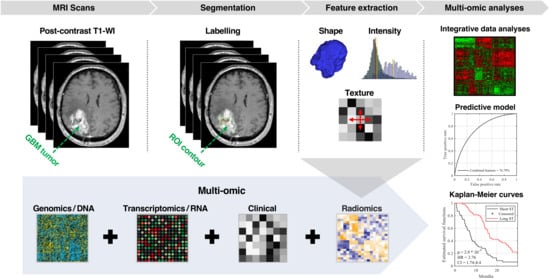
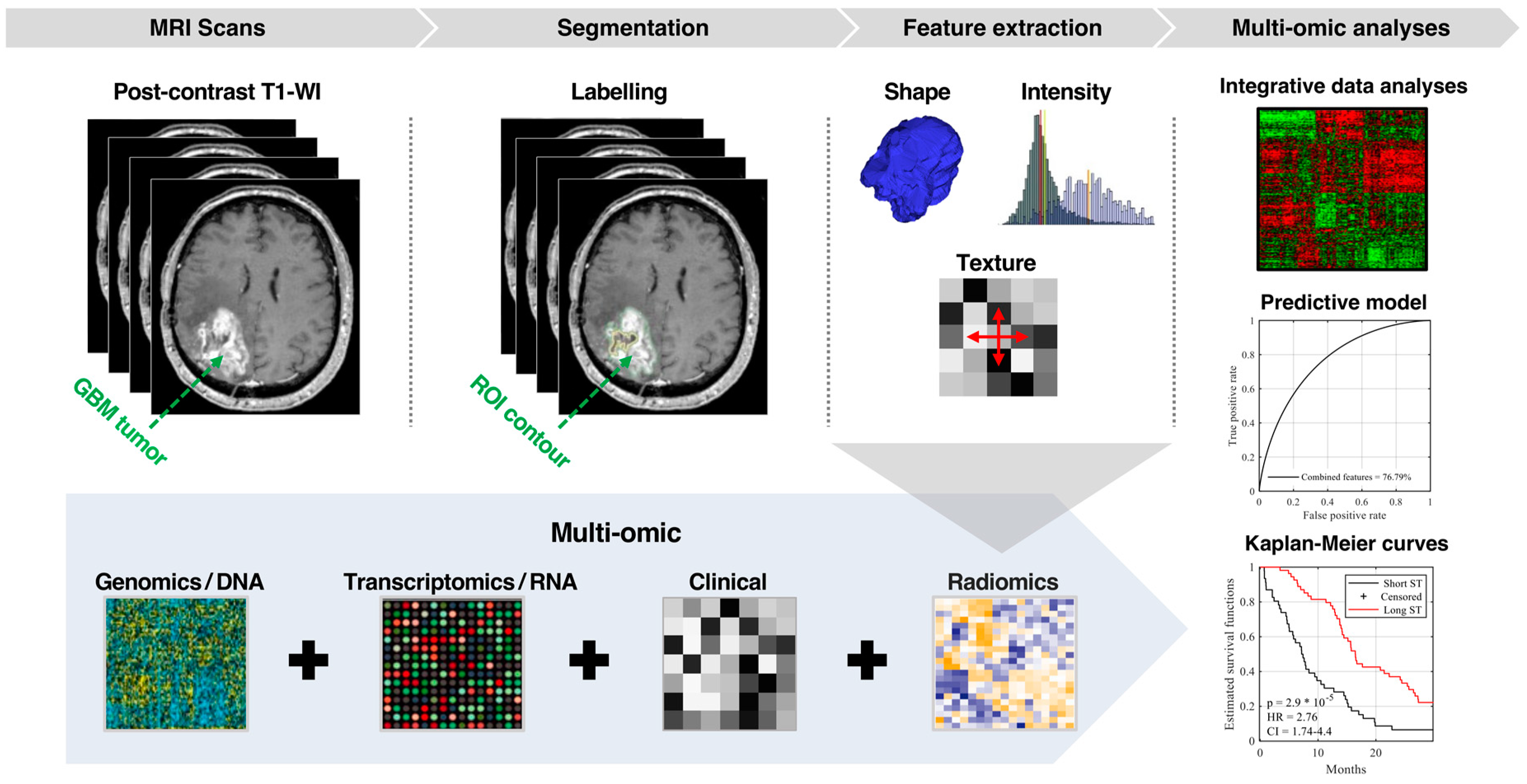
Integration of Radiomic and Multi-omic Analyses Predicts Survival of Newly Diagnosed IDH1 Wild-Type Glioblastoma
Abstract
1. Introduction
2. Results
2.1. Patients Characteristics
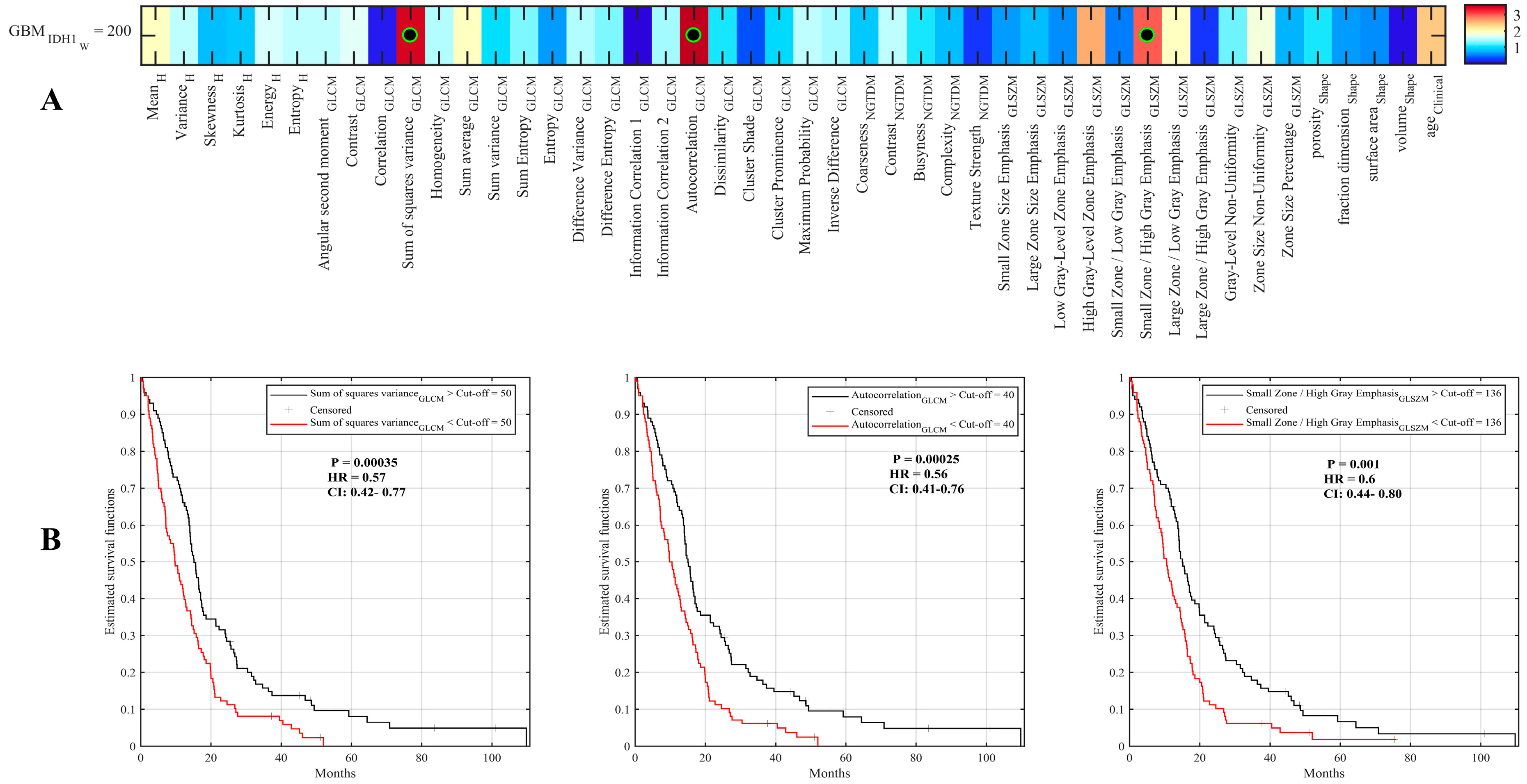
2.2. Assessment of Radiomic Features to Identify Tumor Characteristics
2.3. Radiomic Signature to Predict Survival of IDH1 Wild-Type GBM Patients
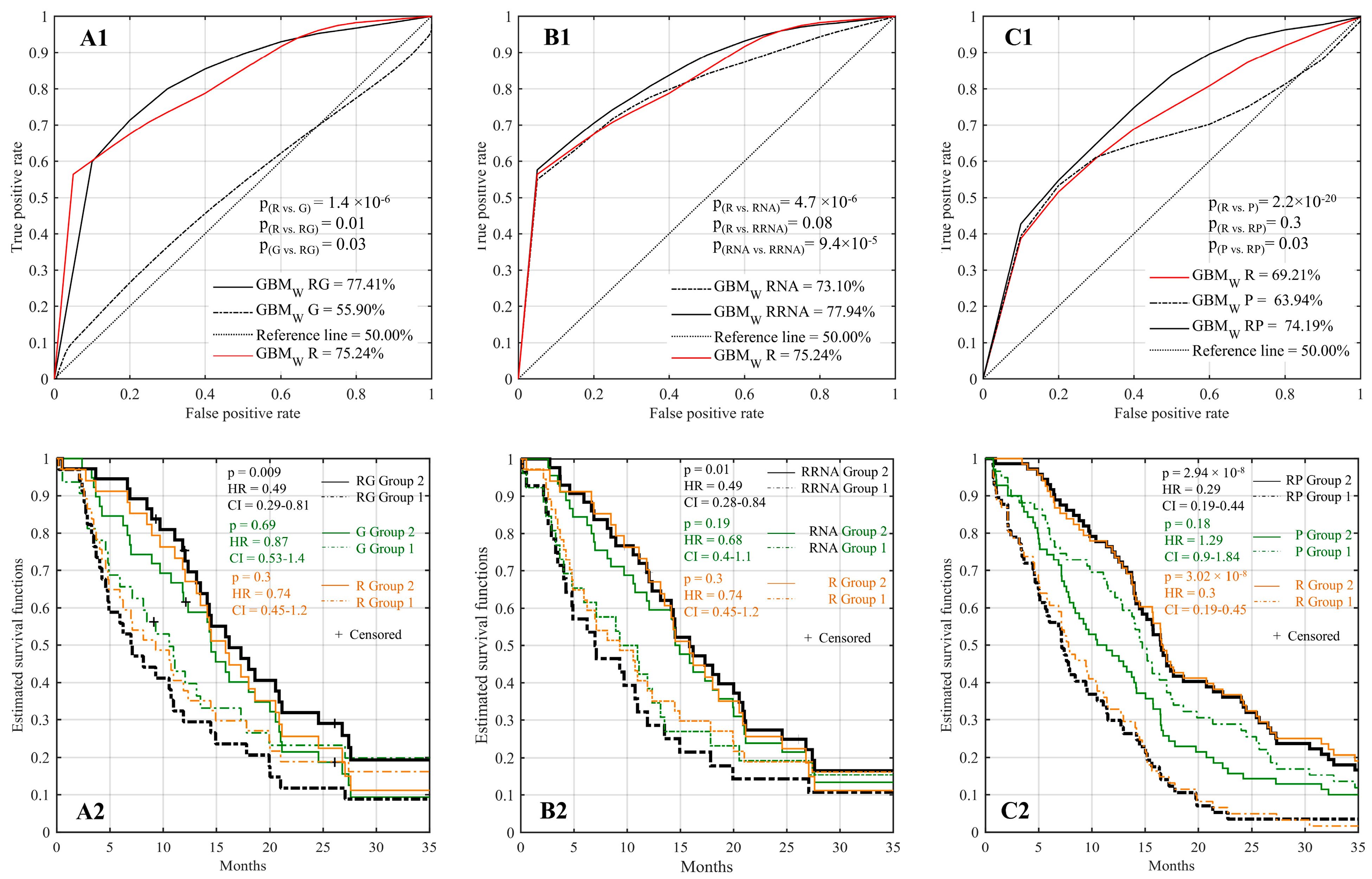
2.4. Improved Prognostic Capacity When Integrating Radiomics with Genomic Features
2.5. Improved Prognostic Capacity When Integrating Radiomics with Transcriptomic Features
2.6. Improved Prognostic Capacity When Integrating Radiomics with Protein Expression
2.7. Multi-Omic Integrative Model and Identification of Predictive Features
3. Discussion
4. Materials and Methods
4.1. Population and Data Acquisition
4.2. ROIs Labeling
4.3. Imaging Feature Extraction
4.4. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Zacharaki, E.I.; Wang, S.; Chawla, S.; Soo Yoo, D.; Wolf, R.; Melhem, E.R.; Davatzikos, C. Classification of brain tumor type and grade using MRI texture and shape in a machine learning scheme. Magn. Reson. Med. 2009, 62, 1609–1618. [Google Scholar] [CrossRef] [PubMed]
- Stupp, R.; Hegi, M.E.; van den Bent, M.J.; Mason, W.P.; Weller, M.; Mirimanoff, R.O.; Cairncross, J.G. Changing Paradigms—An Update on the Multidisciplinary Management of Malignant Glioma. Oncologist 2006, 11, 165–180. [Google Scholar] [CrossRef] [PubMed]
- Stupp, R.; Mason, W.P.; van den Bent, M.J.; Weller, M.; Fisher, B.; Taphoorn, M.J.B.; Belanger, K.; Brandes, A.A.; Marosi, C.; Bogdahn, U.; et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N. Engl. J. Med. 2005, 352, 987–996. [Google Scholar] [CrossRef] [PubMed]
- Verhaak, R.G.W.; Hoadley, K.A.; Purdom, E.; Wang, V.; Qi, Y.; Wilkerson, M.D.; Miller, C.R.; Ding, L.; Golub, T.; Mesirov, J.P.; et al. Integrated Genomic Analysis Identifies Clinically Relevant Subtypes of Glioblastoma Characterized by Abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 2010, 17, 98–110. [Google Scholar] [CrossRef] [PubMed]
- Bleeker, F.E.; Atai, N.A.; Lamba, S.; Jonker, A.; Rijkeboer, D.; Bosch, K.S.; Tigchelaar, W.; Troost, D.; Dertop, W.P.V.; Bardelli, A.; et al. The prognostic IDH1R132 mutation is associated with reduced NADP+-dependent IDH activity in glioblastoma. Acta Neuropathol. 2010, 119, 487–494. [Google Scholar] [CrossRef] [PubMed]
- Guo, C.; Pirozzi, C.J.; Lopez, G.Y.; Yan, H. Isocitrate dehydrogenase mutations in gliomas: mechanisms, biomarkers and therapeutic target. Curr. Opin. Neurol. 2011, 24, 648–652. [Google Scholar] [CrossRef] [PubMed]
- Cancer Genome Atlas Research Network; Brat, D.J.; Verhaak, R.G.W.; Aldape, K.D.; Yung, W.K.A.; Salama, S.R.; Cooper, L.A.D.; Rheinbay, E.; Miller, C.R.; Vitucci, M.; et al. Comprehensive, Integrative Genomic Analysis of Diffuse Lower-Grade Gliomas. N. Engl. J. Med. 2015, 372, 2481–2498. [Google Scholar]
- Reifenberger, G.; Wirsching, H.-G.; Knobbe-Thomsen, C.B.; Weller, M. Advances in the molecular genetics of gliomas—Implications for classification and therapy. Nat. Rev. Clin. Oncol. 2017, 14, 434–452. [Google Scholar] [CrossRef]
- Eckel-Passow, J.E.; Lachance, D.H.; Molinaro, A.M.; Walsh, K.M.; Decker, P.A.; Sicotte, H.; Pekmezci, M.; Rice, T.; Kosel, M.L.; Smirnov, I.V.; et al. Glioma Groups Based on 1p/19q, IDH, and TERT Promoter Mutations in Tumors. New Engl. J. Med. 2015, 372, 2499–2508. [Google Scholar] [CrossRef]
- Chaddad, A.; Desrosiers, C.; Abdulkarim, B.; Niazi, T. Predicting the Gene Status and Survival Outcome of Lower Grade Glioma Patients with Multimodal MRI Features. IEEE Access 2019, 7, 75976–75984. [Google Scholar] [CrossRef]
- Zhou, H.; Vallières, M.; Bai, H.X.; Su, C.; Tang, H.; Oldridge, D.; Zhang, Z.; Xiao, B.; Liao, W.; Tao, Y.; et al. MRI features predict survival and molecular markers in diffuse lower-grade gliomas. Neuro-Oncology 2017, 19, 862–870. [Google Scholar] [CrossRef] [PubMed]
- Chaddad, A.; Tanougast, C. Extracted magnetic resonance texture features discriminate between phenotypes and are associated with overall survival in glioblastoma multiforme patients. Med. Biol. Eng. Comput. 2016, 54, 1707–1718. [Google Scholar] [CrossRef] [PubMed]
- Chaddad, A.; Sabri, S.; Niazi, T.; Abdulkarim, B. Prediction of survival with multi-scale radiomic analysis in glioblastoma patients. Med. Biol. Eng. Comput. 2018, 56, 2287–2300. [Google Scholar] [CrossRef] [PubMed]
- Chaddad, A.; Daniel, P.; Desrosiers, C.; Toews, M.; Abdulkarim, B. Novel Radiomic Features Based on Joint Intensity Matrices for Predicting Glioblastoma Patient Survival Time. IEEE J. Biomed. Health Inform. 2019, 23, 795–804. [Google Scholar] [CrossRef] [PubMed]
- Chaddad, A.; Desrosiers, C.; Hassan, L.; Tanougast, C. A quantitative study of shape descriptors from glioblastoma multiforme phenotypes for predicting survival outcome. Br. J. Radiol. 2016, 89, 20160575. [Google Scholar] [CrossRef] [PubMed]
- Grossmann, P.; Gutman, D.A.; Dunn, W.D.; Holder, C.A.; Aerts, H.J.W.L. Imaging-genomics reveals driving pathways of MRI derived volumetric tumor phenotype features in Glioblastoma. BMC Cancer 2016, 16, 611. [Google Scholar] [CrossRef] [PubMed]
- Gutman, D.A.; Dunn, W.D.; Grossmann, P.; Cooper, L.A.D.; Holder, C.A.; Ligon, K.L.; Alexander, B.M.; Aerts, H.J.W.L. Somatic mutations associated with MRI-derived volumetric features in glioblastoma. Neuroradiology 2015, 57, 1227–1237. [Google Scholar] [CrossRef] [PubMed]
- Drabycz, S.; Roldán, G.; de Robles, P.; Adler, D.; McIntyre, J.B.; Magliocco, A.M.; Cairncross, J.G.; Mitchell, J.R. An analysis of image texture, tumor location, and MGMT promoter methylation in glioblastoma using magnetic resonance imaging. NeuroImage 2010, 49, 1398–1405. [Google Scholar] [CrossRef]
- Chaddad, A.; Tanougast, C. High-Throughput Quantification of Phenotype Heterogeneity Using Statistical Features. Adv. Bioinform. 2015, 2015, e728164. [Google Scholar] [CrossRef]
- Chaddad, A.; Desrosiers, C.; Toews, M. GBM heterogeneity characterization by radiomic analysis of phenotype anatomical planes. In Medical Imaging 2016: Image Processing; International Society for Optics and Photonics; SPIE Medical Imaging: San Diego, CA, USA, 2016; Volume 9784, p. 978424. [Google Scholar]
- Chaddad, A.; Desrosiers, C.; Toews, M. Phenotypic characterization of glioblastoma identified through shape descriptors. In Medical Imaging 2016: Computer-Aided Diagnosis; International Society for Optics and Photonics; SPIE Medical Imaging: San Diego, CA, USA, 2016; Volume 9785, p. 97852M. [Google Scholar]
- Azoulay, M.; Santos, F.; Souhami, L.; Panet-Raymond, V.; Petrecca, K.; Owen, S.; Guiot, M.-C.; Patyka, M.; Sabri, S.; Shenouda, G.; et al. Comparison of radiation regimens in the treatment of Glioblastoma multiforme: results from a single institution. Radiat. Oncol. 2015, 10, 106. [Google Scholar] [CrossRef]
- Holm, S. A Simple Sequentially Rejective Multiple Test Procedure. Scand. J. Stat. 1979, 6, 65–70. [Google Scholar]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative Analysis of Complex Cancer Genomics and Clinical Profiles Using the cBioPortal. Sci. Signal. 2013, 6, pl1. [Google Scholar] [CrossRef]
- Archer, K.J.; Kimes, R.V. Empirical characterization of random forest variable importance measures. Comput. Stat. Data Anal. 2008, 52, 2249–2260. [Google Scholar] [CrossRef]
- Kickingereder, P.; Neuberger, U.; Bonekamp, D.; Piechotta, P.L.; Götz, M.; Wick, A.; Sill, M.; Kratz, A.; Shinohara, R.T.; Jones, D.T. Radiomic subtyping improves disease stratification beyond key molecular, clinical and standard imaging characteristics in patients with glioblastoma. Neuro-Oncology 2017, 20, 848–857. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Chang, K.; Ramkissoon, S.; Tanguturi, S.; Bi, W.L.; Reardon, D.A.; Ligon, K.L.; Alexander, B.M.; Wen, P.Y.; Huang, R.Y. Multimodal MRI features predict isocitrate dehydrogenase genotype in high-grade gliomas. Neuro-Oncology 2017, 19, 109–117. [Google Scholar] [CrossRef]
- Grossmann, P.; Narayan, V.; Chang, K.; Rahman, R.; Abrey, L.; Reardon, D.A.; Schwartz, L.H.; Wen, P.Y.; Alexander, B.M.; Huang, R.; et al. Quantitative imaging biomarkers for risk stratification of patients with recurrent glioblastoma treated with bevacizumab. Neuro-Oncology 2017, 19, 1688–1697. [Google Scholar] [CrossRef]
- Rathore, S.; Akbari, H.; Rozycki, M.; Abdullah, K.G.; Nasrallah, M.P.; Binder, Z.A.; Davuluri, R.V.; Lustig, R.A.; Dahmane, N.; Bilello, M.; et al. Radiomic MRI signature reveals three distinct subtypes of glioblastoma with different clinical and molecular characteristics, offering prognostic value beyond IDH1. Sci. Rep. 2018, 8, 5087. [Google Scholar] [CrossRef] [PubMed]
- Chaddad, A.; Desrosiers, C.; Toews, M. Multi-scale radiomic analysis of sub-cortical regions in MRI related to autism, gender and age. Sci. Rep. 2017, 7, 45639. [Google Scholar] [CrossRef]
- Bakas, S.; Akbari, H.; Sotiras, A.; Bilello, M.; Rozycki, M.; Kirby, J.S.; Freymann, J.B.; Farahani, K.; Davatzikos, C. Advancing the Cancer Genome Atlas glioma MRI collections with expert segmentation labels and radiomic features. Sci. Data 2017, 4, 170117. [Google Scholar] [CrossRef]
- Bakas, S.; Zeng, K.; Sotiras, A.; Rathore, S.; Akbari, H.; Gaonkar, B.; Rozycki, M.; Pati, S.; Davatzikos, C. GLISTRboost: Combining Multimodal MRI Segmentation, Registration, and Biophysical Tumor Growth Modeling with Gradient Boosting Machines for Glioma Segmentation. In Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries; Springer: Cham, Switzerland, 2015; pp. 144–155. [Google Scholar]
- Beig, N.; Patel, J.; Prasanna, P.; Partovi, S.; Varadhan, V.; Madabhushi, A.; Tiwari, P. Radiogenomic analysis of hypoxia pathway reveals computerized MRI descriptors predictive of overall survival in Glioblastoma. In SPIE Medical Imaging; International Society for Optics and Photonics: Bellingham, WA, USA, 2017; p. 101341U. [Google Scholar]
- Prasanna, P.; Patel, J.; Partovi, S.; Madabhushi, A.; Tiwari, P. Radiomic features from the peritumoral brain parenchyma on treatment-naïve multi-parametric MR imaging predict long versus short-term survival in glioblastoma multiforme: Preliminary findings. Eur. Radiol. 2017, 27, 4188–4197. [Google Scholar] [CrossRef]
- Colen, R.R.; Wang, J.; Singh, S.K.; Gutman, D.A.; Zinn, P.O. Glioblastoma: Imaging Genomic Mapping Reveals Sex-specific Oncogenic Associations of Cell Death. Radiology 2014, 275, 215–227. [Google Scholar] [CrossRef] [PubMed]
- Colen, R.R.; Vangel, M.; Wang, J.; Gutman, D.A.; Hwang, S.N.; Wintermark, M.; Jain, R.; Jilwan-Nicolas, M.; Chen, J.Y.; Raghavan, P.; et al. Imaging genomic mapping of an invasive MRI phenotype predicts patient outcome and metabolic dysfunction: A TCGA glioma phenotype research group project. BMC Med. Genom. 2014, 7, 30. [Google Scholar] [CrossRef] [PubMed]
- Akbari, H.; Bakas, S.; Pisapia, J.M.; Nasrallah, M.P.; Rozycki, M.; Martinez-Lage, M.; Morrissette, J.J.D.; Dahmane, N.; O’Rourke, D.M.; Davatzikos, C. In vivo evaluation of EGFRvIII mutation in primary glioblastoma patients via complex multiparametric MRI signature. Neuro-Oncology 2018, 20, 1068–1079. [Google Scholar] [CrossRef] [PubMed]
- Chaddad, A.; Tanougast, C. Quantitative evaluation of robust skull stripping and tumor detection applied to axial MR images. Brain Inf. 2016, 3, 53–61. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, D.; Bandyopadhyay, S.K. Brain tumor detection from MRI image: An approach. IJAR 2017, 3, 1152–1159. [Google Scholar]
- Parmar, C.; Grossmann, P.; Bussink, J.; Lambin, P.; Aerts, H.J.W.L. Machine Learning methods for Quantitative Radiomic Biomarkers. Sci. Rep. 2015, 5, 13087. [Google Scholar] [CrossRef]
- Kickingereder, P.; Götz, M.; Muschelli, J.; Wick, A.; Neuberger, U.; Shinohara, R.T.; Sill, M.; Nowosielski, M.; Schlemmer, H.-P.; Radbruch, A.; et al. Large-scale Radiomic Profiling of Recurrent Glioblastoma Identifies an Imaging Predictor for Stratifying Anti-Angiogenic Treatment Response. Clin. Cancer Res. 2016, 22, 5765–5771. [Google Scholar] [CrossRef]
- Quayle, S.N.; Lee, J.Y.; Cheung, L.W.T.; Ding, L.; Wiedemeyer, R.; Dewan, R.W.; Huang-Hobbs, E.; Zhuang, L.; Wilson, R.K.; Ligon, K.L.; et al. Somatic mutations of PIK3R1 promote gliomagenesis. PLoS ONE 2012, 7, e49466. [Google Scholar] [CrossRef]
- Kraus, J.A.; Glesmann, N.; Beck, M.; Krex, D.; Klockgether, T.; Schackert, G.; Schlegel, U. Molecular analysis of the PTEN, TP53 and CDKN2A tumor suppressor genes in long-term survivors of glioblastoma multiforme. J. Neurooncol. 2000, 48, 89–94. [Google Scholar] [CrossRef]
- Rich, J.N.; Hans, C.; Jones, B.; Iversen, E.S.; McLendon, R.E.; Rasheed, B.K.A.; Dobra, A.; Dressman, H.K.; Bigner, D.D.; Nevins, J.R.; et al. Gene expression profiling and genetic markers in glioblastoma survival. Cancer Res. 2005, 65, 4051–4058. [Google Scholar] [CrossRef]
- Petitjean, A.; Achatz, M.I.W.; Borresen-Dale, A.L.; Hainaut, P.; Olivier, M. TP53 mutations in human cancers: functional selection and impact on cancer prognosis and outcomes. Oncogene 2007, 26, 2157–2165. [Google Scholar] [CrossRef] [PubMed]
- Moon, W.-J.; Choi, J.W.; Roh, H.G.; Lim, S.D.; Koh, Y.-C. Imaging parameters of high grade gliomas in relation to the MGMT promoter methylation status: the CT, diffusion tensor imaging, and perfusion MR imaging. Neuroradiology 2012, 54, 555–563. [Google Scholar] [CrossRef]
- Chaddad, A.; Kucharczyk, M.J.; Daniel, P.; Sabri, S.; Jean-Claude, B.J.; Niazi, T.; Abdulkarim, B. Radiomics in Glioblastoma: Current Status and Challenges Facing Clinical Implementation. Front. Oncol. 2019, 9. [Google Scholar] [CrossRef]
- Prior, F.W.; Clark, K.; Commean, P.; Freymann, J.; Jaffe, C.; Kirby, J.; Moore, S.; Smith, K.; Tarbox, L.; Vendt, B.; et al. TCIA: An information resource to enable open science. Conf. Proc. IEEE Eng. Med. Biol. Soc. 2013, 2013, 1282–1285. [Google Scholar] [PubMed]
- Yang, X.; Beyenal, H.; Harkin, G.; Lewandowski, Z. Quantifying biofilm structure using image analysis. J. Microbiol. Methods 2000, 39, 109–119. [Google Scholar] [CrossRef]
- Legland, D.; Kiêu, K.; Devaux, M.-F. Computation of Minkowski measures on 2D and 3D binary images. Image Anal. Stereol. 2007, 26, 83–92. [Google Scholar] [CrossRef]
- Haralick, R.M. Statistical and structural approaches to texture. Proc. IEEE 1979, 67, 786–804. [Google Scholar] [CrossRef]
- Amadasun, M.; King, R. Textural features corresponding to textural properties. IEEE Trans. Syst. Man Cybern. 1989, 19, 1264–1274. [Google Scholar] [CrossRef]
- Thibault, G.; Fertil, B.; Navarro, C.; Pereira, S.; Cau, P.; Levy, N.; Sequeira, J.; Mari, J.J. Texture indexes and gray level size zone matrix application to cell nuclei classification. Available online: https://pdfs.semanticscholar.org/fec6/bd9b7f5d6a50410109991857494c8d25f290.pdf (accessed on 14 June 2019).
- Chaddad, A.; Desrosiers, C.; Toews, M.; Abdulkarim, B.; Chaddad, A.; Desrosiers, C.; Toews, M.; Abdulkarim, B. Predicting survival time of lung cancer patients using radiomic analysis. Oncotarget 2017, 8, 104393–104407. [Google Scholar] [CrossRef]
- Chaddad, A.; Desrosiers, C.; Hassan, L.; Tanougast, C. Hippocampus and amygdala radiomic biomarkers for the study of autism spectrum disorder. BMC Neurosci. 2017, 18, 52. [Google Scholar] [CrossRef]
- Pratt, J.W. Remarks on Zeros and Ties in the Wilcoxon Signed Rank Procedures. J. Am. Stat. Assoc. 1959, 54, 655–667. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Kleinbaum, D.G.; Klein, M. Kaplan-Meier Survival Curves and the Log-Rank Test. In Survival Analysis; Statistics for Biology and Health; Springer: New York, NY, USA, 2012; pp. 55–96. ISBN 978-1-4419-6645-2. [Google Scholar]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Ishwaran, H.; Gerds, T.A.; Kogalur, U.B.; Moore, R.D.; Gange, S.J.; Lau, B.M. Random survival forests for competing risks. Biostatistics 2014, 15, 757–773. [Google Scholar] [CrossRef]
- Marshall, R.J. The use of classification and regression trees in clinical epidemiology. J. Clin. Epidemiol. 2001, 54, 603–609. [Google Scholar] [CrossRef]
- Hanley, J.A.; McNeil, B.J. The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology 1982, 143, 29–36. [Google Scholar] [CrossRef]
- Sumi, N.S.; Islam, M.A.; Hossain, M.A. Evaluation and Computation of Diagnostic Tests: A Simple Alternative. Bull. Malays. Math. Sci. Soc. 2014, 37, 411–423. [Google Scholar]

| TCGA/TCIA | MUHC Site | |||
|---|---|---|---|---|
| (N = 73) | (N = 132) | |||
| Number | % | Number | % | |
| Age | ||||
| Median (min–max) | 61(18–84) | 62 (22–84) | ||
| ≤65 | 48 | 65.75 | 84 | 63.63 |
| >65 | 25 | 34.25 | 48 | 36.36 |
| Sex | ||||
| Female | 30 | 41.09 | 73 | 55.30 |
| Male | 43 | 58.90 | 59 | 44.69 |
| KPS | ||||
| <70 | 13 | 17.80 | 14 | 10.60 |
| ≥70 | 47 | 64.38 | 95 | 71.96 |
| Unknown | 13 | 17.80 | 23 | 17.42 |
| MGMT methylation | ||||
| Methylated | 24 | 32.87 | 44 | 33.33 |
| Unmethylated | 23 | 31.50 | 58 | 43.93 |
| Unknown | 26 | 35.61 | 30 | 22.72 |
| IDH1 | ||||
| Mutation R132H | 2 | 2.73 | 3 | 2.27 |
| Wild type | 71 | 97.26 | 129 | 97.72 |
| Extent of Surgery | ||||
| Subtotal resection | NA | - | 87 | 65.90 |
| Gross total | NA | - | 45 | 34.09 |
| Radiation treatment | ||||
| Yes | 61 | 83.56 | 116 | 87.87 |
| No | 9 | 12.32 | 15 | 11.36 |
| NA | 3 | 4.10 | 1 | 0.75 |
| Chemotherapy | ||||
| Yes | 53 | 72.60 | 90 | 68.18 |
| No | 20 | 27.39 | 38 | 28.78 |
| NA | 0 | 0 | 4 | 3.03 |
| Survival (censored) | ||||
| Median (months) | 12.06 | 13.75 | ||
| <1year | 37 (3) | 50.68 | 59 (0) | 43.70 |
| 1–4 years | 31 (1) | 42.46 | 67 (4) | 50.75 |
| >4 years | 5 (2) | 6.84 | 6 (3) | 4.54 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chaddad, A.; Daniel, P.; Sabri, S.; Desrosiers, C.; Abdulkarim, B. Integration of Radiomic and Multi-omic Analyses Predicts Survival of Newly Diagnosed IDH1 Wild-Type Glioblastoma. Cancers 2019, 11, 1148. https://doi.org/10.3390/cancers11081148
Chaddad A, Daniel P, Sabri S, Desrosiers C, Abdulkarim B. Integration of Radiomic and Multi-omic Analyses Predicts Survival of Newly Diagnosed IDH1 Wild-Type Glioblastoma. Cancers. 2019; 11(8):1148. https://doi.org/10.3390/cancers11081148
Chicago/Turabian StyleChaddad, Ahmad, Paul Daniel, Siham Sabri, Christian Desrosiers, and Bassam Abdulkarim. 2019. "Integration of Radiomic and Multi-omic Analyses Predicts Survival of Newly Diagnosed IDH1 Wild-Type Glioblastoma" Cancers 11, no. 8: 1148. https://doi.org/10.3390/cancers11081148
APA StyleChaddad, A., Daniel, P., Sabri, S., Desrosiers, C., & Abdulkarim, B. (2019). Integration of Radiomic and Multi-omic Analyses Predicts Survival of Newly Diagnosed IDH1 Wild-Type Glioblastoma. Cancers, 11(8), 1148. https://doi.org/10.3390/cancers11081148