Deleterious and Oncogenic Mutations in the IL7RA
Abstract
1. Introduction
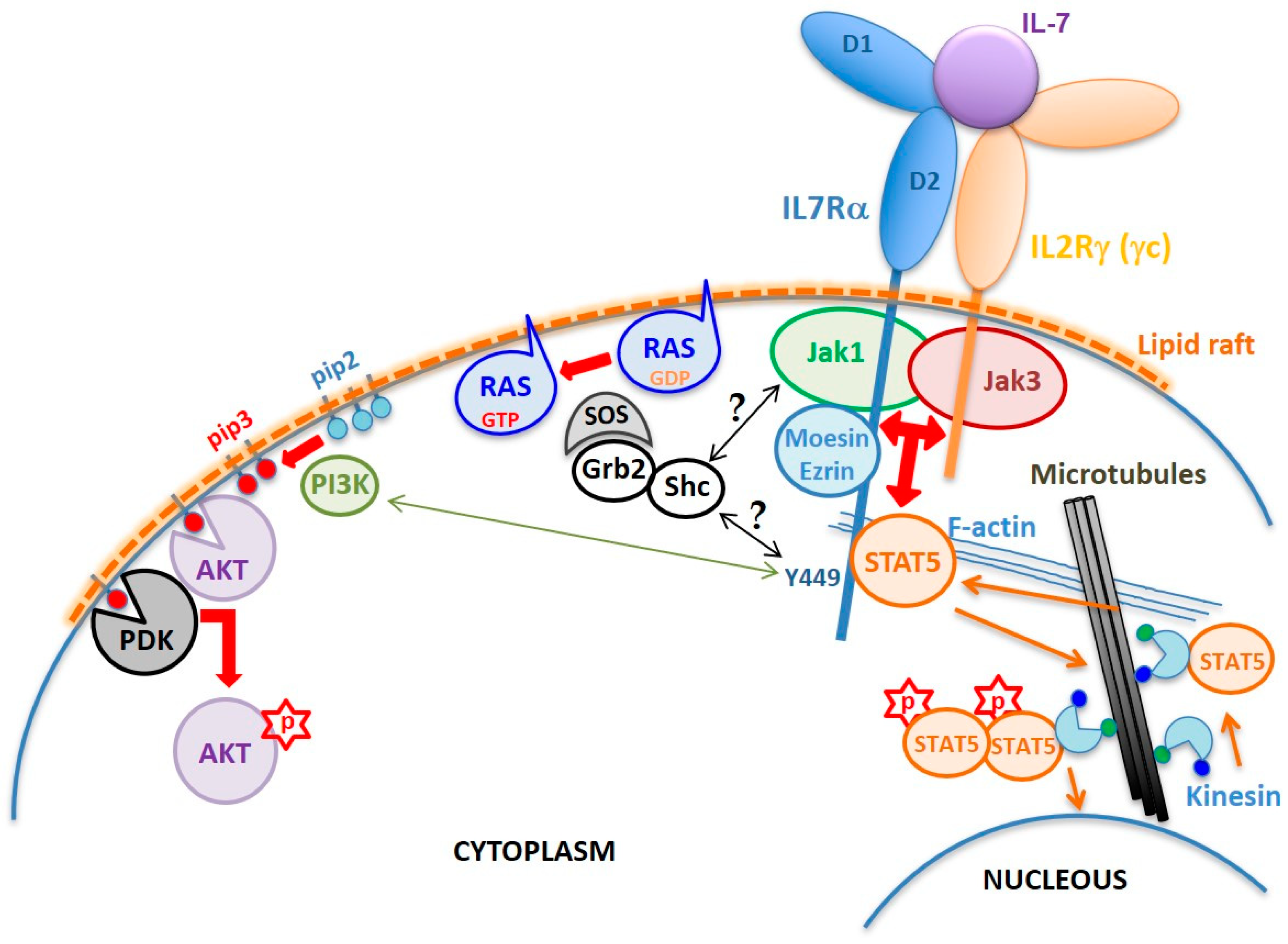
2. Structural Determinants of the IL7Rα Activity
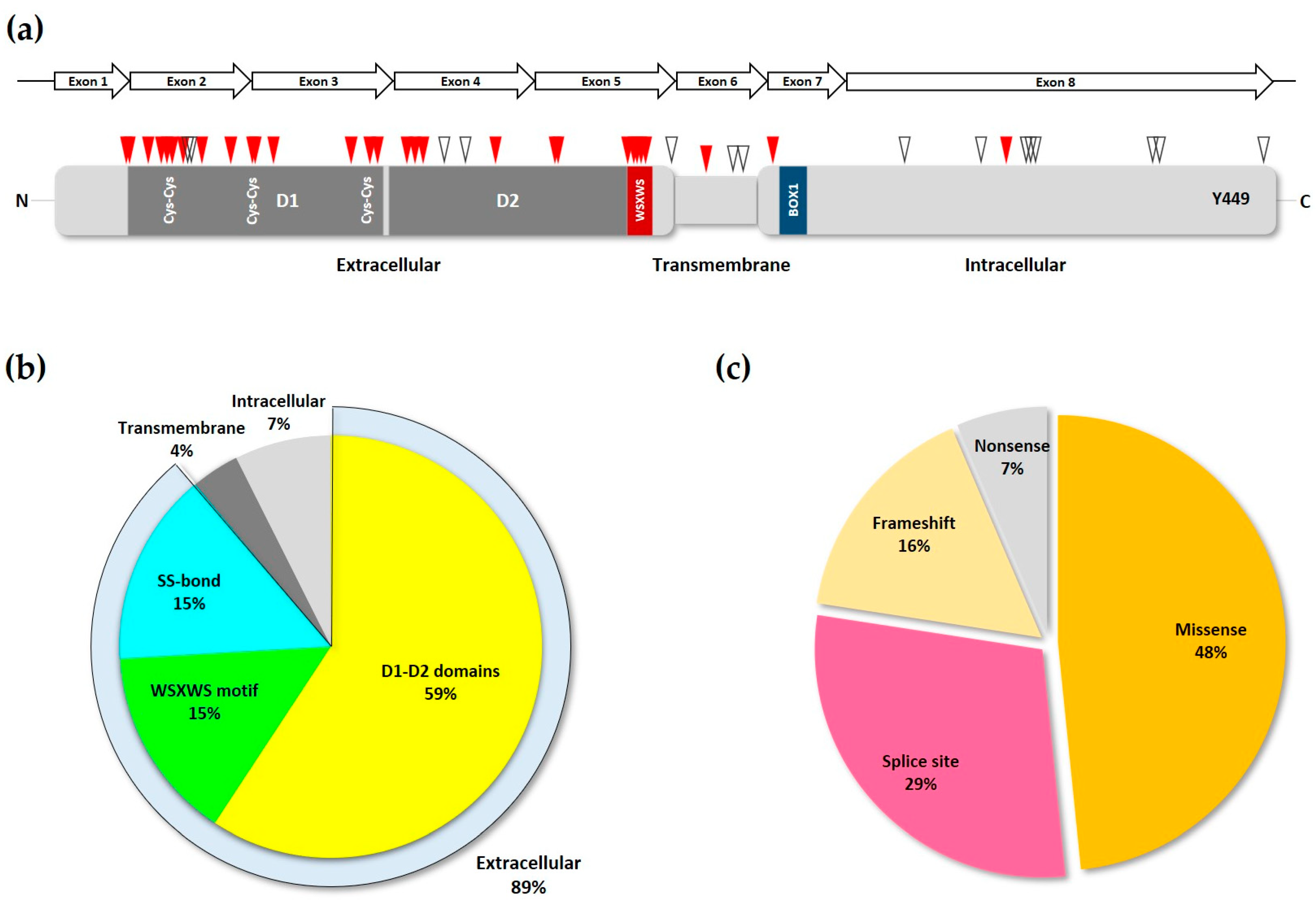
3. Deleterious Mutations in the IL7R
4. Oncogenic Mutations in the IL7R
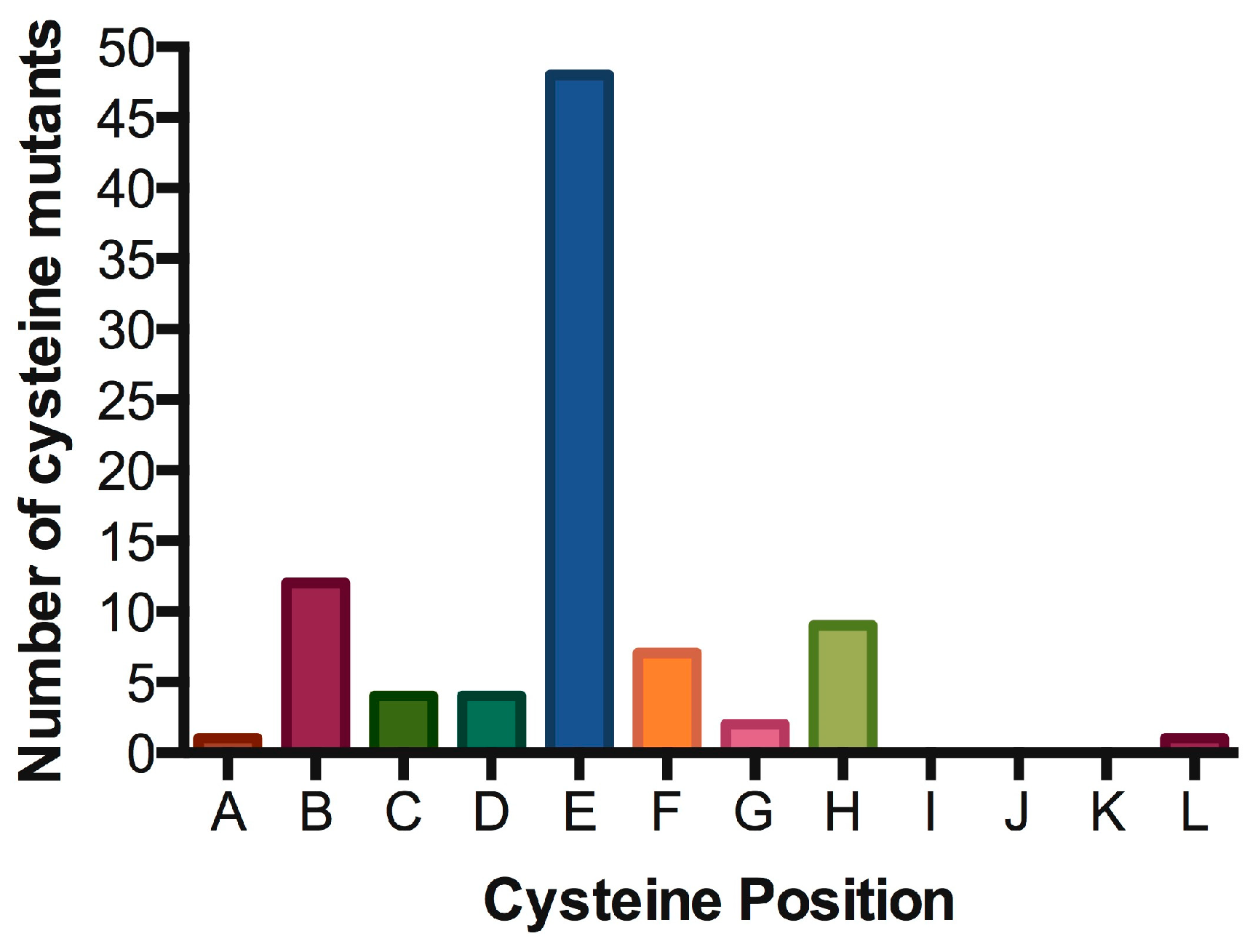
4.1. IL7Rα Cysteine Mutants
4.2. IL7Rα Cysteine-Lacking Mutants
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Cui, L.; Fu, J.; Pang, J.C.-S.; Qiu, Z.-K.; Liu, X.-M.; Chen, F.-R.; Shi, H.-L.; Ng, H.-K.; Chen, Z. Overexpression of IL-7 enhances cisplatin resistance in glioma. Cancer Biol. Ther. 2012, 13, 496–503. [Google Scholar] [CrossRef] [PubMed]
- Al-Rawi, M.A.A.; Rmali, K.; Mansel, R.E.; Jiang, W.G. Interleukin 7 induces the growth of breast cancer cells through a wortmannin-sensitive pathway. Br. J. Surg. 2004, 91, 61–68. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Zeng, Z.; Peng, Y.; Chen, J.; Pan, L.; Pan, D. IL-7 splicing variant IL-7δ5 induces EMT and metastasis of human breast cancer cell lines MCF-7 and BT-20 through activation of PI3K/Akt pathway. Histochem. Cell Biol. 2014, 142, 401–410. [Google Scholar] [CrossRef] [PubMed]
- Ming, J.; Jiang, G.; Zhang, Q.; Qiu, X.; Wang, E. Interleukin-7 up-regulates cyclin D1 via activator protein-1 to promote proliferation of cell in lung cancer. Cancer Immunol. Immunother. 2012, 61, 79–88. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.-H.; Wang, M.-H.; Ren, H.-J.; Qu, W.; Sun, L.-M.; Zhang, Q.-F.; Qiu, X.-S.; Wang, E.-H. Interleukin 7 signaling prevents apoptosis by regulating bcl-2 and bax via the p53 pathway in human non-small cell lung cancer cells. Int. J. Clin. Exp. Pathol. 2014, 7, 870–881. [Google Scholar] [PubMed]
- Suzuki, K.; Kadota, K.; Sima, C.S.; Nitadori, J.; Rusch, V.W.; Travis, W.D.; Sadelain, M.; Adusumilli, P.S. Clinical Impact of Immune Microenvironment in Stage I Lung Adenocarcinoma: Tumor Interleukin-12 Receptor β2 (IL-12Rβ2), IL-7R, and Stromal FoxP3/CD3 Ratio Are Independent Predictors of Recurrence. J. Clin. Oncol. 2013, 31, 490–498. [Google Scholar] [CrossRef]
- Cosenza, L.; Gorgun, G.; Urbano, A.; Foss, F. Interleukin-7 receptor expression and activation in nonhaematopoietic neoplastic cell lines. Cell. Signal. 2002, 14, 317–325. [Google Scholar] [CrossRef]
- Mazzucchelli, R.; Durum, S.K. Interleukin-7 receptor expression: Intelligent design. Nat. Rev. Immunol. 2007, 7, 144–154. [Google Scholar] [CrossRef]
- Carrette, F.; Surh, C.D. IL-7 signaling and CD127 receptor regulation in the control of T cell homeostasis. Semin. Immunol. 2012, 24, 209–217. [Google Scholar] [CrossRef]
- Barata, J.T.; Durum, S.K.; Seddon, B. Flip the coin: IL-7 and IL-7R in health and disease. Nat. Immunol. 2019, 20, 1584–1593. [Google Scholar] [CrossRef]
- Goodwin, R. Cloning of the human and murine interleukin-7 receptors: Demonstration of a soluble form and homology to a new receptor superfamily. Cell 1990, 60, 941–951. [Google Scholar] [CrossRef]
- Lundstrom, W.; Highfill, S.; Walsh, S.T.R.; Beq, S.; Morse, E.; Kockum, I.; Alfredsson, L.; Olsson, T.; Hillert, J.; Mackall, C.L. Soluble IL7R potentiates IL-7 bioactivity and promotes autoimmunity. Proc. Natl. Acad. Sci. USA 2013, 110, E1761–E1770. [Google Scholar] [CrossRef] [PubMed]
- Rose, T.; Pillet, A.-H.; Lavergne, V.; Tamarit, B.; Lenormand, P.; Rousselle, J.-C.; Namane, A.; Thèze, J. Interleukin-7 Compartmentalizes Its Receptor Signaling Complex to Initiate CD4 T Lymphocyte Response. J. Biol. Chem. 2010, 285, 14898–14908. [Google Scholar] [CrossRef] [PubMed]
- McElroy, C.A.; Holland, P.J.; Zhao, P.; Lim, J.-M.; Wells, L.; Eisenstein, E.; Walsh, S.T.R. Structural reorganization of the interleukin-7 signaling complex. Proc. Natl. Acad. Sci. USA 2012, 109, 2503–2508. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Q.; Li, W.Q.; Hofmeister, R.R.; Young, H.A.; Hodge, D.R.; Keller, J.R.; Khaled, A.R.; Durum, S.K. Distinct Regions of the Interleukin-7 Receptor Regulate Different Bcl2 Family Members. Mol. Cell. Biol. 2004, 24, 6501–6513. [Google Scholar] [CrossRef]
- Jiang, Q.; Li, W.Q.; Aiello, F.B.; Mazzucchelli, R.; Asefa, B.; Khaled, A.R.; Durum, S.K. Cell biology of IL-7, a key lymphotrophin. Cytokine Growth Factor Rev. 2005, 16, 513–533. [Google Scholar] [CrossRef]
- Jiang, Q.; Li, W.-Q.; Aiello, F.B.; Klarmann, K.D.; Keller, J.R.; Durum, S.K. Retroviral transduction of IL-7Rα into IL-7Rα−/− bone marrow progenitors: Correction of lymphoid deficiency and induction of neutrophilia. Gene Ther. 2005, 12, 1761–1768. [Google Scholar] [CrossRef][Green Version]
- Palmer, M.J.; Mahajan, V.S.; Trajman, L.C.; Irvine, D.J.; Lauffenburger, D.A.; Chen, J. Interleukin-7 Receptor Signaling Network: An Integrated Systems Perspective. Cell. Mol. Immunol. 2008, 5, 79–89. [Google Scholar] [CrossRef]
- Venkitaraman, A.R.; Cowling, R.J. Interleukin-7 induces the association of phosphatidylinositol 3-kinase with the α chain of the interleukin-7 receptor. Eur. J. Immunol. 1994, 24, 2168–2174. [Google Scholar] [CrossRef]
- Crawley, J.B.; Willcocks, J.; Foxwell, B.M.J. Interleukin-7 induces T cell proliferation in the absence of Erk/MAP kinase activity. Eur. J. Immunol. 1996, 26, 2717–2723. [Google Scholar] [CrossRef]
- Osborne, L.C.; Dhanji, S.; Snow, J.W.; Priatel, J.J.; Ma, M.C.; Miners, M.J.; Teh, H.-S.; Goldsmith, M.A.; Abraham, N. Impaired CD8 T cell memory and CD4 T cell primary responses in IL-7Rα mutant mice. J. Exp. Med. 2007, 204, 619–631. [Google Scholar] [CrossRef]
- Canté-Barrett, K.; Spijkers-Hagelstein, J.A.P.; Buijs-Gladdines, J.G.C.A.M.; Uitdehaag, J.C.M.; Smits, W.K.; van der Zwet, J.; Buijsman, R.C.; Zaman, G.J.R.; Pieters, R.; Meijerink, J.P.P. MEK and PI3K-AKT inhibitors synergistically block activated IL7 receptor signaling in T-cell acute lymphoblastic leukemia. Leukemia 2016, 30, 1832–1843. [Google Scholar] [CrossRef] [PubMed]
- Bousoik, E.; Montazeri Aliabadi, H. “Do We Know Jack” About JAK? A Closer Look at JAK/STAT Signaling Pathway. Front. Oncol. 2018, 8, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Winston, L.A.; Hunter, T. Intracellular signalling: Putting JAKs on the kinase MAP. Curr. Biol. 1996, 6, 668–671. [Google Scholar] [CrossRef]
- Pandey, A.; Ozaki, K.; Baumann, H.; Levin, S.D.; Puel, A.; Farr, A.G.; Ziegler, S.F.; Leonard, W.J.; Lodish, H.F. Cloning of a receptor subunit required for signaling by thymic stromal lymphopoietin. Nat. Immunol. 2000, 1, 59–64. [Google Scholar] [CrossRef]
- Levin, S.D.; Koelling, R.M.; Friend, S.L.; Isaksen, D.E.; Ziegler, S.F.; Perlmutter, R.M.; Farr, A.G. Thymic stromal lymphopoietin: A cytokine that promotes the development of IgM+ B cells in vitro and signals via a novel mechanism. J. Immunol. 1999, 162, 677–683. [Google Scholar]
- Isaksen, D.E.; Baumann, H.; Trobridge, P.A.; Farr, A.G.; Levin, S.D.; Ziegler, S.F. Requirement for stat5 in thymic stromal lymphopoietin-mediated signal transduction. J. Immunol. 1999, 163, 5971–5977. [Google Scholar]
- McElroy, C.A.; Dohm, J.A.; Walsh, S.T.R. Structural and Biophysical Studies of the Human IL-7/IL-7Rα Complex. Structure 2009, 17, 54–65. [Google Scholar] [CrossRef]
- Pillet, A.-H.; Juffroy, O.; Mazard-Pasquier, V.; Moreau, J.-L.; Gesbert, F.; Chastagner, P.; Colle, J.-H.; Thèze, J.; Rose, T. Human IL-Rbeta chains form IL-2 binding homodimers. Eur. Cytokine Netw. 2008, 19, 49–59. [Google Scholar]
- Tamarit, B.; Bugault, F.; Pillet, A.-H.; Lavergne, V.; Bochet, P.; Garin, N.; Schwarz, U.; Thèze, J.; Rose, T. Membrane Microdomains and Cytoskeleton Organization Shape and Regulate the IL-7 Receptor Signalosome in Human CD4 T-cells. J. Biol. Chem. 2013, 288, 8691–8701. [Google Scholar] [CrossRef]
- Henriques, C.M.; Rino, J.; Nibbs, R.J.; Graham, G.J.; Barata, J.T. IL-7 induces rapid clathrin-mediated internalization and JAK3-dependent degradation of IL-7Rα in T cells. Blood 2010, 115, 3269–3277. [Google Scholar] [CrossRef] [PubMed]
- Dooms, H. Interleukin-7: Fuel for the autoimmune attack. J. Autoimmun. 2013, 45, 40–48. [Google Scholar] [CrossRef] [PubMed]
- Giliani, S.; Mori, L.; de Saint Basile, G.; Le Deist, F.; Rodriguez-Perez, C.; Forino, C.; Mazzolari, E.; Dupuis, S.; Elhasid, R.; Kessel, A.; et al. Interleukin-7 receptor alpha (IL-7Ralpha) deficiency: Cellular and molecular bases. Analysis of clinical, immunological, and molecular features in 16 novel patients. Immunol. Rev. 2005, 203, 110–126. [Google Scholar] [CrossRef] [PubMed]
- Ye, S.K.; Maki, K.; Kitamura, T.; Sunaga, S.; Akashi, K.; Domen, J.; Weissman, I.L.; Honjo, T.; Ikuta, K. Induction of germline transcription in the TCRγ, locus by Stat5: Implications for accessibility control by the IL-7 receptor. Immunity 1999, 11, 213–223. [Google Scholar] [CrossRef]
- Puel, A.; Ziegler, S.F.; Buckley, R.H.; Leonard, W.J. Defective IL7R expression in T-B+NK+ severe combined immunodeficiency. Nat. Genet. 1998, 20, 394–397. [Google Scholar] [CrossRef] [PubMed]
- Roifman, C.M.; Zhang, J.; Chitayat, D.; Sharfe, N. A partial deficiency of interleukin-7R alpha is sufficient to abrogate T-cell development and cause severe combined immunodeficiency. Blood 2000, 96, 2803–2807. [Google Scholar] [CrossRef]
- Buckley, R.H. Primary cellular immunodeficiencies. J. Allergy Clin. Immunol. 2002, 109, 747–757. [Google Scholar] [CrossRef]
- Kwan, A.; Abraham, R.S.; Currier, R.; Brower, A.; Andruszewski, K.; Abbott, J.K.; Baker, M.; Ballow, M.; Bartoshesky, L.E.; Bonilla, F.A.; et al. Newborn screening for severe combined immunodeficiency in 11 screening programs in the United States. JAMA J. Am. Med. Assoc. 2014, 312, 729. [Google Scholar] [CrossRef]
- Giliani, S.; Bonfim, C.; de Saint Basile, G.; Lanzi, G.; Brousse, N.; Koliski, A.; Malvezzi, M.; Fischer, A.; Notarangelo, L.D.; Le Deist, F. Omenn syndrome in an infant with IL7RA gene mutation. J. Pediatr. 2006, 148, 272–274. [Google Scholar] [CrossRef]
- Lebet, T.; Chiles, R.; Hsu, A.P.; Mansfield, E.S.; Warrington, J.A.; Puck, J.M. Mutations causing severe combined immunodeficiency: Detection with a custom resequencing microarray. Genet. Med. 2008, 10, 575–585. [Google Scholar] [CrossRef]
- Lev, A.; Simon, A.J.; Barel, O.; Eyal, E.; Glick-Saar, E.; Nayshool, O.; Birk, O.; Stauber, T.; Hochberg, A.; Broides, A.; et al. Reduced Function and Diversity of T Cell Repertoire and Distinct Clinical Course in Patients With IL7RA Mutation. Front. Immunol. 2019, 10, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Engelhardt, K.R.; Xu, Y.; Grainger, A.; Germani Batacchi, M.G.C.; Swan, D.J.; Willet, J.D.P.; Abd Hamid, I.J.; Agyeman, P.; Barge, D.; Bibi, S.; et al. Identification of Heterozygous Single- and Multi-exon Deletions in IL7R by Whole Exome Sequencing. J. Clin. Immunol. 2017, 37, 42–50. [Google Scholar] [CrossRef]
- Lundtoft, C.; Awuah, A.A.-A.; Güler, A.; Harling, K.; Schaal, H.; Mayatepek, E.; Phillips, R.O.; Nausch, N.; Owusu-Dabo, E.; Jacobsen, M. An IL7RA exon 5 polymorphism is associated with impaired IL-7Rα splicing and protection against tuberculosis in Ghana. Genes Immun. 2019, 20, 514–519. [Google Scholar] [CrossRef] [PubMed]
- Hahn, W.-H.; Suh, J.-S.; Park, H.-J.; Cho, B.-S. Interleukin 7 receptor gene polymorphisms and haplotypes are associated with susceptibility to IgA nephropathy in Korean children. Exp. Ther. Med. 2011, 2, 1121–1126. [Google Scholar] [CrossRef] [PubMed]
- Gallego-Bustos, F.; Gotea, V.; Ramos-Amador, J.T.; Rodríguez-Pena, R.; Gil-Herrera, J.; Sastre, A.; Delmiro, A.; Rai, G.; Elnitski, L.; González-Granado, L.I.; et al. A Case of IL-7R Deficiency Caused by a Novel Synonymous Mutation and Implications for Mutation Screening in SCID Diagnosis. Front. Immunol. 2016, 7, 1–7. [Google Scholar] [CrossRef]
- Bayer, D.K.; Martinez, C.A.; Sorte, H.S.; Forbes, L.R.; Demmler-Harrison, G.J.; Hanson, I.C.; Pearson, N.M.; Noroski, L.M.; Zaki, S.R.; Bellini, W.J.; et al. Vaccine-associated varicella and rubella infections in severe combined immunodeficiency with isolated CD4 lymphocytopenia and mutations in IL 7 R detected by tandem whole exome sequencing and chromosomal microarray. Clin. Exp. Immunol. 2014, 178, 459–469. [Google Scholar] [CrossRef]
- Shamim, Z.; Spellman, S.; Haagenson, M.; Wang, T.; Lee, S.J.; Ryder, L.P.; Müller, K. Polymorphism in the Interleukin-7 Receptor-alpha and Outcome after Allogeneic Hematopoietic Cell Transplantation with Matched Unrelated Donor. Scand. J. Immunol. 2013, 78, 214–220. [Google Scholar] [CrossRef]
- Liao, C.-Y.; Yu, H.-W.; Cheng, C.-N.; Chen, J.-S.; Lin, C.-W.; Chen, P.-C.; Shieh, C.-C. A novel pathogenic mutation on Interleukin-7 receptor leading to severe combined immunodeficiency identified with newborn screening and whole exome sequencing. J. Microbiol. Immunol. Infect. 2018, 18, 1–7. [Google Scholar] [CrossRef]
- Lundmark, F.; Duvefelt, K.; Iacobaeus, E.; Kockum, I.; Wallström, E.; Khademi, M.; Oturai, A.; Ryder, L.P.; Saarela, J.; Harbo, H.F.; et al. Variation in interleukin 7 receptor α chain (IL7R) influences risk of multiple sclerosis. Nat. Genet. 2007, 39, 1108–1113. [Google Scholar] [CrossRef]
- Gregory, S.G.; Schmidt, S.; Seth, P.; Oksenberg, J.R.; Hart, J.; Prokop, A.; Caillier, S.J.; Ban, M.; Goris, A.; Barcellos, L.F.; et al. Interleukin 7 receptor α chain ( IL7R ) shows allelic and functional association with multiple sclerosis. Nat. Genet. 2007, 39, 1083–1091. [Google Scholar] [CrossRef]
- Todd, J.A.; Walker, N.M.; Cooper, J.D.; Smyth, D.J.; Downes, K.; Plagnol, V.; Bailey, R.; Nejentsev, S.; Field, S.F.; Payne, F.; et al. Robust associations of four new chromosome regions from genome-wide analyses of type 1 diabetes. Nat. Genet. 2007, 39, 857–864. [Google Scholar] [CrossRef] [PubMed]
- O’Doherty, C.; Alloza, I.; Rooney, M.; Vandenbroeck, K. IL7RA polymorphisms and chronic inflammatory arthropathies. Tissue Antigens 2009, 74, 429–431. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Duvefelt, K.; Svensson, F.; Masterman, T.; Jonasdottir, G.; Salter, H.; Emahazion, T.; Hellgren, D.; Falk, G.; Olsson, T.; et al. Two genes encoding immune-regulatory molecules (LAG3 and IL7R) confer susceptibility to multiple sclerosis. Genes Immun. 2005, 6, 145–152. [Google Scholar] [CrossRef] [PubMed]
- Heron, M.; Grutters, J.C.; van Moorsel, C.H.M.; Ruven, H.J.T.; Huizinga, T.W.J.; van der Helm-van Mil, A.H.M.; Claessen, A.M.E.; van den Bosch, J.M.M. Variation in IL7R predisposes to sarcoid inflammation. Genes Immun. 2009, 10, 647–653. [Google Scholar] [CrossRef][Green Version]
- Bodian, D.L.; McCutcheon, J.N.; Kothiyal, P.; Huddleston, K.C.; Iyer, R.K.; Vockley, J.G.; Niederhuber, J.E. Germline Variation in Cancer-Susceptibility Genes in a Healthy, Ancestrally Diverse Cohort: Implications for Individual Genome Sequencing. PLoS ONE 2014, 9, e94554. [Google Scholar] [CrossRef]
- Genain, C.P.; Cannella, B.; Hauser, S.L.; Raine, C.S. Identification of autoantibodies associated with myelin damage in multiple sclerosis. Nat. Med. 1999, 5, 170–175. [Google Scholar] [CrossRef]
- Hafler, D.A.; Compston, A.; Sawcer, S.; Lander, E.S.; Daly, M.J.; De Jager, P.L.; De Bakker, P.I.W.; Gabriel, S.B.; Mirel, D.B.; Ivinson, A.J.; et al. Risk Alleles for Multiple Sclerosis Identified by a Genomewide Study. N. Engl. J. Med. 2007, 357, 851–862. [Google Scholar]
- Cox, A.L.; Thompson, S.A.J.; Jones, J.L.; Robertson, V.H.; Hale, G.; Waldmann, H.; Compston, D.A.S.; Coles, A.J. Lymphocyte homeostasis following therapeutic lymphocyte depletion in multiple sclerosis. Eur. J. Immunol. 2005, 35, 3332–3342. [Google Scholar] [CrossRef]
- Traggiai, E.; Biagioli, T.; Rosati, E.; Ballerini, C.; Mazzanti, B.; Ben Nun, A.; Massacesi, L.; Vergelli, M. IL-7-enhanced T-cell response to myelin proteins in multiple sclerosis. J. Neuroimmunol. 2001, 121, 111–119. [Google Scholar] [CrossRef]
- Galarza-Muñoz, G.; Briggs, F.B.S.; Evsyukova, I.; Schott-Lerner, G.; Kennedy, E.M.; Nyanhete, T.; Wang, L.; Bergamaschi, L.; Widen, S.G.; Tomaras, G.D.; et al. Human Epistatic Interaction Controls IL7R Splicing and Increases Multiple Sclerosis Risk. Cell 2017, 169, 72–84. [Google Scholar] [CrossRef]
- Zenatti, P.P.; Ribeiro, D.; Li, W.; Zuurbier, L.; Silva, M.C.; Paganin, M.; Tritapoe, J.; Hixon, J.A.; Silveira, A.B.; Cardoso, B.A.; et al. Oncogenic IL7R gain-of-function mutations in childhood T-cell acute lymphoblastic leukemia. Nat. Genet. 2011, 43, 932–939. [Google Scholar] [CrossRef] [PubMed]
- Shochat, C.; Tal, N.; Bandapalli, O.R.; Palmi, C.; Ganmore, I.; te Kronnie, G.; Cario, G.; Cazzaniga, G.; Kulozik, A.E.; Stanulla, M.; et al. Gain-of-function mutations in interleukin-7 receptor -α ( IL7R ) in childhood acute lymphoblastic leukemias. J. Exp. Med. 2011, 208, 1333. [Google Scholar] [CrossRef]
- Shochat, C.; Tal, N.; Gryshkova, V.; Birger, Y.; Bandapalli, O.R.; Cazzaniga, G.; Gershman, N.; Kulozik, A.E.; Biondi, A.; Mansour, M.R.; et al. Novel activating mutations lacking cysteine in type I cytokine receptors in acute lymphoblastic leukemia. Blood 2014, 124, 106–110. [Google Scholar] [CrossRef] [PubMed]
- Huh, H.J.; Lee, S.H.; Yoo, K.H.; Sung, K.W.; Koo, H.H.; Jang, J.H.; Kim, K.; Kim, S.J.; Kim, W.S.; Jung, C.W.; et al. Gene mutation profiles and prognostic implications in Korean patients with T-lymphoblastic leukemia. Ann. Hematol. 2013, 92, 635–644. [Google Scholar] [CrossRef]
- Kim, M.S.; Chung, N.G.; Kim, M.S.; Yoo, N.J.; Lee, S.H. Somatic mutation of IL7R exon 6 in acute leukemias and solid cancers. Hum. Pathol. 2013, 44, 551–555. [Google Scholar] [CrossRef]
- Richter-Pechańska, P.; Kunz, J.B.; Hof, J.; Zimmermann, M.; Rausch, T.; Bandapalli, O.R.; Orlova, E.; Scapinello, G.; Sagi, J.C.; Stanulla, M.; et al. Identification of a genetically defined ultra-high-risk group in relapsed pediatric T-lymphoblastic leukemia. Blood Cancer J. 2017, 7, e523. [Google Scholar] [CrossRef]
- Zhang, J.; Ding, L.; Holmfeldt, L.; Wu, G.; Heatley, S.L.; Payne-Turner, D.; Easton, J.; Chen, X.; Wang, J.; Rusch, M.; et al. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature 2012, 481, 157–163. [Google Scholar] [CrossRef]
- Roberts, K.G.; Li, Y.; Payne-Turner, D.; Harvey, R.C.; Yang, Y.-L.; Pei, D.; McCastlain, K.; Ding, L.; Lu, C.; Song, G.; et al. Targetable Kinase-Activating Lesions in Ph-like Acute Lymphoblastic Leukemia. N. Engl. J. Med. 2014, 371, 1005–1015. [Google Scholar] [CrossRef]
- Roberts, K.G.; Morin, R.D.; Zhang, J.; Hirst, M.; Zhao, Y.; Su, X.; Chen, S.-C.; Payne-Turner, D.; Churchman, M.L.; Harvey, R.C.; et al. Genetic Alterations Activating Kinase and Cytokine Receptor Signaling in High-Risk Acute Lymphoblastic Leukemia. Cancer Cell 2012, 22, 153–166. [Google Scholar] [CrossRef]
- Roberts, K.G.; Yang, Y.-L.; Payne-Turner, D.; Lin, W.; Files, J.K.; Dickerson, K.; Gu, Z.; Taunton, J.; Janke, L.J.; Chen, T.; et al. Oncogenic role and therapeutic targeting of ABL-class and JAK-STAT activating kinase alterations in Ph-like ALL. Blood Adv. 2017, 1, 1657–1671. [Google Scholar]
- Rozovski, U.; Li, P.; Harris, D.; Ohanian, M.; Kantarjian, H.; Estrov, Z. Interleukin-7 receptor- α gene mutations are not detected in adult T-cell acute lymphoblastic leukemia. Cancer Med. 2014, 3, 550–554. [Google Scholar] [CrossRef]
- Hixon, J.A.; Andrews, C.; Kashi, L.; Kohnhorst, C.L.; Senkevitch, E.; Czarra, K.; Barata, J.T.; Li, W.; Schneider, J.P.; Walsh, S.T.R.; et al. New anti-IL-7Rα monoclonal antibodies show efficacy against T cell acute lymphoblastic leukemia in pre-clinical models. Leukemia 2019, 1–15. [Google Scholar] [CrossRef]
- Akkapeddi, P.; Fragoso, R.; Hixon, J.A.; Ramalho, A.S.; Oliveira, M.L.; Carvalho, T.; Gloger, A.; Matasci, M.; Corzana, F.; Durum, S.K.; et al. A fully human anti-IL-7Rα antibody promotes antitumor activity against T-cell acute lymphoblastic leukemia. Leukemia 2019, 33, 2155–2168. [Google Scholar] [CrossRef] [PubMed]
- Mendes, R.D.; Sarmento, L.M.; Cante-Barrett, K.; Zuurbier, L.; Buijs-Gladdines, J.G.C.A.M.; Povoa, V.; Smits, W.K.; Abecasis, M.; Yunes, J.A.; Sonneveld, E.; et al. PTEN microdeletions in T-cell acute lymphoblastic leukemia are caused by illegitimate RAG-mediated recombination events. Blood 2014, 124, 567–578. [Google Scholar] [CrossRef] [PubMed]
- Papaemmanuil, E.; Rapado, I.; Li, Y.; Potter, N.E.; Wedge, D.C.; Tubio, J.; Alexandrov, L.B.; Van Loo, P.; Cooke, S.L.; Marshall, J.; et al. RAG-mediated recombination is the predominant driver of oncogenic rearrangement in ETV6-RUNX1 acute lymphoblastic leukemia. Nat. Genet. 2014, 46, 116–125. [Google Scholar] [CrossRef] [PubMed]
- Kirkham, C.M.; Scott, J.N.F.; Wang, X.; Smith, A.L.; Kupinski, A.P.; Ford, A.M.; Westhead, D.R.; Stockley, P.G.; Tuma, R.; Boyes, J. Cut-and-Run: A Distinct Mechanism by which V(D)J Recombination Causes Genome Instability. Mol. Cell 2019, 74, 584–597. [Google Scholar] [CrossRef]
- Stroud, R.M.; Wells, J.A. Mechanistic Diversity of Cytokine Receptor Signaling Across Cell Membranes. Sci. Signal. 2004, 2004, re7. [Google Scholar] [CrossRef]
- Lu, X.; Gross, A.W.; Lodish, H.F. Active Conformation of the Erythropoietin Receptor. J. Biol. Chem. 2006, 281, 7002–7011. [Google Scholar] [CrossRef]
- Brooks, A.J.; Dai, W.; O’Mara, M.L.; Abankwa, D.; Chhabra, Y.; Pelekanos, R.A.; Gardon, O.; Tunny, K.A.; Blucher, K.M.; Morton, C.J.; et al. Mechanism of Activation of Protein Kinase JAK2 by the Growth Hormone Receptor. Science 2014, 344, 1249783. [Google Scholar] [CrossRef]
- Liu, Y.; Easton, J.; Shao, Y.; Maciaszek, J.; Wang, Z.; Wilkinson, M.R.; McCastlain, K.; Edmonson, M.; Pounds, S.B.; Shi, L.; et al. The genomic landscape of pediatric and young adult T-lineage acute lymphoblastic leukemia. Nat. Genet. 2017, 49, 1211–1218. [Google Scholar] [CrossRef]
- Porcu, M.; Kleppe, M.; Gianfelici, V.; Geerdens, E.; De Keersmaecker, K.; Tartaglia, M.; Foà, R.; Soulier, J.; Cauwelier, B.; Uyttebroeck, A.; et al. Mutation of the receptor tyrosine phosphatase PTPRC (CD45) in T-cell acute lymphoblastic leukemia. Blood 2012, 119, 4476–4479. [Google Scholar] [CrossRef] [PubMed]
- Weijenborg Campos, L.; Pini Zenatti, P.; Granato Pissinato, L.; Libanio Rodrigues, G.O.; Artico, L.L.; Rafael Guimarães, T.; Fröhlich Archangelo, L.; Martínez, L.; Brooks, A.J.; Yunes, J.A. Oncogenic basic amino acid insertions at the extracellular juxtamembrane region of IL7RA cause receptor hypersensitivity. Blood 2019, 133, 1259–1263. [Google Scholar] [CrossRef] [PubMed]
- Fry, T.J.; Mackall, C.L. The Many Faces of IL-7: From Lymphopoiesis to Peripheral T Cell Maintenance. J. Immunol. 2005, 174, 6571–6576. [Google Scholar] [CrossRef] [PubMed]
- Silva, A.; Laranjeira, A.B.A.; Martins, L.R.; Cardoso, B.A.; Demengeot, J.; Yunes, J.A.; Seddon, B.; Barata, J.T. IL-7 Contributes to the Progression of Human T-cell Acute Lymphoblastic Leukemias. Cancer Res. 2011, 71, 4780–4789. [Google Scholar] [CrossRef]


| Mutation/PolyMorphism | Nucleotide Change | Exon | Protein Site | Effect/Possible Effect | Associated Diseases |
|---|---|---|---|---|---|
| p.Q26X | c.76C>T | Exon 1 | Extracellular | Premature stop-codon | SCID [42] |
| p.G28R | c.82G>A | Exon 2 | Extracellular | Structural: Ligand affinity | SCID [33] |
| p.G28fsX35 | c.221+2T>G | Exon 2 | Extracellular | Splicing: Exon skipping | SCID [42] |
| p.G28fsX51 | delExon2-4 | Exon 2 | Extracellular | Frameshift | SCID [42] |
| p.L35Q | c.104T>A | Exon 2 | Extracellular/D1-D2 | Structural: Ligand affinity | SCID [40] |
| p.F40L | c.120C>G | Exon 2 | Extracellular/D1-D2 | Protein thermo-stability | SCID [41] |
| p.C42Y | c.125G>A | Exon 2 | Extracellular/SS-bond | Structural: Ligand affinity | SCID |
| p.S44R | c.132C>A | Exon 2 | Extracellular/D1-D2 | Structural: Ligand affinity | SCID [33] |
| p.V48fsX59 | c.143delTG | Exon 2 | Extracellular/D1-D2 | Frameshift | SCID [33] |
| p.L55Q | c.164T>A | Exon 2 | Extracellular/D1-D2 | Structural: Ligand affinity | SCID [33] |
| p.I66T | c.197T>C | Exon 2 | Extracellular/D1-D2 | Splicing: Exon skipping | SCID [35]/Tuberculosis [43]/IgAN [44] |
| p.C74Y | c.221G>A | Exon 2 | Extracellular/SS-bond | Structural: Ligand affinity | SCID [33] |
| p.G75fsX75 | delExon3 | Exon 3 | Extracellular/D1-D2 | Frameshift | SCID [42] |
| p.C82S | c.244T>A | Exon 3 | Extracellular/SS-bond | Structural: Ligand affinity | SCID |
| p.V111= | c.333T>A | Exon 3 | Extracellular/D1-D2 | Splicing: Exon truncation | SCID [45] |
| p.C118Y | c.353G>A | Exon 3 | Extracellular/SS-bond | Structural: Ligand affinity | OS [39]/SCID [45] |
| p.I121fsX128 | c.361dupA | Exon 3 | Extracellular/D1-D2 | Frameshift | SCID [46] |
| p.P132S | c.394C>T | Exon 4 | Extracellular/D1-D2 | Structural: Ligand affinity | SCID [36] |
| p.L135R | c.404T>G | Exon 4 | Extracellular/D1-D2 | Structural: Ligand affinity | SCID [33] |
| p.V138I | c.412G>A | Exon 4 | Extracellular/D1-D2 | Splicing: Exon truncation | SCID [35]/GvHD [47] |
| p.H165= | c.495C>T | Exon 4 | Extracellular/D1-D2 | Splicing: Exon truncation | SCID |
| p.K187= | c.561G>A | Exon 5 | Extracellular/D1-D2 | Splicing: Exon truncation | Tuberculosis [43] |
| p.L188fsX188 | c.562delC | Exon 5 | Extracellular/D1-D2 | Frameshift | SCID [48] |
| p.G215V | c.644G>T | Exon 5 | Extracellular/D1-D2 | Structural: Ligand affinity | SCID [40] |
| p.W217X | c.651G>A | Exon 5 | Extracellular/WSXWS | Premature stop-codon | SCID [35] |
| p.S218N | c.653G>A | Exon 5 | Extracellular/WSXWS | Structural: Ligand affinity | SCID [33] |
| p.W220C | c.660G>C | Exon 5 | Extracellular/WSXWS | Structural: Ligand affinity | SCID [33] |
| p.S221I | c.662G>T | Exon 5 | Extracellular/WSXWS | Structural: Ligand affinity | SCID [40] |
| p.T244I | c.731C>T | Exon 6 | Transmembrane | Splicing: Exon skipping | MS [49,50]/T1D [51]/RA [52] |
| p.K269fsX269 | c.876+6T>G | Exon 7 | Intracellular | Splicing: Exon skipping | SCID [42] |
| p.I356V | c.1066A>G | Exon 8 | Intracellular | Splicing: Exon truncation | MS [49,53]/T1D [51] |
| - | Int. A>C, T (35857748) | Intron 1 | - | Unknown | Sarcoidosis [54] |
| - | c.83-2A>T,G (35860850) | Intron 1 | - | Splicing defect | SCID [33] |
| Protein Mutation | TM Sequence | Associated Disease |
|---|---|---|
| WT | YFRTPEINNSSGEMDPILLTISILSFFSVALLVILACVLWKKRIK | |
| -------------ABCDEFGHIJKLM------------------- | ||
| p.L242>FCTPVP | EINNSSGEMDPIFCTPVPLTISILSFFSVALLVILACVLWKKRIK | T-ALL [80] |
| p.I241_L242>insCLEG | RTPEINNSSGEMDPCLEGLTISILSFFSVALLVILACVLWKKRIK | T-ALL [61] |
| p.L242_L243insFCRKD | EINNSSGEMDPILFCRKDLTISILSFFSVALLVILACVLWKKRIK | T-ALL [62] |
| p.L242>FDCIGV | EINNSSGEMDPIFDCIGVLTISILSFFSVALLVILACVLWKKRIK | T-ALL [62] |
| p.L242_T243>CGIREI | TPEINNSSGEMDPICGIREIISILSFFSVALLVILACVLWKKRIK | T-ALL [62] |
| p.I241_L242>CRPH | RTPEINNSSGEMDPCRPHLTISILSFFSVALLVILACVLWKKRIK | T-ALL [62] |
| p.L242>CWMK | TPEINNSSGEMDPICWMKLTISILSFFSVALLVILACVLWKKRIK | T-ALL [62] |
| p.P240_I241insCS | RTPEINNSSGEMDPCSILLTISILSFFSVALLVILACVLWKKRIK | T-ALL [65] |
| p.L242>CSQI | TPEINNSSGEMDPICSQILTISILSFFSVALLVILACVLWKKRIK | T-ALL [80] |
| p.L243>PCAQGI | EINNSSGEMDPILPCAQGITISILSFFSVALLVILACVLWKKRIK | T-ALL [80] |
| p.L242delinsLCHRK | PEINNSSGEMDPILCHRKLTISILSFFSVALLVILACVLWKKRIK | T-ALL [66] |
| p.I241>ITLYCKT | INNSSGEMDPITLYCKTLLTISILSFFSVALLVILACVLWKKRIK | T-ALL [67] |
| p.L242>FSCGP | PEINNSSGEMDPIFSCGPLTISILSFFSVALLVILACVLWKKRIK | T-ALL [62] |
| p.L242_L243insCPS | TPEINNSSGEMDPILCPSLTISILSFFSVALLVILACVLWKKRIK | T-ALL [80] |
| p.L243>CPSP | TPEINNSSGEMDPILCPSPTISILSFFSVALLVILACVLWKKRIK | T-ALL [62] |
| p.T244_I245insCPDGR | EINNSSGEMDPILLTCPDGRISILSFFSVALLVILACVLWKKRIK | ph-Like ALL [80] |
| p.L242delinsLTACQP | EINNSSGEMDPILTACQPLTISILSFFSVALLVILACVLWKKRIK | T-ALL [66] |
| p.L243>RCPS | TPEINNSSGEMDPILRCPSTISILSFFSVALLVILACVLWKKRIK | T-ALL [62] |
| p.T244_I245insPPVCSVT | NNSSGEMDPILLTPPVCSVTISILSFFSVALLVILACVLWKKRIK | B-ALL [62] |
| p.IL241-242TC | RTPEINNSSGEMDPITCLLTISILSFFSVALLVILACVLWKKRIK | T-ALL [67] |
| p.L242_L243insNPC | TPEINNSSGEMDPILNPCLTISILSFFSVALLVILACVLWKKRIK | T-ALL [61] |
| p.T244_I245insCPT | TPEINNSSGEMDPILLTCPTISILSFFSVALLVILACVLWKKRIK | T-ALL [61] |
| p.I241_T244>SANCGA | RTPEINNSSGEMDPSANCGAISILSFFSVALLVILACVLWKKRIK | T-ALL [61] |
| p.L243_T244insVSCP | PEINNSSGEMDPILLVSCPTISILSFFSVALLVILACVLWKKRIK | T-ALL [61] |
| p.P240_L242>QSPSC | RTPEINNSSGEMDQSPSCLIISILSFFSVALLVILACVLWKKRIK | T-ALL [61] |
| p.P240_T244>RFCPH | YFRTPEINNSSGEMDRFCPHISILSFFSVALLVILACVLWKKRIK | T-ALL [61] |
| p.L242_T244>FHPFNCGP | EINNSSGEMDPIFHPFNCGPISILSFFSVALLVILACVLWKKRIK | T-ALL [61] |
| p.L243_T244insMCP | TPEINNSSGEMDPILLMCPTISILSFFSVALLVILACVLWKKRIK | T-ALL [61] |
| p.L243>RLECV | PEINNSSGEMDPILRLECVTISILSFFSVALLVILACVLWKKRIK | T-ALL [61] |
| p.L242_L243>WAALLNCE | INNSSGEMDPIWAALLNCETISILSFFSVALLVILACVLWKKRIK | T-ALL [81] |
| p.L242_L243insRC | RTPEINNSSGEMDPILRCLTISILSFFSVALLVILACVLWKKRIK | T-ALL [62] |
| p.L243_T244>PCPL | RTPEINNSSGEMDPILPCPLISILSFFSVALLVILACVLWKKRIK | T-ALL [62] |
| p.244 Ins MPEQDCP +S246T | NNSSGEMDPILLMPEQDCPTITILSFFSVALLVILACVLWKKRIK | T-ALL [62] |
| p.E237_L242>ASWC | SYYFRTPEINNSSGASWCLTISILSFFSVALLVILACVLWKKRIK | T-ALL [62] |
| p.L242_T244>CPP | YFRTPEINNSSGEMDPICPPISILSFFSVALLVILACVLWKKRIK | T-ALL [62] |
| p.L243_T244>PLCSA | TPEINNSSGEMDPILPLCSAISILSFFSVALLVILACVLWKKRIK | T-ALL [62] |
| p.L243_T244>PIYRCVL | EINNSSGEMDPILPIYRCVLISILSFFSVALLVILACVLWKKRIK | T-ALL [62] |
| p.L242>FEC | RTPEINNSSGEMDPIFECLTISILSFFSVALLVILACVLWKKRIK | T-ALL [62] |
| p.L242_T244>FTCPS | RTPEINNSSGEMDPIFTCPSISILSFFSVALLVILACVLWKKRIK | T-ALL [62] |
| p.S249_F250insCSTISILS | NSSGEMDPILLTISILSCSTISILSFFSVALLVILACVLWKKRIK | T-ALL [62] |
| p.243 Ins RCI | RTPEINNSSGEMDPILRCITISILSFFSVALLVILACVLWKKRIK | T-ALL [62] |
| p.L242_L243insGC | RTPEINNSSGEMDPILGCLTISILSFFSVALLVILACVLWKKRIK | T-ALL [62] |
| p.L243>GCI | RTPEINNSSGEMDPILGCITISILSFFSVALLVILACVLWKKRIK | T-ALL [62] |
| p.T244_I245insLPCVY | EINNSSGEMDPILLTLPCVYISILSFFSVALLVILACVLWKKRIK | T-ALL [62] |
| p.T244>KKCTN | PEINNSSGEMDPILLKKCTNISILSFFSVALLVILACVLWKKRIK | T-ALL [62] |
| p.L243_T244insPPCL | PEINNSSGEMDPILLPPCLTISILSFFSVALLVILACVLWKKRIK | B-ALL [62] |
| p.T244_I245insCHL | TPEINNSSGEMDPILLTCHLISILSFFSVALLVILACVLWKKRIK | B-ALL [62] |
| p.L243_T244insSRCL | PEINNSSGEMDPILLSRCLTISILSFFSVALLVILACVLWKKRIK | T-ALL [65] |
| p.M238_L243>PCK | PSYYFRTPEINNSSGEPCKTISILSFFSVALLVILACVLWKKRIK | B-ALL [65] |
| p.L242_L243insLTARGC | INNSSGEMDPILLTARGCLTISILSFFSVALLVILACVLWKKRIK | B-ALL [65] |
| p.T244_I245insNPPCGT | INNSSGEMDPILLTNPPCGTISILSFFSVALLVILACVLWKKRIK | T-ALL [64] |
| P.L243>RCL | RTPEINNSSGEMDPILRCLTISILSFFSVALLVILACVLWKKRIK | T-ALL [80] |
| P.L243>RGCL | TPEINNSSGEMDPILRGCLTISILSFFSVALLVILACVLWKKRIK | T-ALL [80] |
| p.L242_L243SRC | TPEINNSSGEMDPILSRCLTISILSFFSVALLVILACVLWKKRIK | T-ALL [80] |
| p.T244>RRCSS | PEINNSSGEMDPILLRRCSSISILSFFSVALLVILACVLWKKRIK | T-ALL [80] |
| p.L243>LQRCT | PEINNSSGEMDPILLQRCTTISILSFFSVALLVILACVLWKKRIK | T-ALL [80] |
| p.T244>RGFHITCQT | NSSGEMDPILLRGFHITCQTISILSFFSVALLVILACVLWKKRIK | T-ALL [80] |
| p.P240_T244>SCLI | YYFRTPEINNSSGEMDSCLIISILSFFSVALLVILACVLWKKRIK | ph-Like ALL [68] |
| p.L243_T244>CAN | FRTPEINNSSGEMDPILCANISILSFFSVALLVILACVLWKKRIK | ph-Like ALL [68] |
| p.L243_T244>RCPP | RTPEINNSSGEMDPILRCPPISILSFFSVALLVILACVLWKKRIK | ph-Like ALL [68] |
| p.GCinsL243 | RTPEINNSSGEMDPILGCLTISILSFFSVALLVILACVLWKKRIK | ETP-ALL [67] |
| p.L242>DTRVYNSIC | NSSGEMDPIDTRVYNSICLTISILSFFSVALLVILACVLWKKRIK | ETP-ALL [67] |
| p.LL242-243>SPCI | RTPEINNSSGEMDPISPCITISILSFFSVALLVILACVLWKKRIK | ETP-ALL [67] |
| p.L242delinsLPC | RTPEINNSSGEMDPILPCLTISILSFFSVALLVILACVLWKKRIK | T-ALL [66] |
| p.L243delinsLMCP | TPEINNSSGEMDPILLMCPTISILSFFSVALLVILACVLWKKRIK | T-ALL [66] |
| p.L242delinsLSRPC | PEINNSSGEMDPILSRPCLTISILSFFSVALLVILACVLWKKRIK | T-ALL [66] |
| p.P240_L242>SC | YFRTPEINNSSGEMDSCLTISILSFFSVALLVILACVLWKKRIK | ph-Like ALL [69] |
| p.L242>FPGVC | PEINNSSGEMDPIFPGVCLTISILSFFSVALLVILACVLWKKRIK | B-ALL [69] |
| p.L243_T244>RCGA | TPEINNSSGEMDPILLRCGAISILSFFSVALLVILACVLWKKRIK | B-ALL [69] |
| p.L242_L243>FPHQHC | PEINNSSGEMDPIFPHQHCTISILSFFSVALLVILACVLWKKRIK | T-ALL [61] |
| p.T244_I245insRPCG | PEINNSSGEMDPILLTRPCGISILSFFSVALLVILACVLWKKRIK | T-ALL [62] |
| p.T244>SRCG | TPEINNSSGEMDPILLSRCGISILSFFSVALLVILACVLWKKRIK | T-ALL [64] |
| T244>TSPPCG | EINNSSGEMDPILLTSPPCGISILSFFSVALLVILACVLWKKRIK | T-ALL [80] |
| p.I245>TKPCII | EINNSSGEMDPILLTTKPCIISILSFFSVALLVILACVLWKKRIK | T-ALL [80] |
| p.L243_T244>RQGCP | TPEINNSSGEMDPILRQGCPISILSFFSVALLVILACVLWKKRIK | ph-Like ALL [68] |
| p.T244>TGPCF | PEINNSSGEMDPILLTGPCFISILSFFSVALLVILACVLWKKRIK | B-ALL [69] |
| p.T244>NDCS | RTPEINNSSGEMDPILLNDCSSILSFFSVALLVILACVLWKKRIK | T-ALL [77] |
| p.D239_T244>SFC | YFRTPEINNSSGEMSFCISILSFFSVALLVILACVLWKKRIK | ph-Like ALL [68] |
| p.P240_S246>LKC | SPSYYFRTPEINNSSGEMDLKCILSFFSVALLVILACVLWKKRIK | T-ALL [61] |
| p.L242_S246>PQGGC | YFRTPEINNSSGEMDPIPQGGCILSFFSVALLVILACVLWKKRIK | T-ALL [61] |
| p.P240_S246>LQSC | PSYYFRTPEINNSSGEMDLQSCILSFFSVALLVILACVLWKKRIK | T-ALL [61] |
| p.I245_S246>HRGC | RTPEINNSSGEMDPILLTHRGCILSFFSVALLVILACVLWKKRIK | T-ALL [62] |
| p.I245_S246>SHQPC | TPEINNSSGEMDPILLTSHQPCILSFFSVALLVILACVLWKKRIK | T-ALL [62] |
| p.I247>KCH | RTPEINNSSGEMDPILLTISKCHLSFFSVALLVILACVLWKKRIK | T-ALL [62] |
| p.I241_S246>TC | SPSYYFRTPEINNSSGEMDPTCILSFFSVALLVILACVLWKKRIK | ph-Like ALL [68] |
| p.L243_S246>RVPGC | FRTPEINNSSGEMDPILRVPGCILSFFSVALLVILACVLWKKRIK | ph-Like ALL [68] |
| p.P240_S246>RAYC | YFRTPEINNSSGEMDRAYCILSFFSVALLVILACVLWKKRIK | ph-Like ALL [68] |
| p.L248_S251>CQ | SYYFRTPEINNSSGEMDPILLTISICQSVALLVILACVLWKKRIK | T-ALL [62] |
| Protein Mutation | TM Sequence | Associated Disease |
|---|---|---|
| IL7R_WT | FRTPEINNSSGEMDPILLTISILSFFSVALLVILACVLWKKRIK | - |
| TM mutations | ||
| p.I247_L248insQW | TPEINNSSGEMDPILLTISIQWLSFFSVALLVILACVLWKKRIK | T-ALL [61] |
| p.S252_A254>WN | YFRTPEINNSSGEMDPILLTISILSFFWNLLVILACVLWKKRIK | T-ALL [61] |
| p.V253>GPSL | PEINNSSGEMDPILLTISILSFFSGPSLALLVILACVLWKKRIK | T-ALL [61] |
| p.V253_L254insGEA | PEINNSSGEMDPILLTISILSFFSVGEAALLVILACVLWKKRIK | T-ALL [62] |
| p.A254_L255>EKV | RTPEINNSSGEMDPILLTISILSFFSVEKVLVILACVLWKKRIK | T-ALL [62] |
| p.V253G | FRTPEINNSSGEMDPILLTISILSFFSGALLVILACVLWKKRIK | T-ALL [62] |
| p.F250_V253>PLGE | FRTPEINNSSGEMDPILLTISILSPLGEALLVILACVLWKKRIK | T-ALL [81] |
| p.V253>GPLV | PEINNSSGEMDPILLTISILSFFSGPLVALLVILACVLWKKRIK | T-ALL [80] |
| p.L256>FLEL | PEINNSSGEMDPILLTISILSFFSVALFLELVILACVLWKKRIK | T-ALL [80] |
| p.V253>GFSV | PEINNSSGEMDPILLTISILSFFSGFSVALLVILACVLWKKRIK | ETP-ALL [67] |
| EJM mutations | ||
| p.L243>RRI | TPEINNSSGEMDPILRRITISILSFFSVALLVILACVLWKKRIK | T-ALL [65] |
| p.T244>RI | RTPEINNSSGEMDPILLRIISILSFFSVALLVILACVLWKKRIK | T-ALL [64] |
| p.L243>RRL | TPEINNSSGEMDPILRRLTISILSFFSVALLVILACVLWKKRIK | T-ALL [80] |
| p.I241>IH | RTPEINNSSGEMDPIHLLTISILSFFSVALLVILACVLWKKRIK | T-ALL [80] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Campos, L.W.; Pissinato, L.G.; Yunes, J.A. Deleterious and Oncogenic Mutations in the IL7RA. Cancers 2019, 11, 1952. https://doi.org/10.3390/cancers11121952
Campos LW, Pissinato LG, Yunes JA. Deleterious and Oncogenic Mutations in the IL7RA. Cancers. 2019; 11(12):1952. https://doi.org/10.3390/cancers11121952
Chicago/Turabian StyleCampos, Lívia Weijenborg, Leonardo Granato Pissinato, and José Andrés Yunes. 2019. "Deleterious and Oncogenic Mutations in the IL7RA" Cancers 11, no. 12: 1952. https://doi.org/10.3390/cancers11121952
APA StyleCampos, L. W., Pissinato, L. G., & Yunes, J. A. (2019). Deleterious and Oncogenic Mutations in the IL7RA. Cancers, 11(12), 1952. https://doi.org/10.3390/cancers11121952





