Hispidulin Enhances TRAIL-Mediated Apoptosis via CaMKKβ/AMPK/USP51 Axis-Mediated Bim Stabilization
Abstract
1. Introduction
2. Results
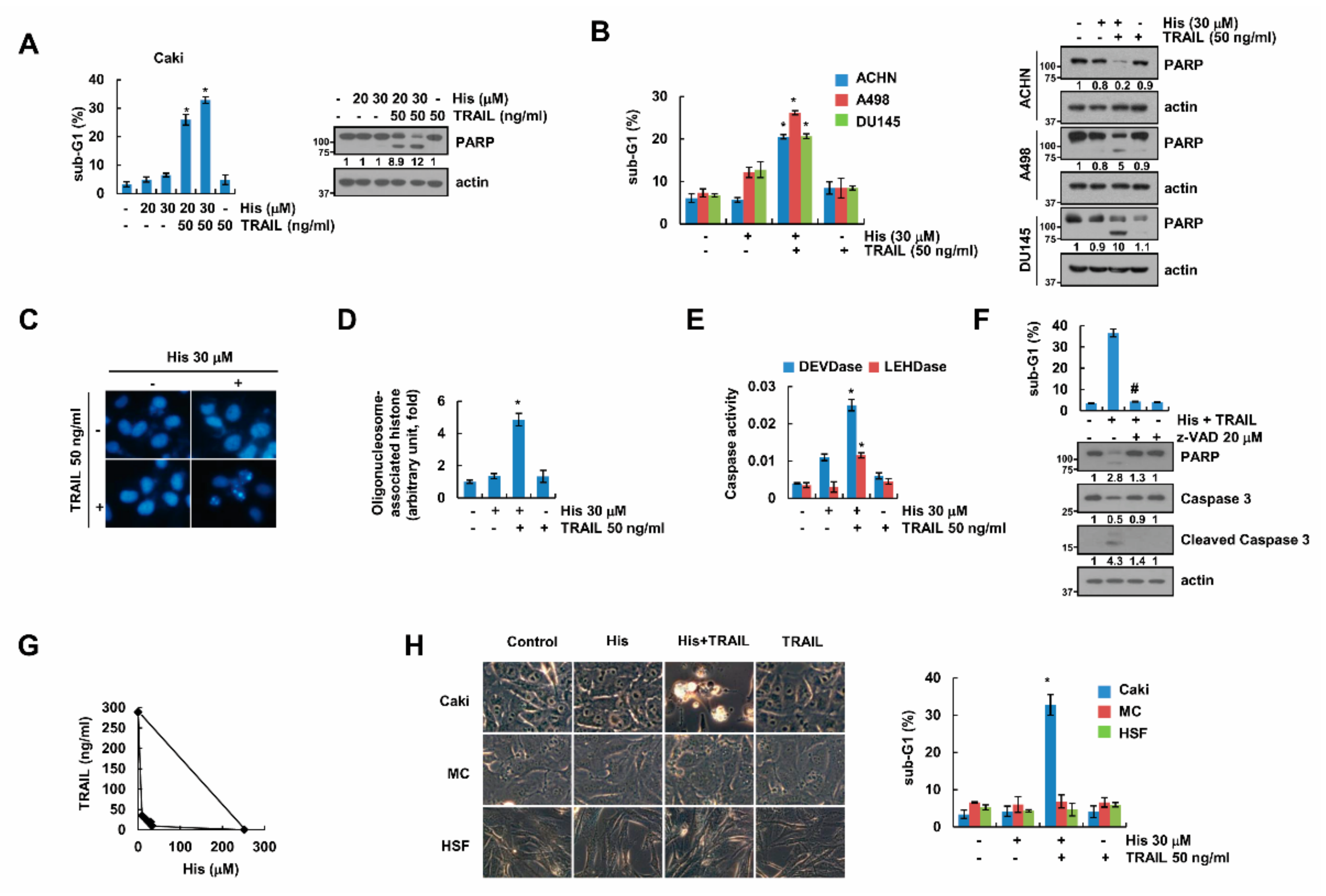
2.1. Hispidulin Enhances TRAIL-Mediated Apoptosis in Cancer Cells
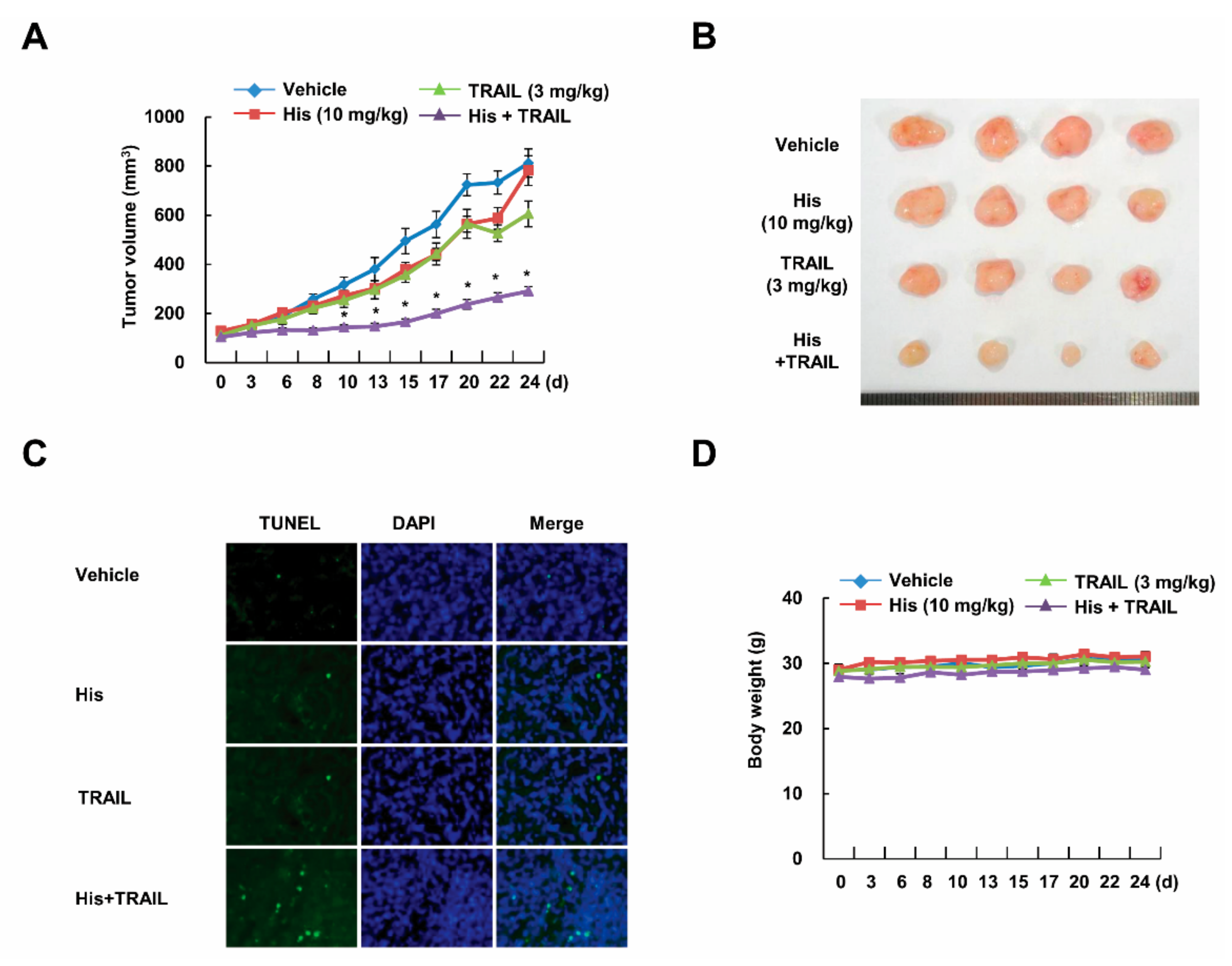
2.2. Co-Treatment with Hispidulin and TRAIL Reduces Tumor Volume In Vivo
2.3. Hispidulin Induces Loss of Mitochondrial Membrane Potential
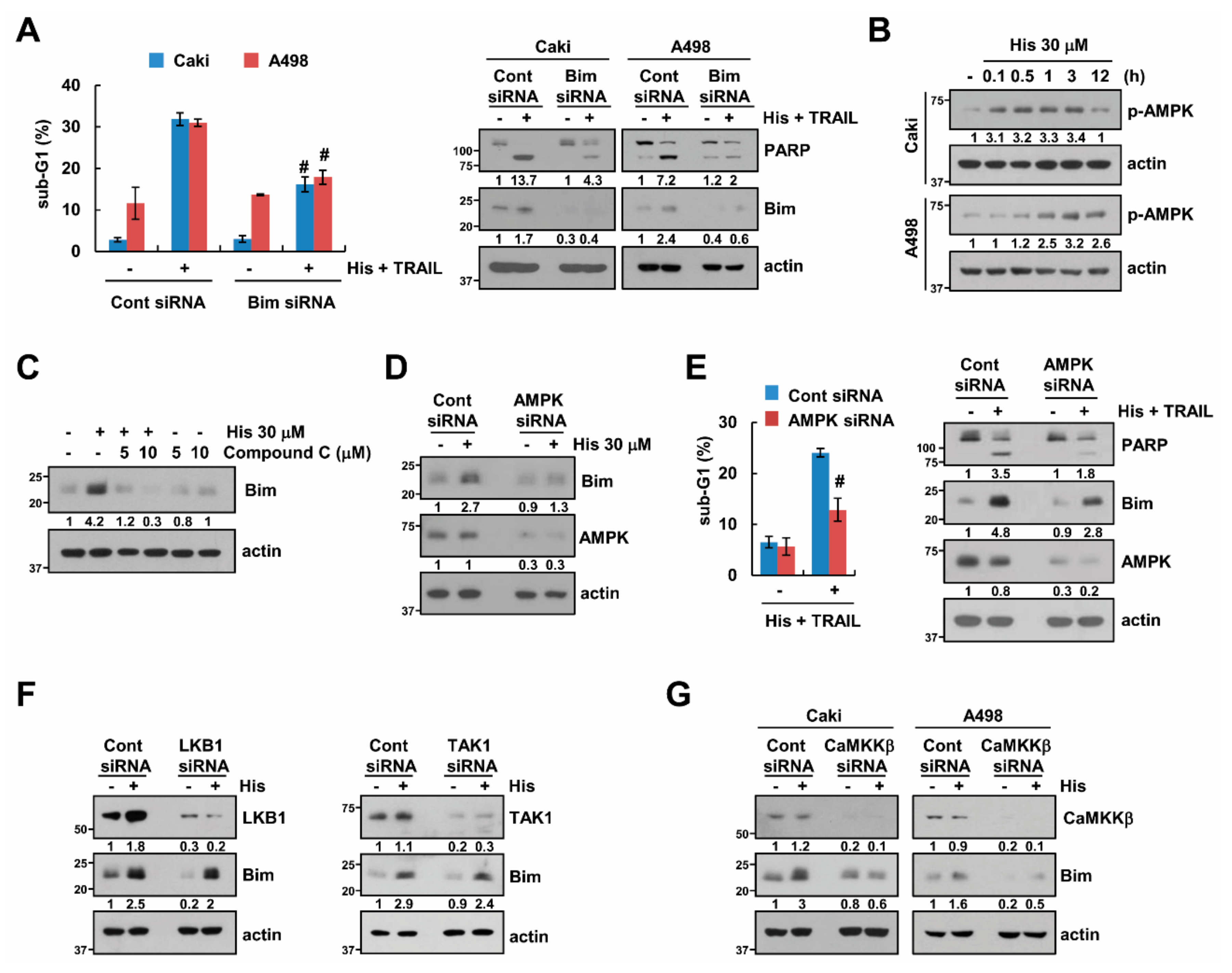
2.4. Stabilization of Bim Is Involved in Combined Treatment-Induced Apoptosis
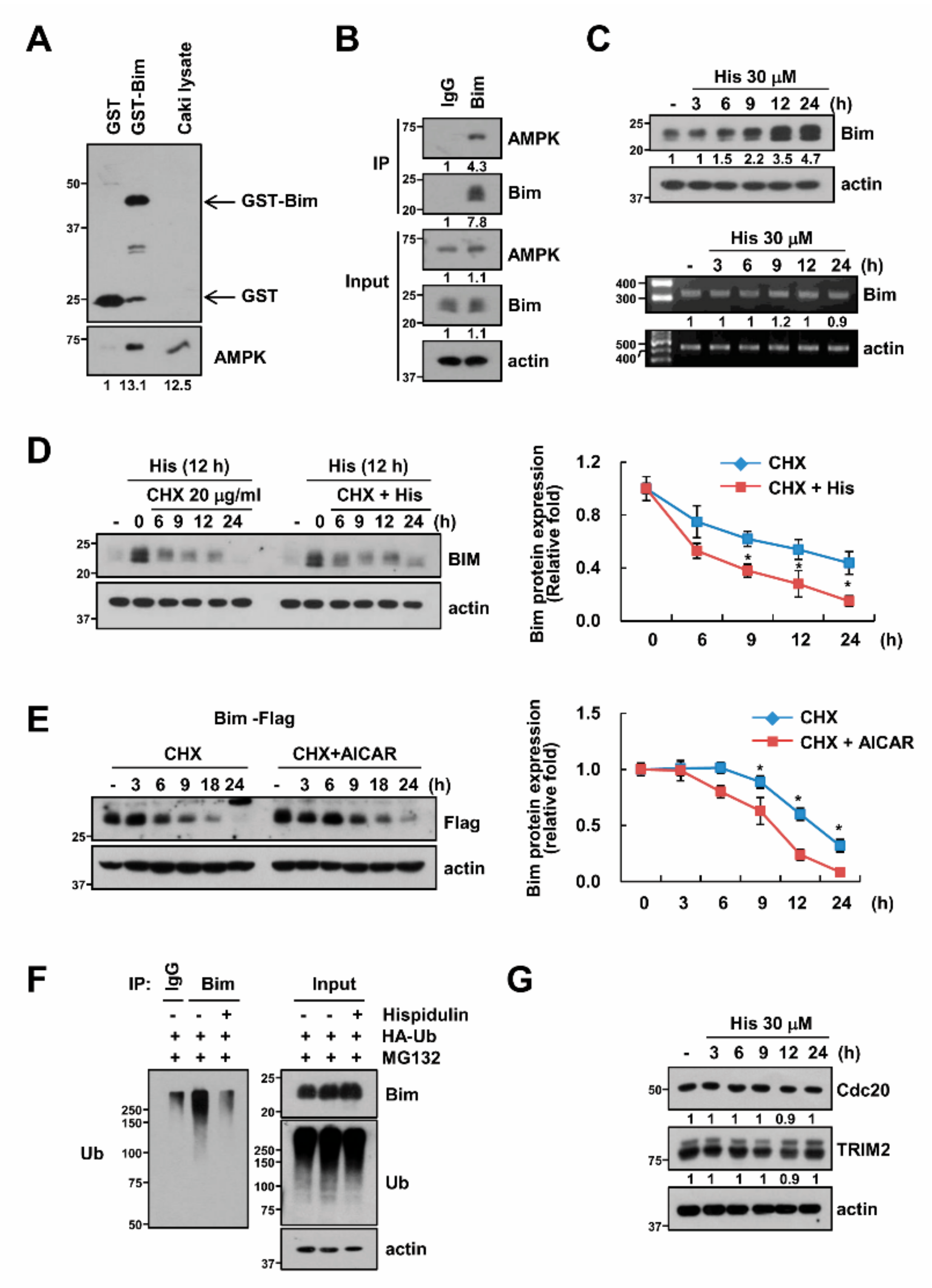
2.5. AMPK Increases Bim Protein Stability
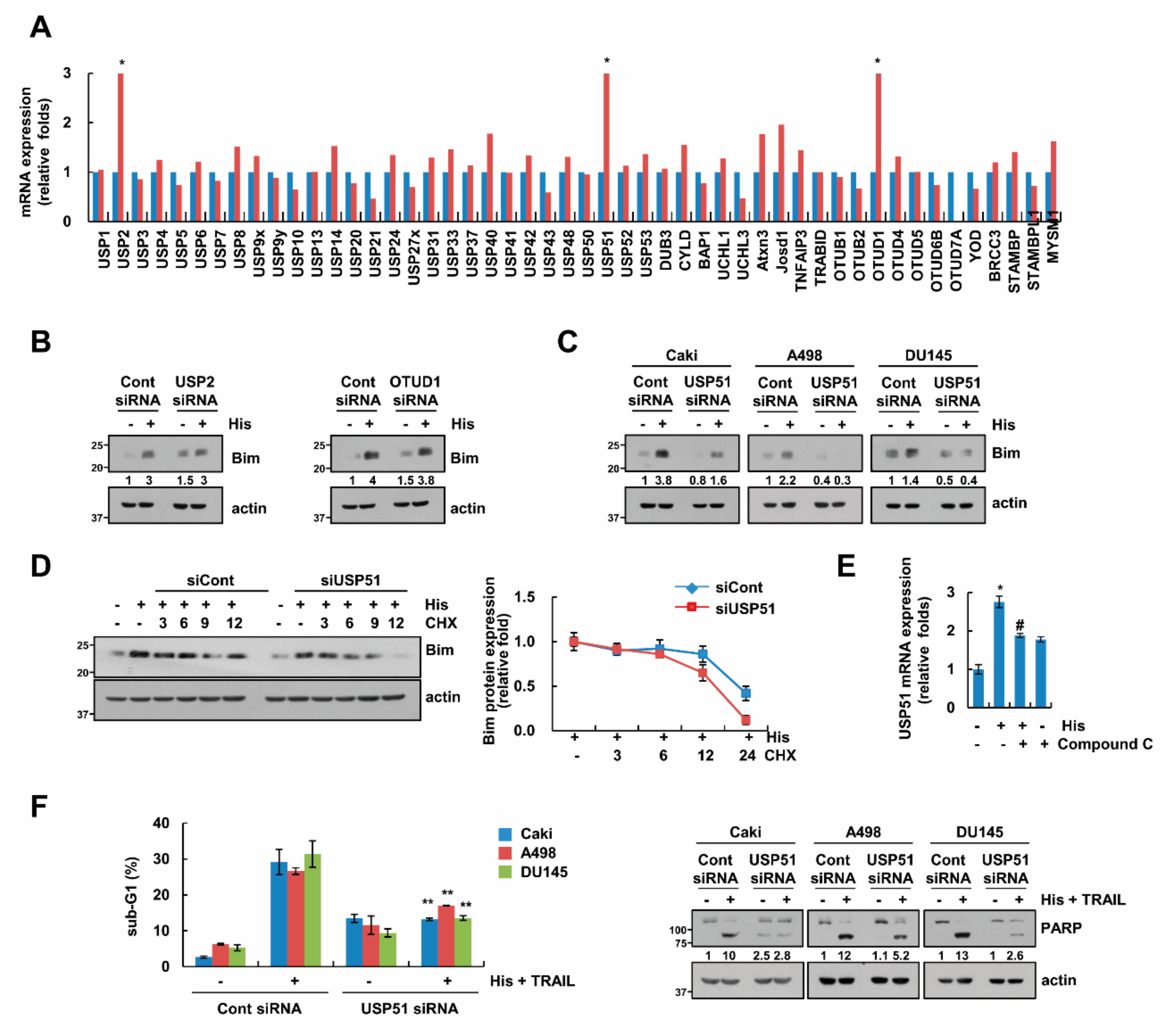
2.6. USP51 Modulates Hispidulin-Induced Bim Stabilization
3. Discussion
4. Materials and Methods
4.1. Cell Cultures and Materials
4.2. Flow Cytometry Analysis
4.3. Western Blot Analysis
4.4. Determination of Synergy
4.5. 4′,6′-Diamidino-2-Phenylindole Staining (DAPI) for Nuclei Condensation and Cell Death Assessment by DNA Fragmentation Assay
4.6. Asp-Glu-Val-Asp-Ase (DEVDase) Activity Assay
4.7. In Vivo Xenograft Model
4.8. Determination of the Mitochondrial Membrane Potential
4.9. Preparation of Cytosolic and Mitochondrial Fractions
4.10. Assay for Bax Activation and Oligomerization
4.11. GST Protein Purification and GST Pull-Down Assay
4.12. Ubiquitination Assay
4.13. Reverse Transcription Polymerase Chain Reaction and Real Time PCR
4.14. Small-Interfering RNAs (siRNAs)
4.15. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| TRAIL | Tumor necrosis factor-related apoptosis-inducing ligand |
| PARP | poly (ADP-ribose) polymerase |
| MMP | Mitochondrial membrane potential |
| FADD | Fas-associated death domain |
| DR | death receptor |
| AMPK | 5’ AMP-activated protein kinase |
| LKB1 | Liver kinase B1 |
| TAK1 | TGF-beta-activated kinase 1 |
| CaMKK | Calcium/calmodulin dependent protein kinase kinase |
| STAT3 | Signal transducer and activator of transcription 3 |
| β-TrCP | β-transducin repeats-containing proteins |
| USP | Ubiquitin specific peptidase |
| OTUD1 | OTU deubiquitinase 1 |
References
- Yin, Y.; Gong, F.Y.; Wu, X.X.; Sun, Y.; Li, Y.H.; Chen, T.; Xu, Q. Anti-inflammatory and immunosuppressive effect of flavones isolated from Artemisia vestita. J. Ethnopharmacol. 2008, 120, 1–6. [Google Scholar] [CrossRef]
- Merfort, I. Methylated Flavonoids from Arnica montana and Arnica chamissonis. Planta Med. 1984, 50, 107–108. [Google Scholar] [CrossRef]
- Lin, T.Y.; Lu, C.W.; Wang, S.J.; Huang, S.K. Protective effect of hispidulin on kainic acid-induced seizures and neurotoxicity in rats. Eur. J. Pharmacol. 2015, 755, 6–15. [Google Scholar] [CrossRef]
- Tan, R.X.; Lu, H.; Wolfender, J.L.; Yu, T.T.; Zheng, W.F.; Yang, L.; Gafner, S.; Hostettmann, K. Mono- and sesquiterpenes and antifungal constituents from Artemisia species. Planta Med. 1999, 65, 64–67. [Google Scholar] [CrossRef]
- Atif, M.; Ali, I.; Hussain, A.; Hyder, S.V.; Niaz, B.; Khan, F.A.; Maalik, A.; Farooq, U. Pharmacological Assessment of Hispidulin—A Natural Bioactive Flavone. Acta Pol. Pharm. 2015, 72, 829–842. [Google Scholar]
- Gao, H.; Wang, H.; Peng, J. Hispidulin induces apoptosis through mitochondrial dysfunction and inhibition of P13k/Akt signalling pathway in HepG2 cancer cells. Cell Biochem. Biophys. 2014, 69, 27–34. [Google Scholar] [CrossRef]
- Gao, H.; Liu, Y.; Li, K.; Wu, T.; Peng, J.; Jing, F. Hispidulin induces mitochondrial apoptosis in acute myeloid leukemia cells by targeting extracellular matrix metalloproteinase inducer. Am. J. Transl. Res. 2016, 8, 1115–1132. [Google Scholar]
- Qi, X.; Disatnik, M.H.; Shen, N.; Sobel, R.A.; Mochly-Rosen, D. Aberrant mitochondrial fission in neurons induced by protein kinase Cδ under oxidative stress conditions in vivo. Mol. Biol. Cell 2011, 22, 256–265. [Google Scholar] [CrossRef]
- Fan, Y.; Lu, H.; Ma, H.; Feng, F.; Hu, X.; Zhang, Q.; Wang, J.; Xu, Y.; Zhao, Q. Bioactive compounds of Eriocaulon sieboldianum blocking proliferation and inducing apoptosis of HepG2 cells might be involved in Aurora kinase inhibition. Food Funct. 2015, 6, 3746–3759. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, W.; He, X.; Fei, Z. Hispidulin enhances the anti-tumor effects of temozolomide in glioblastoma by activating AMPK. Cell Biochem. Biophys. 2015, 71, 701–706. [Google Scholar] [CrossRef]
- Gao, H.; Jiang, Q.; Han, Y.; Peng, J.; Wang, C. Hispidulin potentiates the antitumor effect of sunitinib against human renal cell carcinoma in laboratory models. Cell Biochem. Biophys. 2015, 71, 757–764. [Google Scholar] [CrossRef]
- Yang, J.M.; Hung, C.M.; Fu, C.N.; Lee, J.C.; Huang, C.H.; Yang, M.H.; Lin, C.L.; Kao, J.Y.; Way, T.D. Hispidulin sensitizes human ovarian cancer cells to TRAIL-induced apoptosis by AMPK activation leading to Mcl-1 block in translation. J. Agric. Food Chem. 2010, 58, 10020–10026. [Google Scholar] [CrossRef]
- Kischkel, F.C.; Lawrence, D.A.; Chuntharapai, A.; Schow, P.; Kim, K.J.; Ashkenazi, A. Apo2L/TRAIL-dependent recruitment of endogenous FADD and caspase-8 to death receptors 4 and 5. Immunity 2000, 12, 611–620. [Google Scholar] [CrossRef]
- Ashkenazi, A.; Pai, R.C.; Fong, S.; Leung, S.; Lawrence, D.A.; Marsters, S.A.; Blackie, C.; Chang, L.; McMurtrey, A.E.; Hebert, A.; et al. Safety and antitumor activity of recombinant soluble Apo2 ligand. J. Clin. Investig. 1999, 104, 155–162. [Google Scholar] [CrossRef]
- Jin, Z.; McDonald, E.R., 3rd; Dicker, D.T.; El-Deiry, W.S. Deficient tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) death receptor transport to the cell surface in human colon cancer cells selected for resistance to TRAIL-induced apoptosis. J. Biol. Chem. 2004, 279, 35829–35839. [Google Scholar] [CrossRef]
- Kelly, M.M.; Hoel, B.D.; Voelkel-Johnson, C. Doxorubicin pretreatment sensitizes prostate cancer cell lines to TRAIL induced apoptosis which correlates with the loss of c-FLIP expression. Cancer Biol. Ther. 2002, 1, 520–527. [Google Scholar] [CrossRef]
- Ng, C.P.; Zisman, A.; Bonavida, B. Synergy is achieved by complementation with Apo2L/TRAIL and actinomycin D in Apo2L/TRAIL-mediated apoptosis of prostate cancer cells: Role of XIAP in resistance. Prostate 2002, 53, 286–299. [Google Scholar] [CrossRef]
- Walczak, H.; Bouchon, A.; Stahl, H.; Krammer, P.H. Tumor necrosis factor-related apoptosis-inducing ligand retains its apoptosis-inducing capacity on Bcl-2- or Bcl-xL-overexpressing chemotherapy-resistant tumor cells. Cancer Res. 2000, 60, 3051–3057. [Google Scholar]
- Zhang, Y.; Zhang, B. TRAIL resistance of breast cancer cells is associated with constitutive endocytosis of death receptors 4 and 5. Mol. Cancer Res. MCR 2008, 6, 1861–1871. [Google Scholar] [CrossRef]
- Lin, J.; Wu, L.; Bai, X.; Xie, Y.; Wang, A.; Zhang, H.; Yang, X.; Wan, X.; Lu, X.; Sang, X.; et al. Combination treatment including targeted therapy for advanced hepatocellular carcinoma. Oncotarget 2016, 7, 71036–71051. [Google Scholar] [CrossRef]
- Kroemer, G.; Galluzzi, L.; Brenner, C. Mitochondrial membrane permeabilization in cell death. Physiol. Rev. 2007, 87, 99–163. [Google Scholar] [CrossRef]
- Jurgensmeier, J.M.; Xie, Z.; Deveraux, Q.; Ellerby, L.; Bredesen, D.; Reed, J.C. Bax directly induces release of cytochrome c from isolated mitochondria. Proc. Natl. Acad. Sci. USA 1998, 95, 4997–5002. [Google Scholar] [CrossRef]
- Han, M.A.; Min, K.J.; Woo, S.M.; Seo, B.R.; Kwon, T.K. Eupafolin enhances TRAIL-mediated apoptosis through cathepsin S-induced down-regulation of Mcl-1 expression and AMPK-mediated Bim up-regulation in renal carcinoma Caki cells. Oncotarget 2016, 7, 65707–65720. [Google Scholar] [CrossRef]
- Eom, J.W.; Lee, J.M.; Koh, J.Y.; Kim, Y.H. AMP-activated protein kinase contributes to zinc-induced neuronal death via activation by LKB1 and induction of Bim in mouse cortical cultures. Mol. Brain 2016, 9, 14. [Google Scholar] [CrossRef]
- Cheng, J.; Zhang, T.; Ji, H.; Tao, K.; Guo, J.; Wei, W. Functional characterization of AMP-activated protein kinase signaling in tumorigenesis. Biochim. Biophys. Acta 2016, 1866, 232–251. [Google Scholar] [CrossRef]
- Concannon, C.G.; Tuffy, L.P.; Weisova, P.; Bonner, H.P.; Davila, D.; Bonner, C.; Devocelle, M.C.; Strasser, A.; Ward, M.W.; Prehn, J.H. AMP kinase-mediated activation of the BH3-only protein Bim couples energy depletion to stress-induced apoptosis. J. Cell Biol. 2010, 189, 83–94. [Google Scholar] [CrossRef]
- Weber, A.; Heinlein, M.; Dengjel, J.; Alber, C.; Singh, P.K.; Hacker, G. The deubiquitinase Usp27x stabilizes the BH3-only protein Bim and enhances apoptosis. EMBO Rep. 2016, 17, 724–738. [Google Scholar] [CrossRef]
- Ley, R.; Balmanno, K.; Hadfield, K.; Weston, C.; Cook, S.J. Activation of the ERK1/2 signaling pathway promotes phosphorylation and proteasome-dependent degradation of the BH3-only protein, Bim. J. Biol. Chem. 2003, 278, 18811–18816. [Google Scholar] [CrossRef]
- Lu, C.; Wang, W.; Jia, Y.; Liu, X.; Tong, Z.; Li, B. Inhibition of AMPK/autophagy potentiates parthenolide-induced apoptosis in human breast cancer cells. J. Cell. Biochem. 2014, 115, 1458–1466. [Google Scholar] [CrossRef]
- Thompson, S.; Pearson, A.N.; Ashley, M.D.; Jessick, V.; Murphy, B.M.; Gafken, P.; Henshall, D.C.; Morris, K.T.; Simon, R.P.; Meller, R. Identification of a novel Bcl-2-interacting mediator of cell death (Bim) E3 ligase, tripartite motif-containing protein 2 (TRIM2), and its role in rapid ischemic tolerance-induced neuroprotection. J. Biol. Chem. 2011, 286, 19331–19339. [Google Scholar] [CrossRef]
- Herbst, R.S.; Eckhardt, S.G.; Kurzrock, R.; Ebbinghaus, S.; O’Dwyer, P.J.; Gordon, M.S.; Novotny, W.; Goldwasser, M.A.; Tohnya, T.M.; Lum, B.L.; et al. Phase I dose-escalation study of recombinant human Apo2L/TRAIL, a dual proapoptotic receptor agonist, in patients with advanced cancer. J. Clin. Oncol. 2010, 28, 2839–2846. [Google Scholar] [CrossRef]
- Leong, S.; Cohen, R.B.; Gustafson, D.L.; Langer, C.J.; Camidge, D.R.; Padavic, K.; Gore, L.; Smith, M.; Chow, L.Q.; von Mehren, M.; et al. Mapatumumab, an antibody targeting TRAIL-R1, in combination with paclitaxel and carboplatin in patients with advanced solid malignancies: Results of a phase I and pharmacokinetic study. J. Clin. Oncol. 2009, 27, 4413–4421. [Google Scholar] [CrossRef]
- Forero-Torres, A.; Infante, J.R.; Waterhouse, D.; Wong, L.; Vickers, S.; Arrowsmith, E.; He, A.R.; Hart, L.; Trent, D.; Wade, J.; et al. Phase 2, multicenter, open-label study of tigatuzumab (CS-1008), a humanized monoclonal antibody targeting death receptor 5, in combination with gemcitabine in chemotherapy-naive patients with unresectable or metastatic pancreatic cancer. Cancer Med. 2013, 2, 925–932. [Google Scholar] [CrossRef]
- Lemke, J.; von Karstedt, S.; Zinngrebe, J.; Walczak, H. Getting TRAIL back on track for cancer therapy. Cell Death Differ. 2014, 21, 1350–1364. [Google Scholar] [CrossRef]
- Hardie, D.G. AMP-activated protein kinase: An energy sensor that regulates all aspects of cell function. Genes Dev. 2011, 25, 1895–1908. [Google Scholar] [CrossRef]
- Kamel, W.; Sugihara, E.; Nobusue, H.; Yamaguchi-Iwai, S.; Onishi, N.; Maki, K.; Fukuchi, Y.; Matsuo, K.; Muto, A.; Saya, H.; et al. Simvastatin-induced apoptosis in osteosarcoma cells: A key role of RhoA-AMPK-p38 MAPK signaling in antitumor activity. Mol. Cancer Ther. 2016, 16, 182–192. [Google Scholar] [CrossRef]
- Liu, Z.; Ren, L.; Liu, C.; Xia, T.; Zha, X.; Wang, S. Phenformin Induces Cell Cycle Change, Apoptosis, and Mesenchymal-Epithelial Transition and Regulates the AMPK/mTOR/p70s6k and MAPK/ERK Pathways in Breast Cancer Cells. PLoS ONE 2015, 10, e0131207. [Google Scholar] [CrossRef]
- Chen, M.B.; Shen, W.X.; Yang, Y.; Wu, X.Y.; Gu, J.H.; Lu, P.H. Activation of AMP-activated protein kinase is involved in vincristine-induced cell apoptosis in B16 melanoma cell. J. Cell Physiol. 2011, 226, 1915–1925. [Google Scholar] [CrossRef]
- Ji, C.; Yang, B.; Yang, Y.L.; He, S.H.; Miao, D.S.; He, L.; Bi, Z.G. Exogenous cell-permeable C6 ceramide sensitizes multiple cancer cell lines to Doxorubicin-induced apoptosis by promoting AMPK activation and mTORC1 inhibition. Oncogene 2010, 29, 6557–6568. [Google Scholar] [CrossRef]
- Jang, J.E.; Eom, J.I.; Jeung, H.K.; Cheong, J.W.; Lee, J.Y.; Kim, J.S.; Min, Y.H. AMPK-ULK1-mediated autophagy confers resistance to BET inhibitor JQ1 in acute myeloid leukemia stem cells. Clin. Cancer. Res. 2016, 23, 2781–2794. [Google Scholar] [CrossRef]
- Wang, J.; Qi, Q.; Feng, Z.; Zhang, X.; Huang, B.; Chen, A.; Prestegarden, L.; Li, X.; Wang, J. Berberine induces autophagy in glioblastoma by targeting the AMPK/mTOR/ULK1-pathway. Oncotarget 2016, 7, 66944–66958. [Google Scholar] [CrossRef]
- Liu, Q.; Osterlund, E.J.; Chi, X.; Pogmore, J.; Leber, B.; Andrews, D.W. Bim escapes displacement by BH3-mimetic anti-cancer drugs by double-bolt locking both Bcl-XL and Bcl-2. eLife 2019, 8, e37689. [Google Scholar] [CrossRef]
- Song, K.A.; Niederst, M.J.; Lochmann, T.L.; Hata, A.N.; Kitai, H.; Ham, J.; Floros, K.V.; Hicks, M.A.; Hu, H.; Mulvey, H.E.; et al. Epithelial-to-Mesenchymal Transition Antagonizes Response to Targeted Therapies in Lung Cancer by Suppressing BIM. Clin. Cancer Res. 2018, 24, 197–208. [Google Scholar] [CrossRef]
- Delbridge, A.R.; Grabow, S.; Strasser, A. Loss of BIM augments resistance of ATM-deficient thymocytes to DNA damage-induced apoptosis but does not accelerate lymphoma development. Cell Death Differ. 2017, 24, 1987–1988. [Google Scholar] [CrossRef]
- Dehan, E.; Bassermann, F.; Guardavaccaro, D.; Vasiliver-Shamis, G.; Cohen, M.; Lowes, K.N.; Dustin, M.; Huang, D.C.; Taunton, J.; Pagano, M. betaTrCP- and Rsk1/2-mediated degradation of BimEL inhibits apoptosis. Mol. Cell 2009, 33, 109–116. [Google Scholar] [CrossRef]
- Ittiudomrak, T.; Puthong, S.; Roytrakul, S.; Chanchao, C. Alpha-mangostin and apigenin induced cell cycle arrest and programmed cell death in SKOV-3 ovarian cancer cells. Toxicol. Res. 2019, 35, 167–179. [Google Scholar] [CrossRef]
- Tallarida, R.J. Drug synergism: Its detection and applications. J. Pharmacol. Exp. Ther. 2001, 298, 865–872. [Google Scholar]
- Seo, J.; Lee, E.W.; Shin, J.; Seong, D.; Nam, Y.W.; Jeong, M.; Lee, S.H.; Lee, C.; Song, J. K6 linked polyubiquitylation of FADD by CHIP prevents death inducing signaling complex formation suppressing cell death. Oncogene 2018, 37, 4994–5006. [Google Scholar] [CrossRef]
- Kim, S.; Woo, S.M.; Min, K.J.; Seo, S.U.; Lee, T.J.; Kubatka, P.; Kim, D.E.; Kwon, T.K. WP1130 Enhances TRAIL-Induced Apoptosis through USP9X-Dependent miR-708-Mediated Downregulation of c-FLIP. Cancers 2019, 11, 344. [Google Scholar] [CrossRef]


© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Woo, S.M.; Seo, S.U.; Kim, S.H.; Nam, J.-O.; Kim, S.; Park, J.-W.; Min, K.-j.; Kwon, T.K. Hispidulin Enhances TRAIL-Mediated Apoptosis via CaMKKβ/AMPK/USP51 Axis-Mediated Bim Stabilization. Cancers 2019, 11, 1960. https://doi.org/10.3390/cancers11121960
Woo SM, Seo SU, Kim SH, Nam J-O, Kim S, Park J-W, Min K-j, Kwon TK. Hispidulin Enhances TRAIL-Mediated Apoptosis via CaMKKβ/AMPK/USP51 Axis-Mediated Bim Stabilization. Cancers. 2019; 11(12):1960. https://doi.org/10.3390/cancers11121960
Chicago/Turabian StyleWoo, Seon Min, Seung Un Seo, Sang Hyun Kim, Ju-Ock Nam, Shin Kim, Jong-Wook Park, Kyoung-jin Min, and Taeg Kyu Kwon. 2019. "Hispidulin Enhances TRAIL-Mediated Apoptosis via CaMKKβ/AMPK/USP51 Axis-Mediated Bim Stabilization" Cancers 11, no. 12: 1960. https://doi.org/10.3390/cancers11121960
APA StyleWoo, S. M., Seo, S. U., Kim, S. H., Nam, J.-O., Kim, S., Park, J.-W., Min, K.-j., & Kwon, T. K. (2019). Hispidulin Enhances TRAIL-Mediated Apoptosis via CaMKKβ/AMPK/USP51 Axis-Mediated Bim Stabilization. Cancers, 11(12), 1960. https://doi.org/10.3390/cancers11121960







