Spatial EGFR Dynamics and Metastatic Phenotypes Modulated by Upregulated EphB2 and Src Pathways in Advanced Prostate Cancer
Abstract
1. Introduction
2. Results
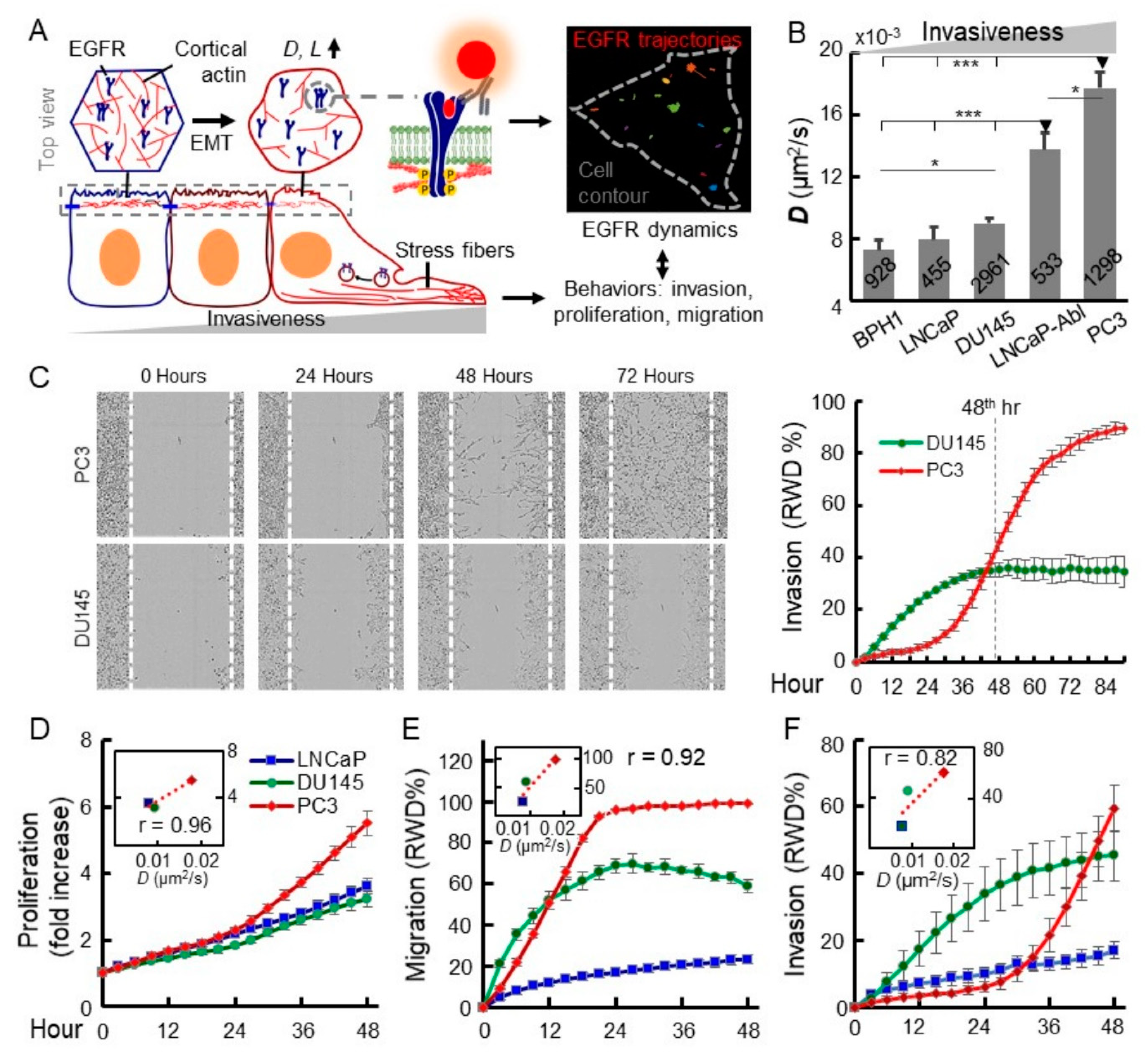
2.1. Increased EGFR Diffusivity is Correlated with Advanced Malignancy
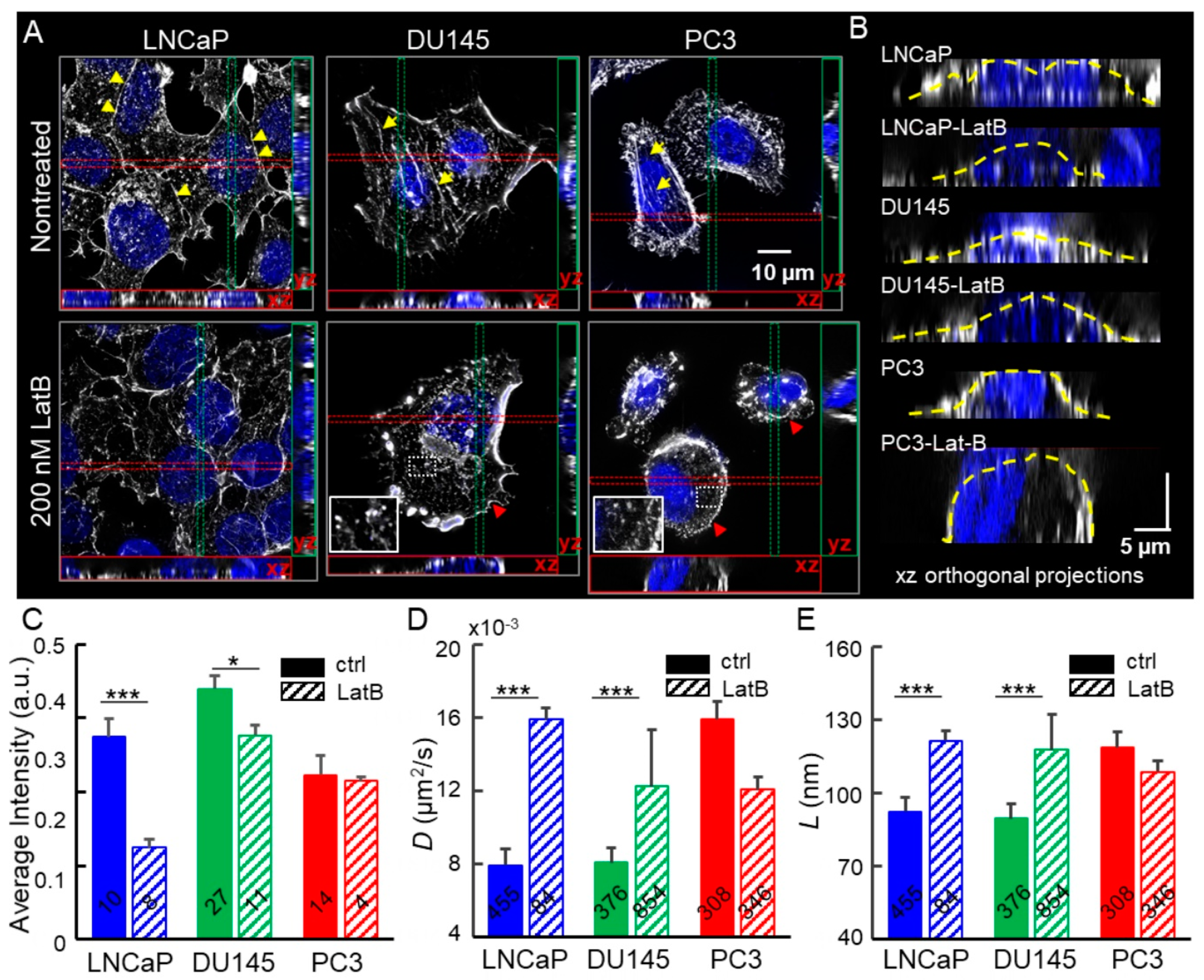
2.2. Depolymerization of Cortical Actin Matrix Increased EGFR Diffusivity and Enlarged Membrane Compartments
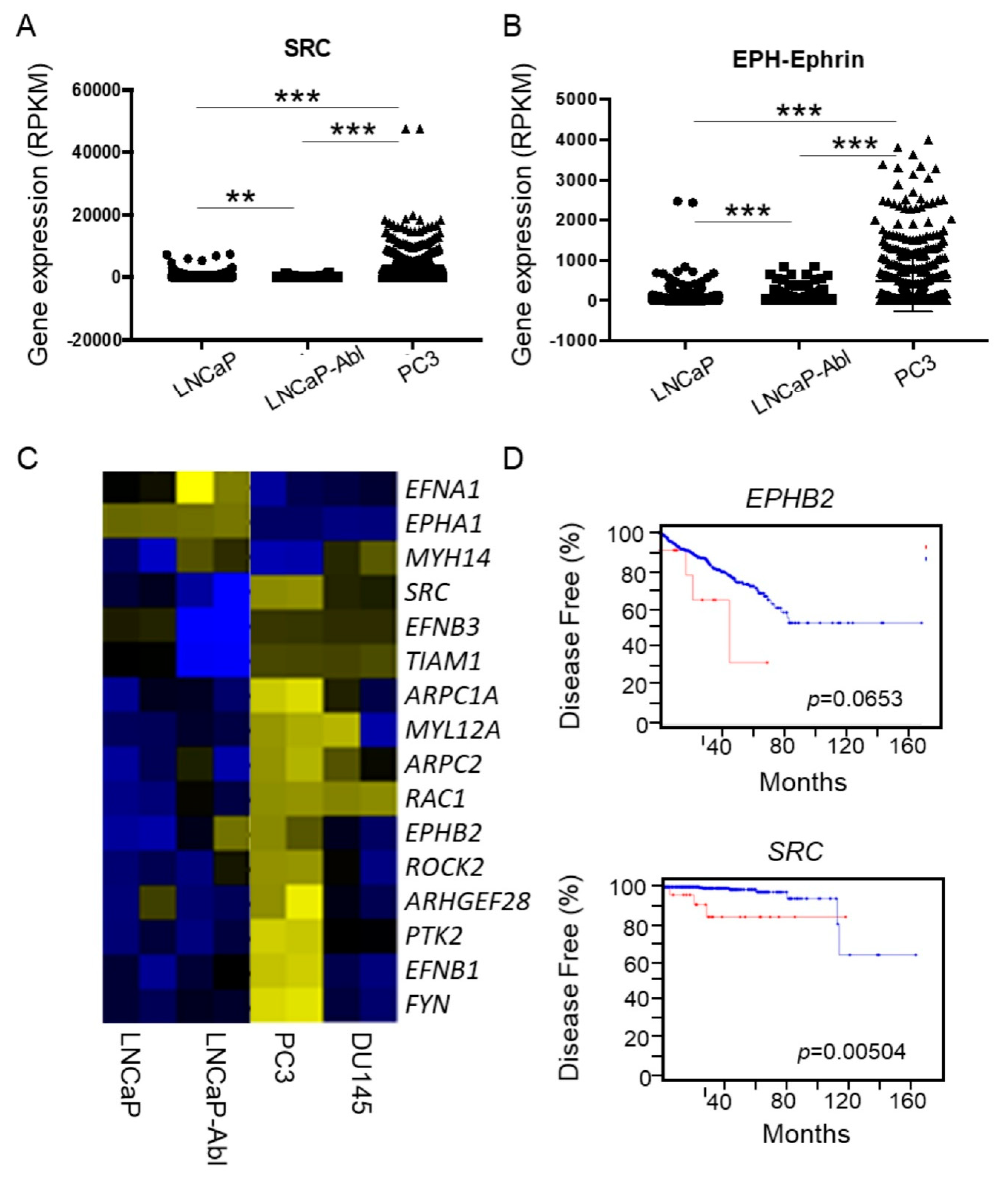
2.3. Upregulated EphB2 and Src Pathways In Cell Lines, CTCs, and Tumors Predicted Actin Reorganization, Metastasis, and Poor Prognosis
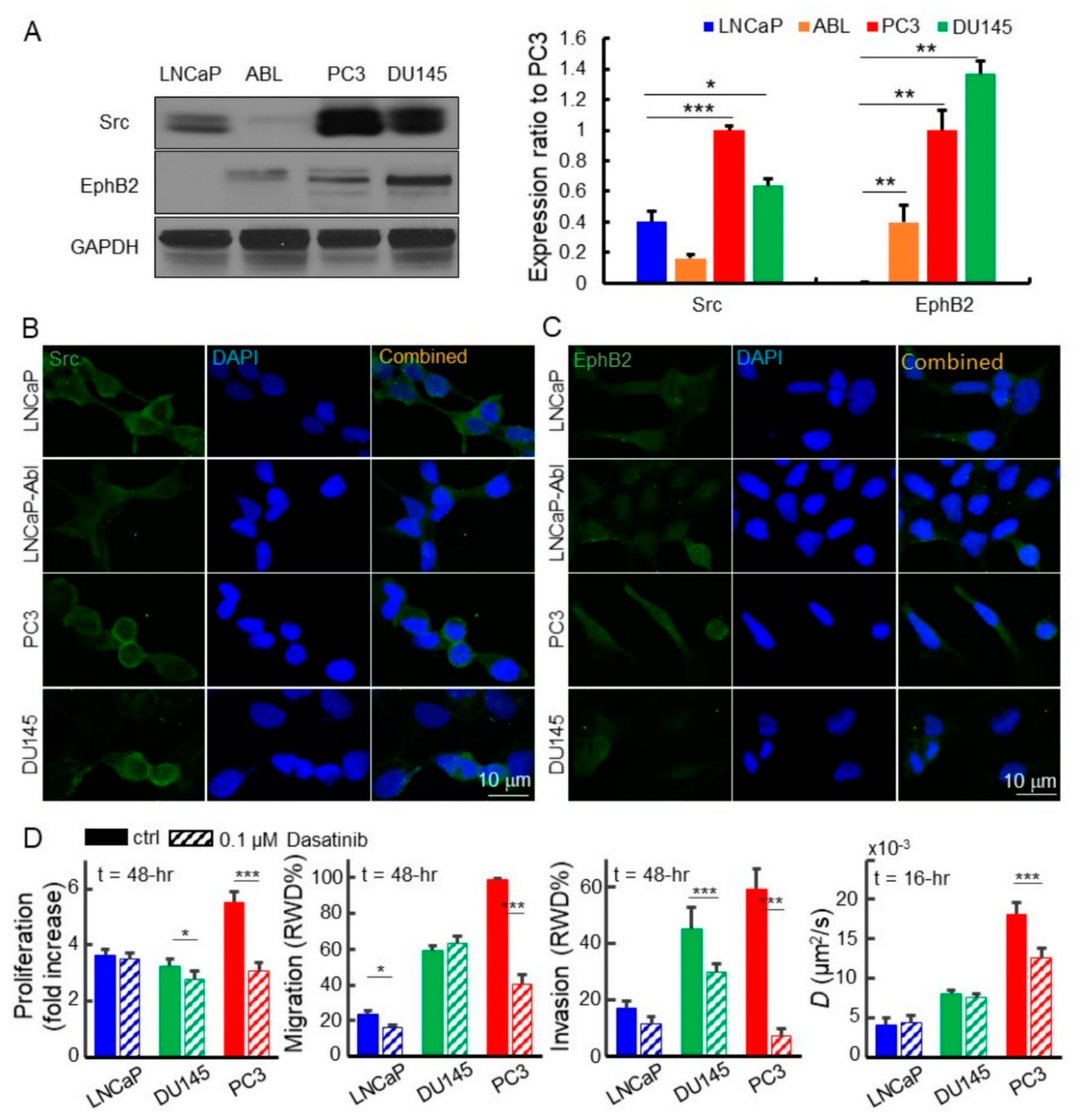
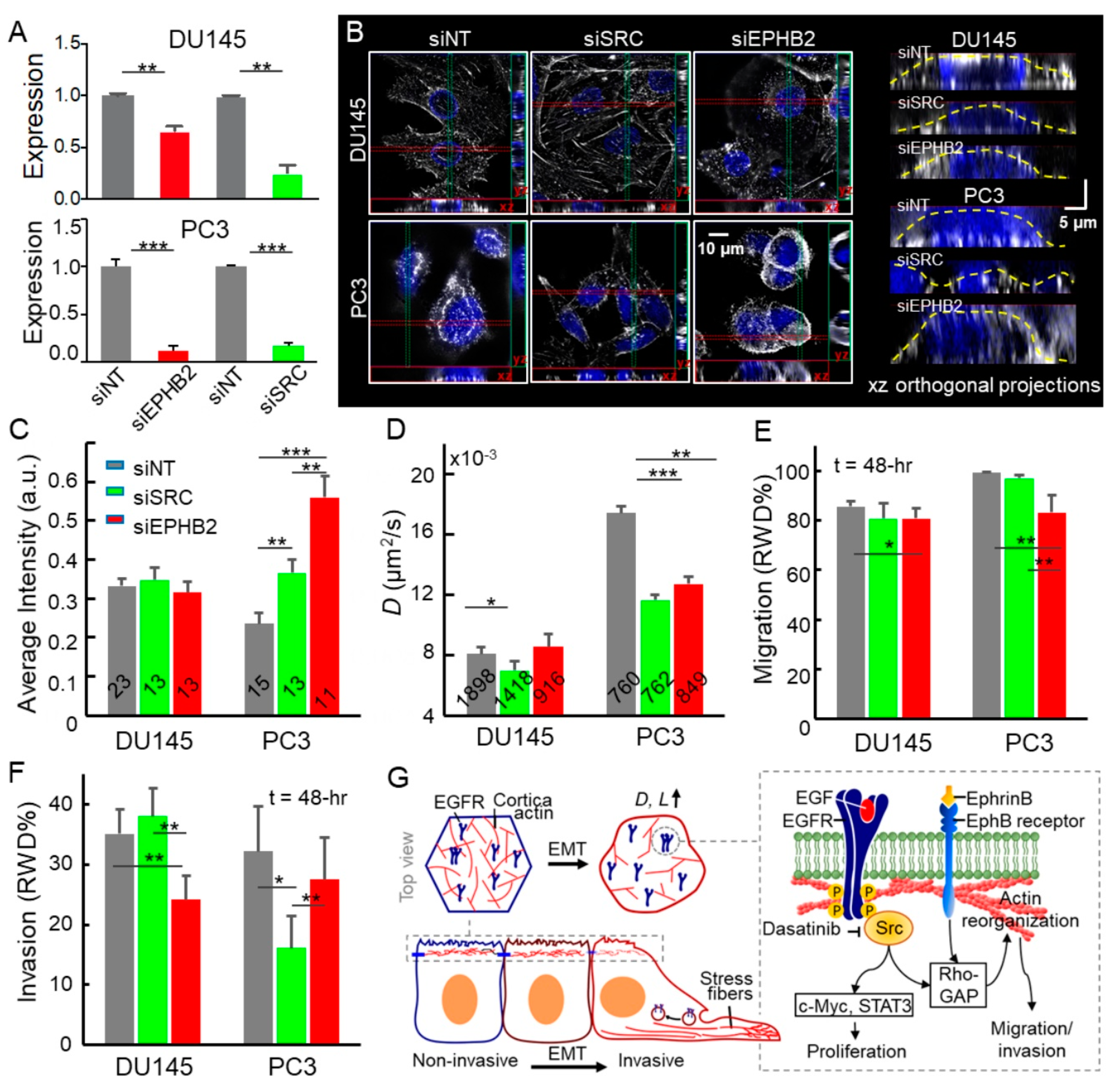
2.4. Disruption of the EphB2 and Src Pathways Led to Decreased EGFR Dynamics, Cell Migration, and Invasion in Advanced Prostate Cancer Cells
2.5. Downregulated EPHB2 and SRC Attenuated Cell Motility, Invasion, and EGFR Diffusivity in Advanced Prostate Cancer Cells
3. Discussion
4. Materials and Methods
4.1. Cells, Antibodies, and Chemicals
4.2. Fluorescent Probe Labeling to EGFR
4.3. D Single-Particle Tracking and Trajectory Analysis
4.4. In Vitro Cell Proliferation, Migration, and Invasion Assays
4.5. Cell Fluorescence Imaging Using Structured Illumination Microscopy
4.6. Single-Cell SMART-seq2 (scSMART-seq2) Gene Profiling and Pathway Analysis
4.7. qRT-PCR
4.8. Western Blot Analysis
4.9. Patients and Ethical Approval
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Lemmon, M.A.; Schlessinger, J. Cell signaling by receptor tyrosine kinases. Cell 2010, 141, 1117–1134. [Google Scholar] [CrossRef] [PubMed]
- Herbst, R.S. Review of epidermal growth factor receptor biology. Int. J. Radiat. Oncol. Biol. Phys. 2004, 59, S21–S26. [Google Scholar] [CrossRef]
- Blume-Jensen, P.; Hunter, T. Oncogenic kinase signalling. Nature 2001, 411, 355–365. [Google Scholar] [CrossRef] [PubMed]
- Yewale, C.; Baradia, D.; Vhora, I.; Patil, S.; Misra, A. Epidermal growth factor receptor targeting in cancer: A review of trends and strategies. Biomaterials 2013, 34, 8690–8707. [Google Scholar] [CrossRef]
- Kris, R.M.; Lax, I.; Gullick, W.; Waterfield, M.D.; Ullrich, A.; Fridkin, M.; Schlessinger, J. Antibodies against a synthetic peptide as a probe for the kinase activity of the avian EGF receptor and v-erbB protein. Cell 1985, 40, 619–625. [Google Scholar] [CrossRef]
- Casaletto, J.B.; McClatchey, A.I. Spatial regulation of receptor tyrosine kinases in development and cancer. Nat. Rev. Cancer 2012, 12, 387–400. [Google Scholar] [CrossRef]
- Tomas, A.; Futter, C.E.; Eden, E.R. EGF receptor trafficking: Consequences for signaling and cancer. Trends Cell Biol. 2014, 24, 26–34. [Google Scholar] [CrossRef]
- Lajoie, P.; Partridge, E.A.; Guay, G.; Goetz, J.G.; Pawling, J.; Lagana, A.; Joshi, B.; Dennis, J.W.; Nabi, I.R. Plasma membrane domain organization regulates EGFR signaling in tumor cells. J. Cell Biol. 2007, 179, 341–356. [Google Scholar] [CrossRef]
- Mosesson, Y.; Mills, G.B.; Yarden, Y. Derailed endocytosis: An emerging feature of cancer. Nat. Rev. Cancer 2008, 8, 835–850. [Google Scholar] [CrossRef]
- Sorkin, A.; von Zastrow, M. Endocytosis and signalling: intertwining molecular networks. Nat. Rev. Mol. Cell Biol. 2009, 10, 609–622. [Google Scholar] [CrossRef]
- Wang, Y.-N.; Hung, M.-C. Nuclear functions and subcellular trafficking mechanisms of the epidermal growth factor receptor family. Cell Biosci. 2012, 2, 13. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.-L.; Perillo, E.P.; Liu, C.; Yu, P.; Chou, C.-K.; Hung, M.-C.; Dunn, A.K.; Yeh, H.-C. Segmentation of 3D Trajectories Acquired by TSUNAMI Microscope: An Application to EGFR Trafficking. Biophys. J. 2016, 111, 2214–2227. [Google Scholar] [CrossRef] [PubMed]
- Perillo, E.P.; Liu, Y.-L.; Huynh, K.; Liu, C.; Chou, C.-K.; Hung, M.-C.; Yeh, H.-C.; Dunn, A.K. Deep and high-resolution three-dimensional tracking of single particles using nonlinear and multiplexed illumination. Nat. Commun. 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- Manzo, C.; Garcia-Parajo, M.F. A review of progress in single particle tracking: From methods to biophysical insights. Rep. Prog. Phys. 2015, 78, 124601. [Google Scholar] [CrossRef] [PubMed]
- Kusumi, A.; Tsunoyama, T.A.; Hirosawa, K.M.; Kasai, R.S.; Fujiwara, T.K. Tracking single molecules at work in living cells. Nat. Chem. Biol. 2014, 10, 524–532. [Google Scholar] [CrossRef] [PubMed]
- Kusumi, A.; Sako, Y.; Yamamoto, M. Confined lateral diffusion of membrane receptors as studied by single particle tracking (nanovid microscopy). Effects of calcium-induced differentiation in cultured epithelial cells. Biophys. J. 1993, 65, 2021–2040. [Google Scholar] [CrossRef]
- Chung, I.; Akita, R.; Vandlen, R.; Toomre, D.; Schlessinger, J.; Mellman, I. Spatial control of EGF receptor activation by reversible dimerization on living cells. Nature 2010, 464, 783–787. [Google Scholar] [CrossRef]
- Low-Nam, S.T.; Lidke, K.A.; Cutler, P.J.; Roovers, R.C.; en Henegouwen, P.M.v.B.; Wilson, B.S.; Lidke, D.S. ErbB1 dimerization is promoted by domain co-confinement and stabilized by ligand binding. Nat. Struct. Mol. Biol. 2011, 18, 1244–1249. [Google Scholar] [CrossRef]
- Clausen, M.P.; Lagerholm, B.C. Visualization of plasma membrane compartmentalization by high-speed quantum dot tracking. Nano Lett. 2013, 13, 2332–2337. [Google Scholar] [CrossRef]
- Kusumi, A.; Nakada, C.; Ritchie, K.; Murase, K.; Suzuki, K.; Murakoshi, H.; Kasai, R.S.; Kondo, J.; Fujiwara, T. Paradigm shift of the plasma membrane concept from the two-dimensional continuum fluid to the partitioned fluid: High-speed single-molecule tracking of membrane molecules. Annu. Rev. Biophys. Biomol. Struct. 2005, 34, 351–378. [Google Scholar] [CrossRef]
- Treanor, B.; Depoil, D.; Gonzalez-Granja, A.; Barral, P.; Weber, M.; Dushek, O.; Bruckbauer, A.; Batista, F.D. The Membrane Skeleton Controls Diffusion Dynamics and Signaling through the B Cell Receptor. Immunity 2010, 32, 187–199. [Google Scholar] [CrossRef] [PubMed]
- Freeman, S.A.; Jaumouillé, V.; Choi, K.; Hsu, B.E.; Wong, H.S.; Abraham, L.; Graves, M.L.; Coombs, D.; Roskelley, C.D.; Das, R.; et al. Toll-like receptor ligands sensitize B-cell receptor signalling by reducing actin-dependent spatial confinement of the receptor. Nat. Communi. 2015, 6, 6168. [Google Scholar] [CrossRef] [PubMed]
- Jaqaman, K.; Grinstein, S. Regulation from within: The cytoskeleton in transmembrane signaling. Trends Cell Biol. 2012, 22, 515–526. [Google Scholar] [CrossRef] [PubMed]
- Salaita, K.; Nair, P.M.; Petit, R.S.; Neve, R.M.; Das, D.; Gray, J.W.; Groves, J.T. Restriction of receptor movement alters cellular response: Physical force sensing by EphA2. Science 2010, 327, 1380–1385. [Google Scholar] [CrossRef]
- Pryor, M.M.; Low-Nam, S.T.; Halász, Á.M.; Lidke, D.S.; Wilson, B.S.; Edwards, J.S. Dynamic Transition States of ErbB1 Phosphorylation Predicted by Spatial Stochastic Modeling. Biophys. J. 2013, 105, 1533–1543. [Google Scholar] [CrossRef][Green Version]
- Ibach, J.; Radon, Y.; Gelléri, M.; Sonntag, M.H.; Brunsveld, L.; Bastiaens, P.I.; Verveer, P.J. Single Particle Tracking Reveals that EGFR Signaling Activity Is Amplified in Clathrin-Coated Pits. PLoS ONE 2015, 10, e0143162. [Google Scholar] [CrossRef]
- Liu, Y.-L.; Chou, C.-K.; Kim, M.; Vasisht, R.; Kuo, Y.-A.; Ang, P.; Liu, C.; Perillo, E.P.; Chen, Y.-A.; Blocher, K.; et al. Assessing metastatic potential of breast cancer cells based on EGFR dynamics. Sci. Rep. 2019, 9, 3395. [Google Scholar] [CrossRef]
- Engelman, J.A.; Zejnullahu, K.; Mitsudomi, T.; Song, Y.; Hyland, C.; Park, J.O.; Lindeman, N.; Gale, C.M.; Zhao, X.; Christensen, J.; et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science 2007, 316, 1039–1043. [Google Scholar] [CrossRef]
- Turke, A.B.; Zejnullahu, K.; Wu, Y.L.; Song, Y.; Dias-Santagata, D.; Lifshits, E.; Toschi, L.; Rogers, A.; Mok, T.; Sequist, L.; et al. Preexistence and clonal selection of MET amplification in EGFR mutant NSCLC. Cancer Cell 2010, 17, 77–88. [Google Scholar] [CrossRef]
- Curto, M.; Cole, B.K.; Lallemand, D.; Liu, C.H.; McClatchey, A.I. Contact-dependent inhibition of EGFR signaling by Nf2/Merlin. J. Cell Biol. 2007, 177, 893–903. [Google Scholar] [CrossRef]
- Lallemand, D.; Curto, M.; Saotome, I.; Giovannini, M.; McClatchey, A.I. NF2 deficiency promotes tumorigenesis and metastasis by destabilizing adherens junctions. Genes Dev. 2003, 17, 1090–1100. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Roche, O.; Xu, C.; Moriyama, E.H.; Heir, P.; Chung, J.; Roos, F.C.; Chen, Y.; Finak, G.; Milosevic, M.; et al. Hypoxia promotes ligand-independent EGF receptor signaling via hypoxia-inducible factor-mediated upregulation of caveolin-1. Proc. Natl. Acad. Sci. USA 2012, 109, 4892–4897. [Google Scholar] [CrossRef] [PubMed]
- Mateus, A.R.; Seruca, R.; Machado, J.C.; Keller, G.; Oliveira, M.J.; Suriano, G.; Luber, B. EGFR regulates RhoA-GTP dependent cell motility in E-cadherin mutant cells. Hum. Mol. Genet. 2007, 16, 1639–1647. [Google Scholar] [CrossRef] [PubMed]
- Bremm, A.; Walch, A.; Fuchs, M.; Mages, J.; Duyster, J.; Keller, G.; Hermannstadter, C.; Becker, K.F.; Rauser, S.; Langer, R.; et al. Enhanced activation of epidermal growth factor receptor caused by tumor-derived E-cadherin mutations. Cancer Res. 2008, 68, 707–714. [Google Scholar] [CrossRef]
- Cortina, C.; Palomo-Ponce, S.; Iglesias, M.; Fernandez-Masip, J.L.; Vivancos, A.; Whissell, G.; Huma, M.; Peiro, N.; Gallego, L.; Jonkheer, S.; et al. EphB-ephrin-B interactions suppress colorectal cancer progression by compartmentalizing tumor cells. Nat. Genet. 2007, 39, 1376–1383. [Google Scholar] [CrossRef]
- Astin, J.W.; Batson, J.; Kadir, S.; Charlet, J.; Persad, R.A.; Gillatt, D.; Oxley, J.D.; Nobes, C.D. Competition amongst Eph receptors regulates contact inhibition of locomotion and invasiveness in prostate cancer cells. Nat. Cell Biol. 2010, 12, 1194–1204. [Google Scholar] [CrossRef]
- Pasquale, E.B. Eph receptors and ephrins in cancer: Bidirectional signalling and beyond. Nat. Rev. Cancer 2010, 10, 165–180. [Google Scholar] [CrossRef]
- Lisabeth, E.M.; Falivelli, G.; Pasquale, E.B. Eph receptor signaling and ephrins. Cold Spring Harb. Perspect. Biol. 2013, 5, a009159. [Google Scholar] [CrossRef]
- Georgakopoulos, A.; Litterst, C.; Ghersi, E.; Baki, L.; Xu, C.; Serban, G.; Robakis, N.K. Metalloproteinase/Presenilin1 processing of ephrinB regulates EphB-induced Src phosphorylation and signaling. EMBO J. 2006, 25, 1242–1252. [Google Scholar] [CrossRef]
- Macrae, M.; Neve, R.M.; Rodriguez-Viciana, P.; Haqq, C.; Yeh, J.; Chen, C.; Gray, J.W.; McCormick, F. A conditional feedback loop regulates Ras activity through EphA2. Cancer Cell 2005, 8, 111–118. [Google Scholar] [CrossRef]
- Saxton, M.J.; Jacobson, K. Single-particle tracking: Applications to membrane dynamics. Ann. Rev. Biophys. Biomol. Struct. 1997, 26, 373–399. [Google Scholar] [CrossRef]
- Martin, D.S.; Forstner, M.B.; Käs, J.A. Apparent subdiffusion inherent to single particle tracking. Biophys. J. 2002, 83, 2109–2117. [Google Scholar] [CrossRef]
- Horning, A.M.; Wang, Y.; Lin, C.K.; Louie, A.D.; Jadhav, R.R.; Hung, C.N.; Wang, C.M.; Lin, C.L.; Kirma, N.B.; Liss, M.A.; et al. Single-Cell RNA-seq reveals a subpopulation of prostate cancer cells with enhanced cell-Cycle–Related transcription and attenuated androgen response. Cancer Res. 2018, 78, 853–864. [Google Scholar] [CrossRef]
- Li, C.-W.; Lim, S.-O.; Chung, E.M.; Kim, Y.-S.; Park, A.H.; Yao, J.; Cha, J.-H.; Xia, W.; Chan, L.-C.; Kim, T.; et al. Eradication of triple-negative breast cancer cells by targeting glycosylated PD-L1. Cancer Cell 2018, 33, 187–201. [Google Scholar] [CrossRef]
- Chen, Z.; Oh, D.; Dubey, A.K.; Yao, M.; Yang, B.; Groves, J.T.; Sheetz, M. EGFR family and Src family kinase interactions: Mechanics matters? Curr. Opin. Cell Biol. 2018, 51, 97–102. [Google Scholar] [CrossRef] [PubMed]
- Paszek, M.J.; DuFort, C.C.; Rossier, O.; Bainer, R.; Mouw, J.K.; Godula, K.; Hudak, J.E.; Lakins, J.N.; Wijekoon, A.C.; Cassereau, L.; et al. The cancer glycocalyx mechanically primes integrin-mediated growth and survival. Nature 2014, 511, 319–325. [Google Scholar] [CrossRef]
- Sadegh, S.; Higgins, J.L.; Mannion, P.C.; Tamkun, M.M.; Krapf, D. Plasma membrane is compartmentalized by a self-similar cortical actin meshwork. Phys. Rev. X 2017, 7, 011031. [Google Scholar] [CrossRef]
- Wakatsuki, T.; Schwab, B.; Thompson, N.C.; Elson, E.L. Effects of cytochalasin D and latrunculin B on mechanical properties of cells. J. Cell Sci. 2001, 114, 1025–1036. [Google Scholar]
- Tehrani, S.; Tomasevic, N.; Weed, S.; Sakowicz, R.; Cooper, J.A. Src phosphorylation of cortactin enhances actin assembly. Proc. Natl. Acad. Sci. USA 2007, 104, 11933–11938. [Google Scholar] [CrossRef]
- Prosperi, M.T.; Lepine, P.; Dingli, F.; Paul-Gilloteaux, P.; Martin, R.; Loew, D.; Knolker, H.J.; Coudrier, E. Myosin 1b functions as an effector of EphB signaling to control cell repulsion. J. Cell Biol. 2015, 210, 347–361. [Google Scholar] [CrossRef]
- Wilkinson, D.G. Eph receptors and ephrins: Regulators of guidance and assembly. Int. Rev. Cytol. 2000, 196, 177–244. [Google Scholar] [PubMed]
- Huang da, W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef]
- Stelzer, G.; Rosen, N.; Plaschkes, I.; Zimmerman, S.; Twik, M.; Fishilevich, S.; Stein, T.I.; Nudel, R.; Lieder, I.; Mazor, Y.; et al. The GeneCards Suite: From Gene Data Mining to Disease Genome Sequence Analyses. Curr. Protoc. Bioinform. 2016, 54, 1–30. [Google Scholar] [CrossRef] [PubMed]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef]
- Xi, H.Q.; Wu, X.S.; Wei, B.; Chen, L. Eph receptors and ephrins as targets for cancer therapy. J. Cell Mol. Med. 2012, 16, 2894–2909. [Google Scholar] [CrossRef]
- Boyd, A.W.; Bartlett, P.F.; Lackmann, M. Therapeutic targeting of EPH receptors and their ligands. Nat. Rev. Drug Discov. 2014, 13, 39–62. [Google Scholar] [CrossRef]
- Park, S.I.; Zhang, J.; Phillips, K.A.; Araujo, J.C.; Najjar, A.M.; Volgin, A.Y.; Gelovani, J.G.; Kim, S.J.; Wang, Z.; Gallick, G.E. Targeting SRC family kinases inhibits growth and lymph node metastases of prostate cancer in an orthotopic nude mouse model. Cancer Res. 2008, 68, 3323–3333. [Google Scholar] [CrossRef]
- Lombardo, L.J.; Lee, F.Y.; Chen, P.; Norris, D.; Barrish, J.C.; Behnia, K.; Castaneda, S.; Cornelius, L.A.; Das, J.; Doweyko, A.M.; et al. Discovery of N-(2-chloro-6-methyl-phenyl)-2-(6-(4-(2-hydroxyethyl)-piperazin-1-yl)-2-methylpyrimidin-4-ylamino) thiazole-5-carboxamide (BMS-354825), a dual Src/Abl kinase inhibitor with potent antitumor activity in preclinical assays. J. Med. Chem. 2004, 47, 6658–6661. [Google Scholar] [CrossRef]
- Katayama, Y.; Burkacky, O.; Meyer, M.; Bräuchle, C.; Gratton, E.; Lamb, D.C. Real-Time Nanomicroscopy via Three-Dimensional Single-Particle Tracking. Chem.Phys. Chem. 2009, 10, 2458–2464. [Google Scholar] [CrossRef]
- Pavani, S.R.P.; Thompson, M.A.; Biteen, J.S.; Lord, S.J.; Liu, N.; Twieg, R.J.; Piestun, R.; Moerner, W. Three-dimensional, single-molecule fluorescence imaging beyond the diffraction limit by using a double-helix point spread function. Proc. Natl. Acad. Sci. USA 2009, 106, 2995–2999. [Google Scholar] [CrossRef]
- Wells, N.P.; Lessard, G.A.; Goodwin, P.M.; Phipps, M.E.; Cutler, P.J.; Lidke, D.S.; Wilson, B.S.; Werner, J.H. Time-resolved three-dimensional molecular tracking in live cells. Nano Lett. 2010, 10, 4732–4737. [Google Scholar] [CrossRef] [PubMed]
- Ram, S.; Kim, D.; Ober, R.J.; Ward, E.S. 3D single molecule tracking with multifocal plane microscopy reveals rapid intercellular transferrin transport at epithelial cell barriers. Biophys. J. 2012, 103, 1594–1603. [Google Scholar] [CrossRef]
- Welsher, K.; Yang, H. Multi-resolution 3D visualization of the early stages of cellular uptake of peptide-coated nanoparticles. Nat. Nanotechnol. 2014, 9, 198–203. [Google Scholar] [CrossRef]
- Suzuki, K.G.; Kasai, R.S.; Hirosawa, K.M.; Nemoto, Y.L.; Ishibashi, M.; Miwa, Y.; Fujiwara, T.K.; Kusumi, A. Transient GPI-anchored protein homodimers are units for raft organization and function. Nat. Chem. Biol. 2012, 8, 774–783. [Google Scholar] [CrossRef]
- Liu, C.; Liu, Y.-L.; Perillo, E.; Jiang, N.; Dunn, A.; Yeh, H.-C. Improving z-tracking accuracy in the two-photon single-particle tracking microscope. Appl. Phys. Lett. 2015, 107, 153701. [Google Scholar] [CrossRef]
- Sherwood, E.R.; Van Dongen, J.L.; Wood, C.G.; Liao, S.; Kozlowski, J.M.; Lee, C. Epidermal growth factor receptor activation in androgen-independent but not androgen-stimulated growth of human prostatic carcinoma cells. Br. J. Cancer 1998, 77, 855–861. [Google Scholar] [CrossRef]
- Unlu, A.; Leake, R.E. The effect of EGFR-related tyrosine kinase activity inhibition on the growth and invasion mechanisms of prostate carcinoma cell lines. Int. J. Biol. Markers 2003, 18, 139–146. [Google Scholar] [CrossRef]
- Chen, C.L.; Mahalingam, D.; Osmulski, P.; Jadhav, R.R.; Wang, C.M.; Leach, R.J.; Chang, T.C.; Weitman, S.D.; Kumar, A.P.; Sun, L.; et al. Single-cell analysis of circulating tumor cells identifies cumulative expression patterns of EMT-related genes in metastatic prostate cancer. Prostate 2013, 73, 813–826. [Google Scholar] [CrossRef]
- Osmulski, P.; Mahalingam, D.; Gaczynska, M.E.; Liu, J.; Huang, S.; Horning, A.M.; Wang, C.-M.; Thompson, I.M.; Huang, T.H.-M.; Chen, C.-L. Nanomechanical biomarkers of single circulating tumor cells for detection of castration resistant prostate cancer. Prostate 2014. [Google Scholar] [CrossRef]
- Agoulnik, I.U.; Vaid, A.; Bingman, W.E.; Erdeme, H.; Frolov, A.; Smith, C.L.; Ayala, G.; Ittmann, M.M.; Weigel, N.L. Role of SRC-1 in the promotion of prostate cancer cell growth and tumor progression. Cancer Res. 2005, 65, 7959–7967. [Google Scholar] [CrossRef]
- Yamaguchi, H.; Condeelis, J. Regulation of the actin cytoskeleton in cancer cell migration and invasion. Biochim. Biophys. Acta 2007, 1773, 642–652. [Google Scholar] [CrossRef] [PubMed]
- Hall, A. Rho family GTPases. Biochem. Soc. Trans. 2012, 40, 1378–1382. [Google Scholar] [CrossRef] [PubMed]
- Chugh, P.; Paluch, E.K. Actin cortex architecture regulates cell surface tension. Nat. Cell Biol. 2017, 19, 689–697. [Google Scholar] [CrossRef]
- Chugh, P.; Paluch, E.K. The actin cortex at a glance. J. Cell Sci. 2018, 131, jcs186254. [Google Scholar] [CrossRef]
- Bovellan, M.; Romeo, Y.; Biro, M.; Boden, A.; Chugh, P.; Yonis, A.; Vaghela, M.; Fritzsche, M.; Moulding, D.; Thorogate, R.; et al. Cellular control of cortical actin nucleation. Curr. Biol. 2014, 24, 1628–1635. [Google Scholar] [CrossRef]
- Calzado-Martín, A.; Encinar, M.; Tamayo, J.; Calleja, M.; San Paulo, A. Effect of actin organization on the stiffness of living breast cancer cells revealed by peak-force modulation atomic force microscopy. ACS Nano 2016, 10, 3365–3374. [Google Scholar] [CrossRef]
- Zhang, G.; Long, M.; Wu, Z.-Z.; Yu, W.-Q. Mechanical properties of hepatocellular carcinoma cells. World J. Gastroenterol. 2002, 8, 243. [Google Scholar] [CrossRef]
- Baker, E.L.; Srivastava, J.; Yu, D.; Bonnecaze, R.T.; Zaman, M.H. Cancer cell migration: Integrated roles of matrix mechanics and transforming potential. PLoS ONE 2011, 6, e20355. [Google Scholar] [CrossRef]
- Li, Z.; Song, J.; Mantini, G.; Lu, M.-Y.; Fang, H.; Falconi, C.; Chen, L.-J.; Wang, Z.L. Quantifying the traction force of a single cell by aligned silicon nanowire array. Nano Lett. 2009, 9, 3575–3580. [Google Scholar] [CrossRef]
- Dahan, M.; Levi, S.; Luccardini, C.; Rostaing, P.; Riveau, B.; Triller, A. Diffusion dynamics of glycine receptors revealed by single-quantum dot tracking. Science 2003, 302, 442–445. [Google Scholar] [CrossRef]
- Andrews, N.L.; Lidke, K.A.; Pfeiffer, J.R.; Burns, A.R.; Wilson, B.S.; Oliver, J.M.; Lidke, D.S. Actin restricts Fcϵ RI diffusion and facilitates antigen-induced receptor immobilization. Nat. Cell Biol. 2008, 10, 955–963. [Google Scholar] [PubMed]
- Hendriks, C.L.; Van Vliet, L.; Rieger, B.; van Kempen, G.; van Ginkel, M. DIPimage: A Scientific Image Processing Toolbox for MATLAB; Quantitative Imaging Group, Faculty of Applied Sciences, Delft University of Technology: Delft, The Netherlands, 1999. [Google Scholar]
- Kamburov, A.; Pentchev, K.; Galicka, H.; Wierling, C.; Lehrach, H.; Herwig, R. ConsensusPathDB: Toward a more complete picture of cell biology. Nucleic Acids Res. 2011, 39, D712-7. [Google Scholar] [CrossRef]
- Kharchenko, P.V.; Silberstein, L.; Scadden, D.T. Bayesian approach to single-cell differential expression analysis. Nat. Methods 2014, 11, 740–742. [Google Scholar] [CrossRef] [PubMed]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]
- Wade, W.F.; Freed, J.H.; Edidin, M. Translational diffusion of class II major histocompatibility complex molecules is constrained by their cytoplasmic domains. J. Cell Biol. 1989, 109, 3325–3331. [Google Scholar] [CrossRef]
- Saxton, M.J. Single-particle tracking: The distribution of diffusion coefficients. Biophys. J. 1997, 72, 1744. [Google Scholar] [CrossRef]
- Casella, G.; Berger, R.L. Statistical Inference; Duxbury: Pacific Grove, CA, USA, 2002; Volume 2. [Google Scholar]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Y.-L.; Horning, A.M.; Lieberman, B.; Kim, M.; Lin, C.-K.; Hung, C.-N.; Chou, C.-W.; Wang, C.-M.; Lin, C.-L.; Kirma, N.B.; et al. Spatial EGFR Dynamics and Metastatic Phenotypes Modulated by Upregulated EphB2 and Src Pathways in Advanced Prostate Cancer. Cancers 2019, 11, 1910. https://doi.org/10.3390/cancers11121910
Liu Y-L, Horning AM, Lieberman B, Kim M, Lin C-K, Hung C-N, Chou C-W, Wang C-M, Lin C-L, Kirma NB, et al. Spatial EGFR Dynamics and Metastatic Phenotypes Modulated by Upregulated EphB2 and Src Pathways in Advanced Prostate Cancer. Cancers. 2019; 11(12):1910. https://doi.org/10.3390/cancers11121910
Chicago/Turabian StyleLiu, Yen-Liang, Aaron M. Horning, Brandon Lieberman, Mirae Kim, Che-Kuang Lin, Chia-Nung Hung, Chih-Wei Chou, Chiou-Miin Wang, Chun-Lin Lin, Nameer B. Kirma, and et al. 2019. "Spatial EGFR Dynamics and Metastatic Phenotypes Modulated by Upregulated EphB2 and Src Pathways in Advanced Prostate Cancer" Cancers 11, no. 12: 1910. https://doi.org/10.3390/cancers11121910
APA StyleLiu, Y.-L., Horning, A. M., Lieberman, B., Kim, M., Lin, C.-K., Hung, C.-N., Chou, C.-W., Wang, C.-M., Lin, C.-L., Kirma, N. B., Liss, M. A., Vasisht, R., Perillo, E. P., Blocher, K., Horng, H., Taverna, J. A., Ruan, J., Yankeelov, T. E., Dunn, A. K., ... Chen, C.-L. (2019). Spatial EGFR Dynamics and Metastatic Phenotypes Modulated by Upregulated EphB2 and Src Pathways in Advanced Prostate Cancer. Cancers, 11(12), 1910. https://doi.org/10.3390/cancers11121910




