BRAF Mutation in Colorectal Rhabdoid and Poorly Differentiated Medullary Carcinomas
Abstract
1. Introduction
2. Results
2.1. Clinicopathological Features
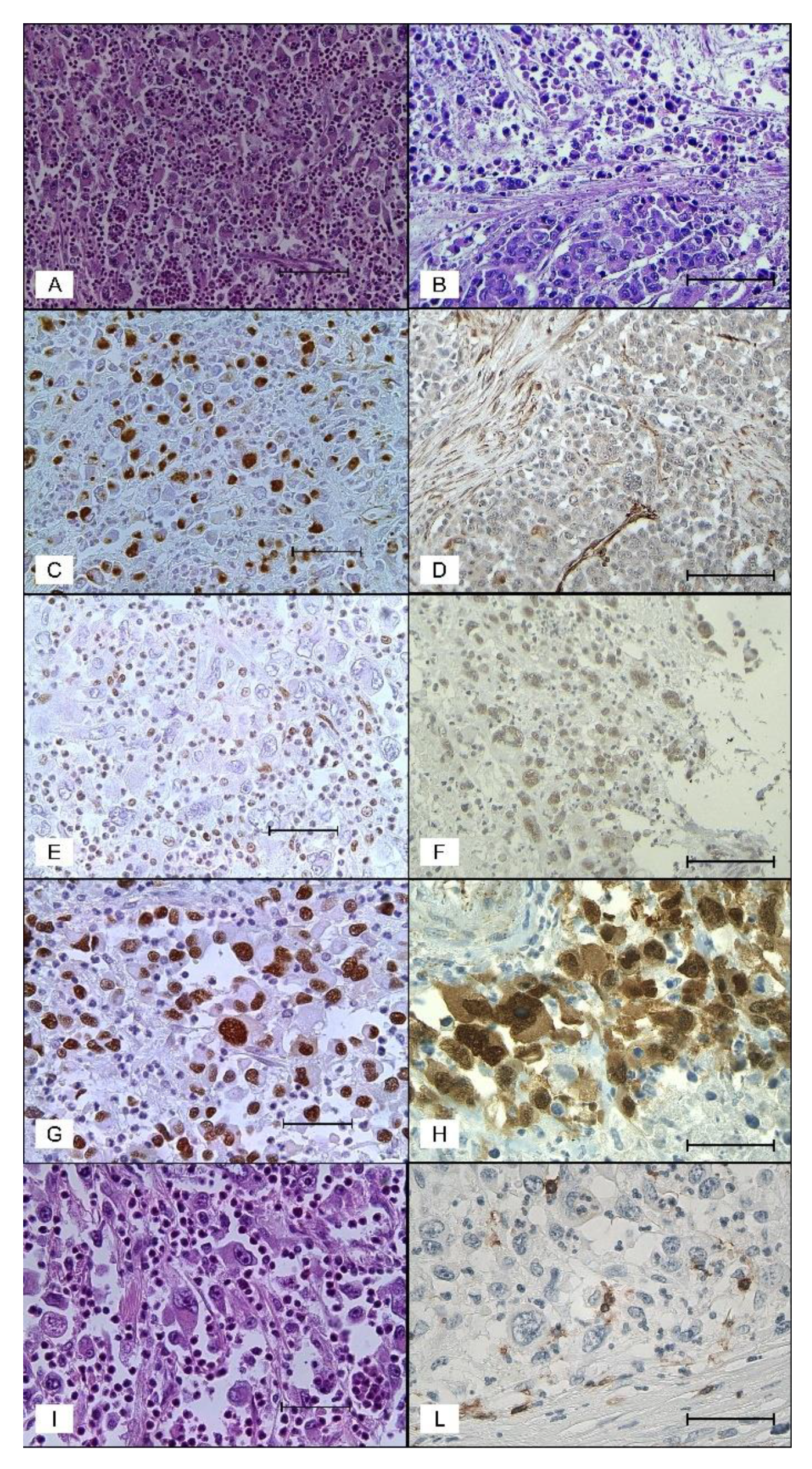
2.2. Pathologic Findings
2.3. Immunohistochemical Findings
2.4. Molecular Findings
2.5. Literature Review
3. Discussion
4. Materials and Methods
4.1. Histopathologic and Immunophenotypical Study
4.2. Molecular Study
4.2.1. MSI and CpG Island Methylator Phenotype (CIMP) Analysis
4.2.2. Targeted Sequencing Libraries and Massively Parallel Sequencing
4.2.3. Next-Generation Sequencing Data Analysis
4.3. Literature Review
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Agaimy, A.; Rau, T.T.; Hartmann, A.; Stoehr, R. SMARCB1 (INI1)-negative rhabdoid carcinomas of the gastrointestinal tract: Clinicopathologic and molecular study of a highly aggressive variant with literature review. Am. J. Surg. Pathol. 2014, 38, 910–920. [Google Scholar] [CrossRef] [PubMed]
- Agaimy, A. The expanding family of SMARCB1(INI1)-deficient neoplasia: Implications of phenotypic, biological, and molecular heterogeneity. Adv. Anat. Pathol. 2014, 21, 394–410. [Google Scholar] [CrossRef] [PubMed]
- Pancione, M.; Remo, A.; Sabatino, L.; Zanella, C.; Votino, C.; Fucci, A.; Di Blasi, A.; Lepore, G.; Daniele, B.; Fenizia, F.; et al. Right-sided rhabdoid colorectal tumors might be related to the serrated pathway. Diagn. Pathol. 2013, 20, 31. [Google Scholar] [CrossRef] [PubMed]
- Remo, A.; Manfrin, E.; Parcesepe, P.; Ferrarini, A.; Han, H.S.; Mickys, U.; Laudanna, C.; Simbolo, M.; Malanga, D.; Oliveira, D.M.; et al. Centrosome Linker-induced Tetraploid Segregation Errors Link Rhabdoid Phenotypes and Lethal Colorectal Cancers. Mol. Cancer Res. 2018, 16, 1385–1395. [Google Scholar] [CrossRef] [PubMed]
- Kono, T.; Imai, Y.; Imura, J.; Ono, Y.; Hagiwara, S.; Taira, K.; Fujimori, T. Cecal adenocarcinoma with prominent rhabdoid feature: Report of a case with immunohistochemical, ultrastructural, and molecular analyses. Int. J. Surg. Pathol. 2007, 15, 414–420. [Google Scholar] [CrossRef] [PubMed]
- Samalavicius, N.E.; Stulpinas, R.; Gasilionis, V.; Baltruskeviciene, E.; Aleknavicius, E.; Mickys, U. Rhabdoid carcinoma of the rectum. Ann. Coloproctol. 2013, 29, 252–255. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lee, S.H.; Seol, H.; Kim, W.Y.; Lim, S.D.; Kim, W.S.; Hwang, T.S.; Han, H.S. Rhabdoid colorectal carcinomas: Reports of two cases. Korean. J. Pathol. 2013, 47, 372–377. [Google Scholar] [CrossRef]
- Moussaly, E.; Atallah, J.P. A rare case of undifferentiated carcinoma of the colon with rhabdoid features: A case report and review of the literature. Case Rep. Oncol. Med. 2015, 2015, 531348. [Google Scholar] [CrossRef][Green Version]
- Kalyan, A.; Pasricha, G.; Monga, D.; Singhi, A.; Bahary, N. Case report of rhabdoid colon cancer and review of literature. Clin. Colorectal. Cancer 2015, 14, e5–e8. [Google Scholar] [CrossRef]
- Wang, J.; Andrici, J.; Sioson, L.; Clarkson, A.; Sheen, A.; Farzin, M.; Toon, C.W.; Turchini, J.; Gill, A.J. Loss of INI1 expression in colorectal carcinoma is associated with high tumor grade, poor survival, BRAFV600E mutation, and mismatch repair deficiency. Hum. Pathol. 2016, 55, 83–90. [Google Scholar] [CrossRef]
- Agaimy, A.; Daum, O.; Märkl, B.; Lichtmannegger, I.; Michal, M.; Hartmann, A. SWI/SNF Complex-deficient Undifferentiated/Rhabdoid Carcinomas of the Gastrointestinal Tract: A Series of 13 Cases Highlighting Mutually Exclusive Loss of SMARCA4 and SMARCA2 and Frequent Co-inactivation of SMARCB1 and SMARCA2. Am. J. Surg. Pathol. 2016, 40, 544–553. [Google Scholar] [CrossRef] [PubMed]
- Pancione, M.; Di Blasi, A.; Sabatino, L.; Fucci, A.; Dalena, A.M.; Palombi, N.; Colantuoni, V. A novel case of rhabdoid colon carcinoma associated with a positive CpG island methylator phenotype and BRAF mutation. Hum. Pathol. 2011, 42, 1047–1052. [Google Scholar] [CrossRef] [PubMed]
- Bak, M.; Teglbjaerg, P.S. Pleomorphic (giant cell) carcinoma of the intestine. An immunohistochemical and electron microscopic study. Cancer 1989, 64, 2557–2564. [Google Scholar] [CrossRef]
- Chetty, R.; Bhathal, P.S. Caecal adenocarcinoma with rhabdoid phenotype: An immunohistochemical and ultrastructural analysis. Virchows Arch. A Pathol. Anat. Histopathol. 1993, 422, 179–182. [Google Scholar] [CrossRef] [PubMed]
- Yang, A.H.; Chen, W.Y.; Chiang, H. Malignant rhabdoid tumour of colon. Histopathology 1994, 24, 89–91. [Google Scholar] [CrossRef] [PubMed]
- Macak, J.; Kodet, R. Rectal adenocarcinoma with rhabdoid phenotype. Pathologica 1995, 87, 696–699. [Google Scholar] [PubMed]
- Marcus, V.A.; Viloria, J.; Owen, D.; Tsao, M.S. Malignant rhabdoid tumor of the colon. Report of a case with molecular analysis. Dis. Colon. Rectum. 1996, 39, 1322–1326. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, I.; Nakano, K.; Nakayama, K.; Ishii, Y.; Ohta, K.; Takahashi, M.; Takenoshita, S. Malignant rhabdoid tumor of the colon: Report of a case. Surg. Today 1999, 29, 1083–1087. [Google Scholar] [CrossRef]
- Oh, H.K.; Cho, C.H.; Kum, Y.S. Adenocarcinoma of the sigmoid colon with prominent rhabdoid features: A case report. Korean. J. Pathol. 2008, 42, 63–65. [Google Scholar]
- Mastoraki, A.; Kotsilianou, O.; Papanikolaou, I.S.; Foukas, P.G.; Sakorafas, G.; Safioleas, M. Malignant rhabdoid tumor of the large intestine. Int. J. Colorectal. Dis. 2009, 24, 1357–1358. [Google Scholar] [CrossRef]
- Han, S.L.; Li, J.L.; Liu, Z.; Cheng, J.; Guo, S.C.; Wu, S.L. Malignant rhabdoid tumor of rectum: Report of a case. Tech. Coloproctol. 2010, 14, 199–200. [Google Scholar] [CrossRef] [PubMed]
- Baba, Y.; Uchiyama, T.; Hamada, K.; Ishihara, Y.; Tanaka, H.; Isono, Y.; Ajioka, Y. A case report of undifferentiated carcinoma of the sigmoid colon with rhabdoid features. Nihon Shokakibyo Gakkai Zasshi 2014, 111, 1384–1390. [Google Scholar] [PubMed]
- Romera Barba, E.; Sanchez Perez, A.; Duque Perez, C.; Garcia Marcilla, J.A.; Vazquez Rojas, J.L. Malignant rhabdoid tumor of the colon: A case report. Cir. Esp. 2014, 92, 638–640. [Google Scholar] [CrossRef] [PubMed]
- Cho, I.J.; Kim, S.S.; Min, Y.D.; Noh, M.W.; Hong, R. Poorly differentiated cecal adenocarcinoma showing prominent rhabdoid feature combined with appendiceal mucinous cystadenoma: A case report and review of the literature. Oncol. Lett. 2015, 9, 1527–1530. [Google Scholar] [CrossRef] [PubMed]
- Dhavaleshwar, D.; Clarke, K. Malignant Extrarenal Rhabdoid Tumor (MERT) of the Colon. Pract. Gastroenterol. 2015, 65, 64–66. [Google Scholar]
- D’Amico, F.; Bertacco, A.; Cesari, M.; Mescoli, C.; Caturegli, G.; Gondolesi, G.; Cillo, U. Extraordinary disease-free survival in a rare malignant extrarenal rhabdoid tumor: A case report and review of the literature. J. Med. Case. Rep. 2018, 12, 39. [Google Scholar] [CrossRef]
- Bond, C.E.; Whitehall, V.L.J. How the BRAF V600E Mutation Defines a Distinct Subgroup of Colorectal Cancer: Molecular and Clinical Implications. Gastroenterol. Res. Pract. 2018, 2018, 9250757. [Google Scholar] [CrossRef]
- Remo, A.; Zanella, C.; Molinari, E.; Talamini, A.; Tollini, F.; Piacentini, P.; Battaglia, P.; Baritono, E.; Bonetti, A.; Lanza, F.; et al. Rhabdoid carcinoma of the colon: A distinct entity with a very aggressive behavior: A case report associated with a polyposis coli and review of the literature. Int. J. Surg. Pathol. 2012, 20, 185–190. [Google Scholar] [CrossRef]
- Geiger, T.R.; Peeper, D.S. Metastasis mechanisms. Biochim. Biophys. Acta 2009, 1796, 293–308. [Google Scholar] [CrossRef]
- Thiery, J.P.; Sleeman, J.P. Complex networks orchestrate epithelial-mesenchymal transitions. Nat. Rev. Mol. Cell Biol. 2006, 7, 131–142. [Google Scholar] [CrossRef]
- Ryu, H.S.; Chung, J.H.; Lee, K.; Shin, E.; Jing, J.; Choe, G.; Kim, H.; Xu, X.; Lee, H.E.; Kim, D.G.; et al. Overexpression of epithelial-mesenchymal transition-related markers according to cell dedifferentiation: Clinical implications as an independent predictor of poor prognosis in cholangiocarcinoma. Hum. Pathol. 2012, 43, 2360–2370. [Google Scholar] [CrossRef] [PubMed]
- Mitselou, A.; Galani, V.; Skoufi, U.; Arvanitis, D.L.; Lampri, E.; Ioachim, E. Syndecan-1, Epithelial-Mesenchymal Transition Markers (E-cadherin/β-catenin) and Neoangiogenesis-related Proteins (PCAM-1 and Endoglin) in Colorectal Cancer. Anticancer Res. 2016, 36, 2271–2280. [Google Scholar] [PubMed]
- Gupta, P.B.; Onder, T.T.; Jiang, G.; Tao, K.; Kuperwasser, C.; Weinberg, R.A.; Lander, E.S. Identification of selective inhibitors of cancer stem cells by high-throughput screening. Cell 2009, 138, 645–659. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.E.; Bae, J.S.; Kang, M.J.; Chung, M.J.; Jang, K.Y.; Park, H.S.; Moon, W.S. Expression of epithelial-mesenchymal transition and cancer stem cell markers in colorectal adenocarcinoma: Clinicopathological significance. Oncol. Rep. 2017, 38, 1695–1705. [Google Scholar] [CrossRef] [PubMed]
- De Smedt, L.; Palmans, S.; Andel, D.; Govaere, O.; Boeckx, B.; Smeets, D.; Galle, E.; Wouters, J.; Barras, D.; Suffiotti, M.; et al. Expression profiling of budding cells incolorectal cancer reveals an EMT-like phenotype and molecular subtype switching. Br. J. Cancer 2017, 116, 58–65. [Google Scholar] [CrossRef]
- Harikrishnan, K.N.; Chow, M.Z.; Baker, E.K.; Pal, S.; Bassal, S.; Brasacchio, D.; Wang, L.; Craig, J.M.; Jones, P.L.; Sif, S.; et al. Brahma links the SWI/SNF chromatin-remodeling complex with MeCP2-dependent transcriptional silencing. Nat. Genet. 2005, 37, 254–264. [Google Scholar]
- Sévenet, N.; Lellouch-Tubiana, A.; Schofield, D.; Hoang-Xuan, K.; Gessler, M.; Birnbaum, D.; Jeanpierre, C.; Jouvet, A.; Delattre, O. Spectrum of hSNF5/INI1 somatic mutations in human cancer and genotype-phenotype correlations. Hum. Mol. Genet. 1999, 8, 2359–2368. [Google Scholar] [CrossRef]
- Landau, M.S.; Kuan, S.F.; Chiosea, S.; Pai, R.K. BRAF-mutated microsatellite stable colorectal carcinoma: An aggressive adenocarcinoma with reduced CDX2 and increased cytokeratin 7 immunohistochemical expression. Hum. Pathol. 2014, 45, 1704–1712. [Google Scholar] [CrossRef]
- Pai, R.K.; Jayachandran, P.; Koong, A.C.; Chang, D.T.; Kwok, S.; Ma, L.; Arber, D.A.; Balise, R.R.; Tubbs, R.R.; Shadrach, B.; et al. BRAF-mutated, microsatellite-stable adenocarcinoma of the proximal colon: An aggressive adenocarcinoma with poor survival, mucinous differentiation, and adverse morphologic features. Am. J. Surg. Pathol. 2012, 36, 744–752. [Google Scholar] [CrossRef]
- Digiacomo, N.; Bolzacchini, E.; Veronesi, G.; Cerutti, R.; Sahnane, N.; Pinotti, G.; Bregni, M.; Artale, S.; Verusio, C.; Crivelli, F.; et al. Neuroendocrine Differentiation, Microsatellite Instability, and Tumor-infiltrating Lymphocytes in Advanced Colorectal Cancer with BRAF Mutation. Clin. Colorectal. Cancer 2019, 18, e251–e260. [Google Scholar] [CrossRef]
- Barras, D.; Missiaglia, E.; Wirapati, P.; Sieber, O.M.; Jorissen, R.N.; Love, C.; Molloy, P.L.; Jones, I.T.; McLaughlin, S.; Gibbs, P.; et al. BRAF V600E Mutant Colorectal Cancer Subtypes Based on GeneExpression. Clin. Cancer Res. 2017, 23, 104–115. [Google Scholar] [CrossRef] [PubMed]
- Mizuno, R.; Kawada, K.; Itatani, Y.; Ogawa, R.; Kiyasu, Y.; Sakai, Y. The Role of Tumor-Associated Neutrophils in Colorectal Cancer. Int. J. Mol. Sci. 2019, 20, 529. [Google Scholar] [CrossRef] [PubMed]
- Rao, H.L.; Chen, J.W.; Li, M.; Xiao, Y.B.; Fu, J.; Zeng, Y.X.; Cai, M.Y.; Xie, D. Increased intratumoral neutrophil in colorectal carcinomas correlates closely with malignant phenotype and predicts patients’ adverse prognosis. PLoS ONE 2012, 7, e30806. [Google Scholar] [CrossRef] [PubMed]
- Bond, C.E.; Umapathy, A.; Ramsnes, I.; Greco, S.A.; Zhao, Z.Z.; Mallitt, K.A.; Buttenshaw, R.L.; Montgomery, G.W.; Leggett, B.A.; Whitehall, V.L. p53 mutation is common in microsatellite stable, BRAF mutant colorectal cancers. Int. J. Cancer 2012, 130, 1567–1576. [Google Scholar] [CrossRef] [PubMed]
- Bond, C.E.; Nancarrow, D.J.; Wockner, L.F.; Wallace, L.; Montgomery, G.W.; Leggett, B.A.; Whitehall, V.L. Microsatellite stable colorectal cancers stratified by the BRAF V600E mutation show distinct patterns of chromosomal instability. PLoS ONE 2014, 9, e91739. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, T.; Mitomi, H.; Kanazawa, H.; Ohkura, Y.; Watanabe, M. Tumor budding as an index to identify high-risk patients with stage II colon cancer. Dis. Colon. Rectum. 2008, 51, 568–572. [Google Scholar] [CrossRef] [PubMed]
- Kasajima, A.; Sers, C.; Sasano, H.; Jöhrens, K.; Stenzinger, A.; Noske, A.; Buckendahl, A.C.; Darb-Esfahani, S.; Müller, B.M.; Budczies, J.; et al. Down-regulation of the antigen processing machinery is linked to a loss of inflammatory response in colorectal cancer. Hum. Pathol. 2010, 41, 1758–1769. [Google Scholar] [CrossRef] [PubMed]
- Sahnane, N.; Furlan, D.; Monti, M.; Romualdi, C.; Vanoli, A.; Vicari, E.; Solcia, E.; Capella, C.; Sessa, F.; La Rosa, S. Microsatellite unstable gastrointestinal neuroendocrine carcinomas: A new clinicopathologic entity. Endocr. Relat. Cancer 2015, 22, 35–45. [Google Scholar] [CrossRef] [PubMed]
- Ogino, S.; Kawasaki, T.; Kirkner, G.J.; Kraft, P.; Loda, M.; Fuchs, C.S. Evaluation of markers for CpG island methylator phenotype (CIMP) in colorectal cancer by anlarge population-based sample. J. Mol. Diagn. 2007, 9, 305–314. [Google Scholar] [CrossRef] [PubMed]
| Studies | MSI Status | BRAF | KRAS | PIK3CA | CROCC | INI-1 § | TP53 | CIMP |
|---|---|---|---|---|---|---|---|---|
| Kono et al. 2007 [5] | MSS | - | WT | - | - | - | - | - |
| Samalavicius et al. 2013 [6] | MSS | V600E | WT | - | - | - | - | - |
| Lee et al. 2013 (case 1) [7] | MSS | WT | WT | - | WT | WT | - | - |
| Lee et al. 2013 (case 2) [7] | MSS | V600E | WT | - | WT | WT | - | - |
| Agaimy et al. 2014 [1] | MSI | V600E | - | - | - | NEG | - | CIMP |
| Moussaly et al. 2015 [8] | MSI | - | - | - | - | - | - | - |
| Kalyan et al. 2015 [9] | MSS | WT | Q61H | WT | - | POS | R273H | - |
| Wang et al. 2016 (case 1) [10] | MSS | V600E | - | - | - | POS | - | - |
| Wang et al. 2016 (case 2) [10] | MSI | V600E | - | - | - | NEG | - | - |
| Wang et al. 2016 (case 3) [10] | MSS | V600E | - | - | - | POS | - | - |
| Agaimy et al. 2016 [11] | MSS | - | - | - | - | POS | - | - |
| Remo et al. 2018 * [4] | MSS | WT | WT | - | - | - | - | - |
| Remo et al. 2018 * [4] | MSS | WT | WT | - | - | - | - | - |
| Remo et al. 2018 * [4] | MSS | WT | MUT | - | - | POS | - | - |
| Remo et al. 2018 * [4] | MSI | V600E | - | - | - | POS | - | - |
| Remo et al. 2018 (RC1) [4] | MSI | V600E | WT | - | A161S | WT | - | CIMP |
| Remo et al. 2018 (RC2) ** [4] | MSI | V600E | - | - | V1885A | WT | - | CIMP |
| Remo et al. 2018 (RC5) [4] | MSS | V600E | WT | - | WT | WT | - | - |
| Remo et al. 2018 (RC6) [4] | MSS | V600E | G12V | - | WT | WT | - | - |
| Remo et al. 2018 (RC7) [4] | MSI | V600E | WT | - | WT | WT | - | - |
| Remo et al. 2018 (RC8) [4] | MSS | WT | WT | - | WT | WT | - | - |
| Remo et al. 2018 (RC9) [4] | MSI | V600E | WT | - | S1320I | WT | - | - |
| Remo et al. 2018 (RC10) [4] | MSS | WT | WT | - | WT | WT | - | - |
| Remo et al. 2018 (RC11) [4] | MSI | WT | WT | - | A1510T | WT | - | - |
| Remo et al. 2018 (RC12) [4] | MSS | WT | WT | - | WT | WT | - | - |
| Cases | Gender | Age | Site | Size (cm) | Type | Metastases | Stage | Treatment | Outcome * |
|---|---|---|---|---|---|---|---|---|---|
| CRbC 1 | F | 63 | Hepatic flexure | 10 | Pure | N | IIIC | Surgery | 2 months |
| CRbC 2 | F | 76 | Sigmoid colon | 4 | Pure | - | - | Surgery + CT | 7 months |
| CRbC 3 | M | 85 | Splenic flexure | 6 | Pure | N, L | IVA | Surgery | 2 months |
| CRbC 4 | M | 65 | Cecum | 6 | Pure | N | IIIB | Surgery | 216 months (alive) |
| CRbC 5 | M | 63 | Rectum | 6 | Pure | N | IIIC | Surgery | 10 months |
| CRbC 6 | M | 64 | Hepatic flexure | 6 | Combined | N | IIIB | Surgery | - |
| CRbC 7 | F | 77 | Ascending colon | 7 | Combined | absence | IIA | Surgery | 187 months (alive) |
| PDMC 1 | M | 79 | Ascending colon | 10 | - | N | IIIC | Surgery | 11 months |
| PDMC 2 | F | 94 | Ascending colon | 8 | - | N | IIIC | Surgery | 5 months |
| PDMC 3 | F | 53 | Sigmoid colon | 13 | - | N | IIIC | Surgery | - |
| PDMC 4 | M | 73 | Cecum | 8 | - | N | IIIC | Surgery | 124 months (alive) |
| ID | Vim | Pancytokeratin | INI-1 | Nuclear | p53 | CD8+ (PLI/ILI) |
|---|---|---|---|---|---|---|
| β-Catenin | ||||||
| CRbC 1 | 3+ | 3+ | 0 | 0 | 3+ | 5/15 |
| CRbC 2 | 3+ | 3+ | 0 | 3+ | 3+ | 10/3 |
| CRbC 3 | 3+ | 1+ | 0 | 2+ | 3+ | 5/6 |
| CRbC 4 | 2+ | 3+ | 0 | 0 | 3+ | 73/29 |
| CRbC 5 | 3+ | 2+ | 0 | 3+ | 2+ | 53/13 |
| CRbC 6 | 3+ | 3+ | 1+ | 2+ | 3+ | 61/38 |
| CRbC 7 | 1+ | 1+ | 1+ | 2+ | 2+ | 15/16 |
| PDMC1 | 0 | 3+ | 3+ | 3+ | 0 | 23/4 |
| PDMC2 | 0 | 3+ | 3+ | 3+ | 3+ | 26/2 |
| PDMC3 | 0 | 3+ | 3+ | 1+ | 0 | 54/5 |
| PDMC4 | 0 | n.a. | 3+ | 3+ | 2+ | 106/15 |
| ID | MSI | CIMP | BRAF (mAF) | KRAS (mAF) | NRAS (mAF) | PIK3CA (mAF) | TP53 (mAF) |
|---|---|---|---|---|---|---|---|
| CRbC 1 | MSS | no CIMP | V600E (7.5) | WT | WT | WT | R273C (11.9) |
| CRbC 2 | MSS | no CIMP | V600E (22.6) | WT | WT | WT | R273C (29.7) |
| CRbC 3 | MSS | no CIMP | G466A (25.1) | WT | G12D (28.2) | WT | G245S (55.7) |
| CRbC 4 | MSS | no CIMP | WT | Q61K (19.7) | WT | WT | R273C (30.9) |
| CRbC 5 | MSS | no CIMP | WT | G13D (86.5) | WT | WT | P278A (56.9) |
| CRbC 6 | MSI | no CIMP | V600E (21.2) | WT | WT | H1047R (29.1) | R273C (12.3) |
| CRbC 7 | MSI | - | V600E * | - | - | - | - |
| PDMC 1 | MSI | CIMP-H | V600E (43.31) | WT | WT | WT | * P152fs, 18 (37.9) * V73fs, 50 (34.3) |
| PDMC 2 | MSI | - | V600E (21.85) | WT | WT | WT | WT |
| PDMC 3 | MSI | CIMP-H | V600E (68.89) | WT | WT | R93Q (33.6) M772I (39.5) | V272M (33.1) |
| PDMC 4 | MSI | CIMP-H | V600E (43.27) | WT | WT | WT | Y163C (41) |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bolzacchini, E.; Digiacomo, N.; Marrazzo, C.; Sahnane, N.; Maragliano, R.; Gill, A.; Albarello, L.; Sessa, F.; Furlan, D.; Capella, C. BRAF Mutation in Colorectal Rhabdoid and Poorly Differentiated Medullary Carcinomas. Cancers 2019, 11, 1252. https://doi.org/10.3390/cancers11091252
Bolzacchini E, Digiacomo N, Marrazzo C, Sahnane N, Maragliano R, Gill A, Albarello L, Sessa F, Furlan D, Capella C. BRAF Mutation in Colorectal Rhabdoid and Poorly Differentiated Medullary Carcinomas. Cancers. 2019; 11(9):1252. https://doi.org/10.3390/cancers11091252
Chicago/Turabian StyleBolzacchini, Elena, Nunzio Digiacomo, Cristina Marrazzo, Nora Sahnane, Roberta Maragliano, Anthony Gill, Luca Albarello, Fausto Sessa, Daniela Furlan, and Carlo Capella. 2019. "BRAF Mutation in Colorectal Rhabdoid and Poorly Differentiated Medullary Carcinomas" Cancers 11, no. 9: 1252. https://doi.org/10.3390/cancers11091252
APA StyleBolzacchini, E., Digiacomo, N., Marrazzo, C., Sahnane, N., Maragliano, R., Gill, A., Albarello, L., Sessa, F., Furlan, D., & Capella, C. (2019). BRAF Mutation in Colorectal Rhabdoid and Poorly Differentiated Medullary Carcinomas. Cancers, 11(9), 1252. https://doi.org/10.3390/cancers11091252





