The Emerging Roles of ATP-Dependent Chromatin Remodeling Complexes in Pancreatic Cancer
Abstract
1. Introduction
2. Epigenetic Dysregulation in Pancreatic Cancer Development and Heterogeneity
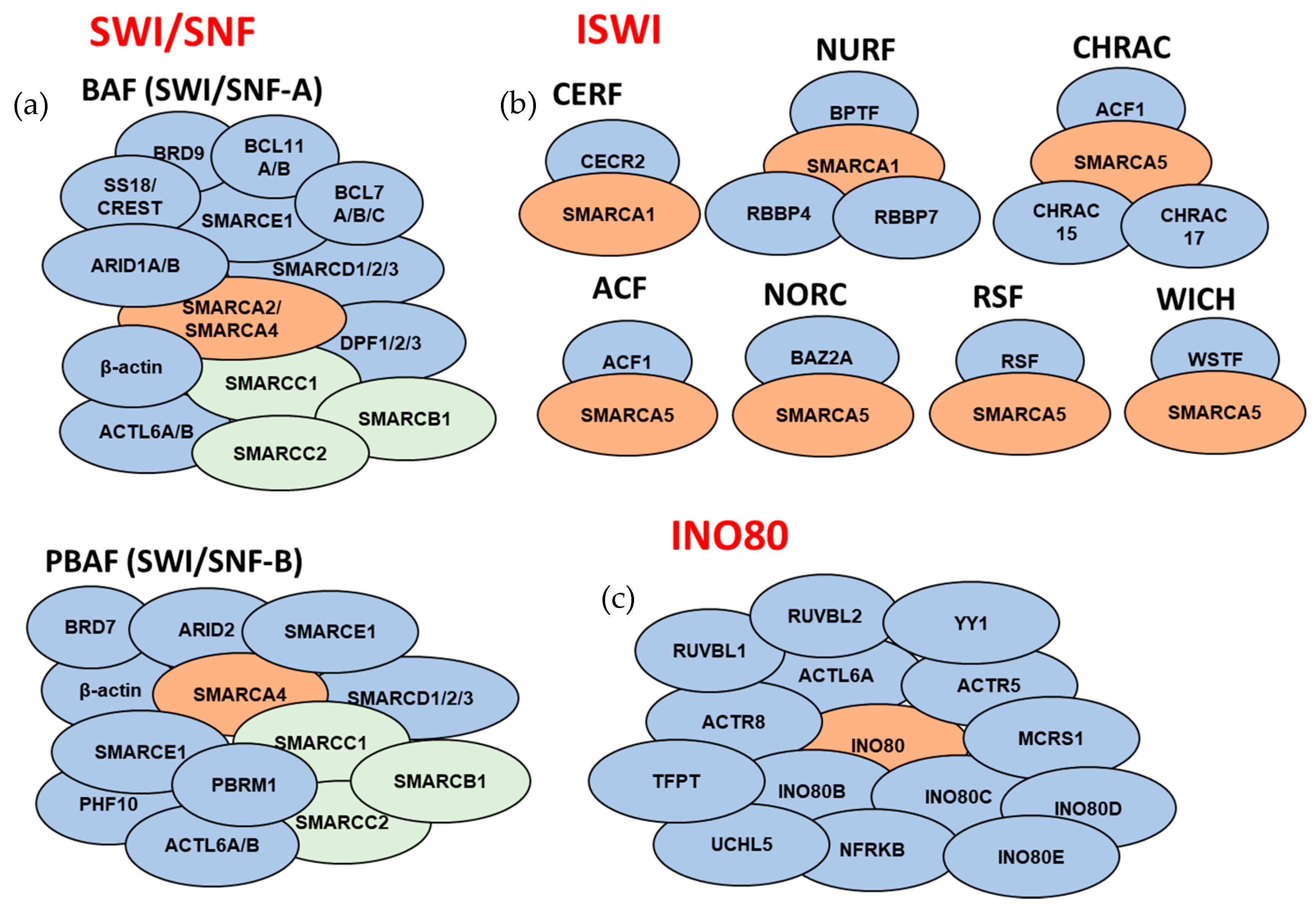
3. ATP-Dependent Chromatin Remodeling Complexes
3.1. SWI/SNF Subfamily
3.2. ISWI Subfamily
3.3. CHD Subfamily
3.4. INO80 Subfamily
4. Mechanistic Studies of the ATP-Dependent Chromatin Remodeling Complexes in PDAC
4.1. SWI/SNF Subfamily
4.1.1. ARID1A
4.1.2. ARID1B
4.1.3. SMARCA2
4.1.4. SMARCA4
4.1.5. SMARCC1
4.1.6. ACTL6B
4.2. ISWI Subfamily
4.2.1. BPTF
4.3. CHD Subfamily
4.3.1. CHD1
4.3.2. CHD5
4.3.3. CHD7
4.4. INO80 Subfamily
4.4.1. INO80
4.4.2. INO80C
4.5. SWI/SNF and INO80 Subfamilies
4.5.1. ACTB
4.5.2. ACTL6A
5. Therapeutic Targeting of Chromatin Remodeling in Pancreatic Cancer
6. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Rahib, L.; Smith, B.D.; Aizenberg, R.; Rosenzweig, A.B.; Fleshman, J.M.; Matrisian, L.M. Projecting cancer incidence and deaths to 2030: The unexpected burden of thyroid, liver, and pancreas cancers in the united states. Cancer Res. 2014, 74, 2913–2921. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Xie, K.; Wolff, R.; Abbruzzese, J.L. Pancreatic cancer. Lancet 2004, 363, 1049–1057. [Google Scholar] [CrossRef]
- Jones, P.A.; Baylin, S.B. The fundamental role of epigenetic events in cancer. Nat. Rev. Genet. 2002, 3, 415–428. [Google Scholar] [CrossRef] [PubMed]
- Jones, P.A.; Baylin, S.B. The epigenomics of cancer. Cell 2007, 128, 683–692. [Google Scholar] [CrossRef]
- Baylin, S.B.; Jones, P.A. Epigenetic determinants of cancer. Cold Spring Harb. Perspect. Biol. 2016, 8, a019505. [Google Scholar] [CrossRef]
- Lomberk, G.; Dusetti, N.; Iovanna, J.; Urrutia, R. Emerging epigenomic landscapes of pancreatic cancer in the era of precision medicine. Nat. Commun. 2019, 10, 3875. [Google Scholar] [CrossRef]
- Yi, J.M.; Guzzetta, A.A.; Bailey, V.J.; Downing, S.R.; Van Neste, L.; Chiappinelli, K.B.; Keeley, B.P.; Stark, A.; Herrera, A.; Wolfgang, C.; et al. Novel methylation biomarker panel for the early detection of pancreatic cancer. Clin. Cancer Res. 2013, 19, 6544–6555. [Google Scholar] [CrossRef]
- Morel, D.; Almouzni, G.; Soria, J.C.; Postel-Vinay, S. Targeting chromatin defects in selected solid tumors based on oncogene addiction, synthetic lethality and epigenetic antagonism. Ann. Oncol. 2017, 28, 254–269. [Google Scholar] [CrossRef]
- Iacobuzio-Donahue, C.A. Epigenetic changes in cancer. Annu. Rev. Pathol. 2009, 4, 229–249. [Google Scholar] [CrossRef]
- Luger, K.; Rechsteiner, T.J.; Flaus, A.J.; Waye, M.M.; Richmond, T.J. Characterization of nucleosome core particles containing histone proteins made in bacteria. J. Mol. Biol. 1997, 272, 301–311. [Google Scholar] [CrossRef]
- Luo, R.X.; Dean, D.C. Chromatin remodeling and transcriptional regulation. J. Natl. Cancer Inst. 1999, 91, 1288–1294. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Torres, K.; Liu, X.; Liu, C.G.; Pollock, R.E. An overview of chromatin-regulating proteins in cells. Curr. Protein Pept. Sci. 2016, 17, 401–410. [Google Scholar] [CrossRef] [PubMed]
- Lawrence, M.; Daujat, S.; Schneider, R. Lateral thinking: How histone modifications regulate gene expression. Trends Genet. 2016, 32, 42–56. [Google Scholar] [CrossRef] [PubMed]
- Audia, J.E.; Campbell, R.M. Histone modifications and cancer. Cold Spring Harb. Perspect. Biol. 2016, 8, a019521. [Google Scholar] [CrossRef] [PubMed]
- Leroy, G.; Dimaggio, P.A.; Chan, E.Y.; Zee, B.M.; Blanco, M.A.; Bryant, B.; Flaniken, I.Z.; Liu, S.; Kang, Y.; Trojer, P.; et al. A quantitative atlas of histone modification signatures from human cancer cells. Epigenetics Chromatin 2013, 6, 20. [Google Scholar] [CrossRef] [PubMed]
- Kadoch, C.; Crabtree, G.R. Mammalian swi/snf chromatin remodeling complexes and cancer: Mechanistic insights gained from human genomics. Sci. Adv. 2015, 1, e1500447. [Google Scholar] [CrossRef] [PubMed]
- Kadoch, C.; Hargreaves, D.C.; Hodges, C.; Elias, L.; Ho, L.; Ranish, J.; Crabtree, G.R. Proteomic and bioinformatic analysis of mammalian swi/snf complexes identifies extensive roles in human malignancy. Nat. Genet. 2013, 45, 592–601. [Google Scholar] [CrossRef]
- Narlikar, G.J.; Sundaramoorthy, R.; Owen-Hughes, T. Mechanisms and functions of atp-dependent chromatin-remodeling enzymes. Cell 2013, 154, 490–503. [Google Scholar] [CrossRef]
- Valencia, A.M.; Kadoch, C. Chromatin regulatory mechanisms and therapeutic opportunities in cancer. Nat. Cell Biol. 2019, 21, 152–161. [Google Scholar] [CrossRef]
- Shain, A.H.; Giacomini, C.P.; Matsukuma, K.; Karikari, C.A.; Bashyam, M.D.; Hidalgo, M.; Maitra, A.; Pollack, J.R. Convergent structural alterations define switch/sucrose nonfermentable (swi/snf) chromatin remodeler as a central tumor suppressive complex in pancreatic cancer. Proc. Natl. Acad. Sci. USA 2012, 109, E252–E259. [Google Scholar] [CrossRef]
- Witkiewicz, A.K.; McMillan, E.A.; Balaji, U.; Baek, G.; Lin, W.C.; Mansour, J.; Mollaee, M.; Wagner, K.U.; Koduru, P.; Yopp, A.; et al. Whole-exome sequencing of pancreatic cancer defines genetic diversity and therapeutic targets. Nat. Commun. 2015, 6, 6744. [Google Scholar] [CrossRef] [PubMed]
- Biankin, A.V.; Waddell, N.; Kassahn, K.S.; Gingras, M.C.; Muthuswamy, L.B.; Johns, A.L.; Miller, D.K.; Wilson, P.J.; Patch, A.M.; Wu, J.; et al. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature 2012, 491, 399–405. [Google Scholar] [CrossRef] [PubMed]
- Klein, A.P.; Brune, K.A.; Petersen, G.M.; Goggins, M.; Tersmette, A.C.; Offerhaus, G.J.; Griffin, C.; Cameron, J.L.; Yeo, C.J.; Kern, S.; et al. Prospective risk of pancreatic cancer in familial pancreatic cancer kindreds. Cancer Res. 2004, 64, 2634–2638. [Google Scholar] [CrossRef] [PubMed]
- Brosens, L.A.; Hackeng, W.M.; Offerhaus, G.J.; Hruban, R.H.; Wood, L.D. Pancreatic adenocarcinoma pathology: Changing “landscape”. J. Gastrointest. Oncol. 2015, 6, 358–374. [Google Scholar]
- Patra, K.C.; Bardeesy, N.; Mizukami, Y. Diversity of precursor lesions for pancreatic cancer: The genetics and biology of intraductal papillary mucinous neoplasm. Clin. Transl. Gastroenterol. 2017, 8, e86. [Google Scholar] [CrossRef]
- Wu, J.; Jiao, Y.; Dal Molin, M.; Maitra, A.; de Wilde, R.F.; Wood, L.D.; Eshleman, J.R.; Goggins, M.G.; Wolfgang, C.L.; Canto, M.I.; et al. Whole-exome sequencing of neoplastic cysts of the pancreas reveals recurrent mutations in components of ubiquitin-dependent pathways. Proc. Natl. Acad. Sci. USA 2011, 108, 21188–21193. [Google Scholar] [CrossRef]
- Bailey, M.H.; Tokheim, C.; Porta-Pardo, E.; Sengupta, S.; Bertrand, D.; Weerasinghe, A.; Colaprico, A.; Wendl, M.C.; Kim, J.; Reardon, B.; et al. Comprehensive characterization of cancer driver genes and mutations. Cell 2018, 174, 1034–1035. [Google Scholar] [CrossRef]
- Collisson, E.A.; Sadanandam, A.; Olson, P.; Gibb, W.J.; Truitt, M.; Gu, S.; Cooc, J.; Weinkle, J.; Kim, G.E.; Jakkula, L.; et al. Subtypes of pancreatic ductal adenocarcinoma and their differing responses to therapy. Nat. Med. 2011, 17, 500–503. [Google Scholar] [CrossRef]
- Moffitt, R.A.; Marayati, R.; Flate, E.L.; Volmar, K.E.; Loeza, S.G.; Hoadley, K.A.; Rashid, N.U.; Williams, L.A.; Eaton, S.C.; Chung, A.H.; et al. Virtual microdissection identifies distinct tumor- and stroma-specific subtypes of pancreatic ductal adenocarcinoma. Nat. Genet. 2015, 47, 1168–1178. [Google Scholar] [CrossRef]
- Puleo, F.; Nicolle, R.; Blum, Y.; Cros, J.; Marisa, L.; Demetter, P.; Quertinmont, E.; Svrcek, M.; Elarouci, N.; Iovanna, J.; et al. Stratification of pancreatic ductal adenocarcinomas based on tumor and microenvironment features. Gastroenterology 2018, 155, 1999–2013. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Research Network. Integrated genomic characterization of pancreatic ductal adenocarcinoma. Cancer Cell 2017, 32, 185–203. [Google Scholar] [CrossRef]
- Lomberk, G.; Blum, Y.; Nicolle, R.; Nair, A.; Gaonkar, K.S.; Marisa, L.; Mathison, A.; Sun, Z.; Yan, H.; Elarouci, N.; et al. Distinct epigenetic landscapes underlie the pathobiology of pancreatic cancer subtypes. Nat. Commun. 2018, 9, 1978. [Google Scholar] [CrossRef]
- Hayashi, A.; Fan, J.; Chen, R.; Ho, Y.; Makohon-Moore, A.P.; Zhong, Y.; Hong, J.; Sakamoto, H.; Attiyeh, M.A.; Kohutek, Z.A.; et al. The genetic basis of transcriptional heterogeneity for basal-like features in pancreatic ductal adenocarcinoma. bioRxiv 2019. [Google Scholar] [CrossRef]
- Nicolle, R.; Blum, Y.; Marisa, L.; Loncle, C.; Gayet, O.; Moutardier, V.; Turrini, O.; Giovannini, M.; Bian, B.; Bigonnet, M.; et al. Pancreatic adenocarcinoma therapeutic targets revealed by tumor-stroma cross-talk analyses in patient-derived xenografts. Cell Rep. 2017, 21, 2458–2470. [Google Scholar] [CrossRef] [PubMed]
- McDonald, O.G.; Li, X.; Saunders, T.; Tryggvadottir, R.; Mentch, S.J.; Warmoes, M.O.; Word, A.E.; Carrer, A.; Salz, T.H.; Natsume, S.; et al. Epigenomic reprogramming during pancreatic cancer progression links anabolic glucose metabolism to distant metastasis. Nat. Genet. 2017, 49, 367–376. [Google Scholar] [CrossRef]
- Natale, F.; Vivo, M.; Falco, G.; Angrisano, T. Deciphering DNA methylation signatures of pancreatic cancer and pancreatitis. Clin. Epigenetics 2019, 11, 132. [Google Scholar] [CrossRef]
- Thompson, M.J.; Rubbi, L.; Dawson, D.W.; Donahue, T.R.; Pellegrini, M. Pancreatic cancer patient survival correlates with DNA methylation of pancreas development genes. PLoS ONE 2015, 10, e0128814. [Google Scholar] [CrossRef]
- Roe, J.S.; Hwang, C.I.; Somerville, T.D.D.; Milazzo, J.P.; Lee, E.J.; Da Silva, B.; Maiorino, L.; Tiriac, H.; Young, C.M.; Miyabayashi, K.; et al. Enhancer reprogramming promotes pancreatic cancer metastasis. Cell 2017, 170, 875–888. [Google Scholar] [CrossRef]
- Kumar, R.; Li, D.Q.; Muller, S.; Knapp, S. Epigenomic regulation of oncogenesis by chromatin remodeling. Oncogene 2016, 35, 4423–4436. [Google Scholar] [CrossRef]
- Hohmann, A.F.; Vakoc, C.R. A rationale to target the swi/snf complex for cancer therapy. Trends Genet. 2014, 30, 356–363. [Google Scholar] [CrossRef]
- Grobner, S.N.; Worst, B.C.; Weischenfeldt, J.; Buchhalter, I.; Kleinheinz, K.; Rudneva, V.A.; Johann, P.D.; Balasubramanian, G.P.; Segura-Wang, M.; Brabetz, S.; et al. The landscape of genomic alterations across childhood cancers. Nature 2018, 555, 321–327. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Liu, Y.; Liu, Y.; Alexandrov, L.B.; Edmonson, M.N.; Gawad, C.; Zhou, X.; Li, Y.; Rusch, M.C.; Easton, J.; et al. Pan-cancer genome and transcriptome analyses of 1699 paediatric leukaemias and solid tumours. Nature 2018, 555, 371–376. [Google Scholar] [CrossRef] [PubMed]
- Koschmann, C.; Nunez, F.J.; Mendez, F.; Brosnan-Cashman, J.A.; Meeker, A.K.; Lowenstein, P.R.; Castro, M.G. Mutated chromatin regulatory factors as tumor drivers in cancer. Cancer Res. 2017, 77, 227–233. [Google Scholar] [CrossRef]
- Ho, L.; Crabtree, G.R. Chromatin remodelling during development. Nature 2010, 463, 474–484. [Google Scholar] [CrossRef] [PubMed]
- Bracken, A.P.; Brien, G.L.; Verrijzer, C.P. Dangerous liaisons: Interplay between swi/snf, nurd, and polycomb in chromatin regulation and cancer. Genes Dev. 2019, 33, 936–959. [Google Scholar] [CrossRef]
- McKenna, B.; Guo, M.; Reynolds, A.; Hara, M.; Stein, R. Dynamic recruitment of functionally distinct swi/snf chromatin remodeling complexes modulates pdx1 activity in islet beta cells. Cell Rep. 2015, 10, 2032–2042. [Google Scholar] [CrossRef]
- Campbell, S.A.; Hoffman, B.G. Chromatin regulators in pancreas development and diabetes. Trends Endocrinol. Metab. 2016, 27, 142–152. [Google Scholar] [CrossRef]
- Spaeth, J.M.; Liu, J.H.; Peters, D.; Guo, M.; Osipovich, A.B.; Mohammadi, F.; Roy, N.; Bhushan, A.; Magnuson, M.A.; Hebrok, M.; et al. The pdx1-bound swi/snf chromatin remodeling complex regulates pancreatic progenitor cell proliferation and mature islet beta-cell function. Diabetes 2019, 68, 1806–1818. [Google Scholar] [CrossRef]
- Wang, W.; Friedland, S.C.; Guo, B.; O’Dell, M.R.; Alexander, W.B.; Whitney-Miller, C.L.; Agostini-Vulaj, D.; Huber, A.R.; Myers, J.R.; Ashton, J.M.; et al. Arid1a, a swi/snf subunit, is critical to acinar cell homeostasis and regeneration and is a barrier to transformation and epithelial-mesenchymal transition in the pancreas. Gut 2019, 68, 1245–1258. [Google Scholar] [CrossRef]
- Wang, S.C.; Nassour, I.; Xiao, S.; Zhang, S.; Luo, X.; Lee, J.; Li, L.; Sun, X.; Nguyen, L.H.; Chuang, J.C.; et al. Swi/snf component arid1a restrains pancreatic neoplasia formation. Gut 2019, 68, 1259–1270. [Google Scholar] [CrossRef]
- Poli, J.; Gasser, S.M.; Papamichos-Chronakis, M. The ino80 remodeller in transcription, replication and repair. Philos. Trans. R. Soc. B Biol. Sci. 2017, 372, 20160290. [Google Scholar] [CrossRef] [PubMed]
- Clapier, C.R.; Iwasa, J.; Cairns, B.R.; Peterson, C.L. Mechanisms of action and regulation of atp-dependent chromatin-remodelling complexes. Nat. Rev. Mol. Cell Biol. 2017, 18, 407–422. [Google Scholar] [CrossRef] [PubMed]
- Aydin, O.Z.; Vermeulen, W.; Lans, H. Iswi chromatin remodeling complexes in the DNA damage response. Cell Cycle 2014, 13, 3016–3025. [Google Scholar] [CrossRef] [PubMed]
- Hargreaves, D.C.; Crabtree, G.R. Atp-dependent chromatin remodeling: Genetics, genomics and mechanisms. Cell Res. 2011, 21, 396–420. [Google Scholar] [CrossRef]
- Michel, B.C.; D’Avino, A.R.; Cassel, S.H.; Mashtalir, N.; McKenzie, Z.M.; McBride, M.J.; Valencia, A.M.; Zhou, Q.; Bocker, M.; Soares, L.M.M.; et al. A non-canonical swi/snf complex is a synthetic lethal target in cancers driven by baf complex perturbation. Nat. Cell Biol. 2018, 20, 1410–1420. [Google Scholar] [CrossRef]
- Lessard, J.; Wu, J.I.; Ranish, J.A.; Wan, M.; Winslow, M.M.; Staahl, B.T.; Wu, H.; Aebersold, R.; Graef, I.A.; Crabtree, G.R. An essential switch in subunit composition of a chromatin remodeling complex during neural development. Neuron 2007, 55, 201–215. [Google Scholar] [CrossRef]
- Lessard, J.A.; Crabtree, G.R. Chromatin regulatory mechanisms in pluripotency. Annu. Rev. Cell Dev. Biol. 2010, 26, 503–532. [Google Scholar] [CrossRef]
- Davidson, J.; Shen, Z.; Gong, X.; Pollack, J.R. Swi/snf aberrations sensitize pancreatic cancer cells to DNA crosslinking agents. Oncotarget 2018, 9, 9608–9617. [Google Scholar] [CrossRef][Green Version]
- Oppikofer, M.; Bai, T.; Gan, Y.; Haley, B.; Liu, P.; Sandoval, W.; Ciferri, C.; Cochran, A.G. Expansion of the iswi chromatin remodeler family with new active complexes. EMBO Rep. 2017, 18, 1697–1706. [Google Scholar] [CrossRef]
- Xiao, H.; Sandaltzopoulos, R.; Wang, H.M.; Hamiche, A.; Ranallo, R.; Lee, K.M.; Fu, D.; Wu, C. Dual functions of largest nurf subunit nurf301 in nucleosome sliding and transcription factor interactions. Mol. Cell 2001, 8, 531–543. [Google Scholar] [CrossRef]
- Mills, A.A. The chromodomain helicase DNA-binding chromatin remodelers: Family traits that protect from and promote cancer. Cold Spring Harb. Perspect. Med. 2017, 7, a026450. [Google Scholar] [CrossRef] [PubMed]
- Basta, J.; Rauchman, M. The nucleosome remodeling and deacetylase complex in development and disease. Transl. Res. 2015, 165, 36–47. [Google Scholar] [CrossRef] [PubMed]
- Cai, Y.; Geutjes, E.J.; de Lint, K.; Roepman, P.; Bruurs, L.; Yu, L.R.; Wang, W.; van Blijswijk, J.; Mohammad, H.; de Rink, I.; et al. The nurd complex cooperates with dnmts to maintain silencing of key colorectal tumor suppressor genes. Oncogene 2014, 33, 2157–2168. [Google Scholar] [CrossRef] [PubMed]
- Xia, L.; Huang, W.; Bellani, M.; Seidman, M.M.; Wu, K.; Fan, D.; Nie, Y.; Cai, Y.; Zhang, Y.W.; Yu, L.R.; et al. Chd4 has oncogenic functions in initiating and maintaining epigenetic suppression of multiple tumor suppressor genes. Cancer Cell 2017, 31, 653–668 e657. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Z.; Elsayed, Z.; Peterkin, V.; Alkatib, S.; Bennett, D.; Landry, J.W. Ino80 is essential for proximal-distal axis asymmetry in part by regulating bmp4 expression. BMC Biol. 2016, 14, 18. [Google Scholar] [CrossRef]
- Rhee, S.; Chung, J.I.; King, D.A.; D’Amato, G.; Paik, D.T.; Duan, A.; Chang, A.; Nagelberg, D.; Sharma, B.; Jeong, Y.; et al. Endothelial deletion of ino80 disrupts coronary angiogenesis and causes congenital heart disease. Nat. Commun. 2018, 9, 368. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Zhou, B.; Wang, L.; Li, P.; Bennett, B.D.; Snyder, R.; Garantziotis, S.; Fargo, D.C.; Cox, A.D.; Chen, L.; et al. Ino80 is required for oncogenic transcription and tumor growth in non-small cell lung cancer. Oncogene 2017, 36, 1430–1439. [Google Scholar] [CrossRef]
- Zhou, B.; Wang, L.; Zhang, S.; Bennett, B.D.; He, F.; Zhang, Y.; Xiong, C.; Han, L.; Diao, L.; Li, P.; et al. Ino80 governs superenhancer-mediated oncogenic transcription and tumor growth in melanoma. Genes Dev. 2016, 30, 1440–1453. [Google Scholar] [CrossRef]
- Lee, S.A.; Lee, H.S.; Hur, S.K.; Kang, S.W.; Oh, G.T.; Lee, D.; Kwon, J. Ino80 haploinsufficiency inhibits colon cancer tumorigenesis via replication stress-induced apoptosis. Oncotarget 2017, 8, 115041–115053. [Google Scholar] [CrossRef][Green Version]
- Wang, L.; Du, Y.; Ward, J.M.; Shimbo, T.; Lackford, B.; Zheng, X.; Miao, Y.L.; Zhou, B.; Han, L.; Fargo, D.C.; et al. Ino80 facilitates pluripotency gene activation in embryonic stem cell self-renewal, reprogramming, and blastocyst development. Cell Stem Cell 2014, 14, 575–591. [Google Scholar] [CrossRef]
- Beckwith, S.L.; Schwartz, E.K.; Garcia-Nieto, P.E.; King, D.A.; Gowans, G.J.; Wong, K.M.; Eckley, T.L.; Paraschuk, A.P.; Peltan, E.L.; Lee, L.R.; et al. The ino80 chromatin remodeler sustains metabolic stability by promoting tor signaling and regulating histone acetylation. PLoS Genet. 2018, 14, e1007216. [Google Scholar] [CrossRef] [PubMed]
- Helming, K.C.; Wang, X.; Roberts, C.W.M. Vulnerabilities of mutant swi/snf complexes in cancer. Cancer Cell 2014, 26, 309–317. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013, 6, pl1. [Google Scholar] [CrossRef] [PubMed]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cbio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef]
- Hoadley, K.A.; Yau, C.; Hinoue, T.; Wolf, D.M.; Lazar, A.J.; Drill, E.; Shen, R.; Taylor, A.M.; Cherniack, A.D.; Thorsson, V.; et al. Cell-of-origin patterns dominate the molecular classification of 10,000 tumors from 33 types of cancer. Cell 2018, 173, 291–304. [Google Scholar] [CrossRef]
- Ellrott, K.; Bailey, M.H.; Saksena, G.; Covington, K.R.; Kandoth, C.; Stewart, C.; Hess, J.; Ma, S.; Chiotti, K.E.; McLellan, M.; et al. Scalable open science approach for mutation calling of tumor exomes using multiple genomic pipelines. Cell Syst. 2018, 6, 271–281. [Google Scholar] [CrossRef]
- Taylor, A.M.; Shih, J.; Ha, G.; Gao, G.F.; Zhang, X.; Berger, A.C.; Schumacher, S.E.; Wang, C.; Hu, H.; Liu, J.; et al. Genomic and functional approaches to understanding cancer aneuploidy. Cancer Cell 2018, 33, 676–689. [Google Scholar] [CrossRef]
- Gao, Q.; Liang, W.W.; Foltz, S.M.; Mutharasu, G.; Jayasinghe, R.G.; Cao, S.; Liao, W.W.; Reynolds, S.M.; Wyczalkowski, M.A.; Yao, L.; et al. Driver fusions and their implications in the development and treatment of human cancers. Cell Rep. 2018, 23, 227–238. [Google Scholar] [CrossRef]
- Liu, J.; Lichtenberg, T.; Hoadley, K.A.; Poisson, L.M.; Lazar, A.J.; Cherniack, A.D.; Kovatich, A.J.; Benz, C.C.; Levine, D.A.; Lee, A.V.; et al. An integrated tcga pan-cancer clinical data resource to drive high-quality survival outcome analytics. Cell 2018, 173, 400–416. [Google Scholar] [CrossRef]
- Sanchez-Vega, F.; Mina, M.; Armenia, J.; Chatila, W.K.; Luna, A.; La, K.C.; Dimitriadoy, S.; Liu, D.L.; Kantheti, H.S.; Saghafinia, S.; et al. Oncogenic signaling pathways in the cancer genome atlas. Cell 2018, 173, 321–337. [Google Scholar] [CrossRef]
- Bailey, P.; Chang, D.K.; Nones, K.; Johns, A.L.; Patch, A.M.; Gingras, M.C.; Miller, D.K.; Christ, A.N.; Bruxner, T.J.; Quinn, M.C.; et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature 2016, 531, 47–52. [Google Scholar] [CrossRef] [PubMed]
- Kimura, Y.; Fukuda, A.; Ogawa, S.; Maruno, T.; Takada, Y.; Tsuda, M.; Hiramatsu, Y.; Araki, O.; Nagao, M.; Yoshikawa, T.; et al. Arid1a maintains differentiation of pancreatic ductal cells and inhibits development of pancreatic ductal adenocarcinoma in mice. Gastroenterology 2018, 155, 194–209. [Google Scholar] [CrossRef] [PubMed]
- Numata, M.; Morinaga, S.; Watanabe, T.; Tamagawa, H.; Yamamoto, N.; Shiozawa, M.; Nakamura, Y.; Kameda, Y.; Okawa, S.; Rino, Y.; et al. The clinical significance of swi/snf complex in pancreatic cancer. Int. J. Oncol. 2013, 42, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Khursheed, M.; Kolla, J.N.; Kotapalli, V.; Gupta, N.; Gowrishankar, S.; Uppin, S.G.; Sastry, R.A.; Koganti, S.; Sundaram, C.; Pollack, J.R.; et al. Arid1b, a member of the human swi/snf chromatin remodeling complex, exhibits tumour-suppressor activities in pancreatic cancer cell lines. Br. J. Cancer 2013, 108, 2056–2062. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Wang, F.; Du, C.; Guo, H.; Ma, L.; Liu, X.; Kornmann, M.; Tian, X.; Yang, Y. Brm/smarca2 promotes the proliferation and chemoresistance of pancreatic cancer cells by targeting jak2/stat3 signaling. Cancer Lett. 2017, 402, 213–224. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Tian, X.; Wang, F.; Ma, Y.; Kornmann, M.; Yang, Y. Brg1 promotes chemoresistance of pancreatic cancer cells through crosstalking with akt signalling. Eur. J. Cancer 2014, 50, 2251–2262. [Google Scholar] [CrossRef] [PubMed]
- Dal Molin, M.; Hong, S.M.; Hebbar, S.; Sharma, R.; Scrimieri, F.; de Wilde, R.F.; Mayo, S.C.; Goggins, M.; Wolfgang, C.L.; Schulick, R.D.; et al. Loss of expression of the swi/snf chromatin remodeling subunit brg1/smarca4 is frequently observed in intraductal papillary mucinous neoplasms of the pancreas. Hum. Pathol. 2012, 43, 585–591. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Roy, N.; Malik, S.; Villanueva, K.E.; Urano, A.; Lu, X.; Von Figura, G.; Seeley, E.S.; Dawson, D.W.; Collisson, E.A.; Hebrok, M. Brg1 promotes both tumor-suppressive and oncogenic activities at distinct stages of pancreatic cancer formation. Genes Dev. 2015, 29, 658–671. [Google Scholar] [CrossRef]
- Von Figura, G.; Fukuda, A.; Roy, N.; Liku, M.E.; Morris Iv, J.P.; Kim, G.E.; Russ, H.A.; Firpo, M.A.; Mulvihill, S.J.; Dawson, D.W.; et al. The chromatin regulator brg1 suppresses formation of intraductal papillary mucinous neoplasm and pancreatic ductal adenocarcinoma. Nat. Cell Biol. 2014, 16, 255–267. [Google Scholar] [CrossRef]
- Iwagami, Y.; Eguchi, H.; Nagano, H.; Akita, H.; Hama, N.; Wada, H.; Kawamoto, K.; Kobayashi, S.; Tomokuni, A.; Tomimaru, Y.; et al. Mir-320c regulates gemcitabine-resistance in pancreatic cancer via smarcc1. Br. J. Cancer 2013, 109, 502–511. [Google Scholar] [CrossRef]
- Taniuchi, K.; Furihata, M.; Naganuma, S.; Dabanaka, K.; Hanazaki, K.; Saibara, T. Bcl7b, a predictor of poor prognosis of pancreatic cancers, promotes cell motility and invasion by influencing creb signaling. Am. J. Cancer Res. 2018, 8, 387–404. [Google Scholar] [PubMed]
- Arpalahti, L.; Saukkonen, K.; Hagstrom, J.; Mustonen, H.; Seppanen, H.; Haglund, C.; Holmberg, C.I. Nuclear ubiquitin c-terminal hydrolase l5 expression associates with increased patient survival in pancreatic ductal adenocarcinoma. Tumour Biol. 2017, 39, 1010428317710411. [Google Scholar] [CrossRef] [PubMed]
- Hall, W.A.; Petrova, A.V.; Colbert, L.E.; Hardy, C.W.; Fisher, S.B.; Saka, B.; Shelton, J.W.; Warren, M.D.; Pantazides, B.G.; Gandhi, K.; et al. Low chd5 expression activates the DNA damage response and predicts poor outcome in patients undergoing adjuvant therapy for resected pancreatic cancer. Oncogene 2014, 33, 5450–5456. [Google Scholar] [CrossRef] [PubMed]
- Livshits, G.; Alonso-Curbelo, D.; Morris, J.P.t.; Koche, R.; Saborowski, M.; Wilkinson, J.E.; Lowe, S.W. Arid1a restrains kras-dependent changes in acinar cell identity. Elife 2018, 7, e35216. [Google Scholar] [CrossRef]
- Helming, K.C.; Wang, X.; Wilson, B.G.; Vazquez, F.; Haswell, J.R.; Manchester, H.E.; Kim, Y.; Kryukov, G.V.; Ghandi, M.; Aguirre, A.J.; et al. Arid1b is a specific vulnerability in arid1a-mutant cancers. Nat. Med. 2014, 20, 251–254. [Google Scholar] [CrossRef]
- Guerrero-Martinez, J.A.; Reyes, J.C. High expression of smarca4 or smarca2 is frequently associated with an opposite prognosis in cancer. Sci. Rep. 2018, 8, 2043. [Google Scholar] [CrossRef]
- Zhang, Z.; Li, J.; Guo, H.; Wang, F.; Ma, L.; Du, C.; Wang, Y.; Wang, Q.; Kornmann, M.; Tian, X.; et al. Brm transcriptionally regulates mir-302a-3p to target socs5/stat3 signaling axis to potentiate pancreatic cancer metastasis. Cancer Lett. 2019, 449, 215–225. [Google Scholar] [CrossRef]
- Marquez-Vilendrer, S.B.; Thompson, K.; Lu, L.; Reisman, D. Mechanism of brg1 silencing in primary cancers. Oncotarget 2016, 7, 56153–56169. [Google Scholar] [CrossRef]
- Strobeck, M.W.; Knudsen, K.E.; Fribourg, A.F.; DeCristofaro, M.F.; Weissman, B.E.; Imbalzano, A.N.; Knudsen, E.S. Brg-1 is required for rb-mediated cell cycle arrest. Proc. Natl. Acad. Sci. USA 2000, 97, 7748–7753. [Google Scholar] [CrossRef]
- Reisman, D.N.; Strobeck, M.W.; Betz, B.L.; Sciariotta, J.; Funkhouser, W., Jr.; Murchardt, C.; Yaniv, M.; Sherman, L.S.; Knudsen, E.S.; Weissman, B.E. Concomitant down-regulation of brm and brg1 in human tumor cell lines: Differential effects on rb-mediated growth arrest vs cd44 expression. Oncogene 2002, 21, 1196–1207. [Google Scholar] [CrossRef][Green Version]
- Hoffman, G.R.; Rahal, R.; Buxton, F.; Xiang, K.; McAllister, G.; Frias, E.; Bagdasarian, L.; Huber, J.; Lindeman, A.; Chen, D.; et al. Functional epigenetics approach identifies brm/smarca2 as a critical synthetic lethal target in brg1-deficient cancers. Proc. Natl. Acad. Sci. USA 2014, 111, 3128–3133. [Google Scholar] [CrossRef] [PubMed]
- Wilson, B.G.; Helming, K.C.; Wang, X.; Kim, Y.; Vazquez, F.; Jagani, Z.; Hahn, W.C.; Roberts, C.W. Residual complexes containing smarca2 (brm) underlie the oncogenic drive of smarca4 (brg1) mutation. Mol. Cell. Biol. 2014, 34, 1136–1144. [Google Scholar] [CrossRef] [PubMed]
- Ehrenhofer-Wolfer, K.; Puchner, T.; Schwarz, C.; Rippka, J.; Blaha-Ostermann, S.; Strobl, U.; Hormann, A.; Bader, G.; Kornigg, S.; Zahn, S.; et al. Smarca2-deficiency confers sensitivity to targeted inhibition of smarca4 in esophageal squamous cell carcinoma cell lines. Sci. Rep. 2019, 9, 11661. [Google Scholar] [CrossRef]
- DelBove, J.; Rosson, G.; Strobeck, M.; Chen, J.; Archer, T.K.; Wang, W.; Knudsen, E.S.; Weissman, B.E. Identification of a core member of the swi/snf complex, baf155/smarcc1, as a human tumor suppressor gene. Epigenetics 2011, 6, 1444–1453. [Google Scholar] [CrossRef]
- Wang, L.; Zhao, Z.; Meyer, M.B.; Saha, S.; Yu, M.; Guo, A.; Wisinski, K.B.; Huang, W.; Cai, W.; Pike, J.W.; et al. Carm1 methylates chromatin remodeling factor baf155 to enhance tumor progression and metastasis. Cancer Cell 2014, 25, 21–36. [Google Scholar] [CrossRef]
- Bell, S.; Rousseau, J.; Peng, H.; Aouabed, Z.; Priam, P.; Theroux, J.F.; Jefri, M.; Tanti, A.; Wu, H.; Kolobova, I.; et al. Mutations in actl6b cause neurodevelopmental deficits and epilepsy and lead to loss of dendrites in human neurons. Am. J. Hum. Genet. 2019, 104, 815–834. [Google Scholar] [CrossRef]
- Zhu, B.; Ueda, A.; Song, X.; Horike, S.I.; Yokota, T.; Akagi, T. Baf53a is involved in survival of mouse es cells, which can be compensated by baf53b. Sci. Rep. 2017, 7, 14059. [Google Scholar] [CrossRef]
- Varela, I.; Tarpey, P.; Raine, K.; Huang, D.; Ong, C.K.; Stephens, P.; Davies, H.; Jones, D.; Lin, M.L.; Teague, J.; et al. Exome sequencing identifies frequent mutation of the swi/snf complex gene pbrm1 in renal carcinoma. Nature 2011, 469, 539–542. [Google Scholar] [CrossRef]
- Miao, D.; Margolis, C.A.; Gao, W.; Voss, M.H.; Li, W.; Martini, D.J.; Norton, C.; Bosse, D.; Wankowicz, S.M.; Cullen, D.; et al. Genomic correlates of response to immune checkpoint therapies in clear cell renal cell carcinoma. Science 2018, 359, 801–806. [Google Scholar] [CrossRef]
- Sato, Y.; Yoshizato, T.; Shiraishi, Y.; Maekawa, S.; Okuno, Y.; Kamura, T.; Shimamura, T.; Sato-Otsubo, A.; Nagae, G.; Suzuki, H.; et al. Integrated molecular analysis of clear-cell renal cell carcinoma. Nat. Genet. 2013, 45, 860–867. [Google Scholar] [CrossRef]
- Porter, E.G.; Dhiman, A.; Chowdhury, B.; Carter, B.C.; Lin, H.; Stewart, J.C.; Kazemian, M.; Wendt, M.K.; Dykhuizen, E.C. Pbrm1 regulates stress response in epithelial cells. iScience 2019, 15, 196–210. [Google Scholar] [CrossRef] [PubMed]
- Kohashi, K.; Oda, Y. Oncogenic roles of smarcb1/ini1 and its deficient tumors. Cancer Sci. 2017, 108, 547–552. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.S.; Kim, M.S.; Yoo, N.J.; Lee, S.H. Frameshift mutations of a chromatin-remodeling gene smarcc2 in gastric and colorectal cancers with microsatellite instability. APMIS 2013, 121, 168–169. [Google Scholar] [CrossRef] [PubMed]
- Fraser, M.; Sabelnykova, V.Y.; Yamaguchi, T.N.; Heisler, L.E.; Livingstone, J.; Huang, V.; Shiah, Y.J.; Yousif, F.; Lin, X.; Masella, A.P.; et al. Genomic hallmarks of localized, non-indolent prostate cancer. Nature 2017, 541, 359–364. [Google Scholar] [CrossRef]
- Nakazato, H.; Takeshima, H.; Kishino, T.; Kubo, E.; Hattori, N.; Nakajima, T.; Yamashita, S.; Igaki, H.; Tachimori, Y.; Kuniyoshi, Y.; et al. Early-stage induction of swi/snf mutations during esophageal squamous cell carcinogenesis. PLoS ONE 2016, 11, e0147372. [Google Scholar] [CrossRef]
- Revill, K.; Wang, T.; Lachenmayer, A.; Kojima, K.; Harrington, A.; Li, J.; Hoshida, Y.; Llovet, J.M.; Powers, S. Genome-wide methylation analysis and epigenetic unmasking identify tumor suppressor genes in hepatocellular carcinoma. Gastroenterology 2013, 145, 1424–1435. [Google Scholar] [CrossRef]
- Hong, C.F.; Lin, S.Y.; Chou, Y.T.; Wu, C.W. Microrna-7 compromises p53 protein-dependent apoptosis by controlling the expression of the chromatin remodeling factor smarcd1. J. Biol. Chem. 2016, 291, 1877–1889. [Google Scholar] [CrossRef]
- Oh, J.; Sohn, D.H.; Ko, M.; Chung, H.; Jeon, S.H.; Seong, R.H. Baf60a interacts with p53 to recruit the swi/snf complex. J. Biol. Chem. 2008, 283, 11924–11934. [Google Scholar] [CrossRef]
- Arts, F.A.; Keogh, L.; Smyth, P.; O’Toole, S.; Ta, R.; Gleeson, N.; O’Leary, J.J.; Flavin, R.; Sheils, O. Mir-223 potentially targets swi/snf complex protein smarcd1 in atypical proliferative serous tumor and high-grade ovarian serous carcinoma. Hum. Pathol. 2017, 70, 98–104. [Google Scholar] [CrossRef]
- Inoue, C.; Zhao, C.; Tsuduki, Y.; Udono, M.; Wang, L.; Nomura, M.; Katakura, Y. Smarcd1 regulates senescence-associated lipid accumulation in hepatocytes. NPJ Aging Mech. Dis. 2017, 3, 11. [Google Scholar] [CrossRef]
- Shen, J.; Xiao, Z.; Wu, W.K.; Wang, M.H.; To, K.F.; Chen, Y.; Yang, W.; Li, M.S.; Shin, V.Y.; Tong, J.H.; et al. Epigenetic silencing of mir-490-3p reactivates the chromatin remodeler smarcd1 to promote helicobacter pylori-induced gastric carcinogenesis. Cancer Res. 2015, 75, 754–765. [Google Scholar] [CrossRef]
- Van de Wijngaart, D.J.; Dubbink, H.J.; Molier, M.; de Vos, C.; Trapman, J.; Jenster, G. Functional screening of fxxlf-like peptide motifs identifies smarcd1/baf60a as an androgen receptor cofactor that modulates tmprss2 expression. Mol. Endocrinol. 2009, 23, 1776–1786. [Google Scholar] [CrossRef] [PubMed]
- Witzel, M.; Petersheim, D.; Fan, Y.; Bahrami, E.; Racek, T.; Rohlfs, M.; Puchalka, J.; Mertes, C.; Gagneur, J.; Ziegenhain, C.; et al. Chromatin-remodeling factor smarcd2 regulates transcriptional networks controlling differentiation of neutrophil granulocytes. Nat. Genet. 2017, 49, 742–752. [Google Scholar] [CrossRef] [PubMed]
- Ji, S.; Zhu, L.; Gao, Y.; Zhang, X.; Yan, Y.; Cen, J.; Li, R.; Zeng, R.; Liao, L.; Hou, C.; et al. Baf60b-mediated atm-p53 activation blocks cell identity conversion by sensing chromatin opening. Cell Res. 2017, 27, 642–656. [Google Scholar] [CrossRef] [PubMed]
- Jordan, N.V.; Prat, A.; Abell, A.N.; Zawistowski, J.S.; Sciaky, N.; Karginova, O.A.; Zhou, B.; Golitz, B.T.; Perou, C.M.; Johnson, G.L. Swi/snf chromatin-remodeling factor smarcd3/baf60c controls epithelial-mesenchymal transition by inducing wnt5a signaling. Mol. Cell. Biol. 2013, 33, 3011–3025. [Google Scholar] [CrossRef]
- Sokol, E.S.; Feng, Y.X.; Jin, D.X.; Tizabi, M.D.; Miller, D.H.; Cohen, M.A.; Sanduja, S.; Reinhardt, F.; Pandey, J.; Superville, D.A.; et al. Smarce1 is required for the invasive progression of in situ cancers. Proc. Natl. Acad. Sci. USA 2017, 114, 4153–4158. [Google Scholar] [CrossRef]
- Link, K.A.; Balasubramaniam, S.; Sharma, A.; Comstock, C.E.; Godoy-Tundidor, S.; Powers, N.; Cao, K.H.; Haelens, A.; Claessens, F.; Revelo, M.P.; et al. Targeting the baf57 swi/snf subunit in prostate cancer: A novel platform to control androgen receptor activity. Cancer Res. 2008, 68, 4551–4558. [Google Scholar] [CrossRef]
- Papadakis, A.I.; Sun, C.; Knijnenburg, T.A.; Xue, Y.; Grernrum, W.; Holzel, M.; Nijkamp, W.; Wessels, L.F.; Beijersbergen, R.L.; Bernards, R.; et al. Smarce1 suppresses egfr expression and controls responses to met and alk inhibitors in lung cancer. Cell Res. 2015, 25, 445–458. [Google Scholar] [CrossRef]
- Yamaguchi, T.; Kurita, T.; Nishio, K.; Tsukada, J.; Hachisuga, T.; Morimoto, Y.; Iwai, Y.; Izumi, H. Expression of baf57 in ovarian cancer cells and drug sensitivity. Cancer Sci. 2015, 106, 359–366. [Google Scholar] [CrossRef]
- Banga, S.S.; Peng, L.; Dasgupta, T.; Palejwala, V.; Ozer, H.L. Phf10 is required for cell proliferation in normal and sv40-immortalized human fibroblast cells. Cytogenet. Genome Res. 2009, 126, 227–242. [Google Scholar] [CrossRef]
- Anbunathan, H.; Verstraten, R.; Singh, A.D.; Harbour, J.W.; Bowcock, A.M. Integrative copy number analysis of uveal melanoma reveals novel candidate genes involved in tumorigenesis including a tumor suppressor role for phf10/baf45a. Clin. Cancer Res. 2019, 25, 5156–5166. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Herlong, F.H.; Stroehlein, J.R.; Mishra, L. Mutations of chromatin structure regulating genes in human malignancies. Curr. Protein Pept. Sci. 2016, 17, 411–437. [Google Scholar] [CrossRef]
- Duan, Y.; Tian, L.; Gao, Q.; Liang, L.; Zhang, W.; Yang, Y.; Zheng, Y.; Pan, E.; Li, S.; Tang, N. Chromatin remodeling gene arid2 targets cyclin d1 and cyclin e1 to suppress hepatoma cell progression. Oncotarget 2016, 7, 45863–45875. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Wang, J.; Han, Y.; Huang, Z.; Ying, J.; Bi, X.; Zhao, J.; Fang, Y.; Zhou, H.; Zhou, J.; et al. Arid2: A new tumor suppressor gene in hepatocellular carcinoma. Oncotarget 2011, 2, 886–891. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Li, Z.; Shen, J. Brd7: A novel tumor suppressor gene in different cancers. Am. J. Transl. Res. 2016, 8, 742–748. [Google Scholar]
- Gatchalian, J.; Malik, S.; Ho, J.; Lee, D.S.; Kelso, T.W.R.; Shokhirev, M.N.; Dixon, J.R.; Hargreaves, D.C. A non-canonical brd9-containing baf chromatin remodeling complex regulates naive pluripotency in mouse embryonic stem cells. Nat. Commun. 2018, 9, 5139. [Google Scholar] [CrossRef]
- Sima, X.; He, J.; Peng, J.; Xu, Y.; Zhang, F.; Deng, L. The genetic alteration spectrum of the swi/snf complex: The oncogenic roles of brd9 and actl6a. PLoS ONE 2019, 14, e0222305. [Google Scholar] [CrossRef]
- Brien, G.L.; Remillard, D.; Shi, J.; Hemming, M.L.; Chabon, J.; Wynne, K.; Dillon, E.T.; Cagney, G.; Van Mierlo, G.; Baltissen, M.P.; et al. Targeted degradation of brd9 reverses oncogenic gene expression in synovial sarcoma. Elife 2018, 7, e41305. [Google Scholar] [CrossRef]
- Uehara, T.; Kage-Nakadai, E.; Yoshina, S.; Imae, R.; Mitani, S. The tumor suppressor bcl7b functions in the wnt signaling pathway. PLoS Genet. 2015, 11, e1004921. [Google Scholar] [CrossRef]
- Lazarus, K.A.; Hadi, F.; Zambon, E.; Bach, K.; Santolla, M.F.; Watson, J.K.; Correia, L.L.; Das, M.; Ugur, R.; Pensa, S.; et al. Bcl11a interacts with sox2 to control the expression of epigenetic regulators in lung squamous carcinoma. Nat. Commun. 2018, 9, 3327. [Google Scholar] [CrossRef]
- Khaled, W.T.; Choon Lee, S.; Stingl, J.; Chen, X.; Raza Ali, H.; Rueda, O.M.; Hadi, F.; Wang, J.; Yu, Y.; Chin, S.F.; et al. Bcl11a is a triple-negative breast cancer gene with critical functions in stem and progenitor cells. Nat. Commun. 2015, 6, 5987. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Luo, N.; Hu, Y.; Li, X.; Zhang, K. Mir-137 suppresses triple-negative breast cancer stemness and tumorigenesis by perturbing bcl11a-dnmt1 interaction. Cell. Physiol. Biochem. 2018, 47, 2147–2158. [Google Scholar] [CrossRef] [PubMed]
- Grabarczyk, P.; Nahse, V.; Delin, M.; Przybylski, G.; Depke, M.; Hildebrandt, P.; Volker, U.; Schmidt, C.A. Increased expression of bcl11b leads to chemoresistance accompanied by g1 accumulation. PLoS ONE 2010, 5, e12532. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez, A.; Kentsis, A.; Sanda, T.; Holmfeldt, L.; Chen, S.C.; Zhang, J.; Protopopov, A.; Chin, L.; Dahlberg, S.E.; Neuberg, D.S.; et al. The bcl11b tumor suppressor is mutated across the major molecular subtypes of t-cell acute lymphoblastic leukemia. Blood 2011, 118, 4169–4173. [Google Scholar] [CrossRef]
- Sakamaki, A.; Katsuragi, Y.; Otsuka, K.; Tomita, M.; Obata, M.; Iwasaki, T.; Abe, M.; Sato, T.; Ochiai, M.; Sakuraba, Y.; et al. Bcl11b swi/snf-complex subunit modulates intestinal adenoma and regeneration after gamma-irradiation through wnt/beta-catenin pathway. Carcinogenesis 2015, 36, 622–631. [Google Scholar] [CrossRef]
- Qiu, Z.; Ghosh, A. A calcium-dependent switch in a crest-brg1 complex regulates activity-dependent gene expression. Neuron 2008, 60, 775–787. [Google Scholar] [CrossRef]
- Pan, Y.; Yuan, F.; Li, Y.; Wang, G.; Lin, Z.; Chen, L. Bromodomain phdfinger transcription factor promotes glioma progression and indicates poor prognosis. Oncol. Rep. 2019, 41, 246–256. [Google Scholar]
- Dar, A.A.; Majid, S.; Bezrookove, V.; Phan, B.; Ursu, S.; Nosrati, M.; De Semir, D.; Sagebiel, R.W.; Miller, J.R., III; Debs, R.; et al. Bptf transduces mitf-driven prosurvival signals in melanoma cells. Proc. Natl. Acad. Sci. USA 2016, 113, 6254–6258. [Google Scholar] [CrossRef]
- Dar, A.A.; Nosrati, M.; Bezrookove, V.; de Semir, D.; Majid, S.; Thummala, S.; Sun, V.; Tong, S.; Leong, S.P.; Minor, D.; et al. The role of bptf in melanoma progression and in response to braf-targeted therapy. J. Natl. Cancer Inst. 2015, 107. [Google Scholar] [CrossRef]
- Xiao, S.; Liu, L.; Lu, X.; Long, J.; Zhou, X.; Fang, M. The prognostic significance of bromodomain phd-finger transcription factor in colorectal carcinoma and association with vimentin and e-cadherin. J. Cancer Res. Clin. Oncol. 2015, 141, 1465–1474. [Google Scholar] [CrossRef]
- Zhao, X.; Zheng, F.; Li, Y.; Hao, J.; Tang, Z.; Tian, C.; Yang, Q.; Zhu, T.; Diao, C.; Zhang, C.; et al. Bptf promotes hepatocellular carcinoma growth by modulating htert signaling and cancer stem cell traits. Redox Biol. 2019, 20, 427–441. [Google Scholar] [CrossRef] [PubMed]
- Balbas-Martinez, C.; Sagrera, A.; Carrillo-de-Santa-Pau, E.; Earl, J.; Marquez, M.; Vazquez, M.; Lapi, E.; Castro-Giner, F.; Beltran, S.; Bayes, M.; et al. Recurrent inactivation of stag2 in bladder cancer is not associated with aneuploidy. Nat. Genet. 2013, 45, 1464–1469. [Google Scholar] [CrossRef] [PubMed]
- Richart, L.; Carrillo-de Santa Pau, E.; Rio-Machin, A.; de Andres, M.P.; Cigudosa, J.C.; Lobo, V.J.S.; Real, F.X. Bptf is required for c-myc transcriptional activity and in vivo tumorigenesis. Nat. Commun. 2016, 7, 10153. [Google Scholar] [CrossRef]
- Eckey, M.; Kuphal, S.; Straub, T.; Rummele, P.; Kremmer, E.; Bosserhoff, A.K.; Becker, P.B. Nucleosome remodeler snf2l suppresses cell proliferation and migration and attenuates wnt signaling. Mol. Cell. Biol. 2012, 32, 2359–2371. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ye, Y.; Xiao, Y.; Wang, W.; Wang, Q.; Yearsley, K.; Wani, A.A.; Yan, Q.; Gao, J.X.; Shetuni, B.S.; Barsky, S.H. Inhibition of expression of the chromatin remodeling gene, snf2l, selectively leads to DNA damage, growth inhibition, and cancer cell death. Mol. Cancer Res. 2009, 7, 1984–1999. [Google Scholar] [CrossRef] [PubMed]
- Gigek, C.O.; Lisboa, L.C.; Leal, M.F.; Silva, P.N.; Lima, E.M.; Khayat, A.S.; Assumpcao, P.P.; Burbano, R.R.; Smith Mde, A. Smarca5 methylation and expression in gastric cancer. Cancer Investig. 2011, 29, 162–166. [Google Scholar] [CrossRef] [PubMed]
- Jin, Q.; Mao, X.; Li, B.; Guan, S.; Yao, F.; Jin, F. Overexpression of smarca5 correlates with cell proliferation and migration in breast cancer. Tumour Biol. 2015, 36, 1895–1902. [Google Scholar] [CrossRef]
- Wang, Y.; Qin, J.; Liu, Q.; Hong, X.; Li, T.; Zhu, Y.; He, L.; Zheng, B.; Li, M. Snf2h promotes hepatocellular carcinoma proliferation by activating the wnt/beta-catenin signaling pathway. Oncol. Lett. 2016, 12, 1329–1336. [Google Scholar] [CrossRef]
- Dluhosova, M.; Curik, N.; Vargova, J.; Jonasova, A.; Zikmund, T.; Stopka, T. Epigenetic control of spi1 gene by ctcf and iswi atpase smarca5. PLoS ONE 2014, 9, e87448. [Google Scholar] [CrossRef]
- Klement, K.; Luijsterburg, M.S.; Pinder, J.B.; Cena, C.S.; Del Nero, V.; Wintersinger, C.M.; Dellaire, G.; van Attikum, H.; Goodarzi, A.A. Opposing iswi- and chd-class chromatin remodeling activities orchestrate heterochromatic DNA repair. J. Cell Biol. 2014, 207, 717–733. [Google Scholar] [CrossRef]
- Toiber, D.; Erdel, F.; Bouazoune, K.; Silberman, D.M.; Zhong, L.; Mulligan, P.; Sebastian, C.; Cosentino, C.; Martinez-Pastor, B.; Giacosa, S.; et al. Sirt6 recruits snf2h to DNA break sites, preventing genomic instability through chromatin remodeling. Mol. Cell 2013, 51, 454–468. [Google Scholar] [CrossRef] [PubMed]
- Kokavec, J.; Zikmund, T.; Savvulidi, F.; Kulvait, V.; Edelmann, W.; Skoultchi, A.I.; Stopka, T. The ISWI atpase smarca5 (snf2h) is required for proliferation and differentiation of hematopoietic stem and progenitor cells. Stem Cells 2017, 35, 1614–1623. [Google Scholar] [CrossRef] [PubMed]
- Bossi, D.; Cicalese, A.; Dellino, G.I.; Luzi, L.; Riva, L.; D’Alesio, C.; Diaferia, G.R.; Carugo, A.; Cavallaro, E.; Piccioni, R.; et al. In vivo genetic screens of patient-derived tumors revealed unexpected frailty of the transformed phenotype. Cancer Discov. 2016, 6, 650–663. [Google Scholar] [CrossRef] [PubMed]
- Meng, J.; Zhang, X.T.; Liu, X.L.; Fan, L.; Li, C.; Sun, Y.; Liang, X.H.; Wang, J.B.; Mei, Q.B.; Zhang, F.; et al. Wstf promotes proliferation and invasion of lung cancer cells by inducing emt via pi3k/akt and il-6/stat3 signaling pathways. Cell. Signal. 2016, 28, 1673–1682. [Google Scholar] [CrossRef] [PubMed]
- Xiao, A.; Li, H.; Shechter, D.; Ahn, S.H.; Fabrizio, L.A.; Erdjument-Bromage, H.; Ishibe-Murakami, S.; Wang, B.; Tempst, P.; Hofmann, K.; et al. Wstf regulates the h2a.X DNA damage response via a novel tyrosine kinase activity. Nature 2009, 457, 57–62. [Google Scholar] [CrossRef]
- Ansari, D.; Andersson, R.; Bauden, M.P.; Andersson, B.; Connolly, J.B.; Welinder, C.; Sasor, A.; Marko-Varga, G. Protein deep sequencing applied to biobank samples from patients with pancreatic cancer. J. Cancer Res. Clin. Oncol. 2015, 141, 369–380. [Google Scholar] [CrossRef]
- Gu, L.; Frommel, S.C.; Oakes, C.C.; Simon, R.; Grupp, K.; Gerig, C.Y.; Bar, D.; Robinson, M.D.; Baer, C.; Weiss, M.; et al. Baz2a (tip5) is involved in epigenetic alterations in prostate cancer and its overexpression predicts disease recurrence. Nat. Genet. 2015, 47, 22–30. [Google Scholar] [CrossRef]
- Postepska-Igielska, A.; Krunic, D.; Schmitt, N.; Greulich-Bode, K.M.; Boukamp, P.; Grummt, I. The chromatin remodelling complex norc safeguards genome stability by heterochromatin formation at telomeres and centromeres. EMBO Rep. 2013, 14, 704–710. [Google Scholar] [CrossRef]
- Sheu, J.J.; Choi, J.H.; Guan, B.; Tsai, F.J.; Hua, C.H.; Lai, M.T.; Wang, T.L.; Shih Ie, M. Rsf-1, a chromatin remodelling protein, interacts with cyclin e1 and promotes tumour development. J. Pathol. 2013, 229, 559–568. [Google Scholar] [CrossRef]
- Sheu, J.J.; Guan, B.; Choi, J.H.; Lin, A.; Lee, C.H.; Hsiao, Y.T.; Wang, T.L.; Tsai, F.J.; Shih Ie, M. Rsf-1, a chromatin remodeling protein, induces DNA damage and promotes genomic instability. J. Biol. Chem. 2010, 285, 38260–38269. [Google Scholar] [CrossRef]
- Wang, X.; Sheu, J.J.; Lai, M.T.; Yin-Yi Chang, C.; Sheng, X.; Wei, L.; Gao, Y.; Wang, X.; Liu, N.; Xie, W.; et al. Rsf-1 overexpression determines cancer progression and drug resistance in cervical cancer. Biomedicine 2018, 8, 4. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.I.; Ahn, J.H.; Lee, K.T.; Shih Ie, M.; Choi, J.H. Rsf1 is a positive regulator of nf-kappab-induced gene expression required for ovarian cancer chemoresistance. Cancer Res. 2014, 74, 2258–2269. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.H.; Sheu, J.J.; Guan, B.; Jinawath, N.; Markowski, P.; Wang, T.L.; Shih Ie, M. Functional analysis of 11q13.5 amplicon identifies rsf-1 (hbxap) as a gene involved in paclitaxel resistance in ovarian cancer. Cancer Res. 2009, 69, 1407–1415. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Garcia, F.; Villagrasa, P.; Matsui, J.; Kotliar, D.; Castro, V.; Akavia, U.D.; Chen, B.J.; Saucedo-Cuevas, L.; Rodriguez Barrueco, R.; Llobet-Navas, D.; et al. Integration of genomic data enables selective discovery of breast cancer drivers. Cell 2014, 159, 1461–1475. [Google Scholar] [CrossRef]
- Min, S.; Kim, K.; Kim, S.G.; Cho, H.; Lee, Y. Chromatin-remodeling factor, rsf1, controls p53-mediated transcription in apoptosis upon DNA strand breaks. Cell Death Dis. 2018, 9, 1079. [Google Scholar] [CrossRef]
- Li, X.; Ding, D.; Yao, J.; Zhou, B.; Shen, T.; Qi, Y.; Ni, T.; Wei, G. Chromatin remodeling factor baz1a regulates cellular senescence in both cancer and normal cells. Life Sci. 2019, 229, 225–232. [Google Scholar] [CrossRef]
- Kostrhon, S.; Kontaxis, G.; Kaufmann, T.; Schirghuber, E.; Kubicek, S.; Konrat, R.; Slade, D. A histone-mimicking interdomain linker in a multidomain protein modulates multivalent histone binding. J. Biol. Chem. 2017, 292, 17643–17657. [Google Scholar] [CrossRef]
- Mahmood, S.F.; Gruel, N.; Chapeaublanc, E.; Lescure, A.; Jones, T.; Reyal, F.; Vincent-Salomon, A.; Raynal, V.; Pierron, G.; Perez, F.; et al. A sirna screen identifies rad21, eif3h, chrac1 and tanc2 as driver genes within the 8q23, 8q24.3 and 17q23 amplicons in breast cancer with effects on cell growth, survival and transformation. Carcinogenesis 2014, 35, 670–682. [Google Scholar] [CrossRef]
- Wang, Y.L.; Faiola, F.; Xu, M.; Pan, S.; Martinez, E. Human atac is a gcn5/pcaf-containing acetylase complex with a novel nc2-like histone fold module that interacts with the tata-binding protein. J. Biol. Chem. 2008, 283, 33808–33815. [Google Scholar] [CrossRef]
- Bolognese, F.; Forni, C.; Caretti, G.; Frontini, M.; Minuzzo, M.; Mantovani, R. The pole3 bidirectional unit is regulated by myc and e2fs. Gene 2006, 366, 109–116. [Google Scholar] [CrossRef]
- Thornley, J.A.; Trask, H.W.; Ringelberg, C.S.; Ridley, C.J.; Wang, S.; Sal-Lari, R.C.; Moore, J.H.; Korc, M.; Tomlinson, C.R. Smad4-dependent polysome rna recruitment in human pancreatic cancer cells. Mol. Carcinog. 2012, 51, 771–782. [Google Scholar] [CrossRef]
- Van Gool, I.C.; Eggink, F.A.; Freeman-Mills, L.; Stelloo, E.; Marchi, E.; de Bruyn, M.; Palles, C.; Nout, R.A.; de Kroon, C.D.; Osse, E.M.; et al. Pole proofreading mutations elicit an antitumor immune response in endometrial cancer. Clin. Cancer Res. 2015, 21, 3347–3355. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Zhou, J.; Jiang, J.; Yuan, J.; Zhang, Y.; Wei, X.; Loo, N.; Wang, Y.; Pan, Y.; Zhang, T.; et al. Genomic characterization of genes encoding histone acetylation modulator proteins identifies therapeutic targets for cancer treatment. Nat. Commun. 2019, 10, 733. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.K.; Park, E.J.; Lee, H.S.; Lee, Y.S.; Kwon, J. Genome-wide screen of human bromodomain-containing proteins identifies cecr2 as a novel DNA damage response protein. Mol. Cells 2012, 34, 85–91. [Google Scholar] [CrossRef] [PubMed]
- Dawe, C.E.; Kooistra, M.K.; Fairbridge, N.A.; Pisio, A.C.; McDermid, H.E. Role of chromatin remodeling gene cecr2 in neurulation and inner ear development. Dev. Dyn. 2011, 240, 372–383. [Google Scholar] [CrossRef] [PubMed]
- Marfella, C.G.; Imbalzano, A.N. The chd family of chromatin remodelers. Mutat. Res. 2007, 618, 30–40. [Google Scholar] [CrossRef]
- Gaspar-Maia, A.; Alajem, A.; Polesso, F.; Sridharan, R.; Mason, M.J.; Heidersbach, A.; Ramalho-Santos, J.; McManus, M.T.; Plath, K.; Meshorer, E.; et al. Chd1 regulates open chromatin and pluripotency of embryonic stem cells. Nature 2009, 460, 863–868. [Google Scholar] [CrossRef]
- Chaudhary, K.; Deb, S.; Moniaux, N.; Ponnusamy, M.P.; Batra, S.K. Human rna polymerase ii-associated factor complex: Dysregulation in cancer. Oncogene 2007, 26, 7499–7507. [Google Scholar] [CrossRef]
- Dey, P.; Ponnusamy, M.P.; Deb, S.; Batra, S.K. Human rna polymerase ii-association factor 1 (hpaf1/pd2) regulates histone methylation and chromatin remodeling in pancreatic cancer. PLoS ONE 2011, 6, e26926. [Google Scholar] [CrossRef]
- Yu, B.; Swatkoski, S.; Holly, A.; Lee, L.C.; Giroux, V.; Lee, C.S.; Hsu, D.; Smith, J.L.; Yuen, G.; Yue, J.; et al. Oncogenesis driven by the ras/raf pathway requires the sumo e2 ligase ubc9. Proc. Natl. Acad. Sci. USA 2015, 112, E1724–E1733. [Google Scholar] [CrossRef]
- Kari, V.; Mansour, W.Y.; Raul, S.K.; Baumgart, S.J.; Mund, A.; Grade, M.; Sirma, H.; Simon, R.; Will, H.; Dobbelstein, M.; et al. Loss of chd1 causes DNA repair defects and enhances prostate cancer therapeutic responsiveness. EMBO Rep. 2016, 17, 1609–1623. [Google Scholar] [CrossRef] [PubMed]
- Shenoy, T.R.; Boysen, G.; Wang, M.Y.; Xu, Q.Z.; Guo, W.; Koh, F.M.; Wang, C.; Zhang, L.Z.; Wang, Y.; Gil, V.; et al. Chd1 loss sensitizes prostate cancer to DNA damaging therapy by promoting error-prone double-strand break repair. Ann. Oncol. 2017, 28, 1495–1507. [Google Scholar] [CrossRef] [PubMed]
- Bagchi, A.; Papazoglu, C.; Wu, Y.; Capurso, D.; Brodt, M.; Francis, D.; Bredel, M.; Vogel, H.; Mills, A.A. Chd5 is a tumor suppressor at human 1p36. Cell 2007, 128, 459–475. [Google Scholar] [CrossRef] [PubMed]
- Fatemi, M.; Paul, T.A.; Brodeur, G.M.; Shokrani, B.; Brim, H.; Ashktorab, H. Epigenetic silencing of chd5, a novel tumor-suppressor gene, occurs in early colorectal cancer stages. Cancer 2014, 120, 172–180. [Google Scholar] [CrossRef]
- Kolla, V.; Naraparaju, K.; Zhuang, T.; Higashi, M.; Kolla, S.; Blobel, G.A.; Brodeur, G.M. The tumour suppressor chd5 forms a nurd-type chromatin remodelling complex. Biochem. J. 2015, 468, 345–352. [Google Scholar] [CrossRef][Green Version]
- Du, Z.; Li, L.; Huang, X.; Jin, J.; Huang, S.; Zhang, Q.; Tao, Q. The epigenetic modifier chd5 functions as a novel tumor suppressor for renal cell carcinoma and is predominantly inactivated by promoter cpg methylation. Oncotarget 2016, 7, 21618–21630. [Google Scholar] [CrossRef]
- Mulero-Navarro, S.; Esteller, M. Chromatin remodeling factor chd5 is silenced by promoter cpg island hypermethylation in human cancer. Epigenetics 2008, 3, 210–215. [Google Scholar] [CrossRef]
- Wang, F.; Zhu, Y.; Huang, Y.; McAvoy, S.; Johnson, W.B.; Cheung, T.H.; Chung, T.K.; Lo, K.W.; Yim, S.F.; Yu, M.M.; et al. Transcriptional repression of wee1 by kruppel-like factor 2 is involved in DNA damage-induced apoptosis. Oncogene 2005, 24, 3875–3885. [Google Scholar] [CrossRef]
- Quan, J.; Adelmant, G.; Marto, J.A.; Look, A.T.; Yusufzai, T. The chromatin remodeling factor chd5 is a transcriptional repressor of wee1. PLoS ONE 2014, 9, e108066. [Google Scholar] [CrossRef]
- Cuneo, K.C.; Morgan, M.A.; Sahai, V.; Schipper, M.J.; Parsels, L.A.; Parsels, J.D.; Devasia, T.; Al-Hawaray, M.; Cho, C.S.; Nathan, H.; et al. Dose escalation trial of the wee1 inhibitor adavosertib (azd1775) in combination with gemcitabine and radiation for patients with locally advanced pancreatic cancer. J. Clin. Oncol. 2019, 37, 2643–2650. [Google Scholar] [CrossRef]
- Bouazoune, K.; Kingston, R.E. Chromatin remodeling by the chd7 protein is impaired by mutations that cause human developmental disorders. Proc. Natl. Acad. Sci. USA 2012, 109, 19238–19243. [Google Scholar] [CrossRef] [PubMed]
- Engelen, E.; Akinci, U.; Bryne, J.C.; Hou, J.; Gontan, C.; Moen, M.; Szumska, D.; Kockx, C.; van Ijcken, W.; Dekkers, D.H.; et al. Sox2 cooperates with chd7 to regulate genes that are mutated in human syndromes. Nat. Genet. 2011, 43, 607–611. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.S.; Chung, N.G.; Kang, M.R.; Yoo, N.J.; Lee, S.H. Genetic and expressional alterations of chd genes in gastric and colorectal cancers. Histopathology 2011, 58, 660–668. [Google Scholar] [CrossRef] [PubMed]
- Tahara, T.; Yamamoto, E.; Madireddi, P.; Suzuki, H.; Maruyama, R.; Chung, W.; Garriga, J.; Jelinek, J.; Yamano, H.O.; Sugai, T.; et al. Colorectal carcinomas with cpg island methylator phenotype 1 frequently contain mutations in chromatin regulators. Gastroenterology 2014, 146, 530–538 e535. [Google Scholar] [CrossRef] [PubMed]
- Chu, X.; Guo, X.; Jiang, Y.; Yu, H.; Liu, L.; Shan, W.; Yang, Z.Q. Genotranscriptomic meta-analysis of the chd family chromatin remodelers in human cancers-initial evidence of an oncogenic role for chd7. Mol. Oncol. 2017, 11, 1348–1360. [Google Scholar] [CrossRef] [PubMed]
- Machado, R.A.C.; Schneider, H.; DeOcesano-Pereira, C.; Lichtenstein, F.; Andrade, F.; Fujita, A.; Trombetta-Lima, M.; Weller, M.; Bowman-Colin, C.; Sogayar, M.C. Chd7 promotes glioblastoma cell motility and invasiveness through transcriptional modulation of an invasion signature. Sci. Rep. 2019, 9, 3952. [Google Scholar] [CrossRef]
- Mishra, N.K.; Guda, C. Genome-wide DNA methylation analysis reveals molecular subtypes of pancreatic cancer. Oncotarget 2017, 8, 28990–29012. [Google Scholar] [CrossRef]
- Colbert, L.E.; Petrova, A.V.; Fisher, S.B.; Pantazides, B.G.; Madden, M.Z.; Hardy, C.W.; Warren, M.D.; Pan, Y.; Nagaraju, G.P.; Liu, E.A.; et al. Chd7 expression predicts survival outcomes in patients with resected pancreatic cancer. Cancer Res. 2014, 74, 2677–2687. [Google Scholar] [CrossRef]
- Rodriguez, D.; Bretones, G.; Quesada, V.; Villamor, N.; Arango, J.R.; Lopez-Guillermo, A.; Ramsay, A.J.; Baumann, T.; Quiros, P.M.; Navarro, A.; et al. Mutations in chd2 cause defective association with active chromatin in chronic lymphocytic leukemia. Blood 2015, 126, 195–202. [Google Scholar] [CrossRef]
- Semba, Y.; Harada, A.; Maehara, K.; Oki, S.; Meno, C.; Ueda, J.; Yamagata, K.; Suzuki, A.; Onimaru, M.; Nogami, J.; et al. Chd2 regulates chromatin for proper gene expression toward differentiation in mouse embryonic stem cells. Nucleic Acids Res. 2017, 45, 8758–8772. [Google Scholar] [CrossRef]
- Moore, S.; Berger, N.D.; Luijsterburg, M.S.; Piett, C.G.; Stanley, F.K.T.; Schrader, C.U.; Fang, S.; Chan, J.A.; Schriemer, D.C.; Nagel, Z.D.; et al. The chd6 chromatin remodeler is an oxidative DNA damage response factor. Nat. Commun. 2019, 10, 241. [Google Scholar] [CrossRef] [PubMed]
- Sawada, G.; Ueo, H.; Matsumura, T.; Uchi, R.; Ishibashi, M.; Mima, K.; Kurashige, J.; Takahashi, Y.; Akiyoshi, S.; Sudo, T.; et al. Chd8 is an independent prognostic indicator that regulates wnt/beta-catenin signaling and the cell cycle in gastric cancer. Oncol. Rep. 2013, 30, 1137–1142. [Google Scholar] [CrossRef] [PubMed]
- Thompson, B.A.; Tremblay, V.; Lin, G.; Bochar, D.A. Chd8 is an atp-dependent chromatin remodeling factor that regulates beta-catenin target genes. Mol. Cell. Biol. 2008, 28, 3894–3904. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Peng, H.; Huang, X.X.; Xia, Y.B.; Hu, K.F.; Zhang, Z.M. Decreased expression of chromodomain helicase DNA-binding protein 9 is a novel independent prognostic biomarker for colorectal cancer. Braz. J. Med. Biol. Res. 2018, 51, e7588. [Google Scholar] [CrossRef]
- Hibi, K.; Kitamura, Y.H.; Mizukami, H.; Goto, T.; Sakuraba, K.; Sakata, M.; Saito, M.; Ishibashi, K.; Kigawa, G.; Nemoto, H.; et al. Frequent cdh3 demethylation in advanced gastric carcinoma. Anticancer Res. 2009, 29, 3945–3947. [Google Scholar]
- Hibi, K.; Goto, T.; Mizukami, H.; Kitamura, Y.H.; Sakuraba, K.; Sakata, M.; Saito, M.; Ishibashi, K.; Kigawa, G.; Nemoto, H.; et al. Demethylation of the cdh3 gene is frequently detected in advanced colorectal cancer. Anticancer Res. 2009, 29, 2215–2217. [Google Scholar]
- Wang, H.C.; Chou, C.L.; Yang, C.C.; Huang, W.L.; Hsu, Y.C.; Luo, C.W.; Chen, T.J.; Li, C.F.; Pan, M.R. Over-expression of chd4 is an independent biomarker of poor prognosis in patients with rectal cancers receiving concurrent chemoradiotherapy. Int. J. Mol. Sci. 2019, 20, 4087. [Google Scholar] [CrossRef]
- Sheng, W.; Chen, Y.; Gong, Y.; Dong, T.; Zhang, B.; Gao, W. Mir-148a inhibits self-renewal of thyroid cancer stem cells via repressing ino80 expression. Oncol. Rep. 2016, 36, 3387–3396. [Google Scholar] [CrossRef][Green Version]
- Chen, L.; Conaway, R.C.; Conaway, J.W. Multiple modes of regulation of the human ino80 snf2 atpase by subunits of the ino80 chromatin-remodeling complex. Proc. Natl. Acad. Sci. USA 2013, 110, 20497–20502. [Google Scholar] [CrossRef]
- Yau, E.H.; Kummetha, I.R.; Lichinchi, G.; Tang, R.; Zhang, Y.; Rana, T.M. Genome-wide crispr screen for essential cell growth mediators in mutant kras colorectal cancers. Cancer Res. 2017, 77, 6330–6339. [Google Scholar] [CrossRef]
- Runge, J.S.; Raab, J.R.; Magnuson, T. Identification of two distinct classes of the human ino80 complex genome-wide. G3 2018, 8, 1095–1102. [Google Scholar] [CrossRef]
- Su, J.; Sui, Y.; Ding, J.; Li, F.; Shen, S.; Yang, Y.; Lu, Z.; Wang, F.; Cao, L.; Liu, X.; et al. Human ino80/yy1 chromatin remodeling complex transcriptionally regulates the brca2- and cdkn1a-interacting protein (bccip) in cells. Protein Cell 2016, 7, 749–760. [Google Scholar] [CrossRef] [PubMed]
- Ayala, R.; Willhoft, O.; Aramayo, R.J.; Wilkinson, M.; McCormack, E.A.; Ocloo, L.; Wigley, D.B.; Zhang, X. Structure and regulation of the human ino80-nucleosome complex. Nature 2018, 556, 391–395. [Google Scholar] [CrossRef] [PubMed]
- Dong, S.; Han, J.; Chen, H.; Liu, T.; Huen, M.S.Y.; Yang, Y.; Guo, C.; Huang, J. The human srcap chromatin remodeling complex promotes DNA-end resection. Curr. Biol. 2014, 24, 2097–2110. [Google Scholar] [CrossRef] [PubMed]
- Morrison, A.J.; Shen, X. Chromatin remodelling beyond transcription: The ino80 and swr1 complexes. Nat. Rev. Mol. Cell Biol. 2009, 10, 373–384. [Google Scholar] [CrossRef]
- Fang, L.T.; Lee, S.; Choi, H.; Kim, H.K.; Jew, G.; Kang, H.C.; Chen, L.; Jablons, D.; Kim, I.J. Comprehensive genomic analyses of a metastatic colon cancer to the lung by whole exome sequencing and gene expression analysis. Int. J. Oncol. 2014, 44, 211–221. [Google Scholar] [CrossRef]
- Thakur, A.; Bollig, A.; Wu, J.; Liao, D.J. Gene expression profiles in primary pancreatic tumors and metastatic lesions of ela-c-myc transgenic mice. Mol. Cancer 2008, 7, 11. [Google Scholar] [CrossRef]
- Shen, X.; Ranallo, R.; Choi, E.; Wu, C. Involvement of actin-related proteins in atp-dependent chromatin remodeling. Mol. Cell 2003, 12, 147–155. [Google Scholar] [CrossRef]
- Willhoft, O.; Bythell-Douglas, R.; McCormack, E.A.; Wigley, D.B. Synergy and antagonism in regulation of recombinant human ino80 chromatin remodeling complex. Nucleic Acids Res. 2016, 44, 8179–8188. [Google Scholar] [CrossRef]
- Gentili, C.; Castor, D.; Kaden, S.; Lauterbach, D.; Gysi, M.; Steigemann, P.; Gerlich, D.W.; Jiricny, J.; Ferrari, S. Chromosome missegregation associated with ruvbl1 deficiency. PLoS ONE 2015, 10, e0133576. [Google Scholar] [CrossRef]
- Tarangelo, A.; Lo, N.; Teng, R.; Kim, E.; Le, L.; Watson, D.; Furth, E.E.; Raman, P.; Ehmer, U.; Viatour, P. Recruitment of pontin/reptin by e2f1 amplifies e2f transcriptional response during cancer progression. Nat. Commun. 2015, 6, 10028. [Google Scholar] [CrossRef] [PubMed]
- Bauer, A.; Chauvet, S.; Huber, O.; Usseglio, F.; Rothbacher, U.; Aragnol, D.; Kemler, R.; Pradel, J. Pontin52 and reptin52 function as antagonistic regulators of beta-catenin signalling activity. EMBO J. 2000, 19, 6121–6130. [Google Scholar] [CrossRef] [PubMed]
- Wood, M.A.; McMahon, S.B.; Cole, M.D. An atpase/helicase complex is an essential cofactor for oncogenic transformation by c-myc. Mol. Cell 2000, 5, 321–330. [Google Scholar] [CrossRef]
- Haurie, V.; Menard, L.; Nicou, A.; Touriol, C.; Metzler, P.; Fernandez, J.; Taras, D.; Lestienne, P.; Balabaud, C.; Bioulac-Sage, P.; et al. Adenosine triphosphatase pontin is overexpressed in hepatocellular carcinoma and coregulated with reptin through a new posttranslational mechanism. Hepatology 2009, 50, 1871–1883. [Google Scholar] [CrossRef]
- Grigoletto, A.; Lestienne, P.; Rosenbaum, J. The multifaceted proteins reptin and pontin as major players in cancer. Biochim. Biophys. Acta 2011, 1815, 147–157. [Google Scholar] [CrossRef]
- Lauscher, J.C.; Elezkurtaj, S.; Dullat, S.; Lipka, S.; Grone, J.; Buhr, H.J.; Huber, O.; Kruschewski, M. Increased pontin expression is a potential predictor for outcome in sporadic colorectal carcinoma. Oncol. Rep. 2012, 28, 1619–1624. [Google Scholar] [CrossRef][Green Version]
- Sun, Q.; Li, F.; Yu, S.; Zhang, X.; Shi, F.; She, J. Pontin acts as a potential biomarker for poor clinical outcome and promotes tumor invasion in hilar cholangiocarcinoma. Biomed. Res. Int. 2018, 2018, 6135016. [Google Scholar] [CrossRef]
- Zhang, X.; Ren, J.; Yan, L.; Tang, Y.; Zhang, W.; Li, D.; Zang, Y.; Kong, F.; Xu, Z. Cytoplasmic expression of pontin in renal cell carcinoma correlates with tumor invasion, metastasis and patients’ survival. PLoS ONE 2015, 10, e0118659. [Google Scholar] [CrossRef]
- Taniuchi, K.; Furihata, M.; Iwasaki, S.; Tanaka, K.; Shimizu, T.; Saito, M.; Saibara, T. Ruvbl1 directly binds actin filaments and induces formation of cell protrusions to promote pancreatic cancer cell invasion. Int. J. Oncol. 2014, 44, 1945–1954. [Google Scholar] [CrossRef]
- Mao, Y.Q.; Houry, W.A. The role of pontin and reptin in cellular physiology and cancer etiology. Front. Mol. Biosci. 2017, 4, 58. [Google Scholar] [CrossRef]
- Grigoletto, A.; Neaud, V.; Allain-Courtois, N.; Lestienne, P.; Rosenbaum, J. The atpase activity of reptin is required for its effects on tumor cell growth and viability in hepatocellular carcinoma. Mol. Cancer Res. 2013, 11, 133–139. [Google Scholar] [CrossRef] [PubMed]
- Menard, L.; Taras, D.; Grigoletto, A.; Haurie, V.; Nicou, A.; Dugot-Senant, N.; Costet, P.; Rousseau, B.; Rosenbaum, J. In vivo silencing of reptin blocks the progression of human hepatocellular carcinoma in xenografts and is associated with replicative senescence. J. Hepatol. 2010, 52, 681–689. [Google Scholar] [CrossRef] [PubMed]
- Rousseau, B.; Menard, L.; Haurie, V.; Taras, D.; Blanc, J.F.; Moreau-Gaudry, F.; Metzler, P.; Hugues, M.; Boyault, S.; Lemiere, S.; et al. Overexpression and role of the atpase and putative DNA helicase ruvb-like 2 in human hepatocellular carcinoma. Hepatology 2007, 46, 1108–1118. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Zhang, C.; Yue, X.; Li, X.; Liu, J.; Yu, H.; Belyi, V.A.; Yang, Q.; Feng, Z.; Hu, W. Pontin, a new mutant p53-binding protein, promotes gain-of-function of mutant p53. Cell Death Differ. 2015, 22, 1824–1836. [Google Scholar] [CrossRef]
- Yuan, P.; He, X.H.; Rong, Y.F.; Cao, J.; Li, Y.; Hu, Y.P.; Liu, Y.; Li, D.; Lou, W.; Liu, M.F. Kras/nf-kappab/yy1/mir-489 signaling axis controls pancreatic cancer metastasis. Cancer Res. 2017, 77, 100–111. [Google Scholar] [CrossRef]
- Jiang, W.; Zhao, S.; Shen, J.; Guo, L.; Sun, Y.; Zhu, Y.; Ma, Z.; Zhang, X.; Hu, Y.; Xiao, W.; et al. The mir-135b-bmal1-yy1 loop disturbs pancreatic clockwork to promote tumourigenesis and chemoresistance. Cell Death Dis. 2018, 9, 149. [Google Scholar] [CrossRef]
- De Nigris, F.; Crudele, V.; Giovane, A.; Casamassimi, A.; Giordano, A.; Garban, H.J.; Cacciatore, F.; Pentimalli, F.; Marquez-Garban, D.C.; Petrillo, A.; et al. Cxcr4/yy1 inhibition impairs vegf network and angiogenesis during malignancy. Proc. Natl. Acad. Sci. USA 2010, 107, 14484–14489. [Google Scholar] [CrossRef]
- Palmer, M.B.; Majumder, P.; Cooper, J.C.; Yoon, H.; Wade, P.A.; Boss, J.M. Yin yang 1 regulates the expression of snail through a distal enhancer. Mol. Cancer Res. 2009, 7, 221–229. [Google Scholar] [CrossRef]
- Liu, D.; Zhang, J.; Wu, Y.; Shi, G.; Yuan, H.; Lu, Z.; Zhu, Q.; Wu, P.; Lu, C.; Guo, F.; et al. Yy1 suppresses proliferation and migration of pancreatic ductal adenocarcinoma by regulating the cdkn3/mdm2/p53/p21 signaling pathway. Int. J. Cancer 2018, 142, 1392–1404. [Google Scholar] [CrossRef]
- Zhang, J.J.; Zhu, Y.; Zhang, X.F.; Liu, D.F.; Wang, Y.; Yang, C.; Shi, G.D.; Peng, Y.P.; Zhang, K.; Tian, L.; et al. Yin yang-1 suppresses pancreatic ductal adenocarcinoma cell proliferation and tumor growth by regulating sox2ot-sox2 axis. Cancer Lett. 2017, 408, 144–154. [Google Scholar] [CrossRef]
- Chen, Q.; Zhang, J.J.; Ge, W.L.; Chen, L.; Yuan, H.; Meng, L.D.; Huang, X.M.; Shen, P.; Miao, Y.; Jiang, K.R. Yy1 inhibits the migration and invasion of pancreatic ductal adenocarcinoma by downregulating the fer/stat3/mmp2 signaling pathway. Cancer Lett. 2019, 463, 37–49. [Google Scholar] [CrossRef] [PubMed]
- Cho, A.A.; Bonavida, B. Targeting the overexpressed yy1 in cancer inhibits emt and metastasis. Crit. Rev. Oncog. 2017, 22, 49–61. [Google Scholar] [CrossRef]
- Khachigian, L.M. The yin and yang of yy1 in tumor growth and suppression. Int. J. Cancer 2018, 143, 460–465. [Google Scholar] [CrossRef] [PubMed]
- Wan, M.; Huang, W.; Kute, T.E.; Miller, L.D.; Zhang, Q.; Hatcher, H.; Wang, J.; Stovall, D.B.; Russell, G.B.; Cao, P.D.; et al. Yin yang 1 plays an essential role in breast cancer and negatively regulates p27. Am. J. Pathol. 2012, 180, 2120–2133. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Stovall, D.B.; Inoue, K.; Sui, G. The oncogenic role of yin yang 1. Crit. Rev. Oncog. 2011, 16, 163–197. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.X.; Zhou, K.C.; Cao, Y. Mcrs1 overexpression, which is specifically inhibited by mir-129*, promotes the epithelial-mesenchymal transition and metastasis in non-small cell lung cancer. Mol. Cancer 2014, 13, 245. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Zhou, K.; Huang, Y.; Cao, Y. The candidate oncogene (mcrs1) promotes the growth of human lung cancer cells via the mir-155-rb1 pathway. J. Exp. Clin. Cancer Res. 2015, 34, 121. [Google Scholar] [CrossRef]
- Shi, H.; Chen, S.; Jin, H.; Xu, C.; Dong, G.; Zhao, Q.; Wang, W.; Zhang, H.; Lin, W.; Zhang, J.; et al. Downregulation of msp58 inhibits growth of human colorectal cancer cells via regulation of the cyclin d1-cyclin-dependent kinase 4-p21 pathway. Cancer Sci. 2009, 100, 1585–1590. [Google Scholar] [CrossRef]
- Fawal, M.A.; Brandt, M.; Djouder, N. Mcrs1 binds and couples rheb to amino acid-dependent mtorc1 activation. Dev. Cell 2015, 33, 67–81. [Google Scholar] [CrossRef]
- Li, C.; Chen, M.; Zhao, P.; Ayana, D.A.; Wang, L.; Jiang, Y. Expression of mcrs1 and mcrs2 and their correlation with serum carcinoembryonic antigen in colorectal cancer. Exp. Ther. Med. 2016, 12, 589–596. [Google Scholar] [CrossRef][Green Version]
- Vander Linden, R.T.; Hemmis, C.W.; Schmitt, B.; Ndoja, A.; Whitby, F.G.; Robinson, H.; Cohen, R.E.; Yao, T.; Hill, C.P. Structural basis for the activation and inhibition of the uch37 deubiquitylase. Mol. Cell 2015, 57, 901–911. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Fu, D.; Tang, W.; Cai, Y.; Ma, D.; Wang, H.; Xue, R.; Liu, T.; Huang, X.; Dong, L.; et al. Ubiquitin c-terminal hydrolase 37, a novel predictor for hepatocellular carcinoma recurrence, promotes cell migration and invasion via interacting and deubiquitinating prp19. Biochim. Biophys. Acta 2013, 1833, 559–572. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Fu, D.; Xi, J.; Ji, Z.; Liu, T.; Ma, Y.; Zhao, Y.; Dong, L.; Wang, Q.; Shen, X. Expression and clinical significance of uch37 in human esophageal squamous cell carcinoma. Dig. Dis. Sci. 2012, 57, 2310–2317. [Google Scholar] [CrossRef] [PubMed]
- Nan, L.; Jacko, A.M.; Tan, J.; Wang, D.; Zhao, J.; Kass, D.J.; Ma, H.; Zhao, Y. Ubiquitin carboxyl-terminal hydrolase-l5 promotes tgfbeta-1 signaling by de-ubiquitinating and stabilizing smad2/smad3 in pulmonary fibrosis. Sci. Rep. 2016, 6, 33116. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Yao, X.; Pang, S.; Chen, P.; Jiang, W.; Shan, Z.; Zhang, Q. The deubiquitinase uchl5/uch37 positively regulates hedgehog signaling by deubiquitinating smoothened. J. Mol. Cell Biol. 2018, 10, 243–257. [Google Scholar] [CrossRef]
- Barber, K.E.; Harrison, C.J.; Broadfield, Z.J.; Stewart, A.R.; Wright, S.L.; Martineau, M.; Strefford, J.C.; Moorman, A.V. Molecular cytogenetic characterization of tcf3 (e2a)/19p13.3 rearrangements in b-cell precursor acute lymphoblastic leukemia. Genes Chromosomes Cancer 2007, 46, 478–486. [Google Scholar] [CrossRef]
- Guo, C.; Liu, S.; Wang, J.; Sun, M.Z.; Greenaway, F.T. Actb in cancer. Clin. Chim. Acta 2013, 417, 39–44. [Google Scholar] [CrossRef]
- Hu, X.; Du, S.; Yu, J.; Yang, X.; Yang, C.; Zhou, D.; Wang, Q.; Qin, S.; Yan, X.; He, L.; et al. Common housekeeping proteins are upregulated in colorectal adenocarcinoma and hepatocellular carcinoma, making the total protein a better “housekeeper”. Oncotarget 2016, 7, 66679–66688. [Google Scholar] [CrossRef]
- Xu, J.; Zhang, Z.; Chen, J.; Liu, F.; Bai, L. Overexpression of beta-actin is closely associated with metastasis of gastric cancer. Hepatogastroenterology 2013, 60, 620–623. [Google Scholar]
- Morris, H.T.; Machesky, L.M. Actin cytoskeletal control during epithelial to mesenchymal transition: Focus on the pancreas and intestinal tract. Br. J. Cancer 2015, 112, 613–620. [Google Scholar] [CrossRef]
- Olson, M.F.; Sahai, E. The actin cytoskeleton in cancer cell motility. Clin. Exp. Metastasis 2009, 26, 273–287. [Google Scholar] [CrossRef] [PubMed]
- Spencer, V.A.; Costes, S.; Inman, J.L.; Xu, R.; Chen, J.; Hendzel, M.J.; Bissell, M.J. Depletion of nuclear actin is a key mediator of quiescence in epithelial cells. J. Cell Sci. 2011, 124, 123–132. [Google Scholar] [CrossRef] [PubMed]
- Fiore, A.; Spencer, V.A.; Mori, H.; Carvalho, H.F.; Bissell, M.J.; Bruni-Cardoso, A. Laminin-111 and the level of nuclear actin regulate epithelial quiescence via exportin-6. Cell Rep. 2017, 19, 2102–2115. [Google Scholar] [CrossRef] [PubMed]
- Pfitzer, L.; Moser, C.; Gegenfurtner, F.; Arner, A.; Foerster, F.; Atzberger, C.; Zisis, T.; Kubisch-Dohmen, R.; Busse, J.; Smith, R.; et al. Targeting actin inhibits repair of doxorubicin-induced DNA damage: A novel therapeutic approach for combination therapy. Cell Death Dis. 2019, 10, 302. [Google Scholar] [CrossRef]
- Chang, L.; Azzolin, L.; Di Biagio, D.; Zanconato, F.; Battilana, G.; Lucon Xiccato, R.; Aragona, M.; Giulitti, S.; Panciera, T.; Gandin, A.; et al. The swi/snf complex is a mechanoregulated inhibitor of yap and taz. Nature 2018, 563, 265–269. [Google Scholar] [CrossRef]
- Nguyen, A.V.; Nyberg, K.D.; Scott, M.B.; Welsh, A.M.; Nguyen, A.H.; Wu, N.; Hohlbauch, S.V.; Geisse, N.A.; Gibb, E.A.; Robertson, A.G.; et al. Stiffness of pancreatic cancer cells is associated with increased invasive potential. Integr. Biol. 2016, 8, 1232–1245. [Google Scholar] [CrossRef]
- Barretina, J.; Caponigro, G.; Stransky, N.; Venkatesan, K.; Margolin, A.A.; Kim, S.; Wilson, C.J.; Lehar, J.; Kryukov, G.V.; Sonkin, D.; et al. The cancer cell line encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 2012, 483, 603–607. [Google Scholar] [CrossRef]
- Lemma, S.; Avnet, S.; Salerno, M.; Chano, T.; Baldini, N. Identification and validation of housekeeping genes for gene expression analysis of cancer stem cells. PLoS ONE 2016, 11, e0149481. [Google Scholar] [CrossRef]
- Deindl, E.; Boengler, K.; van Royen, N.; Schaper, W. Differential expression of gapdh and beta3-actin in growing collateral arteries. Mol. Cell. Biochem. 2002, 236, 139–146. [Google Scholar] [CrossRef]
- Selvey, S.; Thompson, E.W.; Matthaei, K.; Lea, R.A.; Irving, M.G.; Griffiths, L.R. Beta-actin--an unsuitable internal control for rt-pcr. Mol. Cell. Probes 2001, 15, 307–311. [Google Scholar] [CrossRef]
- Saladi, S.V.; Ross, K.; Karaayvaz, M.; Tata, P.R.; Mou, H.; Rajagopal, J.; Ramaswamy, S.; Ellisen, L.W. Actl6a is co-amplified with p63 in squamous cell carcinoma to drive yap activation, regenerative proliferation, and poor prognosis. Cancer Cell 2017, 31, 35–49. [Google Scholar] [CrossRef] [PubMed]
- Zhong, P.Q.; Zhong, L.; Yao, J.J.; Liu, D.D.; Yuan, Z.; Liu, J.M.; Chen, M.; Yao, S.F.; Zhao, Y.; Liu, L.; et al. Actl6a interacts with p53 in acute promyelocytic leukemia cell lines to affect differentiation via the sox2/notch1 signaling pathway. Cell. Signal. 2019, 53, 390–399. [Google Scholar] [CrossRef]
- Xiao, S.; Chang, R.M.; Yang, M.Y.; Lei, X.; Liu, X.; Gao, W.B.; Xiao, J.L.; Yang, L.Y. Actin-like 6a predicts poor prognosis of hepatocellular carcinoma and promotes metastasis and epithelial-mesenchymal transition. Hepatology 2016, 63, 1256–1271. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Z.; Yang, H.; Xiao, S. Actl6a expression promotes invasion, metastasis and epithelial mesenchymal transition of colon cancer. BMC Cancer 2018, 18, 1020. [Google Scholar] [CrossRef]
- Wee, Y.; Liu, Y.; Lu, J.; Li, X.; Zhao, M. Identification of novel prognosis-related genes associated with cancer using integrative network analysis. Sci. Rep. 2018, 8, 3233. [Google Scholar] [CrossRef]
- Lu, W.; Fang, L.; Ouyang, B.; Zhang, X.; Zhan, S.; Feng, X.; Bai, Y.; Han, X.; Kim, H.; He, Q.; et al. Actl6a protects embryonic stem cells from differentiating into primitive endoderm. Stem Cells 2015, 33, 1782–1793. [Google Scholar] [CrossRef]
- Gerhold, C.B.; Winkler, D.D.; Lakomek, K.; Seifert, F.U.; Fenn, S.; Kessler, B.; Witte, G.; Luger, K.; Hopfner, K.P. Structure of actin-related protein 8 and its contribution to nucleosome binding. Nucleic Acids Res. 2012, 40, 11036–11046. [Google Scholar] [CrossRef]
- Nishimoto, N.; Watanabe, M.; Watanabe, S.; Sugimoto, N.; Yugawa, T.; Ikura, T.; Koiwai, O.; Kiyono, T.; Fujita, M. Heterocomplex formation by arp4 and beta-actin is involved in the integrity of the brg1 chromatin remodeling complex. J. Cell Sci. 2012, 125, 3870–3882. [Google Scholar] [CrossRef]
- Xu, J.; Wang, Q.; Leung, E.L.H.; Li, Y.; Fan, X.; Wu, Q.; Yao, X.; Liu, L. Compound c620-0696, a new potent inhibitor targeting bptf, the chromatin-remodeling factor in non-small-cell lung cancer. Front. Med. 2019. [Google Scholar] [CrossRef]
- Bevill, S.M.; Olivares-Quintero, J.F.; Sciaky, N.; Golitz, B.T.; Singh, D.; Beltran, A.S.; Rashid, N.U.; Stuhlmiller, T.J.; Hale, A.; Moorman, N.J.; et al. Gsk2801, a baz2/brd9 bromodomain inhibitor, synergizes with bet inhibitors to induce apoptosis in triple-negative breast cancer. Mol. Cancer Res. 2019, 17, 1503–1518. [Google Scholar] [CrossRef]
- Vangamudi, B.; Paul, T.A.; Shah, P.K.; Kost-Alimova, M.; Nottebaum, L.; Shi, X.; Zhan, Y.; Leo, E.; Mahadeshwar, H.S.; Protopopov, A.; et al. The smarca2/4 atpase domain surpasses the bromodomain as a drug target in swi/snf-mutant cancers: Insights from cdna rescue and pfi-3 inhibitor studies. Cancer Res. 2015, 75, 3865–3878. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Sharma, S.; Cui, H.; LeBlanc, S.E.; Zhang, H.; Muthuswami, R.; Nickerson, J.A.; Imbalzano, A.N. Targeting the chromatin remodeling enzyme brg1 increases the efficacy of chemotherapy drugs in breast cancer cells. Oncotarget 2016, 7, 27158–27175. [Google Scholar] [CrossRef] [PubMed]
- Rakesh, R.; Hussain, S.; Goel, K.; Sharma, S.; Bisht, D.; Chanana, U.B.; Hockensmith, J.W.; Muthuswami, R. Altering mammalian transcription networking with adaadi: An inhibitor of atp-dependent chromatin remodeling. BioRxiv 2019. [Google Scholar] [CrossRef]
- Muthuswami, R.; Mesner, L.D.; Wang, D.; Hill, D.A.; Imbalzano, A.N.; Hockensmith, J.W. Phosphoaminoglycosides inhibit swi2/snf2 family DNA-dependent molecular motor domains. Biochemistry 2000, 39, 4358–4365. [Google Scholar] [CrossRef]
- Farnaby, W.; Koegl, M.; Roy, M.J.; Whitworth, C.; Diers, E.; Trainor, N.; Zollman, D.; Steurer, S.; Karolyi-Oezguer, J.; Riedmueller, C.; et al. Baf complex vulnerabilities in cancer demonstrated via structure-based protac design. Nat. Chem. Biol. 2019, 15, 672–680. [Google Scholar] [CrossRef]


| Gene | Deep Deletions | Amplifications | Fusions | Somatic Mutations | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| UTSW | TCGA | UTSW | TCGA | TCGA | UTSW | QCMG | TCGA | ICGC | ||
| SWI/SNF subfamily | ARID1A | 8.26 | 1.09 | 0.54 | 6.42 | 7.57 | 4.89 | 4.04 | ||
| ARID1B | 3.67 | 0.54 | 0.52 | 1.09 | ||||||
| PBRM1 | 4.59 | 0.54 | 1.83 | 0.92 | 1.83 | 2.72 | 1.01 | |||
| SMARCA2 | 7.34 | 0.54 | 0.92 | 0.54 | 0.54 | 0.78 | 0.54 | 1.01 | ||
| SMARCA4 | 0.92 | 1.09 | 5.55 | 1.84 | 2.87 | 1.09 | 1.01 | |||
| SMARCB1 | 2.75 | 0.92 | 0.52 | 0.54 | ||||||
| SMARCC1 | 4.59 | 0.54 | 3.67 | 0.54 | 0.54 | |||||
| SMARCC2 | 1.83 | 4.59 | 1.63 | 0.26 | 0.54 | |||||
| ACTB * | 15.60 | 0.54 | 2.75 | 0.54 | ||||||
| ACTL6A * | 2.75 | 2.17 | ||||||||
| ACTL6B | 0.92 | 23.85 | 3.26 | |||||||
| SMARCD1 | 1.83 | 0.54 | 0.52 | 0.54 | ||||||
| SMARCD2 | 3.67 | 1.63 | 0.54 | |||||||
| SMARCD3 | 11.01 | 0.54 | 0.54 | |||||||
| SMARCE1 | 0.92 | 4.59 | 1.63 | 0.26 | 1.09 | 1.01 | ||||
| PHF10 | 3.67 | 0.54 | 0.92 | 0.26 | 0.54 | |||||
| DPF1 | 16.51 | 4.89 | 0.54 | |||||||
| DPF2 | 9.17 | 0.54 | ||||||||
| DPF3 | 1.83 | 0.92 | 1.04 | 0.54 | ||||||
| ARID2 | 5.5 | 0.54 | 0.92 | 1.83 | 2.09 | 1.63 | 3.03 | |||
| BRD7 | 2.75 | 1.83 | 0.54 | 0.26 | ||||||
| BCL7A | 1.83 | 0.54 | ||||||||
| BCL7B | 11.93 | 1.09 | 0.26 | |||||||
| BCL7C | 0.92 | 6.42 | 0.26 | |||||||
| BCL11A | 1.83 | 1.83 | 0.92 | 0.78 | 0.54 | 1.01 | ||||
| BCL11B | 2.75 | 0.92 | 1.04 | 1.09 | ||||||
| BRD9 | 13.76 | 0.54 | 1.83 | 0.26 | ||||||
| BICRA | 0.92 | 12.84 | 1.09 | 0.92 | 0.52 | 0.54 | ||||
| BICRAL | 1.83 | 6.42 | 1.63 | 0.52 | 1.09 | |||||
| SS18 | 10.09 | 0.54 | 5.5 | 4.35 | 0.54 | 0.92 | 0.26 | 0.54 | ||
| SS18L1 | 0.92 | 11.01 | 1.09 | |||||||
| ISWI subfamily | SMARCA1 | 0.54 | 1.83 | 0.78 | 1.09 | |||||
| SMARCA5 | 5.5 | 0.92 | 0.26 | |||||||
| BAZ1B | 6.42 | 1.09 | 0.92 | 0.26 | 0.54 | |||||
| BAZ2A | 1.83 | 3.67 | 1.63 | 0.26 | 0.54 | |||||
| RSF1 | 0.92 | 0.92 | 0.26 | |||||||
| BAZ1A | 1.83 | 2.75 | 0.92 | 0.26 | ||||||
| CHRAC1 | 0.92 | 0.54 | 12.84 | 8.7 | 0.54 | 0.54 | ||||
| POLE3 | 0.92 | 0.54 | 1.83 | |||||||
| BPTF | 3.67 | 2.75 | 2.17 | 0.92 | 0.52 | 2.72 | ||||
| CECR2 | 0.92 | 0.78 | 1.09 | |||||||
| RBBP4 | 5.5 | 0.92 | ||||||||
| RBBP7 | 0.54 | 0.54 | 0.78 | 0.54 | 1.01 | |||||
| CHD subfamily | CHD1 | 3.67 | 1.83 | 0.92 | 0.78 | |||||
| CHD2 | 5.5 | 2.17 | 1.83 | 1.04 | 0.54 | |||||
| CHD3 | 5.5 | 0.54 | 0.92 | 0.26 | 1.09 | |||||
| CHD4 | 0.92 | 7.34 | 2.72 | 1.83 | 0.52 | 1.09 | ||||
| CHD5 | 7.34 | 1.09 | 5.5 | 1.63 | 0.78 | 1.09 | ||||
| CHD6 | 0.92 | 4.59 | 0.54 | 1.57 | 2.17 | |||||
| CHD7 | 0.92 | 4.59 | 3.26 | 0.92 | 0.78 | 0.54 | 2.02 | |||
| CHD8 | 0.92 | 0.54 | 0.78 | 1.09 | 1.01 | |||||
| CHD9 | 1.83 | 2.75 | 0.54 | 1.83 | 0.52 | 1.09 | ||||
| INO80 subfamily | INO80 | 4.59 | 0.54 | 1.31 | 0.54 | 1.01 | ||||
| ACTR5 | 0.92 | 3.67 | 0.52 | 0.54 | ||||||
| ACTR8 | 5.5 | 0.92 | 0.26 | 0.54 | ||||||
| INO80B | 0.92 | 4.59 | 0.26 | |||||||
| INO80C | 18.35 | 2.17 | 0.92 | 1.63 | 0.26 | 0.54 | ||||
| INO80D | 0.92 | 1.83 | 1.63 | 0.26 | ||||||
| INO80E | 7.34 | 0.26 | 0.54 | |||||||
| RUVBL1 | 2.75 | 1.09 | ||||||||
| RUVBL2 | 2.75 | 12.84 | 1.09 | 0.26 | 0.54 | |||||
| YY1 | 3.67 | 4.59 | 0.26 | 0.54 | ||||||
| MCRS1 | 2.75 | 0.54 | 2.75 | 0.26 | ||||||
| NFRKB | 0.92 | 2.75 | 0.54 | 0.54 | 0.92 | 0.52 | 0.54 | 1.01 | ||
| UCHL5 | 2.75 | 0.54 | 10.09 | 2.17 | 0.52 | |||||
| TFPT | 0.92 | 11.93 | 1.09 | |||||||
| Protein | Summary of Findings |
|---|---|
| ARID1A | Expression absent in 22% of surgically resected IPMN and in 36% of PDAC samples [82]. ARID1A expression was absent or low in 61% of the gastric and 10% of pancreaticobiliary IPMN subtypes [50]. Deficiency was significantly associated with poor outcome in PDAC [21]. Another study concluded that there was no association between ARID1A expression and clinicopathological features or overall survival [83]. |
| ARID1B | Reduced/nondetectable expression in pancreatic tumor compared to matched normal samples. Reduction in expression was more noticeable in advanced-stage tumors [84]. |
| PBRM1 | High PBRM1 expression was related to smaller pancreatic tumor size. PBRM1high patients had improved 5-year survival rate compared to PBRM1low patients [83]. |
| SMARCA2 | SMARCA2 expression was associated with worse clinicopathological features in pancreatic cancer cases. The survival rate of SMARCA2high patients was significantly worse compared to SMARCA2low patients [83]. SMARCA2 expression correlated significantly with tumor histological grade. SMARCA2high group (56.5%) had significantly worse survival rate compared to the SMARCA2low (43.5%) group [85]. |
| SMARCA4 | SMARCA4 expression was increased in pancreatic cancer tissues [83,86]. Association between SMARCA4 expression, histology, and stage was observed: SMARCA4high correlated with stage IV disease [83]. SMARCA4 has been shown to be expressed heterogeneously in pancreatic cancer tissues. Trend between SMARCA4 expression and tumor grade was observed, and SMARCA4low group had a tendency for higher survival rate [86]. SMARCA4 expression was lost in 8.3% and reduced in 53.3% of the IPMN cases, and decreased SMARCA4 expression correlated with increased dysplasia in IPMN lesions. High-grade IPMNs had more frequent loss (76%) compared to intermediate-grade (52%) and low-grade IPMNs (28%) [87]. SMARCA4 expression was higher in PDAC compared with its precursor IPMN lesions [88,89]. |
| SMARCC1 | Nuclear staining of SMARCC1 was detected in normal pancreatic ductal cells, whereas variable expression was observed in pancreatic cancer lesions (47% had positive staining and 53% had negative staining). SMARCC1 expression did not correlate with patient survival [90]. |
| BCL7B | BCL7B was overexpressed in pancreatic cancer. BCL7Bhigh was associated with shorter survival time. Normal pancreatic ducts did not stain for BCL7B [91]. |
| UCHL5 | Both nuclear and cytoplasmic localization was observed in human PDAC tissues and positive nuclear UCHL5 expression was associated with better prognosis in PDAC patients [92]. |
| CHD5 | CHD5 expression correlated with patient survival. Low CHD5 expression predicted worse survival in patients with resected PDAC receiving adjuvant chemotherapy [93]. |
| SWI/SNF Subfamily | |
|---|---|
| Subunit | Protein Name/Functional Studies |
| ARID1A (BAF250A) | AT-Rich Interaction Domain 1A. Most mutated subunit in pancreatic cancer. Tumor suppressor. See Section 4.1.1. |
| ARID1B (BAF250B) | AT-Rich Interaction Domain 1B. Tumor suppressor. See Section 4.1.2. |
| PBRM1 (BAF180) * | Polybromo 1. Tumor suppressor. High incidence of truncating mutations [108] and association between PBRM1 loss and tumor response to immunotherapy in clear cell renal carcinoma [109]. PBRM1-deficient renal carcinoma tumors have a distinct transcriptional signature linked to hypoxia and other altered signaling pathways [109,110]. PBRM1 has been shown to regulate stress response in normal epithelial cells and its deletion led to increased proliferation and EMT [111]. |
| SMARCA2 (BRM, BAF190B) | SWI/SNF Related, Matrix Associated, Actin Dependent Regulator of Chromatin, Subfamily A, Member 2. Tumor-suppressive role. See Section 4.1.3. |
| SMARCA4 (BRG1, BAF190A) | SWI/SNF Related, Matrix Associated, Actin Dependent Regulator of Chromatin, Subfamily A, Member 4. Tumor-suppressor and oncogenic roles depending on stage of tumor progression. See Section 4.1.4. |
| SMARCB1 (BAF47, INI1, hSNF5) * | SWI/SNF Related, Matrix Associated, Actin Dependent Regulator of Chromatin, Subfamily B, Member 1. Tumor suppressor linked to pathways associated with tumor proliferation and progression [112]. |
| SMARCC1 (BAF155) | SWI/SNF Related, Matrix Associated, Actin-Dependent Regulator of Chromatin Subfamily C Member 1. See Section 4.1.5. |
| SMARCC2 (BAF170) * | SWI/SNF Related, Matrix Associated, Actin-Dependent Regulator of Chromatin Subfamily C Member 2. Frameshift mutations in SMARCC2 in gastric and colorectal cancers with microsatellite instability [113]. |
| ACTB | Actin Beta. See Section 4.5.1. |
| ACTL6A (BAF53A) | Actin Like 6A. See Section 4.5.2. |
| ACTL6B (BAF53B) * | Actin Like 6B. Aberrant promoter methylation observed in esophageal cancer, liver cancer, and prostate cancer [114,115,116]. See Section 4.1.6. |
| SMARCD1 (BAF60A) * | SWI/SNF Related, Matrix Associated, Actin-Dependent Regulator of Chromatin, Subfamily D, Member 1. Interacts with p53 and mostly acts as a tumor suppressor [117,118]. Decreased expression in ovarian cancer [119] and in lung cancer [117]. SMARCD1 sensitized lung cancer cells to cisplatin-induced apoptosis [117], and its reduced expression triggered cellular senescence in hepatocytes [120]. Opposite results in gastric cancer: overexpressed in gastric cancer tissues and correlated with worse survival outcomes [121]. |
| SMARCD2 (BAF60B) * | SWI/SNF Related, Matrix Associated, Actin Dependent Regulator of Chromatin, Subfamily D, Member 2. Highly expressed in pancreas [122], potential tumor suppressor in leukemia [123]. Involved in chromatin opening of hepatic genes and lineage conversion [124]. |
| SMARCD3 (BAF60C) * | SWI/SNF Related, Matrix Associated, Actin Dependent Regulator of Chromatin, Subfamily D, Member 3. Induced Wnt5a signaling and controlled EMT in breast cancer [125]. Amplified in PDAC (Table 1). |
| SMARCE1 (BAF57) * | SWI/SNF Related, Matrix Associated, Actin Dependent Regulator of Chromatin, Subfamily E, Member 1. Promoted invasive and metastatic progression of breast cancer through upregulation of proteases that degrade ECM by forming a SWI/SNF-independent complex [126]. High expression in metastatic prostate cancer [127]. SMARCE1 loss induced EGFR expression, activated AKT and ERK signaling in lung cancer and conferred resistance to MET and ALK inhibitors [128]. Knockdown led to decreased cell growth and increased sensitivity to anticancer agents in ovarian cancer and breast cancer cell lines [129]. |
| PHF10 (BAF45A) * | PHD Finger Protein 10. Might be neuron specific [56], required for cell proliferation in normal fibroblasts [130]. Tumor suppressor role in uveal melanoma [131]. |
| DPF1/3/2 BAF45B/C/D) * | Double PHD Fingers 1/3/2. Rarely mutated in cancers [132]. |
| ARID2 (BAF200) * | AT-Rich Interaction Domain 2. Tumor suppressor in hepatocellular carcinoma [133,134]. |
| BRD7 * | Bromodomain-Containing Protein 7. Tumor suppressor involved in tumor development and progression in multiple cancers [135]. |
| BRD9, BICRA (GLTSCR1), BICRAL (GLTSCR1L) * | Bromodomain Containing 9/ BRD4 Interacting Chromatin Remodeling Complex Associated Protein/ BRD4 Interacting Chromatin Remodeling Complex Associated Protein Like. Components of a newly identified noncanonical SWI/SNF complex involved in maintaining pluripotency in mouse embryonic stem cells [136]. Amplified in several cancers and may act as oncogenic drivers [137,138]. |
| BCL7 (A/B/C) * | BAF Chromatin Remodeling Complex Subunit BCL7A/B/C. Accumulated in the cell protrusions of migrating pancreatic cells, involved in motility and invasiveness through CREB signaling pathway [91]. Tumor suppressor negatively regulating the Wnt-signaling pathway in gastric cancer cells [139]. |
| BCL11 (A/B) * | BAF Chromatin Remodeling Complex Subunit BCL11A/B. BCL11A: Highly expressed in breast cancer and lung cancer, involved in cancer stemness and tumorigenesis [140,141,142]. BCL11B: overexpression led to chemoresistance in T-cell lines [143], acted as a tumor suppressor in T-cell acute lymphoblastic leukemia [144]. Downregulation in intestinal crypt cells increased expression of β-catenin genes, promoting tumor development [145]. |
| SS18/SS18L1 (CREST) * | SS18 Subunit of BAF Chromatin Remodeling Complex/ SS18L1 Subunit of BAF Chromatin Remodeling Complex. Involved in neural development, and links Ca2+ signaling and chromatin reorganization [146]. |
| ISWI Subfamily | |
|---|---|
| Subunit | Protein Name/Functional Studies |
| SMARCA1 (SNF2L) * | SWI/SNF Related, Matrix Associated, Actin-Dependent Regulator of Chromatin, Subfamily A, Member 1. Expression was decreased in malignant melanoma; depletion in HeLa cells led to activated Wnt signaling, increased proliferation and migration [154]. Expression was not detected in normal pancreas [154]. SMARCA1 depletion in cancer cells led to increased apoptosis, DNA damage response and upregulation of genes related to cell-cycle checkpoint arrest [155]. |
| SMARCA5 SNF2H) * | SWI/SNF Related, Matrix Associated, Actin-Dependent Regulator of Chromatin, Subfamily A, Member 5. Expressed in human pancreas [154]. Increased in gastric cancer [156], breast cancer [157], and liver cancer [158]. Activated Wnt/β-catenin signaling [158] and promoted cancer cell proliferation, colony formation and invasion [157,158]. Depletion in HeLa cells led to apoptotic phenotype [154]. Interacts with CCCTC-binding factor (CTCF) and is associated with chromatin to regulate transcription [159]. Involved in DNA repair [160,161]. Required for proliferation and differentiation of hematopoietic stem cells [162]. |
| BAZ1B (WSTF) * | Bromodomain Adjacent to Zinc Finger Domain 1B. Knockdown decreased melanoma tumor growth [163]. In lung cancer models, overexpression promoted proliferation and invasion through activating the PI3K/Akt and IL-6/STAT3 signaling pathways [164]. Involved in DNA damage response [165]. Promoted cell growth and reduced DNA-damage induced cell death in HeLa cells [59]. |
| BAZ2A (TIP5) * | Bromodomain Adjacent to Zinc Finger Domain 2A. Upregulated in the serum of pancreatic cancer patients. Interacts with p53 and is involved in histone acetylation [166]. Overexpressed in prostate cancer and contributed to cell proliferation and viability. Associated with the CIMP molecular subtype and interacted with EZH2 to coordinate epigenetic silencing in prostate cancer cells [167]. It is also a part of the nucleolar remodeling complex (NoRC): involved in heterochromatin formation at telomeres and centromeres, thus maintaining genome stability [166,168]. |
| RSF1 * | Remodeling and Spacing Factor 1. Overexpressed in ovarian cancer and other cancers [169,170,171]. Overexpression has been associated with poor prognosis in ovarian cancer patients. It is a co-activator of NF-kB signaling [172] and is involved in the development of chemoresistance in ovarian cancer cells [172,173]. Identified as a potential oncogene in breast cancer, overexpression led to increase the colony formation ability in vitro and enhanced tumorigenesis and invasion in vivo [174]. Interacts with cyclin E1 and promotes tumorigenesis [169]. Increased RSF1 expression induced chromosomal instability [170]. Involved in DDR and DNA repair [175]. |
| BAZ1A (ACF1) * | Bromodomain Adjacent to Zinc Finger Domain 1A. Promoted cell growth after DNA damage and reduced DNA-damage induced cell death in HeLa cells [59]. Knockdown induced senescence associated phenotype through upregulation of SMAD3 [176]. |
| BAZ2B * | Bromodomain Adjacent to Zinc Finger Domain 2B. Newly added to the ISWI complexes [59]. Paralogue of BAZ2A. Histone binding protein [177]. |
| CHRAC1 (CHRAC15) * | Chromatin Accessibility Complex Subunit 1. Identified as a driver gene in breast cancer regulating proliferation [178]. Amplified in PDAC. |
| POLE3 (CHRAC17) * | DNA Polymerase Epsilon 3. Involved in chromatin remodeling and DNA replication [179], regulated by MYC [180,181]. Polysomes of SMAD4-/- BxPC3 cells had increased level of POLE3, thus it might contribute to the genomic instability in PDAC [181]. POLE3 proofreading mutations in endometrial cancer have been associated with higher T cell content and antitumor response [182]. |
| BPTF * | Bromodomain PHD Finger Transcription Factor. Protumorigenic role. See Section 4.2.1. |
| CECR2 * | CECR2 Histone Acetyl-Lysine Reader. Histone acetylation modulator protein [183]. Identified as a DNA damage response protein [184], involved in neurulation [185]. |
| CHD Subfamily | |
|---|---|
| Subunit | Protein Name/Functional Studies |
| CHD1 * | Chromodomain Helicase DNA Binding Protein 1. See Section 4.3.1. |
| CHD2 * | Chromodomain Helicase DNA Binding Protein 2. Tumor suppressor role in chronic lymphocytic leukemia [209]. Hypomethylated in PDAC [207]. Required to maintain the differentiation potential of mouse ESCs [210]. |
| CHD6 * | Chromodomain Helicase DNA Binding Protein 6. A cancer driver and key regulator of the oxidative DNA damage response [211]. |
| CHD7 * | Chromodomain Helicase DNA Binding Protein 7. See Section 4.3.3. |
| CHD8 * | Chromodomain Helicase DNA Binding Protein 8. Differentially methylated in PDAC [207]. Decreased expression in gastric cancer samples [212]. Negative regulator of the Wnt/β-catenin pathway [212,213], CHD8 knockdown in gastric cancer cells promoted proliferation [212]. |
| CHD9 * | Chromodomain Helicase DNA Binding Protein 9. Decreased expression in CRC patient samples that correlated with worse prognosis [214]. |
| CHD3 * | Chromodomain Helicase DNA Binding Protein 3. Component of the NuRD complex. Aberrant methylation was detected in advanced CRC and gastric cancer [215,216]. Overexpressed in cancers, including PDAC [216]. |
| CHD4 * | Chromodomain Helicase DNA Binding Protein 4. Component of the NuRD complex. High expression was associated with tumor status, metastasis and poor prognosis in rectal cancer [217]. In CRC, CHD4 interacted with oxidative DNA damage sites and double-strand breaks recruiting repressive chromatin proteins that maintained epigenetic silencing of tumor suppressor genes [64]; high levels of CHD4 were associated with poor prognosis [64]. CHD4 was identified as a potential therapeutic target in CRC [63,64] as knockdown of CHD4 sensitized cells to DAC-induced cell death and reactivated tumor suppressor genes [63]. |
| CHD5 | Chromodomain Helicase DNA Binding Protein 5. Component of the NuRD complex. Tumor suppressor. See Section 4.3.2. |
| INO80 Subfamily (INO 80 Complex) | |
|---|---|
| Subunit | Protein Name/Functional Studies |
| INO80 | INO80 Complex ATPase Subunit. See Section 4.4.1. |
| ACTL6A | Actin Like 6A. See Section 4.5.2. |
| ACTR5 (INO80M) * | Actin Related Protein 5. Increased in CRC [226], decreased in pancreatic tumors [227]. ACTR5 facilitates binding of INO80 complex to DNA, INO80 complexes lacking ACTR5 have reduced ATPase and chromatin remodeling activities in vitro [228]. Involved in nucleosome recognition [219]. |
| ACTR8 (INO80N) * | Actin Related Protein 8. ACTR8 facilitates binding of INO80 complex to DNA, INO80 complexes lacking ACTR8 have reduced ATPase and chromatin remodeling activities in vitro [228]. |
| INO80B * | INO80 Complex Subunit B. Regulates INO80 ATPase activity in vitro [219,229]. |
| INO80C * | INO80 Complex Subunit C. Tumor suppressor role. See Section 4.4.2. |
| RUVBL1 (RVB1, Tip49a, pontin)/RUVBL2 (RVB2, Tip49b, reptin) * | RuvB Like AAA ATPase 1/2. RUVBL1: Required for efficient mitosis and proliferation of cells [230]. Expression is increased in HCC, CRC and other cancers, involved in cell invasion and EMT. Interacts with oncogene c-MYC and β-catenin. Roles in cell growth and viability [231,232,233,234,235,236,237,238]. In a mouse model of liver cancer, accumulation of E2f1 recruits the RUVBL1/RUVBL2 complex that opens the chromatin conformation at E2f target genes and amplifies the E2f transcriptional response during cancer progression. Can function as a separate complex, not involved in INO80 subfamilies [231]. Cytoplasmic RUVBL1 interacts with actin filaments at cell protrusions and thus promotes invasiveness and migration of PDAC cells [239], which is a role independent of its chromatin remodeling [240]. No other data in PDAC. RUVBL2: Expression is increased in HCC, CRC. Interacts with oncogene c-MYC and β-catenin. Roles in cell growth and viability [231,232,233,235,240,241,242,243]. Interacts with mutant p53 [244]. |
| YY1 | YY1 Transcription Factor. A zinc finger transcription factor, that can either repress or activate gene transcription by recruiting different cofactors. YY1 expression is increased in PDAC [245,246], higher YY1 levels are associated with oncogenic KRASG12D status in pancreatic cancer cell lines and patient samples [245]. YY1 regulates the expression of Snail1 and VEGF, promoting EMT and angiogenesis [247,248]. Conflicting results reporting its role as a tumor suppressor in inhibiting the migration, invasiveness and proliferation in PDAC cells [249,250,251]. Other studies also report a dual tumor suppressor and oncogenic role [247,252,253,254,255]. |
| MCRS1 (MSP58) * | Microspherule Protein 1. Promoted proliferation, invasion and metastasis of lung cancer cells [256,257] and proliferation and tumor growth of colon carcinoma cells [258]. Increased in CRC [258,259,260]. Involved in mTORC1 activation, thus having an oncogenic role [259]. |
| NFRKB | Nuclear Factor Related to KappaB Binding Protein. NFRKB binds to UCH37, disrupting the active site for ubiquitin binding and inhibiting its function [261]. |
| UCHL5 (UCH37) * | Ubiquitin C-Terminal Hydrolase L5. UCHL5 deubiquitylase-dual roles component of INO80 and 26S proteasome [261]. Implicated in cancer [262,263]. Promotes Hedgehog signaling and TGFb-1 signaling [264,265]. |
| TFPT * | TCF3 Fusion Partner. Translocations are involved in B-cell precursor acute lymphoblastic leukemia [266] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hasan, N.; Ahuja, N. The Emerging Roles of ATP-Dependent Chromatin Remodeling Complexes in Pancreatic Cancer. Cancers 2019, 11, 1859. https://doi.org/10.3390/cancers11121859
Hasan N, Ahuja N. The Emerging Roles of ATP-Dependent Chromatin Remodeling Complexes in Pancreatic Cancer. Cancers. 2019; 11(12):1859. https://doi.org/10.3390/cancers11121859
Chicago/Turabian StyleHasan, Nesrin, and Nita Ahuja. 2019. "The Emerging Roles of ATP-Dependent Chromatin Remodeling Complexes in Pancreatic Cancer" Cancers 11, no. 12: 1859. https://doi.org/10.3390/cancers11121859
APA StyleHasan, N., & Ahuja, N. (2019). The Emerging Roles of ATP-Dependent Chromatin Remodeling Complexes in Pancreatic Cancer. Cancers, 11(12), 1859. https://doi.org/10.3390/cancers11121859





