Targeted-Gene Sequencing to Catch Triple Negative Breast Cancer Heterogeneity before and after Neoadjuvant Chemotherapy
Abstract
1. Introduction
2. Results
2.1. Patient Characteristics
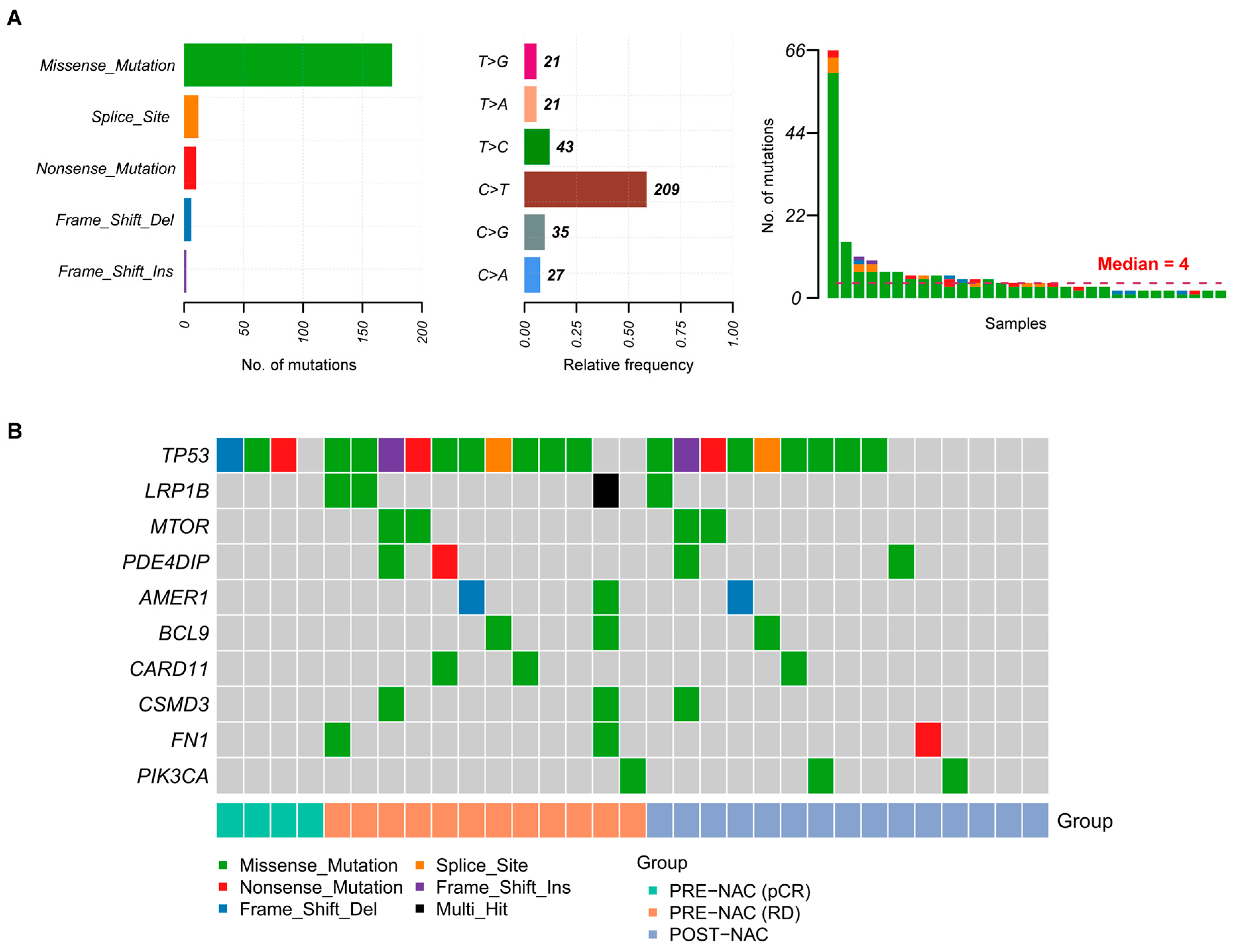
2.2. Somatic Mutations in TNBC Samples
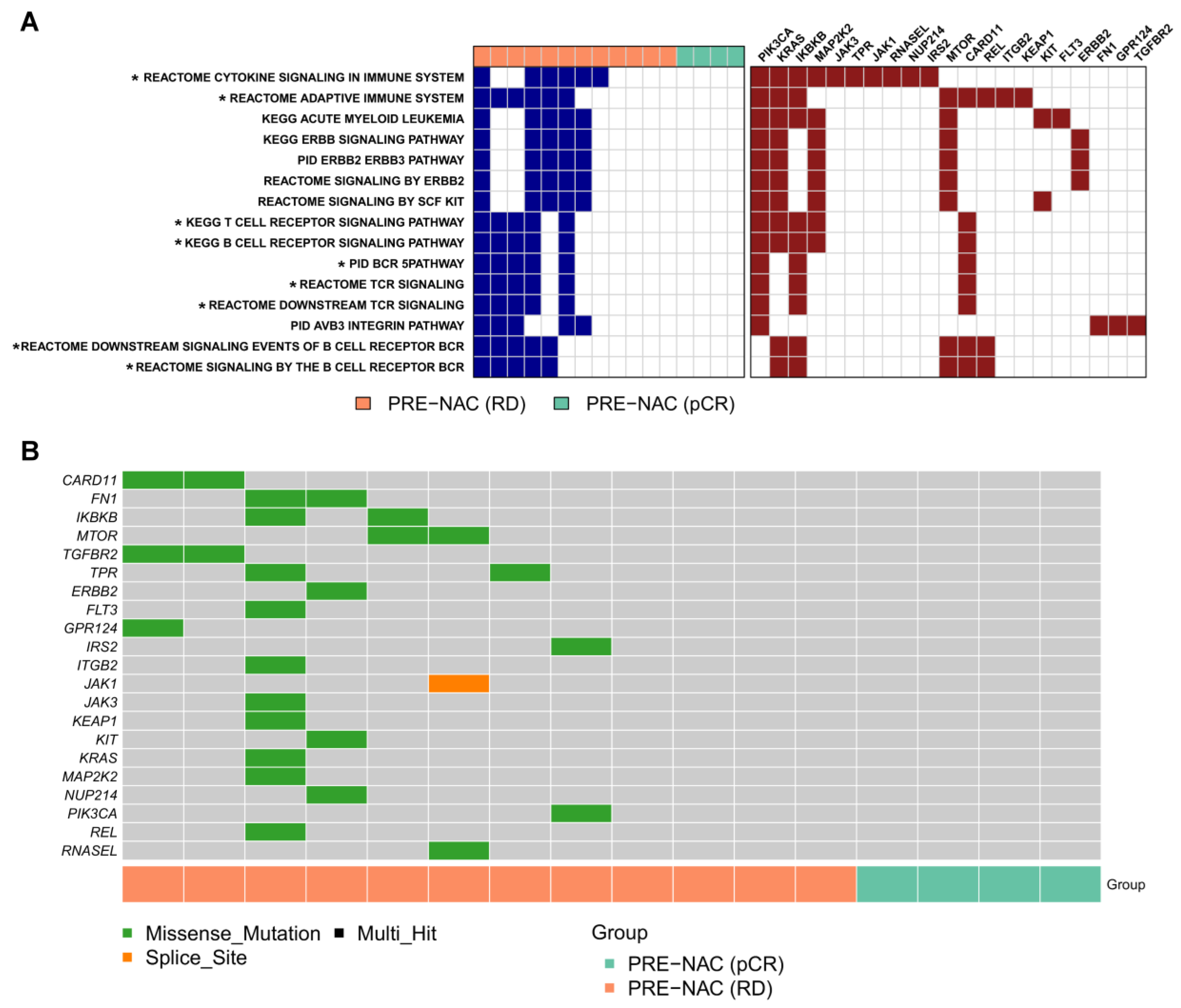
2.3. Genes and Pathways Associated with pCR
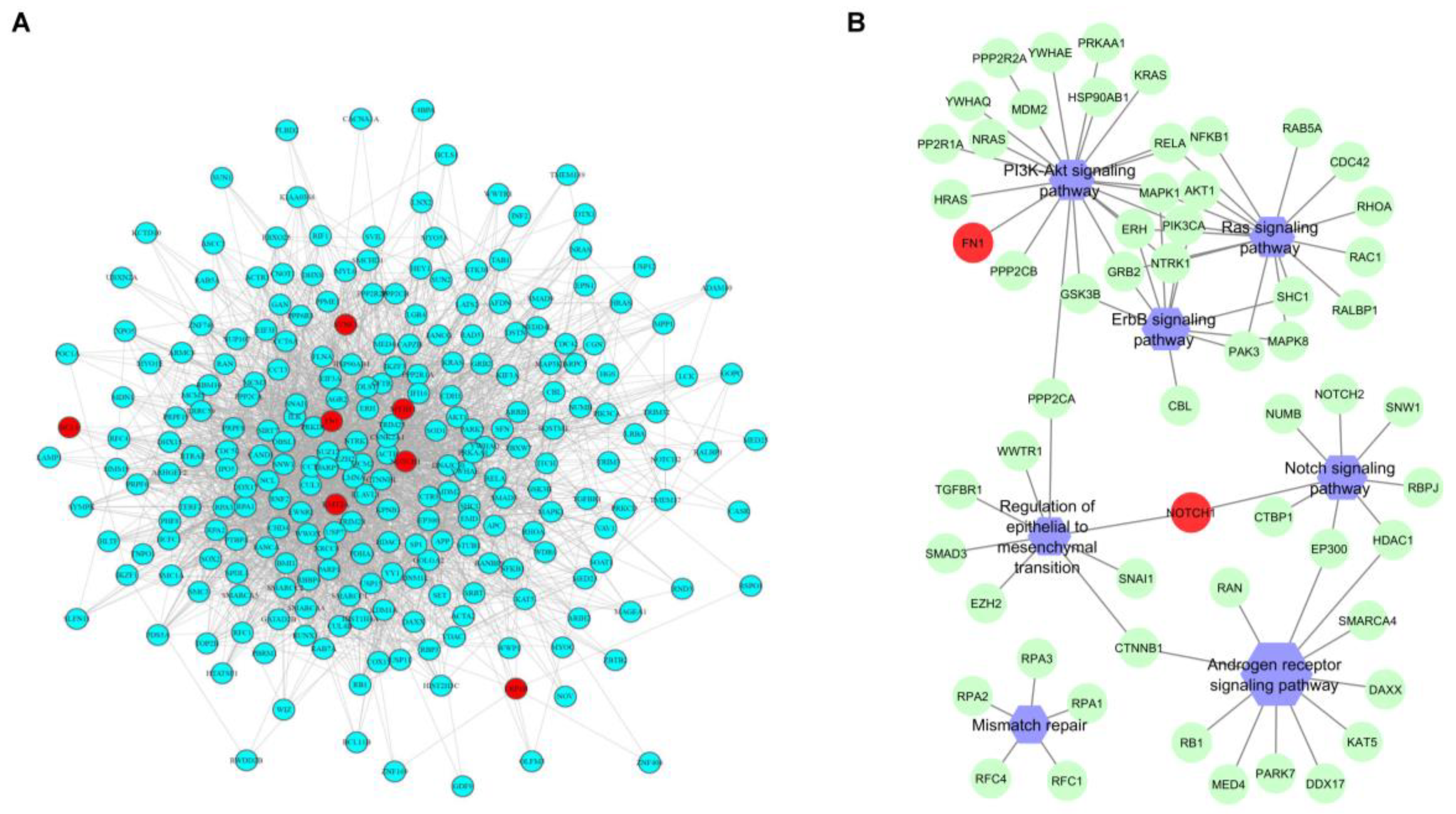
2.4. Comparison between Pre- and Post-NAC Samples
2.5. Tumor-Infiltrating Lymphocytes
3. Discussion
4. Materials and Methods
4.1. Patient Cohort
4.2. Targeted Next Generation Sequencing
4.3. Sequencing and Bio-Informatic Data Analysis
4.4. Statistical Analysis for Association of Genomic and Clinical Data
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hudis, C.A.; Gianni, L. Triple-negative breast cancer: An unmet medical need. Oncologist 2011, 16 (Suppl. 1), 1–11. [Google Scholar] [CrossRef]
- Mustacchi, G.; De Laurentiis, M. The role of taxanes in triple-negative breast cancer: Literature review. Drug Des. Dev. Ther. 2015, 9, 4303–4318. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, L.S.; Mayer, I.A. Platinum agents in the treatment of early-stage triple-negative breast cancer: Is it time to change practice? Clin. Adv. Hematol. Oncol. 2014, 12, 654–658. [Google Scholar] [PubMed]
- Wahba, H.A.; El-Hadaad, H.A. Current approaches in treatment of triple-negative breast cancer. Cancer Biol. Med. 2015, 12, 106–116. [Google Scholar] [PubMed]
- Cortazar, P.; Zhang, L.; Untch, M.; Mehta, K.; Costantino, J.P.; Wolmark, N.; Bonnefoi, H.; Cameron, D.; Gianni, L.; Valagussa, P.; et al. Pathological complete response and long-term clinical benefit in breast cancer: The CTNeoBC pooled analysis. Lancet 2014, 384, 164–172. [Google Scholar] [CrossRef]
- Masuda, N.; Lee, S.J.; Ohtani, S.; Im, Y.H.; Lee, E.S.; Yokota, I.; Kuroi, K.; Im, S.A.; Park, B.W.; Kim, S.B.; et al. Adjuvant Capecitabine for Breast Cancer after Preoperative Chemotherapy. N. Engl. J. Med. 2017, 376, 2147–2159. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.S.; Sundar, R.; Lopez, J. Combining DNA damaging therapeutics with immunotherapy: More haste, less speed. Br. J. Cancer 2018, 118, 312–324. [Google Scholar] [CrossRef]
- Meyerson, M.; Gabriel, S.; Getz, G. Advances in understanding cancer genomes through second-generation sequencing. Nat. Rev. Genet. 2010, 11, 685–696. [Google Scholar] [CrossRef]
- Schmitt, M.W.; Loeb, L.A.; Salk, J.J. The influence of subclonal resistance mutations on targeted cancer therapy. Nat. Rev. Clin. Oncol. 2016, 13, 335–347. [Google Scholar] [CrossRef]
- Gerlinger, M.; Rowan, A.J.; Horswell, S.; Math, M.; Larkin, J.; Endesfelder, D.; Gronroos, E.; Martinez, P.; Matthews, N.; Stewart, A.; et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N. Engl. J. Med. 2012, 366, 883–892. [Google Scholar] [CrossRef]
- Shah, S.P.; Roth, A.; Goya, R.; Oloumi, A.; Ha, G.; Zhao, Y.; Turashvili, G.; Ding, J.; Tse, K.; Haffari, G.; et al. The clonal and mutational evolution spectrum of primary triple-negative breast cancers. Nature 2012, 486, 395–399. [Google Scholar] [CrossRef] [PubMed]
- Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature 2012, 490, 61–70. [Google Scholar] [CrossRef] [PubMed]
- Shlien, A.; Raine, K.; Fuligni, F.; Arnold, R.; Nik-Zainal, S.; Dronov, S.; Mamanova, L.; Rosic, A.; Ju, Y.S.; Cooke, S.L.; et al. Direct Transcriptional Consequences of Somatic Mutation in Breast Cancer. Cell Rep. 2016, 16, 2032–2046. [Google Scholar] [CrossRef] [PubMed]
- Bertucci, F.; Chaffanet, M.; Birnbaum, D. An ICGC major achievement in breast cancer: A comprehensive catalog of mutations and mutational signatures. Chin. Clin. Oncol. 2017, 6, 4. [Google Scholar] [CrossRef] [PubMed]
- Balko, J.M.; Giltnane, J.M.; Wang, K.; Schwarz, L.J.; Young, C.D.; Cook, R.S.; Owens, P.; Sanders, M.E.; Kuba, M.G.; Sánchez, V.; et al. Molecular profiling of the residual disease of triple-negative breast cancers after neoadjuvant chemotherapy identifies actionable therapeutic targets. Cancer Discov. 2014, 4, 232–245. [Google Scholar] [CrossRef]
- Lips, E.H.; Michaut, M.; Hoogstraat, M.; Mulder, L.; Besselink, N.J.; Koudijs, M.J.; Cuppen, E.; Voest, E.E.; Bernards, R.; Nederlof, P.M.; et al. Next generation sequencing of triple negative breast cancer to find predictors for chemotherapy response. Breast Cancer Res. 2015, 17, 134. [Google Scholar] [CrossRef]
- Binnewies, M.; Roberts, E.W.; Kersten, K.; Chan, V.; Fearon, D.F.; Merad, M.; Coussens, L.M.; Gabrilovich, D.I.; Ostrand-Rosenberg, S.; Hedrick, C.C.; et al. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat. Med. 2018, 24, 541–550. [Google Scholar] [CrossRef]
- Shree, T.; Olson, O.C.; Elie, B.T.; Kester, J.C.; Garfall, A.L.; Simpson, K.; Bell-McGuinn, K.M.; Zabor, E.C.; Brogi, E.; Joyce, J.A. Macrophages and cathepsin proteases blunt chemotherapeutic response in breast cancer. Genes Dev. 2011, 25, 2465–2479. [Google Scholar] [CrossRef]
- Rapoport, B.L.; Anderson, R. Realizing the Clinical Potential of Immunogenic Cell Death in Cancer Chemotherapy and Radiotherapy. Int. J. Mol. Sci. 2019, 20, 959. [Google Scholar] [CrossRef]
- Awasthee, N.; Rai, V.; Chava, S.; Nallasamy, P.; Kunnumakkara, A.B.; Bishayee, A.; Chauhan, S.C.; Challagundla, K.B.; Gupta, S.C. Targeting IκappaB kinases for cancer therapy. Semin Cancer Biol. 2019, 56, 12–24. [Google Scholar] [CrossRef]
- Banerjee, S.; Gusho, E.; Gaughan, C.; Dong, B.; Gu, X.; Holvey-Bates, E.; Talukdar, M.; Li, Y.; Weiss, S.R.; Sicheri, F.; et al. OAS-RNase L innate immune pathway mediates the cytotoxicity of a DNA-demethylating drug. Proc. Natl. Acad. Sci. USA 2019, 12, 5071–5076. [Google Scholar] [CrossRef] [PubMed]
- Martelotto, L.G.; Ng, C.K.Y.; Piscuoglio, S.; Weigelt, B.; Reis-Filho, J.S. Breast cancer intra-tumor heterogeneity. Breast Cancer Res. 2014, 16, 210. [Google Scholar] [CrossRef] [PubMed]
- Pribluda, A.; de la Cruz, C.C.; Jackson, E.L. Intratumoral Heterogeneity: From Diversity Comes Resistance. Clin. Cancer Res. 2015, 21, 2916–2923. [Google Scholar] [CrossRef] [PubMed]
- Dieci, M.V.; Criscitiello, C.; Goubar, A.; Viale, G.; Conte, P.; Guarneri, V.; Ficarra, G.; Mathieu, M.C.; Delaloge, S.; Curigliano, G.; et al. Prognostic value of tumor-infiltrating lymphocytes on residual disease after primary chemotherapy for triple-negative breast cancer: A retrospective multicenter study. Ann. Oncol. 2014, 25, 611–618. [Google Scholar] [CrossRef]
- Luen, S.J.; Salagado, M.; Dieci, V.; Vingiani, A.; Curigliano, G.; Gould, R.E.; Castaneda, C.; D’Alfonso, T.; Sanchez, J.; Cheng, E.; et al. Prognostic implications of residual disease tumor-infiltrating lymphocytes and residual cancer burden in triple-negative breast cancer patients after neoadjuvant chemotherapy. Ann. Oncol. 2019, 30, 236–242. [Google Scholar] [CrossRef]
- Lehmann, B.D.; Jovanović, B.; Chen, X.; Estrada, M.V.; Johnson, K.N.; Shyr, Y.; Moses, H.L.; Sanders, M.E.; Pietenpol, J.A. Refinement of Triple-Negative Breast Cancer Molecular Subtypes: Implications for Neoadjuvant Chemotherapy Selection. PLoS ONE 2016, 11, e0157368. [Google Scholar] [CrossRef]
- Baili, P.; Torresani, M.; Agresti, R.; Rosito, G.; Daidone, M.G.; Veneroni, S.; Cavallo, I.; Funaro, F.; Giunco, M.; Turco, A.; et al. A Breast Cancer Clinical Registry in An Italian Comprehensive Cancer Center: An Instrument for Descriptive, Clinical, and Experimental Research. Tumori. 2015, 101, 440–446. [Google Scholar] [CrossRef]
- Edge, S.B.; Compton, C.C. The American Joint Committee on Cancer: The 7th edition of the AJCC cancer staging manual and the future of TNM. Ann. Surg. Oncol. 2010, 17, 1471–1474. [Google Scholar] [CrossRef]
- Hammond, M.E.; Hayes, D.F.; Wolff, A.C.; Mangu, P.B.; Temin, S. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer. J. Oncol. Pract. 2010, 6, 195–197. [Google Scholar] [CrossRef]
- Wolff, A.C.; Hammond, M.E.; Hicks, D.G.; Dowsett, M.; McShane, L.M.; Allison, K.H.; Allred, D.C.; Bartlett, J.M.; Bilous, M.; Fitzgibbons, P.; et al. Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. J. Clin. Oncol. 2013, 25, 118–145. [Google Scholar] [CrossRef]
- Salgado, R.; Denkert, C.; Demaria, S.; Sirtaine, N.; Klauschen, F.; Pruneri, G.; Wienert, S.; Van den Eynden, G.; Baehner, F.L.; Penault-Llorca, F.; et al. The evaluation of tumor-infiltrating lymphocytes (TILs) in breast cancer: Recommendations by an International TILs Working Group 2014. Ann. Oncol. 2015, 26, 259–271. [Google Scholar] [CrossRef] [PubMed]
- Feldman, L.D.; Hortobagyi, G.N.; Buzdar, A.U.; Ames, F.C.; Blumenschein, G.R. Pathological assessment of response to induction chemotherapy in breast cancer. Cancer Res. 1986, 46, 2578–2581. [Google Scholar] [PubMed]
- Ramos, A.H.; Lichtenstein, L.; Gupta, M.; Lawrence, M.S.; Pugh, T.J.; Saksena, G.; Meyerson, M.; Getz, G. Oncotator: Cancer variant annotation tool. Hum. Mutat. 2015, 36, E2423–E2429. [Google Scholar] [CrossRef] [PubMed]
- Mayakonda, A.; Lin, D.C.; Assenov, Y.; Plass, C.; Koeffler, H.P. Maftools: Efficient and comprehensive analysis of somatic variants in cancer. Genome Res. 2018, 28, 1747–1756. [Google Scholar] [CrossRef] [PubMed]
- Csardi, G.; Nepusz, T. The igraph software package for complex network research. InterJ. Complex Syst. 2006, 1695, 1–9. [Google Scholar]
- Diez, D.; Hutchins, A.P.; Miranda-Saavedra, D. Systematic identification of transcriptional regulatory modules from protein-protein interaction networks. Nucleic Acids Res. 2014, 42, e6. [Google Scholar] [CrossRef]

| Characteristics | N (%) |
|---|---|
| Patient age | |
| <50 years | 12 (63) |
| ≥50 years | 7 (37) |
| Clinical T size | |
| 2–5 cm | 13 (68) |
| >5 cm | 6 (32) |
| Clinical nodal status | |
| N0 | 8 (42) |
| N1-3 | 11 (58) |
| Clinical stage | |
| IIA | 5 (26) |
| IIB | 10 (53) |
| IIIA | 2 (11) |
| IIIB | 1 (5) |
| IIIC * | 1 (5) |
| Tumor grade | |
| G2 | 1 (5) |
| G3 | 18 (95) |
| Ki67 | |
| <30% | 3 (16) |
| ≥30% <60% | 3 (16) |
| ≥60% | 12 (63) |
| Missing | 1 (5) |
| Type of NAC | |
| – Doxorubicin/Paclitaxel every 3 weeks followed by CMF1–8 every 4 weeks ^ | 11 (58) |
| – Doxorubicin/Paclitaxel every 3 weeks followed by Eribulin 1–8 every 3 weeks ^^ | 5 (26) |
| – Other | 3 (16) |
| Path Findings | |
| pCR | |
| ypT0N0 | 4 (21) |
| Residual Disease | |
| ypT1N0 | 6 (32) |
| ypT2-3N0 | 7 (37) |
| ypT2N1 | 1 (5) |
| ypT4N3 | 1 (5) |
| Evaluable Cases | |
| Pre-NAC | 16 (84) |
| Post-NAC | 15 (79) |
| Paired pre- and post-NAC | 12 (63) |
| Events | |
| Distant metastases | 7 (37) |
| Patient | Cluster | No. of Genes in Cluster | No. of Genes in Cluster Mutated in POST-NAC Tumor | Enriched Pathways Including Genes Mutated in POST-NAC Tumor |
|---|---|---|---|---|
| p5 | C1 | 42 | 1 | No pathways found |
| C2 | 10 | 1 | No pathways found | |
| C3 | 15 | 0 | NA | |
| p10 | C1 | 3 | 3 | EGFR tyrosine kinase inhibitor resistance (KEGG - hsa01521); Ras signaling pathway (KEGG - hsa04014) |
| C2 | 3 | 0 | NA | |
| p11 | C1 | 3 | 2 | Negative regulation of cell cycle process (GO:0010948); Androgen receptor signaling pathway (GO:0030521) |
| C2 | 5 | 2 | Ras signaling pathway (hsa04014); TOR signaling (GO:0031929); EGFR tyrosine kinase inhibitor resistance (KEGG - hsa01521) | |
| p12 | C1 | 3 | 3 | mTOR signaling pathway (KEGG - hsa04150); PI3K-Akt signaling pathway (KEGG - hsa04151); |
| C2 | 2 | 0 | NA | |
| p13 | C1 | 1 | 1 | PI3K-Akt signaling pathway (KEGG - hsa04151); Negative regulation of cell cycle process (GO:0010948) |
| C2 | 4 | 2 | TORC1 signaling (GO:0038202) | |
| p16 | C1 | 11 | 0 | NA |
| C2 | 3 | 3 | Hippo signaling pathway (KEGG - hsa04390) | |
| C3 | 4 | 0 | NA | |
| p18 | C1 | 2 | 2 | Regulation of cell cycle arrest (GO:0071156); PI3K-Akt signaling pathway (KEGG - hsa04151) |
| C2 | 2 | 2 | Mismatch repair (KEGG - hsa03430); Platinum drug resistance (KEGG - hsa01524) | |
| C3 | 2 | 0 | NA |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Di Cosimo, S.; Appierto, V.; Silvestri, M.; Pruneri, G.; Vingiani, A.; Perrone, F.; Busico, A.; Folli, S.; Scaperrotta, G.; de Braud, F.G.; et al. Targeted-Gene Sequencing to Catch Triple Negative Breast Cancer Heterogeneity before and after Neoadjuvant Chemotherapy. Cancers 2019, 11, 1753. https://doi.org/10.3390/cancers11111753
Di Cosimo S, Appierto V, Silvestri M, Pruneri G, Vingiani A, Perrone F, Busico A, Folli S, Scaperrotta G, de Braud FG, et al. Targeted-Gene Sequencing to Catch Triple Negative Breast Cancer Heterogeneity before and after Neoadjuvant Chemotherapy. Cancers. 2019; 11(11):1753. https://doi.org/10.3390/cancers11111753
Chicago/Turabian StyleDi Cosimo, Serena, Valentina Appierto, Marco Silvestri, Giancarlo Pruneri, Andrea Vingiani, Federica Perrone, Adele Busico, Secondo Folli, Gianfranco Scaperrotta, Filippo Guglielmo de Braud, and et al. 2019. "Targeted-Gene Sequencing to Catch Triple Negative Breast Cancer Heterogeneity before and after Neoadjuvant Chemotherapy" Cancers 11, no. 11: 1753. https://doi.org/10.3390/cancers11111753
APA StyleDi Cosimo, S., Appierto, V., Silvestri, M., Pruneri, G., Vingiani, A., Perrone, F., Busico, A., Folli, S., Scaperrotta, G., de Braud, F. G., Bianchi, G. V., Cavalieri, S., Daidone, M. G., & Dugo, M. (2019). Targeted-Gene Sequencing to Catch Triple Negative Breast Cancer Heterogeneity before and after Neoadjuvant Chemotherapy. Cancers, 11(11), 1753. https://doi.org/10.3390/cancers11111753





