A Radiogenomic Approach for Decoding Molecular Mechanisms Underlying Tumor Progression in Prostate Cancer
Abstract
1. Introduction
2. Results and Discussion
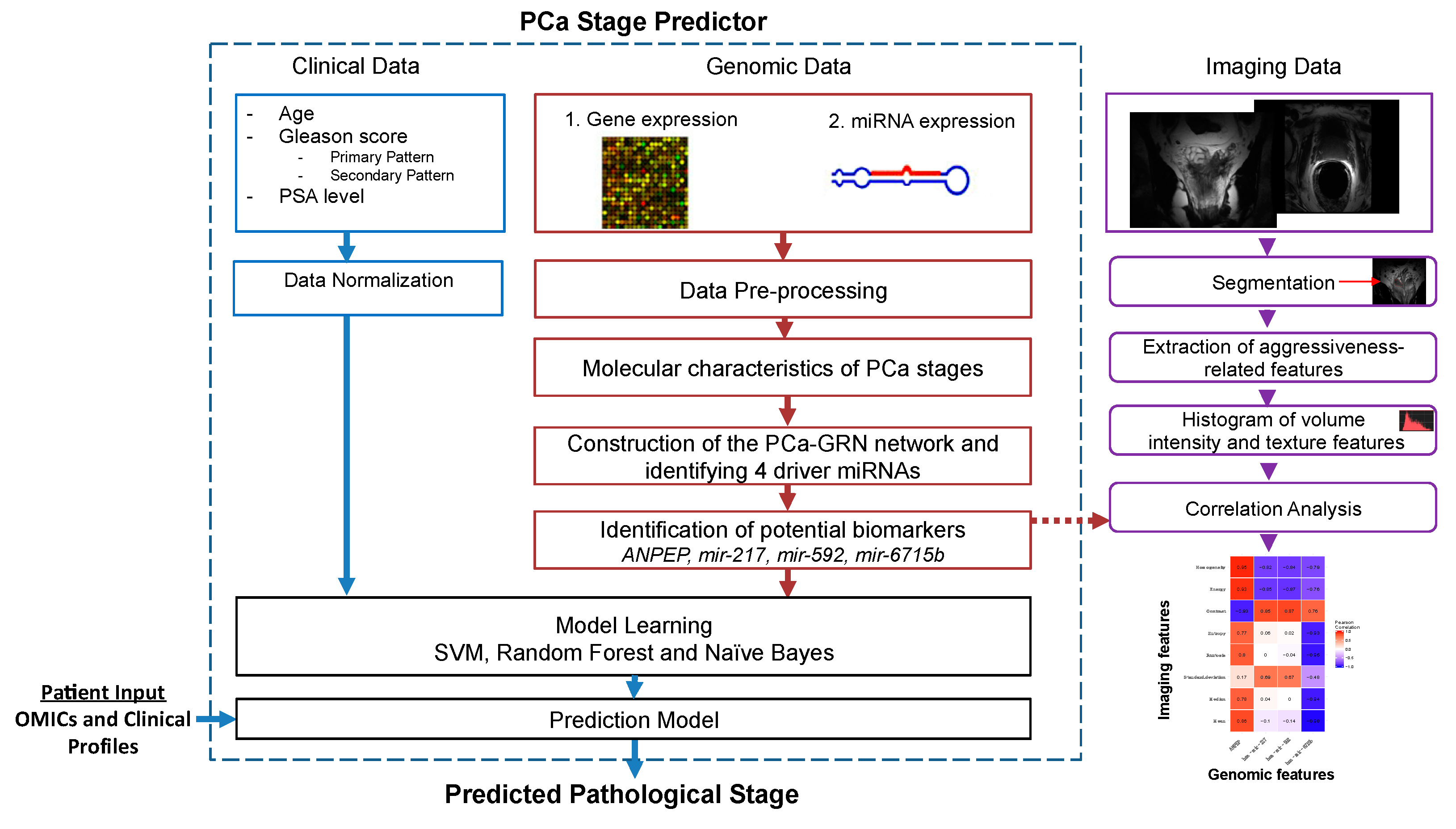
2.1. Description of the Radiogenomic Approach
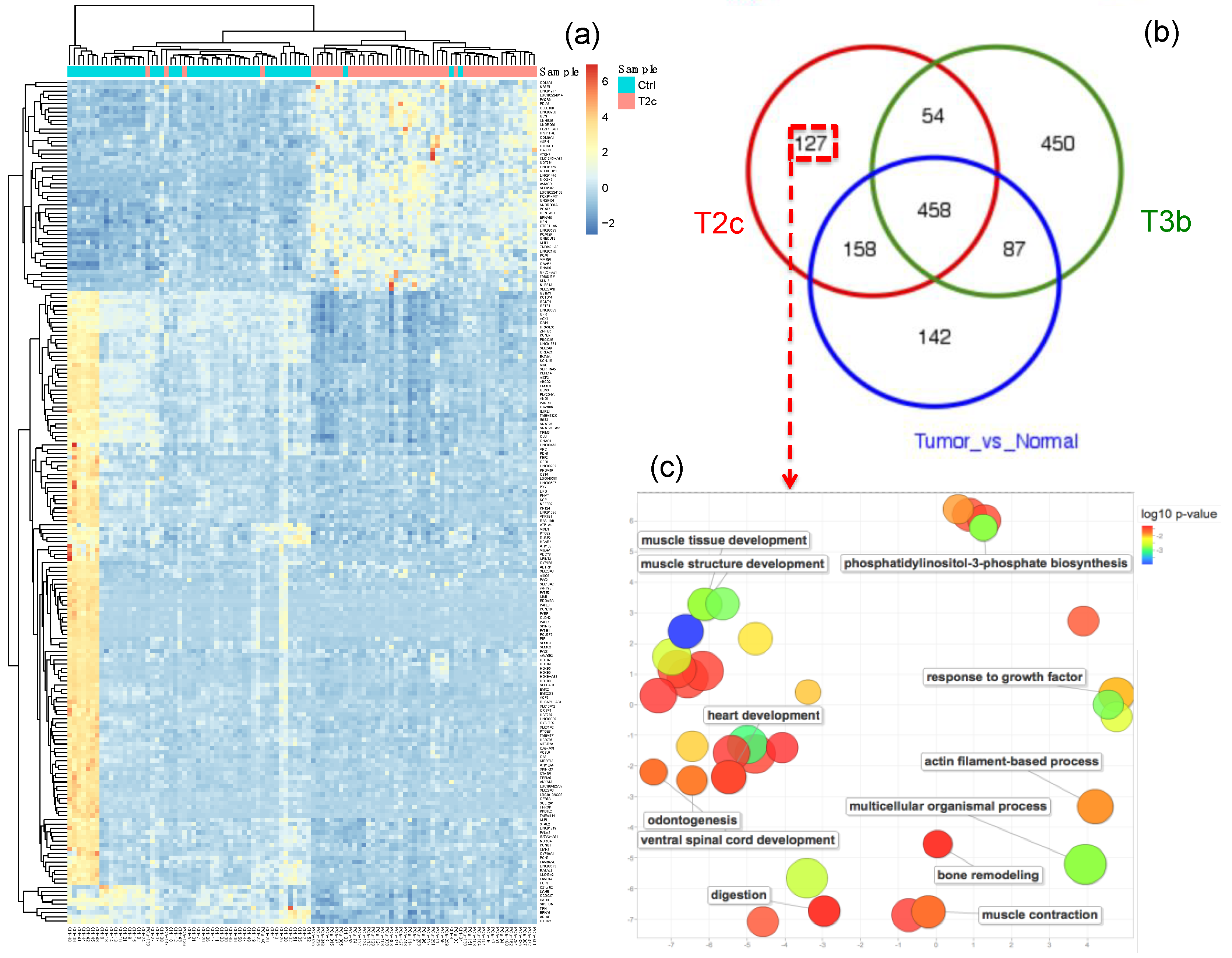
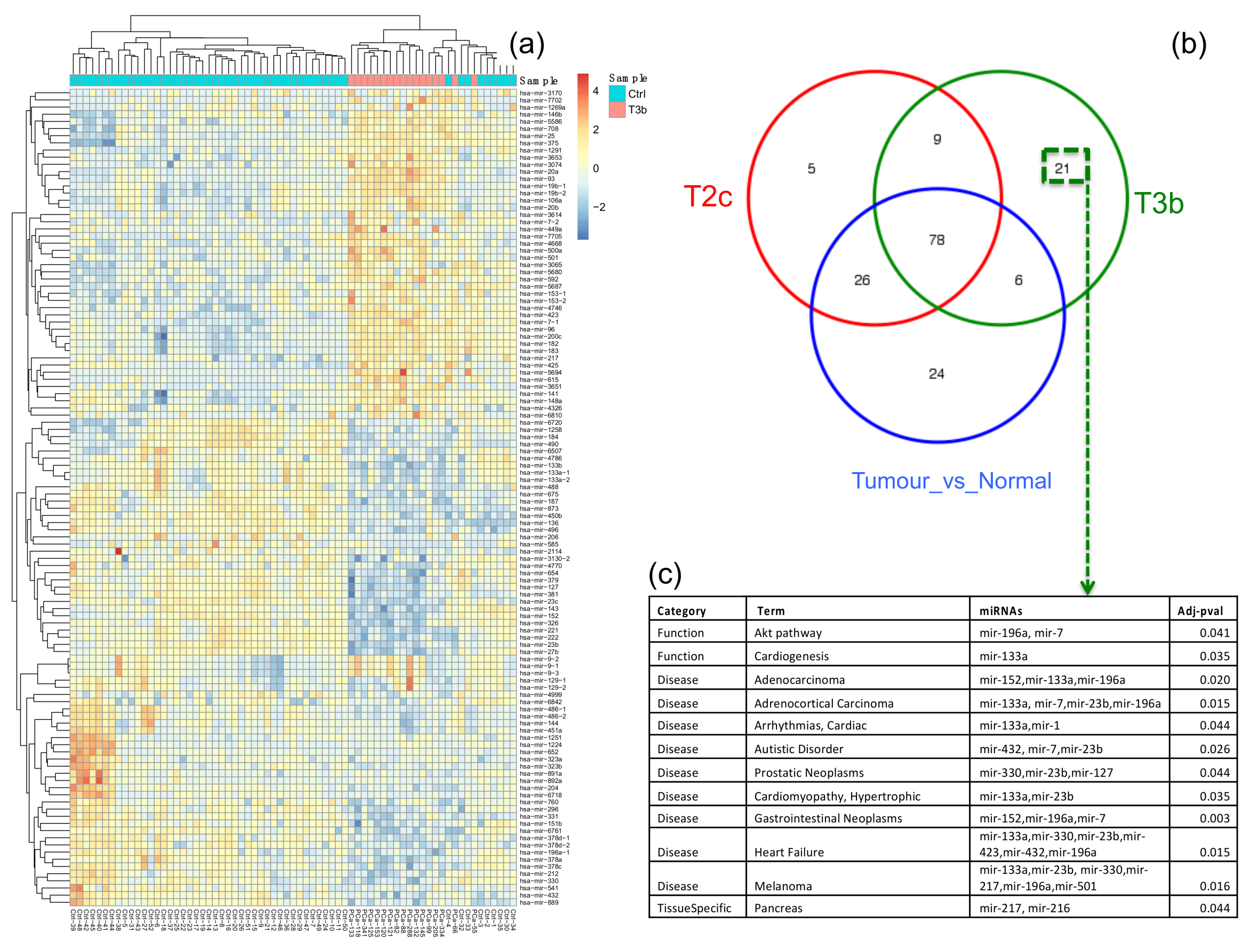
2.2. Functional Characteristics of T2c and T3b Stages
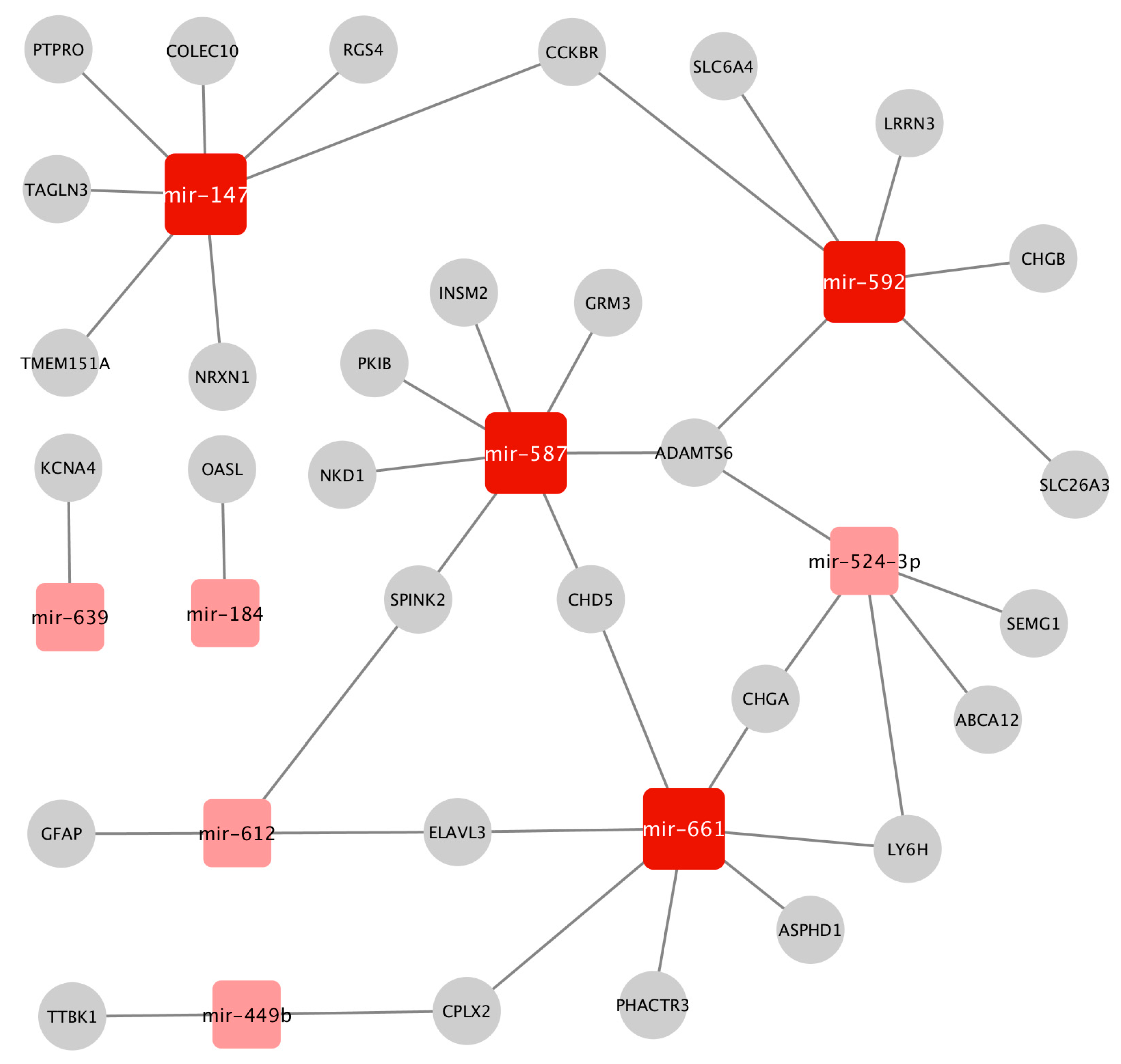
2.3. Construction of the Prostate Cancer (PCa-GRN) Network
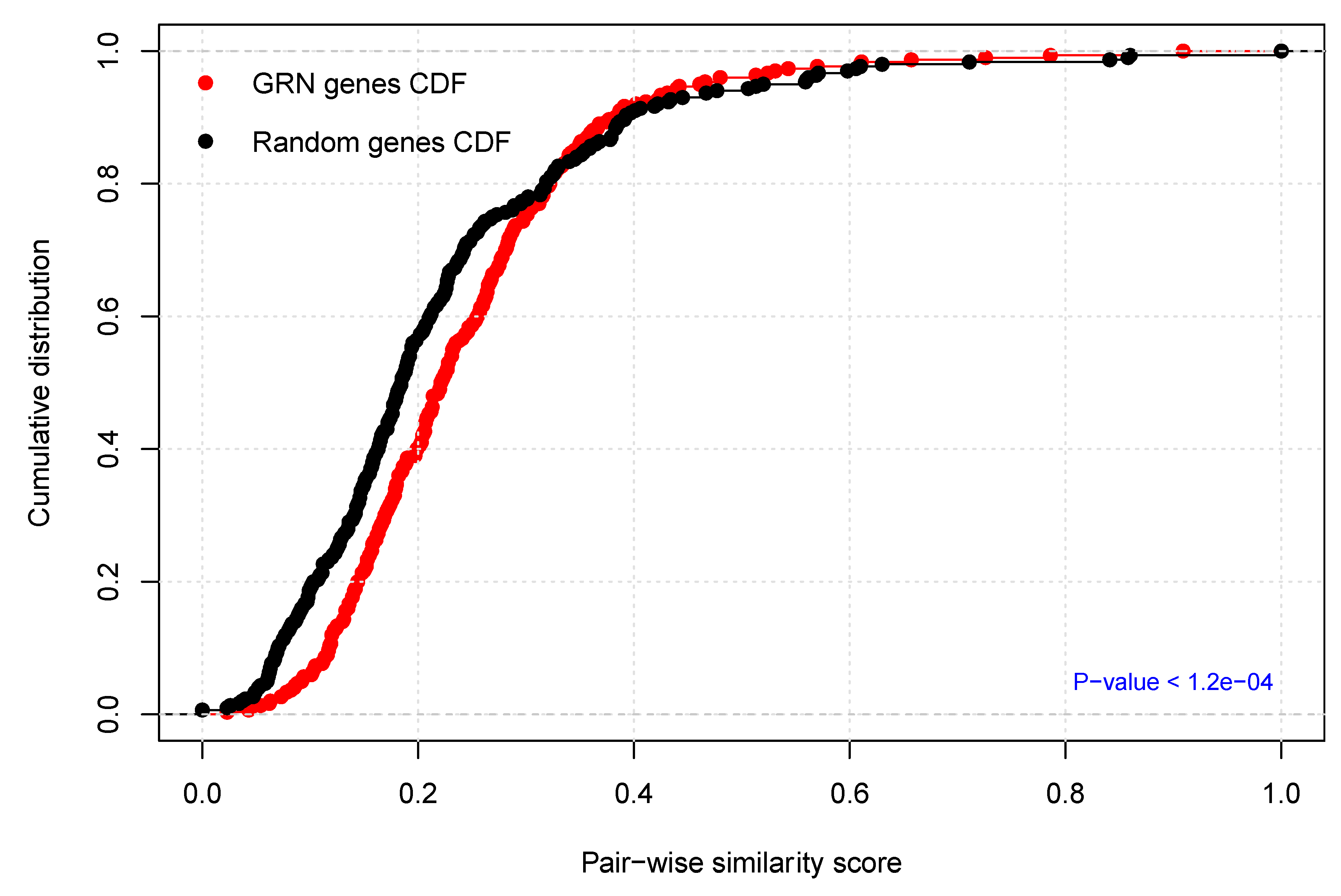
2.4. Functional Homogeneity within the Prostate Cancer (PCa-GRN) Network
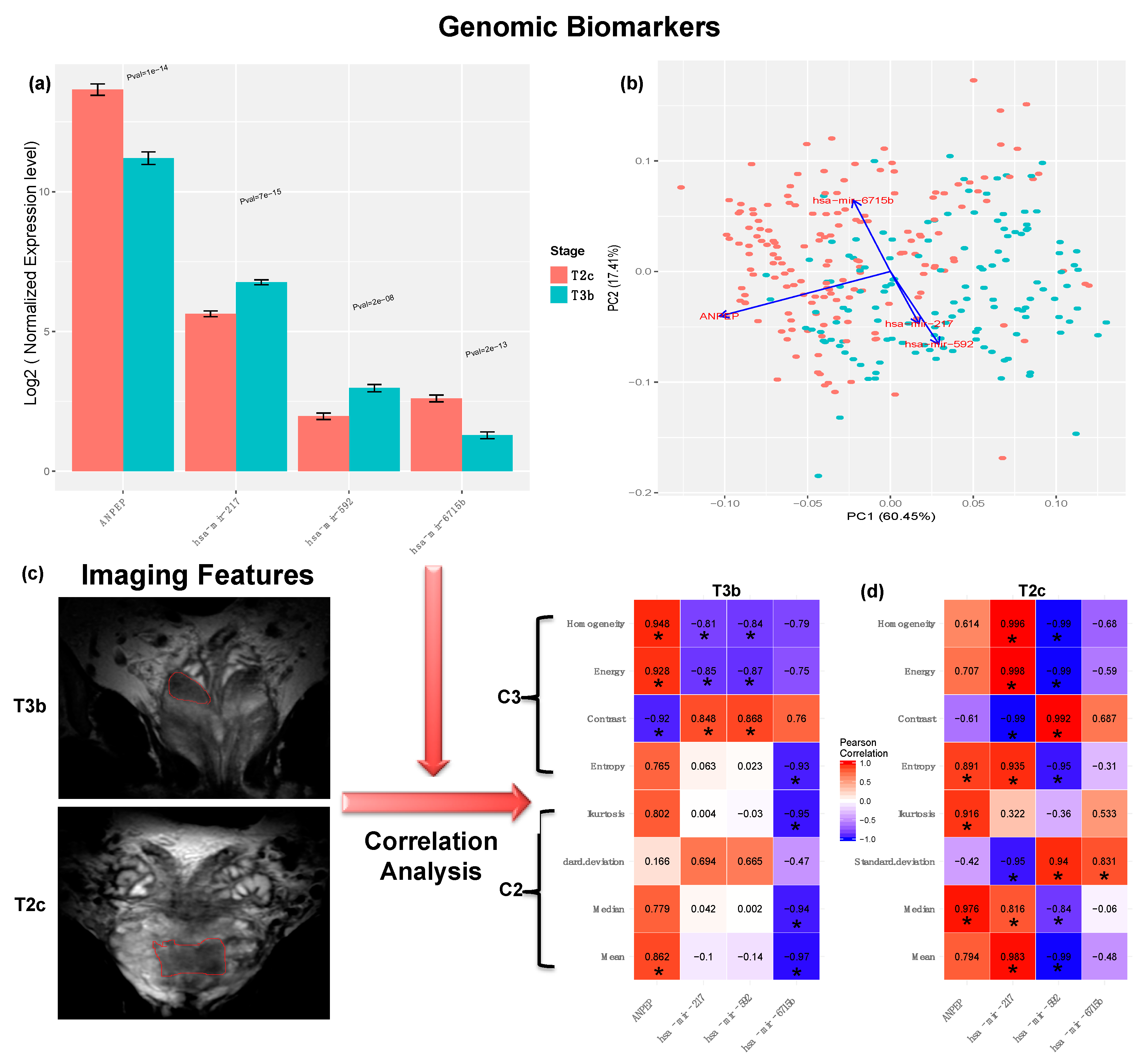
2.5. Biomarker Identification
2.6. Correlation Analysis with Aggressiveness-Related Imaging Features
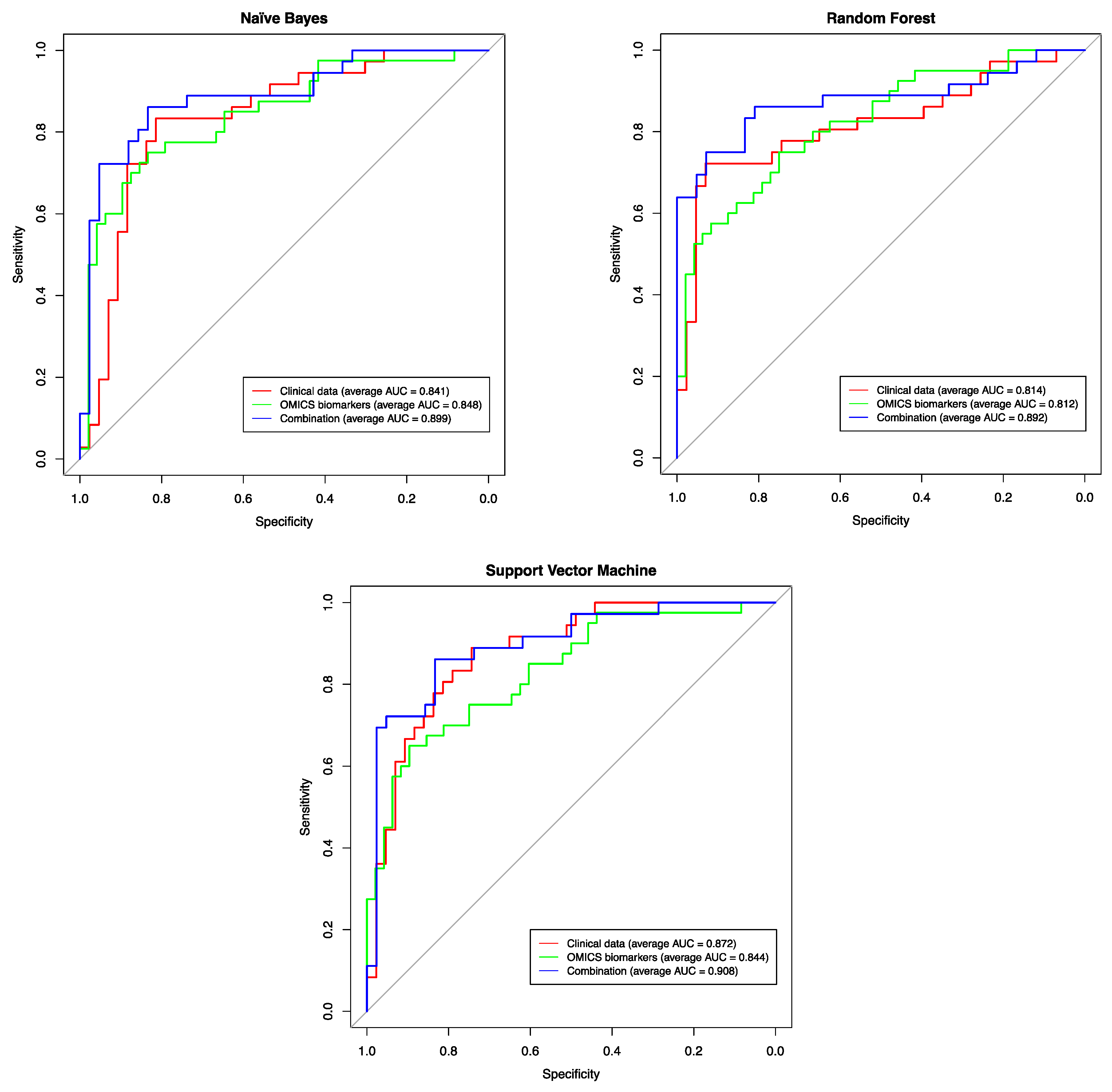
2.7. Assessing the Predictive Power of the Identified Biomarkers
3. Materials and Methods
3.1. Datasets
3.2. Data Pre-Processing
3.2.1. Genomic Data
3.2.2. Clinical Data
3.2.3. Imaging Data
3.3. Differential Expression and Association Analysis
3.4. Construction of Prostate-Specific GRN Network (PRAD-GRN)
3.5. Assessment of the Constructed Prostate Cancer (PCa-GRN) Network
3.5.1. Functional Homogeneity within the PCa-GRN Genes (Semantic Validation)
3.5.2. Enrichment Analysis of Genes and miRNAs
3.6. Correlation Analysis
3.7. Prediction Models
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Abeshouse, A.; Ahn, J.; Akbani, R.; Ally, A.; Amin, S.; Andry, C.D.; Annala, M.; Aprikian, A.; Armenia, J.; Arora, A. The molecular taxonomy of primary prostate cancer. Cell 2015, 163, 1011–1025. [Google Scholar] [CrossRef]
- Ferlay, J.; Soerjomataram, I.; Dikshit, R.; Eser, S.; Mathers, C.; Rebelo, M.; Parkin, D.M.; Forman, D.; Bray, F. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer 2015, 136, E359–E386. [Google Scholar] [CrossRef]
- Stoyanova, R.; Takhar, M.; Tschudi, Y.; Ford, J.C.; Solórzano, G.; Erho, N.; Balagurunathan, Y.; Punnen, S.; Davicioni, E.; Gillies, R.J. Prostate cancer radiomics and the promise of radiogenomics. Transl. Cancer Res. 2016, 5, 432. [Google Scholar] [CrossRef]
- Walz, J.; Joniau, S.; Chun, F.K.; Isbarn, H.; Jeldres, C.; Yossepowitch, O.; Chao-Yu, H.; Klein, E.A.; Scardino, P.T.; Reuther, A.; et al. Pathological results and rates of treatment failure in high-risk prostate cancer patients after radical prostatectomy. BJU Int. 2011, 107, 765–770. [Google Scholar] [CrossRef]
- Incoronato, M.; Aiello, M.; Infante, T.; Cavaliere, C.; Grimaldi, A.M.; Mirabelli, P.; Monti, S.; Salvatore, M. Radiogenomic analysis of oncological data: A technical survey. Int. J. Mol. Sci. 2017, 18, 805. [Google Scholar] [CrossRef]
- Barbieri, C.E.; Demichelis, F.; Rubin, M.A. Molecular genetics of prostate cancer: Emerging appreciation of genetic complexity. Histopathology 2012, 60, 187–198. [Google Scholar] [CrossRef]
- Jansen, F.H.; van Schaik, R.H.; Kurstjens, J.; Horninger, W.; Klocker, H.; Bektic, J.; Wildhagen, M.F.; Roobol, M.J.; Bangma, C.H.; Bartsch, G. Prostate-specific antigen (PSA) isoform p2PSA in combination with total PSA and free PSA improves diagnostic accuracy in prostate cancer detection. Eur. Urol. 2010, 57, 921–927. [Google Scholar] [CrossRef]
- Cookson, M.S.; Fleshner, N.E.; Soloway, S.M.; Fair, W.R. Correlation between Gleason score of needle biopsy and radical prostatectomy specimen: Accuracy and clinical implications. J. Urol. 1997, 157, 559–562. [Google Scholar] [CrossRef]
- Epstein, J.I.; Walsh, P.C.; Carmichael, M.; Brendler, C.B. Pathologic and clinical findings to predict tumor extent of nonpalpable (stage t1 c) prostate cancer. JAMA 1994, 271, 368–374. [Google Scholar] [CrossRef]
- Epstein, J.I.; Pizov, G.; Walsh, P.C. Correlation of pathologic findings with progression after radical retropubic prostatectomy. Cancer 1993, 71, 3582–3593. [Google Scholar] [CrossRef]
- Tosoian, J.J.; Chappidi, M.; Feng, Z.; Humphreys, E.B.; Han, M.; Pavlovich, C.P.; Epstein, J.I.; Partin, A.W.; Trock, B.J. Prediction of pathological stage based on clinical stage, serum prostate-specific antigen, and biopsy Gleason score: Partin Tables in the contemporary era. BJU Int. 2017, 119, 676–683. [Google Scholar] [CrossRef]
- Gospodarowicz, M.K.; Brierley, J.D.; Wittekind, C. TNM Classification of Malignant Tumours; John Wiley & Sons: Hoboken, NJ, USA, 2017. [Google Scholar]
- Mohler, J.L.; Armstrong, A.J.; Bahnson, R.R.; D’Amico, A.V.; Davis, B.J.; Eastham, J.A.; Enke, C.A.; Farrington, T.A.; Higano, C.S.; Horwitz, E.M. Prostate cancer, version 1.2016. J. Natl. Compr. Cancer Netw. 2016, 14, 19–30. [Google Scholar] [CrossRef]
- Dotan, Z.A.; Ramon, J. Nomograms as a tool in predicting prostate cancer prognosis. Eur. Urol. Suppl. 2009, 8, 721–724. [Google Scholar] [CrossRef]
- MSK. Memorial Sloan Kettering Cancer Center. Available online: https://www.mskcc.org/nomograms/prostate (accessed on 15 August 2019).
- Harada, T.; Abe, T.; Kato, F.; Matsumoto, R.; Fujita, H.; Murai, S.; Miyajima, N.; Tsuchiya, K.; Maruyama, S.; Kudo, K.; et al. Five-point Likert scaling on MRI predicts clinically significant prostate carcinoma. BMC Urol. 2015, 15, 91. [Google Scholar] [CrossRef]
- Briganti, A.; Karakiewicz, P.I.; Joniau, S.; Van Poppel, H. The motion: Nomograms should become a routine tool in determining prostate cancer prognosis. Eur. Urol. 2009, 55, 743–747. [Google Scholar] [CrossRef]
- Chun, F.K.H.; Karakiewicz, P.I.; Briganti, A.; Walz, J.; Kattan, M.W.; Huland, H.; Graefen, M. A critical appraisal of logistic regression-based nomograms, artificial neural networks, classification and regression-tree models, look-up tables and risk-group stratification models for prostate cancer. BJU Int. 2007, 99, 794–800. [Google Scholar] [CrossRef]
- Cosma, G.; Acampora, G.; Brown, D.; Rees, R.C.; Khan, M.; Pockley, A.G. Prediction of pathological stage in patients with prostate cancer: A neuro-fuzzy model. PLoS ONE 2016, 11, e0155856. [Google Scholar] [CrossRef]
- Hariri, A.R.; Weinberger, D.R. Imaging genomics. Br. Med. Bull. 2003, 65, 259–270. [Google Scholar] [CrossRef]
- Bai, H.X.; Lee, A.M.; Yang, L.; Zhang, P.; Davatzikos, C.; Maris, J.M.; Diskin, S.J. Imaging genomics in cancer research: Limitations and promises. Br. J. Radiol. 2016, 89, 20151030. [Google Scholar] [CrossRef]
- Bertoli, G.; Cava, C.; Castiglioni, I. MicroRNAs as biomarkers for diagnosis, prognosis and theranostics in prostate cancer. Int. J. Mol. Sci. 2016, 17, 421. [Google Scholar] [CrossRef]
- Bibikova, M.; Chudin, E.; Arsanjani, A.; Zhou, L.; Garcia, E.W.; Modder, J.; Kostelec, M.; Barker, D.; Downs, T.; Fan, J.B. Expression signatures that correlated with Gleason score and relapse in prostate cancer. Genomics 2007, 89, 666–672. [Google Scholar] [CrossRef]
- Agell, L.; Hernández, S.; Nonell, L.; Lorenzo, M.; Puigdecanet, E.; de Muga, S.; Juanpere, N.; Bermudo, R.; Fernández, P.L.; Lorente, J.A. A 12-gene expression signature is associated with aggressive histological in prostate cancer: SEC14L1 and TCEB1 genes are potential markers of progression. Am. J. Pathol. 2012, 181, 1585–1594. [Google Scholar] [CrossRef]
- Bismar, T.A.; Demichelis, F.; Riva, A.; Kim, R.; Varambally, S.; He, L.; Kutok, J.; Aster, J.C.; Tang, J.; Kuefer, R. Defining aggressive prostate cancer using a 12-gene model. Neoplasia 2006, 8, 59–68. [Google Scholar] [CrossRef]
- Cheville, J.C.; Karnes, R.J.; Therneau, T.M.; Kosari, F.; Munz, J.M.; Tillmans, L.; Basal, E.; Rangel, L.J.; Bergstralh, E.; Kovtun, I.V. Gene panel model predictive of outcome in men at high-risk of systemic progression and death from prostate cancer after radical retropubic prostatectomy. J. Clin. Oncol. 2008, 26, 3930. [Google Scholar] [CrossRef]
- Cuzick, J.; Swanson, G.P.; Fisher, G.; Brothman, A.R.; Berney, D.M.; Reid, J.E.; Mesher, D.; Speights, V.; Stankiewicz, E.; Foster, C.S. Prognostic value of an RNA expression signature derived from cell cycle proliferation genes in patients with prostate cancer: A retrospective study. Lancet Oncol. 2011, 12, 245–255. [Google Scholar] [CrossRef]
- Larkin, S.; Holmes, S.; Cree, I.; Walker, T.; Basketter, V.; Bickers, B.; Harris, S.; Garbis, S.D.; Townsend, P.; Aukim-Hastie, C. Identification of markers of prostate cancer progression using candidate gene expression. Br. J. Cancer 2012, 106, 157. [Google Scholar] [CrossRef]
- Singh, D.; Febbo, P.G.; Ross, K.; Jackson, D.G.; Manola, J.; Ladd, C.; Tamayo, P.; Renshaw, A.A.; D’Amico, A.V.; Richie, J.P. Gene expression correlates of clinical prostate cancer behavior. Cancer Cell 2002, 1, 203–209. [Google Scholar] [CrossRef]
- Erho, N.; Crisan, A.; Vergara, I.A.; Mitra, A.P.; Ghadessi, M.; Buerki, C.; Bergstralh, E.J.; Kollmeyer, T.; Fink, S.; Haddad, Z. Discovery and validation of a prostate cancer genomic classifier that predicts early metastasis following radical prostatectomy. PLoS ONE 2013, 8, e66855. [Google Scholar] [CrossRef]
- Al-Kafaji, G.; Said, H.M.; Alam, M.A.; Al Naieb, Z.T. Blood-based microRNAs as diagnostic biomarkers to discriminate localized prostate cancer from benign prostatic hyperplasia and allow cancer-risk stratification. Oncol. Lett. 2018, 16, 1357–1365. [Google Scholar] [CrossRef]
- Stoyanova, R.; Pollack, A.; Takhar, M.; Lynne, C.; Parra, N.; Lam, L.L.; Alshalalfa, M.; Buerki, C.; Castillo, R.; Jorda, M.; et al. Association of multiparametric MRI quantitative imaging features with prostate cancer gene expression in MRI-targeted prostate biopsies. Oncotarget 2016, 7, 53362–53376. [Google Scholar] [CrossRef]
- Corn, P.G.; Wang, F.; McKeehen, W.; Navone, N. Targeting fibroblast growth factor pathways in prostate cancer. Clin. Cancer Res. 2013, 19, 5856–5866. [Google Scholar] [CrossRef]
- Chandran, U.R.; Ma, C.; Dhir, R.; Bisceglia, M.; Lyons-Weiler, M.; Liang, W.; Michalopoulos, G.; Becich, M.; Monzon, F.A. Gene expression profiles of prostate cancer reveal involvement of multiple molecular pathways in the metastatic process. BMC Cancer 2007, 7, 64. [Google Scholar] [CrossRef]
- Mazaris, E.; Tsiotras, A. Molecular pathways in prostate cancer. Nephro Urol. Mon. 2013, 5, 792. [Google Scholar] [CrossRef]
- Vivanco, I.; Sawyers, C.L. The phosphatidylinositol 3-kinase–AKT pathway in human cancer. Nat. Rev. Cancer 2002, 2, 489. [Google Scholar] [CrossRef]
- Supek, F.; Bošnjak, M.; Škunca, N.; Šmuc, T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS ONE 2011, 6, e21800. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, H.E.; Liu, Z. MicroRNA-147 suppresses proliferation, invasion and migration through the AKT/mTOR signaling pathway in breast cancer. Oncol. Lett. 2016, 11, 405–410. [Google Scholar] [CrossRef]
- Zhang, Y.; Talmon, G.; Wang, J. MicroRNA-587 antagonizes 5-FU-induced apoptosis and confers drug resistance by regulating PPP2R1B expression in colorectal cancer. Cell Death Dis. 2016, 7, e2525. [Google Scholar] [CrossRef]
- Liu, A.Y.; Roudier, M.P.; True, L.D. Heterogeneity in primary and metastatic prostate cancer as defined by cell surface CD profile. Am. J. Pathol. 2004, 165, 1543–1556. [Google Scholar] [CrossRef]
- Dall’Era, M.A.; True, L.D.; Siegel, A.F.; Porter, M.P.; Sherertz, T.M.; Liu, A.Y. Differential expression of CD10 in prostate cancer and its clinical implication. BMC Urol. 2007, 7, 3. [Google Scholar] [CrossRef]
- Sørensen, K.D.; Abildgaard, M.O.; Haldrup, C.; Ulhøi, B.P.; Kristensen, H.; Strand, S.; Parker, C.; Høyer, S.; Borre, M.; Ørntoft, T.F. Prognostic significance of aberrantly silenced ANPEP expression in prostate cancer. Br. J. Cancer 2013, 108, 420. [Google Scholar] [CrossRef]
- Lin, H.M.; Nikolic, I.; Yang, J.; Castillo, L.; Deng, N.; Chan, C.L.; Yeung, N.K.; Dodson, E.; Elsworth, B.; Spielman, C.; et al. MicroRNAs as potential therapeutics to enhance chemosensitivity in advanced prostate cancer. Sci. Rep. 2018, 8, 7820. [Google Scholar] [CrossRef]
- Lv, Z.; Rao, P.; Li, W. MiR-592 represses FOXO3 expression and promotes the proliferation of prostate cancer cells. Int. J. Clin. Exp. Med. 2015, 8, 15246–15253. [Google Scholar]
- Giulietti, M.; Occhipinti, G.; Principato, G.; Piva, F. Identification of candidate miRNA biomarkers for pancreatic ductal adenocarcinoma by weighted gene co-expression network analysis. Cell. Oncol. (Dordr.) 2017, 40, 181–192. [Google Scholar] [CrossRef]
- Rosset, A.; Spadola, L.; Ratib, O. OsiriX: An open-source software for navigating in multidimensional DICOM images. J. Digit. Imaging 2004, 17, 205–216. [Google Scholar] [CrossRef]
- Wibmer, A.; Hricak, H.; Gondo, T.; Matsumoto, K.; Veeraraghavan, H.; Fehr, D.; Zheng, J.; Goldman, D.; Moskowitz, C.; Fine, S.W.; et al. Haralick texture analysis of prostate MRI: Utility for differentiating non-cancerous prostate from prostate cancer and differentiating prostate cancers with different Gleason scores. Eur. Radiol. 2015, 25, 2840–2850. [Google Scholar] [CrossRef]
- Shen, J.; Hruby, G.W.; McKiernan, J.M.; Gurvich, I.; Lipsky, M.J.; Benson, M.C.; Santella, R.M. Dysregulation of circulating microRNAs and prediction of aggressive prostate cancer. Prostate 2012, 72, 1469–1477. [Google Scholar] [CrossRef]
- Tsao, C.W.; Liu, C.Y.; Cha, T.L.; Wu, S.T.; Sun, G.H.; Yu, D.S.; Chen, H.I.; Chang, S.Y.; Chen, S.C.; Hsu, C.Y. Artificial neural network for predicting pathological stage of clinically localized prostate cancer in a Taiwanese population. J. Chin. Med. Assoc. 2014, 77, 513–518. [Google Scholar] [CrossRef][Green Version]
- Clark, K.; Vendt, B.; Smith, K.; Freymann, J.; Kirby, J.; Koppel, P.; Moore, S.; Phillips, S.; Maffitt, D.; Pringle, M. The Cancer Imaging Archive (TCIA): Maintaining and operating a public information repository. J. Digit. Imaging 2013, 26, 1045–1057. [Google Scholar] [CrossRef]
- The Cancer Genome Atlas Prostate Adenocarcinoma (TCGA-PRAD) Data Collection. Available online: https://portal.gdc.cancer.gov/projects/TCGA-PRAD (accessed on 11 January 2019).
- Gentleman, R.C.; Carey, V.J.; Bates, D.M.; Bolstad, B.; Dettling, M.; Dudoit, S.; Ellis, B.; Gautier, L.; Ge, Y.; Gentry, J. Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol. 2004, 5, R80. [Google Scholar] [CrossRef]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Genome Biol. 2010, 11, R106. [Google Scholar] [CrossRef]
- Ihaka, R.; Gentleman, R. R: A language for data analysis and graphics. J. Comput. Graph. Stat. 1996, 5, 299–314. [Google Scholar]
- Gallón, S.; Loubes, J.M.; Maza, E. Statistical properties of the quantile normalization method for density curve alignment. Math. Biosci. 2013, 242, 129–142. [Google Scholar] [CrossRef][Green Version]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671. [Google Scholar] [CrossRef]
- Hochberg, Y.; Benjamini, Y. More powerful procedures for multiple significance testing. Stat. Med. 1990, 9, 811–818. [Google Scholar] [CrossRef]
- Hamed, M.; Spaniol, C.; Nazarieh, M.; Helms, V. TFmiR: A web server for constructing and analyzing disease-specific transcription factor and miRNA co-regulatory networks. Nucleic Acids Res. 2015, 43, W283–W288. [Google Scholar] [CrossRef]
- Li, Y.; Qiu, C.; Tu, J.; Geng, B.; Yang, J.; Jiang, T.; Cui, Q. HMDD v2. 0: A database for experimentally supported human microRNA and disease associations. Nucleic Acids Res. 2013, 42, D1070–D1074. [Google Scholar] [CrossRef]
- Piñero, J.; Queralt-Rosinach, N.; Bravo, À.; Deu-Pons, J.; Bauer-Mehren, A.; Baron, M.; Sanz, F.; Furlong, L.I. DisGeNET: A discovery platform for the dynamical exploration of human diseases and their genes. Database 2015, 2015, bav028. [Google Scholar] [CrossRef]
- Piñero, J.; Bravo, À.; Queralt-Rosinach, N.; Gutiérrez-Sacristán, A.; Deu-Pons, J.; Centeno, E.; García-García, J.; Sanz, F.; Furlong, L.I. DisGeNET: A comprehensive platform integrating information on human disease-associated genes and variants. Nucleic Acids Res. 2016, gkw943. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Yu, G.; Li, F.; Qin, Y.; Bo, X.; Wu, Y.; Wang, S. GOSemSim: An R package for measuring semantic similarity among GO terms and gene products. Bioinformatics 2010, 26, 976–978. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2008, 4, 44. [Google Scholar] [CrossRef]
- Lu, M.; Shi, B.; Wang, J.; Cao, Q.; Cui, Q. TAM: A method for enrichment and depletion analysis of a microRNA category in a list of microRNAs. BMC Bioinform. 2010, 11, 419. [Google Scholar] [CrossRef]
- Hamed, M.; Trumm, J.; Spaniol, C.; Sethi, R.; Irhimeh, M.R.; Fuellen, G.; Paulsen, M.; Helms, V. Linking Hematopoietic Differentiation to Co-Expressed Sets of Pluripotency-Associated and Imprinted Genes and to Regulatory microRNA-Transcription Factor Motifs. PLoS ONE 2017, 12, e0166852. [Google Scholar] [CrossRef]
- Kuhn, M. Caret: Classification and Regression Training. Astrophys. Source Code Library. 2015. Available online: https://rdrr.io/rforge/caret/ (accessed on 23 March 2019).
- Robin, X.; Turck, N.; Hainard, A.; Tiberti, N.; Lisacek, F.; Sanchez, J.C.; Müller, M. pROC: An open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinform. 2011, 12, 77. [Google Scholar] [CrossRef]
| Pathological STAGE | Count | Age Median (Min–Max) | PSA-Value Median (Min–Max) | Gleason Score | Count | Clinical Stage | Count | Biochemical Recurrence | Count | Ethnicity | Count |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Primary + Secondary | Stage | ||||||||||
| T2c | 164 | 59 (41–77) | 0.1 (0.01–14.69) | 3 + 3 | 25 | T1b | 0 | Yes | 6 | Black or African American | 3 |
| 3 + 4 | 84 | T1c | 84 | No | 131 | White | 58 | ||||
| 4 + 3 | 31 | T2 | 4 | Not available | 27 | Not available | 103 | ||||
| ≥8 | 24 | T2a | 16 | ||||||||
| T2b | 10 | ||||||||||
| T2c | 23 | ||||||||||
| T3a | 2 | ||||||||||
| T3b | 0 | ||||||||||
| T4 | 0 | ||||||||||
| Not available | 25 | ||||||||||
| T3b | 134 | 62 (46–78) | 0.1 (0.01–37.36) | 3 + 3 | 1 | T1b | 2 | Yes | 29 | White | 27 |
| 3 + 4 | 8 | T1c | 23 | No | 94 | Not available | 107 | ||||
| 4 + 3 | 21 | T2 | 6 | Not available | 11 | ||||||
| ≥8 | 104 | T2a | 14 | ||||||||
| T2b | 12 | ||||||||||
| T2c | 16 | ||||||||||
| T3a | 14 | ||||||||||
| T3b | 16 | ||||||||||
| T4 | 1 | ||||||||||
| Not available | 30 |
| Enriched Pathways Which Characterize the T2c Stage but Not the T3b Stage | ||
| Pathways | Involved Genes | Adj-Pval |
| hsa05218: Melanoma | FGF6, FGF8, FGF23, FGF3 | 3.00 × 10−3 |
| hsa04010: MAPK signaling pathway | FGF6, DUSP4, FGF8, FGF23, FGF3, PLA2G4D | 3.00 × 10−3 |
| hsa04810: Regulation of actin cytoskeleton | FGF6, FGF8, FGF23, MYLPF, FGF3 | 9.00 × 10−3 |
| hsa04151: PI3K-Akt signaling pathway | FGF6, FGF8, COL6A5, FGF23, FGF3, EIF4E1B | 1.00 × 10−2 |
| hsa04014: Ras signaling pathway | FGF6, FGF8, FGF23, FGF3, PLA2G4D | 1.1 × 10−2 |
| hsa04015: Rap1 signaling pathway | FGF6, FGF8, FGF23, FGF3 | 4.7 × 10−2 |
| Enriched Pathways Which Characterize the T3b Stage But Not the T2c Stage | ||
| Pathways | Involved Genes | Adj-Pval |
| hsa04080: Neuroactive ligand-receptor interaction | GABRD, MCHR1, GABRA2, GABRA3, GABRB2, ADCYAP1R1, GRIA3, NTSR2, GHRHR, HRH3, PRLR, GALR1, HRH2, P2RX2, NPFFR1, CHRNA1, ADRA1D, GABRQ | 2.23 × 10−5 |
| hsa05033: Nicotine addiction | GABRD, GABRA2, GABRB2, GABRA3, GRIA3, GABRQ | 9.70 × 10−4 |
| hsa04972: Pancreatic secretion | KCNMA1, CD38, ATP2B4, PLA2G2A, PLA2G2C, CPA1, ATP1A2, PRKCB | 2.09 × 10−3 |
| hsa05143: African trypanosomiasis | IL6, HBA2, HBB, SELE, PRKCB | 3.58 × 10−3 |
| hsa04727: GABAergic synapse | GABRD, PLCL1, GABRA2, GABRB2, GABRA3, GABRQ, PRKCB | 5.94 × 10−3 |
| hsa04510: Focal adhesion | CAV3, CAV1, RASGRF1, PAK3, RAC3, ACTN2, ITGB3, FLNC, COL4A6, PRKCB, FN1 | 6.53 × 10−3 |
| hsa04723: Retrograde endocannabinoid signaling | GABRD, GABRA2, GABRB2, GABRA3, GRIA3, GABRQ, PRKCB | 1.34 × 10−2 |
| hsa05144: Malaria | IL6, CXCL8, HBA2, HBB, SELE | 1.46 × 10−2 |
| hsa05146: Amoebiasis | GNAL, IL6, CXCL8, ACTN2, COL4A6, PRKCB, FN1 | 1.67 × 10−2 |
| hsa04020: Calcium signaling pathway | GNAL, CD38, ATP2B4, ERBB4, HRH2, PLN, P2RX2, ADRA1D, PRKCB | 2.27 × 10−2 |
| hsa04970: Salivary secretion | KCNMA1, CD38, ATP2B4, ATP1A2, ADRA1D, PRKCB | 2.53 × 10−2 |
| hsa04270: Vascular smooth muscle contraction | KCNMA1, ACTG2, PLA2G2A, PLA2G2C, ADRA1D, KCNMB1, PRKCB | 2.77 × 10−2 |
| hsa05032: Morphine addiction | GABRD, GABRA2, GABRB2, GABRA3, GABRQ, PRKCB | 3.14 × 10−2 |
| hsa05205: Proteoglycans in cancer | CAV3, MIR10B, WNT16, CAV1, ERBB4, ITGB3, FLNC, PRKCB, FN1 | 4.02 × 10−2 |
| hsa05412: Arrhythmogenic right ventricular cardiomyopathy (ARVC) | SGCG, DMD, ACTN2, ITGB3, CTNNA3 | 4.85 × 10−2 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fischer, S.; Tahoun, M.; Klaan, B.; Thierfelder, K.M.; Weber, M.-A.; Krause, B.J.; Hakenberg, O.; Fuellen, G.; Hamed, M. A Radiogenomic Approach for Decoding Molecular Mechanisms Underlying Tumor Progression in Prostate Cancer. Cancers 2019, 11, 1293. https://doi.org/10.3390/cancers11091293
Fischer S, Tahoun M, Klaan B, Thierfelder KM, Weber M-A, Krause BJ, Hakenberg O, Fuellen G, Hamed M. A Radiogenomic Approach for Decoding Molecular Mechanisms Underlying Tumor Progression in Prostate Cancer. Cancers. 2019; 11(9):1293. https://doi.org/10.3390/cancers11091293
Chicago/Turabian StyleFischer, Sarah, Mohamed Tahoun, Bastian Klaan, Kolja M. Thierfelder, Marc-André Weber, Bernd J. Krause, Oliver Hakenberg, Georg Fuellen, and Mohamed Hamed. 2019. "A Radiogenomic Approach for Decoding Molecular Mechanisms Underlying Tumor Progression in Prostate Cancer" Cancers 11, no. 9: 1293. https://doi.org/10.3390/cancers11091293
APA StyleFischer, S., Tahoun, M., Klaan, B., Thierfelder, K. M., Weber, M.-A., Krause, B. J., Hakenberg, O., Fuellen, G., & Hamed, M. (2019). A Radiogenomic Approach for Decoding Molecular Mechanisms Underlying Tumor Progression in Prostate Cancer. Cancers, 11(9), 1293. https://doi.org/10.3390/cancers11091293






