The mRNA-binding Protein TTP/ZFP36 in Hepatocarcinogenesis and Hepatocellular Carcinoma
Abstract
:1. Introduction
2. Results
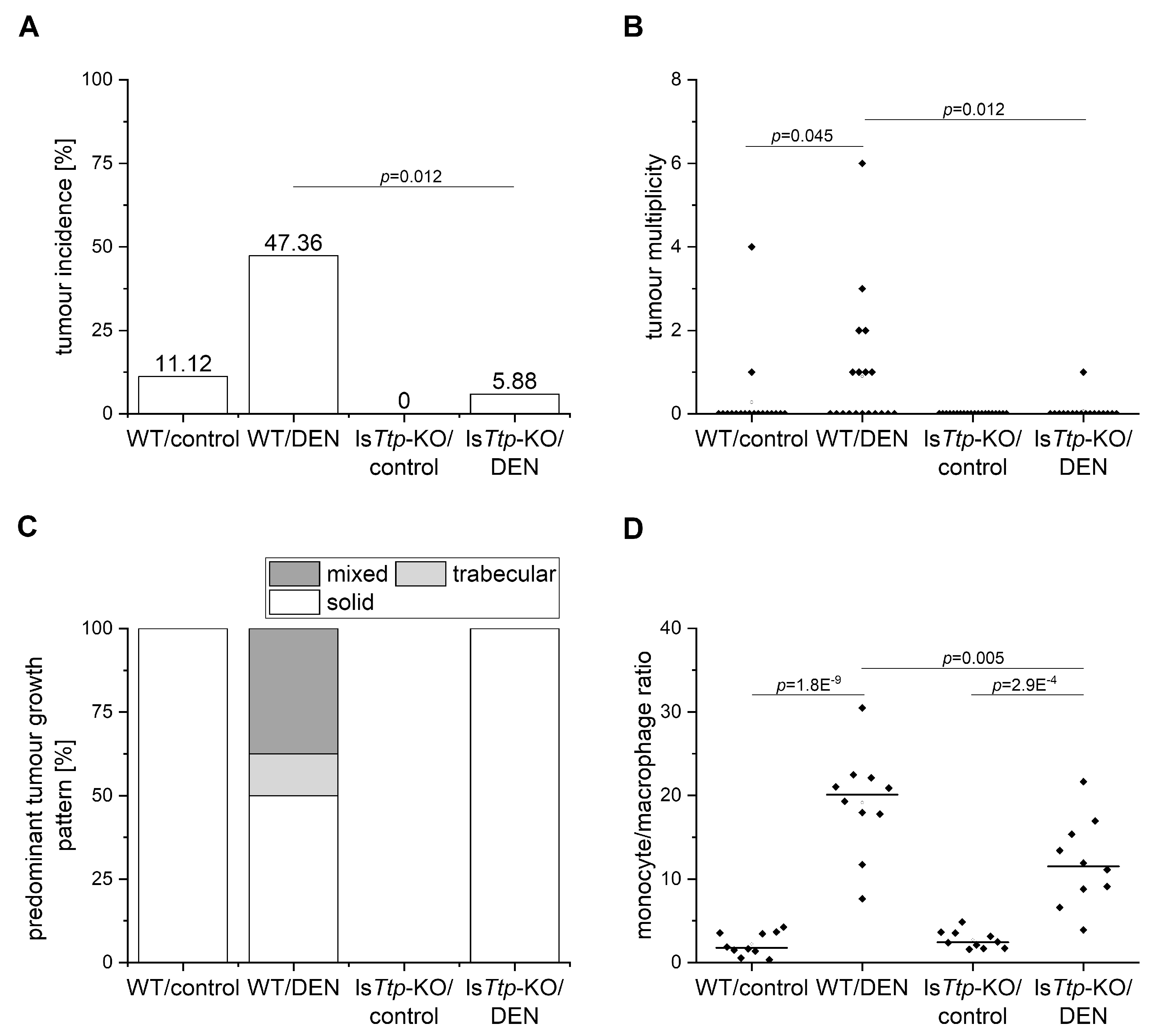
2.1. TTP and Tumor Initiation
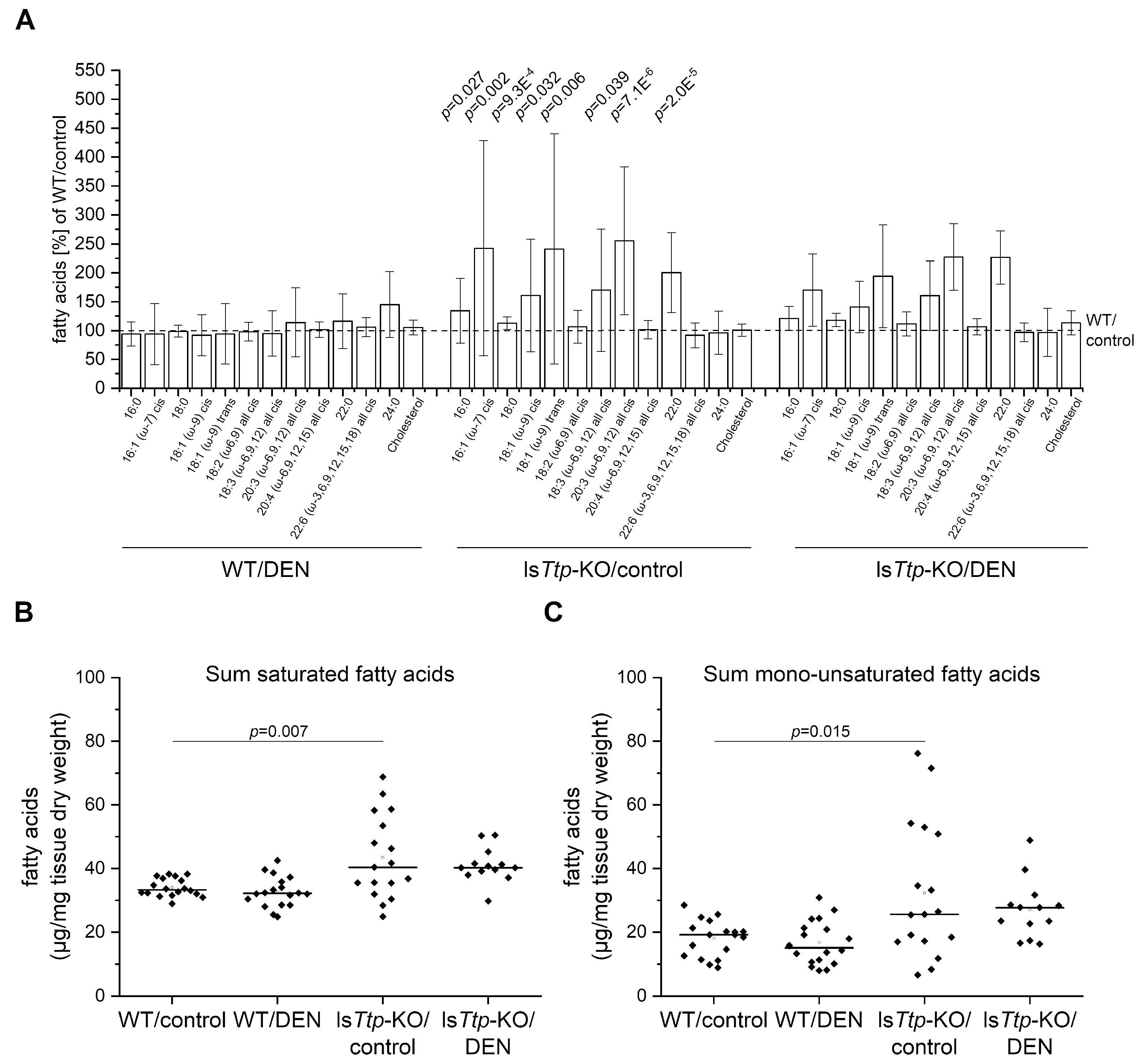
2.2. DEN-Induced Leukocyte Recruitment and Hepatic Lipids
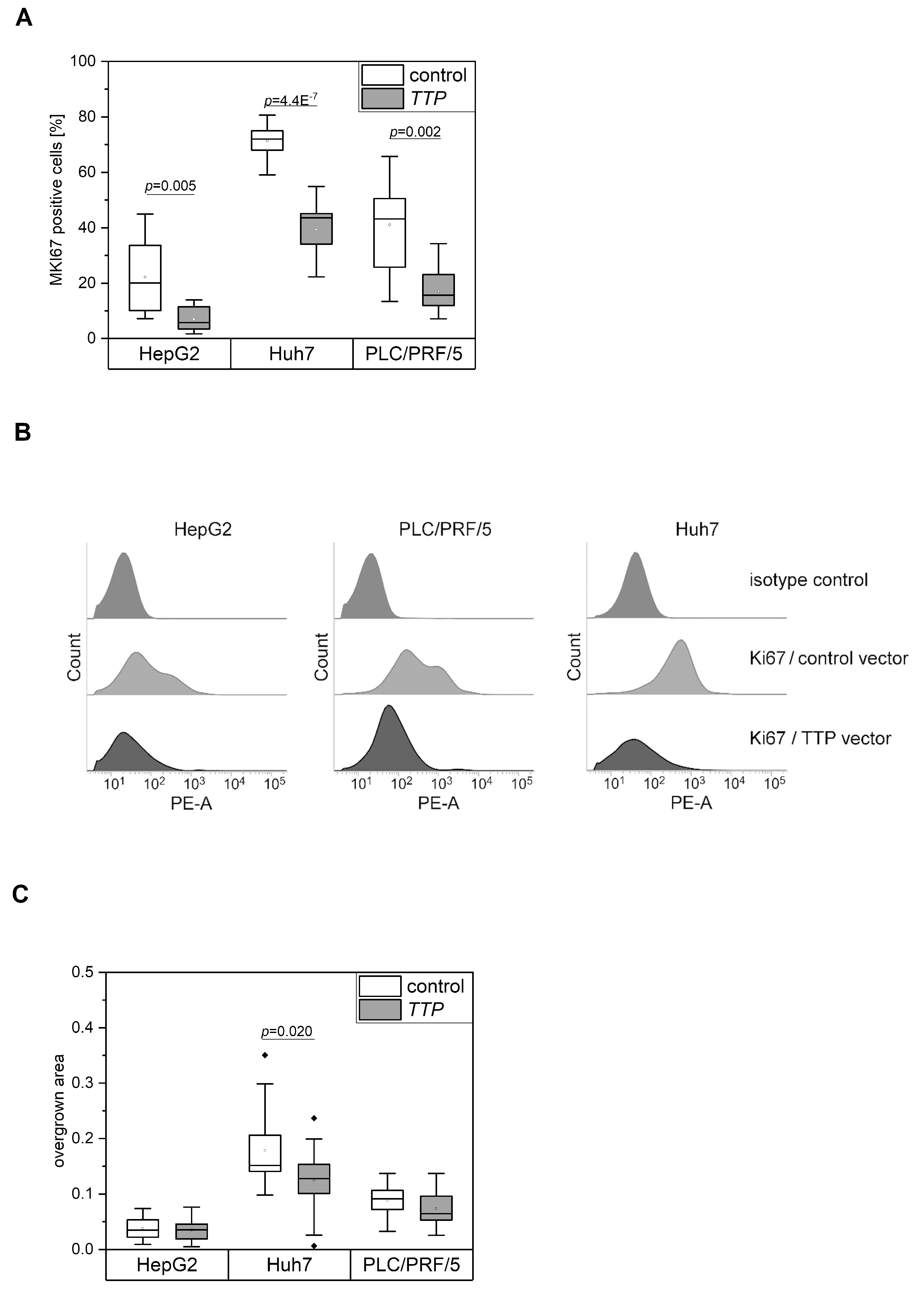
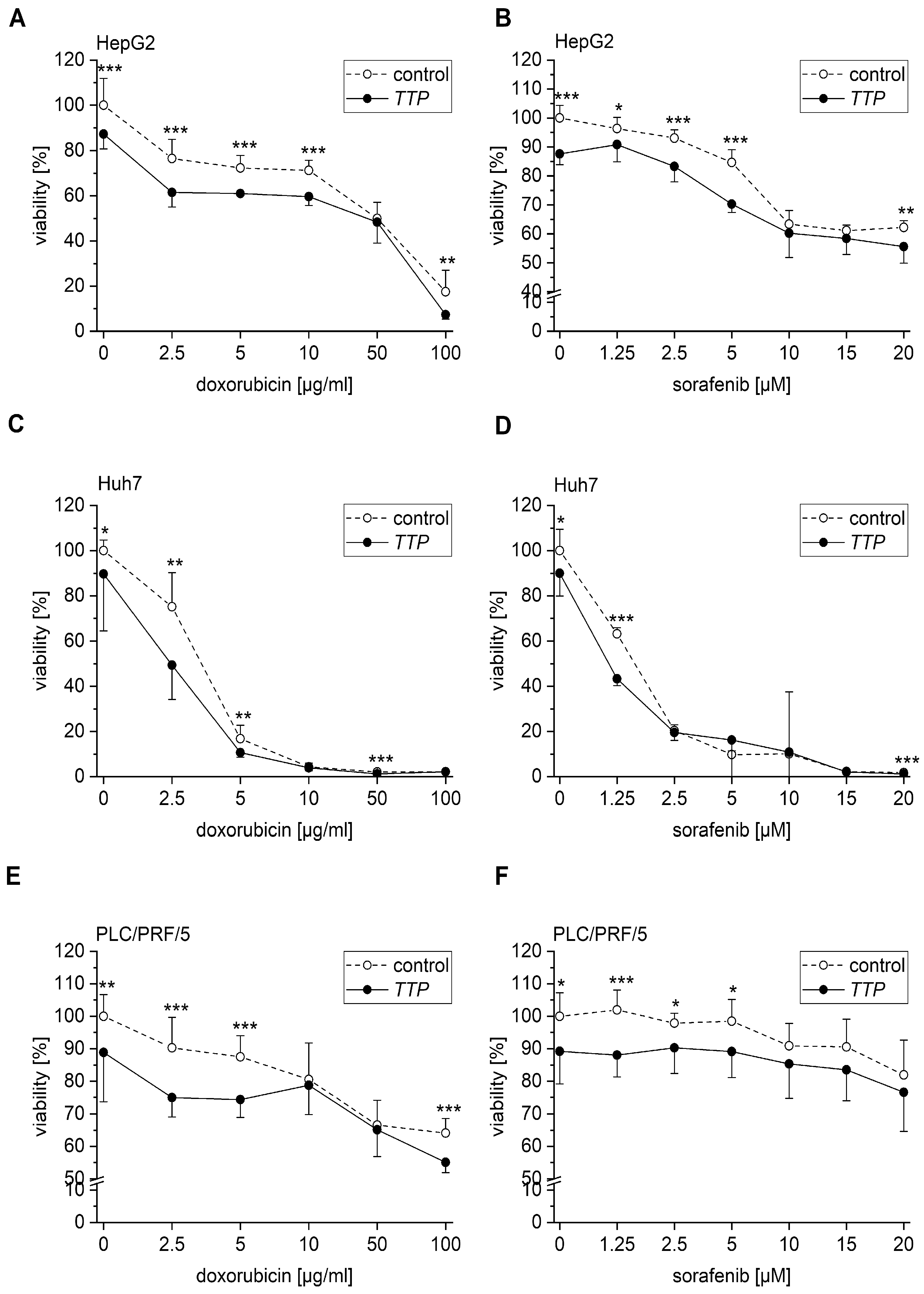
2.3. Effects of TTP on Hallmarks of Cancer
2.4. Expression Changes of Potential TTP Targets
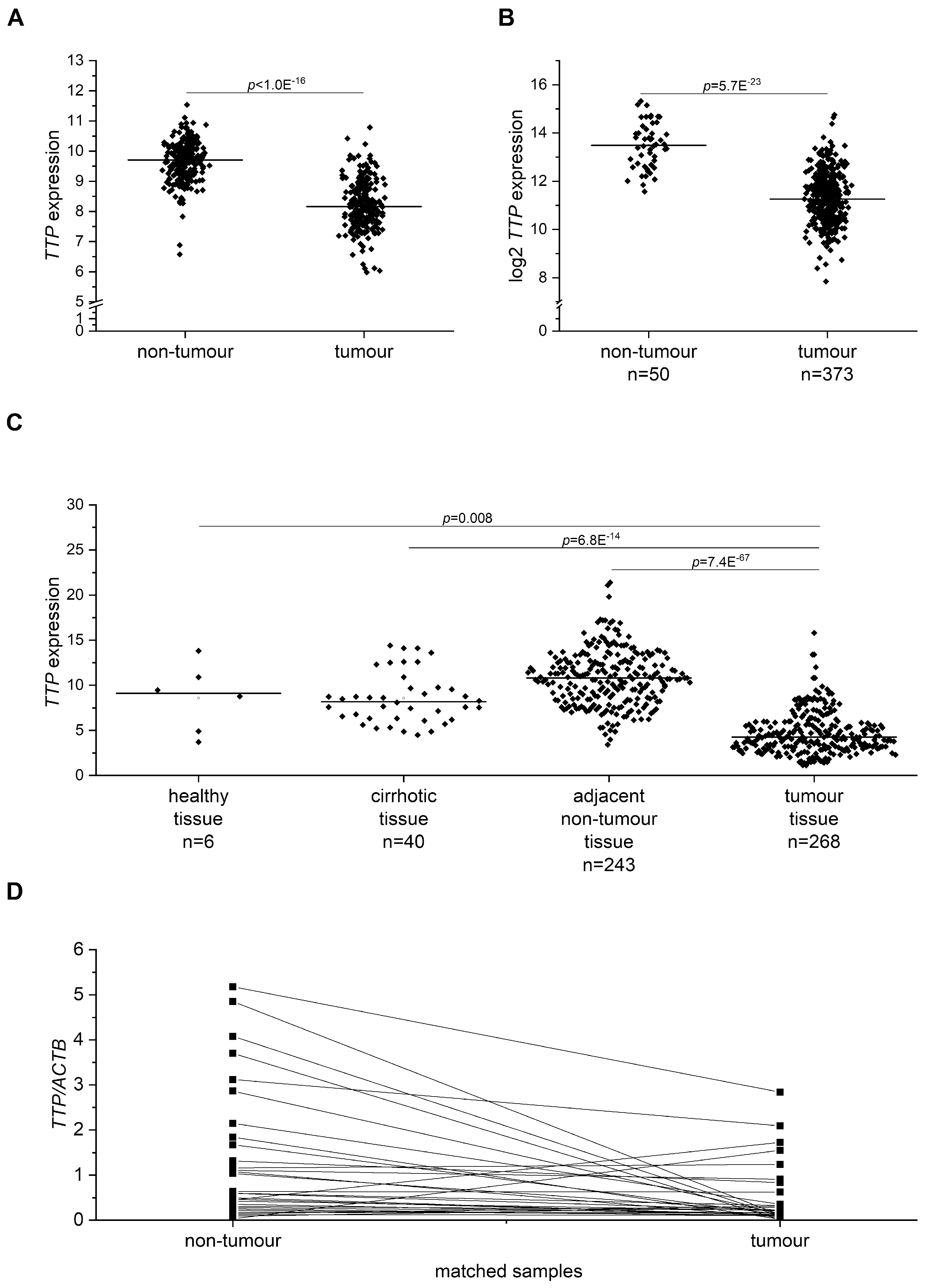
2.5. TTP (gene name ZFP36) Expression in Human HCC Tissue
3. Discussion
4. Materials and Methods
4.1. Animals
4.2. Histology
4.3. Determination of Hepatic Fatty Acid Profile in Mice
4.4. Cell Culture
4.5. Transient TTP Overexpression
4.6. Cytotoxicity Assay (MTT Assay)
4.7. Scratch Assay
4.8. Human HCC
4.9. qPCR
4.10. Flow Cytometry
4.11. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bruix, J.; Han, K.H.; Gores, G.; Llovet, J.M.; Mazzaferro, V. Liver cancer: Approaching a personalized care. J. Hepatol. 2015, 62, S144–S156. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, A.; Hallouch, O.; Chernyak, V.; Kamaya, A.; Sirlin, C.B. Epidemiology of hepatocellular carcinoma: Target population for surveillance and diagnosis. Abdom. Radiol. (NY) 2017, 43, 13–25. [Google Scholar] [CrossRef] [PubMed]
- Farazi, P.A.; DePinho, R.A. Hepatocellular carcinoma pathogenesis: From genes to environment. Nat. Rev. Cancer 2006, 6, 674–687. [Google Scholar] [CrossRef] [PubMed]
- El-Serag, H.B. Surveillance for hepatocellular carcinoma: Long way to achieve effectiveness. Dig. Dis. Sci. 2012, 57, 3050–3051. [Google Scholar] [CrossRef]
- Reeves, H.L.; Zaki, M.Y.; Day, C.P. Hepatocellular carcinoma in obesity, type 2 diabetes, and NAFLD. Dig. Dis. Sci. 2016, 61, 1234–1245. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646. [Google Scholar] [CrossRef]
- Dang, H.; Takai, A.; Forgues, M.; Pomyen, Y.; Mou, H.; Xue, W.; Ray, D.; Ha, K.C.H.; Morris, Q.D.; Hughes, T.R.; et al. Oncogenic activation of the RNA binding protein NELFE and MYC signaling in hepatocellular carcinoma. Cancer Cell 2017, 32, 101–114.e8. [Google Scholar] [CrossRef]
- Gutschner, T.; Hammerle, M.; Pazaitis, N.; Bley, N.; Fiskin, E.; Uckelmann, H.; Heim, A.; Grobeta, M.; Hofmann, N.; Geffers, R.; et al. Insulin-like growth factor 2 mRNA-binding protein 1 (IGF2BP1) is an important protumorigenic factor in hepatocellular carcinoma. Hepatology 2014, 59, 1900–1911. [Google Scholar] [CrossRef]
- Kessler, S.M.; Laggai, S.; Barghash, A.; Schultheiss, C.S.; Lederer, E.; Artl, M.; Helms, V.; Haybaeck, J.; Kiemer, A.K. Imp2/p62 induces genomic instability and an aggressive hepatocellular carcinoma phenotype. Cell Death Dis. 2015, 6, e1894. [Google Scholar] [CrossRef]
- Li, T.; Li, S.; Chen, D.; Chen, B.; Yu, T.; Zhao, F.; Wang, Q.; Yao, M.; Huang, S.; Chen, Z.; et al. Transcriptomic analyses of RNA-binding proteins reveal eif3c promotes cell proliferation in hepatocellular carcinoma. Cancer Sci. 2017, 108, 877–885. [Google Scholar] [CrossRef]
- Guhaniyogi, J.; Brewer, G. Regulation of mRNA stability in mammalian cells. Gene 2001, 265, 11–23. [Google Scholar] [CrossRef] [Green Version]
- Blackshear, P.J. Tristetraprolin and other CCCH tandem zinc-finger proteins in the regulation of mRNA turnover. Biochem. Soc. Trans. 2002, 30, 945–952. [Google Scholar] [CrossRef] [PubMed]
- Hitti, E.; Bakheet, T.; Al-Souhibani, N.; Moghrabi, W.; Al-Yahya, S.; Al-Ghamdi, M.; Al-Saif, M.; Shoukri, M.M.; Lanczky, A.; Grepin, R.; et al. Systematic analysis of AU-rich element expression in cancer reveals common functional clusters regulated by key RNA-binding proteins. Cancer Res. 2016, 76, 4068–4080. [Google Scholar] [CrossRef] [PubMed]
- Sanduja, S.; Blanco, F.F.; Young, L.E.; Kaza, V.; Dixon, D.A. The role of tristetraprolin in cancer and inflammation. Front. Biosci. (Landmark Ed.) 2012, 17, 174–188. [Google Scholar] [CrossRef] [PubMed]
- Brennan, S.E.; Kuwano, Y.; Alkharouf, N.; Blackshear, P.J.; Gorospe, M.; Wilson, G.M. The mRNA-destabilizing protein tristetraprolin is suppressed in many cancers, altering tumorigenic phenotypes and patient prognosis. Cancer Res. 2009, 69, 5168–5176. [Google Scholar] [CrossRef] [PubMed]
- Sawicki, K.T.; Chang, H.-C.; Shapiro, J.S.; Bayeva, M.; De Jesus, A.; Finck, B.N.; Wertheim, J.A.; Blackshear, P.J.; Ardehali, H. Hepatic tristetraprolin promotes insulin resistance through RNA destabilization of FGF21. JCI Insight 2018, 3, e95948. [Google Scholar] [CrossRef] [PubMed]
- Kessler, S.M.; Laggai, S.; Barghash, A.; Helms, V.; Kiemer, A.K. Lipid metabolism signatures in nash-associated hcc-letter. Cancer Res. 2014, 74, 2903. [Google Scholar] [CrossRef]
- Kessler, S.M.; Simon, Y.; Gemperlein, K.; Gianmoena, K.; Cadenas, C.; Zimmer, V.; Pokorny, J.; Barghash, A.; Helms, V.; van Rooijen, N.; et al. Fatty acid elongation in non-alcoholic steatohepatitis and hepatocellular carcinoma. Int. J. Mol. Sci. 2014, 15, 5762–5773. [Google Scholar] [CrossRef]
- Naugler, W.E.; Sakurai, T.; Kim, S.; Maeda, S.; Kim, K.; Elsharkawy, A.M.; Karin, M. Gender disparity in liver cancer due to sex differences in MyD88-dependent Il-6 production. Science 2007, 317, 121–124. [Google Scholar] [CrossRef]
- Kessler, S.M.; Hoppstädter, J.; Hosseini, K.; Laggai, S.; Haybaeck, J.; Kiemer, A.K. Lack of kupffer cell depletion in diethylnitrosamine-induced hepatic inflammation. J. Hepatol. 2019, 70, 813–815. [Google Scholar] [CrossRef]
- Dembek, A.; Laggai, S.; Kessler, S.M.; Czepukojc, B.; Simon, Y.; Kiemer, A.K.; Hoppstädter, J. Hepatic interleukin-6 production is maintained during endotoxin tolerance and facilitates lipid accumulation. Immunobiology 2017, 222, 786–796. [Google Scholar] [CrossRef] [PubMed]
- Horie, Y.; Suzuki, A.; Kataoka, E.; Sasaki, T.; Hamada, K.; Sasaki, J.; Mizuno, K.; Hasegawa, G.; Kishimoto, H.; Iizuka, M.; et al. Hepatocyte-specific Pten deficiency results in steatohepatitis and hepatocellular carcinomas. J. Clin. Investig. 2004, 113, 1774. [Google Scholar] [CrossRef] [PubMed]
- Selmi, T.; Alecci, C.; dell’ Aquila, M.; Montorsi, L.; Martello, A.; Guizzetti, F.; Volpi, N.; Parenti, S.; Ferrari, S.; Salomoni, P.; et al. Zfp36 stabilizes RIP1 via degradation of XIAP and cIAP2 thereby promoting ripoptosome assembly. BMC Cancer 2015, 15, 357. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Ding, N.; Guo, J.; Xia, J.; Ruan, Y. Dysregulation of TTP and HuR plays an important role in cancers. Tumour Biol. 2016, 37, 14451–14461. [Google Scholar] [CrossRef]
- Mukherjee, N.; Jacobs, N.C.; Hafner, M.; Kennington, E.A.; Nusbaum, J.D.; Tuschl, T.; Blackshear, P.J.; Ohler, U. Global target mRNA specification and regulation by the RNA-binding protein ZFP36. Genome Biol. 2014, 15, R12. [Google Scholar] [CrossRef]
- Jeng, Y.M.; Chang, C.C.; Hu, F.C.; Chou, H.Y.; Kao, H.L.; Wang, T.H.; Hsu, H.C. RNA-binding protein insulin-like growth factor II mRNA-binding protein 3 expression promotes tumor invasion and predicts early recurrence and poor prognosis in hepatocellular carcinoma. Hepatology 2008, 48, 1118. [Google Scholar] [CrossRef]
- Li, X.M.; Tang, Z.Y.; Zhou, G.; Lui, Y.K.; Ye, S.L. Significance of vascular endothelial growth factor mRNA expression in invasion and metastasis of hepatocellular carcinoma. J. Exp. Clin. Cancer Res. 1998, 17, 13–17. [Google Scholar]
- Carballo, E.; Lai, W.S.; Blackshear, P.J. Feedback inhibition of macrophage tumor necrosis factor-alpha production by tristetraprolin. Science 1998, 281, 1001–1005. [Google Scholar] [CrossRef]
- Qiu, L.-Q.; Stumpo, D.J.; Blackshear, P.J. Myeloid-specific tristetraprolin deficiency in mice results in extreme lipopolysaccharide sensitivity in an otherwise minimal phenotype. J. Immunol. 2012, 188, 5150–5159. [Google Scholar] [CrossRef]
- Milke, L.; Schulz, K.; Weigert, A.; Sha, W.; Schmid, T.; Brüne, B. Depletion of tristetraprolin in breast cancer cells increases interleukin-16 expression and promotes tumor infiltration with monocytes/macrophages. Carcinogenesis 2013, 34, 850–857. [Google Scholar] [CrossRef]
- Karagozian, R.; Derdák, Z.; Baffy, G. Obesity-associated mechanisms of hepatocarcinogenesis. Metabolism 2014, 63, 607–617. [Google Scholar] [CrossRef] [PubMed]
- Beloribi-Djefaflia, S.; Vasseur, S.; Guillaumond, F. Lipid metabolic reprogramming in cancer cells. Oncogenesis 2016, 5, e189. [Google Scholar] [CrossRef] [PubMed]
- Matsuzaka, T.; Atsumi, A.; Matsumori, R.; Nie, T.; Shinozaki, H.; Suzuki-Kemuriyama, N.; Kuba, M.; Nakagawa, Y.; Ishii, K.; Shimada, M.; et al. Elovl6 promotes nonalcoholic steatohepatitis. Hepatology 2012, 56, 2199. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Pang, Y.; Xiang, X.; Du, J.; Mai, K.; Ai, Q. Molecular cloning, characterization, and nutritional regulation of Elovl6 in large yellow croaker (Larimichthys crocea). Int. J. Mol. Sci. 2019, 20, 1801. [Google Scholar] [CrossRef] [PubMed]
- Sohn, B.H.; Park, I.Y.; Lee, J.J.; Yang, S.J.; Jang, Y.J.; Park, K.C.; Kim, D.J.; Lee, D.C.; Sohn, H.A.; Kim, T.W.; et al. Functional switching of TGF-β1 signaling in liver cancer via epigenetic modulation of a single CpG site in TTP promoter. Gastroenterology 2010, 138, 1898–1908.e1812. [Google Scholar] [CrossRef] [PubMed]
- Vicente-Manzanares, M.; Horwitz, A.R. Cell migration: An overview. Methods Mol. Biol. 2011, 769, 1–24. [Google Scholar] [CrossRef]
- Balic, M.; Lin, H.; Young, L.; Hawes, D.; Giuliano, A.; McNamara, G.; Datar, R.H.; Cote, R.J. Most early disseminated cancer cells detected in bone marrow of breast cancer patients have a putative breast cancer stem cell phenotype. Clin. Cancer Res. 2006, 12, 5615–5621. [Google Scholar] [CrossRef]
- Li, F.; Tiede, B.; Massague, J.; Kang, Y. Beyond tumorigenesis: Cancer stem cells in metastasis. Cell Res. 2007, 17, 3–14. [Google Scholar] [CrossRef]
- Lee, H.H.; Lee, S.R.; Leem, S.H. Tristetraprolin regulates prostate cancer cell growth through suppression of e2f1. J. Microbiol. Biotechnol. 2014, 24, 287–294. [Google Scholar] [CrossRef]
- Van Tubergen, E.; Vander Broek, R.; Lee, J.; Wolf, G.; Carey, T.; Bradford, C.; Prince, M.; Kirkwood, K.L.; D’Silva, N.J. Tristetraprolin regulates interleukin-6, which is correlated with tumor progression in patients with head and neck squamous cell carcinoma. Cancer 2011, 117, 2677–2689. [Google Scholar] [CrossRef]
- Yoon, N.A.; Jo, H.G.; Lee, U.H.; Park, J.H.; Yoon, J.E.; Ryu, J.; Kang, S.S.; Min, Y.J.; Ju, S.A.; Seo, E.H.; et al. Tristetraprolin suppresses the EMT through the down-regulation of Twist1 and Snail1 in cancer cells. Oncotarget 2016, 7, 8931–8943. [Google Scholar] [CrossRef] [PubMed]
- Al-Souhibani, N.; Al-Ahmadi, W.; Hesketh, J.E.; Blackshear, P.J.; Khabar, K.S. The RNA-binding zinc-finger protein tristetraprolin regulates au-rich mRNAs involved in breast cancer-related processes. Oncogene 2010, 29, 4205–4215. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.H.; Son, Y.J.; Lee, W.H.; Park, Y.W.; Chae, S.W.; Cho, W.J.; Kim, Y.M.; Choi, H.-J.; Choi, D.H.; Jung, S.W.; et al. Tristetraprolin regulates expression of VEGF and tumorigenesis in human colon cancer. Int. J. Cancer 2010, 126, 1817–1827. [Google Scholar] [CrossRef] [PubMed]
- Park, S.B.; Lee, J.H.; Jeong, W.W.; Kim, Y.H.; Cha, H.J.; Joe, Y.; Chung, H.T.; Cho, W.J.; Do, J.W.; Lee, B.J.; et al. Ttp mediates cisplatin-induced apoptosis of head and neck cancer cells by down-regulating the expression of Bcl-2. J. Chemother. 2015, 27, 174–180. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Wang, H. Heterogeneity of liver cancer and personalized therapy. Cancer Lett. 2016, 379, 191–197. [Google Scholar] [CrossRef] [PubMed]
- Ghasemi, R.; Ghaffari, S.H.; Momeny, M.; Pirouzpanah, S.; Yousefi, M.; Malehmir, M.; Alimoghaddam, K.; Ghavamzadeh, A. Multitargeting and antimetastatic potentials of silibinin in human HepG-2 and PLC/PRF/5 hepatoma cells. Nutr. Cancer 2013, 65, 590–599. [Google Scholar] [CrossRef] [PubMed]
- Hsu, I.C.; Tokiwa, T.; Bennett, W.; Metcalf, R.A.; Welsh, J.A.; Sun, T.; Harris, C.C. p53 gene mutation and integrated hepatitis b viral DNA sequences in human liver cancer cell lines. Carcinogenesis 1993, 14, 987–992. [Google Scholar] [CrossRef]
- Kanno, S.; Kurauchi, K.; Tomizawa, A.; Yomogida, S.; Ishikawa, M. Pifithrin-alpha has a p53-independent cytoprotective effect on docosahexaenoic acid-induced cytotoxicity in human hepatocellular carcinoma HepG2 cells. Toxicol. Lett. 2015, 232, 393–402. [Google Scholar] [CrossRef] [Green Version]
- Worns, M.A.; Weinmann, A.; Schuchmann, M.; Galle, P.R. Systemic therapies in hepatocellular carcinoma. Dig. Dis. 2009, 27, 175–188. [Google Scholar] [CrossRef]
- Adriaens, C.; Standaert, L.; Barra, J.; Latil, M.; Verfaillie, A.; Kalev, P.; Boeckx, B.; Wijnhoven, P.W.G.; Radaelli, E.; Vermi, W.; et al. p53 induces formation of NEAT1 lncRNA-containing paraspeckles that modulate replication stress response and chemosensitivity. Nat. Med. 2016, 22, 861–868. [Google Scholar] [CrossRef]
- Kessler, S.M.; Hosseini, K.; Hussein, U.K.; Kim, K.M.; List, M.; Schultheiß, C.S.; Schulz, M.H.; Laggai, S.; Jang, K.Y.; Kiemer, A.K. Hepatocellular carcinoma and nuclear paraspeckles: Induction in chemoresistance and prediction for poor survival. Cell. Physiol. Biochem. 2019, 52, 787–801. [Google Scholar] [CrossRef] [PubMed]
- Clark, M.B.; Johnston, R.L.; Inostroza-Ponta, M.; Fox, A.H.; Fortini, E.; Moscato, P.; Dinger, M.E.; Mattick, J.S. Genome-wide analysis of long noncoding RNA stability. Genome Res. 2012, 22, 885–898. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, S.; Chen, W.; Luo, Y.; Ren, F.; Zhong, T.; Rong, M.; Dang, Y.; Feng, Z.; Chen, G. Clinical implication of long non-coding RNA NEAT1 expression in hepatocellular carcinoma patients. Int. J. Clin. Exp. Pathol. 2015, 8, 5395–5402. [Google Scholar] [PubMed]
- Dhanasekaran, R.; Kooby, D.A.; Staley, C.A.; Kauh, J.S.; Khanna, V.; Kim, H.S. Comparison of conventional transarterial chemoembolization (TACE) and chemoembolization with doxorubicin drug eluting beads (DEB) for unresectable hepatocelluar carcinoma (HCC). J. Surg. Oncol. 2010, 101, 476–480. [Google Scholar] [CrossRef]
- Lee, J.Y.; Kim, H.J.; Yoon, N.A.; Lee, W.H.; Min, Y.J.; Ko, B.K.; Lee, B.J.; Lee, A.; Cha, H.J.; Cho, W.J.; et al. Tumor suppressor p53 plays a key role in induction of both tristetraprolin and let-7 in human cancer cells. Nucleic Acids Res. 2013, 41, 5614–5625. [Google Scholar] [CrossRef] [Green Version]
- Schultheiss, C.S.; Laggai, S.; Hussein, U.K.; Golob-Schwarzl, N.; Czepukojc, B.; Tierling, S.; List, M.; Schulz, M.; Barghash, A.; Zimmer, V.; et al. The long non-coding RNA H19 suppresses carcinogenesis and chemoresistance in hepatocellular carcinoma. Cell Stress 2017, 1, 37–54. [Google Scholar] [CrossRef] [Green Version]
- Vesselinovitch, S.D. Perinatal mouse liver carcinogenesis as a sensitive carcinogenesis model and the role of the sex hormonal environment in tumor development. Prog. Clin. Biol. Res. 1990, 331, 53–68. [Google Scholar]
- Tybl, E.; Shi, F.-D.; Kessler, S.M.; Tierling, S.; Walter, J.; Bohle, R.M.; Wieland, S.; Zhang, J.; Tan, E.M.; Kiemer, A.K. Overexpression of the IGF2-mRNA binding protein p62 in transgenic mice induces a steatotic phenotype. J. Hepatol. 2011, 54, 994–1001. [Google Scholar] [CrossRef] [Green Version]
- Fengler, V.H.I.; Macheiner, T.; Kessler, S.M.; Czepukojc, B.; Gemperlein, K.; Müller, R.; Kiemer, A.K.; Magnes, C.; Haybaeck, J.; Lackner, C.; et al. Susceptibility of different mouse wild type strains to develop diet-induced NAFLD/AFLD-associated liver disease. PLoS ONE 2016, 11, e0155163. [Google Scholar] [CrossRef]
- Laggai, S.; Kessler, S.M.; Boettcher, S.; Lebrun, V.; Gemperlein, K.; Lederer, E.; Leclercq, I.A.; Mueller, R.; Hartmann, R.W.; Haybaeck, J.; et al. The IGF2 mRNA binding protein p62/IGF2BP2-2 induces fatty acid elongation as a critical feature of steatosis. J. Lipid Res. 2014, 55, 1087–1097. [Google Scholar] [CrossRef] [Green Version]
- Simon, Y.; Kessler, S.M.; Gemperlein, K.; Bohle, R.M.; Müller, R.; Haybaeck, J.; Kiemer, A.K. Elevated free cholesterol as a hallmark of non-alcoholic steatohepatitis in p62/insulin-like growth factor 2 mRNA binding protein 2-2 transgenic animals. World J. Gastroenterol. 2014, 20, 17839–17850. [Google Scholar] [CrossRef] [PubMed]
- Bode, H.B.; Ring, M.W.; Schwär, G.; Kroppenstedt, R.M.; Kaiser, D.; Müller, R. 3-Hydroxy-3-methylglutaryl-coenzyme a (CoA) synthase is involved in biosynthesis of isovaleryl-CoA in the myxobacterium Myxococcus xanthus during fruiting body formation. J. Bacteriol. 2006, 188, 6524. [Google Scholar] [CrossRef] [PubMed]
- Hoppstädter, J.; Diesel, B.; Eifler, L.K.; Schmid, T.; Brüne, B.; Kiemer, P.A.K. Glucocorticoid-induced leucine zipper is downregulated in human alveolar macrophages upon toll-like receptor activation. Eur. J. Immunol. 2012, 42, 1282. [Google Scholar] [CrossRef] [PubMed]
- Fechir, M.; Linker, K.; Pautz, A.; Hubrich, T.; Forstermann, U.; Rodriguez-Pascual, F.; Kleinert, H. Tristetraprolin regulates the expression of the human inducible nitric-oxide synthase gene. Mol. Pharmacol. 2005, 67, 2148–2161. [Google Scholar] [CrossRef]
- Kessler, S.M.; Pokorny, J.; Zimmer, V.; Laggai, S.; Lammert, F.; Bohle, R.M.; Kiemer, A.K. IGF2 mRNA binding protein p62/IMP2-2 in hepatocellular carcinoma: Antiapoptotic action is independent of IGF2/PI3K signaling. Am. J. Physiol. Gastrointest. Liver Physiol. 2013, 304, G328–G336. [Google Scholar] [CrossRef]
- Roessler, S.; Jia, H.-L.; Budhu, A.; Forgues, M.; Ye, Q.-H.; Lee, J.-S.; Thorgeirsson, S.S.; Sun, Z.; Tang, Z.-Y.; Qin, L.-X.; et al. A unique metastasis gene signature enables prediction of tumor relapse in early-stage hepatocellular carcinoma patients. Cancer Res. 2010, 70, 10202–10212. [Google Scholar] [CrossRef] [PubMed]
- Tung, E.K.-K.; Mak, C.K.-M.; Fatima, S.; Lo, R.C.-L.; Zhao, H.; Zhang, C.; Dai, H.; Poon, R.T.-P.; Yuen, M.-F.; Lai, C.-L.; et al. Clinicopathological and prognostic significance of serum and tissue Dickkopf-1 levels in human hepatocellular carcinoma. Liver Int. 2011, 31, 1494–1504. [Google Scholar] [CrossRef]
- Mínguez, B.; Hoshida, Y.; Villanueva, A.; Toffanin, S.; Cabellos, L.; Thung, S.; Mandeli, J.; Sia, D.; April, C.; Fan, J.-B.; et al. Gene-expression signature of vascular invasion in hepatocellular carcinoma. J. Hepatol. 2011, 55, 1325. [Google Scholar] [CrossRef]
- Vivian, J.; Rao, A.A.; Nothaft, F.A.; Ketchum, C.; Armstrong, J.; Novak, A.; Pfeil, J.; Narkizian, J.; Deran, A.D.; Musselman-Brown, A.; et al. Toil enables reproducible, open source, big biomedical data analyses. Nat. Biotechnol. 2017, 35, 314–316. [Google Scholar] [CrossRef] [Green Version]
- Li, B.; Dewey, C.N. Rsem: Accurate transcript quantification from RNA-seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef]
- Hoppstädter, J.; Kessler, S.M.; Bruscoli, S.; Huwer, H.; Riccardi, C.; Kiemer, A.K. Glucocorticoid-induced leucine zipper: A critical factor in macrophage endotoxin tolerance. J. Immunol. 2015, 194, 6057–6067. [Google Scholar] [CrossRef] [PubMed]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andersen, C.L.; Jensen, J.L.; Ørntoft, T.F. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Strugnell, R.; Wijburg, O.; Brodnicki, T. Measuring bacterial load and immune responses in mice infected with Listeria monocytogenes. J. Vis. Exp. 2011, 54, e3076. [Google Scholar] [CrossRef] [PubMed]
- Blériot, C.; Dupuis, T.; Jouvion, G.; Eberl, G.; Disson, O.; Lecuit, M. Liver-resident macrophage necroptosis orchestrates type 1 microbicidal inflammation and type-2-mediated tissue repair during bacterial infection. Immunity 2015, 42, 145–158. [Google Scholar] [CrossRef]
- Ramachandran, P.; Pellicoro, A.; Vernon, M.A.; Boulter, L.; Aucott, R.L.; Ali, A.; Hartland, S.N.; Snowdon, V.K.; Cappon, A.; Gordon-Walker, T.T.; et al. Differential Ly-6c expression identifies the recruited macrophage phenotype, which orchestrates the regression of murine liver fibrosis. Proc. Natl. Acad. Sci. USA 2012, 109, E3186–E3195. [Google Scholar] [CrossRef]


© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kröhler, T.; Kessler, S.M.; Hosseini, K.; List, M.; Barghash, A.; Patial, S.; Laggai, S.; Gemperlein, K.; Haybaeck, J.; Müller, R.; et al. The mRNA-binding Protein TTP/ZFP36 in Hepatocarcinogenesis and Hepatocellular Carcinoma. Cancers 2019, 11, 1754. https://doi.org/10.3390/cancers11111754
Kröhler T, Kessler SM, Hosseini K, List M, Barghash A, Patial S, Laggai S, Gemperlein K, Haybaeck J, Müller R, et al. The mRNA-binding Protein TTP/ZFP36 in Hepatocarcinogenesis and Hepatocellular Carcinoma. Cancers. 2019; 11(11):1754. https://doi.org/10.3390/cancers11111754
Chicago/Turabian StyleKröhler, Tarek, Sonja M. Kessler, Kevan Hosseini, Markus List, Ahmad Barghash, Sonika Patial, Stephan Laggai, Katja Gemperlein, Johannes Haybaeck, Rolf Müller, and et al. 2019. "The mRNA-binding Protein TTP/ZFP36 in Hepatocarcinogenesis and Hepatocellular Carcinoma" Cancers 11, no. 11: 1754. https://doi.org/10.3390/cancers11111754
APA StyleKröhler, T., Kessler, S. M., Hosseini, K., List, M., Barghash, A., Patial, S., Laggai, S., Gemperlein, K., Haybaeck, J., Müller, R., Helms, V., Schulz, M. H., Hoppstädter, J., Blackshear, P. J., & Kiemer, A. K. (2019). The mRNA-binding Protein TTP/ZFP36 in Hepatocarcinogenesis and Hepatocellular Carcinoma. Cancers, 11(11), 1754. https://doi.org/10.3390/cancers11111754







