Therapeutic Targeting of Stat3 Using Lipopolyplex Nanoparticle-Formulated siRNA in a Syngeneic Orthotopic Mouse Glioma Model
Abstract
1. Introduction
2. Results
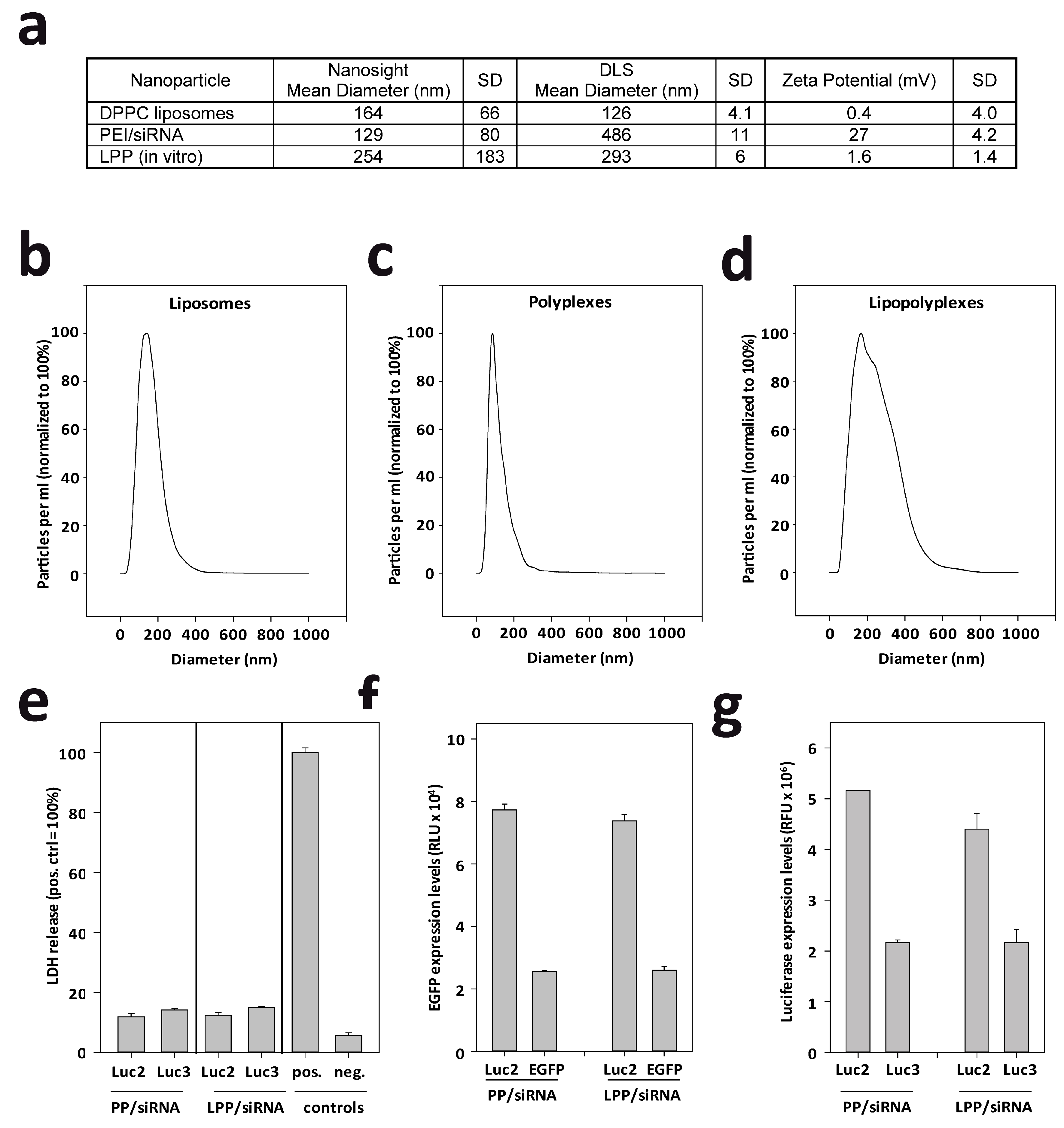
2.1. LPP Nanoparticles Provide a Stable and Efficient siRNA Formulation
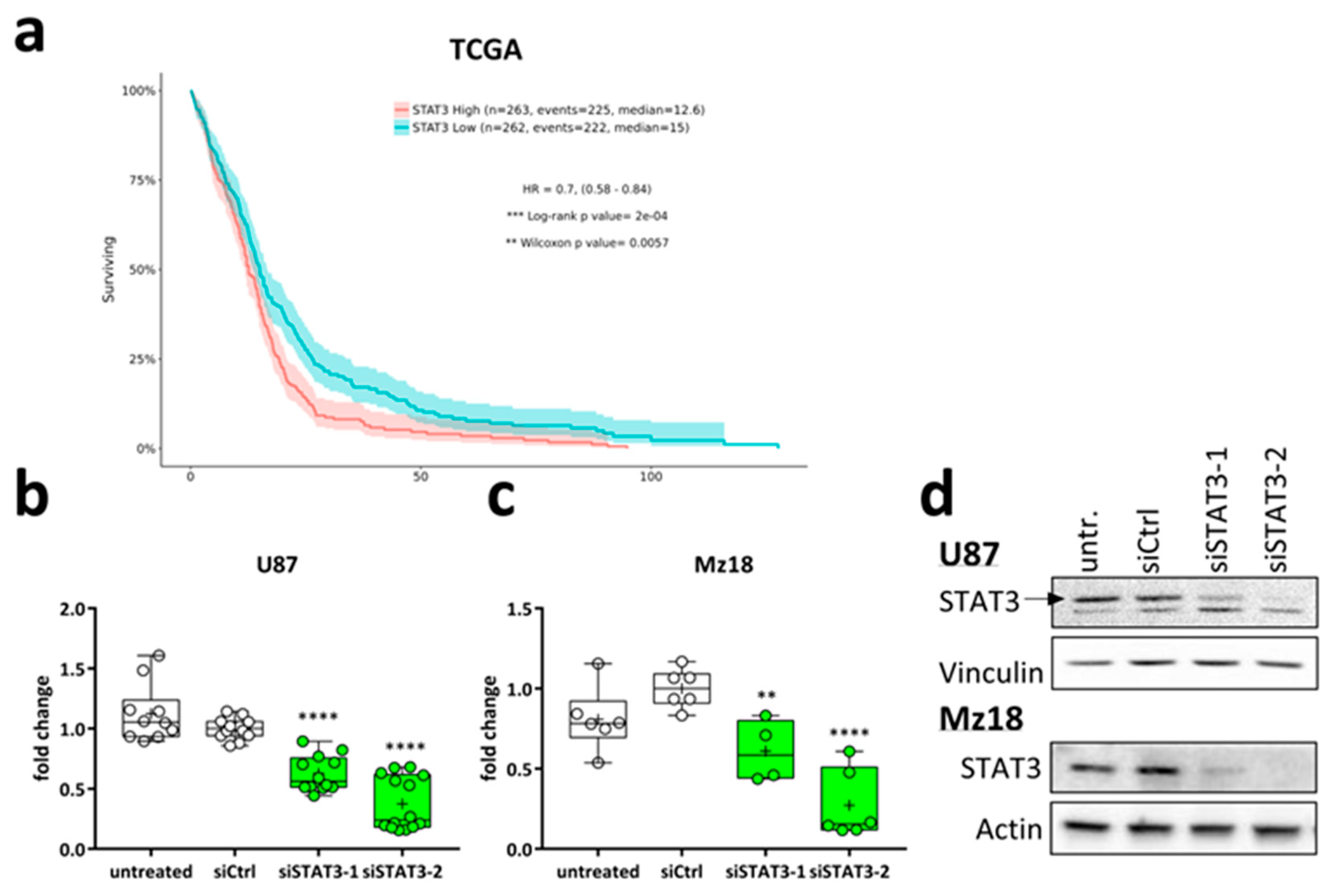
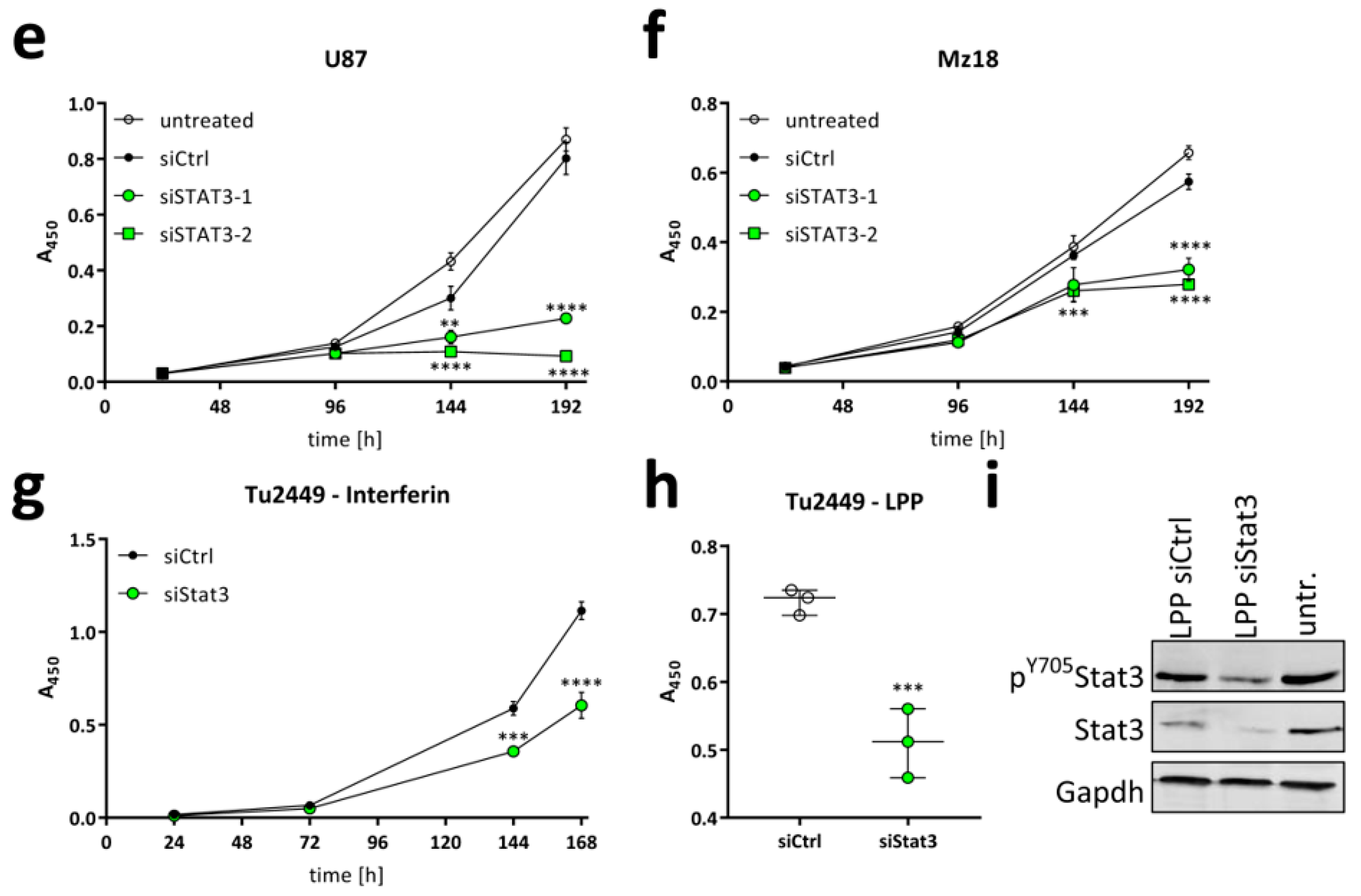
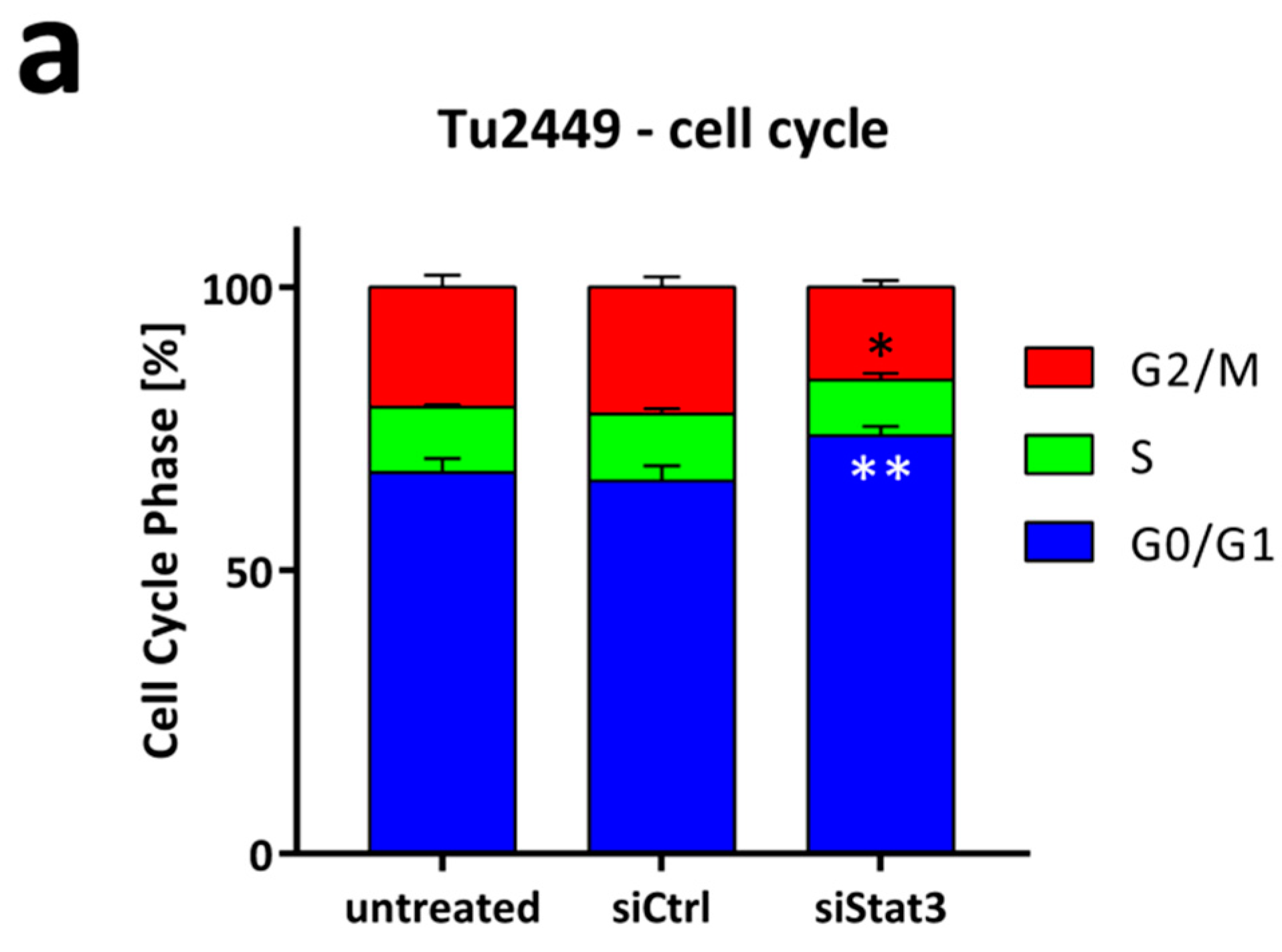
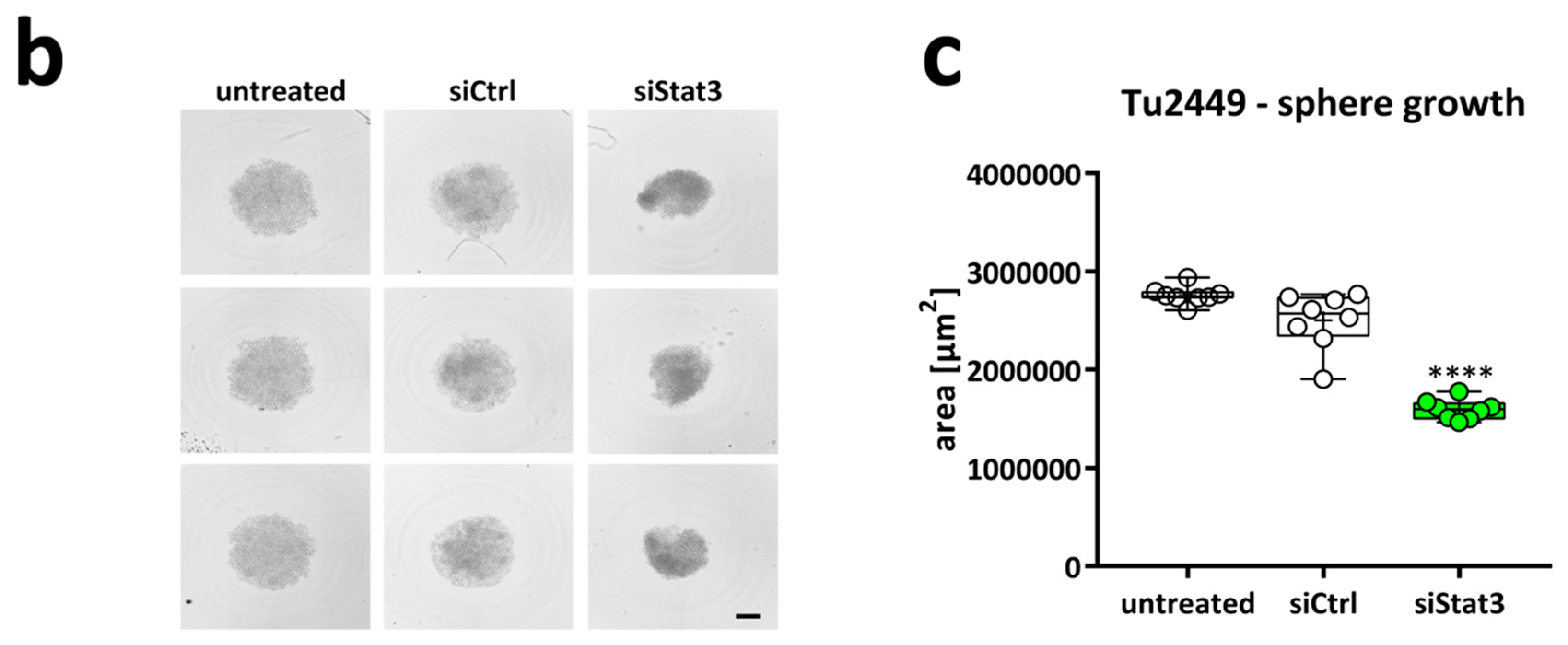
2.2. Targeting STAT3 Reduces Cancer Cell Proliferation In Vitro
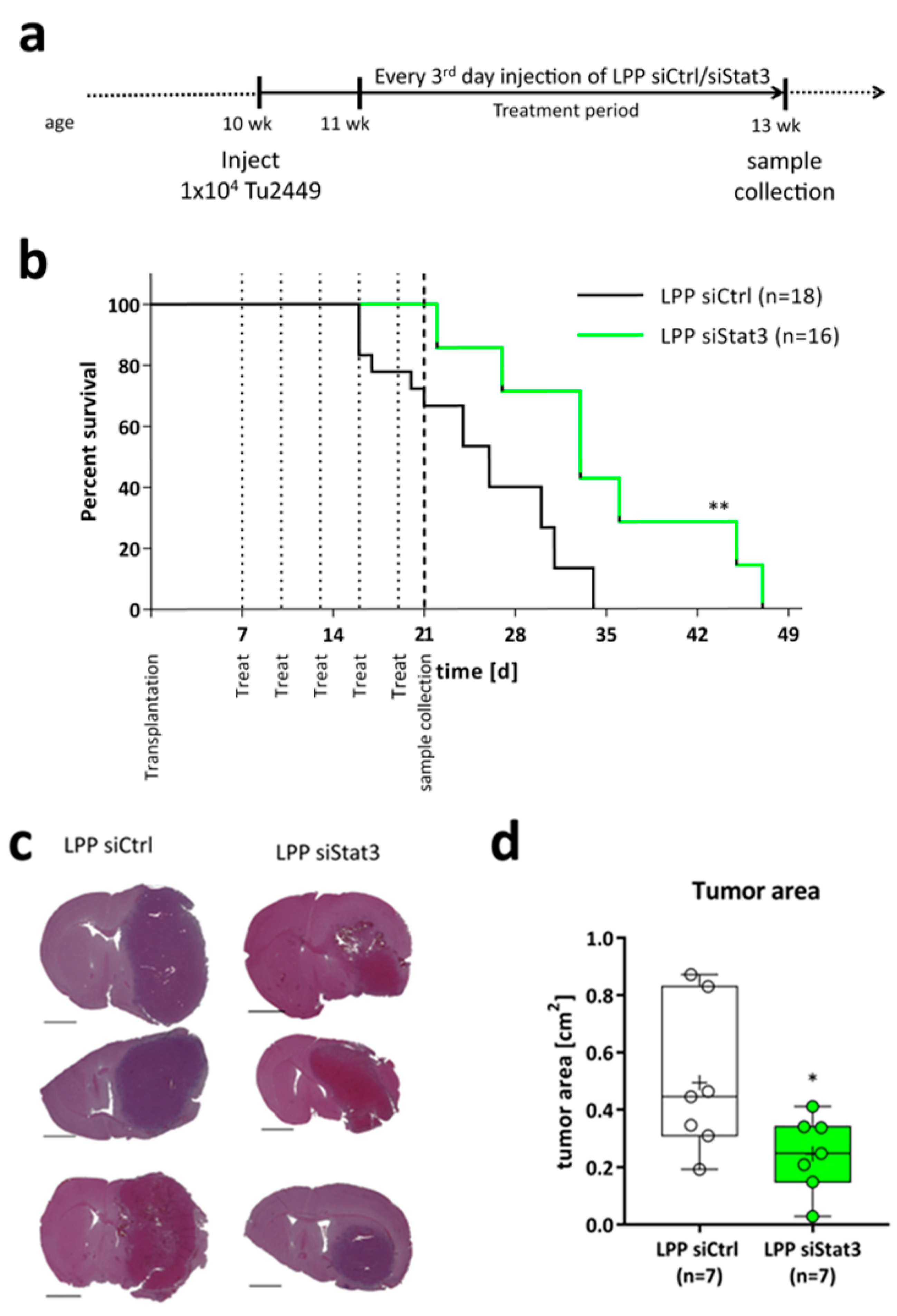
2.3. LPP siStat3 Prolongs Overall Survival and Reduces Tumor Size in Glioma-Bearing Mice
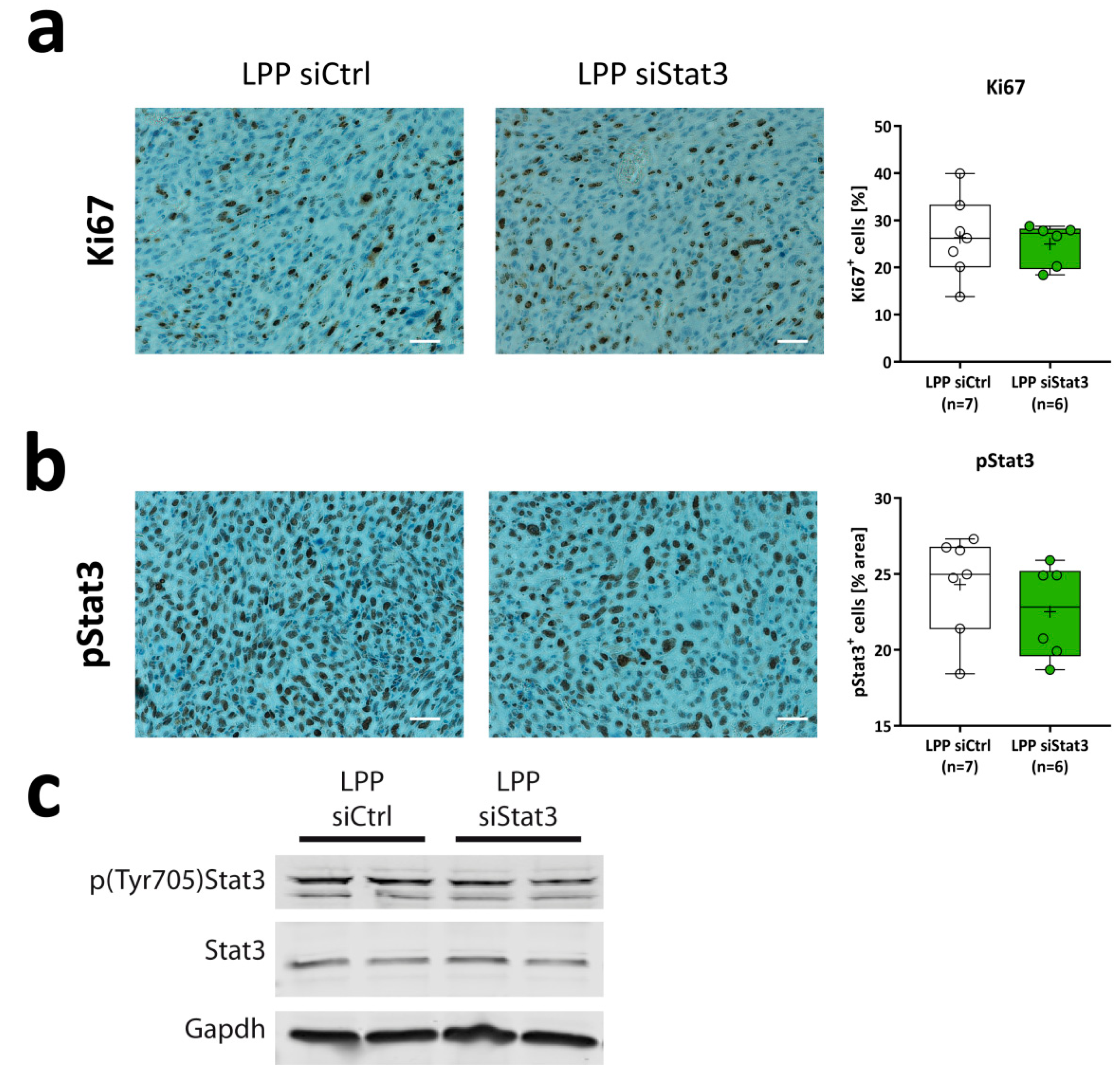
2.4. LPP siStat3 Slightly Reduces Stat3 Activation, But Does Not Affect Gross Tumor Proliferation Rates In Vivo
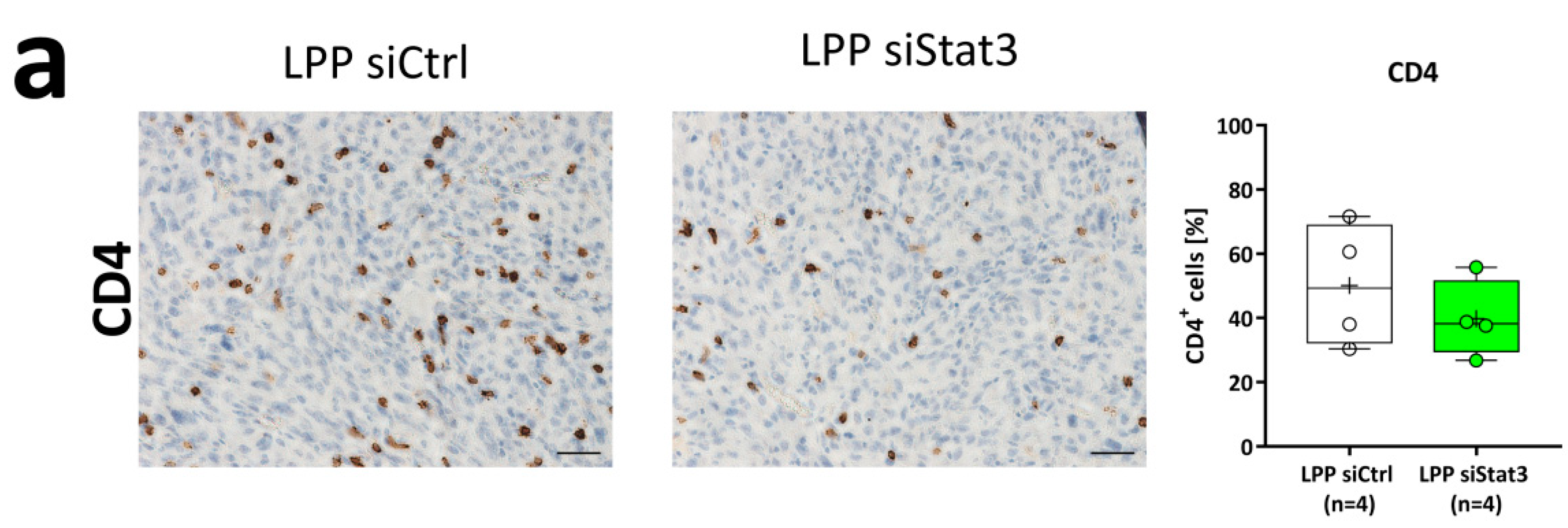
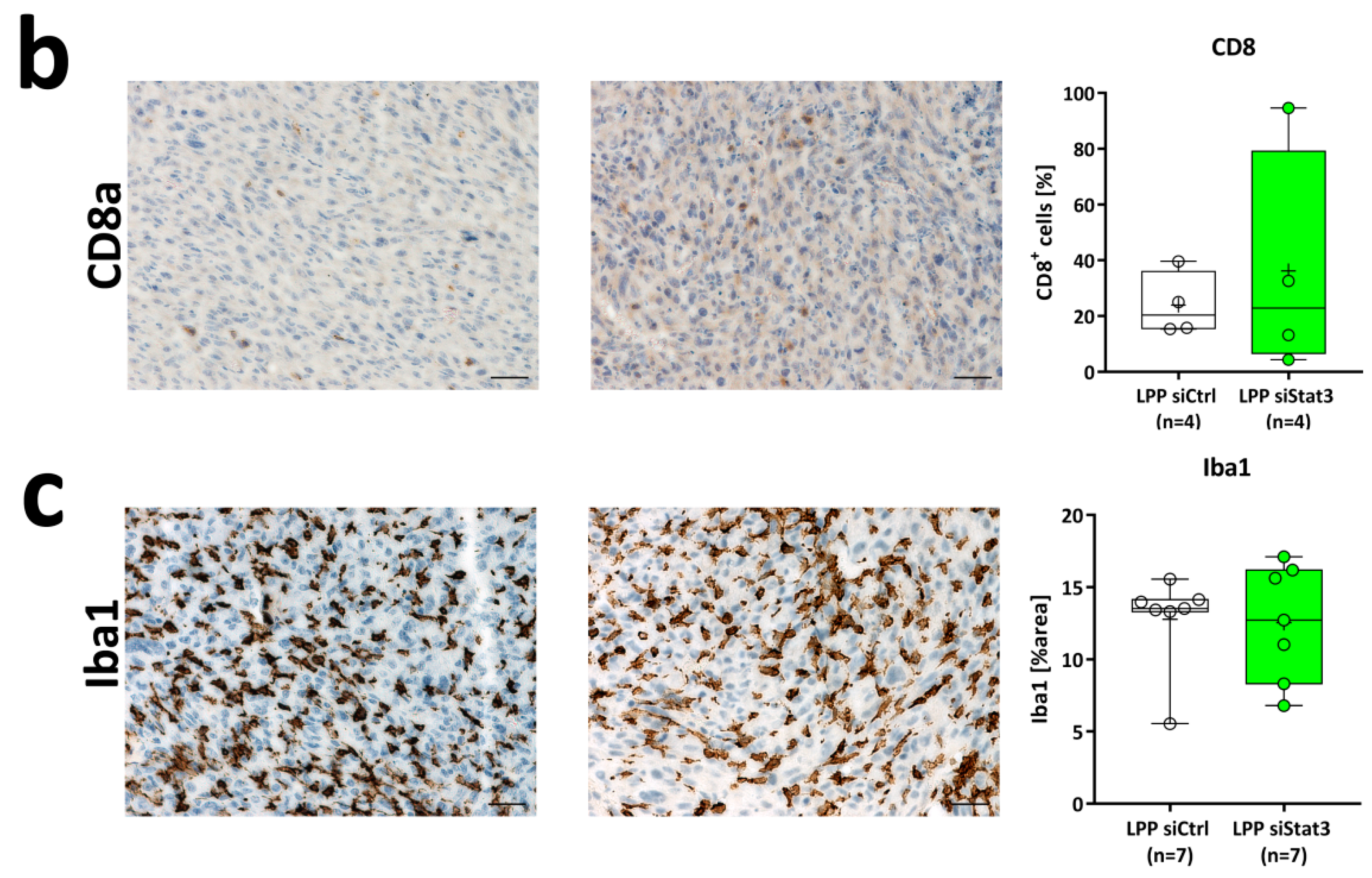
2.5. LPP siStat3 Reduces Stat3 Expression in the Core Region of the Tumor
3. Discussion
4. Materials and Methods
4.1. Analysis of TCGA Data Sets
4.2. Cells and Compounds
4.3. siRNAs + Sequences
4.4. Polyplex and Lipopolyplex Preparation
4.5. qRT-PCR
4.6. Western Blotting
4.7. Antibodies
4.8. Nanoparticle Characterization
4.9. Cell Transfection
4.10. Determination of Knockdown Efficacies
4.11. Cell Proliferation and Viability Assays
4.12. Cell Cycle Analysis
4.13. Spheroid Assay
4.14. Animal Experiments
4.15. Determination of Tumor Size
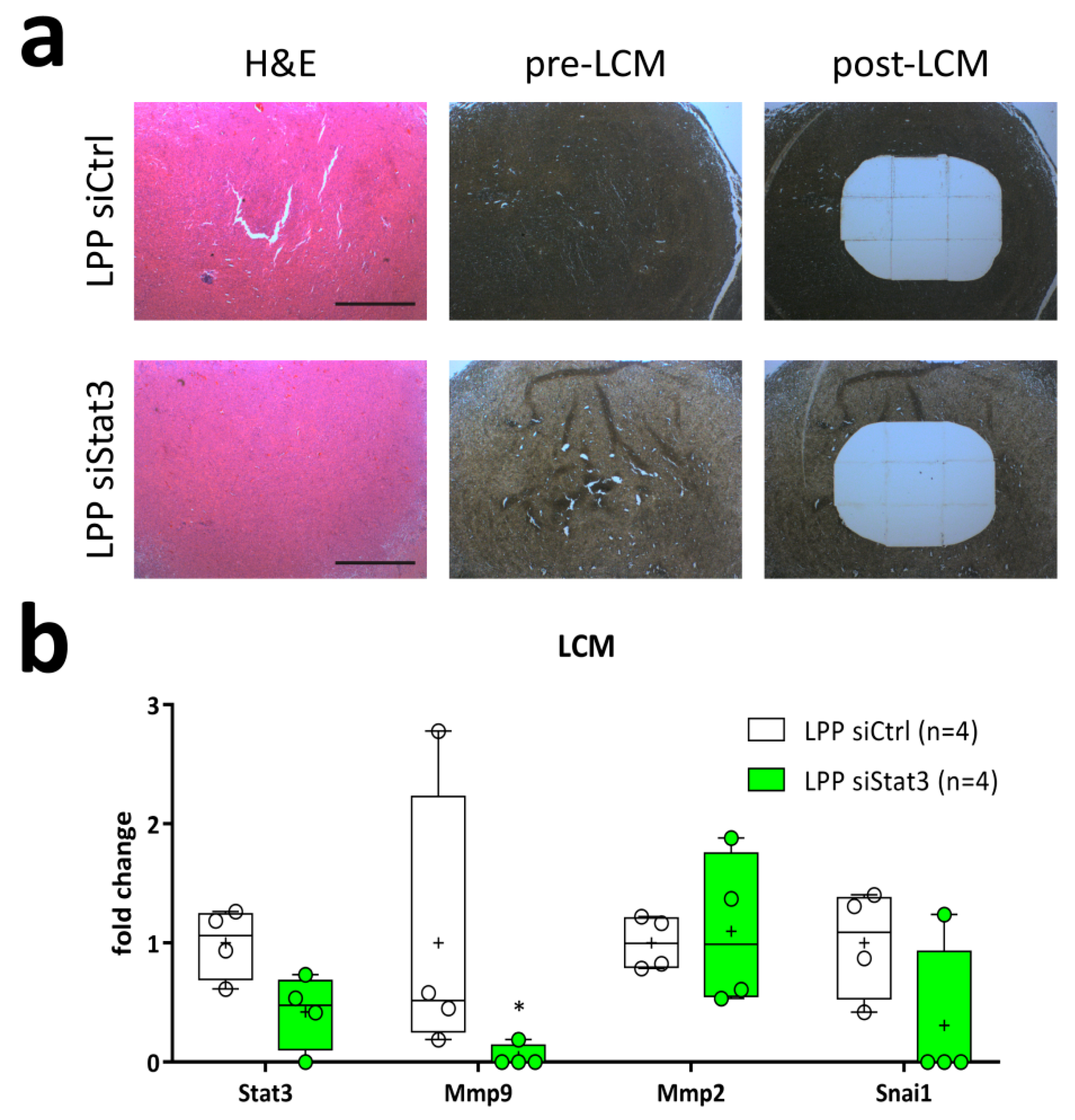
4.16. Laser-Capture Microdissection
4.17. Statistics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Louis, D.N.; Perry, A.; Reifenberger, G.; von Deimling, A.; Figarella-Branger, D.; Cavenee, W.K.; Ohgaki, H.; Wiestler, O.D.; Kleihues, P.; Ellison, D.W. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: A summary. Acta Neuropathol. 2016, 131, 803–820. [Google Scholar] [CrossRef]
- Becker, K.P.; Yu, J. Status quo–standard-of-care medical and radiation therapy for glioblastoma. Cancer J. 2012, 18, 12–19. [Google Scholar] [CrossRef]
- Verhaak, R.G.; Hoadley, K.A.; Purdom, E.; Wang, V.; Qi, Y.; Wilkerson, M.D.; Miller, C.R.; Ding, L.; Golub, T.; Mesirov, J.P.; et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 2010, 17, 98–110. [Google Scholar] [CrossRef]
- Olar, A.; Aldape, K.D. Using the molecular classification of glioblastoma to inform personalized treatment. J. Pathol. 2014, 232, 165–177. [Google Scholar] [CrossRef]
- Carro, M.S.; Lim, W.K.; Alvarez, M.J.; Bollo, R.J.; Zhao, X.; Snyder, E.Y.; Sulman, E.P.; Anne, S.L.; Doetsch, F.; Colman, H.; et al. The transcriptional network for mesenchymal transformation of brain tumours. Nature 2010, 463, 318–325. [Google Scholar] [CrossRef]
- Hossain, A.; Gumin, J.; Gao, F.; Figueroa, J.; Shinojima, N.; Takezaki, T.; Priebe, W.; Villarreal, D.; Kang, S.G.; Joyce, C.; et al. Mesenchymal Stem Cells Isolated From Human Gliomas Increase Proliferation and Maintain Stemness of Glioma Stem Cells Through the IL-6/gp130/STAT3 Pathway. Stem. Cells 2015, 33, 2400–2415. [Google Scholar] [CrossRef]
- Yu, H.; Lee, H.; Herrmann, A.; Buettner, R.; Jove, R. Revisiting STAT3 signalling in cancer: New and unexpected biological functions. Nat. Rev. Cancer 2014, 14, 736–746. [Google Scholar] [CrossRef]
- Galoczova, M.; Coates, P.; Vojtesek, B. STAT3, stem cells, cancer stem cells and p63. Cell. Mol. Biol. Lett. 2018, 23, 12. [Google Scholar] [CrossRef]
- Kim, J.E.; Patel, M.; Ruzevick, J.; Jackson, C.M.; Lim, M. STAT3 Activation in Glioblastoma: Biochemical and Therapeutic Implications. Cancers (Basel) 2014, 6, 376–395. [Google Scholar] [CrossRef]
- Brantley, E.C.; Benveniste, E.N. Signal transducer and activator of transcription-3: A molecular hub for signaling pathways in gliomas. Mol. Cancer Res. 2008, 6, 675–684. [Google Scholar] [CrossRef]
- Ganguly, D.; Fan, M.; Yang, C.H.; Zbytek, B.; Finkelstein, D.; Roussel, M.F.; Pfeffer, L.M. The critical role that STAT3 plays in glioma-initiating cells: STAT3 addiction in glioma. Oncotarget 2018, 9, 22095–22112. [Google Scholar] [CrossRef]
- Luwor, R.B.; Baradaran, B.; Taylor, L.E.; Iaria, J.; Nheu, T.V.; Amiry, N.; Hovens, C.M.; Wang, B.; Kaye, A.H.; Zhu, H.J. Targeting Stat3 and Smad7 to restore TGF-beta cytostatic regulation of tumor cells in vitro and in vivo. Oncogene 2013, 32, 2433–2441. [Google Scholar] [CrossRef]
- Westermarck, J.; Kahari, V.M. Regulation of matrix metalloproteinase expression in tumor invasion. FASEB J. 1999, 13, 781–792. [Google Scholar] [CrossRef]
- Wendt, M.K.; Balanis, N.; Carlin, C.R.; Schiemann, W.P. STAT3 and epithelial-mesenchymal transitions in carcinomas. JAKSTAT 2014, 3, e28975. [Google Scholar] [CrossRef]
- Kudo-Saito, C.; Shirako, H.; Takeuchi, T.; Kawakami, Y. Cancer metastasis is accelerated through immunosuppression during Snail-induced EMT of cancer cells. Cancer Cell 2009, 15, 195–206. [Google Scholar] [CrossRef]
- Avalle, L.; Camporeale, A.; Camperi, A.; Poli, V. STAT3 in cancer: A double edged sword. Cytokine 2017, 98, 42–50. [Google Scholar] [CrossRef]
- Liu, Y.; Li, C.; Lin, J. STAT3 as a Therapeutic Target for Glioblastoma. Anticancer Agents Med. Chem 2010, 10, 512–519. [Google Scholar] [CrossRef]
- Li, W.; Graeber, M.B. The molecular profile of microglia under the influence of glioma. Neuro Oncol. 2012, 14, 958–978. [Google Scholar] [CrossRef]
- Esposito, C.L.; Nuzzo, S.; Catuogno, S.; Romano, S.; de Nigris, F.; de Franciscis, V. STAT3 Gene Silencing by Aptamer-siRNA Chimera as Selective Therapeutic for Glioblastoma. Mol. Nucleic Acids 2018, 10, 398–411. [Google Scholar] [CrossRef]
- Kiprianova, I.; Remy, J.; Milosch, N.; Mohrenz, I.V.; Seifert, V.; Aigner, A.; Kogel, D. Sorafenib Sensitizes Glioma Cells to the BH3 Mimetic ABT-737 by Targeting MCL1 in a STAT3-Dependent Manner. Neoplasia 2015, 17, 564–573. [Google Scholar] [CrossRef]
- Priester, M.; Copanaki, E.; Vafaizadeh, V.; Hensel, S.; Bernreuther, C.; Glatzel, M.; Seifert, V.; Groner, B.; Kogel, D.; Weissenberger, J. STAT3 silencing inhibits glioma single cell infiltration and tumor growth. Neuro Oncol. 2013, 15, 840–852. [Google Scholar] [CrossRef]
- Senft, C.; Priester, M.; Polacin, M.; Schroder, K.; Seifert, V.; Kogel, D.; Weissenberger, J. Inhibition of the JAK-2/STAT3 signaling pathway impedes the migratory and invasive potential of human glioblastoma cells. J. Neurooncol. 2011, 101, 393–403. [Google Scholar] [CrossRef]
- Sherry, M.M.; Reeves, A.; Wu, J.L.K.; Cochran, B.H. STAT3 Is Required for Proliferation and Maintenance of Multipotency in Glioblastoma Stem Cells. Stem. Cells 2009, 27, 2383–2392. [Google Scholar] [CrossRef]
- Takeda, K.; Kaisho, T.; Yoshida, N.; Takeda, J.; Kishimoto, T.; Akira, S. Stat3 activation is responsible for IL-6-dependent T cell proliferation through preventing apoptosis: Generation and characterization of T cell-specific Stat3-deficient mice. J. Immunol. 1998, 161, 4652–4660. [Google Scholar] [CrossRef]
- Aigner, A. Applications of RNA interference: Current state and prospects for siRNA-based strategies in vivo. Appl. Microbiol. Biotechnol. 2007, 76, 9–21. [Google Scholar] [CrossRef]
- Aigner, A.; Kogel, D. Nanoparticle/siRNA-based therapy strategies in glioma: Which nanoparticles, which siRNAs? Nanomedicine (London) 2018, 13, 89–103. [Google Scholar] [CrossRef]
- Hobel, S.; Koburger, I.; John, M.; Czubayko, F.; Hadwiger, P.; Vornlocher, H.P.; Aigner, A. Polyethylenimine/small interfering RNA-mediated knockdown of vascular endothelial growth factor in vivo exerts anti-tumor effects synergistically with Bevacizumab. J. Gene Med. 2010, 12, 287–300. [Google Scholar]
- Hendruschk, S.; Wiedemuth, R.; Aigner, A.; Topfer, K.; Cartellieri, M.; Martin, D.; Kirsch, M.; Ikonomidou, C.; Schackert, G.; Temme, A. RNA interference targeting survivin exerts antitumoral effects in vitro and in established glioma xenografts in vivo. Neuro Oncol. 2011, 13, 1074–1089. [Google Scholar] [CrossRef]
- Ibrahim, A.F.; Weirauch, U.; Thomas, M.; Grunweller, A.; Hartmann, R.K.; Aigner, A. MicroRNA replacement therapy for miR-145 and miR-33a is efficacious in a model of colon carcinoma. Cancer Res. 2011, 71, 5214–5224. [Google Scholar] [CrossRef]
- Hampl, V.; Martin, C.; Aigner, A.; Hoebel, S.; Singer, S.; Frank, N.; Sarikas, A.; Ebert, O.; Prywes, R.; Gudermann, T.; et al. Depletion of the transcriptional coactivators megakaryoblastic leukaemia 1 and 2 abolishes hepatocellular carcinoma xenograft growth by inducing oncogene-induced senescence. EMBO Mol. Med. 2013, 5, 1367–1382. [Google Scholar] [CrossRef]
- Hobel, S.; Aigner, A. Polyethylenimines for siRNA and miRNA delivery in vivo. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2013, 5, 484–501. [Google Scholar] [CrossRef]
- Ewe, A.; Panchal, O.; Pinnapireddy, S.R.; Bakowsky, U.; Przybylski, S.; Temme, A.; Aigner, A. Liposome-polyethylenimine complexes (DPPC-PEI lipopolyplexes) for therapeutic siRNA delivery in vivo. Nanomedicine 2017, 13, 209–218. [Google Scholar] [CrossRef]
- Heyes, J.; Palmer, L.; Chan, K.; Giesbrecht, C.; Jeffs, L.; MacLachlan, I. Lipid encapsulation enables the effective systemic delivery of polyplex plasmid DNA. Molcure 2007, 15, 713–720. [Google Scholar] [CrossRef]
- Ko, Y.T.; Kale, A.; Hartner, W.C.; Papahadjopoulos-Sternberg, B.; Torchilin, V.P. Self-assembling micelle-like nanoparticles based on phospholipid-polyethyleneimine conjugates for systemic gene delivery. J. Control. Release 2009, 133, 132–138. [Google Scholar] [CrossRef]
- Ewe, A.; Schaper, A.; Barnert, S.; Schubert, R.; Temme, A.; Bakowsky, U.; Aigner, A. Storage stability of optimal liposome-polyethylenimine complexes (lipopolyplexes) for DNA or siRNA delivery. Acta Biomater. 2014, 10, 2663–2673. [Google Scholar] [CrossRef]
- Werth, S.; Urban-Klein, B.; Dai, L.; Hobel, S.; Grzelinski, M.; Bakowsky, U.; Czubayko, F.; Aigner, A. A low molecular weight fraction of polyethylenimine (PEI) displays increased transfection efficiency of DNA and siRNA in fresh or lyophilized complexes. J. Control. Release 2006, 112, 257–270. [Google Scholar] [CrossRef]
- Schafer, J.; Hobel, S.; Bakowsky, U.; Aigner, A. Liposome-polyethylenimine complexes for enhanced DNA and siRNA delivery. Biomaterials 2010, 31, 6892–6900. [Google Scholar] [CrossRef]
- Ewe, A.; Aigner, A. Cationic Lipid-Coated Polyplexes (Lipopolyplexes) for DNA and Small RNA Delivery. Methods Mol. Biol. 2016, 1445, 187–200. [Google Scholar]
- Dasgupta, A.; Raychaudhuri, B.; Haqqi, T.; Prayson, R.; Van Meir, E.G.; Vogelbaum, M.; Haque, S.J. Stat3 activation is required for the growth of U87 cell-derived tumours in mice. Eur. J. Cancer 2009, 45, 677–684. [Google Scholar] [CrossRef]
- The Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 2008, 455, 1061–1068. [Google Scholar]
- Smilowitz, H.M.; Weissenberger, J.; Weis, J.; Brown, J.D.; O’Neill, R.J.; Laissue, J.A. Orthotopic transplantation of v-src-expressing glioma cell lines into immunocompetent mice: Establishment of a new transplantable in vivo model for malignant glioma. J. Neurosurg. 2007, 106, 652–659. [Google Scholar] [CrossRef] [PubMed]
- Stechishin, O.D.; Luchman, H.A.; Ruan, Y.; Blough, M.D.; Nguyen, S.A.; Kelly, J.J.; Cairncross, J.G.; Weiss, S. On-target JAK2/STAT3 inhibition slows disease progression in orthotopic xenografts of human glioblastoma brain tumor stem cells. Neuro Oncol. 2013, 15, 198–207. [Google Scholar] [CrossRef]
- Doucette, T.A.; Kong, L.Y.; Yang, Y.; Ferguson, S.D.; Yang, J.; Wei, J.; Qiao, W.; Fuller, G.N.; Bhat, K.P.; Aldape, K.; et al. Signal transducer and activator of transcription 3 promotes angiogenesis and drives malignant progression in glioma. Neuro Oncol. 2012, 14, 1136–1145. [Google Scholar] [CrossRef] [PubMed]
- Weissenberger, J.; Priester, M.; Bernreuther, C.; Rakel, S.; Glatzel, M.; Seifert, V.; Kogel, D. Dietary curcumin attenuates glioma growth in a syngeneic mouse model by inhibition of the JAK1,2/STAT3 signaling pathway. Clin. Cancer Res. 2010, 16, 5781–5795. [Google Scholar] [CrossRef] [PubMed]
- Ewe, A.; Aigner, A. Nebulization of liposome–polyethylenimine complexes (lipopolyplexes) for DNA or siRNA delivery: Physicochemical properties and biological activity. Eur. J. Lipid Sci. Technol. 2014, 116, 1195–1204. [Google Scholar] [CrossRef]
- Planas, A.M.; Soriano, M.A.; Berruezo, M.; Justicia, C.; Estrada, A.; Pitarch, S.; Ferrer, I. Induction of Stat3, a signal transducer and transcription factor, in reactive microglia following transient focal cerebral ischaemia. Eur. J. Neurosci. 1996, 8, 2612–2618. [Google Scholar] [CrossRef]
- Rodriguez, M.; Lapierre, J.; Ojha, C.R.; Kaushik, A.; Batrakova, E.; Kashanchi, F.; Dever, S.M.; Nair, M.; El-Hage, N. Intranasal drug delivery of small interfering RNA targeting Beclin1 encapsulated with polyethylenimine (PEI) in mouse brain to achieve HIV attenuation. Sci. Rep. 2017, 7, 1862. [Google Scholar] [CrossRef]
- Matsuda, M.; Yamamoto, T.; Matsumura, A.; Kaneda, Y. Highly efficient eradication of intracranial glioblastoma using Eg5 siRNA combined with HVJ envelope. Gene 2009, 16, 1465–1476. [Google Scholar] [CrossRef]
- Gillespie, D.L.; Aguirre, M.T.; Ravichandran, S.; Leishman, L.L.; Berrondo, C.; Gamboa, J.T.; Wang, L.; King, R.; Wang, X.; Tan, M.; et al. RNA interference targeting hypoxia-inducible factor 1alpha via a novel multifunctional surfactant attenuates glioma growth in an intracranial mouse model. J. Neurosurg. 2015, 122, 331–341. [Google Scholar] [CrossRef]
- Michiue, H.; Eguchi, A.; Scadeng, M.; Dowdy, S.F. Induction of in vivo synthetic lethal RNAi responses to treat glioblastoma. Cancer Biol. 2009, 8, 2306–2313. [Google Scholar] [CrossRef]
- Merz, L.; Hobel, S.; Kallendrusch, S.; Ewe, A.; Bechmann, I.; Franke, H.; Merz, F.; Aigner, A. Tumor tissue slice cultures as a platform for analyzing tissue-penetration and biological activities of nanoparticles. Eur. J. Pharm. Biopharm. 2017, 112, 45–50. [Google Scholar] [CrossRef]
- Helmschrodt, C.; Hobel, S.; Schoniger, S.; Bauer, A.; Bonicelli, J.; Gringmuth, M.; Fietz, S.A.; Aigner, A.; Richter, A.; Richter, F. Polyethylenimine Nanoparticle-Mediated siRNA Delivery to Reduce alpha-Synuclein Expression in a Model of Parkinson’s Disease. Mol. Nucleic Acids 2017, 9, 57–68. [Google Scholar] [CrossRef] [PubMed]
- Mathieson, T.; Franken, H.; Kosinski, J.; Kurzawa, N.; Zinn, N.; Sweetman, G.; Poeckel, D.; Ratnu, V.S.; Schramm, M.; Becher, I.; et al. Systematic analysis of protein turnover in primary cells. Nat. Commun. 2018, 9, 689. [Google Scholar] [CrossRef] [PubMed]
- Bocchini, C.E.; Nahmod, K.; Katsonis, P.; Kim, S.; Kasembeli, M.M.; Freeman, A.; Lichtarge, O.; Makedonas, G.; Tweardy, D.J. Protein stabilization improves STAT3 function in autosomal dominant hyper-IgE syndrome. Blood 2016, 128, 3061–3072. [Google Scholar] [CrossRef] [PubMed]
- Boisvert, F.M.; Ahmad, Y.; Gierlinski, M.; Charriere, F.; Lamont, D.; Scott, M.; Barton, G.; Lamond, A.I. A quantitative spatial proteomics analysis of proteome turnover in human cells. Mol. Cell. Proteom. 2012, 11, M111.011429. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.Y.; Ozawa, T.; Drummond, D.C.; Kalra, A.; Fitzgerald, J.B.; Kirpotin, D.B.; Wei, K.C.; Butowski, N.; Prados, M.D.; Berger, M.S.; et al. Comparing routes of delivery for nanoliposomal irinotecan shows superior anti-tumor activity of local administration in treating intracranial glioblastoma xenografts. Neuro Oncol. 2013, 15, 189–197. [Google Scholar] [CrossRef] [PubMed]
- Bruce, J.N.; Fine, R.L.; Canoll, P.; Yun, J.; Kennedy, B.C.; Rosenfeld, S.S.; Sands, S.A.; Surapaneni, K.; Lai, R.; Yanes, C.L.; et al. Regression of recurrent malignant gliomas with convection-enhanced delivery of topotecan. Neurosurgery 2011, 69, 1272–1279, discussion 1279–1280. [Google Scholar] [CrossRef] [PubMed]
- Jaschinski, F.; Rothhammer, T.; Jachimczak, P.; Seitz, C.; Schneider, A.; Schlingensiepen, K.H. The antisense oligonucleotide trabedersen (AP 12009) for the targeted inhibition of TGF-beta2. Curr. Pharm. Biotechnol. 2011, 12, 2203–2213. [Google Scholar] [CrossRef] [PubMed]
- Voges, J.; Reszka, R.; Gossmann, A.; Dittmar, C.; Richter, R.; Garlip, G.; Kracht, L.; Coenen, H.H.; Sturm, V.; Wienhard, K.; et al. Imaging-guided convection-enhanced delivery and gene therapy of glioblastoma. Ann. Neurol. 2003, 54, 479–487. [Google Scholar] [CrossRef] [PubMed]
- Yu, D.; Khan, O.F.; Suva, M.L.; Dong, B.; Panek, W.K.; Xiao, T.; Wu, M.; Han, Y.; Ahmed, A.U.; Balyasnikova, I.V.; et al. Multiplexed RNAi therapy against brain tumor-initiating cells via lipopolymeric nanoparticle infusion delays glioblastoma progression. Proc. Natl. Acad. Sci. USA 2017, 114, E6147–E6156. [Google Scholar] [CrossRef] [PubMed]
- Vogelbaum, M.A.; Brewer, C.; Barnett, G.H.; Mohammadi, A.M.; Peereboom, D.M.; Ahluwalia, M.S.; Gao, S. First-in-human evaluation of the Cleveland Multiport Catheter for convection-enhanced delivery of topotecan in recurrent high-grade glioma: Results of pilot trial 1. J. Neurosurg. 2018. [Google Scholar] [CrossRef] [PubMed]
- Ceccarelli, M.; Barthel, F.P.; Malta, T.M.; Sabedot, T.S.; Salama, S.R.; Murray, B.A.; Morozova, O.; Newton, Y.; Radenbaugh, A.; Pagnotta, S.M.; et al. Molecular Profiling Reveals Biologically Discrete Subsets and Pathways of Progression in Diffuse Glioma. Cell 2016, 164, 550–563. [Google Scholar] [CrossRef] [PubMed]
- Bowman, R.L.; Wang, Q.; Carro, A.; Verhaak, R.G.; Squatrito, M. GlioVis data portal for visualization and analysis of brain tumor expression datasets. Neuro Oncol. 2017, 19, 139–141. [Google Scholar] [CrossRef] [PubMed]
- Pohl, U.; Wick, W.; Weissenberger, J.; Steinbach, J.P.; Dichgans, J.; Aguzzi, A.; Weller, M. Characterization of Tu-2449, a glioma cell line derived from a spontaneous tumor in GFAP-v-src-transgenic mice: Comparison with established murine glioma cell lines. Int. J. Oncol. 1999, 15, 829–834. [Google Scholar] [CrossRef] [PubMed]
- Hetschko, H.; Voss, V.; Horn, S.; Seifert, V.; Prehn, J.H.; Kogel, D. Pharmacological inhibition of Bcl-2 family members reactivates TRAIL-induced apoptosis in malignant glioma. J. Neurooncol. 2008, 86, 265–272. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Antonietti, P.; Linder, B.; Hehlgans, S.; Mildenberger, I.C.; Burger, M.C.; Fulda, S.; Steinbach, J.P.; Gessler, F.; Rodel, F.; Mittelbronn, M.; et al. Interference with the HSF1/HSP70/BAG3 Pathway Primes Glioma Cells to Matrix Detachment and BH3 Mimetic-Induced Apoptosis. Mol. Cancer 2017, 16, 156–168. [Google Scholar] [CrossRef]
- Weissenberger, J.; Loeffler, S.; Kappeler, A.; Kopf, M.; Lukes, A.; Afanasieva, T.A.; Aguzzi, A.; Weis, J. IL-6 is required for glioma development in a mouse model. Oncogene 2004, 23, 3308–3316. [Google Scholar] [CrossRef]
- Schulze, J.; Hendrikx, S.; Schulz-Siegmund, M.; Aigner, A. Microparticulate poly(vinyl alcohol) hydrogel formulations for embedding and controlled release of polyethylenimine (PEI)-based nanoparticles. Acta Biomater. 2016, 45, 210–222. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Linder, B.; Weirauch, U.; Ewe, A.; Uhmann, A.; Seifert, V.; Mittelbronn, M.; Harter, P.N.; Aigner, A.; Kögel, D. Therapeutic Targeting of Stat3 Using Lipopolyplex Nanoparticle-Formulated siRNA in a Syngeneic Orthotopic Mouse Glioma Model. Cancers 2019, 11, 333. https://doi.org/10.3390/cancers11030333
Linder B, Weirauch U, Ewe A, Uhmann A, Seifert V, Mittelbronn M, Harter PN, Aigner A, Kögel D. Therapeutic Targeting of Stat3 Using Lipopolyplex Nanoparticle-Formulated siRNA in a Syngeneic Orthotopic Mouse Glioma Model. Cancers. 2019; 11(3):333. https://doi.org/10.3390/cancers11030333
Chicago/Turabian StyleLinder, Benedikt, Ulrike Weirauch, Alexander Ewe, Anja Uhmann, Volker Seifert, Michel Mittelbronn, Patrick N. Harter, Achim Aigner, and Donat Kögel. 2019. "Therapeutic Targeting of Stat3 Using Lipopolyplex Nanoparticle-Formulated siRNA in a Syngeneic Orthotopic Mouse Glioma Model" Cancers 11, no. 3: 333. https://doi.org/10.3390/cancers11030333
APA StyleLinder, B., Weirauch, U., Ewe, A., Uhmann, A., Seifert, V., Mittelbronn, M., Harter, P. N., Aigner, A., & Kögel, D. (2019). Therapeutic Targeting of Stat3 Using Lipopolyplex Nanoparticle-Formulated siRNA in a Syngeneic Orthotopic Mouse Glioma Model. Cancers, 11(3), 333. https://doi.org/10.3390/cancers11030333






