Microfluidic Droplet Digital PCR Is a Powerful Tool for Detection of BRAF and TERT Mutations in Papillary Thyroid Carcinomas
Abstract
1. Introduction
2. Results
2.1. Optimization of dPCR for Detection of BRAFV600E and TERT Promoter Mutations
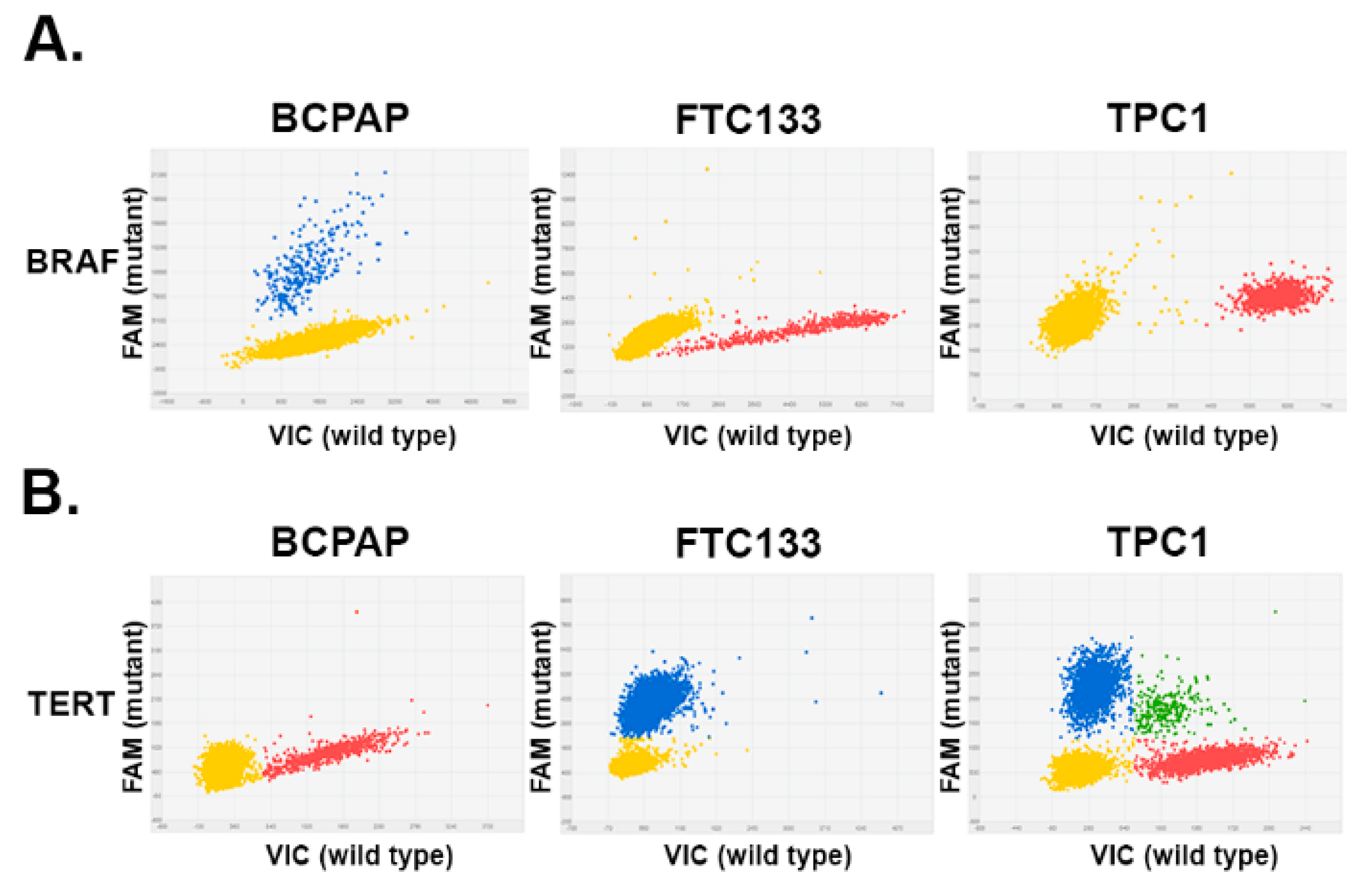
2.2. Detection of BRAFV600 and TERT Promoter Mutations in Thyroid Cancer Cell Lines
2.3. Detection of BRAFV600 and TERT228 in Follicular Adenomas, Follicular Cancers, and Medullary Thyroid Cancers
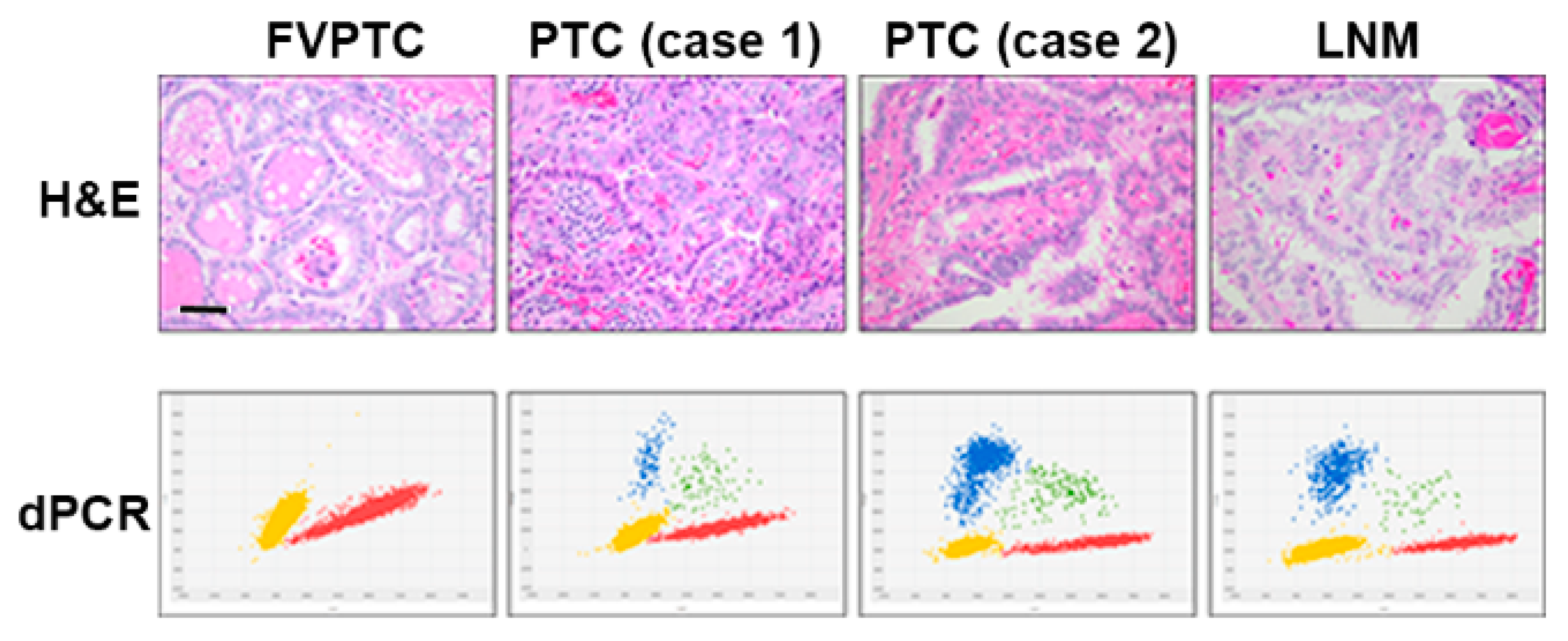
2.4. Detection of BRAFV600 by Digital PCR in Papillary Thyroid Cancers
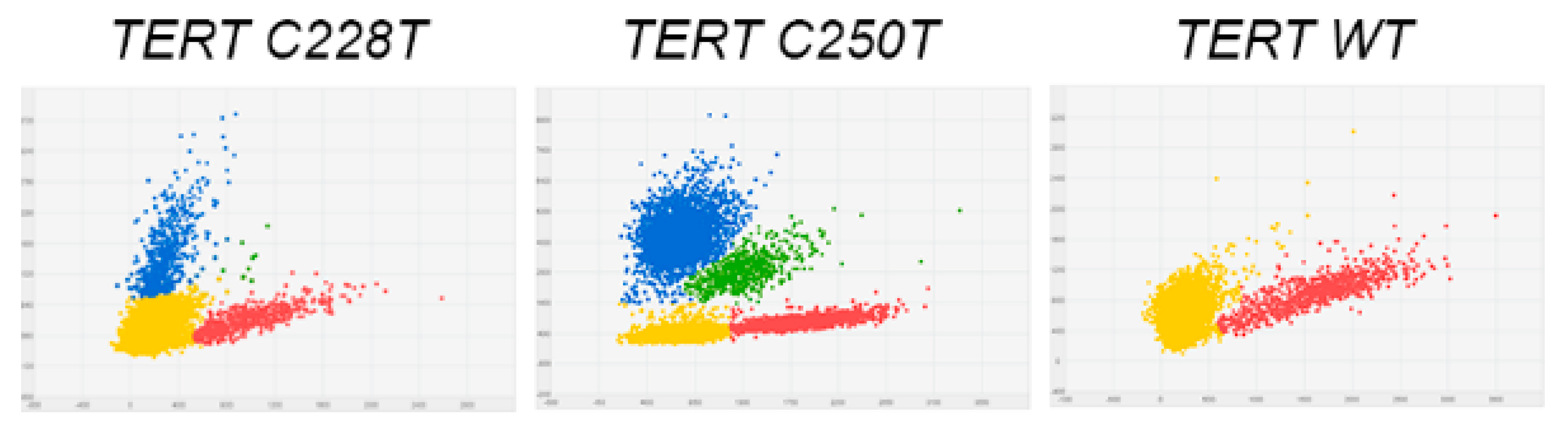
2.5. Detection of TERT228 and TERTC250T by Digital PCR in Papillary Thyroid Cancers
3. Discussion
4. Material and Methods
4.1. Human Tumor Specimens
4.2. Thyroid Cancer Cell Lines
4.3. DNA/RNA Extraction
4.4. Sanger Sequencing
4.5. Microfluidic Digital PCR
4.6. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Disclaimer
References
- Morris, L.G.; Tuttle, R.M.; Davies, L. Changing Trends in the Incidence of Thyroid Cancer in the United States. JAMA Otolaryngol. Head Neck Surg. 2016, 142, 709–711. [Google Scholar] [CrossRef] [PubMed]
- Cancer Genome Atlas Research Network. Integrated genomic characterization of papillary thyroid carcinoma. Cell 2014, 159, 676–690. [Google Scholar] [CrossRef] [PubMed]
- Haugen, B.R.; Sherman, S.I. Evolving approaches to patients with advanced differentiated thyroid cancer. Endocr. Rev. 2013, 34, 439–455. [Google Scholar] [CrossRef] [PubMed]
- Fagin, J.A.; Wells, S.A., Jr. Biologic and Clinical Perspectives on Thyroid Cancer. N. Engl. J. Med. 2016, 375, 1054–1067. [Google Scholar] [CrossRef]
- Knauf, J.A.; Fagin, J.A. Role of MAPK pathway oncoproteins in thyroid cancer pathogenesis and as drug targets. Curr. Opin. Cell Biol. 2009, 21, 296–303. [Google Scholar] [CrossRef]
- Mercer, K.; Giblett, S.; Green, S.; Lloyd, D.; DaRocha Dias, S.; Plumb, M.; Marais, R.; Pritchard, C. Expression of endogenous oncogenic V600EB-raf induces proliferation and developmental defects in mice and transformation of primary fibroblasts. Cancer Res. 2005, 65, 11493–11500. [Google Scholar] [CrossRef]
- Sithanandam, G.; Kolch, W.; Duh, F.M.; Rapp, U.R. Complete coding sequence of a human B-raf cDNA and detection of B-raf protein kinase with isozyme specific antibodies. Oncogene 1990, 5, 1775–1780. [Google Scholar]
- Wan, P.T.; Garnett, M.J.; Roe, S.M.; Lee, S.; Niculescu-Duvaz, D.; Good, V.M.; Jones, C.M.; Marshall, C.J.; Springer, C.J.; Barford, D.; et al. Mechanism of activation of the RAF-ERK signaling pathway by oncogenic mutations of B-RAF. Cell 2004, 116, 855–867. [Google Scholar] [CrossRef]
- Mitsutake, N.; Knauf, J.A.; Mitsutake, S.; Mesa, C., Jr.; Zhang, L.; Fagin, J.A. Conditional BRAFV600E expression induces DNA synthesis, apoptosis, dedifferentiation, and chromosomal instability in thyroid PCCL3 cells. Cancer Res. 2005, 65, 2465–2473. [Google Scholar] [CrossRef]
- Namba, H.; Nakashima, M.; Hayashi, T.; Hayashida, N.; Maeda, S.; Rogounovitch, T.I.; Ohtsuru, A.; Saenko, V.A.; Kanematsu, T.; Yamashita, S. Clinical implication of hot spot BRAF mutation, V599E, in papillary thyroid cancers. J. Clin. Endocrinol. Metab. 2003, 88, 4393–4397. [Google Scholar] [CrossRef]
- Nikiforova, M.N.; Kimura, E.T.; Gandhi, M.; Biddinger, P.W.; Knauf, J.A.; Basolo, F.; Zhu, Z.; Giannini, R.; Salvatore, G.; Fusco, A.; et al. BRAF mutations in thyroid tumors are restricted to papillary carcinomas and anaplastic or poorly differentiated carcinomas arising from papillary carcinomas. J. Clin. Endocrinol. Metab. 2003, 88, 5399–5404. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Lee, E.S.; Kim, Y.S. Clinicopathologic significance of BRAF V600E mutation in papillary carcinomas of the thyroid: A meta-analysis. Cancer 2007, 110, 38–46. [Google Scholar] [CrossRef] [PubMed]
- Xing, M. BRAF mutation in papillary thyroid cancer: Pathogenic role, molecular bases, and clinical implications. Endocr. Rev. 2007, 28, 742–762. [Google Scholar] [CrossRef] [PubMed]
- Xing, M. BRAF mutation in thyroid cancer. Endocr. Relat. Cancer 2005, 12, 245–262. [Google Scholar] [CrossRef] [PubMed]
- Bonhomme, B.; Godbert, Y.; Perot, G.; Al Ghuzlan, A.; Bardet, S.; Belleannee, G.; Criniere, L.; Do Cao, C.; Fouilloux, G.; Guyetant, S.; et al. Molecular Pathology of Anaplastic Thyroid Carcinomas: A Retrospective Study of 144 Cases. Thyroid 2017, 27, 682–692. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Yang, T.; Zhao, T.; Liang, J.; Lin, Y.S. Clinical Outcome of Radioiodine Therapy in Low-intermediate Risk Papillary Thyroid Carcinoma with BRAF(V600E) Mutation. Zhongguo Yi Xue Ke Xue Yuan Xue Bao Acta Acad. Med. Sin. 2016, 38, 346–350. [Google Scholar] [CrossRef]
- Lin, K.L.; Wang, O.C.; Zhang, X.H.; Dai, X.X.; Hu, X.Q.; Qu, J.M. The BRAF mutation is predictive of aggressive clinicopathological characteristics in papillary thyroid microcarcinoma. Ann. Surg. Oncol. 2010, 17, 3294–3300. [Google Scholar] [CrossRef]
- Cabanillas, M.E.; McFadden, D.G.; Durante, C. Thyroid cancer. Lancet 2016, 388, 2783–2795. [Google Scholar] [CrossRef]
- Xing, M.; Alzahrani, A.S.; Carson, K.A.; Shong, Y.K.; Kim, T.Y.; Viola, D.; Elisei, R.; Bendlova, B.; Yip, L.; Mian, C.; et al. Association between BRAF V600E mutation and recurrence of papillary thyroid cancer. J. Clin. Oncol. 2015, 33, 42–50. [Google Scholar] [CrossRef]
- Yang, K.; Wang, H.; Liang, Z.; Liang, J.; Li, F.; Lin, Y. BRAFV600E mutation associated with non-radioiodine-avid status in distant metastatic papillary thyroid carcinoma. Clin. Nucl. Med. 2014, 39, 675–679. [Google Scholar] [CrossRef]
- Xing, M. BRAF V600E mutation and papillary thyroid cancer. Jama 2013, 310, e535. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Qu, S.; Liu, R.; Sheng, C.; Shi, X.; Zhu, G.; Murugan, A.K.; Guan, H.; Yu, H.; Wang, Y.; et al. TERT promoter mutations and their association with BRAF V600E mutation and aggressive clinicopathological characteristics of thyroid cancer. J. Clin. Endocrinol. Metab. 2014, 99, 1130–1136. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Bishop, J.; Shan, Y.; Pai, S.; Liu, D.; Murugan, A.K.; Sun, H.; El-Naggar, A.K.; Xing, M. Highly prevalent TERT promoter mutations in aggressive thyroid cancers. Endocr. Relat. Cancer 2013, 20, 603–610. [Google Scholar] [CrossRef] [PubMed]
- Jin, L.; Chen, E.; Dong, S.; Cai, Y.; Zhang, X.; Zhou, Y.; Zeng, R.; Yang, F.; Pan, C.; Liu, Y.; et al. BRAF and TERT promoter mutations in the aggressiveness of papillary thyroid carcinoma: A study of 653 patients. Oncotarget 2016, 7, 18346–18355. [Google Scholar] [CrossRef] [PubMed]
- Vinagre, J.; Almeida, A.; Populo, H.; Batista, R.; Lyra, J.; Pinto, V.; Coelho, R.; Celestino, R.; Prazeres, H.; Lima, L.; et al. Frequency of TERT promoter mutations in human cancers. Nat. Commun. 2013, 4, e2185. [Google Scholar] [CrossRef]
- Pestana, A.; Vinagre, J.; Sobrinho-Simoes, M.; Soares, P. TERT biology and function in cancer: Beyond immortalisation. J. Mol. Endocrinol. 2017, 58, 129–146. [Google Scholar] [CrossRef]
- Arcila, M.; Lau, C.; Nafa, K.; Ladanyi, M. Detection of KRAS and BRAF mutations in colorectal carcinoma roles for high-sensitivity locked nucleic acid-PCR sequencing and broad-spectrum mass spectrometry genotyping. J. Mol. Diagn. 2011, 13, 64–73. [Google Scholar] [CrossRef]
- Omholt, K.; Platz, A.; Kanter, L.; Ringborg, U.; Hansson, J. NRAS and BRAF mutations arise early during melanoma pathogenesis and are preserved throughout tumor progression. Clin. Cancer Res. 2003, 9, 6483–6488. [Google Scholar]
- Lamy, P.J.; Castan, F.; Lozano, N.; Montelion, C.; Audran, P.; Bibeau, F.; Roques, S.; Montels, F.; Laberenne, A.C. Next-Generation Genotyping by Digital PCR to Detect and Quantify the BRAF V600E Mutation in Melanoma Biopsies. J. Mol. Diagn. 2015, 17, 366–373. [Google Scholar] [CrossRef]
- Kowalik, A.; Kowalska, A.; Walczyk, A.; Chodurska, R.; Kopczynski, J.; Chrapek, M.; Wypiorkiewicz, E.; Chlopek, M.; Pieciak, L.; Gasior-Perczak, D.; et al. Evaluation of molecular diagnostic approaches for the detection of BRAF p.V600E mutations in papillary thyroid cancer: Clinical implications. PLoS ONE 2017, 12, e0179691. [Google Scholar] [CrossRef]
- Volik, S.; Alcaide, M.; Morin, R.D.; Collins, C. Cell-free DNA (cfDNA): Clinical Significance and Utility in Cancer Shaped By Emerging Technologies. Mol. Cancer Res. 2016, 14, 898–908. [Google Scholar] [CrossRef] [PubMed]
- Azuara, D.; Santos, C.; Lopez-Doriga, A.; Grasselli, J.; Nadal, M.; Sanjuan, X.; Marin, F.; Vidal, J.; Montal, R.; Moreno, V.; et al. Nanofluidic Digital PCR and Extended Genotyping of RAS and BRAF for Improved Selection of Metastatic Colorectal Cancer Patients for Anti-EGFR Therapies. Mol. Cancer Ther. 2016, 15, 1106–1112. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Ma, X.; Zhang, X.; Cao, G.; Tang, Y.; Deng, X.; Kang, Z.; Li, M.; Guan, M. Detection of BRAF V600E mutation in fine-needle aspiration fluid of papillary thyroid carcinoma by droplet digital PCR. Clin. Chim. Acta 2019, 491, 91–96. [Google Scholar] [CrossRef] [PubMed]
- Huggett, J.F.; Foy, C.A.; Benes, V.; Emslie, K.; Garson, J.A.; Haynes, R.; Hellemans, J.; Kubista, M.; Mueller, R.D.; Nolan, T.; et al. The digital MIQE guidelines: Minimum Information for Publication of Quantitative Digital PCR Experiments. Clin. Chem. 2013, 59, 892–902. [Google Scholar] [CrossRef]
- Armbruster, D.A.; Pry, T. Limit of blank, limit of detection and limit of quantitation. Clin. Biochem. Rev. 2008, 29, 49–52. [Google Scholar]
- Falchook, G.S.; Millward, M.; Hong, D.; Naing, A.; Piha-Paul, S.; Waguespack, S.G.; Cabanillas, M.E.; Sherman, S.I.; Ma, B.; Curtis, M.; et al. BRAF inhibitor dabrafenib in patients with metastatic BRAF-mutant thyroid cancer. Thyroid 2015, 25, 71–77. [Google Scholar] [CrossRef]
- Rothenberg, S.M.; Daniels, G.H.; Wirth, L.J. Redifferentiation of Iodine-Refractory BRAF V600E-Mutant Metastatic Papillary Thyroid Cancer with Dabrafenib-Response. Clin. Cancer Res. 2015, 21, 5640–5641. [Google Scholar] [CrossRef]
- Brose, M.S.; Cabanillas, M.E.; Cohen, E.E.; Wirth, L.J.; Riehl, T.; Yue, H.; Sherman, S.I.; Sherman, E.J. Vemurafenib in patients with BRAF(V600E)-positive metastatic or unresectable papillary thyroid cancer refractory to radioactive iodine: A non-randomised, multicentre, open-label, phase 2 trial. Lancet Oncol. 2016, 17, 1272–1282. [Google Scholar] [CrossRef]
- Moon, S.; Song, Y.S.; Kim, Y.A.; Lim, J.A.; Cho, S.W.; Moon, J.H.; Hahn, S.; Park, D.J.; Park, Y.J. Effects of Coexistent BRAF(V600E) and TERT Promoter Mutations on Poor Clinical Outcomes in Papillary Thyroid Cancer: A Meta-Analysis. Thyroid 2017, 27, 651–660. [Google Scholar] [CrossRef]
- Liu, R.; Bishop, J.; Zhu, G.; Zhang, T.; Ladenson, P.W.; Xing, M. Mortality Risk Stratification by Combining BRAF V600E and TERT Promoter Mutations in Papillary Thyroid Cancer: Genetic Duet of BRAF and TERT Promoter Mutations in Thyroid Cancer Mortality. JAMA Oncol. 2017, 3, 202–208. [Google Scholar] [CrossRef]
- Tsao, S.C.; Weiss, J.; Hudson, C.; Christophi, C.; Cebon, J.; Behren, A.; Dobrovic, A. Monitoring response to therapy in melanoma by quantifying circulating tumour DNA with droplet digital PCR for BRAF and NRAS mutations. Sci. Rep. 2015, 5, e11198. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.H.; Bae, J.S.; Lim, D.J.; Lee, H.; Jeon, S.R.; Park, G.S.; Jung, C.K. Quantification of BRAF V600E alleles predicts papillary thyroid cancer progression. Endocr. Relat. Cancer 2014, 21, 891–902. [Google Scholar] [CrossRef] [PubMed]
- De Biase, D.; Cesari, V.; Visani, M.; Casadei, G.P.; Cremonini, N.; Gandolfi, G.; Sancisi, V.; Ragazzi, M.; Pession, A.; Ciarrocchi, A.; et al. High-sensitivity BRAF mutation analysis: BRAF V600E is acquired early during tumor development but is heterogeneously distributed in a subset of papillary thyroid carcinomas. J. Clin. Endocrinol. Metab. 2014, 99, 1530–1538. [Google Scholar] [CrossRef] [PubMed]
- Kimbrell, H.Z.; Sholl, A.B.; Ratnayaka, S.; Japa, S.; Lacey, M.; Carpio, G.; Bhatia, P.; Kandil, E. BRAF Testing in Multifocal Papillary Thyroid Carcinoma. BioMed Res. Int. 2015, 2015, e486391. [Google Scholar] [CrossRef]
- Guerra, A.; Sapio, M.R.; Marotta, V.; Campanile, E.; Rossi, S.; Forno, I.; Fugazzola, L.; Budillon, A.; Moccia, T.; Fenzi, G.; et al. The primary occurrence of BRAF(V600E) is a rare clonal event in papillary thyroid carcinoma. J. Clin. Endocrinol. Metab. 2012, 97, 517–524. [Google Scholar] [CrossRef]
- Kouba, E.; Ford, A.; Brown, C.G.; Yeh, C.; Siegal, G.P.; Manne, U.; Eltoum, I.E. Detection of BRAF V600E Mutations With Next-Generation Sequencing in Infarcted Thyroid Carcinomas After Fine-Needle Aspiration. Am. J. Clin. Pathol. 2018, 150, 177–185. [Google Scholar] [CrossRef]
- Ye, W.; Hannigan, B.; Zalles, S.; Mehrotra, M.; Barkoh, B.A.; Williams, M.D.; Cabanillas, M.E.; Edeiken-Monroe, B.; Hu, P.; Duose, D.; et al. Centrifuged supernatants from FNA provide a liquid biopsy option for clinical next-generation sequencing of thyroid nodules. Cancer Cytopathol. 2019, 127, 146–160. [Google Scholar] [CrossRef]
- Zhang, H.; Liu, R.; Yan, C.; Liu, L.; Tong, Z.; Jiang, W.; Yao, M.; Fang, W.; Chen, Z. Advantage of Next-Generation Sequencing in Dynamic Monitoring of Circulating Tumor DNA over Droplet Digital PCR in Cetuximab Treated Colorectal Cancer Patients. Transl. Oncol. 2019, 12, 426–431. [Google Scholar] [CrossRef]
- Dong, L.; Wang, S.; Fu, B.; Wang, J. Evaluation of droplet digital PCR and next generation sequencing for characterizing DNA reference material for KRAS mutation detection. Sci. Rep. 2018, 8, e9650. [Google Scholar] [CrossRef]
- Abd Elmageed, Z.Y.; Sholl, A.B.; Tsumagari, K.; Al-Qurayshi, Z.; Basolo, F.; Moroz, K.; Boulares, A.H.; Friedlander, P.; Miccoli, P.; Kandil, E. Immunohistochemistry as an accurate tool for evaluating BRAF-V600E mutation in 130 samples of papillary thyroid cancer. Surgery 2017, 161, 1122–1128. [Google Scholar] [CrossRef]
- Martinuzzi, C.; Pastorino, L.; Andreotti, V.; Garuti, A.; Minuto, M.; Fiocca, R.; Bianchi-Scarra, G.; Ghiorzo, P.; Grillo, F.; Mastracci, L. A combination of immunohistochemistry and molecular approaches improves highly sensitive detection of BRAF mutations in papillary thyroid cancer. Endocrine 2016, 53, 672–680. [Google Scholar] [CrossRef] [PubMed]
- Szymonek, M.; Kowalik, A.; Kopczynski, J.; Gasior-Perczak, D.; Palyga, I.; Walczyk, A.; Gadawska-Juszczyk, K.; Plusa, A.; Mezyk, R.; Chrapek, M.; et al. Immunohistochemistry cannot replace DNA analysis for evaluation of BRAF V600E mutations in papillary thyroid carcinoma. Oncotarget 2017, 8, 74897–74909. [Google Scholar] [CrossRef] [PubMed]
- Hindson, B.J.; Ness, K.D.; Masquelier, D.A.; Belgrader, P.; Heredia, N.J.; Makarewicz, A.J.; Bright, I.J.; Lucero, M.Y.; Hiddessen, A.L.; Legler, T.C.; et al. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal. Chem. 2011, 83, 8604–8610. [Google Scholar] [CrossRef] [PubMed]
- Garcia, J.; Dusserre, E.; Cheynet, V.; Bringuier, P.P.; Brengle-Pesce, K.; Wozny, A.S.; Rodriguez-Lafrasse, C.; Freyer, G.; Brevet, M.; Payen, L.; et al. Evaluation of pre-analytical conditions and comparison of the performance of several digital PCR assays for the detection of major EGFR mutations in circulating DNA from non-small cell lung cancers: The CIRCAN_0 study. Oncotarget 2017, 8, 87980–87996. [Google Scholar] [CrossRef]
- Dobnik, D.; Stebih, D.; Blejec, A.; Morisset, D.; Zel, J. Multiplex quantification of four DNA targets in one reaction with Bio-Rad droplet digital PCR system for GMO detection. Sci. Rep. 2016, 6, e35451. [Google Scholar] [CrossRef]
- Hui, R.; Pearson, A.; Cortes, J.; Campbell, C.; Poirot, C.; Azim, H.A.; Fumagalli, D.; Lambertini, M.; Daly, F.; Arahmani, A.; et al. Lucitanib for the treatment of HR(+)/ HER2(-) metastatic breast cancer: Results from the multicohort phase II FINESSE study. Clin. Cancer Res. 2019. [Google Scholar] [CrossRef]
- Dalal, A.A.; Guerin, A.; Mutebi, A.; Culver, K.W. Economic analysis of BRAF gene mutation testing in real world practice using claims data: Costs of single gene versus panel tests in patients with lung cancer. J. Med. Econ. 2018, 21, 649–655. [Google Scholar] [CrossRef]
- Nishino, M. Molecular cytopathology for thyroid nodules: A review of methodology and test performance. Cancer Cytopathol. 2016, 124, 14–27. [Google Scholar] [CrossRef]
| Clinical Characteristics | BRAF Positive (n = 42) | BRAF Negative (n = 33) | p Value |
|---|---|---|---|
| Age (mean ± SD) | 38.64 ± 16 | 38.91 ± 14.7 | 0.996 |
| Size (cm) (mean ± SD) | 2.02 ± 1.3 | 2.1 ± 1.3 | 0.523 |
| Histology FVPTC * | 7.1% | 39.3% | 0.001 |
| Gender male | 19% | 21.2% | 0.816 |
| Multifocality | 42.8% | 33.3% | 0.401 |
| Presence of invasion | 48.7% | 25% | 0.048 |
| Presence of lymph node metastasis | 75% | 35.4% | 0.001 |
| Pathological Characteristics | BRAF % | |
|---|---|---|
| Mean ± SD | p Value | |
| Central tumor with positive lymph nodes metastasis | 27.65 ± 11.5 | 0.03 |
| Central tumor without lymph nodes metastasis | 18.4 ± 12.2 | |
| Multifocality | 26.83 ± 11.8 | 0.68 |
| Non multifocality | 25.2 ± 12.9 | |
| Invasion | 24.04 ± 14.6 | 0.34 |
| Non invasion | 27.79 ± 9.8 | |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ylli, D.; Patel, A.; Jensen, K.; Li, Z.-Z.; Mendonca-Torres, M.C.; Costello, J.; Gomes-Lima, C.J.; Wartofsky, L.; Burman, K.D.; Vasko, V.V. Microfluidic Droplet Digital PCR Is a Powerful Tool for Detection of BRAF and TERT Mutations in Papillary Thyroid Carcinomas. Cancers 2019, 11, 1916. https://doi.org/10.3390/cancers11121916
Ylli D, Patel A, Jensen K, Li Z-Z, Mendonca-Torres MC, Costello J, Gomes-Lima CJ, Wartofsky L, Burman KD, Vasko VV. Microfluidic Droplet Digital PCR Is a Powerful Tool for Detection of BRAF and TERT Mutations in Papillary Thyroid Carcinomas. Cancers. 2019; 11(12):1916. https://doi.org/10.3390/cancers11121916
Chicago/Turabian StyleYlli, Dorina, Aneeta Patel, Kirk Jensen, Zhao-Zhang Li, Maria Cecilia Mendonca-Torres, John Costello, Cristiane Jeyce Gomes-Lima, Leonard Wartofsky, Kenneth Dale Burman, and Vasyl V. Vasko. 2019. "Microfluidic Droplet Digital PCR Is a Powerful Tool for Detection of BRAF and TERT Mutations in Papillary Thyroid Carcinomas" Cancers 11, no. 12: 1916. https://doi.org/10.3390/cancers11121916
APA StyleYlli, D., Patel, A., Jensen, K., Li, Z.-Z., Mendonca-Torres, M. C., Costello, J., Gomes-Lima, C. J., Wartofsky, L., Burman, K. D., & Vasko, V. V. (2019). Microfluidic Droplet Digital PCR Is a Powerful Tool for Detection of BRAF and TERT Mutations in Papillary Thyroid Carcinomas. Cancers, 11(12), 1916. https://doi.org/10.3390/cancers11121916






