|---|
| AFAP1-NTRK3 | 4p16.1 | t(4;15)(p16;q25) | Glioblastoma [84] |
| AKAP13-NTRK3 | 15q25.3 | t(15;15)(q25;q25) | Lung adenocarcinoma [102], Low-grade glioma [94] |
| BTBD1-NTRK3 | 15q25.2 | t(15;15)(q25;q25) | Glioma [85] |
| CPEB1-NTRK3 | 15q25 | t(15;15)(q25;q25) | Glioma [85] |
| EML4-NTRK3 | 2p21 | t(2;15)(p21;q25) | Uterine and vaginal sarcomas [95], Dermatofibrosarcoma [96], Infantile fibrosarcoma and Congenital mesoblastic nephroma [97,99], Infantile fibrosarcomas [98], Glioblastoma [84]. |
| ETV6-NTRK3 | 12p13.2 | t(12;15)(p13;q25) | Congenital fibrosarcomas [74,75], Acute myeloid leukemia [76,77], Cellular mesoblastic nephroma [75,77,78], Secretory breast carcinoma [81,82], Colorectal cancer [83,84], Glioma [85,86], Spitz tumor [87], Lung adenocarcinoma [88], Infantile fibrosarcoma [88,89], Gastrointestinal stromal tumor [88,90], Thyroid carcinoma [103], Uterine sarcoma [86], Sinonasal adenocarcinoma [93], thyroid carcinomas [91,92], Mammary analog secretory carcinoma [104] |
| FAT1-NTRK3 | 4q35.2 | t(4;15)(q35;q25) | Cervical squamous cell carcinoma (TCGA Dataset), [105] |
| HNRNPA2B1-NTRK3 | 7p15.2 | t(7;15)(p15;q25) | Multiple myeloma [106] |
| KHDRBS1-NTRK3 | 1p35.2 | t(1;15)(p35;q25) | Pediatric cutaneous congenital skin cancer [107] |
| LYN-NTRK3 | 8q12 | t(8;15)(q12;q25) | Head and Neck squamous cell carcinoma (TCGA Dataset), [87,108] |
| MYH9-NTRK3 | 22q12.3 | t(22;15)(q12;q25) | Spitz tumor [87] |
| MYO5A-NTRK3 | 15q21.2 | t(15;15)(q21;q25) | Spitzoid tumor [87], Epithelioid melanocytic tumor [109] |
| NTRK3-HOMER2 | 15q25.2 | t(15;15)(q25;q25) | Soft tissue sarcoma [110] |
| NTRK3-SCAPER | 15q24.3 | t(15;15)(q24;q25) | Epithelioid melanocytoma [111] |
| TPM4-NTRK3 | 19p13.12 | t(19;15)(p13;q25) | Sarcoma [84] |
| ZNF710-NTRK3 | 15q26 | t(15;15)(q26;q25) | Glioblastoma [84] |
| RBPMS-NTRK3 | 8p12 | t(8;15)(p12;q25) | Glioma [112], Uterine Sarcoma [113], Thyroid carcinoma [66] |
| SPECC1L-NTRK3 | 22q11.23 | t(22;15)(q11;q25) | Uterine sarcoma [86] |
| SQSTM1-NTRK3 | 5q35.3 | t(5;15)(q35;q25) | Thyroid Cancer [114,115], Non-small-cell lung cancer [86,116] |
| STRN-NTRK3 | 2p22.2 | t(2;15)(p22;q25) | Fibrosarcoma [117], Uterine sarcoma [118] |
| STRN3-NTRK3 | 14q12 | t(14;15)(q12;q25) | Fibrosarcoma [117] |
| TFG-NTKR3 | 3q12.2 | t(3;15)(q12;q25) | Solitary fibrous tumor [119,120] |
| UBE2R2-NTRK3 | 9p13.3 | t(9;15)(p13;q25) | Multiple myeloma [106,121] |
| VIM-NTRK3 | 10q13 | t(10;15)(q13;q25) | Thyroid carcinoma [86] |
| VPS18-NTRK3 | 15q15 | t(15;15)(q15;q25) | Colon Adenocarcinoma (TCGA Dataset) [105] |
© 2020 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jin, W. Roles of TrkC Signaling in the Regulation of Tumorigenicity and Metastasis of Cancer. Cancers 2020, 12, 147. https://doi.org/10.3390/cancers12010147
Jin W. Roles of TrkC Signaling in the Regulation of Tumorigenicity and Metastasis of Cancer. Cancers. 2020; 12(1):147. https://doi.org/10.3390/cancers12010147
Chicago/Turabian StyleJin, Wook. 2020. "Roles of TrkC Signaling in the Regulation of Tumorigenicity and Metastasis of Cancer" Cancers 12, no. 1: 147. https://doi.org/10.3390/cancers12010147
APA StyleJin, W. (2020). Roles of TrkC Signaling in the Regulation of Tumorigenicity and Metastasis of Cancer. Cancers, 12(1), 147. https://doi.org/10.3390/cancers12010147





