Role of miRNAs in Melanoma Metastasis
Abstract
1. Introduction
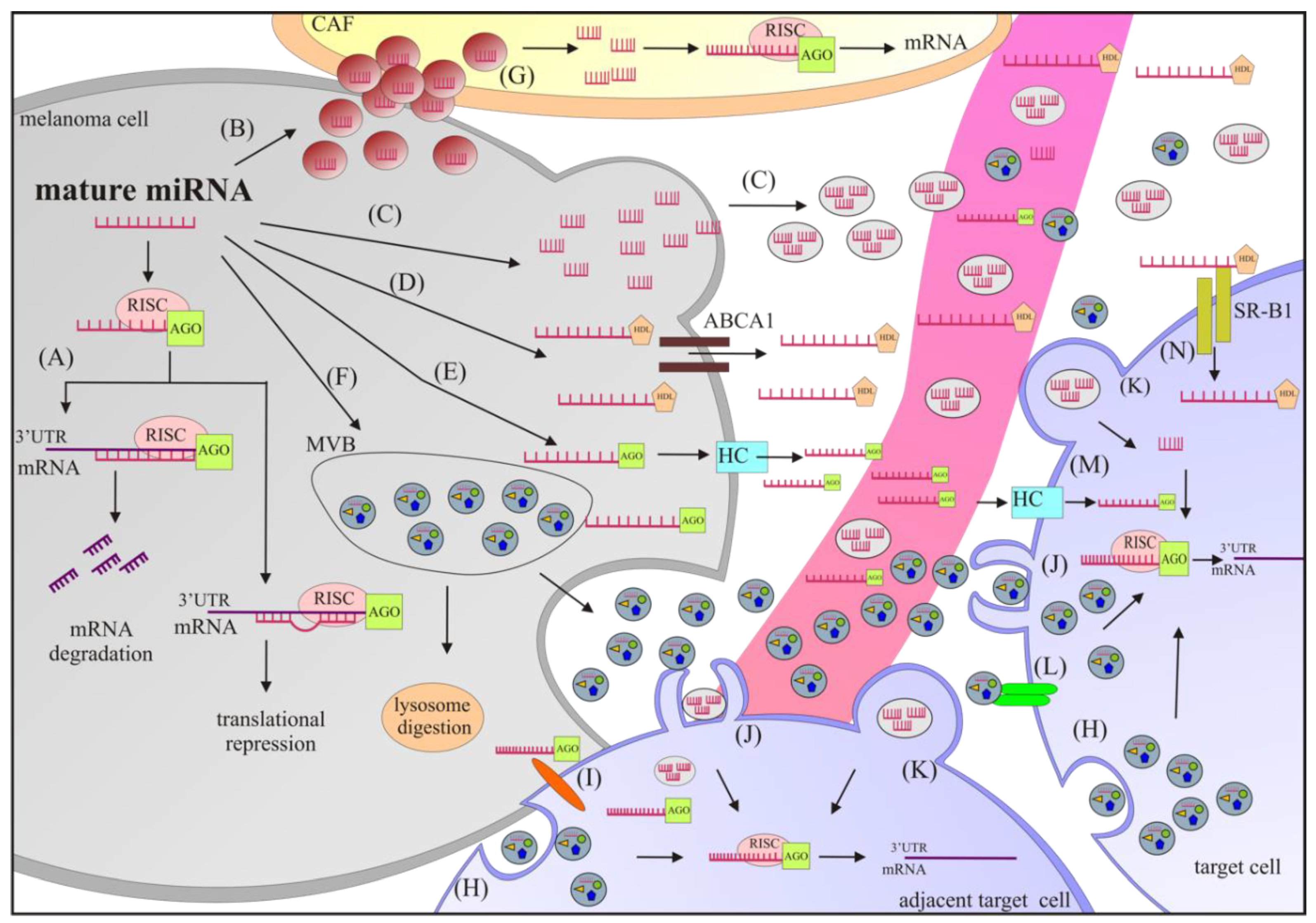
2. Biogenesis, Function, and Extracellular Transport of miRNAs
3. miRNAs Modulate the Tumour Niche within the Dermis
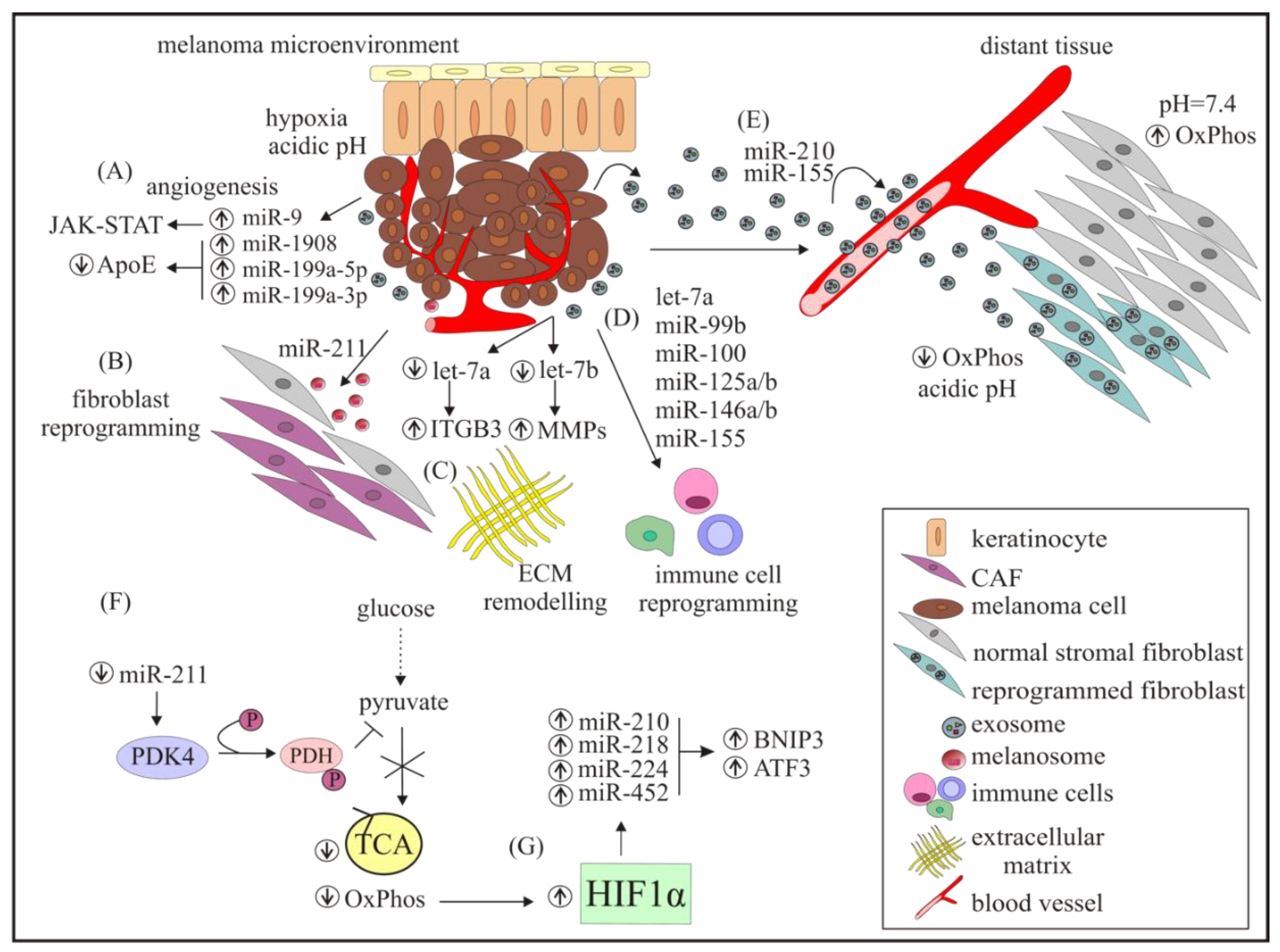
4. miRNAs Modulate Angiogenesis and Immune Response, Intravasation, and Extravasation of Melanoma Cells
5. miRNAs Modulate Melanoma Cell Invasion
6. miRNAs Modulate the Melanoma Metastatic Niche
7. Alterations in miRNAs Expression in Response to Microenvironmental Changes
8. miRNAs in Drug Resistance to Targeted Therapy and Immunotherapy
9. miRNAs as Biomarkers and Therapeutics
10. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Adler, N.R.; Haydon, A.; McLean, C.A.; Kelly, J.W.; Mar, V.J. Metastatic pathways in patients with cutaneous melanoma. Pigment Cell Melanoma Res. 2017, 30, 13–27. [Google Scholar] [CrossRef] [PubMed]
- Hayward, N.K.; Wilmott, J.S.; Waddell, N.; Johansson, P.A.; Field, M.A.; Nones, K.; Patch, A.M.; Kakavand, H.; Alexandrov, L.B.; Burke, H.; et al. Whole-genome landscapes of major melanoma subtypes. Nature 2017, 545, 175–180. [Google Scholar] [CrossRef] [PubMed]
- Alexandrov, L.B.; Nik-Zainal, S.; Wedge, D.C.; Aparicio, S.A.; Behjati, S.; Biankin, A.V.; Bignell, G.R.; Bolli, N.; Borg, A.; Børresen-Dale, A.L.; et al. Signatures of mutational processes in human cancer. Nature 2013, 500, 415–421. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Kelly, J.W.; Trivett, M.; Murray, W.K.; Dowling, J.P.; Wolfe, R.; Mason, G.; Magee, J.; Angel, C.; Dobrovic, A.; et al. Distinct clinical and pathological features are associated with the BRAF(T1799A(V600E)) mutation in primary melanoma. J. Investig. Dermatol. 2007, 127, 900–905. [Google Scholar] [CrossRef] [PubMed]
- Long, G.V.; Stroyakovskiy, D.; Gogas, H.; Levchenko, E.; de Braud, F.; Larkin, J.; Garbe, C.; Jouary, T.; Hauschild, A.; Grob, J.J.; et al. Combined BRAF and MEK inhibition versus BRAF inhibition alone in melanoma. N. Engl. J. Med. 2014, 371, 1877–1888. [Google Scholar] [CrossRef] [PubMed]
- Rizos, H.; Menzies, A.M.; Pupo, G.M.; Carlino, M.S.; Fung, C.; Hyman, J.; Haydu, L.E.; Mijatov, B.; Becker, T.M.; Boyd, S.C.; et al. BRAF inhibitor resistance mechanisms in metastatic melanoma: Spectrum and clinical impact. Clin. Cancer Res. 2014, 20, 1965–1977. [Google Scholar] [CrossRef] [PubMed]
- Gatzka, M.V. Targeted Tumor Therapy Remixed-An Update on the Use of Small-Molecule Drugs in Combination Therapies. Cancers (Basel) 2018, 10, 155. [Google Scholar] [CrossRef] [PubMed]
- Larkin, J.; Hodi, F.S.; Wolchok, J.D. Combined Nivolumab and Ipilimumab or Monotherapy in Untreated Melanoma. N. Engl. J. Med. 2015, 373, 1270–1271. [Google Scholar] [CrossRef] [PubMed]
- Robert, C.; Schachter, J.; Long, G.V.; Arance, A.; Grob, J.J.; Mortier, L.; Daud, A.; Carlino, M.S.; McNeil, C.; Lotem, M.; et al. KEYNOTE-006 investigators. Pembrolizumab versus Ipilimumab in Advanced Melanoma. N. Engl. J. Med. 2015, 372, 2521–2532. [Google Scholar] [CrossRef] [PubMed]
- Tucci, M.; Passarelli, A.; Mannavola, F.; Stucci, L.S.; Ascierto, P.A.; Capone, M.; Madonna, G.; Lopalco, P.; Silvestris, F. Serum exosomes as predictors of clinical response to ipilimumab in metastatic melanoma. Oncoimmunology 2017, 7, e1387706. [Google Scholar] [CrossRef] [PubMed]
- Luke, J.J.; Flaherty, K.T.; Ribas, A.; Long, G.V. Targeted agents and immunotherapies: Optimizing outcomes in melanoma. Nat. Rev. Clin. Oncol. 2017, 14, 463–482. [Google Scholar] [CrossRef] [PubMed]
- Kaufman, H.L.; Margolin, K.; Sullivan, R. Management of Metastatic Melanoma in 2018. JAMA Oncol. 2018, 4, 857–858. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, F.; Haass, N.K. Microenvironment-driven dynamic heterogeneity and phenotypic plasticity as a mechanism of melanoma therapy resistance. Front. Oncol. 2018, 8, 173. [Google Scholar] [CrossRef] [PubMed]
- Croce, C.M. Causes and consequences of microRNA dysregulation in cancer. Nat. Rev. Genet. 2009, 10, 704–714. [Google Scholar] [CrossRef] [PubMed]
- Chan, S.H.; Wang, L.H. Regulation of cancer metastasis by microRNAs. J. Biomed. Sci. 2015, 22, 29. [Google Scholar] [CrossRef] [PubMed]
- Jing, Z.; Han, W.; Sui, X.; Xie, J.; Pan, H. Interaction of autophagy with microRNAs and their potential therapeutic implications in human cancers. Cancer Lett. 2015, 356, 332–338. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.; Li, X.; Ji, W.; Sun, B.; Xu, C.; Li, Z.; Qian, G.; Su, C. Small molecule with big role: MicroRNAs in cancer metastatic microenvironments. Cancer Lett. 2014, 344, 147–156. [Google Scholar] [CrossRef] [PubMed]
- Chan, B.; Manley, J.; Lee, J.; Singh, S.R. The emerging roles of microRNAs in cancer metabolism. Cancer Lett. 2015, 356, 301–308. [Google Scholar] [CrossRef] [PubMed]
- Melnik, B.C. MiR-21: An environmental driver of malignant melanoma? J. Transl. Med. 2015, 13, 202. [Google Scholar] [CrossRef] [PubMed]
- Bennett, P.E.; Bemis, L.; Norris, D.A.; Shellman, Y.G. MiR in melanoma development: MiRNAs and acquired hallmarks of cancer in melanoma. Physiol. Genom. 2013, 45, 1049–1059. [Google Scholar] [CrossRef] [PubMed]
- Varamo, C.; Occelli, M.; Vivenza, D.; Merlano, M.; Lo Nigro, C. MicroRNAs role as potential biomarkers and key regulators in melanoma. Genes Chromosomes Cancer 2017, 56, 3–10. [Google Scholar] [CrossRef] [PubMed]
- Mirzaei, H.; Gholamin, S.; Shahidsales, S.; Sahebkar, A.; Jaafari, M.R.; Mirzaei, H.R.; Hassanian, S.M.; Avan, A. MicroRNAs as potential diagnostic and prognostic biomarkers in melanoma. Eur. J. Cancer 2016, 53, 25–32. [Google Scholar] [CrossRef] [PubMed]
- Wozniak, M.; Mielczarek, A.; Czyz, M. MiRNAs in melanoma: Tumor suppressors and oncogenes with prognostic potential. Curr. Med. Chem. 2016, 23, 3136–3153. [Google Scholar] [CrossRef] [PubMed]
- Mione, M.; Bosserhoff, A. MicroRNAs in melanocyte and melanoma biology. Pigment Cell Melanoma Res. 2015, 28, 340–354. [Google Scholar] [CrossRef] [PubMed]
- Mannavola, F.; Tucci, M.; Felici, C.; Stucci, S.; Silvestris, F. MiRNAs in melanoma: A defined role in tumor progression and metastasis. Expert Rev. Clin. Immunol. 2016, 12, 79–89. [Google Scholar] [CrossRef] [PubMed]
- Mueller, D.W.; Rehli, M.; Bosserhoff, A.K. MiRNA expression profiling in melanocytes and melanoma cell lines reveals miRNAs associated with formation and progression of malignant melanoma. J. Investig. Dermatol. 2009, 129, 1740–1751. [Google Scholar] [CrossRef] [PubMed]
- Li, J.Y.; Zheng, L.L.; Wang, T.T.; Hu, M. Functional annotation of metastasis-associated microRNAs of melanoma: A meta-analysis of expression profiles. Chin. Med. J. (Engl.) 2016, 129, 2484–2490. [Google Scholar] [CrossRef] [PubMed]
- Caramuta, S.; Egyházi, S.; Rodolfo, M.; Witten, D.; Hansson, J.; Larsson, C.; Lui, W.O. MicroRNA expression profiles associated with mutational status and survival in malignant melanoma. J. Investig. Dermatol. 2010, 130, 2062–2070. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.S.; Su, J.L.; Hung, M.C. Dysregulation of microRNAs in cancer. J. Biomed. Sci. 2012, 19, 90. [Google Scholar] [CrossRef] [PubMed]
- Couts, K.L.; Anderson, E.M.; Gross, M.M.; Sullivan, K.; Ahn, N.G. Oncogenic B-Raf signaling in melanoma cells controls a network of microRNAs with combinatorial functions. Oncogene 2013, 32, 1959–1970. [Google Scholar] [CrossRef] [PubMed]
- Latchana, N.; Abrams, Z.B.; Howard, J.H.; Regan, K.; Jacob, N.; Fadda, P.; Terando, A.; Markowitz, J.; Agnese, D.; Payne, P.; et al. Plasma microRNA levels following resection of metastatic melanoma. Bioinform. Biol. Insights 2017, 11, 1177932217694837. [Google Scholar] [CrossRef] [PubMed]
- Mo, M.H.; Chen, L.; Fu, Y.; Wang, W.; Fu, S.W. Cell-free Circulating miRNA Biomarkers in Cancer. J. Cancer 2012, 3, 432–448. [Google Scholar] [CrossRef] [PubMed]
- Dror, S.; Sander, L.; Schwartz, H.; Sheinboim, D.; Barzilai, A.; Dishon, Y.; Apcher, S.; Golan, T.; Greenberger, S.; Barshack, I.; et al. Melanoma miRNA trafficking controls tumour primary niche formation. Nat. Cell Biol. 2016, 18, 1006–1017. [Google Scholar] [CrossRef] [PubMed]
- La Shu, S.; Yang, Y.; Allen, C.L.; Maguire, O.; Minderman, H.; Sen, A.; Ciesielski, M.J.; Collins, K.A.; Bush, P.J.; Singh, P.; et al. Metabolic reprogramming of stromal fibroblasts by melanoma exosome microRNA favours a pre-metastatic microenvironment. Sci. Rep. 2018, 8, 12905. [Google Scholar] [CrossRef] [PubMed]
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The, C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef]
- Bhaskaran, M.; Mohan, M. MicroRNAs: History, biogenesis, and their evolving role in animal development and disease. Vet. Pathol. 2014, 51, 759–774. [Google Scholar] [CrossRef] [PubMed]
- Davalos, V.; Esteller, M. MicroRNAs and cancer epigenetics: A macrorevolution. Curr. Opin. Oncol. 2010, 22, 35–45. [Google Scholar] [CrossRef] [PubMed]
- Fabbri, M.; Calore, F.; Paone, A.; Galli, R.; Calin, G.A. Epigenetic regulation of miRNAs in cancer. Adv. Exp. Med. Biol. 2013, 754, 137–148. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Xia, W.; Khotskaya, Y.B.; Huo, L.; Nakanishi, K.; Lim, S.O.; Du, Y.; Wang, Y.; Chang, W.C.; Chen, C.H.; et al. EGFR modulates microRNA maturation in response to hypoxia through phosphorylation of AGO2. Nature 2013, 497, 383–387. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Target recognition and regulatory functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Sung, Y.M.; Park, J.; Kim, S.; Kim, J.; Park, J.; Ha, H.; Bae, J.Y.; Kim, S.; Baek, D. General rules for functional microRNA targeting. Nat. Genet. 2016, 48, 1517–1526. [Google Scholar] [CrossRef] [PubMed]
- Thomson, D.W.; Dinger, M.E. Endogenous microRNA sponges: Evidence and controversy. Nat. Rev. Genet. 2016, 17, 272–283. [Google Scholar] [CrossRef] [PubMed]
- Hansen, T.B.; Jensen, T.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.; Damgaard, C.K.; Kjems, J. Natural RNA circles function as efficient microRNA sponges. Nature 2013, 495, 384–388. [Google Scholar] [CrossRef] [PubMed]
- Poliseno, L.; Salmena, L.; Zhang, J.; Carver, B.; Haveman, W.J.; Pandolfi, P.P. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature 2010, 465, 1033–1038. [Google Scholar] [CrossRef] [PubMed]
- Gilot, D.; Migault, M.; Bachelot, L.; Journé, F.; Rogiers, A.; Donnou-Fournet, E.; Mogha, A.; Mouchet, N.; Pinel-Marie, M.L.; Mari, B.; et al. A non-coding function of TYRP1 mRNA promotes melanoma growth. Nat. Cell Biol. 2017, 19, 1348–1357. [Google Scholar] [CrossRef] [PubMed]
- Hartman, M.L.; Czyz, M. TYRP1 mRNA level is stable and MITF-M-independent in drug-naïve and vemurafenib-resistant melanoma cells. Melanoma Res. 2019. under review. [Google Scholar]
- Maffioletti, E.; Tardito, D.; Gennarelli, M.; Bocchio-Chiavetto, L. Micro spies from the brain to the periphery: New clues from studies on microRNAs in neuropsychiatric disorders. Front. Cell. Neurosci. 2014, 8, 75. [Google Scholar] [CrossRef] [PubMed]
- Sheervalilou, R.; Shirvaliloo, S.; Aval, S.F.; Khamaneh, A.M.; Sharifi, A.; Ansarin, K.; Zarghami, N. A new insight on reciprocal relationship between microRNA expression and epigenetic modifications in human lung cancer. Tumor Biol. 2017, 39. [Google Scholar] [CrossRef] [PubMed]
- Jahagirdar, D.; Purohit, S.; Jain, A.; Sharma, N.K. Export of microRNAs: A Bridge between Breast Carcinoma and Their Neighboring Cells. Front. Oncol. 2016, 6, 147. [Google Scholar] [CrossRef] [PubMed]
- Minciacchi, V.R.; Freeman, M.R.; Di Vizio, D. Extracellular vesicles in cancer: Exosomes, microvesicles and the emerging role of large oncosomes. Semin. Cell Dev. Biol. 2015, 40, 41–51. [Google Scholar] [CrossRef] [PubMed]
- Becker, A.; Thakur, B.K.; Weiss, J.M.; Kim, H.S.; Peinado, H.; Lyden, D. Extracellular Vesicles in Cancer: Cell-to-Cell Mediators of Metastasis. Cancer Cell 2016, 30, 836–848. [Google Scholar] [CrossRef] [PubMed]
- Di Vizio, D.; Morello, M.; Dudley, A.C.; Schow, P.W.; Adam, R.M.; Morley, S.; Mulholland, D.; Rotinen, M.; Hager, M.H.; Insabato, L.; et al. Large oncosomes in human prostate cancer tissues and in the circulation of mice with metastatic disease. Am. J. Pathol. 2012, 181, 1573–1584. [Google Scholar] [CrossRef] [PubMed]
- Plotkin, L.I.; Pacheco-Costa, R.; Davis, H.M. microRNAs and connexins in bone: Interaction and mechanisms of delivery. Curr. Mol. Biol. Rep. 2017, 3, 63–70. [Google Scholar] [CrossRef] [PubMed]
- Kosaka, N.; Iguchi, H.; Ochiya, T. Circulating microRNA in body fluid: A new potential biomarker for cancer diagnosis and prognosis. Cancer Sci. 2010, 101, 2087–2092. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Yuan, T.; Tschannen, M.; Sun, Z.; Jacob, H.; Du, M.; Liang, M.; Dittmar, R.L.; Liu, Y.; Liang, M.; et al. Characterization of human plasma-derived exosomal RNAs by deep sequencing. BMC Genom. 2013, 14, 319. [Google Scholar] [CrossRef] [PubMed]
- Al-Nedawi, K.; Meehan, B.; Micallef, J.; Lhotak, V.; May, L.; Guha, A.; Rak, J. Intercellular transfer of the oncogenic receptor EGFRvIII by microvesicles derived from tumour cells. Nat. Cell Biol. 2008, 10, 619–624. [Google Scholar] [CrossRef] [PubMed]
- Pan, B.T.; Johnstone, R.M. Fate of the transferrin receptor during maturation of sheep reticulocytes in vitro: Selective externalization of the receptor. Cell 1983, 33, 967–978. [Google Scholar] [CrossRef]
- Raposo, G.; Stoorvogel, W. Extracellular vesicles: Exosomes, microvesicles, and friends. J. Cell Biol. 2013, 200, 373–383. [Google Scholar] [CrossRef] [PubMed]
- Colombo, M.; Moita, C.; van Niel, G.; Kowal, J.; Vigneron, J.; Benaroch, P.; Manel, N.; Moita, L.F.; Théry, C.; Raposo, G. Analysis of ESCRT functions in exosome biogenesis, composition and secretion highlights the heterogeneity of extracellular vesicles. J. Cell Sci. 2013, 126, 5553–5565. [Google Scholar] [CrossRef] [PubMed]
- Shimaoka, M.; Kawamoto, E.; Gaowa, A.; Okamoto, T.; Park, E.J. Connexins and Integrins in Exosomes. Cancers 2019, 11, 106. [Google Scholar] [CrossRef] [PubMed]
- Jella, K.K.; Nasti, T.H.; Li, Z.; Malla, S.R.; Buchwald, Z.S.; Khan, M.K. Exosomes, Their Biogenesis and Role in Inter-Cellular Communication, Tumor Microenvironment and Cancer Immunotherapy. Vaccines (Basel) 2018, 6, 69. [Google Scholar] [CrossRef] [PubMed]
- Gajos-Michniewicz, A.; Duechler, M.; Czyz, M. MiRNA in melanoma-derived exosomes. Cancer Lett. 2014, 347, 29–37. [Google Scholar] [CrossRef] [PubMed]
- Tucci, M.; Mannavola, F.; Passarelli, A.; Stucci, L.S.; Cives, M.; Silvestris, F. Exosomes in melanoma: A role in tumor progression, metastasis and impaired immune system activity. Oncotarget 2018, 9, 20826–20837. [Google Scholar] [CrossRef] [PubMed]
- Grasedieck, S.; Sorrentino, A.; Langer, C.; Buske, C.; Döhner, H.; Mertens, D.; Kuchenbauer, F. Circulating microRNAs in hematological diseases: Principles, challenges, and perspectives. Blood 2013, 121, 4977–4984. [Google Scholar] [CrossRef] [PubMed]
- Boissy, R.E. Melanosome transfer to and translocation in the keratinocyte. Exp. Dermatol. 2003, 12, 5–12. [Google Scholar] [CrossRef] [PubMed]
- Golan, T.; Messer, A.R.; Amitai-Lange, A.; Melamed, Z.; Ohana, R.; Bell, R.E.; Kapitansky, O.; Lerman, G.; Greenberger, S.; Khaled, M.; et al. Interactions of melanoma cells with distal keratinocytes trigger metastasis via Notch signaling inhibition of MITF. Mol. Cell 2015, 59, 664–676. [Google Scholar] [CrossRef] [PubMed]
- Felli, N.; Errico, M.C.; Pedini, F.; Petrini, M.; Puglisi, R.; Bellenghi, M.; Boe, A.; Felicetti, F.; Mattia, G.; De Feo, A.; et al. AP2α controls the dynamic balance between miR-126&126* and miR-221&222 during melanoma progression. Oncogene 2016, 35, 3016–3026. [Google Scholar] [CrossRef] [PubMed]
- Felicetti, F.; Errico, M.C.; Bottero, L.; Segnalini, P.; Stoppacciaro, A.; Biffoni, M.; Felli, N.; Mattia, G.; Petrini, M.; Colombo, M.P.; et al. The promyelocytic leukemia zinc finger-microRNA-221/-222 pathway controls melanoma progression through multiple oncogenic mechanisms. Cancer Res. 2008, 68, 2745–2754. [Google Scholar] [CrossRef] [PubMed]
- Felicetti, F.; De Feo, A.; Coscia, C.; Puglisi, R.; Pedini, F.; Pasquini, L.; Bellenghi, M.; Errico, M.C.; Pagani, E.; Carè, A. Exosome-mediated transfer of miR-222 is sufficient to increase tumor malignancy in melanoma. J. Transl. Med. 2016, 14, 56. [Google Scholar] [CrossRef] [PubMed]
- Igoucheva, O.; Alexeev, V. MicroRNA-dependent regulation of cKit in cutaneous melanoma. Biochem. Biophys. Res. Commun. 2009, 379, 790–794. [Google Scholar] [CrossRef] [PubMed]
- Lohcharoenkal, W.; Das Mahapatra, K.; Pasquali, L.; Crudden, C.; Kular, L.; Akkaya Ulum, Y.Z.; Zhang, L.; Xu Landén, N.; Girnita, L.; Jagodic, M.; et al. Genome-Wide Screen for MicroRNAs Reveals a Role for miR-203 in Melanoma Metastasis. J. Investig. Dermatol. 2018, 138, 882–892. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.K.; Varney, M.L. IL-8 expression in malignant melanoma: Implications in growth and metastasis. Histol. Histopathol. 2000, 15, 843–849. [Google Scholar] [CrossRef] [PubMed]
- Pencheva, N.; Tran, H.; Buss, C.; Huh, D.; Drobnjak, M.; Busam, K.; Tavazoie, S.F. Convergent multi-miRNA targeting of ApoE drives LRP1/LRP8 dependent melanoma metastasis and angiogenesis. Cell 2012, 151, 1068–1082. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, G.; Wu, X.; Jiang, Z.; Kasman, I.; Yao, J.; Guan, Y.; Oeh, J.; Modrusan, Z.; Bais, C.; Sampath, D.; et al. Tumour-secreted miR-9 promotes endothelial cell migration and angiogenesis by activating the JAK-STAT pathway. EMBO J. 2012, 31, 3513–3523. [Google Scholar] [CrossRef] [PubMed]
- Penna, E.; Orso, F.; Cimino, D.; Tenaglia, E.; Lembo, A.; Quaglino, E.; Poliseno, L.; Haimovic, A.; Osella-Abate, S.; De Pittà, C.; et al. MicroRNA-214 contributes to melanoma tumour progression through suppression of TFAP2C. EMBO J. 2011, 30, 1990–2007. [Google Scholar] [CrossRef] [PubMed]
- Crawford, Y.; Kasman, I.; Yu, L.; Zhong, C.; Wu, X.; Modrusan, Z.; Kaminker, J.; Ferrara, N. PDGF-C mediates the angiogenic and tumorigenic properties of fibroblasts associated with tumors refractory to anti-VEGF treatment. Cancer Cell 2009, 15, 21–34. [Google Scholar] [CrossRef] [PubMed]
- Anderberg, C.; Li, H.; Fredriksson, L.; Andrae, J.; Betsholtz, C.; Li, X.; Eriksson, U.; Pietras, K. Paracrine signaling by platelet-derived growth factor-CC promotes tumor growth by recruitment of cancer-associated fibroblasts. Cancer Res. 2009, 69, 369–378. [Google Scholar] [CrossRef] [PubMed]
- Di Tomaso, E.; London, N.; Fuja, D.; Logie, J.; Tyrrell, J.A.; Kamoun, W.; Munn, L.L.; Jain, R.K. PDGF-C induces maturation of blood vessels in a model of glioblastoma and attenuates the response to anti-VEGF treatment. PLoS ONE 2009, 4, e5123. [Google Scholar] [CrossRef] [PubMed]
- Passarelli, A.; Mannavola, F.; Stucci, L.S.; Tucci, M.; Silvestris, F. Immune system and melanoma biology: A balance between immunosurveillance and immune escape. Oncotarget 2017, 8, 106132–106142. [Google Scholar] [CrossRef] [PubMed]
- Fanini, F.; Fabbri, M. Cancer-derived exosomic microRNAs shape the immune system within the tumor microenvironment: State of the art. Semin. Cell Dev. Biol. 2017, 67, 23–28. [Google Scholar] [CrossRef] [PubMed]
- Graner, M.W.; Schnell, S.; Olin, M.R. Tumor-derived exosomes, microRNAs, and cancer immune suppression. Semin. Immunopathol. 2018, 40, 505–515. [Google Scholar] [CrossRef] [PubMed]
- Alfonsi, R.; Grassi, L.; Signore, M.; Bonci, D. The Double Face of Exosome-Carried MicroRNAs in Cancer Immunomodulation. Int. J. Mol. Sci. 2018, 19, 1183. [Google Scholar] [CrossRef] [PubMed]
- Vallacchi, V.; Camisaschi, C.; Dugo, M.; Vergani, E.; Deho, P.; Gualeni, A.; Huber, V.; Gloghini, A.; Maurichi, A.; Santinami, M.; et al. MicroRNA Expression in Sentinel Nodes from Progressing Melanoma Patients Identifies Networks Associated with Dysfunctional Immune Response. Genes (Basel) 2016, 7, 124. [Google Scholar] [CrossRef] [PubMed]
- Gaziel-Sovran, A.; Segura, M.F.; Di Micco, R.; Collins, M.K.; Hanniford, D.; Vega-Saenz de Miera, E.; Rakus, J.F.; Dankert, J.F.; Shang, S.; Kerbel, R.S.; et al. MiR30b/30d regulation of GalNAc transferases enhances invasion and immunosuppression during metastasis. Cancer Cell 2011, 20, 104–118. [Google Scholar] [CrossRef] [PubMed]
- Ding, H.; Yang, X.; Wei, Y. Fusion Proteins of NKG2D/NKG2DL in Cancer Immunotherapy. Int. J. Mol. Sci. 2018, 19, 177. [Google Scholar] [CrossRef] [PubMed]
- Heinemann, A.; Zhao, F.; Pechlivanis, S.; Eberle, J.; Steinle, A.; Diederichs, S.; Schadendorf, D.; Paschen, A. Tumor suppressive microRNAs miR-34a/c control cancer cell expression of ULBP2, a stress-induced ligand of the natural killer cell receptor NKG2D. Cancer Res. 2012, 72, 460–471. [Google Scholar] [CrossRef] [PubMed]
- Paschen, A.; Sucker, A.; Hill, B.; Moll, I.; Zapatka, M.; Nguyen, X.D.; Sim, G.C.; Gutmann, I.; Hassel, J.; Becker, J.C.; et al. Differential clinical significance of individual NKG2D ligands in melanoma: Soluble ULBP2 as an indicator of poor prognosis superior to S100B. Clin. Cancer Res. 2009, 15, 5208–5215. [Google Scholar] [CrossRef] [PubMed]
- Garraway, L.A.; Widlund, H.R.; Rubin, M.A.; Getz, G.; Berger, A.J.; Ramaswamy, S.; Beroukhim, R.; Milner, D.A.; Granter, S.R.; Du, J.; et al. Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature 2005, 436, 117–222. [Google Scholar] [CrossRef] [PubMed]
- Carreira, S.; Goodall, J.; Denat, L.; Rodriguez, M.; Nuciforo, P.; Hoek, K.S.; Testori, A.; Larue, L.; Goding, C.R. Mitf regulation of Dia1 controls melanoma proliferation and invasiveness. Genes Dev. 2006, 20, 3426–3439. [Google Scholar] [CrossRef] [PubMed]
- Bell, R.E.; Levy, C. The three M’s: Melanoma, microphthalmia-associated transcription factor and microRNA. Pigment Cell Melanoma Res. 2011, 24, 1088–1106. [Google Scholar] [CrossRef] [PubMed]
- Hartman, M.L.; Czyz, M. MITF in melanoma: Mechanisms behind its expression and activity. Cell Mol. Life Sci. 2015, 72, 1249–1260. [Google Scholar] [CrossRef] [PubMed]
- Bemis, L.T.; Chen, R.; Amato, C.M.; Classen, E.H.; Robinson, S.E.; Coffey, D.G.; Erickson, P.F.; Shellman, Y.G.; Robinson, W.A.F. MicroRNA-137 targets microphthalmia-associated transcription factor in melanoma cell lines. Cancer Res. 2008, 68, 1362–1368. [Google Scholar] [CrossRef] [PubMed]
- Luo, C.; Tetteh, P.W.; Merz, P.R.; Dickes, E.; Abukiwan, A.; Hotz-Wagenblatt, A.; Holland-Cunz, S.; Sinnberg, T.; Schittek, B.; Schadendorf, D.; et al. MiR-137 inhibits the invasion of melanoma cells through downregulation of multiple oncogenic target genes. J. Investig. Dermatol. 2013, 133, 768–775. [Google Scholar] [CrossRef] [PubMed]
- Qi, J.; Wang, W.W.; Chen, W.; Lu, W.Y.; Shang, A.Q. Mechanism of miR-137 regulating migration and invasion of melanoma cells by targeting PIK3R3gene. J. Cell Biochem. 2018. [Google Scholar] [CrossRef] [PubMed]
- Haflidadóttir, B.S.; Bergsteinsdóttir, K.; Praetorius, C.; Steingrímsson, E. MiR-148 regulates Mitf in melanoma cells. PLoS ONE 2010, 5, e11574. [Google Scholar] [CrossRef] [PubMed]
- Segura, M.F.; Hanniford, D.; Menendez, S.; Reavie, L.; Zou, X.; Alvarez-Diaz, S.; Zakrzewski, J.; Blochin, E.; Rose, A.; Bogunovic, D.; et al. Aberrant miR-182 expression promotes melanoma metastasis by repressing FOXO3 and microphthalmia-associated transcription factor. Proc. Natl. Acad. Sci. USA 2009, 106, 1814–1819. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Howell, P.M.; Riker, A.I. Up-regulation of miR-182 expression after epigenetic modulation of human melanoma cells. Ann. Surg. Oncol. 2013, 20, 1745–1752. [Google Scholar] [CrossRef] [PubMed]
- Jukic, D.M.; Rao, U.N.; Kelly, L.; Skaf, J.S.; Drogowski, L.M.; Kirkwood, J.M.; Panelli, M.C. Microrna profiling analysis of differences between the melanoma of young adults and older adults. J. Transl. Med. 2010, 19, 8–27. [Google Scholar] [CrossRef] [PubMed]
- Levy, C.; Khaled, M.; Iliopoulos, D.; Janas, M.M.; Schubert, S.; Pinner, S.; Chen, P.H.; Li, S.; Fletcher, A.L.; Yokoyama, S.; et al. Intronic miR-211 assumes the tumor suppressive function of its host gene in melanoma. Mol. Cell 2010, 40, 841–849. [Google Scholar] [CrossRef] [PubMed]
- Mazar, J.; DeYoung, K.; Khaitan, D.; Meister, E.; Almodovar, A.; Goydos, J.; Ray, A.; Perera, R.J. The regulation of miRNA-211 expression and its role in melanoma cell invasiveness. PLoS ONE 2010, 5, e13779. [Google Scholar] [CrossRef] [PubMed]
- Boyle, G.M.; Woods, S.L.; Bonazzi, V.F.; Stark, M.S.; Hacker, E.; Aoude, L.G.; Dutton-Regester, K.; Cook, A.L.; Sturm, R.A.; Hayward, N.K. Melanoma cell invasiveness is regulated by miR-211 suppression of the BRN2 transcription factor. Pigment Cell Melanoma Res. 2011, 24, 525–537. [Google Scholar] [CrossRef] [PubMed]
- Bell, R.E.; Khaled, M.; Netanely, D.; Schubert, S.; Golan, T.; Buxbaum, A.; Janas, M.M.; Postolsky, B.; Goldberg, M.S.; Shamir, R.; et al. Transcription factor/microRNA axis blocks melanoma invasion program by miR-211 targeting NUAK1. J. Investig. Dermatol. 2014, 134, 441–451. [Google Scholar] [CrossRef] [PubMed]
- Eccles, M.R.; He, S.; Ahn, A.; Slobbe, L.; Jeffs, A.R.; Yoon, H.S.; Baguley, B.C. MITF and PAX3 Play Distinct Roles in Melanoma Cell Migration; Outline of a “Genetic Switch” Theory Involving MITF and PAX3 in Proliferative and Invasive Phenotypes of Melanoma. Front. Oncol. 2013, 3, 229. [Google Scholar] [CrossRef] [PubMed]
- Arozarena, I.; Sanchez-Laorden, B.; Packer, L.; Hidalgo-Carcedo, C.; Hayward, R.; Viros, A.; Sahai, E.; Marais, R. Oncogenic BRAF induces melanoma cell invasion by downregulating the cGMP-specific phosphodiesterase PDE5A. Cancer Cell 2011, 19, 45–57. [Google Scholar] [CrossRef] [PubMed]
- Goswami, S.; Tarapore, R.S.; Poenitzsch Strong, A.M.; TeSlaa, J.J.; Grinblat, Y.; Setaluri, V.; Spiegelman, V.S. MicroRNA-340-mediated degradation of microphthalmia-associated transcription factor (MITF) mRNA is inhibited by coding region determinant-binding protein (CRD-BP). J. Biol. Chem. 2015, 290, 384–395. [Google Scholar] [CrossRef] [PubMed]
- Cohen, R.; Greenberg, E.; Nemlich, Y.; Schachter, J.; Markel, G. MiR17 regulates melanoma cell motility by inhibiting the translation of ETV1. Oncotarget 2015, 6, 19006–19016. [Google Scholar] [CrossRef] [PubMed]
- Ohira, T.; Naohiro, S.; Nakayama, Y.; Osaki, M.; Okada, F.; Oshimura, M.; Kugoh, H. miR-19b regulates hTERT mRNA expression through targeting PITX1 mRNA in melanoma cells. Sci. Rep. 2015, 5, 8201. [Google Scholar] [CrossRef] [PubMed]
- Martin del Campo, S.E.; Latchana, N.; Levine, K.M.; Grignol, V.P.; Fairchild, E.T.; Jaime-Ramirez, A.C.; Dao, T.V.; Karpa, V.I.; Carson, M.; Ganju, A.; et al. MiR-21 enhances melanoma invasiveness via inhibition of tissue inhibitor of metalloproteinases 3 expression: In vivo effects of MiR-21 inhibitor. PLoS ONE 2015, 10, e0115919. [Google Scholar] [CrossRef]
- Saldanha, G.; Potter, L.; Lee, Y.S.; Watson, S.; Shendge, P.; Pringle, J.H. MicroRNA-21 expression and its pathogenetic significance in cutaneous melanoma. Melanoma Res. 2016, 26, 21–28. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.H.; Pfeffer, S.R.; Sims, M.; Yue, J.; Wang, Y.; Linga, V.G.; Paulus, E.; Davidoff, A.M.; Pfeffer, L.M. The oncogenic microRNA-21 inhibits the tumor suppressive activity of FBXO11 to promote tumorigenesis. J. Biol. Chem. 2015, 290, 6037–6046. [Google Scholar] [CrossRef] [PubMed]
- Huo, J.; Zhang, Y.; Li, R.; Wang, Y.; Wu, J.; Zhang, D. Upregulated microRNA-25 mediates the migration of melanoma cells by targeting DKK3 through the WNT/β-catenin pathway. Int. J. Mol. Sci. 2016, 17, 1124. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Q.Q.; Liu, W.B. MiR-25 promotes melanoma progression by regulating RNA binding motif protein 47. Med. Sci. (Paris) 2018, 34, 59–65. [Google Scholar] [CrossRef] [PubMed]
- Rambow, F.; Bechadergue, A.; Luciani, F.; Gros, G.; Domingues, M.; Bonaventure, J.; Meurice, G.; Marine, J.C.; Larue, L. Regulation of Melanoma Progression through the TCF4/miR-125b/NEDD9 Cascade. J. Investig. Dermatol. 2016, 136, 1229–1237. [Google Scholar] [CrossRef] [PubMed]
- Forloni, M.; Dogra, S.K.; Dong, Y.; Conte, D., Jr.; Ou, J.; Zhu, L.J.; Deng, A.; Mahalingam, M.; Green, M.R.; Wajapeyee, N. MiR-146a promotes the initiation and progression of melanoma by activating Notch signaling. eLife 2014, 3, e01460. [Google Scholar] [CrossRef] [PubMed]
- Ozsolak, F.; Poling, L.L.; Wang, Z.; Liu, H.; Liu, X.S.; Roeder, R.G.; Zhang, X.; Song, J.S.; Fisher, D.E. Chromatin structure analyses identify miRNA promoters. Genes Dev. 2008, 22, 3172–3183. [Google Scholar] [CrossRef] [PubMed]
- Knoll, S.; Fürst, K.; Kowtharapu, B.; Schmitz, U.; Marquardt, S.; Wolkenhauer, O.; Martin, H.; Pützer, B.M. E2F1 induces miR 224/452 expression to drive EMT through TXNIP downregulation. EMBO Rep. 2014, 15, 1315–1329. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, A.; Schmitz, U.; Raatz, Y.; Schönherr, M.; Kottek, T.; Schauer, M.; Franz, S.; Saalbach, A.; Anderegg, U.; Wolkenhauer, O.; et al. MiR-638 promotes melanoma metastasis and protects melanoma cells from apoptosis and autophagy. Oncotarget 2015, 6, 2966–2980. [Google Scholar] [CrossRef] [PubMed]
- Müller, D.W.; Bosserhoff, A.K. Integrin beta 3 expression is regulated by let-7a miRNA in malignant melanoma. Oncogene 2008, 27, 6698–6706. [Google Scholar] [CrossRef] [PubMed]
- Fu, T.Y.; Chang, C.C.; Lin, C.T.; Lai, C.H.; Peng, S.Y.; Ko, Y.J.; Tang, P.C. Let-7b-mediated suppression of basigin expression and metastasis in mouse melanoma cells. Exp. Cell Res. 2011, 317, 445–451. [Google Scholar] [CrossRef] [PubMed]
- Migliore, C.; Petrelli, A.; Ghiso, E.; Corso, S.; Capparuccia, L.; Eramo, A.; Comoglio, P.M.; Giordano, S. MicroRNAs impair MET-mediated invasive growth. Cancer Res. 2008, 68, 10128–10136. [Google Scholar] [CrossRef] [PubMed]
- Mazar, J.; Khaitan, D.; DeBlasio, D.; Zhong, C.; Govindarajan, S.S.; Kopanathi, S.; Zhang, S.; Ray, A.; Perera, R.J. Epigenetic regulation of microRNA genes and the role of miR-34b in cell invasion and motility in human melanoma. PLoS ONE 2011, 6, e24922. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Tetzlaff, M.T.; Wang, T.; Yang, R.; Xie, L.; Zhang, G.; Krepler, C.; Xiao, M.; Beqiri, M.; Xu, W.; et al. MiR-200c/Bmi1 axis and epithelial-mesenchymal transition contribute to acquired resistance to BRAF inhibitor treatment. Pigment Cell Melanoma Res. 2015, 28, 431–441. [Google Scholar] [CrossRef] [PubMed]
- Noguchi, S.; Kumazaki, M.; Mori, T.; Baba, K.; Okuda, M.; Mizuno, T.; Akao, Y. Analysis of microRNA-203 function in CREB/MITF/RAB27a pathway: Comparison between canine and human melanoma cells. Vet. Comp. Oncol. 2016, 14, 384–394. [Google Scholar] [CrossRef] [PubMed]
- Mazar, J.; Qi, F.; Lee, B.; Marchica, J.; Govindarajan, S.; Shelley, J.; Li, J.L.; Ray, A.; Perera, R.J. MicroRNA 211 Functions as a Metabolic Switch in Human Melanoma Cells. Mol. Cell Biol. 2016, 36, 1090–1108. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Du, Y.; Chen, X.; Li, P.; Wang, Y.; Zang, W.; Zhao, L.; Li, Z.; Zhao, G. Expression patterns of microRNA-218 and its potential functions by targeting CIP2A and BMI1 genes in melanoma. Tumor Biol. 2014, 35, 8007–8015. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Jin, J.; Rong, K.; Zhuo, L.; Li, P. MicroRNA-675 inhibits cell proliferation and invasion in melanoma by directly targeting metadherin. Mol. Med. Rep. 2018, 17, 3372–3379. [Google Scholar] [CrossRef] [PubMed]
- Segura, M.F.; Greenwald, H.S.; Hanniford, D.; Osman, I.; Hernando, E. MicroRNA and cutaneous melanoma: From discovery to prognosis and therapy. Carcinogenesis 2012, 33, 1823–1832. [Google Scholar] [CrossRef] [PubMed]
- Gokhale, A.; Kunder, R.; Goel, A.; Sarin, R.; Moiyadi, A.; Shenoy, A.; Mamidipally, C.; Noronha, S.; Kannan, S.; Shirsat, N.V. Distinctive microRNA signature of medulloblastomas associated with the WNT signaling pathway. J. Cancer Res. Ther. 2010, 6, 521–529. [Google Scholar] [CrossRef] [PubMed]
- Nishinaka, Y.; Nishiyama, A.; Masutani, H.; Oka, S.; Ahsan, K.M.; Nakayama, Y.; Ishii, Y.; Nakamura, H.; Maeda, M.; Yodoi, J. Loss of thioredoxin-bind- ing protein-2/vitamin D3 up-regulated protein 1 in human T-cell leukemia virus type I-dependent T-cell transformation: Implications for adult T-cell leukemia leukemogenesis. Cancer Res. 2004, 64, 1287–1292. [Google Scholar] [CrossRef] [PubMed]
- Adam, L.; Zhong, M.; Choi, W.; Qi, W.; Nicoloso, M.; Arora, A.; Calin, G.; Wang, H.; Siefker-Radtke, A.; McConkey, D.; et al. MiR-200 expression regulates epithelial-to-mesenchymal transition in bladder cancer cells and reverses resistance to epidermal growth factor receptor therapy. Clin. Cancer Res. 2009, 15, 5060–5072. [Google Scholar] [CrossRef] [PubMed]
- Elson-Schwab, I.; Lorentzen, A.; Marshall, C.J. MicroRNA-200 family members differentially regulate morphological plasticity and mode of melanoma cell invasion. PLoS ONE 2010, 5, e13176. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Li, Y.; Ahmad, A.; Azmi, A.S.; Kong, D.; Banerjee, S.; Sarkar, F.H. Targeting miRNAs involved in cancer stem cell and EMT regulation: An emerging concept in overcoming drug resistance. Drug Resist. Updat. 2010, 13, 109–118. [Google Scholar] [CrossRef] [PubMed]
- Wellner, U.; Schubert, J.; Burk, U.C.; Schmalhofer, O.; Zhu, F.; Sonntag, A.; Waldvogel, B.; Vannier, C.; Darling, D.; zur Hausen, A.; et al. The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. Nat. Cell Biol. 2009, 11, 1487–1495. [Google Scholar] [CrossRef] [PubMed]
- Da Cruz, A.T.; Jasiulionis, M.G. MiRNAs and Melanoma: How Are They Connected? Dermatol. Res. Pract. 2012, 2012, 528345. [Google Scholar] [CrossRef] [PubMed]
- Romano, G.; Kwong, L.N. MiRNAs, Melanoma and Microenvironment: An Intricate Network. Int. J. Mol. Sci. 2017, 18, 2354. [Google Scholar] [CrossRef] [PubMed]
- Domingues, M.J.; Rambow, F.; Job, B.; Papon, L.; Liu, W.; Larue, L.; Bonaventure, J. Beta-catenin inhibitor ICAT modulates the invasive motility of melanoma cells. Cancer Res. 2014, 74, 1983–1995. [Google Scholar] [CrossRef] [PubMed]
- Ge, X.; Lv, X.; Feng, L.; Liu, X.; Gao, J.; Chen, N.; Wang, X. Metadherin contributes to the pathogenesis of diffuse large B-cell lymphoma. PLoS ONE 2012, 7, e39449. [Google Scholar] [CrossRef] [PubMed]
- Mazar, J.; DeBlasio, D.; Govindarajan, S.S.; Zhang, S.; Perera, R.J. Epigenetic regulation of microRNA-375 and its role in melanoma development in humans. FEBS Lett. 2011, 585, 2467–2476. [Google Scholar] [CrossRef] [PubMed]
- Nogués, L.; Benito-Martin, A.; Hergueta-Redondo, M.; Peinado, H. The influence of tumour-derived extracellular vesicles on local and distal metastatic dissemination. Mol. Asp. Med. 2018, 60, 15–26. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Cao, X. Characteristics and Significance of the Pre-metastatic Niche. Cancer Cell 2016, 30, 668–681. [Google Scholar] [CrossRef] [PubMed]
- Peinado, H.; Alečković, M.; Lavotshkin, S.; Matei, I.; Costa-Silva, B.; Moreno-Bueno, G.; Hergueta-Redondo, M.; Williams, C.; García-Santos, G.; Ghajar, C.; et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat. Med. 2012, 18, 883–891. [Google Scholar] [CrossRef] [PubMed]
- Czyz, M. HGF/c-MET signaling in melanocytes and melanoma. Int. J. Mol. Sci. 2018, 19, 3844. [Google Scholar] [CrossRef] [PubMed]
- Hanna, S.C.; Krishnan, B.; Bailey, S.T.; Moschos, S.J.; Kuan, P.F.; Shimamura, T.; Osborne, L.D.; Siegel, M.B.; Duncan, L.M.; O′Brien, E.T., 3rd; et al. HIF1α and HIF2α independently activate SRC to promote melanoma metastases. J. Clin. Investig. 2013, 123, 2078–9203. [Google Scholar] [CrossRef] [PubMed]
- Nouraee, N.; Mowla, S.J.; Calin, G.A. Tracking miRNAs’ footprints in tumor-microenvironment interactions: Insights and implications for targeted cancer therapy. Genes Chromosomes Cancer 2015, 54, 335–352. [Google Scholar] [CrossRef] [PubMed]
- Wozniak, M.; Sztiller-Sikorska, M.; Czyz, M. Diminution of miR-340-5p levels is responsible for increased expression of ABCB5 in melanoma cells under oxygen-deprived conditions. Exp. Mol. Pathol. 2015, 99, 707–716. [Google Scholar] [CrossRef] [PubMed]
- Majmundar, A.J.; Wong, W.J.; Simon, M.C. Hypoxia-inducible factors and the response to hypoxic stress. Mol. Cell 2010, 40, 294–309. [Google Scholar] [CrossRef] [PubMed]
- Hwang, H.W.; Baxter, L.L.; Loftus, S.K.; Cronin, J.C.; Trivedi, N.S.; Borate, B.; Pavan, W.J. Distinct microRNA expression signatures are associated with melanoma subtypes and are regulated by HIF1A. Pigment Cell Melanoma Res. 2014, 27, 777–787. [Google Scholar] [CrossRef] [PubMed]
- Maes, H.; Van Eygen, S.; Krysko, D.V.; Vandenabeele, P.; Nys, K.; Rillaerts, K.; Garg, A.D.; Verfaillie, T.; Agostinis, P. BNIP3 supports melanoma cell migration and vasculogenic mimicry by orchestrating the actin cytoskeleton. Cell Death Dis. 2014, 5, e1127. [Google Scholar] [CrossRef] [PubMed]
- Vara-Perez, M.; Maes, H.; Van Dingenen, S.; Agostinis, P. BNIP3 contributes to the glutamine-driven aggressive behavior of melanoma cells. Biol. Chem. 2019, 400, 187–193. [Google Scholar] [CrossRef] [PubMed]
- Thompson, M.R.; Xu, D.; Williams, B.R.G. ATF3 transcription factor and its emerging roles in immunity and cancer. J. Mol. Med. 2009, 87, 1053–1060. [Google Scholar] [CrossRef] [PubMed]
- King, H.W.; Michael, M.Z.; Gleadle, J.M. Hypoxic enhancement of exosome release by breast cancer cells. BMC Cancer 2012, 12, 421. [Google Scholar] [CrossRef] [PubMed]
- Wozniak, M.; Peczek, L.; Czernek, L.; Düchler, M. Analysis of the miRNA profiles of melanoma exosomes derived under normoxic and hypoxic culture conditions. Anticancer Res. 2017, 37, 6779–6789. [Google Scholar] [CrossRef] [PubMed]
- Rofstad, E.K.; Mathiesen, B.; Kindem, K.; Galappathi, K. Acidic extracellular pH promotes experimental metastasis of human melanoma cells in athymic nude mice. Cancer Res. 2006, 66, 6699–6707. [Google Scholar] [CrossRef] [PubMed]
- Logozzi, M.; Mizzoni, D.; Angelini, D.F.; Di Raimo, R.; Falchi, M.; Battistini, L.; Fais, S. Microenvironmental pH and Exosome Levels Interplay in Human Cancer Cell Lines of Different Histotypes. Cancers (Basel) 2018, 10, 370. [Google Scholar] [CrossRef] [PubMed]
- Wagle, N.; Van Allen, E.M.; Treacy, D.J.; Frederick, D.T.; Cooper, Z.A.; Taylor-Weiner, A.; Rosenberg, M.; Goetz, E.M.; Sullivan, R.J.; Farlow, D.N.; et al. MAP kinase pathway alterations in BRAF-mutant melanoma patients with acquired resistance to combined RAF/MEK inhibition. Cancer Discov. 2014, 4, 61–68. [Google Scholar] [CrossRef] [PubMed]
- Amaral, T.; Sinnberg, T.; Meier, F.; Krepler, C.; Levesque, M.; Niessner, H.; Garbe, C. The mitogen-activated protein kinase pathway in melanoma part I—Activation and primary resistance mechanisms to BRAF inhibition. Eur. J. Cancer 2017, 73, 85–92. [Google Scholar] [CrossRef] [PubMed]
- Larribère, L.; Kuphal, S.; Sachpekidis, C.; Sachindra; Hüser, L.; Bosserhoff, A.; Utikal, J. Targeted Therapy-Resistant Melanoma Cells Acquire Transcriptomic Similarities with Human Melanoblasts. Cancers (Basel) 2018, 10, 451. [Google Scholar] [CrossRef]
- Migliore, C.; Giordano, S. Resistance to targeted therapies: A role for microRNAs? Trends Mol. Med. 2013, 19, 633–642. [Google Scholar] [CrossRef] [PubMed]
- Fattore, L.; Mancini, R.; Acunzo, M.; Romano, G.; Laganà, A.; Pisanu, M.E.; Malpicci, D.; Madonna, G.; Mallardo, D.; Capone, M.; et al. MiR-579-3p controls melanoma progression and resistance to target therapy. Proc. Natl. Acad. Sci. USA 2016, 113, E5005–E5013. [Google Scholar] [CrossRef] [PubMed]
- Díaz-Martínez, M.; Benito-Jardón, L.; Alonso, L.; Koetz-Ploch, L.; Hernando, E.; Teixidó, J. MiR-204-5p and miR-211-5p Contribute to BRAF Inhibitor Resistance in Melanoma. Cancer Res. 2018, 78, 1017–1030. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Li, J.; Sun, Y.; Zhang, Y.; Dong, L.; Shen, C.; Yang, L.; Yang, M.; Li, Y.; Shen, G.; et al. MiR-7 reverses the resistance to BRAFi in melanoma by targeting EGFR/IGF-1R/CRAF and inhibiting the MAPK and PI3K/AKT signaling pathways. Oncotarget 2016, 7, 53558–53570. [Google Scholar] [CrossRef] [PubMed]
- Stark, M.S.; Bonazzi, V.F.; Boyle, G.M.; Palmer, J.M.; Symmons, J.; Lanagan, C.M.; Schmidt, C.W.; Herington, A.C.; Ballotti, R.; Pollock, P.M.; et al. MiR-514a regulates the tumour suppressor NF1 and modulates BRAFi sensitivity in melanoma. Oncotarget 2015, 6, 17753–17763. [Google Scholar] [CrossRef] [PubMed]
- Vergani, E.; Di Guardo, L.; Dugo, M.; Rigoletto, S.; Tragni, G.; Ruggeri, R.; Perrone, F.; Tamborini, E.; Gloghini, A.; Arienti, F.; et al. Overcoming melanoma resistance to vemurafenib by targeting CCL2-induced miR-34a, miR-100 and miR-125b. Oncotarget 2016, 7, 4428–4441. [Google Scholar] [CrossRef] [PubMed]
- Luan, W.; Qian, Y.; Ni, X.; Bu, X.; Xia, Y.; Wang, J.; Ruan, H.; Ma, S.; Xu, B. MiR-204-5p acts as a tumor suppressor by targeting matrix metalloproteinases-9 and B-cell lymphoma-2 in malignant melanoma. OncoTargets Ther. 2017, 10, 1237–1246. [Google Scholar] [CrossRef] [PubMed]
- Huber, V.; Vallacchi, V.; Fleming, V.; Hu, X.; Cova, A.; Dugo, M.; Shahaj, E.; Sulsenti, R.; Vergani, E.; Filipazzi, P.; et al. Tumor-derived microRNAs induce myeloid suppressor cells and predict immunotherapy resistance in melanoma. J. Clin. Investig. 2018, 128, 5505–5516. [Google Scholar] [CrossRef] [PubMed]
- Cobos Jiménez, V.; Bradley, E.J.; Willemsen, A.M.; van Kampen, A.H.; Baas, F.; Kootstra, N.A. Next-generation sequencing of microRNAs uncovers expression signatures in polarized macrophages. Physiol. Genom. 2014, 46, 91–103. [Google Scholar] [CrossRef] [PubMed]
- Eigsti, R.L.; Sudan, B.; Wilson, M.E.; Graff, J.W. Regulation of activation-associated microRNA accumulation rates during monocyte-to-macrophage differentiation. J. Biol. Chem. 2014, 289, 28433–28447. [Google Scholar] [CrossRef] [PubMed]
- Kaneda, M.M.; Messer, K.S.; Ralainirina, N.; Li, H.; Leem, C.J.; Gorjestani, S.; Woo, G.; Nguyen, A.V.; Figueiredo, C.C.; Foubert, P.; et al. PI3Kγ is a molecular switch that controls immune suppression. Nature 2016, 539, 437–442. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.M.; Kim, T.S.; Jo, E.K. MiR-146 and miR-125 in the regulation of innate immunity and inflammation. BMB Rep. 2016, 49, 311–318. [Google Scholar] [CrossRef] [PubMed]
- Taganov, K.D.; Boldin, M.P.; Chang, K.J.; Baltimore, D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc. Natl. Acad. Sci. USA 2006, 103, 12481–12486. [Google Scholar] [CrossRef] [PubMed]
- Hildebrand, D.; Eberle, M.E.; Wölfle, S.M.; Egler, F.; Sahin, D.; Sähr, A.; Bode, K.A.; Heeg, K. Hsa-miR-99b/let-7e/miR-125a Cluster Regulates Pathogen Recognition Receptor-Stimulated Suppressive Antigen-Presenting Cells. Front. Immunol. 2018, 9, 1224. [Google Scholar] [CrossRef] [PubMed]
- Fleming, N.H.; Zhong, J.; da Silva, I.P.; Vega-Saenz de Miera, E.; Brady, B.; Han, S.W.; Hanniford, D.; Wang, J.; Shapiro, R.L.; Hernando, E.; et al. Serum-based miRNAs in the prediction and detection of recurrence in melanoma patients. Cancer 2015, 121, 51–59. [Google Scholar] [CrossRef] [PubMed]
- Shiiyama, R.; Fukushima, S.; Jinnin, M.; Yamashita, J.; Miyashita, A.; Nakahara, S.; Kogi, A.; Aoi, J.; Masuguchi, S.; Inoue, Y.; et al. Sensitive detection of melanoma metastasis using circulating microRNA expression profiles. Melanoma Res. 2013, 23, 366–372. [Google Scholar] [CrossRef] [PubMed]
- Margue, C.; Reinsbach, S.; Philippidou, D.; Beaume, N.; Walters, C.; Schneider, J.G.; Nashan, D.; Behrmann, I.; Kreis, S. Comparison of a healthy miRNome with melanoma patient miRNomes: Are microRNAs suitable serum biomarkers for cancer? Oncotarget 2015, 6, 12110–12127. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; He, Q.Y.; Luo, C.Q.; Qian, L.Y. Circulating miR-221 expression level and prognosis of cutaneous malignant melanoma. Med. Sci. Monit. 2014, 20, 2472–2477. [Google Scholar] [CrossRef] [PubMed]
- Kanemaru, H.; Fukushima, S.; Yamashita, J.; Honda, N.; Oyama, R.; Kakimoto, A.; Masuguchi, S.; Ishihara, T.; Inoue, Y.; Jinnin, M.; et al. The circulating microRNA-221 level in patients with malignant melanoma as a new tumor marker. J. Dermatol. Sci. 2011, 61, 187–193. [Google Scholar] [CrossRef] [PubMed]
- Lawrie, C.H.; Gal, S.; Dunlop, H.M.; Pushkaaran, B.; Liggins, A.P.; Pulford, K.; Banham, A.H.; Pezzella, F.; Boultwood, J.; Wainscoat, J.S.; et al. Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large B-cell lymphoma. Br. J. Haematol. 2008, 141, 672–675. [Google Scholar] [CrossRef] [PubMed]
- Xin, Y.; Li, Z.; Chan, M.T.; Wu, W.K. Circulating epigenetic biomarkers in melanoma. Tumor Biol. 2016, 37, 1487–1492. [Google Scholar] [CrossRef] [PubMed]
- Solé, C.; Tramonti, D.; Schramm, M.; Goicoechea, I.; Armesto, M.; Hernandez, L.I.; Manterola, L.; Fernandez-Mercado, M.; Mujika, K.; Tuneu, A.; et al. The circulating transcriptome as a source of biomarkers for melanoma. Cancers (Basel) 2019, 11, 70. [Google Scholar] [CrossRef] [PubMed]
- Saldanha, G.; Potter, L.; Shendge, P.; Osborne, J.; Nicholson, S.; Yii, N.; Varma, S.; Aslam, M.I.; Elshaw, S.; Papadogeorgakis, E.; et al. Plasma microRNA-21 is associated with tumor burden in cutaneous melanoma. J. Investig. Dermatol. 2013, 133, 1381–1384. [Google Scholar] [CrossRef] [PubMed]
- Sempere, L.F.; Keto, J.; Fabbri, M. Exosomal MicroRNAs in Breast Cancer towards Diagnostic and Therapeutic Applications. Cancers (Basel) 2017, 9, 71. [Google Scholar] [CrossRef] [PubMed]
- Nagesh, P.K.B.; Chowdhury, P.; Hatami, E.; Boya, V.K.N.; Kashyap, V.K.; Khan, S.; Hafeez, B.B.; Chauhan, S.C.; Jaggi, M.; Yallapu, M.M. MiRNA-205 Nanoformulation Sensitizes Prostate Cancer Cells to Chemotherapy. Cancers (Basel) 2018, 10, 289. [Google Scholar] [CrossRef] [PubMed]
- Osaki, M.; Okada, F.; Ochiya, T. MiRNA therapy targeting cancer stem cells: A new paradigm for cancer treatment and prevention of tumor recurrence. Ther. Deliv. 2015, 6, 323–337. [Google Scholar] [CrossRef] [PubMed]
- Hosseinahli, N.; Aghapour, M.; Duijf, P.H.G.; Baradaran, B. Treating cancer with microRNA replacement therapy: A literature review. J. Cell Physiol. 2018, 233, 5574–5588. [Google Scholar] [CrossRef] [PubMed]
| Upregulated miRNAs | ||||
| miRNA | Mechanism of miRNA Regulation Recognized/Confirmed in Melanoma | Target mRNA | Type of Experiment | References |
| miR-17 | ND | ETV1 | in vitro | [106] |
| miR-19 | ND | PITX1 | in vitro | [107] |
| clinical specimens | ||||
| miR-21 | AP-1/c-Jun | TIMP3 | in vitro | [108] |
| in vivo | ||||
| PTEN | in vitro | [109] | ||
| clinical specimens | ||||
| PDCD4 FBXO11 | in vitro | [110] | ||
| in vivo | ||||
| clinical specimens | ||||
| miR-25 | ND | DKK3 | in vitro | [111] |
| in vivo | ||||
| clinical specimens | ||||
| RBM47 | in vitro | [112] | ||
| miR-30d/30b | ND | GALNT7 | in vitro | [84] |
| in vivo | ||||
| clinical specimens | ||||
| miR-125b | TCF-4 | NEDD9 | in vitro | [113] |
| clinical specimens | ||||
| miR-146a | MYC | NUMB | in vitro | [114] |
| in vivo | ||||
| clinical specimens | ||||
| miR-182 | methylation [97] | MITF-M FOXO3 | in vitro | [96] |
| in vivo | ||||
| clinical specimens | ||||
| APC | in vitro | [14] | ||
| in vivo | ||||
| clinical specimens | ||||
| miR-214 | ND | AP-2 γ | in vitro | [75] |
| in vivo | ||||
| ITGA3 | clinical specimens | |||
| miR-221/222 | PLZF | c-KIT | in vitro | [68,69] |
| P27 | in vivo | |||
| in vitro | [70] | |||
| MITF | in silico | [115] | ||
| miR-224 miR-452 | E2F1 | TXNIP | in vitro | [116] |
| in vivo | ||||
| clinical specimens | ||||
| miR-340 | CRD-BP | MITF | in vitro | [105] |
| miR-638 | AP-2α | TP53INP2 | in vitro | [117] |
| in vivo | ||||
| clinical specimens | ||||
| miR-1908 miR-199a-5p miR-199a-3p | LNAs | ApoE DNAJA4 | in vitro | [73] |
| in vivo | ||||
| clinical specimens | ||||
| Downregulated miRNAs | ||||
| miRNA | Mechanism of miRNA Regulation Recognized/Confirmed in Melanoma | Target mRNA | Type of Experiment | References |
| let-7a | ND | ITGB3 | in vitro | [118] |
| clinical specimens | ||||
| let-7b | ND | BSG | in vitro | [119] |
| in vivo | ||||
| miR-34b miR-34c | methylation | MET | in vitro | [120,121] |
| miR-137 | α-MSH | PIK3R3 | in vitro | [94] |
| clinical specimens | ||||
| MITF | in vitro | [92] | ||
| miR-200c | ND | BMI-1 | in vitro | [122] |
| in vivo | ||||
| clinical specimens | ||||
| miR-203 | methylation | SLUG IL-8 | in vitro | [71] |
| in vivo | ||||
| clinical specimens | ||||
| CREB1 | in vitro | [123] | ||
| miR-211 | methylation | IGF2R TGFBR2 NFAT5 | in vitro | [99] |
| POU3F2/BRN2 | in vitro | [101] | ||
| PDK4 | in vitro | [124] | ||
| clinical specimens | ||||
| miR-218 | ND | CIP2A | in vitro | [125] |
| BMI1 | clinical specimens | |||
| miR-199a-3p | LNAs | MET | in vitro | [120] |
| miR-675 | ND | MTDH | in vitro | [126] |
| in vivo | ||||
| clinical specimens | ||||
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gajos-Michniewicz, A.; Czyz, M. Role of miRNAs in Melanoma Metastasis. Cancers 2019, 11, 326. https://doi.org/10.3390/cancers11030326
Gajos-Michniewicz A, Czyz M. Role of miRNAs in Melanoma Metastasis. Cancers. 2019; 11(3):326. https://doi.org/10.3390/cancers11030326
Chicago/Turabian StyleGajos-Michniewicz, Anna, and Malgorzata Czyz. 2019. "Role of miRNAs in Melanoma Metastasis" Cancers 11, no. 3: 326. https://doi.org/10.3390/cancers11030326
APA StyleGajos-Michniewicz, A., & Czyz, M. (2019). Role of miRNAs in Melanoma Metastasis. Cancers, 11(3), 326. https://doi.org/10.3390/cancers11030326





