A Circulating miRNA Signature for Stratification of Breast Lesions among Women with Abnormal Screening Mammograms
Abstract
1. Introduction
2. Results
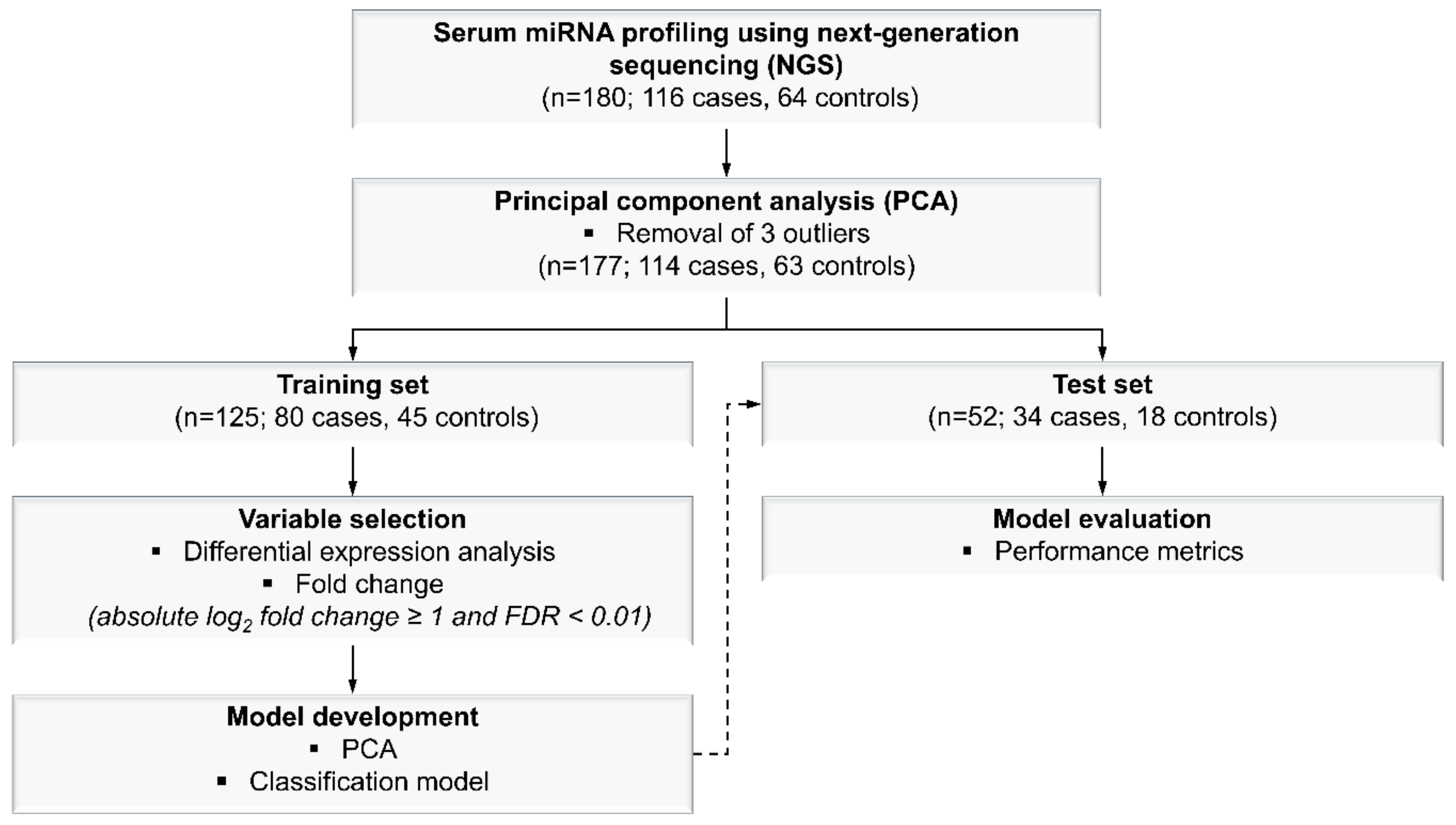
2.1. Study Subject Characteristics
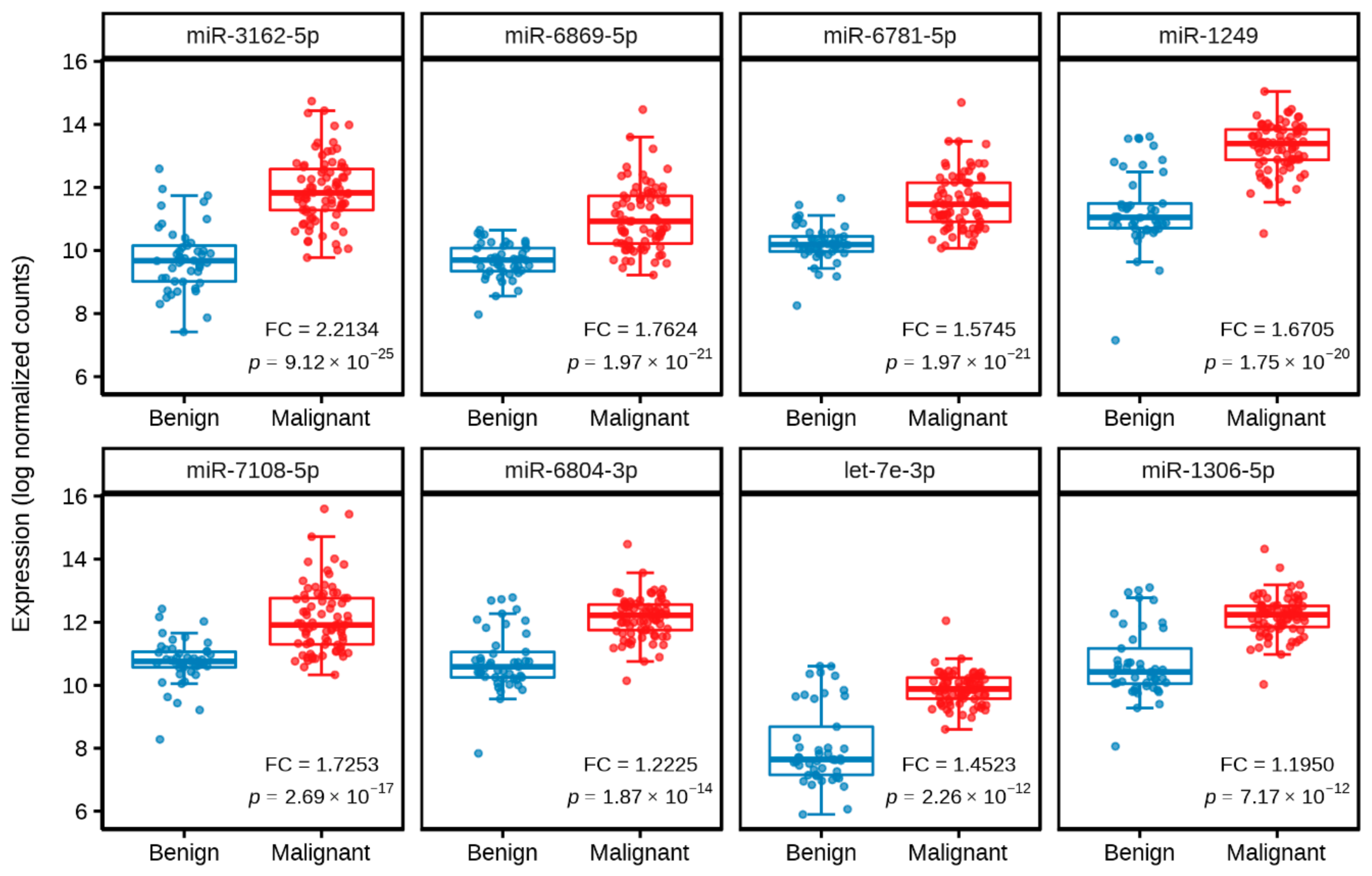
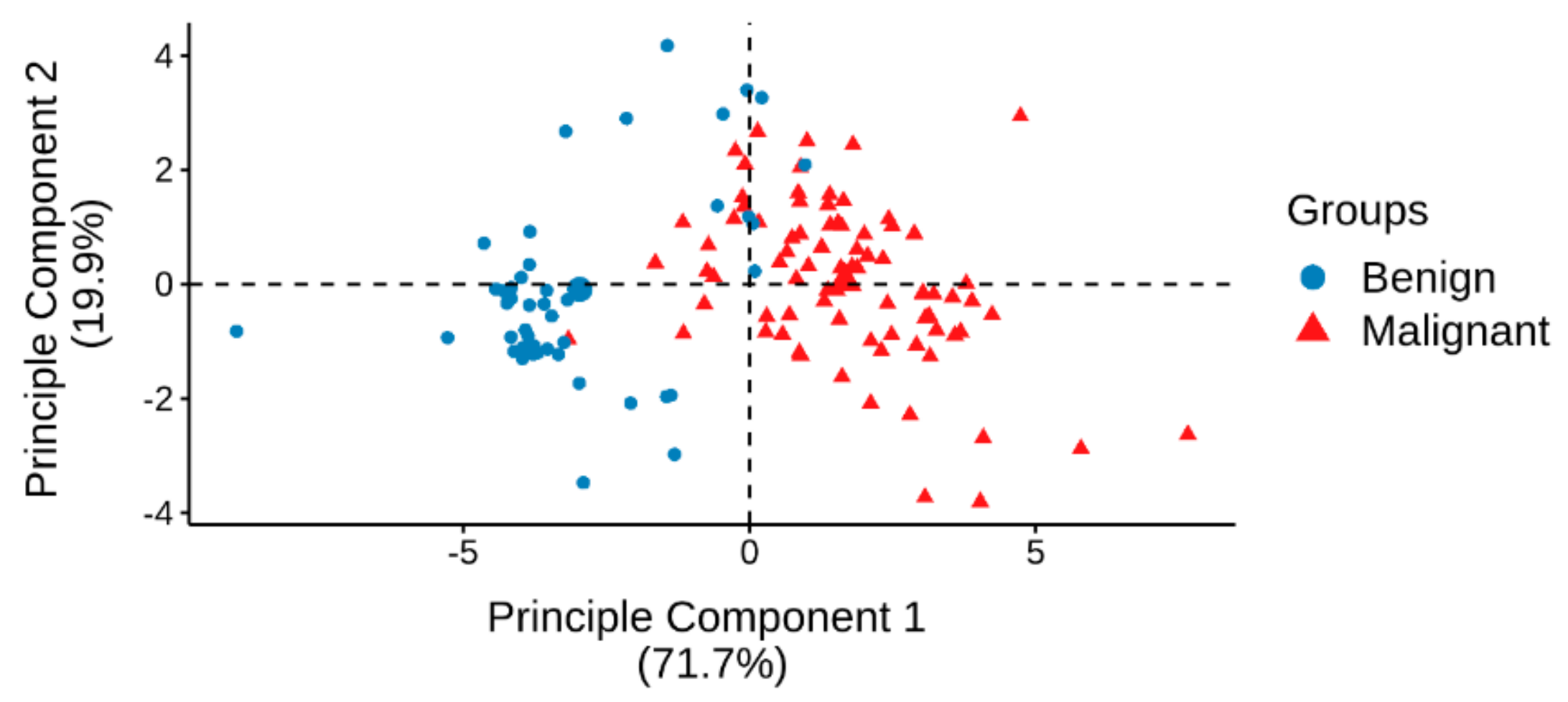
2.2. Identification of Significantly Differentially Expressed Serum Circulating MiRNAs from NGS Profiling
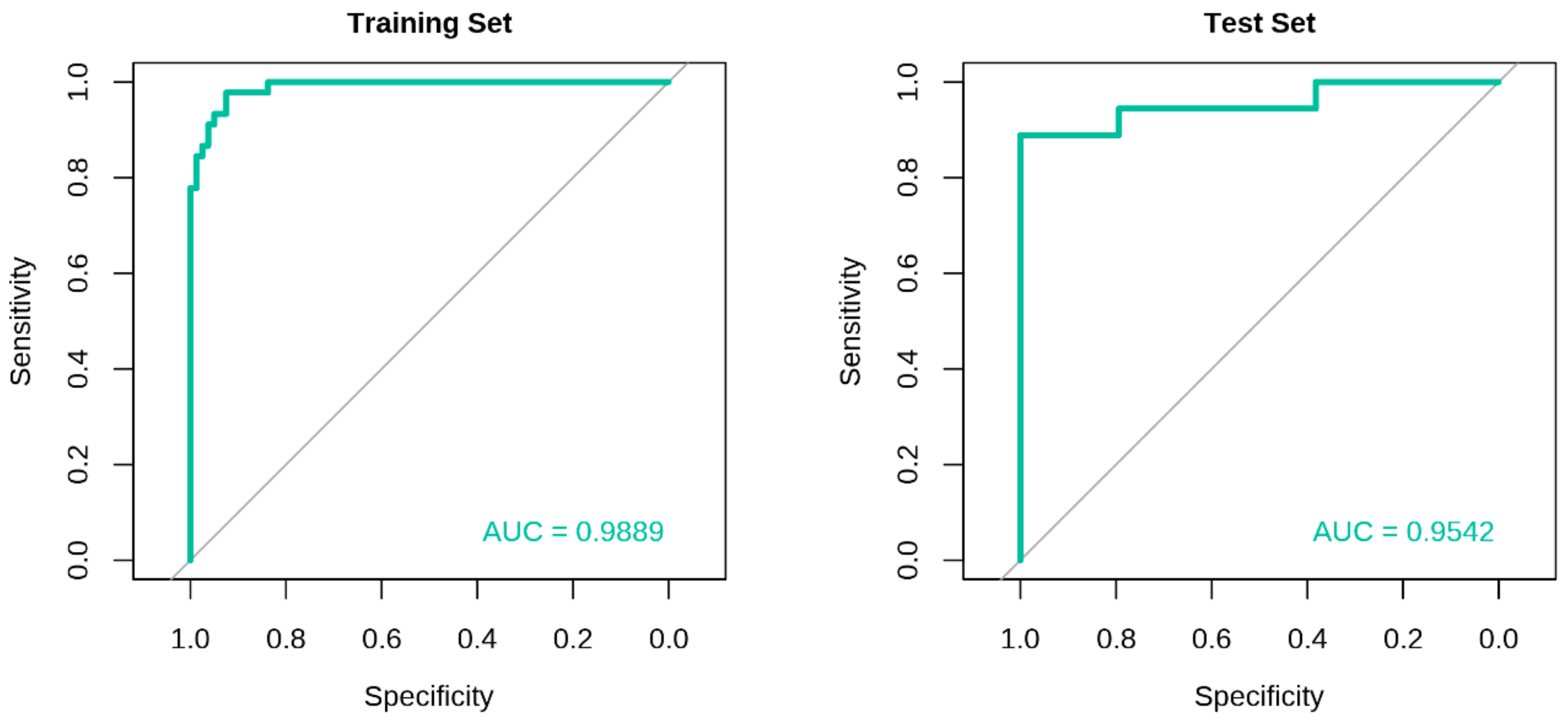
2.3. Development of Classification Model and Receiver Operating Characteristic (ROC) Curve Analysis
3. Discussion
4. Materials and Methods
4.1. Study Subjects
4.2. Serum Samples Processing
4.3. Serum Samples for NGS
4.4. Model Development
4.4.1. MiRNA Selection
4.4.2. Classification Model Generation for Malignant and Benign Breast Lesions
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ong, M.S.; Mandl, K.D. National expenditure for false-positive mammograms and breast cancer overdiagnoses estimated at $4 billion a year. Health Aff. (Millwood) 2015, 34, 576–583. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Martinez, A.; De Miguel-Perez, D.; Ortega, F.G.; Garcia-Puche, J.L.; Robles-Fernandez, I.; Exposito, J.; Martorell-Marugan, J.; Carmona-Saez, P.; Garrido-Navas, M.D.C.; Rolfo, C.; et al. Exosomal mirna profile as complementary tool in the diagnostic and prediction of treatment response in localized breast cancer under neoadjuvant chemotherapy. Breast Cancer Re. 2019, 21, 21. [Google Scholar] [CrossRef] [PubMed]
- Huynh, P.T.; Jarolimek, A.M.; Daye, S. The false-negative mammogram. Radiographics 1998, 18, 1137–1154. [Google Scholar] [CrossRef] [PubMed]
- Kolb, T.M.; Lichy, J.; Newhouse, J.H. Comparison of the performance of screening mammography, physical examination, and breast us and evaluation of factors that influence them: An analysis of 27,825 patient evaluations. Radiology 2002, 225, 165–175. [Google Scholar] [CrossRef]
- Nelson, H.D.; O’Meara, E.S.; Kerlikowske, K.; Balch, S.; Miglioretti, D. Factors associated with rates of false-positive and false-negative results from digital mammography screening: An analysis of registry data. Ann. Intern. Med. 2016, 164, 226–235. [Google Scholar] [CrossRef]
- Hubbard, R.A.; Kerlikowske, K.; Flowers, C.I.; Yankaskas, B.C.; Zhu, W.; Miglioretti, D.L. Cumulative probability of false-positive recall or biopsy recommendation after 10 years of screening mammography: A cohort study. Ann. Intern. Med. 2011, 155, 481–492. [Google Scholar] [CrossRef]
- Heitzer, E.; Perakis, S.; Geigl, J.B.; Speicher, M.R. The potential of liquid biopsies for the early detection of cancer. NPJ Precis. Oncol. 2017, 1, 36. [Google Scholar] [CrossRef]
- Molina, R.; Barak, V.; Van Dalen, A.; Duffy, M.J.; Einarsson, R.; Gion, M.; Goike, H.; Lamerz, R.; Nap, M.; Soletormos, G.; et al. Tumor markers in breast cancer—european group on tumor markers recommendations. Tumour Biol. 2005, 26, 281–293. [Google Scholar] [CrossRef]
- Harris, L.; Fritsche, H.; Mennel, R.; Norton, L.; Ravdin, P.; Taube, S.; Somerfield, M.R.; Hayes, D.F.; Bast, R.C., Jr. American Society of Clinical Oncology 2007 update of recommendations for the use of tumor markers in breast cancer. J. Oncol. Pract. 2007, 3, 336–339. [Google Scholar]
- Loke, S.Y.; Lee, A.S.G. The future of blood-based biomarkers for the early detection of breast cancer. Eur. J. Cancer 2018, 92, 54–68. [Google Scholar] [CrossRef]
- Schwarzenbach, H.; Nishida, N.; Calin, G.A.; Pantel, K. Clinical relevance of circulating cell-free micrornas in cancer. Nat. Rev. Clin. Oncol. 2014, 11, 145–156. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Croce, C.M. The role of micrornas in human cancer. Signal. Transduct Target Ther. 2016, 1, 15004. [Google Scholar] [CrossRef] [PubMed]
- Hayes, J.; Peruzzi, P.P.; Lawler, S. Micrornas in cancer: Biomarkers, functions and therapy. Trends Mol. Med. 2014, 20, 460–469. [Google Scholar] [CrossRef] [PubMed]
- Kent, O.A.; Mendell, J.T. A small piece in the cancer puzzle: Micrornas as tumor suppressors and oncogenes. Oncogene 2006, 25, 6188–6196. [Google Scholar] [CrossRef]
- Motawi, T.M.; Sadik, N.A.; Shaker, O.G.; El Masry, M.R.; Mohareb, F. Study of micrornas-21/221 as potential breast cancer biomarkers in egyptian women. Gene 2016, 590, 210–219. [Google Scholar] [CrossRef]
- Schwarzenbach, H.; Milde-Langosch, K.; Steinbach, B.; Muller, V.; Pantel, K. Diagnostic potential of pten-targeting mir-214 in the blood of breast cancer patients. Breast Cancer Res. Treat. 2012, 134, 933–941. [Google Scholar] [CrossRef]
- Cuk, K.; Zucknick, M.; Madhavan, D.; Schott, S.; Golatta, M.; Heil, J.; Marme, F.; Turchinovich, A.; Sinn, P.; Sohn, C.; et al. Plasma microrna panel for minimally invasive detection of breast cancer. PLoS ONE 2013, 8, e76729. [Google Scholar] [CrossRef]
- Cohen, J.D.; Li, L.; Wang, Y.; Thoburn, C.; Afsari, B.; Danilova, L.; Douville, C.; Javed, A.A.; Wong, F.; Mattox, A.; et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science 2018, 359, 926–930. [Google Scholar] [CrossRef]
- Klein, E.A.; Hubbell, E.; Maddala, T.; Aravanis, A.; Beausang, J.F.; Filippova, D.; Gross, S.; Jamshidi, A.; Kurtzman, K.; Shen, L.; et al. Development of a comprehensive cell-free DNA (cfdna) assay for early detection of multiple tumor types: The circulating cell-free genome atlas (ccga) study. J. Clin. Oncol. 2018, 36, 12021. [Google Scholar] [CrossRef]
- Liu, M.C.; Maddala, T.; Aravanis, A.; Hubbell, E.; Beausang, J.F.; Filippova, D.; Gross, S.; Jamshidi, A.; Kurtzman, K.; Shen, L.; et al. Breast cancer cell-free DNA (cfdna) profiles reflect underlying tumor biology: The circulating cell-free genome atlas (ccga) study. J. Clin. Oncol. 2018, 36, 536. [Google Scholar] [CrossRef]
- Attallah, A.M.; El-Far, M.; Omran, M.M.; Abdallah, S.O.; El-Desouky, M.A.; El-Dosoky, I.; Abdelrazek, M.A.; Attallah, A.A.; Elweresh, M.A.; Abdel Hameed, G.E.; et al. Circulating levels and clinical implications of epithelial membrane antigen and cytokeratin-1 in women with breast cancer: Can their ratio improve the results? Tumour Biol. 2014, 35, 10737–10745. [Google Scholar] [CrossRef] [PubMed]
- Moon, P.G.; Lee, J.E.; Cho, Y.E.; Lee, S.J.; Jung, J.H.; Chae, Y.S.; Bae, H.I.; Kim, Y.B.; Kim, I.S.; Park, H.Y.; et al. Identification of developmental endothelial locus-1 on circulating extracellular vesicles as a novel biomarker for early breast cancer detection. Clin. Cancer Res. 2016, 22, 1757–1766. [Google Scholar] [CrossRef] [PubMed]
- Swellam, M.; Abdelmaksoud, M.D.; Sayed Mahmoud, M.; Ramadan, A.; Abdel-Moneem, W.; Hefny, M.M. Aberrant methylation of apc and rarbeta2 genes in breast cancer patients. IUBMB Life 2015, 67, 61–68. [Google Scholar] [CrossRef] [PubMed]
- Shimomura, A.; Shiino, S.; Kawauchi, J.; Takizawa, S.; Sakamoto, H.; Matsuzaki, J.; Ono, M.; Takeshita, F.; Niida, S.; Shimizu, C.; et al. Novel combination of serum microrna for detecting breast cancer in the early stage. Cancer Sci. 2016, 107, 326–334. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, C.C.; Cheng, H.H.; Tewari, M. Microrna profiling: Approaches and considerations. Nat. Rev. Genet. 2012, 13, 358–369. [Google Scholar] [CrossRef] [PubMed]
- Pezuk, J.A.; Miller, T.L.A.; Bevilacqua, J.L.B.; De Barros, A.; De Andrade, F.E.M.; LFA, E.M.; Aguilar, V.; Claro, A.N.M.; Camargo, A.A.; Galante, P.A.F.; et al. Measuring plasma levels of three micrornas can improve the accuracy for identification of malignant breast lesions in women with bi-rads 4 mammography. Oncotarget 2017, 8, 83940–83948. [Google Scholar] [CrossRef]
- Swellam, M.; Ramadan, A.; El-Hussieny, E.A.; Bakr, N.M.; Hassan, N.M.; Sobeih, M.E.; EzzElArab, L.R. Clinical significance of blood-based mirnas as diagnostic and prognostic nucleic acid markers in breast cancer: Comparative to conventional tumor markers. J. Cell Biochem. 2019, 120, 12321–12330. [Google Scholar] [CrossRef]
- Wang, Q.; Sun, T.; Cao, Y.; Gao, P.; Dong, J.; Fang, Y.; Fang, Z.; Sun, X.; Zhu, Z. A dried blood spot mass spectrometry metabolomic approach for rapid breast cancer detection. Onco. Targets Ther. 2016, 9, 1389–1398. [Google Scholar]
- Chen, X.; Chen, H.; Dai, M.; Ai, J.; Li, Y.; Mahon, B.; Dai, S.; Deng, Y. Plasma lipidomics profiling identified lipid biomarkers in distinguishing early-stage breast cancer from benign lesions. Oncotarget 2016, 7, 36622–36631. [Google Scholar] [CrossRef]
- Farre, P.L.; Scalise, G.D.; Duca, R.B.; Dalton, G.N.; Massillo, C.; Porretti, J.; Grana, K.; Gardner, K.; De Luca, P.; De Siervi, A. Ctbp1 and metabolic syndrome induce an mrna and mirna expression profile critical for breast cancer progression and metastasis. Oncotarget 2018, 9, 13848–13858. [Google Scholar] [CrossRef]
- Aure, M.R.; Leivonen, S.K.; Fleischer, T.; Zhu, Q.; Overgaard, J.; Alsner, J.; Tramm, T.; Louhimo, R.; Alnaes, G.I.; Perala, M.; et al. Individual and combined effects of DNA methylation and copy number alterations on mirna expression in breast tumors. Genome Biol. 2013, 14, R126. [Google Scholar] [CrossRef] [PubMed]
- Ding, J.; Wu, W.; Yang, J.; Wu, M. Long non-coding rna mif-as1 promotes breast cancer cell proliferation, migration and emt process through regulating mir-1249-3p/hoxb8 axis. Pathol. Res. Pract. 2019, 215, 152376. [Google Scholar] [CrossRef] [PubMed]
- Fang, B.; Li, G.; Xu, C.; Hui, Y.; Li, G. Microrna mir-1249 downregulates adenomatous polyposis coli 2 expression and promotes glioma cells proliferation. Am. J. Transl. Res. 2018, 10, 1324–1336. [Google Scholar] [PubMed]
- Chen, X.; Zeng, K.; Xu, M.; Liu, X.; Hu, X.; Xu, T.; He, B.; Pan, Y.; Sun, H.; Wang, S. P53-induced mir-1249 inhibits tumor growth, metastasis, and angiogenesis by targeting vegfa and hmga2. Cell Death Dis. 2019, 10, 131. [Google Scholar] [CrossRef]
- Okumura, T.; Shimada, Y.; Omura, T.; Hirano, K.; Nagata, T.; Tsukada, K. Microrna profiles to predict postoperative prognosis in patients with small cell carcinoma of the esophagus. Anticancer Res. 2015, 35, 719–727. [Google Scholar]
- Katayama, Y.; Maeda, M.; Miyaguchi, K.; Nemoto, S.; Yasen, M.; Tanaka, S.; Mizushima, H.; Fukuoka, Y.; Arii, S.; Tanaka, H. Identification of pathogenesis-related micrornas in hepatocellular carcinoma by expression profiling. Oncol. Lett. 2012, 4, 817–823. [Google Scholar] [CrossRef][Green Version]
- Ye, Y.; Wei, Y.; Xu, Y.; Li, Y.; Wang, R.; Chen, J.; Zhou, Y.; Fu, Z.; Chen, Y.; Wang, X.; et al. Induced mir-1249 expression by aberrant activation of hedegehog signaling pathway in hepatocellular carcinoma. Exp. Cell Res. 2017, 355, 9–17. [Google Scholar] [CrossRef]
- Yan, S.; Liu, G.; Jin, C.; Wang, Z.; Duan, Q.; Xu, J.; Xu, D. Microrna-6869-5p acts as a tumor suppressor via targeting tlr4/nf-kappab signaling pathway in colorectal cancer. J. Cell Physiol. 2018, 233, 6660–6668. [Google Scholar] [CrossRef]
- Chen, J.; Yao, D.; Li, Y.; Chen, H.; He, C.; Ding, N.; Lu, Y.; Ou, T.; Zhao, S.; Li, L.; et al. Serum microrna expression levels can predict lymph node metastasis in patients with early-stage cervical squamous cell carcinoma. Int. J. Mol. Med. 2013, 32, 557–567. [Google Scholar] [CrossRef]
- Stegeman, S.; Amankwah, E.; Klein, K.; O’Mara, T.A.; Kim, D.; Lin, H.Y.; Permuth-Wey, J.; Sellers, T.A.; Srinivasan, S.; Eeles, R.; et al. A large-scale analysis of genetic variants within putative mirna binding sites in prostate cancer. Cancer Discov. 2015, 5, 368–379. [Google Scholar] [CrossRef]
- Matin, F.; Jeet, V.; Srinivasan, S.; Cristino, A.S.; Panchadsaram, J.; Clements, J.A.; Batra, J.; Australian Prostate Cancer, B. Microrna-3162-5p-mediated crosstalk between kallikrein family members including prostate-specific antigen in prostate cancer. Clin. Chem. 2019, 65, 771–780. [Google Scholar] [CrossRef] [PubMed]
- Van Boven, N.; Kardys, I.; Van Vark, L.C.; Akkerhuis, K.M.; De Ronde, M.W.J.; Khan, M.A.F.; Merkus, D.; Liu, Z.; Voors, A.A.; Asselbergs, F.W.; et al. Serially measured circulating micrornas and adverse clinical outcomes in patients with acute heart failure. Eur. J. Heart Fail. 2018, 20, 89–96. [Google Scholar] [CrossRef] [PubMed]
- Bayes-Genis, A.; Lanfear, D.E.; De Ronde, M.W.J.; Lupon, J.; Leenders, J.J.; Liu, Z.; Zuithoff, N.P.A.; Eijkemans, M.J.C.; Zamora, E.; De Antonio, M.; et al. Prognostic value of circulating micrornas on heart failure-related morbidity and mortality in two large diverse cohorts of general heart failure patients. Eur. J. Heart Fail. 2018, 20, 67–75. [Google Scholar] [CrossRef]
- Hindle, A.G.; Thoonen, R.; Jasien, J.V.; Grange, R.M.H.; Amin, K.; Wise, J.; Ozaki, M.; Ritch, R.; Malhotra, R.; Buys, E.S. Identification of candidate mirna biomarkers for glaucoma. Invest. Ophthalmol. Vis. Sci. 2019, 60, 134–146. [Google Scholar] [CrossRef]
- Yan, S.; Zhang, H.; Xie, W.; Meng, F.; Zhang, K.; Jiang, Y.; Zhang, X.; Zhang, J. Altered microrna profiles in plasma exosomes from mesial temporal lobe epilepsy with hippocampal sclerosis. Oncotarget 2017, 8, 4136–4146. [Google Scholar] [CrossRef]
- Hasan, M.M.; Akter, R.; Ullah, M.S.; Abedin, M.J.; Ullah, G.M.; Hossain, M.Z. A computational approach for predicting role of human micrornas in mers-cov genome. Adv. Bioinformatics 2014, 2014, 967946. [Google Scholar] [CrossRef]
- Elias, K.M.; Fendler, W.; Stawiski, K.; Fiascone, S.J.; Vitonis, A.F.; Berkowitz, R.S.; Frendl, G.; Konstantinopoulos, P.; Crum, C.P.; Kedzierska, M.; et al. Diagnostic potential for a serum mirna neural network for detection of ovarian cancer. Elife 2017, 6. [Google Scholar] [CrossRef]
- Rodriguez, M.; Bajo-Santos, C.; Hessvik, N.P.; Lorenz, S.; Fromm, B.; Berge, V.; Sandvig, K.; Line, A.; Llorente, A. Identification of non-invasive mirnas biomarkers for prostate cancer by deep sequencing analysis of urinary exosomes. Mol. Cancer 2017, 16, 156. [Google Scholar] [CrossRef]
- Tiberio, P.; Callari, M.; Angeloni, V.; Daidone, M.G.; Appierto, V. Challenges in using circulating mirnas as cancer biomarkers. BioMed Res. Int. 2015, 2015, 731479. [Google Scholar] [CrossRef]
- Leidner, R.S.; Li, L.; Thompson, C.L. Dampening enthusiasm for circulating microrna in breast cancer. PLoS ONE 2013, 8, e57841. [Google Scholar] [CrossRef]
- Witwer, K.W. Circulating microrna biomarker studies: Pitfalls and potential solutions. Clin. Chem. 2015, 61, 56–63. [Google Scholar] [CrossRef]
- Singh, R.; Ramasubramanian, B.; Kanji, S.; Chakraborty, A.R.; Haque, S.J.; Chakravarti, A. Circulating micrornas in cancer: Hope or hype? Cancer Lett. 2016, 381, 113–121. [Google Scholar] [CrossRef]
- Merker, J.D.; Oxnard, G.R.; Compton, C.; Diehn, M.; Hurley, P.; Lazar, A.J.; Lindeman, N.; Lockwood, C.M.; Rai, A.J.; Schilsky, R.L.; et al. Circulating tumor DNA analysis in patients with cancer: American society of clinical oncology and college of american pathologists joint review. Arch. Pathol. Lab. Med. 2018, 142, 1242–1253. [Google Scholar] [CrossRef]
- Allred, D.C. Ductal carcinoma in situ: Terminology, classification, and natural history. J. Natl. Cancer Inst. Monogr. 2010, 2010, 134–138. [Google Scholar] [CrossRef]
- Guray, M.; Sahin, A.A. Benign breast diseases: Classification, diagnosis, and management. Oncologist 2006, 11, 435–449. [Google Scholar] [CrossRef]
- Arpino, G.; Laucirica, R.; Elledge, R.M. Premalignant and in situ breast disease: Biology and clinical implications. Ann. Intern. Med. 2005, 143, 446–457. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for rna-seq data with deseq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
| Clinico-Pathological Characteristics | Training Set (n = 125) | Test Set (n = 52) |
|---|---|---|
| Age a | ||
| Mean | 56.0 | 55.2 |
| Median | 55.0 | 53.5 |
| Range | 34–87 | 39–79 |
| Race | ||
| Chinese | 108 (86.4%) | 42 (80.8%) |
| Malay | 8 (6.4%) | 1 (1.9%) |
| Indian | 7 (5.6%) | 6 (11.5%) |
| Others | 2 (1.6%) | 3 (5.8%) |
| Malignant Cases | 80 | 34 |
| Age at diagnosis | ||
| Mean | 57.6 | 58.2 |
| Median | 57.5 | 58.0 |
| Range | 34–87 | 41–79 |
| Histological type | ||
| Invasive ductal carcinoma (IDC) | 37 (46.3%) | 18 (52.9%) |
| Invasive lobular carcinoma (ILC) | 3 (3.8%) | 1 (2.9%) |
| Ductal carcinoma in-situ (DCIS) | 26 (32.5%) | 8 (23.5%) |
| Others b | 6 (7.5%) | 3 (8.8%) |
| Mixed | ||
| IDC with another histological type | 7 (8.8%) | 4 (11.8%) |
| Others c | 1 (1.3%) | nil |
| Receptor status | ||
| Estrogen receptor (ER) | ||
| Positive | 70 (87.5%) | 28 (82.4%) |
| Negative | 10 (12.5%) | 5 (14.7%) |
| Unknown | nil | 1 (2.9%) |
| Progesterone receptor (PR) | ||
| Positive | 54 (67.5%) | 23 (67.6%) |
| Negative | 25 (31.3%) | 10 (29.4%) |
| Unknown | 1 (1.3%) | 1 (2.9%) |
| Human epidermal growth factor receptor 2 (HER2) | ||
| Positive | 13 (16.3%) | 4 (11.8%) |
| Negative | 33 (41.3%) | 12 (35.3%) |
| Equivocal | 10 (12.5%) | 10 (29.4%) |
| Not tested (DCIS) d | 20 (25.0%) | 8 (23.5%) |
| Unknown | 4 (5.0%) | nil |
| Tumor size | ||
| <20 mm | 44 (55.0%) | 17 (50.0%) |
| 20 mm to 50 mm | 30 (37.5%) | 15 (44.1%) |
| >50 mm | 4 (5.0%) | 2 (5.9%) |
| Unknown | 2 (2.5%) | nil |
| Tumor grade | ||
| Invasive carcinomas | ||
| Grade 1 | 7 (8.8%) | 7 (20.6%) |
| Grade 2 | 27 (33.8%) | 9 (26.5%) |
| Grade 3 | 18 (22.5%) | 10 (29.4%) |
| Unknown | 2 (2.5%) | nil |
| DCIS | ||
| High nuclear grade | 10 (12.5%) | 1 (2.9%) |
| Intermediate nuclear grade | 12 (15.0%) | 4 (11.8%) |
| Low nuclear grade | 4 (5.0%) | 3 (8.8%) |
| Lymph node status | ||
| Positive | 20 (25.0%) | 9 (26.5%) |
| Negative | 44 (55.0%) | 21 (61.8%) |
| Not tested (DCIS) d | 11 (13.8%) | 3 (8.8%) |
| Unknown | 5 (6.3%) | 1 (2.9%) |
| Benign Controls | 45 | 18 |
| Age at diagnosis | ||
| Mean | 53.1 | 49.5 |
| Median | 52.0 | 47.0 |
| Range | 40–82 | 39–66 |
| Type of lesion e | ||
| Atypical ductal hyperplasia (ADH) | 2 (4.4%) | nil |
| Lobular carcinoma in-situ (LCIS) | 1 (2.2%) | 1 (5.6%) |
| Fibrocystic changes | 17 (37.8%) | 6 (33.3%) |
| Sclerosing adenosis | 2 (4.4%) | 2 (11.1%) |
| Moderate or florid ductal hyperplasia of the usual type | 2 (4.4%) | nil |
| Radial scar | 1 (2.2%) | nil |
| Fibroadenoma (FA) | 21 (46.7%) | 11 (61.1%) |
| Others | 10 (22.2%) | 3 (16.7%) |
| MiRNA | Fold Change (log2) | Adjusted p-Value | Expression |
|---|---|---|---|
| miR-3162-5p | 2.2134 | 9.12 × 10−25 | Upregulation |
| miR-6869-5p | 1.7624 | 1.97 × 10−21 | Upregulation |
| miR-6781-5p | 1.5745 | 1.97 × 10−21 | Upregulation |
| miR-1249 | 1.6705 | 1.75 × 10−20 | Upregulation |
| miR-7108-5p | 1.7253 | 2.69 × 10−17 | Upregulation |
| miR-6804-3p | 1.2225 | 1.87 × 10−14 | Upregulation |
| let-7e-3p | 1.4523 | 2.26 × 10−12 | Upregulation |
| miR-1306-5p | 1.1950 | 7.17 × 10−12 | Upregulation |
| Performance Metrics | Eight-MiRNA Signature Model | |
|---|---|---|
| Training Set | Test Set | |
| AUC (95% CI) | 0.9889 (0.9772, 1.0000) | 0.9542 (0.8832, 1.0000) |
| Recall | 0.9625 | 0.9412 |
| Precision | 0.9506 | 0.9412 |
| Balanced Accuracy | 0.9368 | 0.9150 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Loke, S.Y.; Munusamy, P.; Koh, G.L.; Chan, C.H.T.; Madhukumar, P.; Thung, J.L.; Tan, K.T.B.; Ong, K.W.; Yong, W.S.; Sim, Y.; et al. A Circulating miRNA Signature for Stratification of Breast Lesions among Women with Abnormal Screening Mammograms. Cancers 2019, 11, 1872. https://doi.org/10.3390/cancers11121872
Loke SY, Munusamy P, Koh GL, Chan CHT, Madhukumar P, Thung JL, Tan KTB, Ong KW, Yong WS, Sim Y, et al. A Circulating miRNA Signature for Stratification of Breast Lesions among Women with Abnormal Screening Mammograms. Cancers. 2019; 11(12):1872. https://doi.org/10.3390/cancers11121872
Chicago/Turabian StyleLoke, Sau Yeen, Prabhakaran Munusamy, Geok Ling Koh, Claire Hian Tzer Chan, Preetha Madhukumar, Jee Liang Thung, Kiat Tee Benita Tan, Kong Wee Ong, Wei Sean Yong, Yirong Sim, and et al. 2019. "A Circulating miRNA Signature for Stratification of Breast Lesions among Women with Abnormal Screening Mammograms" Cancers 11, no. 12: 1872. https://doi.org/10.3390/cancers11121872
APA StyleLoke, S. Y., Munusamy, P., Koh, G. L., Chan, C. H. T., Madhukumar, P., Thung, J. L., Tan, K. T. B., Ong, K. W., Yong, W. S., Sim, Y., Oey, C. L., Lim, S. Z., Chan, M. Y. P., Ho, T. S. J., Khoo, B. K. J., Wong, S. L. J., Thng, C. H., Chong, B. K., Tan, E. Y., ... Lee, A. S. G. (2019). A Circulating miRNA Signature for Stratification of Breast Lesions among Women with Abnormal Screening Mammograms. Cancers, 11(12), 1872. https://doi.org/10.3390/cancers11121872





