Panoptic View of Prognostic Models for Personalized Breast Cancer Management
Abstract
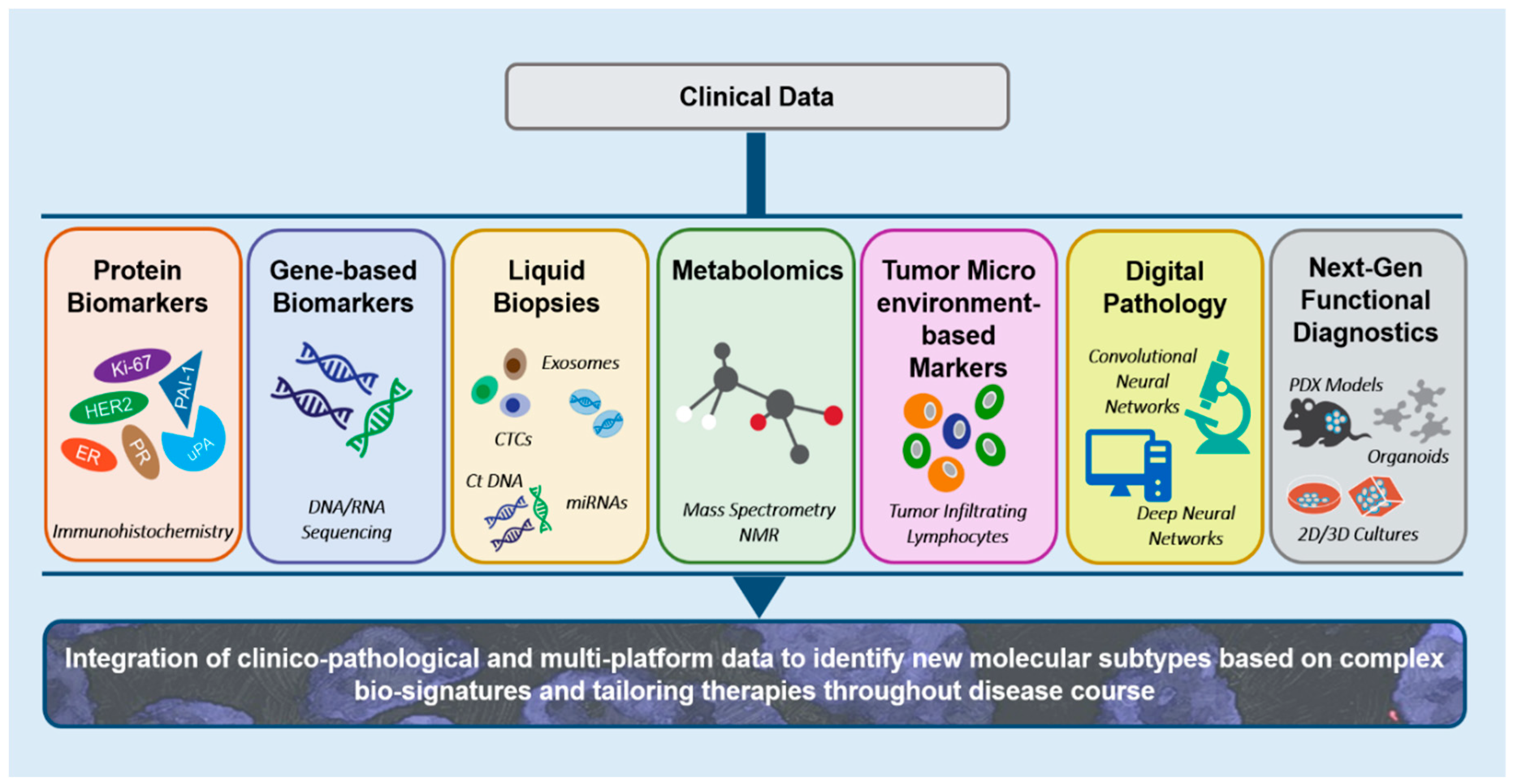
1. Introduction
2. Integrating Clinico-Pathological Variables to Frame Breast Cancer Prognostic Models
3. Immunohistochemistry-Based Prognostic Assays for Breast Cancer
4. Gene-Centered Biomarkers: Translating Molecular Complexity of Tumors by Gene Expression-Based Assays
4.1. Number of Risk Categories: An Ongoing Debate
4.2. Gene-Based Prognostic Assays: The Major Takeaways
5. Liquid Biopsy Holds Promise for Guiding Breast Cancer Management
5.1. Serum Protein Markers
5.2. Circulating microRNAs
6. Metabolomics in Breast Cancer Prognosis
7. Tumor Microenvironment-Based Biomarkers: Tumor Infiltrating Lymphocytes as Prognostic and Predictive Variables in Breast Cancer
8. Neoantigens as Biomarkers of Treatment Response
9. Digital Pathology and Tissue Phenome Analysis
10. Scoring Centrosome Amplification: An Emerging Prognostic Marker
10.1. Immunofluorescence-Based Three-Dimensional Image Analysis Yields a CA Score for Ductal Carcinoma In Situ Stratification
10.2. CA20: A Transcriptomic Signature
11. Prognostic Breast Cancer Staging
12. Future Perspectives
13. Conclusions
Funding
Conflicts of Interest
References
- Haybittle, J.L.; Blamey, R.W.; Elston, C.W.; Johnson, J.; Doyle, P.J.; Campbell, F.C.; Nicholson, R.I.; Griffiths, K. A prognostic index in primary breast cancer. Br. J. Cancer 1982, 45, 361–366. [Google Scholar] [CrossRef] [PubMed]
- Green, A.R.; Soria, D.; Powe, D.G.; Nolan, C.C.; Aleskandarany, M.; Szasz, M.A.; Tokes, A.M.; Ball, G.R.; Garibaldi, J.M.; Rakha, E.A.; et al. Nottingham prognostic index plus (NPI+) predicts risk of distant metastases in primary breast cancer. Breast Cancer Res. Treat. 2016, 157, 65–75. [Google Scholar] [CrossRef] [PubMed]
- Kwon, J.; Eom, K.Y.; Koo, T.R.; Kim, B.H.; Kang, E.; Kim, S.W.; Kim, Y.J.; Park, S.Y.; Kim, I.A. A Prognostic Model for Patients with Triple-Negative Breast Cancer: Importance of the Modified Nottingham Prognostic Index and Age. J. Breast Cancer 2017, 20, 65–73. [Google Scholar] [CrossRef] [PubMed]
- Silverstein, J.M.; Lagios, M.D. Treatment selection for patients with ductal carcinoma in situ (DCIS) of the breast using the University of Southern California/Van Nuys (USC/VNPI) prognostic index. Breast J. 2015, 21, 127–132. [Google Scholar] [CrossRef] [PubMed]
- Dowsett, M.; Sestak, I.; Regan, M.M.; Dodson, A.; Viale, G.; Thurlimann, B.; Colleoni, M.; Cuzick, J. Integration of Clinical Variables for the Prediction of Late Distant Recurrence in Patients With Estrogen Receptor-Positive Breast Cancer Treated with 5 Years of Endocrine Therapy: CTS5. J. Clin. Oncol. 2018, 36, 1941–1948. [Google Scholar] [CrossRef] [PubMed]
- Wazir, U.; Mokbel, K.; Carmichael, A.; Mokbel, K. Are online prediction tools a valid alternative to genomic profiling in the context of systemic treatment of ER-positive breast cancer? Cell. Mol. Biol. Lett. 2017, 22, 20. [Google Scholar] [CrossRef]
- Cuzick, J.; Dowsett, M.; Pineda, S.; Wale, C.; Salter, J.; Quinn, E.; Zabaglo, L.; Mallon, E.; Green, A.R.; Ellis, I.O.; et al. Prognostic value of a combined estrogen receptor, progesterone receptor, Ki-67, and human epidermal growth factor receptor 2 immunohistochemical score and comparison with the Genomic Health recurrence score in early breast cancer. J. Clin. Oncol. 2011, 29, 4273–4278. [Google Scholar] [CrossRef]
- Vieira, A.F.; Schmitt, F. An Update on Breast Cancer Multigene Prognostic Tests-Emergent Clinical Biomarkers. Front. Med. 2018, 5, 248. [Google Scholar] [CrossRef]
- Lakhanpal, R.; Sestak, I.; Shadbolt, B.; Bennett, G.M.; Brown, M.; Phillips, T.; Zhang, Y.; Bullman, A.; Rezo, A. IHC4 score plus clinical treatment score predicts locoregional recurrence in early breast cancer. Breast 2016, 29, 147–152. [Google Scholar] [CrossRef]
- Look, M.P.; van Putten, W.L.; Duffy, M.J.; Harbeck, N.; Christensen, I.J.; Thomssen, C.; Kates, R.; Spyratos, F.; Ferno, M.; Eppenberger-Castori, S.; et al. Pooled analysis of prognostic impact of urokinase-type plasminogen activator and its inhibitor PAI-1 in 8377 breast cancer patients. J. Natl. Cancer Inst. 2002, 94, 116–128. [Google Scholar] [CrossRef]
- Janicke, F.; Prechtl, A.; Thomssen, C.; Harbeck, N.; Meisner, C.; Untch, M.; Sweep, C.G.; Selbmann, H.K.; Graeff, H.; Schmitt, M.; et al. Randomized adjuvant chemotherapy trial in high-risk, lymph node-negative breast cancer patients identified by urokinase-type plasminogen activator and plasminogen activator inhibitor type 1. J. Natl. Cancer Inst. 2001, 93, 913–920. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Harbeck, N.; Schmitt, M.; Meisner, C.; Friedel, C.; Untch, M.; Schmidt, M.; Sweep, C.G.; Lisboa, B.W.; Lux, M.P.; Beck, T.; et al. Ten-year analysis of the prospective multicentre Chemo-N0 trial validates American Society of Clinical Oncology (ASCO)-recommended biomarkers uPA and PAI-1 for therapy decision making in node-negative breast cancer patients. Eur. J. Cancer 2013, 49, 1825–1835. [Google Scholar] [CrossRef] [PubMed]
- Foekens, J.A.; Peters, H.A.; Look, M.P.; Portengen, H.; Schmitt, M.; Kramer, M.D.; Brunner, N.; Janicke, F.; Gelder, M.E.; Henzen-Logmans, S.C.; et al. The urokinase system of plasminogen activation and prognosis in 2780 breast cancer patients. Cancer Res. 2000, 60, 636–643. [Google Scholar] [PubMed]
- Harris, L.; Fritsche, H.; Mennel, R.; Norton, L.; Ravdin, P.; Taube, S.; Somerfield, M.R.; Hayes, D.F.; Bast, R.C., Jr.; American Society of Clinical Oncology. American Society of Clinical Oncology 2007 update of recommendations for the use of tumor markers in breast cancer. J. Clin. Oncol. 2007, 25, 5287–5312. [Google Scholar] [CrossRef] [PubMed]
- Bellocq, J.P.; Luporsi, E.; Barriere, J.; Bonastre, J.; Chetritt, J.; le Corroller, A.G.; de Cremoux, P.; Fina, F.; Gauchez, A.S.; Kassab-Chahmi, D.; et al. uPA/PAI-1, Oncotype DX, MammaPrint((R)). Prognosis and predictive values for clinical utility in breast cancer management. Ann. Pathol. 2014, 34, 349–351. [Google Scholar] [CrossRef] [PubMed]
- Nicolini, A.; Ferrari, P.; Duffy, M.J. Prognostic and predictive biomarkers in breast cancer: Past, present and future. Semin. Cancer Biol. 2018, 52, 56–73. [Google Scholar] [CrossRef]
- Volker, H.U.; Weigel, M.; Strehl, A.; Frey, L. Levels of uPA and PAI-1 in breast cancer and its correlation to Ki67-index and results of a 21-multigene-array. Diagn. Pathol. 2018, 13, 67. [Google Scholar] [CrossRef]
- Dixon, A.R.; Bathany, C.; Tsuei, M.; White, J.; Barald, K.F.; Takayama, S. Recent developments in multiplexing techniques for immunohistochemistry. Expert. Rev. Mol. Diagn. 2015, 15, 1171–1186. [Google Scholar] [CrossRef]
- Ross, J.S.; Hatzis, C.; Symmans, W.F.; Pusztai, L.; Hortobagyi, G.N. Commercialized multigene predictors of clinical outcome for breast cancer. Oncologist 2008, 13, 477–493. [Google Scholar] [CrossRef]
- Paik, S.; Shak, S.; Tang, G.; Kim, C.; Baker, J.; Cronin, M.; Baehner, F.L.; Walker, M.G.; Watson, D.; Park, T.; et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N. Engl. J. Med. 2004, 351, 2817–2826. [Google Scholar] [CrossRef]
- Esteban, J.; Baker, J.; Cronin, M.; Liu, M.-L.; Llamas, M.; Walker, M.; Mena, R.; Shak, S. Tumor gene expression and prognosis in breast cancer: Multi-gene RT-PCR assay of paraffin-embedded tissue. Proc. Am. Soc. Clin. Oncol. 2003, 22, A3416. [Google Scholar]
- Cobleigh, M.; Bitterman, P.; Baker, J.; Cronin, M.; Liu, M.-L.; Borchik, R.; Tabesh, B.; Mosquera, J.-M.; Walker, M.; Shak, S. Tumor gene expression predicts distant disease-free survival (DDFS) in breast cancer patients with 10 or more positive nodes: High throughput RT-PCR assay of paraffin-embedded tumor tissues. Proc. Am. Soc. Clin. Oncol. 2003, 22, A3415. [Google Scholar]
- Habel, L.A.; Shak, S.; Jacobs, M.K.; Capra, A.; Alexander, C.; Pho, M.; Baker, J.; Walker, M.; Watson, D.; Hackett, J.; et al. A population-based study of tumor gene expression and risk of breast cancer death among lymph node-negative patients. Breast Cancer Res. 2006, 8, R25. [Google Scholar] [CrossRef] [PubMed]
- Cronin, M.; Pho, M.; Dutta, D.; Stephans, J.C.; Shak, S.; Kiefer, M.C.; Esteban, J.M.; Baker, J.B. Measurement of gene expression in archival paraffin-embedded tissues: Development and performance of a 92-gene reverse transcriptase-polymerase chain reaction assay. Am. J. Pathol. 2004, 164, 35–42. [Google Scholar] [CrossRef]
- Cobleigh, M.A.; Tabesh, B.; Bitterman, P.; Baker, J.; Cronin, M.; Liu, M.L.; Borchik, R.; Mosquera, J.M.; Walker, M.G.; Shak, S. Tumor gene expression and prognosis in breast cancer patients with 10 or more positive lymph nodes. Clin. Cancer Res. 2005, 11, 8623–8631. [Google Scholar] [CrossRef] [PubMed]
- Dowsett, M.; Cuzick, J.; Wale, C.; Forbes, J.; Mallon, E.A.; Salter, J.; Quinn, E.; Dunbier, A.; Baum, M.; Buzdar, A.; et al. Prediction of risk of distant recurrence using the 21-gene recurrence score in node-negative and node-positive postmenopausal patients with breast cancer treated with anastrozole or tamoxifen: A TransATAC study. J. Clin. Oncol. 2010, 28, 1829–1834. [Google Scholar] [CrossRef]
- Sparano, J.A.; Gray, R.J.; Makower, D.F.; Pritchard, K.I.; Albain, K.S.; Hayes, D.F.; Geyer, C.E., Jr.; Dees, E.C.; Perez, E.A.; Olson, J.A., Jr.; et al. Prospective Validation of a 21-Gene Expression Assay in Breast Cancer. N. Engl. J. Med. 2015, 373, 2005–2014. [Google Scholar] [CrossRef]
- Toi, M.; Iwata, H.; Yamanaka, T.; Masuda, N.; Ohno, S.; Nakamura, S.; Nakayama, T.; Kashiwaba, M.; Kamigaki, S.; Kuroi, K.; et al. Clinical significance of the 21-gene signature (Oncotype DX) in hormone receptor-positive early stage primary breast cancer in the Japanese population. Cancer 2010, 116, 3112–3118. [Google Scholar] [CrossRef]
- Paik, S.; Tang, G.; Shak, S.; Kim, C.; Baker, J.; Kim, W.; Cronin, M.; Baehner, F.L.; Watson, D.; Bryant, J.; et al. Gene expression and benefit of chemotherapy in women with node-negative, estrogen receptor-positive breast cancer. J. Clin. Oncol. 2006, 24, 3726–3734. [Google Scholar] [CrossRef]
- Cronin, M.; Sangli, C.; Liu, M.L.; Pho, M.; Dutta, D.; Nguyen, A.; Jeong, J.; Wu, J.; Langone, K.C.; Watson, D. Analytical validation of the Oncotype DX genomic diagnostic test for recurrence prognosis and therapeutic response prediction in node-negative, estrogen receptor-positive breast cancer. Clin. Chem. 2007, 53, 1084–1091. [Google Scholar] [CrossRef]
- Barcenas, C.H.; Raghavendra, A.; Sinha, A.K.; Syed, M.P.; Hsu, L.; Patangan, M.G., Jr.; Chavez-MacGregor, M.; Shen, Y.; Hortobagyi, G.H.; Valero, V.; et al. Outcomes in patients with early-stage breast cancer who underwent a 21-gene expression assay. Cancer 2017, 123, 2422–2431. [Google Scholar] [CrossRef] [PubMed]
- Stemmer, S.M.; Steiner, M.; Rizel, S.; Soussan-Gutman, L.; Ben-Baruch, N.; Bareket-Samish, A.; Geffen, D.B.; Nisenbaum, B.; Isaacs, K.; Fried, G.; et al. Clinical outcomes in patients with node-negative breast cancer treated based on the recurrence score results: Evidence from a large prospectively designed registry. NPJ Breast Cancer 2017, 3, 33. [Google Scholar] [CrossRef] [PubMed]
- Fayanju, O.M.; Park, K.U.; Lucci, A. Molecular Genomic Testing for Breast Cancer: Utility for Surgeons. Ann. Surg. Oncol. 2018, 25, 512–519. [Google Scholar] [CrossRef] [PubMed]
- Solin, L.J.; Gray, R.; Baehner, F.L.; Butler, S.M.; Hughes, L.L.; Yoshizawa, C.; Cherbavaz, D.B.; Shak, S.; Page, D.L.; Sledge, G.W., Jr.; et al. A multigene expression assay to predict local recurrence risk for ductal carcinoma in situ of the breast. J. Natl. Cancer. Inst. 2013, 105, 701–710. [Google Scholar] [CrossRef] [PubMed]
- Solin, L.J.; Gray, R.; Hughes, L.L.; Wood, W.C.; Lowen, M.A.; Badve, S.S.; Baehner, F.L.; Ingle, J.N.; Perez, E.A.; Recht, A.; et al. Surgical Excision Without Radiation for Ductal Carcinoma in Situ of the Breast: 12-Year Results From the ECOG-ACRIN E5194 Study. J. Clin. Oncol. 2015, 33, 3938–3944. [Google Scholar] [CrossRef] [PubMed]
- Rakovitch, E.; Nofech-Mozes, S.; Narod, S.A.; Hanna, W.; Thiruchelvam, D.; Saskin, R.; Taylor, C.; Tuck, A.; Sengupta, S.; Elavathil, L.; et al. Can we select individuals with low risk ductal carcinoma in situ (DCIS)? A population-based outcomes analysis. Breast Cancer Res. Treat. 2013, 138, 581–590. [Google Scholar] [CrossRef] [PubMed]
- Rakovitch, E.; Nofech-Mozes, S.; Hanna, W.; Baehner, F.L.; Saskin, R.; Butler, S.M.; Tuck, A.; Sengupta, S.; Elavathil, L.; Jani, P.A.; et al. A population-based validation study of the DCIS Score predicting recurrence risk in individuals treated by breast-conserving surgery alone. Breast Cancer Res. Treat. 2015, 152, 389–398. [Google Scholar] [CrossRef]
- Hughes, L.L.; Wang, M.; Page, D.L.; Gray, R.; Solin, L.J.; Davidson, N.E.; Lowen, M.A.; Ingle, J.N.; Recht, A.; Wood, W.C. Local excision alone without irradiation for ductal carcinoma in situ of the breast: A trial of the Eastern Cooperative Oncology Group. J. Clin. Oncol. 2009, 27, 5319–5324. [Google Scholar] [CrossRef]
- Acs, G.; Esposito, N.N.; Kiluk, J.; Loftus, L.; Laronga, C. A mitotically active, cellular tumor stroma and/or inflammatory cells associated with tumor cells may contribute to intermediate or high Oncotype DX Recurrence Scores in low-grade invasive breast carcinomas. Mod. Pathol. 2012, 25, 556–566. [Google Scholar] [CrossRef][Green Version]
- Acs, G.; Kiluk, J.; Loftus, L.; Laronga, C. Comparison of Oncotype DX and Mammostrat risk estimations and correlations with histologic tumor features in low-grade, estrogen receptor-positive invasive breast carcinomas. Mod. Pathol. 2013, 26, 1451–1460. [Google Scholar] [CrossRef][Green Version]
- Grimes, M.; Coad, J.; Oliviero, B. Comparison of Oncotype DX Recurrence Score and standard immunohistochemical prognostic markers (abstract). Mod. Pathol. 2007, 20, 33A. [Google Scholar]
- Kittaneh, M.; Montero, A.J.; Gluck, S. Molecular profiling for breast cancer: A comprehensive review. Biomark. Cancer 2013, 5, 61–70. [Google Scholar] [CrossRef] [PubMed]
- Mook, S.; Schmidt, M.K.; Viale, G.; Pruneri, G.; Eekhout, I.; Floore, A.; Glas, A.M.; Bogaerts, J.; Cardoso, F.; Piccart-Gebhart, M.J.; et al. The 70-gene prognosis-signature predicts disease outcome in breast cancer patients with 1-3 positive lymph nodes in an independent validation study. Breast Cancer Res. Treat. 2009, 116, 295–302. [Google Scholar] [CrossRef] [PubMed]
- Van ‘t Veer, L.J.; Dai, H.; van de Vijver, M.J.; He, Y.D.; Hart, A.A.; Mao, M.; Peterse, H.L.; van der Kooy, K.; Marton, M.J.; Witteveen, A.T.; et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature 2002, 415, 530–536. [Google Scholar] [CrossRef] [PubMed]
- Glas, A.M.; Floore, A.; Delahaye, L.J.; Witteveen, A.T.; Pover, R.C.; Bakx, N.; Lahti-Domenici, J.S.; Bruinsma, T.J.; Warmoes, M.O.; Bernards, R.; et al. Converting a breast cancer microarray signature into a high-throughput diagnostic test. BMC Genomics 2006, 7, 278. [Google Scholar] [CrossRef] [PubMed]
- Cardoso, F.; Piccart-Gebhart, M.; Veer, L.V.; Rutgers, E.; Consortium, T. The MINDACT trial: The first prospective clinical validation of a genomic tool. Mol. Oncol. 2007, 1, 246–251. [Google Scholar] [CrossRef] [PubMed]
- Mook, S.; Veer, L.J.V.; Rutgers, E.J.; Piccart-Gebhart, M.J.; Cardoso, F. Individualization of therapy using Mammaprint: From development to the MINDACT Trial. Cancer Genomics Proteomics 2007, 4, 147–155. [Google Scholar] [PubMed]
- Knauer, M.; Mook, S.; Rutgers, E.J.; Bender, R.A.; Hauptmann, M.; van de Vijver, M.J.; Koornstra, R.H.; Bueno-de-Mesquita, J.M.; Linn, S.C.; Van ‘t Veer, L.J. The predictive value of the 70-gene signature for adjuvant chemotherapy in early breast cancer. Breast Cancer Res. Treat. 2010, 120, 655–661. [Google Scholar] [CrossRef] [PubMed]
- Drukker, C.A.; Bueno-de-Mesquita, J.M.; Retel, V.P.; van Harten, W.H.; van Tinteren, H.; Wesseling, J.; Roumen, R.M.; Knauer, M.; van ‘t Veer, L.J.; Sonke, G.S.; et al. A prospective evaluation of a breast cancer prognosis signature in the observational RASTER study. Int. J. Cancer 2013, 133, 929–936. [Google Scholar] [CrossRef] [PubMed]
- Drukker, C.A.; van Tinteren, H.; Schmidt, M.K.; Rutgers, E.J.; Bernards, R.; van de Vijver, M.J.; Van’t Veer, L.J. Long-term impact of the 70-gene signature on breast cancer outcome. Breast Cancer Res. Treat. 2014, 143, 587–592. [Google Scholar] [CrossRef] [PubMed]
- Cardoso, F.; van’t Veer, L.J.; Bogaerts, J.; Slaets, L.; Viale, G.; Delaloge, S.; Pierga, J.Y.; Brain, E.; Causeret, S.; DeLorenzi, M.; et al. 70-Gene Signature as an Aid to Treatment Decisions in Early-Stage Breast Cancer. N. Engl. J. Med. 2016, 375, 717–729. [Google Scholar] [CrossRef] [PubMed]
- Gluck, S.; de Snoo, F.; Peeters, J.; Stork-Sloots, L.; Somlo, G. Molecular subtyping of early-stage breast cancer identifies a group of patients who do not benefit from neoadjuvant chemotherapy. Breast Cancer Res. Treat. 2013, 139, 759–767. [Google Scholar] [CrossRef]
- Geiss, G.K.; Bumgarner, R.E.; Birditt, B.; Dahl, T.; Dowidar, N.; Dunaway, D.L.; Fell, H.P.; Ferree, S.; George, R.D.; Grogan, T.; et al. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat. Biotechnol. 2008, 26, 317–325. [Google Scholar] [CrossRef] [PubMed]
- Parker, J.S.; Mullins, M.; Cheang, M.C.; Leung, S.; Voduc, D.; Vickery, T.; Davies, S.; Fauron, C.; He, X.; Hu, Z.; et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J. Clin. Oncol. 2009, 27, 1160–1167. [Google Scholar] [CrossRef] [PubMed]
- Bastien, R.R.; Rodriguez-Lescure, A.; Ebbert, M.T.; Prat, A.; Munarriz, B.; Rowe, L.; Miller, P.; Ruiz-Borrego, M.; Anderson, D.; Lyons, B.; et al. PAM50 breast cancer subtyping by RT-qPCR and concordance with standard clinical molecular markers. BMC Med. Genomics 2012, 5, 44. [Google Scholar] [CrossRef] [PubMed]
- Filipits, M.; Nielsen, T.O.; Rudas, M.; Greil, R.; Stoger, H.; Jakesz, R.; Bago-Horvath, Z.; Dietze, O.; Regitnig, P.; Gruber-Rossipal, C.; et al. The PAM50 risk-of-recurrence score predicts risk for late distant recurrence after endocrine therapy in postmenopausal women with endocrine-responsive early breast cancer. Clin. Cancer Res. 2014, 20, 1298–1305. [Google Scholar] [CrossRef] [PubMed]
- Park, J.L.; Park, S.M.; Kim, J.H.; Lee, H.C.; Lee, S.H.; Woo, K.M.; Kim, S.Y. Forensic Body Fluid Identification by Analysis of Multiple RNA Markers Using NanoString Technology. Genomics Inform. 2013, 11, 277–281. [Google Scholar] [CrossRef] [PubMed]
- Dowsett, M.; Sestak, I.; Lopez-Knowles, E.; Sidhu, K.; Dunbier, A.K.; Cowens, J.W.; Ferree, S.; Storhoff, J.; Schaper, C.; Cuzick, J. Comparison of PAM50 risk of recurrence score with oncotype DX and IHC4 for predicting risk of distant recurrence after endocrine therapy. J. Clin. Oncol. 2013, 31, 2783–2790. [Google Scholar] [CrossRef]
- Sestak, I.; Dowsett, M.; Zabaglo, L.; Lopez-Knowles, E.; Ferree, S.; Cowens, J.W.; Cuzick, J. Factors predicting late recurrence for estrogen receptor-positive breast cancer. J. Natl. Cancer Inst. 2013, 105, 1504–1511. [Google Scholar] [CrossRef]
- Nielsen, T.; Wallden, B.; Schaper, C.; Ferree, S.; Liu, S.; Gao, D.; Barry, G.; Dowidar, N.; Maysuria, M.; Storhoff, J. Analytical validation of the PAM50-based Prosigna Breast Cancer Prognostic Gene Signature Assay and nCounter Analysis System using formalin-fixed paraffin-embedded breast tumor specimens. BMC Cancer 2014, 14, 177. [Google Scholar] [CrossRef]
- Goncalves, R.; Bose, R. Using multigene tests to select treatment for early-stage breast cancer. J. Natl. Compr. Canc. Netw. 2013, 11, 174–182. [Google Scholar] [CrossRef]
- Filipits, M.; Rudas, M.; Jakesz, R.; Dubsky, P.; Fitzal, F.; Singer, C.F.; Dietze, O.; Greil, R.; Jelen, A.; Sevelda, P.; et al. A new molecular predictor of distant recurrence in ER-positive, HER2-negative breast cancer adds independent information to conventional clinical risk factors. Clin. Cancer Res. 2011, 17, 6012–6020. [Google Scholar] [CrossRef]
- Dubsky, P.; Brase, J.C.; Jakesz, R.; Rudas, M.; Singer, C.F.; Greil, R.; Dietze, O.; Luisser, I.; Klug, E.; Sedivy, R.; et al. The EndoPredict score provides prognostic information on late distant metastases in ER+/HER2- breast cancer patients. Br. J. Cancer 2013, 109, 2959–2964. [Google Scholar] [CrossRef] [PubMed]
- Dubsky, P.; Filipits, M.; Jakesz, R.; Rudas, M.; Singer, C.F.; Greil, R.; Dietze, O.; Luisser, I.; Klug, E.; Sedivy, R.; et al. EndoPredict improves the prognostic classification derived from common clinical guidelines in ER-positive, HER2-negative early breast cancer. Ann. Oncol. 2013, 24, 640–647. [Google Scholar] [CrossRef] [PubMed]
- Martin, M.; Brase, J.C.; Calvo, L.; Krappmann, K.; Ruiz-Borrego, M.; Fisch, K.; Ruiz, A.; Weber, K.E.; Munarriz, B.; Petry, C.; et al. Clinical validation of the EndoPredict test in node-positive, chemotherapy-treated ER+/HER2- breast cancer patients: Results from the GEICAM 9906 trial. Breast Cancer Res. 2014, 16, R38. [Google Scholar] [CrossRef]
- Ma, X.J.; Salunga, R.; Dahiya, S.; Wang, W.; Carney, E.; Durbecq, V.; Harris, A.; Goss, P.; Sotiriou, C.; Erlander, M.; et al. A five-gene molecular grade index and HOXB13:IL17BR are complementary prognostic factors in early stage breast cancer. Clin. Cancer Res. 2008, 14, 2601–2608. [Google Scholar] [CrossRef] [PubMed]
- Sgroi, D.C.; Sestak, I.; Cuzick, J.; Zhang, Y.; Schnabel, C.A.; Schroeder, B.; Erlander, M.G.; Dunbier, A.; Sidhu, K.; Lopez-Knowles, E.; et al. Prediction of late distant recurrence in patients with oestrogen-receptor-positive breast cancer: A prospective comparison of the breast-cancer index (BCI) assay, 21-gene recurrence score, and IHC4 in the TransATAC study population. Lancet Oncol. 2013, 14, 1067–1076. [Google Scholar] [CrossRef]
- Jerevall, P.L.; Ma, X.J.; Li, H.; Salunga, R.; Kesty, N.C.; Erlander, M.G.; Sgroi, D.C.; Holmlund, B.; Skoog, L.; Fornander, T.; et al. Prognostic utility of HOXB13:IL17BR and molecular grade index in early-stage breast cancer patients from the Stockholm trial. Br. J. Cancer 2011, 104, 1762–1769. [Google Scholar] [CrossRef]
- Sgroi, D.C.; Carney, E.; Zarrella, E.; Steffel, L.; Binns, S.N.; Finkelstein, D.M.; Szymonifka, J.; Bhan, A.K.; Shepherd, L.E.; Zhang, Y.; et al. Prediction of late disease recurrence and extended adjuvant letrozole benefit by the HOXB13/IL17BR biomarker. J. Natl. Cancer Inst. 2013, 105, 1036–1042. [Google Scholar] [CrossRef]
- Zhang, Y.; Schnabel, C.A.; Schroeder, B.E.; Jerevall, P.L.; Jankowitz, R.C.; Fornander, T.; Stal, O.; Brufsky, A.M.; Sgroi, D.; Erlander, M.G. Breast cancer index identifies early-stage estrogen receptor-positive breast cancer patients at risk for early- and late-distant recurrence. Clin. Cancer Res. 2013, 19, 4196–4205. [Google Scholar] [CrossRef]
- Harris, L.N.; Ismaila, N.; McShane, L.M.; Andre, F.; Collyar, D.E.; Gonzalez-Angulo, A.M.; Hammond, E.H.; Kuderer, N.M.; Liu, M.C.; Mennel, R.G.; et al. Use of Biomarkers to Guide Decisions on Adjuvant Systemic Therapy for Women With Early-Stage Invasive Breast Cancer: American Society of Clinical Oncology Clinical Practice Guideline. J. Clin. Oncol. 2016, 34, 1134–1150. [Google Scholar] [CrossRef]
- Sestak, I.; Buus, R.; Cuzick, J.; Dubsky, P.; Kronenwett, R.; Denkert, C.; Ferree, S.; Sgroi, D.; Schnabel, C.; Baehner, F.L.; et al. Comparison of the Performance of 6 Prognostic Signatures for Estrogen Receptor-Positive Breast Cancer: A Secondary Analysis of a Randomized Clinical Trial. JAMA Oncol. 2018, 4, 545–553. [Google Scholar] [CrossRef]
- Guacci, A.; Cordella, A.; Rocco, T.; Giurato, G.; Nassa, G.; Rizzo, F.; Carlomagno, C.; Pepe, S.; Tarallo, R.; Weisz, A. Identification of a novel truncating mutation in PALB2 gene by a multigene sequencing panel for mutational screening of breast cancer risk-associated and related genes. J. Clin. Lab. Anal. 2018, 32, e22418. [Google Scholar] [CrossRef]
- Heeke, A.L.; Pishvaian, M.J.; Lynce, F.; Xiu, J.; Brody, J.R.; Chen, W.J.; Baker, T.M.; Marshall, J.L.; Isaacs, C. Prevalence of Homologous Recombination-Related Gene Mutations Across Multiple Cancer Types. JCO Precis. Oncol. 2018, 2018, 1–13. [Google Scholar] [CrossRef]
- Hodgson, D.R.; Dougherty, B.A.; Lai, Z.; Fielding, A.; Grinsted, L.; Spencer, S.; O’Connor, M.J.; Ho, T.W.; Robertson, J.D.; Lanchbury, J.S.; et al. Candidate biomarkers of PARP inhibitor sensitivity in ovarian cancer beyond the BRCA genes. Br. J. Cancer 2018, 119, 1401–1409. [Google Scholar] [CrossRef]
- Sparano, J.A.; Gray, R.J.; Makower, D.F.; Pritchard, K.I.; Albain, K.S.; Hayes, D.F.; Geyer, C.E., Jr.; Dees, E.C.; Goetz, M.P.; Olson, J.A., Jr.; et al. Adjuvant Chemotherapy Guided by a 21-Gene Expression Assay in Breast Cancer. N. Engl. J. Med. 2018, 379, 111–121. [Google Scholar] [CrossRef]
- Sparano, J.A.; Gray, R.J.; Ravdin, P.M.; Makower, D.F.; Pritchard, K.I.; Albain, K.S.; Hayes, D.F.; Geyer, C.E., Jr.; Dees, E.C.; Goetz, M.P.; et al. Clinical and Genomic Risk to Guide the Use of Adjuvant Therapy for Breast Cancer. N. Engl. J. Med. 2019, 380, 2395–2405. [Google Scholar] [CrossRef]
- Baselga, J.; Im, S.A.; Iwata, H.; Cortes, J.; de Laurentiis, M.; Jiang, Z.; Arteaga, C.L.; Jonat, W.; Clemons, M.; Ito, Y.; et al. Buparlisib plus fulvestrant versus placebo plus fulvestrant in postmenopausal, hormone receptor-positive, HER2-negative, advanced breast cancer (BELLE-2): A randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol. 2017, 18, 904–916. [Google Scholar] [CrossRef]
- Fribbens, C.; O’Leary, B.; Kilburn, L.; Hrebien, S.; Garcia-Murillas, I.; Beaney, M.; Cristofanilli, M.; Andre, F.; Loi, S.; Loibl, S.; et al. Plasma ESR1 Mutations and the Treatment of Estrogen Receptor-Positive Advanced Breast Cancer. J. Clin. Oncol. 2016, 34, 2961–2968. [Google Scholar] [CrossRef]
- Rothe, F.; Silva, M.J.; Venet, D.; Campbell, C.; Bradburry, I.; Rouas, G.; de Azambuja, E.; Maetens, M.; Fumagalli, D.; Rodrik-Outmezguine, V.; et al. Circulating Tumor DNA in HER2-Amplified Breast Cancer: A Translational Research Substudy of the NeoALTTO Phase III Trial. Clin. Cancer Res. 2019, 25, 3581–3588. [Google Scholar] [CrossRef]
- Murtaza, M.; Dawson, S.J.; Pogrebniak, K.; Rueda, O.M.; Provenzano, E.; Grant, J.; Chin, S.F.; Tsui, D.W.; Marass, F.; Gale, D.; et al. Multifocal clonal evolution characterized using circulating tumour DNA in a case of metastatic breast cancer. Nat. Commun. 2015, 6, 8760. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.H.; Hancock, B.A.; Solzak, J.P.; Brinza, D.; Scafe, C.; Miller, K.D.; Radovich, M. Next-generation sequencing of circulating tumor DNA to predict recurrence in triple-negative breast cancer patients with residual disease after neoadjuvant chemotherapy. NPJ Breast Cancer 2017, 3, 24. [Google Scholar] [CrossRef] [PubMed]
- Sparano, J.; O’Neill, A.; Alpaugh, K.; Wolff, A.C.; Northfelt, D.W.; Dang, C.T.; Sledge, G.W.; Miller, K.D. Association of Circulating Tumor Cells with Late Recurrence of Estrogen Receptor-Positive Breast Cancer: A Secondary Analysis of a Randomized Clinical Trial. JAMA Oncol. 2018, 4, 1700–1706. [Google Scholar] [CrossRef] [PubMed]
- Pantel, K.; Brakenhoff, R.H.; Brandt, B. Detection, clinical relevance and specific biological properties of disseminating tumour cells. Nat. Rev. Cancer 2008, 8, 329–340. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, M.M.; Ramani, V.C.; Jeffrey, S.S. Circulating tumor cell technologies. Mol. Oncol. 2016, 10, 374–394. [Google Scholar] [CrossRef] [PubMed]
- Bidard, F.C.; Proudhon, C.; Pierga, J.Y. Circulating tumor cells in breast cancer. Mol. Oncol. 2016, 10, 418–430. [Google Scholar] [CrossRef] [PubMed]
- Cristofanilli, M.; Budd, G.T.; Ellis, M.J.; Stopeck, A.; Matera, J.; Miller, M.C.; Reuben, J.M.; Doyle, G.V.; Allard, W.J.; Terstappen, L.W.; et al. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N. Engl. J. Med. 2004, 351, 781–791. [Google Scholar] [CrossRef]
- Smerage, J.B.; Barlow, W.E.; Hortobagyi, G.N.; Winer, E.P.; Leyland-Jones, B.; Srkalovic, G.; Tejwani, S.; Schott, A.F.; O’Rourke, M.A.; Lew, D.L.; et al. Circulating tumor cells and response to chemotherapy in metastatic breast cancer: SWOG S0500. J. Clin. Oncol. 2014, 32, 3483–3489. [Google Scholar] [CrossRef]
- Budd, G.T.; Cristofanilli, M.; Ellis, M.J.; Stopeck, A.; Borden, E.; Miller, M.C.; Matera, J.; Repollet, M.; Doyle, G.V.; Terstappen, L.W.; et al. Circulating tumor cells versus imaging--predicting overall survival in metastatic breast cancer. Clin. Cancer Res. 2006, 12, 6403–6409. [Google Scholar] [CrossRef]
- Lucci, A.; Hall, C.S.; Lodhi, A.K.; Bhattacharyya, A.; Anderson, A.E.; Xiao, L.; Bedrosian, I.; Kuerer, H.M.; Krishnamurthy, S. Circulating tumour cells in non-metastatic breast cancer: A prospective study. Lancet Oncol. 2012, 13, 688–695. [Google Scholar] [CrossRef]
- Kowalik, A.; Kowalewska, M.; Gozdz, S. Current approaches for avoiding the limitations of circulating tumor cells detection methods-implications for diagnosis and treatment of patients with solid tumors. Transl. Res. 2017, 185, 58–84.e15. [Google Scholar] [CrossRef] [PubMed]
- Alix-Panabieres, C.; Vendrell, J.P.; Pelle, O.; Rebillard, X.; Riethdorf, S.; Muller, V.; Fabbro, M.; Pantel, K. Detection and characterization of putative metastatic precursor cells in cancer patients. Clin. Chem. 2007, 53, 537–539. [Google Scholar] [CrossRef] [PubMed]
- Alix-Panabieres, C. EPISPOT assay: Detection of viable DTCs/CTCs in solid tumor patients. Recent Results Cancer Res. 2012, 195, 69–76. [Google Scholar] [PubMed]
- Ramirez, J.M.; Fehm, T.; Orsini, M.; Cayrefourcq, L.; Maudelonde, T.; Pantel, K.; Alix-Panabieres, C. Prognostic relevance of viable circulating tumor cells detected by EPISPOT in metastatic breast cancer patients. Clin. Chem. 2014, 60, 214–221. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.H.; Wang, Y.; Oliver, C.R.; Thamm, D.H.; Cooling, L.; Paoletti, C.; Smith, K.J.; Nagrath, S.; Hayes, D.F. A temporary indwelling intravascular aphaeretic system for in vivo enrichment of circulating tumor cells. Nat. Commun. 2019, 10, 1478. [Google Scholar] [CrossRef]
- Ignatiadis, M.; Lee, M.; Jeffrey, S.S. Circulating Tumor Cells and Circulating Tumor DNA: Challenges and Opportunities on the Path to Clinical Utility. Clin. Cancer Res. 2015, 21, 4786–4800. [Google Scholar] [CrossRef]
- Reese, D.E.; Henderson, M.C.; Silver, M.; Mulpuri, R.; Letsios, E.; Tran, Q.; Wolf, J.K. Breast density does not impact the ability of Videssa(R) Breast to detect breast cancer in women under age 50. PLoS ONE 2017, 12, e0186198. [Google Scholar] [CrossRef]
- Lourenco, A.P.; Benson, K.L.; Henderson, M.C.; Silver, M.; Letsios, E.; Tran, Q.; Gordon, K.J.; Borman, S.; Corn, C.; Mulpuri, R.; et al. A Noninvasive Blood-based Combinatorial Proteomic Biomarker Assay to Detect Breast Cancer in Women Under the Age of 50 Years. Clin. Breast Cancer 2017, 17, 516–525.e6. [Google Scholar] [CrossRef]
- Sauter, E.R. Reliable Biomarkers to Identify New and Recurrent Cancer. Eur. J. Breast Health 2017, 13, 162–167. [Google Scholar] [CrossRef]
- Van Poznak, C.; Harris, L.N.; Somerfield, M.R. Use of Biomarkers to Guide Decisions on Systemic Therapy for Women with Metastatic Breast Cancer: American Society of Clinical Oncology Clinical Practice Guideline. J. Oncol. Pract. 2015, 11, 514–516. [Google Scholar] [CrossRef]
- Witwer, K.W. Circulating microRNA biomarker studies: Pitfalls and potential solutions. Clin. Chem. 2015, 61, 56–63. [Google Scholar] [CrossRef] [PubMed]
- Duffy, M.J.; Walsh, S.; McDermott, E.W.; Crown, J. Biomarkers in Breast Cancer: Where Are We and Where Are We Going? Adv. Clin. Chem. 2015, 71, 1–23. [Google Scholar] [PubMed]
- Hamam, R.; Hamam, D.; Alsaleh, K.A.; Kassem, M.; Zaher, W.; Alfayez, M.; Aldahmash, A.; Alajez, N.M. Circulating microRNAs in breast cancer: Novel diagnostic and prognostic biomarkers. Cell Death Dis. 2017, 8, e3045. [Google Scholar] [CrossRef] [PubMed]
- Barh, D. (Ed.) OMICS Approaches in Breast Cancer: Towards Next-Generation Diagnosis, Prognosis, and Therapy; Springer: New York, NY, USA, 2014. [Google Scholar]
- Asiago, V.M.; Alvarado, L.Z.; Shanaiah, N.; Gowda, G.A.; Owusu-Sarfo, K.; Ballas, R.A.; Raftery, D. Early detection of recurrent breast cancer using metabolite profiling. Cancer Res. 2010, 70, 8309–8318. [Google Scholar] [CrossRef] [PubMed]
- Giskeodegard, G.F.; Grinde, M.T.; Sitter, B.; Axelson, D.E.; Lundgren, S.; Fjosne, H.E.; Dahl, S.; Gribbestad, I.S.; Bathen, T.F. Multivariate modeling and prediction of breast cancer prognostic factors using MR metabolomics. J. Proteome Res. 2010, 9, 972–979. [Google Scholar] [CrossRef] [PubMed]
- Hadi, N.I.; Jamal, Q.; Iqbal, A.; Shaikh, F.; Somroo, S.; Musharraf, S.G. Serum Metabolomic Profiles for Breast Cancer Diagnosis, Grading and Staging by Gas Chromatography-Mass Spectrometry. Sci. Rep. 2017, 7, 1715. [Google Scholar] [CrossRef] [PubMed]
- Oakman, C.; Tenori, L.; Biganzoli, L.; Santarpia, L.; Cappadona, S.; Luchinat, C.; di Leo, A. Uncovering the metabolomic fingerprint of breast cancer. Int. J. Biochem. Cell Biol. 2011, 43, 1010–1020. [Google Scholar] [CrossRef] [PubMed]
- Tenori, L.; Oakman, C.; Morris, P.G.; Gralka, E.; Turner, N.; Cappadona, S.; Fornier, M.; Hudis, C.; Norton, L.; Luchinat, C.; et al. Serum metabolomic profiles evaluated after surgery may identify patients with oestrogen receptor negative early breast cancer at increased risk of disease recurrence. Results from a retrospective study. Mol. Oncol. 2015, 9, 128–139. [Google Scholar] [CrossRef]
- Wei, S.; Liu, L.; Zhang, J.; Bowers, J.; Gowda, G.A.; Seeger, H.; Fehm, T.; Neubauer, H.J.; Vogel, U.; Clare, S.E.; et al. Metabolomics approach for predicting response to neoadjuvant chemotherapy for breast cancer. Mol. Oncol. 2013, 7, 297–307. [Google Scholar] [CrossRef]
- Hart, C.D.; Vignoli, A.; Tenori, L.; Uy, G.L.; van To, T.; Adebamowo, C.; Hossain, S.M.; Biganzoli, L.; Risi, E.; Love, R.R.; et al. Serum Metabolomic Profiles Identify ER-Positive Early Breast Cancer Patients at Increased Risk of Disease Recurrence in a Multicenter Population. Clin. Cancer Res. 2017, 23, 1422–1431. [Google Scholar] [CrossRef]
- Helland, T.; Henne, N.; Bifulco, E.; Naume, B.; Borgen, E.; Kristensen, V.N.; Kvaloy, J.T.; Lash, T.L.; Alnaes, G.I.G.; van Schaik, R.H.; et al. Serum concentrations of active tamoxifen metabolites predict long-term survival in adjuvantly treated breast cancer patients. Breast Cancer Res. 2017, 19, 125. [Google Scholar] [CrossRef] [PubMed]
- Spratlin, J.L.; Serkova, N.J.; Eckhardt, S.G. Clinical applications of metabolomics in oncology: A review. Clin. Cancer Res. 2009, 15, 431–440. [Google Scholar] [CrossRef] [PubMed]
- Aaltomaa, S.; Lipponen, P.; Eskelinen, M.; Kosma, V.M.; Marin, S.; Alhava, E.; Syrjanen, K. Lymphocyte infiltrates as a prognostic variable in female breast cancer. Eur. J. Cancer 1992, 28A, 859–864. [Google Scholar] [CrossRef]
- Adams, S.; Gray, R.J.; Demaria, S.; Goldstein, L.; Perez, E.A.; Shulman, L.N.; Martino, S.; Wang, M.; Jones, V.E.; Saphner, T.J.; et al. Prognostic value of tumor-infiltrating lymphocytes in triple-negative breast cancers from two phase III randomized adjuvant breast cancer trials: ECOG 2197 and ECOG 1199. J. Clin. Oncol. 2014, 32, 2959–2966. [Google Scholar] [CrossRef] [PubMed]
- Dieci, M.V.; Mathieu, M.C.; Guarneri, V.; Conte, P.; Delaloge, S.; Andre, F.; Goubar, A. Prognostic and predictive value of tumor-infiltrating lymphocytes in two phase III randomized adjuvant breast cancer trials. Ann. Oncol. 2015, 26, 1698–1704. [Google Scholar] [CrossRef] [PubMed]
- Salgado, R.; Denkert, C.; Campbell, C.; Savas, P.; Nuciforo, P.; Aura, C.; de Azambuja, E.; Eidtmann, H.; Ellis, C.E.; Baselga, J.; et al. Tumor-Infiltrating Lymphocytes and Associations With Pathological Complete Response and Event-Free Survival in HER2-Positive Early-Stage Breast Cancer Treated With Lapatinib and Trastuzumab: A Secondary Analysis of the NeoALTTO Trial. JAMA Oncol. 2015, 1, 448–454. [Google Scholar] [CrossRef] [PubMed]
- Denkert, C.; Loibl, S.; Noske, A.; Roller, M.; Muller, B.M.; Komor, M.; Budczies, J.; Darb-Esfahani, S.; Kronenwett, R.; Hanusch, C.; et al. Tumor-associated lymphocytes as an independent predictor of response to neoadjuvant chemotherapy in breast cancer. J. Clin. Oncol. 2010, 28, 105–113. [Google Scholar] [CrossRef]
- Denkert, C.; von Minckwitz, G.; Brase, J.C.; Sinn, B.V.; Gade, S.; Kronenwett, R.; Pfitzner, B.M.; Salat, C.; Loi, S.; Schmitt, W.D.; et al. Tumor-infiltrating lymphocytes and response to neoadjuvant chemotherapy with or without carboplatin in human epidermal growth factor receptor 2-positive and triple-negative primary breast cancers. J. Clin. Oncol. 2015, 33, 983–991. [Google Scholar] [CrossRef]
- Ibrahim, E.M.; Al-Foheidi, M.E.; Al-Mansour, M.M.; Kazkaz, G.A. The prognostic value of tumor-infiltrating lymphocytes in triple-negative breast cancer: A meta-analysis. Breast Cancer Res. Treat. 2014, 148, 467–476. [Google Scholar] [CrossRef]
- Issa-Nummer, Y.; Darb-Esfahani, S.; Loibl, S.; Kunz, G.; Nekljudova, V.; Schrader, I.; Sinn, B.V.; Ulmer, H.U.; Kronenwett, R.; Just, M.; et al. Prospective validation of immunological infiltrate for prediction of response to neoadjuvant chemotherapy in HER2-negative breast cancer—A substudy of the neoadjuvant GeparQuinto trial. PLoS ONE 2013, 8, e79775. [Google Scholar] [CrossRef]
- Loi, S.; Michiels, S.; Salgado, R.; Sirtaine, N.; Jose, V.; Fumagalli, D.; Kellokumpu-Lehtinen, P.L.; Bono, P.; Kataja, V.; Desmedt, C.; et al. Tumor infiltrating lymphocytes are prognostic in triple negative breast cancer and predictive for trastuzumab benefit in early breast cancer: Results from the FinHER trial. Ann. Oncol. 2014, 25, 1544–1550. [Google Scholar] [CrossRef] [PubMed]
- Loi, S.; Sirtaine, N.; Piette, F.; Salgado, R.; Viale, G.; van Eenoo, F.; Rouas, G.; Francis, P.; Crown, J.P.; Hitre, E.; et al. Prognostic and predictive value of tumor-infiltrating lymphocytes in a phase III randomized adjuvant breast cancer trial in node-positive breast cancer comparing the addition of docetaxel to doxorubicin with doxorubicin-based chemotherapy: BIG 02-98. J. Clin. Oncol. 2013, 31, 860–867. [Google Scholar] [CrossRef] [PubMed]
- Mahmoud, S.M.; Paish, E.C.; Powe, D.G.; Macmillan, R.D.; Grainge, M.J.; Lee, A.H.; Ellis, I.O.; Green, A.R. Tumor-infiltrating CD8+ lymphocytes predict clinical outcome in breast cancer. J. Clin. Oncol. 2011, 29, 1949–1955. [Google Scholar] [CrossRef] [PubMed]
- Mao, Y.; Qu, Q.; Zhang, Y.; Liu, J.; Chen, X.; Shen, K. The value of tumor infiltrating lymphocytes (TILs) for predicting response to neoadjuvant chemotherapy in breast cancer: A systematic review and meta-analysis. PLoS ONE 2014, 9, e115103. [Google Scholar] [CrossRef] [PubMed]
- Menard, S.; Tomasic, G.; Casalini, P.; Balsari, A.; Pilotti, S.; Cascinelli, N.; Salvadori, B.; Colnaghi, M.I.; Rilke, F. Lymphoid infiltration as a prognostic variable for early-onset breast carcinomas. Clin. Cancer Res. 1997, 3, 817–819. [Google Scholar] [PubMed]
- Mohammed, Z.M.; Going, J.J.; Edwards, J.; Elsberger, B.; Doughty, J.C.; McMillan, D.C. The relationship between components of tumour inflammatory cell infiltrate and clinicopathological factors and survival in patients with primary operable invasive ductal breast cancer. Br. J. Cancer 2012, 107, 864–873. [Google Scholar] [CrossRef] [PubMed]
- Salgado, R.; Denkert, C.; Demaria, S.; Sirtaine, N.; Klauschen, F.; Pruneri, G.; Wienert, S.; van den Eynden, G.; Baehner, F.L.; Penault-Llorca, F.; et al. The evaluation of tumor-infiltrating lymphocytes (TILs) in breast cancer: Recommendations by an International TILs Working Group 2014. Ann. Oncol. 2015, 26, 259–271. [Google Scholar] [CrossRef] [PubMed]
- Hida, A.I.; Sagara, Y.; Yotsumoto, D.; Kanemitsu, S.; Kawano, J.; Baba, S.; Rai, Y.; Oshiro, Y.; Aogi, K.; Sagara, Y.; et al. Prognostic and predictive impacts of tumor-infiltrating lymphocytes differ between Triple-negative and HER2-positive breast cancers treated with standard systemic therapies. Breast Cancer Res. Treat. 2016, 158, 1–9. [Google Scholar] [CrossRef]
- Gnant, M.; Harbeck, N.; Thomssen, C. Gallen/Vienna 2017: A Brief Summary of the Consensus Discussion about Escalation and De-Escalation of Primary Breast Cancer Treatment. Breast Care 2017, 12, 102–107. [Google Scholar] [CrossRef]
- Kurozumi, S.; Inoue, K.; Matsumoto, H.; Fujii, T.; Horiguchi, J.; Oyama, T.; Kurosumi, M.; Shirabe, K. Prognostic utility of tumor-infiltrating lymphocytes in residual tumor after neoadjuvant chemotherapy with trastuzumab for HER2-positive breast cancer. Sci. Rep. 2019, 9, 1583. [Google Scholar] [CrossRef]
- Mao, Y.; Qu, Q.; Chen, X.; Huang, O.; Wu, J.; Shen, K. The Prognostic Value of Tumor-Infiltrating Lymphocytes in Breast Cancer: A Systematic Review and Meta-Analysis. PLoS ONE 2016, 11, e0152500. [Google Scholar] [CrossRef] [PubMed]
- Schumacher, T.N.; Schreiber, R.D. Neoantigens in cancer immunotherapy. Science 2015, 348, 69–74. [Google Scholar] [CrossRef] [PubMed]
- Snyder, A.; Makarov, V.; Merghoub, T.; Yuan, J.; Zaretsky, J.M.; Desrichard, A.; Walsh, L.A.; Postow, M.A.; Wong, P.; Ho, T.S.; et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N. Engl. J. Med. 2014, 371, 2189–2199. [Google Scholar] [CrossRef] [PubMed]
- Turk, J.L. Rudolf Virchow—Father of cellular pathology. J. R. Soc. Med. 1993, 86, 688–689. [Google Scholar] [PubMed]
- Dong, F.; Irshad, H.; Oh, E.Y.; Lerwill, M.F.; Brachtel, E.F.; Jones, N.C.; Knoblauch, N.W.; Montaser-Kouhsari, L.; Johnson, N.B.; Rao, L.K.; et al. Computational pathology to discriminate benign from malignant intraductal proliferations of the breast. PLoS ONE 2014, 9, e114885. [Google Scholar] [CrossRef] [PubMed]
- Janowczyk, A.; Madabhushi, A. Deep learning for digital pathology image analysis: A comprehensive tutorial with selected use cases. J. Pathol. Inform. 2016, 7, 29. [Google Scholar] [CrossRef] [PubMed]
- Litjens, G.; Sanchez, C.I.; Timofeeva, N.; Hermsen, M.; Nagtegaal, I.; Kovacs, I.; Hulsbergen-Van de Kaa, C.; Bult, P.; van Ginneken, B.; van der Laak, J. Deep learning as a tool for increased accuracy and efficiency of histopathological diagnosis. Sci. Rep. 2016, 6, 26286. [Google Scholar] [CrossRef] [PubMed]
- Madabhushi, A.; Lee, G. Image analysis and machine learning in digital pathology: Challenges and opportunities. Med. Image Anal. 2016, 33, 170–175. [Google Scholar] [CrossRef]
- Ehteshami Bejnordi, B.; Veta, M.; van Diest, P.J.; van Ginneken, B.; Karssemeijer, N.; Litjens, G.; van der Laak, J.; the, C.C.; Hermsen, M.; Manson, Q.F.; et al. Diagnostic Assessment of Deep Learning Algorithms for Detection of Lymph Node Metastases in Women With Breast Cancer. JAMA 2017, 318, 2199–2210. [Google Scholar] [CrossRef]
- Bejnordi, B.E.; Zuidhof, G.; Balkenhol, M.; Hermsen, M.; Bult, P.; van Ginneken, B.; Karssemeijer, N.; Litjens, G.; van der Laak, J. Context-aware stacked convolutional neural networks for classification of breast carcinomas in whole-slide histopathology images. J. Med. Imaging 2017, 4, 044504. [Google Scholar] [CrossRef]
- Klimov, S.; Miligy, I.M.; Gertych, A.; Jiang, Y.; Toss, M.S.; Rida, P.; Ellis, I.O.; Green, A.; Krishnamurti, U.; Rakha, E.A.; et al. A whole slide image-based machine learning approach to predict ductal carcinoma in situ (DCIS) recurrence risk. Breast Cancer Res. 2019, 21, 83. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Gadepalli, K.; Norouzi, M.; Dahl, G.E.; Kohlberger, T.; Boyko, A.; Venugopalan, S.; Timofeev, A.; Nelson, P.Q.; Corrado, G.S.; et al. Detecting cancer metastases on gigapixel pathology images. arXiv 2017, arXiv:1703.02442. [Google Scholar]
- Djuric, U.; Zadeh, G.; Aldape, K.; Diamandis, P. Precision histology: How deep learning is poised to revitalize histomorphology for personalized cancer care. NPJ Precis. Oncol. 2017, 1, 22. [Google Scholar] [CrossRef] [PubMed]
- Pannu, V.; Mittal, K.; Cantuaria, G.; Reid, M.D.; Li, X.; Donthamsetty, S.; McBride, M.; Klimov, S.; Osan, R.; Gupta, M.V.; et al. Rampant centrosome amplification underlies more aggressive disease course of triple negative breast cancers. Oncotarget 2015, 6, 10487–10497. [Google Scholar] [CrossRef] [PubMed]
- Denu, R.A.; Zasadil, L.M.; Kanugh, C.; Laffin, J.; Weaver, B.A.; Burkard, M.E. Centrosome amplification induces high grade features and is prognostic of worse outcomes in breast cancer. BMC Cancer 2016, 16, 47. [Google Scholar] [CrossRef] [PubMed]
- Mittal, K.; Kaur, J.; Wei, G.; Toss, M.S.; Osan, R.M.; Janssen, E.A.; Søiland, H.; Rakha, E.A.; Rida, P.C.; Aneja, R. A quantitative centrosomal amplification score (CAS) predicts local recurrence in ductal carcinoma in situ. In Proceedings of the 2018 San Antonio Breast Cancer Symposium, San Antonio, TX, USA, 4–8 December 2018. Abstract P5-18-02. [Google Scholar] [CrossRef]
- Ogden, A.; Rida, P.C.; Aneja, R. Prognostic value of CA20, a score based on centrosome amplification-associated genes, in breast tumors. Sci. Rep. 2017, 7, 262. [Google Scholar] [CrossRef] [PubMed]
- De Almeida, B.P.; Vieira, A.F.; Paredes, J.; Bettencourt-Dias, M.; Barbosa-Morais, N.L. Pan-cancer association of a centrosome amplification gene expression signature with genomic alterations and clinical outcome. PLoS Comput. Biol. 2019, 15, e1006832. [Google Scholar] [CrossRef]
- Mittendorf, E.A.; Vila, J.; Tucker, S.L.; Chavez-MacGregor, M.; Smith, B.D.; Symmans, W.F.; Sahin, A.A.; Hortobagyi, G.N.; Hunt, K.K. The Neo-Bioscore Update for Staging Breast Cancer Treated with Neoadjuvant Chemotherapy: Incorporation of Prognostic Biologic Factors into Staging After Treatment. JAMA Oncol. 2016, 2, 929–936. [Google Scholar] [CrossRef]
- Friedman, A.A.; Letai, A.; Fisher, D.E.; Flaherty, K.T. Precision medicine for cancer with next-generation functional diagnostics. Nat. Rev. Cancer 2015, 15, 747–756. [Google Scholar] [CrossRef]
- Ridky, T.W.; Chow, J.M.; Wong, D.J.; Khavari, P.A. Invasive three-dimensional organotypic neoplasia from multiple normal human epithelia. Nat. Med. 2010, 16, 1450–1455. [Google Scholar] [CrossRef]
- Sachs, N.; Clevers, H. Organoid cultures for the analysis of cancer phenotypes. Curr. Opin. Genet. Dev. 2014, 24, 68–73. [Google Scholar] [CrossRef] [PubMed]
- Sato, T.; Stange, D.E.; Ferrante, M.; Vries, R.G.; van Es, J.H.; van den Brink, S.; van Houdt, W.J.; Pronk, A.; van Gorp, J.; Siersema, P.D.; et al. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium. Gastroenterology 2011, 141, 1762–1772. [Google Scholar] [CrossRef] [PubMed]
- Siolas, D.; Hannon, G.J. Patient-derived tumor xenografts: Transforming clinical samples into mouse models. Cancer Res. 2013, 73, 5315–5319. [Google Scholar] [CrossRef] [PubMed]
- Jonas, O.; Landry, H.M.; Fuller, J.E.; Santini, J.T., Jr.; Baselga, J.; Tepper, R.I.; Cima, M.J.; Langer, R. An implantable microdevice to perform high-throughput in vivo drug sensitivity testing in tumors. Sci. Transl. Med. 2015, 7, 284ra57. [Google Scholar] [CrossRef] [PubMed]
- Klinghoffer, R.A.; Bahrami, S.B.; Hatton, B.A.; Frazier, J.P.; Moreno-Gonzalez, A.; Strand, A.D.; Kerwin, W.S.; Casalini, J.R.; Thirstrup, D.J.; You, S.; et al. A technology platform to assess multiple cancer agents simultaneously within a patient’s tumor. Sci. Transl. Med. 2015, 7, 284ra58. [Google Scholar] [CrossRef] [PubMed]
| MGT/IHC Assay and Provider | Tissue Type, Technique, Facility | Endorsement | Clinical Indications | Prognostic/Predictive Value | Risk Groups/Stratification and Implications | Trials and Validation Studies | Comparative Advantage |
|---|---|---|---|---|---|---|---|
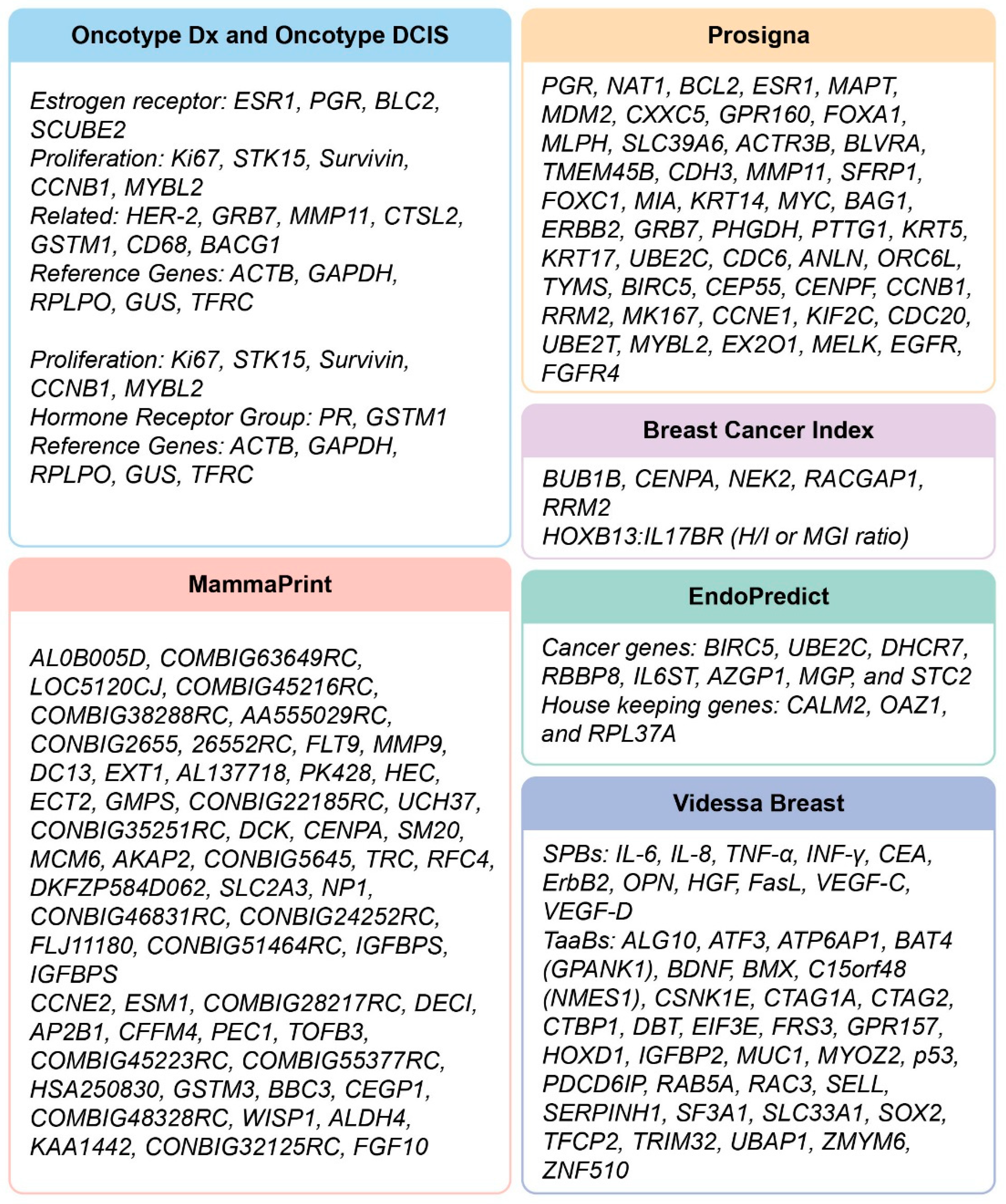
| Oncotype DX Genomic Health [20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41] | FFPE, qRT-PCR, Centralized | NCCN, ASCO, St Gallen | ER+, 0–3 node+, Stage I–II invasive, Treatment decision with tamoxifen or aromatase inhibitors | Prognostic for distant recurrence (5–10 years). Predictive for chemo and radiation sensitive in high recurrence score group. Oncotype DX DCIS is predictive of DCIS recurrence. Benefits women who have had surgery for DCIS, whether additional adjuvant treatment (radiotherapy or tamoxifen) is needed based on their risk score. | Continuous Recurrence Score (formerly triple risk stratification; intermediate score discarded on basis of TAILORx trial results): Low risk (RS 0–25; no additional benefit with chemotherapy), High Risk (RS 26–100; substantial chemotherapy benefit). Risk score for ipsilateral recurrence (invasive or DCIS); Low risk < 39, Intermediate risk 39–54, High risk ≥ 55. | TRANS ATAC, NASBP B 14/B 20, RxPONDER, TAILORx, ECOG-ACRIN, Ontario study | Considered the gold standard in MGTs with high amplification efficiency, precision and linearity. |
| MammaPrint Agendia [42,43,44,45,46,47,48,49,50,51,52] | Fresh/frozen or FFPE, Microarray, Centralized | FDA, St Gallen | Stage I–II, 0–3 node+, ER+ | Prognostic for short-term distant recurrence (0–5 years). Predictive for chemoresponse in high risk group, ER+ cancer. Strong predictor of 10-year metastasis-free survival. | Binary risk classification (MP low risk or MP high risk) for recurrence without adjuvant chemotherapy. Combined with BluePrint (a molecular subtyping test) stratifies patients into four subgroups: Luminal-type/MP Low Risk; Luminal-type/MP High Risk; HER2-type and Basal-type. | TRANSBIG, MINDACT | In contrast to Oncotype DX, test was devised from patients with no hormonal (tamoxifen) or chemo-therapy and thus its robust prognostic ability. The test is endorsed for the clinical high risk group (OncotypeDx is endorsed for clinical low risk group). |
| Prosigna (PAM 50) NanoString Technologies [53,54,55,56,57,58,59,60,61] | FFPE, nCounter, Decentralized; kit compatible with other pathology labs | FDA, NCCN, ASCO, St Gallen | Stage I–III, HR+ | Prognostic for 10 year recurrence in stage I–III. Prognostic and predictive for adjuvant tamoxifen. | Continuous Rate of Recurrence (ROR) score: Low risk (0–40), Intermediate risk (41–60), High risk (>61). | Trans ATAC, ABCSG8, RxPONDER | Its Prediction Analysis of Microarrays (PAM) is an almost fully automated platform technology. RNA is extracted and hybridized (by hand) from FFPE tissue in much smaller quantity than other MGTs. |
| EndoPredict Myriad Genetics [62,63,64,65] | FFPE, RT-PCR, Decentralized | ASCO, St Gallen,NCCN | Early stage, ER+,Her2− | Prognostic for early (0–5 years) and late (5–15 years) distant recurrence. Predictive for benefit from both adjuvant chemotherapy as well as which patients can safely forgo extended endocrine therapy beyond five years. | The multi-gene EP test (Figure 2) and clinical factors (nodal status and tumor size) are combined into an EPClin score stratifying patients into low- or high-risk groups: EP low-risk (<5), EP high-risk (≥5); EPclin low-risk (<3.3), EPclin high-risk (≥3.3). | GEICAM 9906, ABCSG6 and ABCSG8 | EndoPredict is a second-generation, multigene prognostic test. |
| Breast Cancer Index Biotheranostics [66,67,68,69,70] | FPET, Real time RT-PCR, Centralized | ASCO, St Gallen | Stage I–III, HR+, Her2−, Node– | Predictive for adjuvant aromatase inhibitor. Predictive for hormonal therapy for 5 additional years for total of 10 years. Prognostic for late distant recurrence (post-five years). | 0–10 year recurrence risk score is continuous: Low risk BCI < 5.0825, Intermediate risk BCI ≥ 5.0825 to 6.5025 and High risk BCI > 6.5025. Bimodal score informs late distant recurrence: Low risk BCI <5.0825, and High risk BCI ≥ 5.0825. BCI index for predictive utility (to direct adjuvant aromatase inhibitor treatment) is determined with the H/I ratio (Figure 2) and is just a High and Low qualification. | Trans ATAC,Stockholm trial | Outperformed both OncotypeDx and Mammostrat in its 5–10 years’ prognostic ability. |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Saini, G.; Mittal, K.; Rida, P.; Janssen, E.A.M.; Gogineni, K.; Aneja, R. Panoptic View of Prognostic Models for Personalized Breast Cancer Management. Cancers 2019, 11, 1325. https://doi.org/10.3390/cancers11091325
Saini G, Mittal K, Rida P, Janssen EAM, Gogineni K, Aneja R. Panoptic View of Prognostic Models for Personalized Breast Cancer Management. Cancers. 2019; 11(9):1325. https://doi.org/10.3390/cancers11091325
Chicago/Turabian StyleSaini, Geetanjali, Karuna Mittal, Padmashree Rida, Emiel A. M. Janssen, Keerthi Gogineni, and Ritu Aneja. 2019. "Panoptic View of Prognostic Models for Personalized Breast Cancer Management" Cancers 11, no. 9: 1325. https://doi.org/10.3390/cancers11091325
APA StyleSaini, G., Mittal, K., Rida, P., Janssen, E. A. M., Gogineni, K., & Aneja, R. (2019). Panoptic View of Prognostic Models for Personalized Breast Cancer Management. Cancers, 11(9), 1325. https://doi.org/10.3390/cancers11091325






